Abstract
Background
The crown-of-thorns starfish, Acanthaster planci (L.), has been blamed for coral mortality in a large number of coral reef systems situated in the Indo-Pacific region. Because of its high fecundity and the long duration of the pelagic larval stage, the mechanism of outbreaks may be related to its meta-population dynamics, which should be examined by larval sampling and population genetic analysis. However, A. planci larvae have undistinguished morphological features compared with other asteroid larvae, hence it has been difficult to discriminate A. planci larvae in plankton samples without species-specific markers. Also, no tools are available to reveal the dispersal pathway of A. planci larvae. Therefore the development of highly polymorphic genetic markers has the potential to overcome these difficulties. To obtain genomic information for these purposes, the complete nucleotide sequences of the mitochondrial genome of A. planci and its putative sibling species, A. brevispinus were determined and their characteristics discussed.
Results
The complete mtDNA of A. planci and A. brevispinus are 16,234 bp and 16,254 bp in size, respectively. These values fall within the length variation range reported for other metazoan mitochondrial genomes. They contain 13 proteins, 2 rRNA, and 22 tRNA genes and the putative control region in the same order as the asteroid, Asterina pectinifera. The A + T contents of A. planci and A. brevispinus on their L strands that encode the majority of protein-coding genes are 56.3% and 56.4% respectively and are lower than that of A. pectinifera (61.2%). The percent similarity of nucleotide sequences between A. planci and A. brevispinus is found to be highest in the CO2 and CO3 regions (both 90.6%) and lowest in ND2 gene (84.2%) among the 13 protein-coding genes. In the deduced putative amino acid sequences, CO1 is highly conserved (99.2%), and ATP8 apparently evolves faster any of the other protein-coding gene (85.2%).
Conclusion
The gene arrangement, base composition, codon usage and tRNA structure of A. planci are similar to those of A. brevispinus. However, there are significant variations between A. planci and A. brevispinus. Complete mtDNA sequences are useful for the study of phylogeny, larval detection and population genetics.
Background
The crown-of-thorns starfish, Acanthaster planci (L.), is a typical reef coral predator, which sometimes causes population outbreaks and destroys coral reef communities. The activities of this starfish have been responsible for causing extensive coral mortality in a large number of coral reef systems throughout the Indo-Pacific region [1]. Major outbreaks involve large numbers of starfish and widespread coral destruction and consequently they can change the fauna and topographic nature of coral reef communities [1,2].
In Australia, Kenchington [3] hypothesized that outbreaks (termed primary) started and in turn initiated a cascade of additional outbreaks (termed secondary). Similarly, the Great Barrier Reef (GBR) in Australia and the Ryukyu Islands in Japan have experienced two distinct series of outbreaks of the crown-of-thorns starfish [2]. Once the unpredictable primary outbreak occurs, this large population of adult A. planci that reproduces enormous amounts of eggs can lead to secondary outbreaks.
The high fecundity and long pelagic larval stage [4] of A. planci imply that the dispersal and recruitment of larvae play an important role in the occurrence of primary and secondary outbreaks [5]. The lack of knowledge of its larval dispersal has limited the understanding of its population dynamics. Some hydrodynamic models have been developed to describe larval dispersal around the GBR [6,7]. These authors suggested that larval dispersal was not random and provided probabilistic routes for dispersal through the GBR. They also claimed that some larvae might be retained on the natal reef. Verifying the larval dispersal route predicted by these hydrodynamic models is essential. However, there are two difficulties in the study of larval dispersal and recruitment of A. planci from a biological perspective: 1) the morphological appearance of A. planci larvae is so similar to those of other asteroid species [4], hence, it is extremely difficult to differentiated A. planci larvae from other asteroid larvae. Recent concern about pelagic larvae dispersal emphasizes the need for accurate larval identification. 2) There have been no useful tools to reveal the reef-connectivity among reefs and autochthonous recruitment of A. planci larvae. Therefore, the development of highly polymorphic genetic markers such as mitochondrial sequences or nucleic microsatellites has been eagerly desired. To resolve the former problem, Evans [8] developed a Polymerase Chain Reaction (PCR)-based identification tool for other asteroid species larvae (excluding A. planci) and showed the PCR-RFLP assay is a useful tool for larval identification of asteroid larvae. The complete or partial mtDNA sequences of many metazoans are now available [9]. Having a relatively rapid evolutionary rate and lacking intermolecular genetic recombination, these data on mtDNA sequences have been utilized to identify invertebrate larvae [10] or conduct the population genetic study at various taxonomic levels [11]. The genetic information can be applied to overcome both these difficulties in the study of larval dispersal of A. planci.
In this study, we determined the complete sequences of the mitochondrial genome of A. planci and its sibling species, Acanthaster brevispinus and compared in detail the full sequences of both species and a third asteroid species, Asterina pectinifera. Also, we describe some characteristics of both mitochondrial genomes of A. planci and A. brevispinus.
Results and discussion
General Features
The complete mtDNA sequences of A. planci and A. brevispinus are 16,234 bp and 16,254 bp in size, respectively; both fit within the length variation range reported for other metazoan mitochondrial genomes [9]. They were submitted to the DDBJ with Accession No. AB231475 (A. planci) and AB231476 (A. brevispinus). They contain 13 proteins, 2 rRNA, and 22 tRNA genes [see additional file 1] in the same order as another asteroid species, Asterina pectinifera [12] [NCBI: NC-001627]. Both in A. planci and A. brevispinus, the L strand encodes most of the proteins including 10 proteins, 11 tRNA and 12S rRNA genes, whereas the H strand includes the remaining three proteins (ND1, ND2, and ND6), 11 tRNA and 16S rRNA genes [12].
