Abstract
Background
An organ such as the bladder consists of complex, interacting set of tissues and cells. Inflammation has been implicated in every major disease of the bladder, including cancer, interstitial cystitis, and infection. However, scanty is the information about individual detrusor and urothelium transcriptomes in response to inflammation. Here, we used suppression subtractive hybridizations (SSH) to determine bladder tissue- and disease-specific genes and transcriptional regulatory elements (TRE)s. Unique TREs and genes were assembled into putative networks.
Results
It was found that the control bladder mucosa presented regulatory elements driving genes such as myosin light chain phosphatase and calponin 1 that influence the smooth muscle phenotype. In the control detrusor network the Pax-3 TRE was significantly over-represented. During development, the Pax-3 transcription factor (TF) maintains progenitor cells in an undifferentiated state whereas, during inflammation, Pax-3 was suppressed and genes involved in neuronal development (synapsin I) were up-regulated. Therefore, during inflammation, an increased maturation of neural progenitor cells in the muscle may underlie detrusor instability. NF-κB was specifically over-represented in the inflamed mucosa regulatory network. When the inflamed detrusor was compared to control, two major pathways were found, one encoding synapsin I, a neuron-specific phosphoprotein, and the other an important apoptotic protein, siva. In response to LPS-induced inflammation, the liver X receptor was over-represented in both mucosa and detrusor regulatory networks confirming a role for this nuclear receptor in LPS-induced gene expression.
Conclusion
A new approach for understanding bladder muscle-urothelium interaction was developed by assembling SSH, real time PCR, and TRE analysis results into regulatory networks. Interestingly, some of the TREs and their downstream transcripts originally involved in organogenesis and oncogenesis were also activated during inflammation. The latter represents an additional link between inflammation and cancer. The regulatory networks represent key targets for development of novel drugs targeting bladder diseases.
Background
The lower urinary tract is subject to a number of functional disorders in which a cross-communication between urothelium and detrusor muscle is a factor. Bladder overactivity has been attributed to detrusor muscle dysfunction, and several in vitro and in vivo methodologies have been developed to better understand its pathophysiology [1]. Although the detrusor muscle participates intensively in the inflammatory response, practically every major disease of the urinary bladder, including cancer, interstitial cystitis, and infection [2], involves the urothelium [3].
The urinary bladder develops as a result of indispensable epithelial-mesenchymal interactions responsible for directing urothelial differentiation and for normal smooth muscle development [4,5]. However, in adulthood, the urinary bladder is a highly heterogeneous organ, consisting of a large variety of cell types, and this complexity presents challenges to the study of physiological and cellular processes in health and disease. This was evident in studies determining gene regulation [6-11] and target validation [12]. In the latter study, the expression of protease-activating receptors (PAR) was differentially distributed between bladder mucosa, detrusor smooth muscle, and nerve elements. Moreover, during inflammation, PAR expression was up-regulated in the mucosa contrasting with its down-regulation in the detrusor muscle [12]. These results suggested a possible differential distribution of proteins between bladder mucosa and detrusor muscle and indicated the need for reduction of the whole tissue into specific layers.
The present work was undertaken to elucidate the transcriptional complexity of inflammation, as modeled in the mouse urinary bladder. The first step towards the study of individual layers was the separation of the mucosa and submucosa layers away from the detrusor smooth muscle and adventitia (See Additional file 1). We went further to determine low abundant transcripts, using suppression subtractive hybridization (SSH) and selected ones were confirmed by real time PCR. Next, the SSH-originated transcripts were annotated and analyzed for significantly enriched transcriptional regulatory elements (TRE) using PAINT [Promoter Analysis and Interaction Network Toolset; [13]]. PAINT utilizes TRANSFAC database [14] containing eukaryotic cis-acting regulatory DNA elements and trans-acting factors. The pattern search tool MATCH in TRANSFAC suite is employed to identify the TREs on cognate 5' upstream regulatory sequences [15]. Putative regulatory networks were assembled using known interactions among genes and their coded proteins as well as information about TREs that were significantly over-represented in the genes comprising inflammatory and control transcriptomes. Together, these results constitute the first demonstration of the transcriptional complexity underlying the different layers of the urinary bladder and their contribution to the early phases of bladder inflammation.
Results
SSH. From each library, at least 50 clones were further sequenced and annotated, for a total of 300 clones. The present work reports only tissue-specific transcripts (mucosa vs detrusor) and treatment-specific transcripts (saline vs LPS). 120 transcripts were considered to be unique. These indicate that each of these 120 transcript occurred in a specific subtraction. Transcripts that appeared in more than one subtraction were not considered unique and therefore, not included. Selection of clones ceased when a substantial number of additional sequences [approximately 180] did not reveal any additional unique transcript. Next, 120 clones were sequenced and analyzed for homology in the GenBank and EMBL databases.
Seventy six cDNA fragments (Table 2) were further annotated using the Mouse Genome Information [16] according to Gene Ontology and presented in Table 3. Among the 76 fragments selected for futher annotation, 21 fragments had homology with expressed sequence tags (ESTs) and cDNA clones for which information about tissue specificity, biologycal or molecular function is not available. Interestingly, one of these clones (ambladder; RIKEN clone:9530014P05) was originally isolated from an adult male bladder cDNA library [17].
Table 2.
Mouse bladder transcripts isolated by SSHs
| Abbrev | Name | Accession | Library | QPCR |
| 2310015N07 | RIKEN cDNA 2310015N07 gene | NM_025515 | MIDI | N |
| Actg1 | actin, gamma 1, cytoplasmic | NM_009609 | MIC | N |
| Actg2 | actin, gamma 2, smooth muscle, enteric | NM_009610 | MIDI | N |
| Actr3 | ARP3 actin-related protein 3 homolog (yeast) | NM_023735 | MIC | N |
| Ambladder | adult male urinary bladder cDNA, RIKEN clone:9530014P05 | AK020558 | MC | N |
| Amcq | adult male corpora quadrigemina cDNA, RIKEN clone:B230340L02 | AK080832 | MIC | Y |
| Aplp2 | amyloid beta (A4) precursor-like protein 2 isoform 751 | U15571 | MIDI | N |
| Bcap31 | B-cell receptor-associated protein 31 | NM_012060 | MIDI | Y |
| Calm2 | calmodulin 2 | NM_007589 | MC | N |
| Catenin | similar to catenin src | BC043108 | MIC | N |
| Cnn1 | calponin 1 | NM_009922 | MC | N |
| Col3a1 | collagen, type III, alpha 1, RIKEN 3200002K15 | AK019448 | MIDI | N |
| Cox7b | cytochrome c oxidase subunit VIIb | NM_025379 | MIC | N |
| CoxI | mitochondrial gene for subunit I of cytochrome c oxidase | X57780 | MIC | N |
| Cpe | carboxypeptidase E (Cpe), | NM_013494 | MIC | N |
| Cts e | cathepsin E (Ctse) | NM_007799 | MIC | N |
| Cts h | cathepsin H | NM_007801 | DC | N |
| Cts l | cathepsin L | NM_009984 | MIC | Y |
| Ddx3 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3 | NM_010028 | MIDI | Y |
| DnaJ | Hsp40 homolog, subfamily A, member 2 | NM_019794 | MIDI | N |
| Eif4ebp2 | eukaryotic translation initiation factor 4E binding protein 2 | NM_010124 | DIC | N |
| Elp3 | elongation protein 3 homolog, RIKEN clone:2610507P14 | AK012072 | MIDI | Y |
| Endomuc1 | endomucin-1 | BC003706 | MIC | N |
| Fth1 | ferritin heavy chain 1 | NM_010239 | MIC | N |
| Gpam/GPAT | glycerol-3-phosphate acyltransferase | NM_008149 | MIC | Y |
| Grn | epithelin 1 and 2 (granulin) | X62321 | MC | N |
| Grp58 | glucose regulated protein | BC003285 | MIC | Y |
| Gst a4 | glutathione S-transferase, alpha 4 | NM_010357 | MIDI | N |
| Gst m1 | glutathione S-transferase, mu 1 | NM_010358 | DC | Y |
| Gst o1 | glutathione S-transferase omega 1 | NM_010362 | DC | Y |
| Gus | beta-glucuronidase | NM_010368 | MIDI | N |
| IFIT3 | interferon-induced protein with tetratricopeptide repeats 3 | BC003804 | MIC | Y |
| Ifrg15 | interferon alpha responsive gene | NM_022329 | DC | N |
| Lamp2 | lysosomal membrane glycoprotein 2 | NM_010685 | MIC | N |
| Lrrfip2 | leucine rich repeat (in FLII) interacting protein 2 | XM_284541 | MIDI | Y |
| Ly6d | mRNA for THB / lymphocyte antigen 6 complex, locus D | X63782 | MIDI | N |
| Mafb | transcription factor MAFB | AF180338 | MIC | N |
| Mgs1-182e11 | clone mgs1-182e11 strain 129/SvJ | AC096622 | MIDI | Y |
| MLZE | adult female vagina cDNA, RIKEN clone 9930109F21 | AK037079 | MIDI | N |
| Mpp1 | palmytoylated protein p55 | U38196 | DIC | N |
| My h11 | myosin heavy chain 11, smooth muscle | XM_147228 | MIDI | Y |
| My lc2b/MRLC2 | myosin light chain, regulatory B | NM_023402 | MC | Y |
| My19 | myosin, light polypeptide 9, regulatory | XM_283793 | MIDI | N |
| Pls3 | similar to plastin 3 precursor (T-isoform) | NM_145629 | MIC | Y |
| Prdx1 | peroxiredoxin 1 | NM_011034 | MIDI | N |
| Prnp | prion protein | NM_011170 | DC | Y |
| Prss11 | protease, serine, 11 (Igf binding) HtrA1 | NM_019564 | MIC | N |
| RP23-452N23 | clone RP23-452N23 on chromosome 4 | AL928645 | MIC | N |
| RP23-70M6 | chromosome 18 clone RP23-70M6 | AC114820 | MIC | N |
| RPCI23 | Strain C57BL6/J Chromosome 2, clone RP23-111A22 | AC078911 | MIDI | Y |
| RPs21 | ribosomal protein S21 / RIKEN cDNA 2410030A14 gene | BC027563 | MIC | N |
| RPs7 | ribosomal protein S7 | NM_011300 | MIDI | Y |
| SAC1 | suppressor of actin mutations | AJ245720 | MC | Y |
| SC1 | extracellular matrix protein precursor | U77330 | MC | N |
| SCP-1 | SCP-1 mRNA for stromal cell derived protein-1 | D16847 | MIC | Y |
| Sdfr1 | stromal cell derived factor receptor 1 | NM_009145 | DC | N |
| Sepp1/SePP | selenoprotein P, plasma,1 glycoprotein | NM_009155 | MIC | Y |
| Serpina3n | serine (or cysteine) proteinase inhibitor (clade A, member 3N) | NM_009252 | DC | Y |
| Sf 3b2 | splicing factor 3b, subunit 2, clone MGC:61326 IMAGE:6812422 | BC049118 | MIDI | N |
| Sf rs6 | splicing factor, arginine/serine-rich 6 | NM_026499 | MIC | N |
| Sf sc35 | splicing factor SC35 | AF077858 | MIDI | N |
| Sirt1 | Sir1 alpha protein | AF214646 | MIDI | N |
| Siva | Cd27 binding proapoptotic protein | NM_013929 | DIC | Y |
| Smoc2 | SPARC related modular calcium binding 2 | NM_022315 | MIC | N |
| Sparc | secreted acidic cysteine rich glycoprotein | NM_009242 | MIDI | N |
| Sprr 2A | small proline-rich protein 2A | BC010818 | MC | Y |
| Syn1 | synapsin I or ribosomal protein S15a | NM_013680 | DIC | Y |
| Tcra-V13.1 | T-cell receptor alpha/delta locus | AE008684 | MIC | N |
| Thsd6 | thrombospondin, type I domain containing 6 | NM_025629 | DC | Y |
| TMPRSS2 | transmembrane protease, serine 2 | NM_015775 | MIDI | N |
| Tnnt3 | troponin T3, skeletal, fast | NM_011620 | MIDI | N |
| UDP-gluco | UDP-glucuronosyltransferase 1 family, member 2 | BC019434 | MIC | N |
| Upk1a | uroplakin 1a | AF262335 | DC | N |
| Upk1b | uroplakin 1b | NM_178924 | DC | N |
| Wdr1 | WD repeat domain 1 | NM_011715 | MIDI | N |
| Zfp364 | zinc finger protein 364/Rab7 | NM_026406 | MIDI | N |
Table 3.
ANNOTATION (GENE ONTOLOGY) OF SSH-ISOLATED TRANSCRIPT FROM MOUSE URINARY BLADDER
| Biological Process | Abbrev | Molecular Function | Cellular Component | Accession |
| actin dynamics | Wdr1 | actin binding | actin cytoskeleton | NM_011715 |
| apoptosis | Sirt1 | NAD-dependent histone deacetylase | chromatin silencing complex | AF214646 |
| apoptosis | Bcap31 | receptor binding | Golgi membrane | NM_012060 |
| apoptosis | Siva | CD27 receptor binding | cytoplasm | NM_013929 |
| calcium ion binding | SCP-1 | calcium ion binding | integral to membrane | D16847 |
| calcium ion binding | Sparc | calcium ion binding | basement membrane | NM_009242 |
| calcium ion binding | Smoc2 | calcium ion binding | extracellular space | NM_022315 |
| calcium ion binding | Pls3 | calcium ion binding | unknown | NM_145629 |
| calcium ion binding | SC1 | calcium ion binding | extracellular space | U77330 |
| cell adhesion | Col3a1 | extracellular matrix structural constituent | collagen | AK019448 |
| cell adhesion | Catenin | protein binding | cytoskeleton | BC043108 |
| cell cycle/G-protein coupled receptor | Calm2 | protein binding | plasma membrane | NM_007589 |
| cell growth (regulation) | Prss11 | insulin-like growth factor binding | extracellular region | NM_019564 |
| cell motility | Actr3 | structural molecule | actin cytoskeleton | NM_023735 |
| cytoskeleton organization and biogenesis | Actg1 | motor activity | actin cytoskeleton | NM_009609 |
| cytoskeleton organization and biogenesis | Actg2 | motor activity | actin cytoskeleton | NM_009610 |
| cytoskeleton organization and biogenesis | My lc2b | unknown | cytoskeleton | NM_023402 |
| defense response | Ly6d | unknown | plasma membrane | X63782 |
| electron transport | Grp58 | electron transporter | endoplasmic reticulum | BC003285 |
| electron transport | Cox7b | oxidoreductase activity | mitochondrial electron transport chain | NM_025379 |
| electron transport | CoxI | ubiquinol-cytochrome-c reductase activity | mitochondrion | X57780 |
| epithelial cell proliferation | Grn | phospholipase A2 | mitochondrion | X62321 |
| immune response | IFIT3 | unknown | unknown | BC003804 |
| insulin processing | Cpe | carboxypeptidase A and E activity | extracellular space | NM_013494 |
| iron ion transport | Fth1 | ferric iron binding | unknown | NM_010239 |
| metabolism | Gst a4 | glutathione transferase | unknown | NM_010357 |
| metabolism | Gst m1 | glutathione transferase | unknown | NM_010358 |
| metabolism | Gst o1 | glutathione transferase | cytoplasm | NM_010362 |
| negative regulation of translational initiation | Eif4ebp2 | insulin receptor signaling pathway | unknown | NM_010124 |
| nuclear mRNA splicing, via spliceosome | Sf sc35 | DNA binding | spliceosome complex | AF077858 |
| nuclear mRNA splicing, via spliceosome | Sf 3b2 | unknown | nucleus | BC049118 |
| nuclear mRNA splicing, via spliceosome | Sf rs6 | pre-mRNA splicing factor | nucleus | NM_026499 |
| phospholipid biosynthesis | Gpam | acyltransferase | mitochondrion | NM_008149 |
| positive regulation of transcription | Mafb | DNA binding | transcription factor complex | AF180338 |
| post-embryonic development | Sepp1 | selenium binding | extracellular space | NM_009155 |
| protein biosynthesis | RPs21 | structural constituent of ribosome | ribosome | BC027563 |
| protein biosynthesis | RPs7 | RNA binding | ribosome | NM_011300 |
| protein folding | DnaJ | unfolded protein binding | membrane | NM_019794 |
| protein ubiquitination | Zfp364 | ubiquitin-protein ligase activity | ubiquitin ligase complex | NM_026406 |
| proteolysis and peptidolysis | Cts e | neutrophil collagenase activity | extracellular space | NM_007799 |
| proteolysis and peptidolysis | Cts h | cysteine-type endopeptidase activity | lysosome | NM_007801 |
| proteolysis and peptidolysis | Cts l | cysteine-type endopeptidase activity | lysosome | NM_009984 |
| proteolysis and peptidolysis | TMPRSS2 | trypsin activity | integral to membrane | NM_015775 |
| regulation of cell shape | Sprr 2A | constituent of cytoskeleton | cornified envelope | BC010818 |
| regulation of muscle contraction | Cnn1 | calmodulin binding | unknown | NM_009922 |
| regulation of muscle contraction | Myl19 | calcium ion binding | myosin | XM_283793 |
| response to oxidative stress | Prdx1 | antioxidant activity | unknown | NM_011034 |
| response to oxidative stress | Prnp | copper ion binding | Golgi apparatus | NM_011170 |
| synaptic transmission | Syn1 | protein dimerization | synaptic vesicle membrane | NM_013680 |
| tRNA aminoacylation | Lamp2 | tRNA ligase | platelet dense granule membrane | NM_010685 |
| unknown | RPCI23 | unknown | unknown | AC078911 |
| unknown | Mgs1-182e11 | unknown | unknown | AC096622 |
| unknown | RP23-70M6 | unknown | unknown | AC114820 |
| unknown | Tcra-V13.1 | unknown | unknown | AE008684 |
| unknown | Upk1a | unknown | integral to membrane | AF262335 |
| unknown | SAC1 | unknown | integral to membrane | AJ245720 |
| unknown | Elp3 | N-acetyltransferase activity | mitochondrion | AK012072 |
| unknown | Ambladder | unknown | unknown | AK020558 |
| unknown | MLZE | unknown | unknown | AK037079 |
| unknown | Amcq | unknown | unknown | AK080832 |
| unknown | RP23-452N23 | unknown | unknown | AL928645 |
| unknown | Endomuc1 | unknown | integral to membrane | BC003706 |
| unknown | UDP-gluco | unknown | unknown | BC019434 |
| unknown | Sdfr1 | receptor activity | integral to membrane | NM_009145 |
| unknown | Serpina3n | endopeptidase inhibitor | extracellular space | NM_009252 |
| unknown | Ddx3 | ATP-dependent helicase activity | intracellular | NM_010028 |
| unknown | Gus | unknown | unknown | NM_010368 |
| unknown | Tnnt3 | unknown | unknown | NM_011620 |
| unknown | Ifrg15 | unknown | unknown | NM_022329 |
| unknown | 2310015N07 | unknown | unknown | NM_025515 |
| unknown | Thsd6 | unknown | extracellular space | NM_025629 |
| unknown | Upk1b | unknown | integral to membrane | NM_178924 |
| unknown | Aplp2 | serine-type endopeptidase inhibitor activity | integral to membrane | U15571 |
| unknown | Mpp1 | protein binding | membrane | U38196 |
| unknown | My h11 | unknown | unknown | XM_147228 |
| unknown | Lrrfip2 | unknown | unknown | XM_284541 |
SSH-selected transcripts
Table 3 summarizes the isolated transcripts that in general are involved in: actin dynamics (wdr1); apoptosis (sirt1, siva, and bcap31); calcium ion binding (pls3, sc1, scp-1, sparc, and smoc2); cell adhesion (col3a1 and catenin); cell cycle/ G-protein coupled receptor (cam2); cell growth (prss11); cell motility (actr3); cytoskeleton organization and biogenesis (actg1, actg2, and mylc2b);defense response (ly6d); electron transport (cox7b, grp58, and coxI); epithelial cell proliferation (grn); immune response (ifit3); insulin processing (cpe); iron ion transport (fth1); metabolism/ glutathione transferase activity (gsto1, gsta4, and gstm1); negative regulation of translational initiation (eif4ebp2); nuclear mRNA splicing (sfsc35 and sfrs6); phospholipid biosynthesis (gpam); positive regulation of transcription (mafb); post-embryonic development (sepp1); protein biosynthesis (rps7 and rps21); protein folding (dnaJ); protein ubiquitination (zfp364); proteolysis (ctse, ctsh, ctsl, Aplp2, serpina3n, and tmprss2); regulation of cell shape (sprr2A); regulation of muscle contraction (my19 and cnn1); response to oxidative stress (prdx1 and prnp); synaptic transmission (syn1); and tRNA aminoacylation (lamp2).
Target validation by quantitative real-time polymerase chain reaction (QRT-PCR)
From the annotated transcripts (Table 2), twenty six were selected for further analysis by QRT-PCR. The results are summarized in Table 4. In tissues isolated from saline-treated mice the following transcripts were expressed preferentially in the detrusor muscle when compared to the mucosa layer: ctsh, eif4ebp2, gstm1, gsto1, serpina3n, sprr2A, upk1a, and upk1b. With the exception of serpina3n, all detrusor-specific transcripts were also preferentially up regulated in the inflamed detrusor. In contrast, calm2, cnn1, and smoc2 were preferentially expressed in the bladder mucosa of control mice. With the exception of calm2, all other mucosa-specific transcripts were up-regulated during inflammation. Table 4 also segregates transcripts that were represented in both layers of control mice and that were up-regulated during the inflammatory process. The latter include: bcap31, catenin, pls3, mafb, prss11, mpp1, syn1, lamp2, and sepp1.
Table 4.