Base composition and codon usage
Animal mitochondrial genomes sometimes deviate from a random nucleotide usage, which may be due to the directional mutation pressure and the non-asymmetrical base composition of both the strands [13]. The A + T contents of A. planci and A. brevispinus on the L strand are 56.3% and 56.4%, respectively, and both are lower than that of A. pectinifera (61.2%). This result comes from the fact that the third codon position of the protein-coding gene in A. pectinifera prefers T more than those of A. planci and A. brevispinus, particularly in the synonymous codons of Phe (UUU), Leu (CUU), Ile (AUU), Ser (UCU) and Asn (AAU) [see additional file 2]. The mtDNA sequences of A. planci and A. brevispinus have A-bias on the first and second codon positions of protein-coding gene and rRNA genes. There is no significant difference in the base composition of tRNA genes between the Acanthaster species and A. pectinifera [see additional file 3].
Asakawa et al. [13] demonstrated the bias of a strand-specific nucleotide composition in the mtDNA of A. pectinifera that the L strand prefers to use A or C while the H strand prefers to use G or T in the third codons. This asymmetric distribution of G and C between the two complementary DNA strands can be easily measured with the following formulae [14]; GC-skew = (G-C)/(G+C); AT-skew = (A-T)/(A+T) where C, G, A and T are the occurrences of the four bases. The skew is likely to be most pronounced at the third codon positions of each strand, where any mutational change is synonymous and not subject to selection pressure [15]. Thus we calculated the GC-skew and AT-Skew using the third codon positions of each strand. The L strand prefers to use A or C and H strand prefers to use G or T in both Acanthaster species, clearly showing strand-specific bias of the nucleotide compositions. The three asteroid species show almost the same tendency of skew, the skew in other five echinoderms (Cucumaria miniata (holothuroid), Arbacia lixula (echinoid), Ophiopholis aculeata (ophiuroid) and Ophiura lutkeni (ophiuroid)) showed similar bias of skew, although the degree of skew is slightly different among species examined except for Florometra serratissima (crinoid) (data not shown). In F. serratissima mtDNA, the L strand prefers to use G or T and the H strand prefers to use A or C. The pattern of the bias of the skew is likely to be conserved in asteroid species, although the strand-specific bias is not a general rule in echinoderms [16].
The strand specific bias of the A + C content of protein-coding genes is also seen. In short, the A + C content of the H strands in A. planci and A. brevispinus are 31.4% and 30.9 %, respectively, whereas those in the L strands are 57.3% and 57.6 %, respectively. These values are similar to those of A. pectinifera (34.1% for the L strand and 56.5 % for the H strand).
Nucleotide and amino acid differences in protein genes
As previously mentioned, the identified protein-coding genes of the A. planci and A. brevispinus mtDNA are similar with those of A. pectinifera [12] and of other metazoan mtDNA genomes [9]. Most of the protein-coding genes in these species are identified by sequence comparison with A. pectinifera except for ND5 and ND3 genes. Alignment is difficult to perform because size variations occur near their 5' ends. The 5' ends of ND3 and ND5 genes in these two species are inferred from the eligible termination codons without overlapping the preceding gene. In the sequence comparison between A. pectinifera and A. planci/brevispinus, significant changes in the number of codons are observed in ND3 and ND5 genes between the Acanthaster species and Asterina. The nucleotide sequence of ND3 gene in A. pectinifera is 18 bp shorter than those of both Acanthaster counterparts. On the other hand, both Acanthaster ND5 gene nucleotide sequences are 10 bp shorter than that of A. pectinifera. In both of these cases, most of the deletions are observed near 3' end of each gene.
For the given genes, nucleotide and amino acid differences are compared in the protein-coding genes [see additional file 4]. The percent similarity of nucleotide sequences between A. planci and A. brevispinus is found to be highest in the CO2 and CO3 regions (both 90.6%) and lowest in ND2 gene (84.2%) among the 13 protein-coding genes. In the amino acid sequences, CO1 gene is highly conserved (99.2%), and it seems that ATP8 has evolved faster than any of the other protein-coding genes (85.2%). Amino acid sequences are generally more conserved than nucleotide sequences because of the functional constraints. Therefore, most of the identity values of amino acid sequences are higher than those of nucleotide sequences or almost the same except for ATP8 gene. The percentage similarity of amino acid sequences in ATP8 gene is significantly lower than those of nucleotide sequences between Acanthaster species and A. pectinifera and also slightly lower even between Acanthaster species. The pairwise comparison of the ATP8 gene among A. planci, A. brevispinus and A. pectinifera has revealed that the nucleotide substitution rates of nonsynonymous sites (first + second codon positions) are almost the same as those of synonymous sites (third position) (data not shown), suggesting the cause of this improbable phenomenon. A similar result in ATP genes was reported in sea urchins [17] and they showed that the nucleotide substitution rates of nonsynonymous sites are not significantly lower than that of synonymous sites. These data clearly indicate a relaxation of the functional constraints at the amino acid level. They found that the same sequence motif (V/M)PQL(X4)(W/F) is evolutionarily conserved from yeast to mammals at the N-termini of the proteins. When the deduced amino acid sequences were analysed with a computer aided method, the predicted hydrophatic profiles for the corresponding proteins are completely super-imposable. De Giorgi et al. [17] stated that in spite of the divergence observed in the primary structure of the genes, the role played may be the same. On the other hand, the loss of ATP8 has occurred several times in the evolutional process of metazoans [18]. As De Giorgi et al. [17] suggested, the C-terminal of ATP8 might evolve extremely rapidly and in a phylum-specific manner.
The ATP8 and ATP6 genes of starfish overlap each other by 16 nucleotides with a -1 frame shift. A similar overlap is found among metazoan mitochondria, although the extent of overlap varies [19]. Thus, it is easily expected that the functional constraints of ATP8 gene are not so large and the gene could evolve faster than other protein-coding genes. However, further work is required to clarify the causal relationship in the lower percent similarity of amino acid sequence with that of nucleotide sequence in the Acanthaster species and the evolution of ATP8 genes in asteroid species.
Gene initiation and termination
In the case of A. planci, 10 of the 13 protein-coding genes use ATG as an initiation codon. Merely ND3 and ND4L genes use ATT whereas ND1 gene uses GTG. All initiation codons of A. brevispinus are the same as those of A. planci. The initiation codon ATG of ND5 gene in the Acanthaster species mtDNA changes to GTG in Asterina pectinifera. Most of the A. planci and A. brevispinus mtDNA genes terminate with a TAA codon, however ND1, ND2 and ND6 genes terminate with ATG. ND4 gene of A. brevispinus terminates with ATG, however, those of A. planci and A. pectinifera terminate with TAA. There is only one T residue between CO2 or Cytb genes and their downstream tRNA genes in the Acanthaster species and A. pectinifera mtDNA genomes. Furthermore, CO1 gene in the Acanthaster species terminates only with TA residue, whereas the A. pectinifera counterpart has a triplet codon (TAA), although CO1 gene is the most conserved gene among the protein-coding genes [see additional file 4]. Mitochondrial genes commonly employ several alternatives to ATG as an initiation codon and to TAA or TAG as a termination codon [13,20]. However, the mechanism by which these variations occur is not yet well understood [21]. As demonstrated in the human mtDNA genome [20], we speculate that incomplete termination codons such as T and TA are converted to canonical termination codon TAA after the cleavage of the tRNA gene by ribonuclease.
tRNA and rRNA genes
Twenty-two tRNA genes are identified in A. planci and A. brevispinus mtDNA genomes by sequence comparison with known echinoderm tRNA genes and the deduced potential cloverleaf structures are shown in Figure 1.
Figure 1.
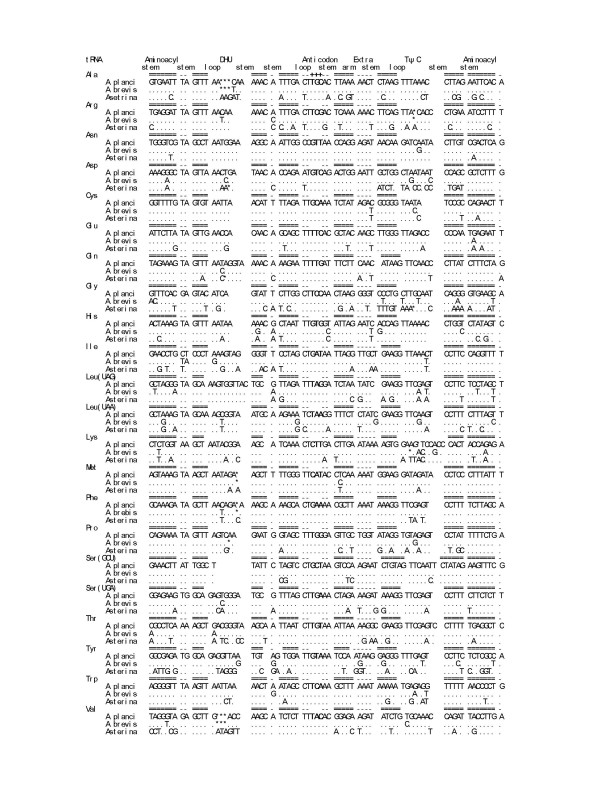
Alignment of the twenty-two tRNA form mitochondrial genome of A. planci, A. brevispinus and Asterina pectinifera. * Data from Asakawa et al. 199510. A. planci: Acanthaster planci; A. brevis: Acanthaster brevispinus; Asterina: Asterina pectinifera, Each anticodon is located at the middle of anticodon loop (+++). Bars above alignments indicate unambiguously aligned stem regions. Asterisks indicate deletions and dots indicate the same nucleotides as A. planci. The last one letter is one nucleotide protrusion of 3' amino-acid accepter stem.
The tRNA genes ranged in size from 67 (tRNACys) to 74 (tRNAThr) and they are large enough for encoded tRNAs to fold into cloverleaf structures. As often found in other metazoan mtDNA genomes, tRNASer (GCU) lacks the DHU loop. The potential secondary structures of tRNAs are quite similar between A. planci and A. brevispinus, whereas the comparison between the Acanthaster species and A. pectinifera reveals that several size variations have occurred at the DHU and TψC loops. The comparison between the DHU and TψX loops shows that size variations are more likely to be found in the DHU loop than the TψX loop.