Differential expression of selected SSH transcripts by quantitative real-time polymerase chain reaction (QRT-PCR) ***
| Genes | Normalized CT values | log2 CT X1000000 | Fold Change (Delta CT Values)* | ||||||||||||||
| Average (n = 3) | SEM | ||||||||||||||||
| LD | LM | CD | CM | LD | LM | CD | CM | LD | LM | CD | CM | CD/CM | CM/CD | LD/LM | LD/CD | LM/CM | |
| Cts h | 26.5 | 24.2 | 26.1 | 24.1 | 0.03 | 0.01 | 0.05 | 0.01 | 96 | 19 | 70 | 18 | 3.9 | 0.3 | 5.0 | 1.4 | 1.1 |
| Eif4ebp2 | 36.5 | 30.4 | 33.7 | 30.9 | 0.62 | 0.07 | 0.07 | 0.05 | 95252 | 1430 | 13833 | 1974 | 7.0 | 0.1 | 66.6 | 6.9 | 0.7 |
| Gst m1 | 23.4 | 19.1 | 23.2 | 18.2 | 0.32 | 0.02 | 0.00 | 0.06 | 11 | 1 | 9 | 0 | 29.9 | 0.0 | 19.7 | 1.2 | 1.8 |
| Gst o1 | 36.0 | 26.9 | 35.5 | 29.8 | 0.14 | 0.17 | 0.56 | 0.06 | 68660 | 125 | 47023 | 909 | 51.7 | 0.0 | 550.9 | 1.5 | 0.1 |
| Serpina3n | 21.8 | 21.1 | 26.1 | 23.5 | 0.03 | 0.02 | 0.09 | 0.05 | 4 | 2 | 72 | 12 | 6.1 | 0.2 | 1.6 | 0.1 | 0.2 |
| Sprr 2A | 29.4 | 21.8 | 25.4 | 19.8 | 0.06 | 0.14 | 0.03 | 0.00 | 697 | 4 | 45 | 1 | 51.0 | 0.0 | 185.1 | 15.3 | 4.2 |
| Upk1a | 32.5 | 24.4 | 30.0 | 23.7 | 0.18 | 0.01 | 0.02 | 0.13 | 6162 | 23 | 1050 | 14 | 76.4 | 0.0 | 271.4 | 5.9 | 1.7 |
| Upk1b | 26.5 | 22.2 | 26.7 | 21.4 | 0.04 | 0.02 | 0.12 | 0.05 | 98 | 5 | 112 | 3 | 39.3 | 0.0 | 20.6 | 0.9 | 1.7 |
| Calm2 | 23.2 | 24.0 | 22.2 | 23.8 | 0.11 | 0.09 | 0.04 | 0.11 | 9 | 17 | 5 | 14 | 0.3 | 3.0 | 0.6 | 2.0 | 1.2 |
| Cnn1 | 32.1 | 33.1 | 27.2 | 31.0 | 0.50 | 0.09 | 0.15 | 0.05 | 4547 | 9400 | 149 | 2177 | 0.1 | 14.6 | 0.5 | 30.5 | 4.3 |
| Smoc2 | 33.9 | 34.7 | 26.0 | 28.4 | 0.06 | 0.27 | 0.04 | 0.06 | 15953 | 27402 | 68 | 352 | 0.2 | 5.2 | 0.6 | 234.8 | 77.7 |
| Bcap31 | 33.3 | 29.2 | 26.7 | 26.9 | 0.40 | 0.07 | 0.08 | 0.01 | 10331 | 627 | 106 | 127 | 0.8 | 1.2 | 16.5 | 97.6 | 4.9 |
| Catenin | 36.9 | 33.0 | 27.1 | 28.0 | 0.23 | 0.07 | 0.42 | 0.09 | 130085 | 8592 | 143 | 273 | 0.5 | 1.9 | 15.1 | 912.8 | 31.5 |
| Mafb | 32.1 | 30.1 | 31.8 | 30.5 | 0.65 | 0.10 | 0.34 | 0.44 | 4703 | 1154 | 3615 | 1570 | 2.3 | 0.4 | 4.1 | 1.3 | 0.7 |
| Pls3 | 37.4 | 35.3 | 28.6 | 29.3 | 0.10 | 0.18 | 0.14 | 0.15 | 179012 | 41906 | 402 | 661 | 0.6 | 1.6 | 4.3 | 444.9 | 63.4 |
| Prss11 | 29.9 | 27.9 | 25.8 | 24.5 | 0.05 | 0.03 | 0.06 | 0.00 | 999 | 259 | 59 | 23 | 2.5 | 0.4 | 3.9 | 17.1 | 11.0 |
| Syn1 | 30.1 | 25.3 | 24.1 | 22.8 | 0.22 | 0.04 | 0.21 | 0.13 | 1148 | 42 | 18 | 7 | 2.4 | 0.4 | 27.3 | 65.6 | 5.9 |
| Lamp2 | 31.6 | 31.1 | 28.1 | 27.8 | 0.21 | 0.14 | 0.05 | 0.01 | 3342 | 2379 | 285 | 230 | 1.2 | 0.8 | 1.4 | 11.7 | 10.3 |
| Mpp1 | 31.0 | 30.0 | 29.2 | 29.5 | 0.44 | 0.11 | 0.11 | 0.19 | 2114 | 1090 | 632 | 753 | 0.8 | 1.2 | 1.9 | 3.3 | 1.4 |
| Sepp1 | 23.0 | 23.9 | 22.0 | 21.6 | 0.06 | 0.03 | 0.08 | 0.05 | 8 | 15 | 4 | 3 | 1.4 | 0.7 | 0.6 | 2.0 | 4.8 |
| Thsd6 | 28.1 | 28.3 | 28.3 | 28.0 | 0.13 | 0.08 | 0.03 | 0.04 | 293 | 329 | 338 | 272 | 1.2 | 0.8 | 0.9 | 0.9 | 1.2 |
| IFIT3 | 30.4 | 31.2 | 30.8 | 30.9 | 0.08 | 0.00 | 0.12 | 0.05 | 1455 | 2396 | 1885 | 1988 | 0.9 | 1.1 | 0.6 | 0.8 | 1.2 |
| Ifrg15 | 27.9 | 27.9 | 28.2 | 27.6 | 0.03 | 0.05 | 0.09 | 0.07 | 254 | 245 | 307 | 210 | 1.5 | 0.7 | 1.0 | 0.8 | 1.2 |
| Sdfr1 | 24.4 | 25.6 | 24.1 | 24.8 | 0.06 | 0.09 | 0.10 | 0.09 | 23 | 52 | 18 | 29 | 0.6 | 1.6 | 0.4 | 1.3 | 1.8 |
| Siva | 26.7 | 25.8 | 27.1 | 25.8 | 0.03 | 0.07 | 0.06 | 0.07 | 110 | 59 | 147 | 59 | 2.5 | 0.4 | 1.9 | 0.7 | 1.0 |
| Prnp | 24.1 | 25.7 | 23.3 | 24.6 | 0.02 | 0.07 | 0.02 | 0.00 | 18 | 53 | 11 | 26 | 0.4 | 2.5 | 0.3 | 1.7 | 2.0 |
*(ratio of antilog2 of cycle threshold values)
** (bolded cells indicate values greater than 3.0)
***Female C57BL/6J mice were instilled with saline (n = 20) or LPS (n = 20). Twenty four hours after LPS instillation, mice were euthanized, the bladder was removed and placed in RNAlater™ (Ambion) for separation of the mucosa and submucosa from the detrusor smooth muscle, as described in Material and Methods. Four sample groups were obtained as follows: control mucosa (CM), control detrusor (CD), LPS-treated mucosa (LM), and LPS-treated detrusor (LD). The QRT-PCR amplifications were accomplished on an ABIPRISM 7700 using SYBRGreen I dye assay chemistry
All samples were run in triplicate with the appropriate single QRT-PCR controls (no reverse transcriptase and no template). From the QRT-PCR data, an average cycle threshold (Ct) value was calculated from the triplicate reactions. Averaged Ct values were then normalized (to adjust for different amounts of cDNA within each reaction) to the exongenous control gene, RCA. The relative expression level of each transcript within each sample group (CD, CM, LD, and LM) was determined by calculating the ratio of the antilog2 of the delta Ct values.
TREs
Figure 2 contains over-represented TREs and the downstream transcripts that were found primarily in the bladder mucosa when compared to detrusor muscle isolated from control mice (MC). In contrast, figure 5 contains over-represented TFs and transcripts that were found primarily in the control detrusor muscle (DC). TFs and downstream transcripts specifically expressed in bladder mucosa during inflammation were compared to control mucosa (Figure 3) or to the inflamed detrusor (Figure 4). In contrast, the response of detrusor muscle to inflammation was determined in comparison to detrusor control (Figure 6) or inflamed mucosa (Figure 7). The results presented here demonstrate the concept that combining SSH methodology with PAINT-guided transcriptional regulatory element analysis permitted the generation of testable hypotheses regarding differences between mucosa and detrusor regulatory networks in health and disease states.
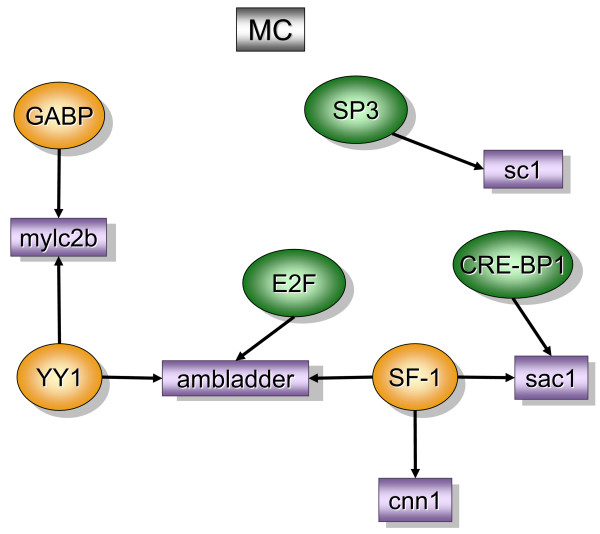
Figure 2.
Transcripts and TREs over-represented in the control mucosa when compared to control detrusor (MC). The regulatory network was determined by a combination of SSH-selected transcripts (green) and PAINT 3.3 query of transcription factor database (TRANSFAC). PAINT 3.3 was employed to examine 2000 base pairs of regulatory region upstream of the transcriptional start site of each differentially expressed transcript detected with the SSH. Genbank accession numbers were used as the gene identifiers in PAINT test files. Individual elements of the matrix are colored by the significance p-values. Over-representation in the matrix when compared to the reference (all TFs in the PAINT database) is indicated in orange (0 < p < = 0.01) or green (0.01 < p < 0.05). For a detailed origin of each library please see Figure 1 and Material and methods.
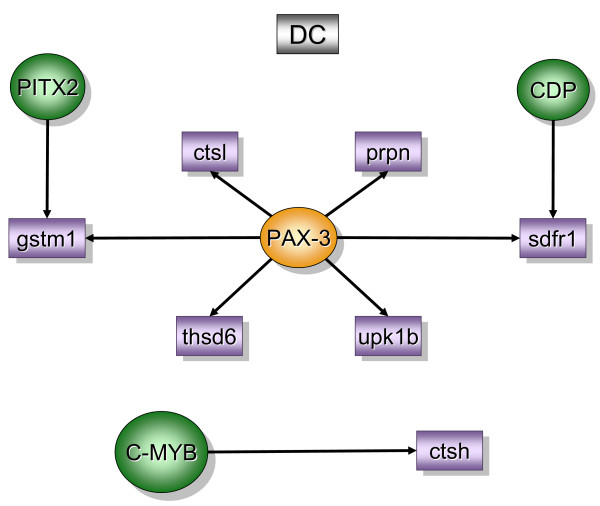
Figure 5.
Transcripts and TREs over-represented in the control detrusor when compared to control mucosa (DC). The regulatory network was determined by a combination of SSH-selected transcripts (green) and PAINT 3.3 query of transcription factor database (TRANSFAC). PAINT 3.3 was employed to examine 2000 base pairs of regulatory region upstream of the transcriptional start site of each differentially expressed transcript detected with the SSH. Genbank accession numbers were used as the gene identifiers in PAINT test files. Individual elements of the matrix are colored by the significance p-values. Over-representation in the matrix when compared to the reference (all TFs in the PAINT database) is indicated in orange (0 < p < = 0.01) or green (0.01 < p < 0.05). For a detailed origin of each library please see Figure 1 and Material and methods.
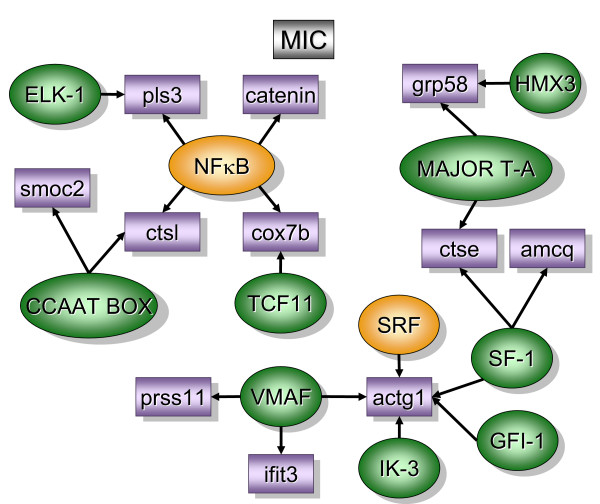
Figure 3.
Transcripts and TREs over-represented in the mucosa inflamed when compared to control mucosa (MIC). The regulatory network was determined by a combination of SSH-selected transcripts (green) and PAINT 3.3 query of transcription factor database (TRANSFAC). PAINT 3.3 was employed to examine 2000 base pairs of regulatory region upstream of the transcriptional start site of each differentially expressed transcript detected with the SSH. Genbank accession numbers were used as the gene identifiers in PAINT test files. Individual elements of the matrix are colored by the significance p-values. Over-representation in the matrix when compared to the reference (all TFs in the PAINT database) is indicated in orange (0 < p < = 0.01) or green (0.01 < p < 0.05). For a detailed origin of each library please see Figure 1 and Material and methods.