The echinoderm mitochondrial rRNA genes are located separately from each other [22], and this arrangement appears also in A. planci and A. brevispinus mtDNA genomes. The 5' and 3' end of 12S rRNA genes are defined as immediately adjacent to the ends of the flanking genes tRNAPhe and tRNAGlu, in the two Acanthaster mtDNA genomes. However, an alignment of A. pectinifera does not support their 3' end boundaries; the 12S rRNA sequences deduced from the sequence comparison with A. pectinifera overlapped with the beginning of tRNAGlu by two nucleotides.
Although the precise ends of the 16S rRNA transcripts cannot be determined without experimentation, the inferred boundaries were determined by sequence comparison with Pisaster ochraceus [22] [NCBI: NC-004610]. In contrast to a rather uniform distribution of nucleotide differences over the entire protein-coding genes, although the substitution was mainly seen in the third positions, replacement differences of rRNA genes are dispersed in clusters. This variability is due to the secondary structures of rRNA genes; the stem regions are relatively conservative, whereas the loop sequences are more variable as is the case of tRNAs. Comparing with 12S rRNA genes, 16S rRNA genes of asteroid species seem to have more variable sites. This variability may be due to the potential loop structures that have evolved much faster than other regions in rRNAs.
Unassigned Sequence
Intergenetic regions represent 4.6% of A. planci mtDNA genome and 4.7% of A. brevispinus mtDNA genome. The largest unassigned regions (the putative control region) are adjacent to 16S rRNA and tRNAThr. The inferred boundaries determined by sequence comparison with P. ochraceus suggest that this region of A. planci spanned 532 bp and that of A. brevispinus spanned 552 bp. This region contains a stem-loop structure, which is the putative origin of light-strand replication. This structure is observed close to tRNAThr, and the lengths are 29 bp for A. planci and 34 bp for A. brevispinus. There is a 12 bp stem (TAACCCCCCCCC) and a 5 bp loop (CCTCC) in A. planci, and a 12 bp stem (ATTACCCCCCCC) and a slightly longer 10 bp loop (CCCCCCCTCC) in A. brevispinus. The second largest unassigned region in length is 51 bp in A. planci and 58 bp in A. brevispinus and it is located between the ND5 and ND6 genes. There are neither stem and loop structures nor promoter-like sequences in these short sequences. However, these sequences are as variable as the largest unassigned sequences. In A. pectinifera mtDNA genome, the second largest region is found between ND4 and tRNAHis, which extends to 140 bp long, whereas in the two Acanthaster species it is only 8 bp long.
Aside from the large two regions, there are 4 TATATATATAA-like sequences found between the following genes in both Acanthaster species: tRNALeu and tRNAGly, tRNAPro and CO1, CO3 and tRNAPro, ND6 and Cytb. Except for the region between the genes for tRNALeu and tRNAGly, all other TATATATATAA-like sequences are located in the boundaries where the coding strand shifts to the complimentary one. As already pointed out by Asakawa et al. [13], these sequences are the possible bi-directional promoters, which play the role for replication and/or transcription. However, the sequence located between 16S rRNA and ND2 that were reported in A. pectinifera and sea urchin species, Strongylocentrotus purpuratus [23] and Paracentrotus lividus [24] are not found in the two Acanthaster species.
Phylogenetic relationship
The complete CO1 sequences of A. planci and A. brevispinus were subjected to the ML, MP and NJ analyses together with the representative asteroid sequences [see additional file 5] available in GenBank (Figure 2). These three analyses produced almost the same tree topologies. The CO1 phylogeny indicated that Pisaster ochraceus (Forcipulatida, Asteriidae) was the most primitive taxon among asteroids. In these trees, the close affinities of species belonging to the same family (except for Asterinidae), showing monophyly, were well supported. In contrast, the confidence values for the branches linking different families were not so higher. The close affinity between Patiriella and Asterina species was found and the Asterinidae mixed together in the same group, suggesting a non-monophyletic relationship. Acanthaster species were positioned closer to Oreaster species than any other members in Valvatida, suggesting the close genetic affiliation, although the bootstrap values were not so high and these were separated from Patiriella and Asterina species by relatively high bootstrap supports in the ML method. A. planci and A. brevispinus were the closest species showing 89.2% homology between the two sequences, but these were consistently separated by 100% bootstrap supports in the tree. Luidia quinalia and Astropecten polyacanthus were grouped together and separated from Acanthaster, Oreaster, Patiriella and Asterina species. In the asteroid phylogeny using mitochondrial 12S rRNA and 16S rRNA also, the monophyly of each family was well supported [25,26] but the monophyly of Asterinidae was not clearly indicated from their ML and MP analyses as well as our data. In this study, P. ochraceus (Forcipulatida, Asteriidae) was the most primitive taxon among the asteroids. On the other hand, the mitochondrial 12S rRNA and 16S rRNA phylogenetic trees [26] do not support the early divergence of Asteriidae such as Asterias amurensis and Coscinasterias acutispina. Our data suggested the close affinity between Luidia and Astropecten species (both are Paxillosida), however, [26] reported a contradictory result that Astropecten species were not positioned close to Luidia species but grouped Leiaster leachii and Certonardoa semiregularis (Valvatida). The reason, why the different topology between CO1 and mitochondrial 12S rRNA and 16S rRNA sequences was obtained, is unclear. The phylogenetic relationships among asteroids remain extremely controversial in spite of many morphological and molecular studies that have been applied to this issue [26].
Figure 2.
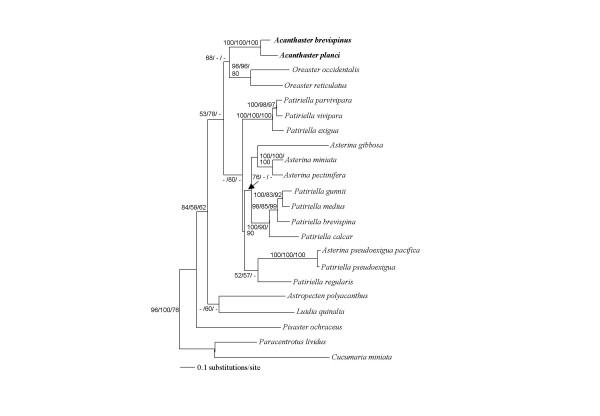
The molecular phylogeny of asteroid species based on the CO1 sequence data. Cucumaria miniata (Cucumariidae) and Paracentrotus lividus (Echinidae) were used as the out-groups. The ML tree was reconstructed with the TrN + I + G model using PAUP 4.0b10. Only bootstrap values of 50% or higher are shown to the left of internal nodes. The first numbers are from neighbor-joining analyses (1000 replications), the second numbers are from maximum likelihood analyses (100 replications) and the last numbers are from Parsimony analyses (1000 replications; characters were unweighted).
Applications of mtDNA sequence of A. planci
A. brevispinus is a very rare species and was once considered as only as a short-spined individual of A. planci [27]. However, Birkeland & Lucas [1] and Lucas & Johns [28] reported that A. planci and A. brevispinus are genetically compatible to the extent that fertile F1 hybrids were produced. However, F2 hybrids and backcrosses were of poor viability and some of them showed morphological abnormalities, suggesting that there are barriers to introgression of genetic material between the species.
Protein and DNA technologies have provided strategies for identification when morphological features are of limited value [29,30]. A method to identify A. planci larvae using monoclonal antibodies provided by Hanna et al. (see Birkeland & Lucas 1990[1]) is not perfect because the monoclonal antibodies do not ensure discrimination among coral reef asteroids other than Culsita novaeguineae. As mentioned in the background of this study, a species-specific genetic marker of A. planci is essential to reveal the larval ecology in the field study. Recent investigations have suggested the feasibility of creating identification systems reliant on the analysis of sequence in small segments of DNA [31]. Hebert et al. [32] focused this discussion by proposing that a DNA barcoding system for animal life could be based upon sequence diversity in cytochrome c oxidase subunit 1 (CO1). They summarized CO1 sequence divergences for 13,320 congeneric pairs of animal species belonging to 11 phyla. Most pairs (79%) showed greater than 8% sequence divergence and more than 98% of species pairs showed greater than 2% sequence divergence in their study (10.9 ± 4.9% (mean ± s.d.%, n = 86) in Echinodamata). Therefore, most congeneric species pairs of animals showed enough sequence divergence to ensure their easy diagnosis, because intraspecific divergences are rarely greater than 2% and most are less than 1% [33]. In the comparison of a complete CO1 sequence of A. planci with the genetically closest species A. brevispinus, the sequence convergence was 10.8%. The phylogenetic tree inferred from the complete CO1 sequences also clearly showed the high divergence among asteroid species. In addition, no sequence variation of the partial CO1 gene (from 4691 bp to 5401 bp) was confirmed among the 3 A. planci specimens collected from different localities (data not shown). Accordingly these results indicate that the identification system for asteroid species based on the CO1 gene will be highly effective and it will easily provide the effective regions for species-specific markers. Moreover, universal primers for metazoan invertebrates in the CO1 genes were already provided [34], therefore, PCR-RFLP (Restriction Fragment Length Polymorphism) assay can also be applied as another tool for species identification of A. planci.
For polymorphic marker using mtDNA variation, the desirable characteristics are as follows: evolutional rate is rapid enough to distinguish among local populations and substitutions are neutral. In A. planci mtDNA sequence, the largest unassigned region between tRNAThe and 16S rRNA is most variable. In this region, size variations ranging from 538 to 546 bp are observed in the 23 individuals. The variation includes 62 base substitutions and 5 deletions/insertions ranging from 1 to 5 bp (data not shown). These sequences can be useful in the analysis of intraspecific variation of A. planci collected from several localities.
Conclusion
The complete mitochondrial genome sequences of Acanthaster planci and its sibling, rare species, A. brevispinus were determined and compared with another known asteroid species, Asterina pectinifera. The genome contains 37 genes most commonly found in metazoan mtDNA and they are in the same order as in A. pectinifera. Characteristics of the two Acanthaster species are quite similar when compared with A. pectinifera, though there exist some significant differences between them especially in the unassigned regions. The sequence information will be useful for molecular ecological studies of A. planci in future.