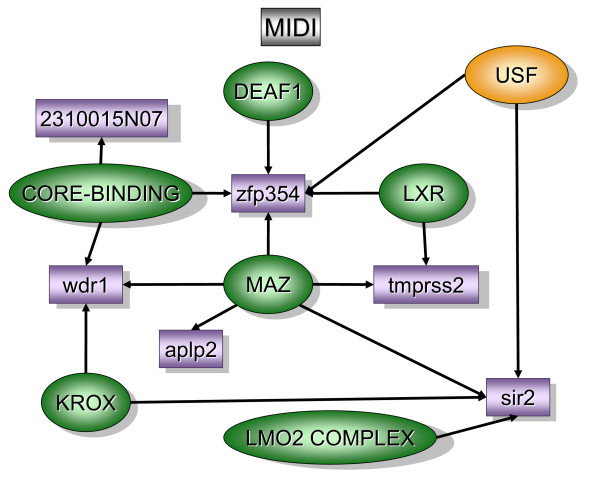
Figure 4.
Transcripts and TREs over-represented in the mucosa inflamed when compared to detrusor inflamed (MIDI). The regulatory network was determined by a combination of SSH-selected transcripts (green) and PAINT 3.3 query of transcription factor database (TRANSFAC). PAINT 3.3 was employed to examine 2000 base pairs of regulatory region upstream of the transcriptional start site of each differentially expressed transcript detected with the SSH. Genbank accession numbers were used as the gene identifiers in PAINT test files. Individual elements of the matrix are colored by the significance p-values. Over-representation in the matrix when compared to the reference (all TFs in the PAINT database) is indicated in orange (0 < p < = 0.01) or green (0.01<p < 0.05).
Figure 6.
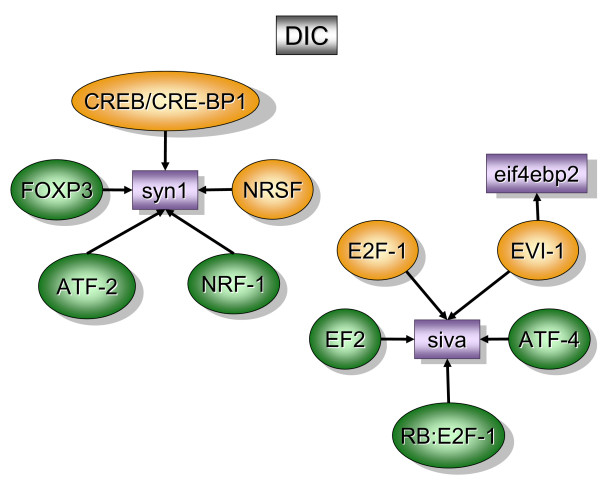
Transcripts and TREs over-represented in the detrusor inflamed when compared to detrusor control (DIC). The regulatory network was determined by a combination of SSH-selected transcripts (green) and PAINT 3.3 query of transcription factor database (TRANSFAC). PAINT 3.3 was employed to examine 2000 base pairs of regulatory region upstream of the transcriptional start site of each differentially expressed transcript detected with the SSH. Genbank accession numbers were used as the gene identifiers in PAINT test files. Individual elements of the matrix are colored by the significance p-values. Over-representation in the matrix when compared to the reference (all TFs in the PAINT database) is indicated in orange (0 < p < = 0.01) or green (0.01 < p < 0.05). For a detailed origin of each library please see Figure 1 and Material and methods.
Figure 7.
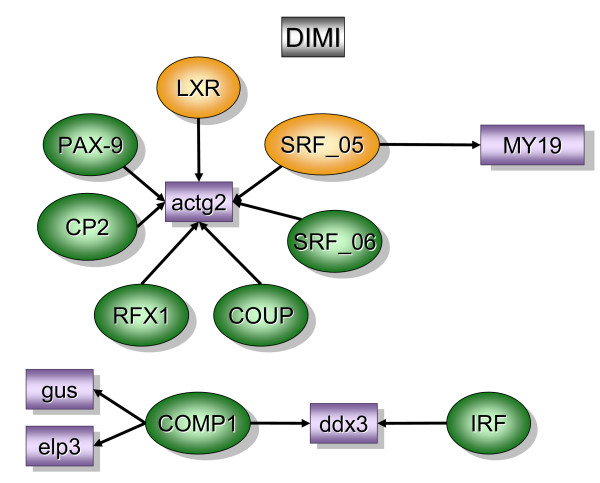
Transcripts and TREs over-represented in the detrusor inflamed when compared to mucosa inflamed (DIMI). The regulatory network was determined by a combination of SSH-selected transcripts (green) and PAINT 3.3 query of transcription factor database (TRANSFAC). PAINT 3.3 was employed to examine 2000 base pairs of regulatory region upstream of the transcriptional start site of each differentially expressed transcript detected with the SSH. Genbank accession numbers were used as the gene identifiers in PAINT test files. Individual elements of the matrix are colored by the significance p-values. Over-representation in the matrix when compared to the reference (all TFs in the PAINT database) is indicated in orange (0 < p < = 0.01) or green (0.01 < p < 0.05). For a detailed origin of each library please see Figure 1 and Material and methods.
Discussion
We used a highly effective method for differential gene analysis, termed suppression subtractive hybridization (SSH), which has been developed for the generation of subtracted cDNA libraries. It is based primarily on suppression PCR and combines normalization and subtraction in a single procedure [18]. The normalization step equalizes the abundance of cDNAs within the target population and the subtraction step excludes the common sequences between the target and driver populations. In a model system, the SSH technique enriched for rare sequences over 1,000-fold in one round of subtractive hybridization [18]. Unlike microarrays, which mainly identify moderate to high abundant genes, SSH identifies clones that are expressed at very low levels. It is possible that some of the extremely low-level gene expression is not biologically significant, as it might arise from 'random transcription' [19]. Therefore, we confirmed the differential expression of twenty six SSH-selected transcripts by QRT-PCR and the results were highly correlated. In addition, we introduced a redundancy factor by comparing the data generated from the analysis of multiple SSH libraries (MC, DC, MIC, MIDI, DID, and DIMI). Nevertheles, the question of validation certainly is an important one, and it is one we have considered. For one, we examined properties of the candidate genes that were independent of expression, such as the presence of promoter sequences, to increase the probability that mechanisms suggested by the expression data were not simply statistical anomalies. Moreover, the finding that mechanisms known to be operant emerged from the analysis also increases confidence that novel ones are valid as well.
This study was carried out at a single time point. The 24 hour time point was chosen because it coincides with the peak of acute inflammation [8,9]. This point characterizes the acute inflammatory responses to LPS and in addition to edema and vasodilation, the peak response of neutrophil infiltration occurred at 24 h in the mucosa and submucosa as well as in the detrusor smooth muscle [8,9]. The rationale for a single time point was to keep the number of variables within a reasonable limit. We understand that 24 h time point may not identify preceding or proceeding events as we indicated previously [7-9]. Therefore, the results of regulatory network here presented should be viewed as a snapshot of the inflammatory transcriptome at the point of maximal inflammatory response. Nevertheless, genes identified as important at this time point can now be followed over time by other techniques.
By using cDNA subtractions between mucosa and detrusor smooth muscle layers isolated from control and inflamed bladders, several clones identified transcripts that were further annotated. It has to be taken in consideration that the mucosal layer contains the urothelial layer and the lamina propria which involves other cell types (fibroblasts, myofibroblasts, etc.) in addition to urothelial cells that may underlie the transcriptomes identified. In addition, during the inflammation, inflammatory cells are present in the detrusor as well as in the mucosa and may contribute to the inflammatory bladder transcriptome.
Some of the transcripts are RIKEN sequences and therefore, have no biological process attributed. Interestingly, some of these transcripts with unknown function, such as ambladder, have been reported in the adult male urinary bladder by others using SSH [17]. However, the present work indicates that the same transcript was isolated from female urinary bladder as well.
Central to elucidation of hypothetical cis-regulatory networks is the identification and classification of naturally occurring transcription factor-binding sites in subtracted libraries. The combination of SSH-derived transcripts with PAINT-guided query of the TRANSFAC database permitted the generation of regulatory networks containing upstream TREs connecting corresponding TFs to the respective transcripts. Interestingly, many of the genes identified by SSH are transcription factors which emphasize the use of SSH to identify low expression transcripts.
Mucosa control [MC], (figure 2)
The regulatory network for the control bladder mucosa (MC) was obtained in comparison to detrusor control and contained the following over-represented TREs: GA-binding protein (GABP), YY1 (yin yang 1), SF-1 (steroidogenic factor-1), EF2, CRE-BP1, and Sp-3 (trans-acting transcription factor 3).
GABP, also known as nuclear respiratory factor 2 (NRF-2), is a transcriptional coordinator of mitochondrial and nuclear-encoded subunits of cytochrome oxidase genes [20]. NRF-2 responds to increased neuronal activity by translocating from the cytoplasm to the nucleus, where it engages in transcriptional activation of target genes [21]. GABP is abundant in the kidney [22], however, the information about GABP is scanty in the rest lower urinary tract.
YY1 is a zinc finger TF which is thought to regulate cell growth and differentiation. YY1 normally antagonizes serum response factor (SRF) [23]. Interestingly, in the inflamed mucosa YY1 was not expressed and SRF was considered over-represented (Figure 2). YY1 along with GAPB drives the expression of myosin light chain phosphatase (mylc2b) which can be developmentally regulated in mammalian urinary bladders [24] and it is involved in the bladder response to obstruction [25].
SF-1 is a zinc finger motif of nuclear receptors [26] essential for steroidogenesis as well as for the development of the reproductive axis [27]. CRE-BP1, which is an ubiquitous basic-leucine zipper, is required for normal skeletal development.
YY1, SF-1, and EF2 were found upstream of ambladder (RIKEN clone 9530014P05 or prothymosin alpha). Prothymosin alpha is an oxidative stress-protecting gene [28] and transgenic mice over-expressing this transcript develop polycystic kidney disease PKD [29]. Another gene downstream of SF-1 was calponin 1 (cnn1). Cnn1 encodes for a multifunctional protein whose expression is tightly restricted to differentiated smooth muscle cell lineages during embryonic and post-natal life [30].
SF-1 and CRE-BP1 were found upstream of sac1 (a murine homolog of S. cerevisiae suppressor of actin mutations). Finally, an additional TRE, Sp-3 was found upstream of sc1 which is a calcium binging protein and an extracellular matrix protein precursor.
Epithelial-stromal interactions in bladder development have been extensively studied [5]. However, the present results depicted in the MC regulatory network raises the hypothesis that the bladder mucosa exhibits TREs and genes whose proteins may regulate the smooth muscle phenotype (Figure 2).
Detrusor control [DC] (figure 5)
The regulatory network for the control detrusor smooth muscle suggests that Pax-3 plays a central role. In addition to Pax-3, other TREs such as: PITX2, CDP, and c-Myb were also found over-represented (figure 5).
Pax-3 was found upstream of the following transcripts: prion protein (prnp), cathepsin L (ctsl), stromal cell derived factor receptor 1 (sdfr1) [31], thrombospondin, type I domain containing 6 (thsd6), and uroplakin 1b (upk1b). Prnp seems to be involved in cell-to-cell interaction and its expression has been observed in germ cell differentiation during spermatogenesis [32]. Ctsl is a lysosomal proteolytic enzyme and an imbalance between ctsl and its inhibitors is believed to correlate with bladder tumor progression [33].
In addition to ctsl, the detrusor also presents ctsh under the control of c-Myb. Interestingly, thsd is involved in bladder cancer development [34] and is a target for methylation [35]. In addition, thsd has been described as inhibitor of angiogenesis. Finally, upk1b gene is highly expressed in normal human urothelium and its mRNA was undetectable or markedly reduced in bladder carcinoma [36].
Interestingly, the control detrusor normally expresses genes in the Pax-3 pathway that maintain neural progenitor cells [37] and myoblasts [38] undifferentiated. Therefore, our data suggests that Pax-3-regulated suppression of neural development in control detrusor changed substantially during inflammation and genes involved in neuronal development such as syn were found to be up-regulated. The later implies that detrusor instability may be a consequence of alterations in Pax-3 pathway leading to increased maturation of neural progenitor cells within the bladder smooth muscle.
Mucosa inflamed versus control [MIC]. (figure 3)
Of all regulatory networks (Figures 2, 3, 4, 5, 6, 7), MIC was the only one presenting NF-κB (p < 0.01), (Figure 3). These results confirmed our previous observation that it is the bladder mucosa and in particular, the urothelium, that responds to LPS with NF-κB translocation [39]. This independent identification of NF-κB [39], which is known to play a key role in bladder inflammation [40] strengthens confidence in the identification of novel pathways.
Three transcripts were found downstream of NF-κB: catenin (cadherin-associated protein, delta 1), cox7b, and pIs3 (plastin 3 T-isoform). Regarding catenin, others have described its presence in the bladder mucosa and in particular in the urothelium [41]. Indeed, alpha1-catenin was reported to be reduced in bladder urothelial cells treated with anti-proliferative factor [42] which make this transcript a possible target in cystitis. Moreover, p120-catenin is frequently altered and/or lost in tumors of bladder [43]. Finally, bladder cancers harboring a beta-catenin mutation may represent aggressive biological behavior with enhanced proliferating activity [44]. Cox7b encodes cytochrome C oxidase, subunit VIIb which is the terminal component of the mitochondrial respiratory chain and catalyzes the electron transfer from reduced cytochrome C to oxygen. Finally, pls3 was found by differential display to have increased expression in cisplatin-resistant human cancer cells [45].
In addition to NF-κB, MIC also had SRF that along with several zinc finger TFs (SF-1 [Steroidogenic factor 1]; Gfi-1 (growth factor independent 1); ik-3 (Ikaros 3); and vMaf [basic region leucine zipper]), drive the activity of actg1. Actg1 is a highly conserved protein involved in various types of cell motility, and maintenance of the cytoskeleton.
The MIC regulatory network also includes the following unique transcripts: smoc2 (secreted modular calcium-binding protein 2), amcq (adult male corpora quadrigemina cDNA; RIKEN clone:B230340L02), prss11 (Serine protease 11), and ifit3 (Interferon-induced protein with tetratricopeptide repeats 3). Smoc2 is a widespread glycoprotein with a calcium-dependent conformation [46]. Prss11 encodes a secreted trypsin (HtrA1) that regulates the availability of insulin-like growth factors (IGFs) by cleaving IGF-binding proteins and therefore, may function as a regulator of cell growth [47]. HtrA is involved in stress response pathways [48]. Interestingly, down-regulation of prss11 expression may be an indicator of melanoma progression [49].
Ifit3 was originally cloned from a cDNA library prepared from the murine cell line, RAW 264.7, after bacterial LPS stimulation [50]. Although it is a transcript induced by IFN [51], its function is still unknown.
Grp58 encodes a ubiquitously expressed chaperone protein that resides in the endoplasmic reticulum and is part of the protein folding machinery [52]. It is likely that grp58 is involved in the oncogenic transformation[53] since expression analysis revealed an up-regulation of grp58 in breast, uterus, lung, and stomach tumors [54].
Together these results confirmed that the bladder mucosa expresses unique transcripts involved in cell growth, motility, and cytoskeleton, protein folding, and proteases. Some of these transcripts have been described to be altered in cystitis as well as in LPS-induced bladder inflammation. However, the most striking result was that of 6 different networks, the MIC library was the only one to have over-representation of NF-κB.
Mucosa inflamed versus detrusor inflamed [MIDI], (figure 4)
The question being answered by this experiment was whether or not the bladder mucosa sets the stage for LPS-induced inflammatory responses by up-regulating a unique set of genes and TFs distinct from the detrusor muscle.
In terms of TFs, the upstream stimulatory factor (USF) was the most significantly over-represented (p < 0.001) and driving unique transcripts (figure 4). USF dimerizes to regulate transcription through E-box motifs in target genes. Although widely expressed, they can mediate tissue-specific transcripts. USF is stimulated by glucose in murine mesangial cells, binds to TGF-β1 promoter, contributes to TGF-β1 expression, and may play a role in diabetes-related gene regulation in the kidney [55]. Others have shown that USF binding activity is enhanced in response to LPS [56].
Another interesting TF was the liver X-activated receptors (LXR) found upstream of several transcripts bearing USF sequence. LXR is a member of the nuclear receptor superfamily [57] is a negative regulator of macrophage inflammatory gene expression [58], and a putative therapeutic agent for the treatment of inflammation [58], diabetes [59], and neurodegenerative diseases [57,58,60].
LIM-only proteins (LMO), which consist of LMO1, LMO2, LMO3, and LMO4, are involved in cell fate determination and differentiation during embryonic development [61]. LMO2 was originally identified through its involvement in T-cell leukemia and subsequently shown to be critical for normal hematopoietic and endothelial development [61]. Accumulating evidence suggests that LMO1 and LMO2 act as oncogenic proteins in T-cell acute lymphoblastic leukemia, whereas LMO4 has recently been implicated in the genesis of breast cancer[61].
MAZ (Myc-associated zinc finger protein), also known as serum amyloid A-activating transcription factor-1 (SAF-1), plays a major role in regulating transcription of several inflammation-responsive genes, including matrix metalloproteinase-1 (81), the mouse mast cell protease (mMCP)-6 [62], and function as growth suppressor in fibroblasts [63]. SAF-1 transgenic mice are prone to develop a severe form of inflammation-induced arthritis [64]. MAZ is upstream of Aplp2 which is a key regulator of structure and function of developing neuromuscular synapses [65].
In terms of unique transcripts isolated from the MIDI library, our results indicate that transcripts such as zfp364 (zinc finger protein 364, also known as rab7) and sir2 are downstream of series of TFs including: USF, LXR, DEAF1, MAZ, and Lmo2 complex (figure 4). Sir2 gene encodes a member of the sirtuin family of proteins which are NAD-dependent histone/protein deacetylases [66]. Zfp364 is a member of the Rab family of small G proteins, and regulates intracellular vesicle traffic to late endosomes [67]. Another unique transcript was the androgen-regulated tmprss2 protease [68] known to be expressed in urogenital tissues [69]. Tmprss2 has gained interest owing to its highly localized expression in the prostate and its over-expression in neoplastic prostate epithelium. Once activated, the serine protease domain of tmprss2 is released from the cell surface into the extracellular space and activates PAR (protease-activated receptor)-2 that has a role in prostate cancer and tumor metastasis [70]. Among the proteins correlating with cytoskeleton dynamics, our SSH identified a transcript (wdr1) encoding a 67-kDa WD40 repeat protein 1 which is the vertebrate homologue of actin-interacting protein 1 [71]. Wdr1 is involved in actin dynamics and seems to be required to induce cell morphologic changes, especially mitotic cell rounding [72]. Others have shown that wdr1 was found upregulated in the lung [73] and cell lines [74] following exposure to nickel oxide-induced carinogenesis. Finally, MIDI library also contained a RIKEN cDNA 2310015N07 gene that was described to be isolated from developing mouse libraries but no function yet has been attributed [75].
Interestingly, a comparison between MIDI and DIMI libraries indicates that both share LXR as a TRE. The major difference between these two libraries was found downstream LXR activation. In the inflamed mucosa, LXR preferentially activates zfp364 and tmprrs2 whereas in the inflamed detrusor LXR was found as a co-modulator of actg2.
In conclusion, the mucosa regulatory network presents USF in a central position raising the hypothesis that USF-target promoters such as the TGF-β1 promoter are involved in the mucosal response to inflammation and whether mucosa inflammation follows similar diabetes- related mucosal gene expression.
Detrusor inflamed versus detrusor control [DIC], (figure 6)
The regulatory network of the detrusor muscle, inflamed versus control, selected the following transcripts: syn1 (Synapsin I or ribosomal protein S15a), siva (CD27-binding protein), and eif4ebp2 (negative regulation of translational initiation), (figure 6).
Syn1 is a member of the synapsin gene family which is a neuron-specific phosphoprotein of small synaptic vesicles.Syn1 has been mapped to an evolutionarily conserved linkage group composed of: araf1, syn1, timp, and properdin located at human chromosome Xp11.2 [76] and mouse chromosome X [77]. Of interest, araf1 is a proto-oncogene which is predominantly expressed in mouse urogenital tissues [77]. In contrast, siva has an important role in the apoptotic pathway induced by the CD27 antigen. Others have described that siva is a direct transcriptional target for both tumor suppressors, p53 and E2F1 [78]. Finally, the eukaryotic initiation factor eIF4E and eIF4E-binding proteins (4E-BPs) control the initiation of protein synthesis and are part of a translational signaling pathway sensitive to insulin [79] and rapamycin [80]. Changes in the state of phosphorylation of eIF4E and 4E-BPs occur at an early stage of apoptosis [81]. Interestingly, eIF4E selectively enhances the translation of powerful angiogenic factors such as FGF-2 and VEGF [82] and therefore may have a role in oncogenesis [82] as well as inflammation.
Over represented TFs in DIC regulatory network were NRSF (Kruppel-type zinc-finger transcriptional repressor RE1-silencing transcription factor [REST]; also known as the neuron-restrictive silencing factor), CREB/CRE-BP1, E2F-1, and Evi-1.
CREB/CRE-BP1, also called transcription factor ATF-2, binds to the cAMP response element and its activity is enhanced after phosphorylation by stress-activated protein kinases such as c-Jun N-terminal kinase and p38. ATF-2 plays a central role in TGFβ signaling by acting as a common nuclear target of both Smad and TAK1 pathways [83].
Nrf-1 (nuclear respiratory factor 1) regulates expression of nuclear-encoded mitochondrial genes and it was shown to be part of the response to LPS in rats [84].
FOXp3 belongs to the forkhead gene family which comprises a diverse group of "winged-helix" TFs with important roles in development, metabolism, cancer and aging [85]. Recently, several forkhead genes have been demonstrated to play critical roles in lymphocyte development and effector function [85]. FoxP3 is a potential target for treatment of experimental chronic inflammatory renal disease [86] and type I diabetes [87]. In addition, both FOXp3 and NRSF seems to be downstream of Wnt-Frizzled signaling [88] which was recently proposed to participate in the pathogenesis of interstitial cystitis [89].
E2F, E2F-1, and Rb-E2F-1 belong to a family of TFs implicated in the regulation of cell proliferation and their binding sites are present in the promoters of several growth-regulating genes. E2F family members are functionally regulated, in part, by complex formation with one or more members of the nuclear pocket protein family such as the retinoblastoma protein (Rb) and play a role in neuronal development [90] by acting as negative regulator of cell proliferation. The interplay between Rb and E2F is critical for proper cell cycle progression [91]. Of interest, E2F-1 has a growth-promoting effect in bladder superficial TCC [92].
ATF-3 (activating transcription factor 3) is transcriptional repressor involved in survival and regeneration of sensory neurons [93,94] that responds to insulin [95]. ATF3 is also a novel stress-activated regulator of p53 protein stability/function providing the cell with a means of responding to a wide range of environmental insults [96]. In addition, ATF3 represents a novel mechanism in which anti-inflammatory drugs exert their anti-invasive activity [97].
The proposed role of nrsf/rest is that of a transcriptional silencer that restricts neuronal gene expression to the nervous system by silencing their expression in non-neural tissues [98]. Interestingly, loss of nrst function in human prostate carcinoma cells is associated with neuroendocrine phenotype, tumor progression, and androgen independence [99]. Others investigators indicated that nrst also modulates the cholinergic gene locus [100,101] which may have some implication in detrusor instability. Recently, it was proposed that activation of the rest/nrsf target genes overrides muscle differentiation pathways and converted myoblasts to a physiologically active neuronal phenotype [102]. It remains to be determined whether nrst promotes the same transformation in the inflamed detrusor muscle. The latter would explain the hyperactivity of detrusor muscle observed in over-active bladder disorders such as obstruction, incontinence, and inflammation.
In conclusion two major networks are proposed to be active in the detrusor inflamed when compared to control. One containing a neuron-specific phosphoprotein of small synaptic vesicles (syn) and the other an important protein of apoptotic pathway (siva). In both cases, analysis of the intense upstream promoter network leads us to the hypothesis that both genes represent a common downstream target of several pro-inflammatory stimuli.
Detrusor inflamed versus mucosa inflamed [DIMI], (figure 7)
The question being answered by this experiment was whether or not the inflamed detrusor muscle expresses unique transcripts and TFs distinct from the bladder mucosa.