Methods
Animal samples and DNA Extraction
A specimen of Acanthaster planci for complete mtDNA sequencing was collected at Akajima Island, Okinawa Prefecture, Japan in August 2003 by snorkeling. A specimen of A. brevispinus was collected at an offshore site at Kushimoto, Wakayama Prefecture, Japan in October 2003. Twenty-three specimens of A. planci to observe the variability of the largest unassigned sequence were collected from Ishigaki Island (n = 4), Tokashiki Island (n = 10) in Okinawa Prefecture and Kinko Bay (n = 9) in Kagoshima Prefecture, Japan. The total genomic DNA of Acanthaster species was extracted from the tube feet using a DNeasy kit (Qiagen) following the manufacturer's procedure.
Long PCR and DNA sequencing
Oligonucleotide primers were designed in accordance with sequences reported in Asterina pectinifera [12] and other species in order to amplify the intervening regions. Sequences were completely determined from the ends of these PCR fragments, then internally by primer walking.
Three partial sequences of A. planci and A. brevispinus mtDNA were obtained. They were amplified to a total volume of 100 μl containing 5 U of LaTaq (TaKaRa), 50 μl of 2 × GC buffer I, 2.5 mM of MgCl2, 16 μl of dNTPs mixture (2.5 mM each), 0.5 μM of each oligonucleotide primers, and 10 to 50 ng of the genomic DNA. The mixture was subjected to 40 cycles of amplification in a thermal cycler (PC-800, ASTEC). The oligonucleotide primers of A. planci and A. brevispinus are summarized in Table 6 [see additional file 6]. The first cycle was preceded by initial denaturation for 1 min at 94°C. Each cycle consisted of denaturation for 30 sec at 94°C, annealing for 30 sec at 50–55°C, and extension for 7 min. The last cycle was followed by a final extension step for 10 min at 72°C.
All PCR products were electrophoresed in 1% agarose gels and stained with ethidium bromide. The agarose gel slices containing the PCR-product band were excised under UV, and DNA was purified from the agarose plug by using GenEluteTM Agarose Spin Columns (SIGMA), followed by an ethanol precipitation. Amplified products were either sequenced directly or cloned into pGEM-T® easy vector in the pGEM-T® easy system (Promega) and TopoXL® vector using the TopoXL (Invitrogen). Recombinant plasmids were extracted by the QIAprep Spin Miniprep kit (Qiagen).
All DNA sequence data were determined using ABI PRISM 3100 Genetic Analyzer (Applied Biosystems) DYEnamic ET Terminator Cycle Sequencing Kit and DYEnamic Terminator Dilution Buffer (Amersham Bioscience).
Phylogeny
Complete CO1 sequences, excluding primer regions, of A. planci and A. brevispinus were aligned with 18 other asteroid sequences obtained from GenBank using the ClustalX package [35] and edited manually. Cucumaria miniata (Cucumariidae) and Paracentrotus lividus (Echinidae) were used as the out-groups [see additional file 5]. A total of 1551 bp sequence were analyzed using maximum likelihood (ML), maximum parsimony (MP) and neighbour-joining (NJ) methods in PAUP* 4.0b10 [36]. In order to find the best model for the CO1 sequences we applied Modeltest (version 3.07) [37]. The best-fit model was TrN + I + G [38] with assumed proportion of invariable site = 0.5180, gamma distribution shape parameter = 0.6099 and base substitution categories (AC = 1.0000, AG = 10.3590, AT = 1.0000, CG = 1.0000, CT = 8.7274, GT = 1.0000). Base frequencies were set at A = 0.3725, C = 0.2639, G = 0.0884 and T = 0.2753. ML analyses were performed with the heuristic search option starting with NJ and TBR branch swapping. One hundred replicates were performed in ML bootstrap analysis. In MP 100 random additions were done using the heuristic search option and TBR branch swapping. Characters were unordered and weighted equally. Bootstrap analyses with 1000 replicates were conducted. The best-fit model suggested by Modeltest (see above) was also used to compute dissimilarity values. The distance matrix obtained was used to build a tree with NJ. One thousand replicates were performed in NJ bootstrap analysis.
Abbreviations
CO1, CO2, CO3, cytochrome oxidase subunit I, II and III protein genes; Cytb, cytochrome b gene; ATP6, ATP8, ATP synthase subunit 6 and 8 genes; ND1, ND2, ND3, ND4, ND4L, ND5 and ND6, NDAH dehydrogenase subunit 1–6, 4L genes; 12S rRNA and 16S rRNA, small and large ribosomal RNA genes; tRNA, transfer RNA genes with standard 3-letter 22 amino acid abbreviations are used. PCR-RFLP, Polymerase Chain Reaction based – Restriction Fragment Length Polymorphism.
Authors' contributions
KN and MH were primarily responsible for the design, coordination and conducting the study. NY and MS were responsible for determining and assembling the mtDNA sequences. NY drafted the manuscript and figures, and MH contributed suggested some valuable comments. KN extensively revised the manuscript. SN conducted phylogenetic analysis. This research has been financially supported by JSPS (The Japan Society for the Promotion of Science), Grand-in-Aid for Scientific Research (A) (No. 14205071 and No. 15254002), and by The Cabinet Office of Japan through Research Institute of Subtropics. KN is the project leader for these grants. All authors have read and approved the final manuscript.
Supplementary Material
Table 1: Localization of genes and features in the mitochondrial genome of Acanthaster planci and A. brevispinus. Genes with asterisks are coded on the complimentary H strand.
Table 2: Comparison of base composition of the mitochondrial genomes of Acanthaster planci, A. brevispinus and Asterina pectinifera. * Data from Asakawa et al. [10].
Table 3: Total codon usage in the 13 protein-coding genes of Acanthaster planci, A. brevispinus and Asterina pectinifera. *Data from Asakawa et al. [10].
Table 4: Comparison of nucleotide and amino acid sequences in 13 protein-coding genes of Acanthaster planci, A. brevispinus and Asterina pectinifera.Genes that showed size variations are indicated as bold.
Table 5: Primer sequences for long PCR to amplify the mitochondrial DNA in A. planci and A. brevispinus.
Table 6: List of specimens used for phylogenetic analysis.
Acknowledgments
Acknowledgements
We would like to express our deep thanks to Dr. Saki Harii, Centre for Marine Studies, The University of Queensland, for valuable encouragement and constructive criticism on the manuscript and her encouragement during this study. We are grateful to Takuya Mori, Masaharu Mizukami and Kazusa Isozumi, Susami Crustacean Aquarium for their help with the A. brevispinus sample collection and also thanks to Hiroki Taniguchi, of Akajima Marine Science Laboratory for help with the A. planci sample collection. We also thank Dr. Kenji Saito, of Tohoku National Research Fisheries Institute, for his assistance with the alignment of tRNA genes. Maria Cecilia Rubio helped us to revise the manuscript.
Contributor Information
Nina Yasuda, Email: yasuda@wv.mei.titech.ac.jp.
Masami Hamaguchi, Email: masami@fra.affrc.go.jp.
Miho Sasaki, Email: tb1214@affrc.go.jp.
Satoshi Nagai, Email: snagai@affrc.go.jp.
Masaki Saba, Email: saba-m@mct.ne.jp.
Kazuo Nadaoka, Email: nadaoka@mei.titech.ac.jp.
References
- Birkeland C, Lucas JS. Acanthaster planci. Major Management Problem of Coral Reefs. CRC Press; 1990. [Google Scholar]
- Moran PJ. The Acanthaster Phenomenon. Oceanogr Mar Biol Ann Rev. 1986;24:379–480. [Google Scholar]
- Kenchington RA. Growth and recruitment of Acanthaster planci (L.) on the Great Barrier Reef. Biol Conserv. 1977;11:103–118. doi: 10.1016/0006-3207(77)90032-5. [DOI] [Google Scholar]
- Yamaguchi M. Early life history of coral reef asteroids, with special reference to Acanthaster planci (L.) In: Jones OA, Endean R, editor. Biology and Geology of Coral Reefs Biology 1. II. New York and London: Academic Press; 1973. pp. 369–389. [Google Scholar]
- Lucas JS. Proceedings Crown-of-Thorns starfish seminar, University of Queensland August 25 1972; Brisbane. The Advisory Committee on Research into the Crown-of-thorns Starfish: AGPS Canberra; 1972. Acanthaster planci : Before it eats coral polyps; pp. 25–36. [Google Scholar]
- Black KP, Moran PJ. Influence of hydrodynamics on the passive dispersal and initial recruitment of larvae of Acanthaster planci (Echinodermata: Asteroidea) on the Great Barrier Reef. Mar Ecol Prog Ser. 1991;69:55–65. [Google Scholar]
- Dight IJ, James MK, Bode L. Modelling the larval dispersal of Acanthaster planci II. Patterns of reef connectivity. Coral Reefs. 1990;9:125–34. doi: 10.1007/BF00258224. [DOI] [Google Scholar]
- Evans BS, White RWG, Ward RD. Genetic identification of asteroid larvae from Tasmania, Australia, by PCR-RFLP. Mol Ecol. 1998;7:1077–1082. doi: 10.1046/j.1365-294x.1998.00395.x. [DOI] [Google Scholar]
- Boore LJ. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu IF, Slater JW, Carney BF. Identification of Mytilus spp. and Pecten maximus in Irish waters by standard PCR of the 18S rDNA gene and multiplex PCR of the 16S rDNA gene. Mar Biotechnol doi: 10.1007/s10126-004-0124-y. [DOI] [PubMed] [Google Scholar]
- Silva EP, Russo CAM. Techniques and statistical data analysis in molecular population genetics. Hydrobiologia. 2000;420:119–135. doi: 10.1023/A:1003993824352. [DOI] [Google Scholar]
- Asakawa S, Himeno H, Miura K, Watanabe K. Nucleotide sequence and gene organization of the starfish Asterina pectinifera mitochondrial genome. Genetics. 1995;140:1047–1060. doi: 10.1093/genetics/140.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa S, Kumazawa Y, Araki T, Himeno H, Mimura K, Watanabe K. Strand-specific nucleotide composition bias in echinoderm and vertebrate mitochondrial genomes. J Mol Evol. 1991;32:511–520. doi: 10.1007/BF02102653. [DOI] [PubMed] [Google Scholar]
- Perna NT, Kocher TD. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 1995;41:353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- Le TH, McManus DP, Blair D. Codon usage and bias in mitochondrial genomes of parasitic platyhelminthes. Korean J Parasitol. 2004;42:159–167. doi: 10.3347/kjp.2004.42.4.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Giorgi C, Martiradonna A, Lanave C, Saccone C. Complete sequence of the mitochondrial DNA inthe sea urchin Arbacia lixula: Conserved features of the echinoid mitochondrial genome. Mol Phyl Evol. 1995;5:323–332. doi: 10.1006/mpev.1996.0027. [DOI] [PubMed] [Google Scholar]