Two major pathways could be constructed with the combination of SSH and PAINT results. The first involves key smooth muscle proteins, a myosin light chain encoded by my19 and gamma actin encoded by actg2. Gamma actins are highly conserved proteins that are involved in various types of cell motility, and maintenance of the cytoskeleton. In addition, a role for smooth muscle alpha actin in force generation by the urinary bladder has been suggested [103]. Several TREs upstream of actg 2 and my19 were over-represented in MIDI, including LXR which was described above, SRF, COUP, Pax-9, CP2, and RFX1.
A second pathway involved the transcripts elp3, gus, and ddx3 and two TFs COMP1 and IRF. Ddx3 is a putative RNA helicase and a member of a highly conserved DEAD box subclass. RNA helicases are highly conserved enzymes involved in transcription, splicing, and translation [104]. There are several examples of the involvement of RNA helicases in differentiation of germ cells, particularly in spermatogenesis. Upstream of ddx3, PAINT selected a TF that cooperates with myogenic proteins (COMP1) and Interferon Regulatory Factor (IRF). Transcription of IRF is synergistically activated by products of inflammation such as IFNγ and TNFα [105].
Two other transcripts were found downstream of COMP1: Gus (beta-glucoronidase) and elp3. Gus is a sensitive indicator LPS activation of macrophages. Elp3 is one of the sub units of the elongator complex, an acetyltransferase important for normal histone acetylation involved in elongation of RNA polymerase II transcription.
By comparing the inflamed detrusor and mucosa (DIMI), the fundamental difference observed was the up-regulation in the detrusor of genes and TFs related to smooth muscle function. It is fair to propose that this network could underlie detrusor instability during inflammation.
Conclusion
We here present a novel approach to understanding the bladder response to inflammation as a system. By using SSH, low abundance, differentially expressed transcripts could be detected that probably would have been lost in the background "noise" of a microarray study. That these genes were, in fact, key players was shown by the remarkable concordance in the transcriptional regulatory elements identified and by target validation with QRT-PCR. We suggest that the results identified key players governing the normal growth and differentiation of bladder mucosa and urothelium as well as the cross-communication of these layers during inflammation resulting from a number of pathologic processes.
As genes encoding DNA-binding TFs are the largest class of genes involved in human oncogenesis, it was obvious that in several instances the vast amount of information was related to cancer, in general and to bladder carcinoma, in particular. Interestingly, some of the TFs and their correlated downstream transcripts originally described to be involved in organogenesis were also activated during inflammation. The implications of these findings may represent one more link between inflammation and cancer.
The networks here described could well represent key targets for development of novel drugs for treatment of bladder diseases.
Methods
Animals
Ten to twelve-week old female C57BL/6J mice were used in these experiments that were performed in conformity with the "Guiding Principles for Research Involving Animals and Human Beings (OUHSC Animal Care & Use Committee protocol #002-109).
Induction of inflammation
Acute inflammation was induced by instillation of LPS into the mouse bladder, as described previously [8,9,106]. Female mice were anesthetized (ketamine 200 mg/kg and xylazine 2.5 mg/kg, i.p.), then transurethrally catheterized (24 Ga.; 3/4 in; Angiocath, Becton Dickson, Sandy, Utah), and the urine was drained by applying slight digital pressure to the lower abdomen. Because the bladder of 10-week old mice has an average capacity of 250 μl, the urinary bladders were instilled with 200 μl of one of the following substances: pyrogen-free saline (control) or Escherichia coli LPS strain 055:B5 (Sigma, St. Louis, MO; 100 μg/ml),) (figure 1a). Substances were infused at a slow rate to avoid trauma and vesicoureteral reflux. To ensure consistent contact of substances with the bladder, infusion was repeated twice within a 1-hour interval and a 1-ml syringe was maintained in the catheter end during this period. The catheter was removed, and mice were allowed to void normally. Twenty-four hours after instillation, mice were euthanized with pentobarbital (100 mg/kg, i.p.) and bladders were removed rapidly.
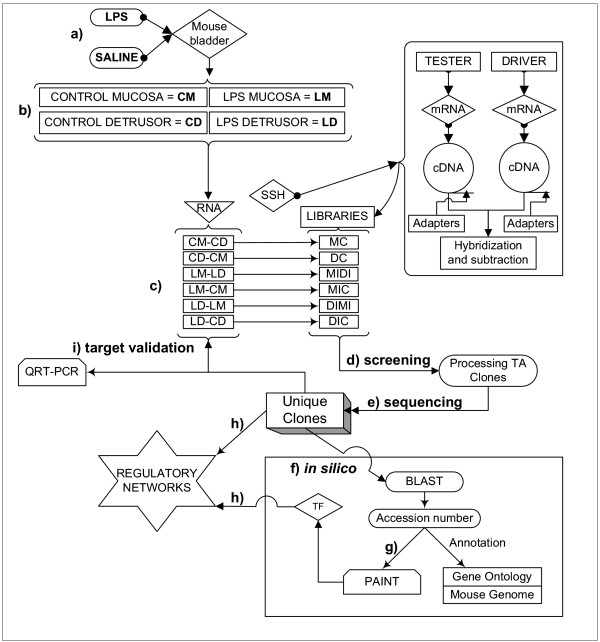
Figure 1.
Overall experimental design. a) A well established animal model of LPS- induced bladder inflammation was used. b) RNA was extracted from isolated detrusor muscle and mucosa layers. c) Extracted RNA was used to generate 6 different libraries by suppressive subtraction hybridization (SSH) in order to determine bladder tissue- and treatment- dependent transcripts. d) Transcripts were then screened and e) Sequenced f) All unique transcripts were fully annotated by querying public (PubMed, Gene Ontology, Mouse Genome [108]) and private (Transfac professional [109]) databases. g) The accession number of each SSH-selected transcript was uploaded into the PAINT 3.3 feasnet builder [110] to query the Transfac database [109]. h) A regulatory network for each library was originated by a combination of SSH-selected transcripts and over-represented TF (0 < p < 0.05) in the matrix when compared to the PAINT database reference equivalent to the all the genes in the Ensembl annotated genome (Figures 2, 3, 4, 5, 6, 7). i) Unique clones were validated by QRT-PCR.
Tissue layers – separating the mucosa from detrusor
Immediately after removal from the animal, bladders were placed in RNAlater™ (Ambion) and visualized under a dissecting microscope (Nikon SMZ 1500). The detrusor smooth muscle was separated by blunt dissection away from the mucosa which contained the epithelium and sub-epithelial layers, (see Additional file 1).
Suppression subtractive hybridization (SSH) (9)
A total of six libraries were obtained by SSH (figure 1c). In order to standardize the names of the groups and to correlate with delta CT values (see below), the SSH libraries were named after the tester minus driver. The first library (MC) was obtained by using the mucosa removed from control saline-treated mice (CM) as tester and the respective detrusor smooth muscle (CD) as driver. The resultant subtraction MC was supposed to contain genes preferentially expressed in the control mucosa. The second library was the reverse of MC and therefore, CD was used as tester and CM as driver and will contain genes preferentially expressed in the detrusor smooth muscle of control mice (DC). The other 4 libraries were obtained to investigate genes whose expression was altered during LPS-induced inflammation (figure 1c). The samples used for each SSH were obtained by pooling RNA from 20 individual mice. The pooling was necessary in order to obtain enough RNA from each layer without amplification.
Construction of subtractive cDNA libraries
mRNA was isolated from total RNA using Poly(A) Quick mRNA Isolation Kit (Stratagene, La Jolla, CA) according to the manufacture's protocol. To compare the two populations of resulting cDNA the method of SSH was performed using PCR-Select cDNA Subtraction Kit (BD Biosciences – Clontech, Palo Alto, CA), as described by Diatchenko and colleagues [18]. This method selectively amplifies differentially expressed sequences, and the generation of high- and low-abundance sequences is equalized during the first hybridization. The PCR allows amplification of equalized differentially expressed sequences. Each step of the cDNA synthesis and subtractive hybridization procedure was monitored using the positive control samples provided by the manufacturer. We verified the efficiency of subtraction by PCR analysis by comparing GAPDH levels in subtracted and un-subtracted cDNA using the method and GAPDH primers provided by the manufacturer. For analysis of efficiency, please see Additional file 2. For analysis of ligation, please see Additional file 3. For the analysis of PCR products, please see Additional file 4. For PCR analysis and subtraction efficiency, please see Additional file 5.
Next, cDNAs from the testers and drivers were digested with RsaI. To select tissue- and treatment-specific transcripts, PCR adapters were ligated to the tester pool population. The tester cDNA pool was then hybridized with excess cDNAs from the driver pool. After hybridization suppression, PCR using primers specific for the tester PCR adapters selectively amplified differentially expressed transcripts.
Screening the clones (plating out, growing up and analyzing the PCR clones), (figure 1d)
After the PCR subtraction, the amplification products were cloned into the pCR 2.1 plasmid of the TA cloning kit (Invitrogen). Ligated DNA was transformed by heat shock in 100 μl of INVαF competent E. coli cells. Colonies were grown overnight at 37°C on Luria broth agar plates containing ampicillin, X-gal, and isopropyl-B-D-thiogalactopyranoside for blue/white colony selection. White colonies were isolated and grown individually in 2 ml of LB medium containing ampicillin for 16 h. After plasmid DNA isolation (Wizard® Plus SV Minipreps DNA Purification System – Promega), digestion was performed using Xba I and BamH I, and the products analyzed in 1% agarose gels. Positive clones (representing fragments larger than the original polylinker in the cloning vector) were sent to a sequencing service (figure 1e), and sequences were submitted for a BLAST analysis in GenBank for identification and annotation was done by searching the gene ontology[16], figure 1f.
Tissue- and treatment-specific transcription factors
After annotation, a bioinformatics approach to identify functionally relevant putative transcriptional regulatory elements (TREs) for all SSH-selected transcripts was used (figure 1g). We used the Promoter Analysis and Interaction Network Toolset [PAINT [13]], available online [107], to integrate functional genomics information from SSH-derived gene expression data with the genomic sequence and TRE data to derive hypotheses about relevant transcriptional regulatory networks. PAINT uses the TRANSFAC® database [14] of transcription factors and position weight matrix descriptions of cis-acting sequences and an associated pattern matching tool MATCH [15] to identify statistically over-represented regulatory sites in 5' upstream sequences of related genes. This information provides a substantially pruned list of TFs regulating tissue- and treatment-specific genes that were identified by SSH. Briefly, the accession numbers of all the SSH-selected transcripts were used as an input gene list in PAINT. Up to 2000 base pairs of 5' upstream sequences were analyzed for the presence of TREs using a MATCH/TRANSFAC setting to minimize false positives and filtering the results further in PAINT to consider only those hits with 100% match to the 5 bp core TRE sequence. The interaction matrix contained 79 genes and 162 TREs.
PAINT can analyze the interaction matrix for over-represented TREs in subsets/clusters of related genes, typically grouped based on gene expression data. The library for each transcript (MC, DC, MI, MIDI, DI and DIMI) was considered as the cluster label and provided as the Gene-Cluster information in the PAINT analysis and visualization step. The TRE over-representation in PAINT is calculated as the hyper-geometric probability of the observed number of TREs in a given cluster as compared to that in randomly selected gene clusters from a 'reference' list. In this study, all the genes in the genome as annotated in Ensembl were used to construct the interaction matrix for use as the reference. To construct the hypothesized regulatory networks, TFs were chosen based on the probability of over-representation (p < 0.05) in any of the six gene groups as compared to all the genes in the genome (or equivalently in PAINT promoter database).
Target validation by QRT-PCR (figure 1i)
RNA isolation and cDNA synthesis
An additional 40 C57BL/6J female mice, ten to twelve-weeks old underwent the same intravesical treatment as described above [LPS, (n = 20 mice) and saline (n = 20 mice)]. Twenty four hours after LPS instillation, mice were euthanized, the bladder was removed and placed in RNAlater™ (Ambion) for separation of the mucosa and submucosa from the detrusor smooth muscle, as described above. Four sample groups were obtained as follows: control mucosa (CM), control detrusor (CD), LPS-treated mucosa (LM), and LPS-treated detrusor (LD). Bladders were pooled and homogenized in Ultraspec RNA solution (Biotecx Laboratories, Houston, TX) for isolation and purification of total RNA used for QRT-PCR. High RNA quality from each of these four groups was verified by capillary gel electrophoresis using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc, Palo Alto, CA). RNA concentration was determined by spectrophotometry using a NanoDrop® ND-1000 UV-Vis Spectrophotometer (Nano Drop Technologies, Wilmington, DE). Subsequently, this total RNA was used as a template for each of the first-strand cDNA syntheses. Prior to cDNA synthesis, an exogenous standard of A. thaliana (RCA) mRNA (Stratagene, L Jolla, CA, USA) was added (0.1 ng) to each CM, CD, LM, and LD total RNA (2 μg) sample for normalization of succeeding gene expression data. RNA was reverse-transcribed according to the Omniscript RT™ kit (Qiagen, Valencia, CA) instructions and subsequently purified using the Montage PCR 96-well cleanup plate (Millipore, Billerica, MA). Prior to the PCR, primer pairs were designed utilizing both Primer Express® (ABI, Foster City, CA) and NetPrimer (PREMIER Biosoft International, Palo Alto, CA) software. Primers were designed according to the general guidelines outlined in the Primer Express® User Bulletin. Details of the primers and the Genebank accession numbers are given in Table 1. The designed primers shared 100% homology with the target sequence but no significant homology with other sequences.
Table 1.
Primers for real time PCR
| Gene | GenBank ID | 5' primer sequence | 3' primer sequence | localization |
| Bcap31 | NM_009922 | AAAGAATATGACCGCCTGCTAGA | AAGCCTTTACTCCTCCTTCTTGACT | 761–847 |
| Catenin | BC043108 | CCTCCCCCACCCACATCTA | ACCCACACCCCACCGAgaa | 4957–5049 |
| Cnn1 | NM_009922 | AGTTGTTTGCTGCCAAGTCTGA | GGTGGAAGGCAGTTTAATGGAGT | 2081–2168 |
| Dlk1 | D16847 | CTTTTTGTGGTGGAGTTTGCTCTAT | gCGTGGTAGCATGGCACACA | 1859–1950 |
| eiF-4E | NM_010124 | TGTGCTTGGCTGCTGAGAGA | ACGGACAGACGGACGATGA | 1446–1531 |
| Grn | X62321 | CTACCTAAAGGGTGTCTGCTGTAGA | AGGAATCTTCTTTCGCAAACACTT | 1630–1729 |
| Grp58 | BC003285 | AAAACCAGAGAGGACAGAATGGATAA | TGTATTTTCAAACAGTGCAGCTAAGAA | 1627–1712 |
| GSTM1 | NM_010358 | TCTCCTTCCCGCTCCCTT | GAGAATGAAGGCTGTGTGGACTT | 965–1047 |
| GSTO1 | NM_010362 | GGCAAGAGCCCTCAGCAA | TGAGAAAGGAGCCAGTGAGAATACT | 826–912 |
| IFIT3 | BC003804 | TGTGGTGGATTCTTGGCAGTT | CTGCCTGTGCCCCAAAGT | 1279–1366 |
| LAMP2 | NM_010685 | TGGCACTGGCTTAATGCTGTT | GTGCTTTGGAGGTATCTCAATATGAA | 3132–3218 |
| Mafb | AF180338 | CCATCTTGAGAAGGTAGCAGCAA | AAAGTTGGGCTTGGTGGGTT | 3731–3840 |
| Mpp1 | U38196 | AGATTGCCATCCTTGACATTGA | GTAGGTGCGATGAACACAATGAA | 1301–1388 |
| Pls3 | NM_145629 | CACACCCAGGCTCAAAGGA | TTGTGATAAAGATTTCCAAACAAACAA | 2816–2902 |
| Prss11 | NM_019564 | AGTCAACATTTGTCCCTTCCCTTA | GGCTGCGAGGACCTTCCT | 1752–1837 |
| Sirt1 | AF214646 | CCTGCATAGATCTTCACCACAaat | ACACTCTCCCCAGTAGAAGTACCATT | 3016–3108 |
| Smoc2 | NM_022315 | CCACTATGGGATGAAGGTTATGA | AGAAAGTGACAGCCAGCCATACA | 2377–2465 |
| SPRR2A | BC010818 | ATAGCAACACTTCCATCCTCCTTT | TGAGGAGCCATCATAAGCACAT | 379–471 |
| SYN1 | NM_013680 | GTTCTAAAGTCATCGTTCGGTTCTTAA | TTCCCAGCTCTGTGATCATCAA | 3334–3424 |
| TMPRSS2 | AF199362 | AATCACACCAGCCATGATCTGT | AATCAGCCACCAGATCCCATT | 1364–1473 |
| Upk1a | AF262335 | GGCAACTTCATCCCCATCAA | AGCAACCCTTGGTAAACAGGTAGT | 580–652 |
The QRT-PCR amplifications were accomplished on an ABI®PRISM 7700 using SYBR®Green I dye assay chemistry. A 15 μL PCR assay for each gene of interest consisted of 7.5 μL of 2X SYBR®Green PCR mix (Applied Biosystems Inc., Foster City, CA), 4.9 μl of H20, 0.6 μl (30 pmoles) of gene-specific forward and reverse primers, and 2 μl (1 ng) of cDNA template. All samples were run in triplicate with the appropriate single QRT-PCR controls (no reverse transcriptase and no template). Cycling conditions used for all amplifications were one cycle of 95°C for 10 minutes and 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Following the QRT-PCR, dissociation curve analysis was performed to confirm the desired single gene product.
From the QRT-PCR data, an average cycle threshold (Ct) value was calculated from the triplicate reactions. Averaged Ct values were then normalized (to adjust for different amounts of cDNA within each reaction) to the exongenous control gene, RCA. The relative expression level of each transcript within each sample group (CD, CM, LD, and LM) was determined by calculating the ratio of the antilog2 of the delta Ct values. The resultant fold-change data is presented in Table 4.
Authors' contributions
MRS participated in its design, carried out the animal experiments, removed the tissues, performed suppression subtractive hybridizations, and performed sequence alignments. NBN helped MRS on the suppression subtractive hybridizations experiments. HLH participated in the design of the study and trained both MRS and NBN to develop SSHs. MT and MC developed Q-PCR.RV developed the PAINT program and guided RS to perform TF analysis. DWD's laboratory sequenced all transcripts. REH participated in its design and helped to draft the manuscript. RS conceived of the study, developed annotation and TF analysis, and draft the manuscript.
Supplementary Material
Separation of bladder layers (mucosa and detrusor).
Detailed methodology for SSH and analysis of the experimental procedures.
Analysis of ligation.
Analysis of PCR products.
The reduced message of a known housekeeping gene between the subtracted and un-subtracted cDNA over the same number of PCR cycles.
Acknowledgments
Acknowledgements
Supported by National Institutes of Health grants 5 R01 DK066101-02(RS) and 5 R01 DK055828-05 (RS).
Contributor Information
Marcia R Saban, Email: marcia-saban@ouhsc.edu.
Helen L Hellmich, Email: hhellmic@utmb.edu.
Mary Turner, Email: Mary-turner@omrf.ouhsc.edu.
Ngoc-Bich Nguyen, Email: NGUYENN2@UTHSCSA.EDU.
Rajanikanth Vadigepalli, Email: raj@mail.dbi.tju.edu.
David W Dyer, Email: david-dyer@ouhsc.edu.
Robert E Hurst, Email: Robert-hurst@ouhsc.edu.
Michael Centola, Email: Mike-Centola@omrf.ouhsc.edu.
Ricardo Saban, Email: Ricardo-saban@ouhsc.edu.
References
- Fry CH. Experimental models to study the physiology, pathophysiology, and pharmacology of the lower urinary tract. J Pharmacol Toxicol Methods. 2004;49:201–210. doi: 10.1016/j.vascn.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- OVERCOMING BLADDER DISEASE. A Strategic Plan for Research. A REPORT OF THE BLADDER RESEARCH PROGRESS REVIEW GROUP. 2002. http://www.niddk.nih.gov/fund/other/archived-conferences/2001/brprg_book.pdf
- Baskin LS, Hayward SW, Sutherland RA, DiSandro MS, Thomson AA. Cellular Signaling in the Bladder. Front Biosci. 1997;2:592–595. doi: 10.2741/a215. [DOI] [PubMed] [Google Scholar]
- Cunha GR, Lung B. The importance of stroma in morphogenesis and functional activity of urogenital epithelium. In Vitro. 1979;15:50–71. doi: 10.1007/BF02627079. [DOI] [PubMed] [Google Scholar]
- Saban M, Nguyen N-B, Hammond T, Saban R. Gene Expression profiling of mouse bladder inflammatory responses to LPS, substance P, and antigen-stimulation. Am J Pathol. 2002;160:2095–2110. doi: 10.1016/S0002-9440(10)61159-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saban MR, Hellmich H, Nguyen NB, Winston J, Hammond TG, Saban R. Time course of LPS-induced gene expression in a mouse model of genitourinary inflammation. Physiol Genomics. 2001;5:147–160. doi: 10.1152/physiolgenomics.2001.5.3.147. [DOI] [PubMed] [Google Scholar]
- Saban MR, Nguyen NB, Hammond TG, Saban R. Gene expression profiling of mouse bladder inflammatory responses to LPS, substance P, and antigen-stimulation. Am J Pathol. 2002;160:2095–2110. doi: 10.1016/S0002-9440(10)61159-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saban MR, Nguyen NB, Hurst RE, Saban R. Gene expression profiling of inflammatory bladder disorders. Expert Rev Mol Diagn. 2003;3:217–235. doi: 10.1586/14737159.3.2.217. [DOI] [PubMed] [Google Scholar]
- Saban R, Gerard NP, Saban MR, Nguyen NB, DeBoer DJ, Wershil BK. Mast cells mediate substance P-induced bladder inflammation through an NK(1) receptor-independent mechanism. Am J Physiol Renal Physiol. 2002;283:F616–629. doi: 10.1152/ajprenal.00096.2002. [DOI] [PubMed] [Google Scholar]
- Saban R, Saban MR, Nguyen NB, Hammond TG, Wershil BK. Mast cell regulation of inflammation and gene expression during antigen-induced bladder inflammation in mice. Physiol Genomics. 2001;7:35–43. doi: 10.1152/physiolgenomics.00044.2001. [DOI] [PubMed] [Google Scholar]
- D'Andrea MR, Saban MR, Nguyen NB, Andrade-Gordon P, Saban R. Expression of protease-activated receptor-1, -2, -3, and -4 in control and experimentally inflamed mouse bladder. Am J Pathol. 2003;162:907–923. doi: 10.1016/S0002-9440(10)63886-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadigepalli R, Chakravarthula P, Zak DE, Schwaber JS, Gonye GE. PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. Omics. 2003;7:235–252. doi: 10.1089/153623103322452378. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.informatics.jax.org/
- Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 2000;10:1617–1630. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lau Y-FC, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, et al. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. PNAS. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Wright MB, Cai L, Flament A, Lindpaintner K. Efficacy of SSH PCR in isolating differentially expressed genes. BMC Genomics. 2002;3:12. doi: 10.1186/1471-2164-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong-Riley MT, Yang SJ, Liang HL, Ning G, Jacobs P. Quantitative immuno-electron microscopic analysis of nuclear respiratory factor 2 alpha and beta subunits: Normal distribution and activity-dependent regulation in mammalian visual cortex. Vis Neurosci. 2005;22:1–18. doi: 10.1017/S0952523805221016. [DOI] [PubMed] [Google Scholar]
- Yang SJ, Liang HL, Ning G, Wong-Riley MT. Ultrastructural study of depolarization-induced translocation of NRF-2 transcription factor in cultured rat visual cortical neurons. Eur J Neurosci. 2004;19:1153–1162. doi: 10.1111/j.1460-9568.2004.03250.x. [DOI] [PubMed] [Google Scholar]