- De Giorgi C, Martiradonna A, Pesole G, Saccone C. Lineage-specific evolution of echinoderm mitochondrial ATP synthase subunit 8. J Bioenerg Biomembr. 1997;29:233–239. doi: 10.1023/A:1022406026196. [DOI] [PubMed] [Google Scholar]
- Dreyer H, Steiner G. The complete nucleotide sequence and gene organization of the mitochondrial genome of the gadilid scaphopod Siphonondentalium lobatum (Mollusca) Mol Phylogenet Evol. 2004;31:605–617. doi: 10.1016/j.ympev.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992;141:173–216. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, De Bruijin M, Coulson A, Drouin J, Eperon I, Nierlich D, Roe B, Sanger F, Schreier P, Smith A, Staden R, Young I. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Lavrov DV, Boore JL, Brown WM. The complete mitochondrial DNA sequence of the horseshoe crab Limulus polyphemus. Mol Biol Evol. 2000;17:813–824. doi: 10.1093/oxfordjournals.molbev.a026360. [DOI] [PubMed] [Google Scholar]
- Scouras A, Beckenbach K, Arndt A, Smith MJ. Complete mitochondrial genome DNA sequence for two ophiuroids and a holothuroid: the utility of protein gene sequence and gene maps in the analyses of deep deuterostome phylogeny. Mol Phylogenet Evol. 2004;31:50–65. doi: 10.1016/j.ympev.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Jacobs HT, Elliott DJ, Math VB, Farquharson A. Nucleotide sequence and gene organization of sea urchin mitochondrial DNA. J Mol Biol. 1988;202:185–217. doi: 10.1016/0022-2836(88)90452-4. [DOI] [PubMed] [Google Scholar]
- Cantatore O, Roberti M, Rainaldi G, Gadaleta MN, Saccone C. The complete nucleotide sequence, gene organization, genetic code of the mitochondrial genome of Paracentrotus lividus. J Biol Chem. 1989;264:10965–10975. [PubMed] [Google Scholar]
- Wada H, Komatsu M, Satoh N. Mitochondrial rDNA phylogeny of the Asteroidea suggests the primitiveness of the Paxillosida. Mol Phylogenet Evol. 1996;6:97–106. doi: 10.1006/mpev.1996.0062. [DOI] [PubMed] [Google Scholar]
- Matsubara M, Komatsu M, Wada H. Close relationship between Asterina and Solasteridae (Asteroidea) supported by both nuclear and mitochondrial gene molecular phylogenies. Zool Sci. 2004;21:785–793. doi: 10.2108/zsj.21.785. [DOI] [PubMed] [Google Scholar]
- Madsen FJ. A note on the sea star genus Acanthaster. Vidensk Medd Dansk Naturhist Foren Kbh. 1955;117:179–192. [Google Scholar]
- Lucas JS, Jones MM. Hybrid crown-of-thorns starfish (Acanthaster planci X A.brevispinus) reared to maturity in the laboratory. Nature. 1976;263:409–412. doi: 10.1038/263409a0. [DOI] [PubMed] [Google Scholar]
- Oberst RD, Hays MP, Bohra LK, Phebus RK, Yamashiro CT, Pasxko-Kolva C, Flood SJA, Sargeant JM, Gillespie JR. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5' nuclease (Taq Man) assay. Appl Environ Microbiol. 1998;64:3389–3396. doi: 10.1128/aem.64.9.3389-3396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Nolte FS, Holloway BP, Morrison CJ. Rapid identification of up to three Candida species in a single reaction tube by a 5' exonuclease assay using fluorescent DNA probes. J Clin Microbiol. 1999;37:165–170. doi: 10.1128/jcm.37.1.165-170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Arctander P, Minelli A, Thomas RH, Vogler AP. A plea For DNA taxonomy. Trends Ecol Evol. 2003;18:70–74. doi: 10.1016/S0169-5347(02)00041-1. [DOI] [Google Scholar]
- Hebert PDN, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B. 2003. (DOI 10.1098/rsbl.2003.0025) [DOI] [PMC free article] [PubMed]
- Johns GC, Avise JC. A comparative summary of genetic distances in the vertebrates from the mitochondrial cytochrome b gene. Mol Biol Evol. 1998;15:1481–1490. doi: 10.1093/oxfordjournals.molbev.a025875. [DOI] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol & Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. Phylogenetic analysis using parsimony (and other methods), version 4. Sinauer Associates, Sunderland, MA, USA; 2002. [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei N. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1: Localization of genes and features in the mitochondrial genome of Acanthaster planci and A. brevispinus. Genes with asterisks are coded on the complimentary H strand.
Table 2: Comparison of base composition of the mitochondrial genomes of Acanthaster planci, A. brevispinus and Asterina pectinifera. * Data from Asakawa et al. [10].
Table 3: Total codon usage in the 13 protein-coding genes of Acanthaster planci, A. brevispinus and Asterina pectinifera. *Data from Asakawa et al. [10].
Table 4: Comparison of nucleotide and amino acid sequences in 13 protein-coding genes of Acanthaster planci, A. brevispinus and Asterina pectinifera.Genes that showed size variations are indicated as bold.
Table 5: Primer sequences for long PCR to amplify the mitochondrial DNA in A. planci and A. brevispinus.
Table 6: List of specimens used for phylogenetic analysis.