- LaMarco K, Thompson CC, Byers BP, Walton EM, McKnight SL. Identification of Ets- and notch-related subunits in GA binding protein. Science. 1991;253:789–792. doi: 10.1126/science.1876836. [DOI] [PubMed] [Google Scholar]
- Gualberto A, LePage D, Pons G, Mader SL, Park K, Atchison ML, Walsh K. Functional antagonism between YY1 and the serum response factor. Mol Cell Biol. 1992;12:4209–4214. doi: 10.1128/mcb.12.9.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman M, Fagher K, Wede M, Stakeberg K, Arner A. Decreased phosphatase activity, increased Ca2+ sensitivity, and Myosin light chain phosphorylation in urinary bladder smooth muscle of newborn mice. J Gen Physiol. 2005;125:187–196. doi: 10.1085/jgp.200409212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSanto ME, Stein R, Chang S, Hypolite JA, Zheng Y, Zderic S, Wein AJ, Chacko S. Alteration in expression of myosin isoforms in detrusor smooth muscle following bladder outlet obstruction. Am J Physiol Cell Physiol. 2003;285:C1397–1410. doi: 10.1152/ajpcell.00513.2002. [DOI] [PubMed] [Google Scholar]
- Ito M, Achermann JC, Jameson JL. A naturally occurring steroidogenic factor-1 mutation exhibits differential binding and activation of target genes. J Biol Chem. 2000;275:31708–31714. doi: 10.1074/jbc.M002892200. [DOI] [PubMed] [Google Scholar]
- Achermann JC, Ito M, Ito M, Hindmarsh PC, Jameson JL. A mutation in the gene encoding steroidogenic factor-1 causes XY sex reversal and adrenal failure in humans. Nat Genet. 1999;22:125–126. doi: 10.1038/9629. [DOI] [PubMed] [Google Scholar]
- Karapetian RN, Evstafieva AG, Abaeva IS, Chichkova NV, Filonov GS, Rubtsov YP, Sukhacheva EA, Melnikov SV, Schneider U, Wanker EE, et al. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: implications for expression of oxidative stress-protecting genes. Mol Cell Biol. 2005;25:1089–1099. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KJ, Shiau AL, Chiou YY, Yo YT, Wu CL. Transgenic overexpression of prothymosin alpha induces development of polycystic kidney disease. Kidney Int. 2005;67:1710–1722. doi: 10.1111/j.1523-1755.2005.00268.x. [DOI] [PubMed] [Google Scholar]
- Miano JM, Thomas S, Disteche CM. Expression and chromosomal mapping of the mouse smooth muscle calponin gene. Mamm Genome. 2001;12:187–191. doi: 10.1007/s003350010266. [DOI] [PubMed] [Google Scholar]
- Shirozu M, Tada H, Tashiro K, Nakamura T, Lopez ND, Nazarea M, Hamada T, Sato T, Nakano T, Honjo T. Characterization of novel secreted and membrane proteins isolated by the signal sequence trap method. Genomics. 1996;37:273–280. doi: 10.1006/geno.1996.0560. [DOI] [PubMed] [Google Scholar]
- Fujisawa M, Kanai Y, Nam SY, Maeda S, Nakamuta N, Kano K, Kurohmaru M, Hayashi Y. Expression of Prnp mRNA (prion protein gene) in mouse spermatogenic cells. J Reprod Dev. 2004;50:565–570. doi: 10.1262/jrd.50.565. [DOI] [PubMed] [Google Scholar]
- Staack A, Koenig F, Daniltchenko D, Hauptmann S, Loening SA, Schnorr D, Jung K. Cathepsins B, H, and L activities in urine of patients with transitional cell carcinoma of the bladder. Urology. 2002;59:308–312. doi: 10.1016/S0090-4295(01)01517-5. [DOI] [PubMed] [Google Scholar]
- Raghavan D. Molecular targeting and pharmacogenomics in the management of advanced bladder cancer. Cancer. 2003;97:2083–2089. doi: 10.1002/cncr.11281. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gomez P, Bello MJ, Alonso ME, Aminoso C, Lopez-Marin I, De Campos JM, Isla A, Gutierrez M, Rey JA. Promoter methylation status of multiple genes in brain metastases of solid tumors. Int J Mol Med. 2004;13:93–98. [PubMed] [Google Scholar]
- Finch JL, Miller J, Aspinall JO, Cowled PA. Cloning of the human uroplakin 1B cDNA and analysis of its expression in urothelial-tumor cell lines and bladder-carcinoma tissue. Int J Cancer. 1999;80:533–538. doi: 10.1002/(SICI)1097-0215(19990209)80:4<533::AID-IJC9>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Reeves FC, Fredericks WJ, Rauscher FJ, 3rd, Lillycrop KA. The DNA binding activity of the paired box transcription factor Pax-3 is rapidly downregulated during neuronal cell differentiation. FEBS Lett. 1998;422:118–122. doi: 10.1016/S0014-5793(97)01598-6. [DOI] [PubMed] [Google Scholar]
- Epstein JA, Lam P, Jepeal L, Maas RL, Shapiro DN. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J Biol Chem. 1995;270:11719–11722. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
- Saban MR, Memet S, Jackson DG, Ash J, Roig AA, Israel A, Saban R. Visualization of lymphatic vessels through NF-{kappa}B activity. Blood. 2004;104:3228–3230. doi: 10.1182/blood-2004-04-1428. [DOI] [PubMed] [Google Scholar]
- Wang XC, Saban R, Kaysen JH, Saban MR, Allen PL, Benes EN, Hammond TG. Nuclear factor kappa B mediates lipopolysaccharide-induced inflammation in the urinary bladder. J Urol. 2000;163:993–998. doi: 10.1016/S0022-5347(05)67870-6. [DOI] [PubMed] [Google Scholar]
- Bryan RT, Nicholls JH, Harrison RF, Jankowski JA, Wallace DM. The role of beta-catenin signaling in the malignant potential of cystitis glandularis. J Urol. 2003;170:1892–1896. doi: 10.1097/01.ju.0000092740.51330.39. [DOI] [PubMed] [Google Scholar]
- Keay S, Seillier-Moiseiwitsch F, Zhang CO, Chai TC, Zhang J. Changes in human bladder epithelial cell gene expression associated with interstitial cystitis or antiproliferative factor treatment. Physiol Genomics. 2003;14:107–115. doi: 10.1152/physiolgenomics.00055.2003. [DOI] [PubMed] [Google Scholar]
- Thoreson MA, Reynolds AB. Altered expression of the catenin p120 in human cancer: implications for tumor progression. Differentiation. 2002;70:583–589. doi: 10.1046/j.1432-0436.2002.700911.x. [DOI] [PubMed] [Google Scholar]
- Shiina H, Igawa M, Shigeno K, Terashima M, Deguchi M, Yamanaka M, Ribeiro-Filho L, Kane CJ, Dahiya R. Beta-catenin mutations correlate with over expression of C-myc and cyclin D1 Genes in bladder cancer. J Urol. 2002;168:2220–2226. doi: 10.1016/S0022-5347(05)64359-5. [DOI] [PubMed] [Google Scholar]
- Hisano T, Ono M, Nakayama M, Naito S, Kuwano M, Wada M. Increased expression of T-plastin gene in cisplatin-resistant human cancer cells: identification by mRNA differential display. FEBS Lett. 1996;397:101–107. doi: 10.1016/S0014-5793(96)01150-7. [DOI] [PubMed] [Google Scholar]
- Vannahme C, Gosling S, Paulsson M, Maurer P, Hartmann U. Characterization of SMOC-2, a modular extracellular calcium-binding protein. Biochem J. 2003;373:805–814. doi: 10.1042/BJ20030532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbrunn J, Trueb B. Primary structure of a putative serine protease specific for IGF-binding proteins. FEBS Lett. 1996;398:187–192. doi: 10.1016/S0014-5793(96)01229-X. [DOI] [PubMed] [Google Scholar]
- Gray CW, Ward RV, Karran E, Turconi S, Rowles A, Viglienghi D, Southan C, Barton A, Fantom KG, West A, et al. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur J Biochem. 2000;267:5699–5710. doi: 10.1046/j.1432-1327.2000.01589.x. [DOI] [PubMed] [Google Scholar]
- Baldi A, De Luca A, Morini M, Battista T, Felsani A, Baldi F, Catricala C, Amantea A, Noonan DM, Albini A, et al. The HtrA1 serine protease is down-regulated during human melanoma progression and represses growth of metastatic melanoma cells. Oncogene. 2002;21:6684–6688. doi: 10.1038/sj.onc.1205911. [DOI] [PubMed] [Google Scholar]
- Lee CG, Demarquoy J, Jackson MJ, O'Brien WE. Molecular cloning and characterization of a murine LPS-inducible cDNA. J Immunol. 1994;152:5758–5767. [PubMed] [Google Scholar]
- Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
- van der Vlies D, Pap EH, Post JA, Celis JE, Wirtz KW. Endoplasmic reticulum resident proteins of normal human dermal fibroblasts are the major targets for oxidative stress induced by hydrogen peroxide. Biochem J. 2002;366:825–830. doi: 10.1042/BJ20020618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano N, Shibasaki F, Sakai R, Tanaka T, Nishida J, Yazaki Y, Takenawa T, Hirai H. Molecular cloning of the human glucose-regulated protein ERp57/GRP58, a thiol-dependent reductase. Identification of its secretory form and inducible expression by the oncogenic transformation. Eur J Biochem. 1995;234:336–342. doi: 10.1111/j.1432-1033.1995.336_c.x. [DOI] [PubMed] [Google Scholar]
- Celli CM, Jaiswal AK. Role of GRP58 in mitomycin C-induced DNA cross-linking. Cancer Res. 2003;63:6016–6025. [PubMed] [Google Scholar]
- Zhu Y, Casado M, Vaulont S, Sharma K. Role of Upstream Stimulatory Factors in Regulation of Renal Transforming Growth Factor-{beta}1. Diabetes. 2005;54:1976–1984. doi: 10.2337/diabetes.54.7.1976. [DOI] [PubMed] [Google Scholar]
- Potter JJ, Rennie-Tankersley L, Mezey E. Endotoxin enhances liver alcohol dehydrogenase by action through upstream stimulatory factor but not by nuclear factor-kappa B. J Biol Chem. 2003;278:4353–4357. doi: 10.1074/jbc.M210097200. [DOI] [PubMed] [Google Scholar]
- Steffensen KR, Neo SY, Stulnig TM, Vega VB, Rahman SS, Schuster GU, Gustafsson JA, Liu ET. Genome-wide expression profiling; a panel of mouse tissues discloses novel biological functions of liver X receptors in adrenals. J Mol Endocrinol. 2004;33:609–622. doi: 10.1677/jme.1.01508. [DOI] [PubMed] [Google Scholar]
- Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- Michael LF, Schkeryantz JM, Burris TP. The pharmacology of LXR. Mini Rev Med Chem. 2005;5:729–740. doi: 10.2174/1389557054553767. [DOI] [PubMed] [Google Scholar]
- Collins JL. Therapeutic opportunities for liver X receptor modulators. Curr Opin Drug Discov Devel. 2004;7:692–702. [PubMed] [Google Scholar]
- Aoyama M, Ozaki T, Inuzuka H, Tomotsune D, Hirato J, Okamoto Y, Tokita H, Ohira M, Nakagawara A. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005;65:4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- Morii E, Oboki K, Kataoka TR, Igarashi K, Kitamura Y. Interaction and cooperation of mi transcription factor (MITF) and myc-associated zinc-finger protein-related factor (MAZR) for transcription of mouse mast cell protease 6 gene. J Biol Chem. 2002;277:8566–8571. doi: 10.1074/jbc.M110392200. [DOI] [PubMed] [Google Scholar]
- Stubbs MC, Min I, Izzo MW, Rallapalli R, Derfoul A, Hall DJ. The ZF87/MAZ transcription factor functions as a growth suppressor in fibroblasts. Biochem Cell Biol. 2000;78:477–485. doi: 10.1139/bcb-78-4-477. [DOI] [PubMed] [Google Scholar]
- Ray A, Kumar D, Shakya A, Brown CR, Cook JL, Ray BK. Serum amyloid A-activating factor-1 (SAF-1) transgenic mice are prone to develop a severe form of inflammation-induced arthritis. J Immunol. 2004;173:4684–4691. doi: 10.4049/jimmunol.173.7.4684. [DOI] [PubMed] [Google Scholar]
- Wang P, Yang G, Mosier DR, Chang P, Zaidi T, Gong YD, Zhao NM, Dominguez B, Lee KF, Gan WB, et al. Defective neuromuscular synapses in mice lacking amyloid precursor protein (APP) and APP-Like protein 2. J Neurosci. 2005;25:1219–1225. doi: 10.1523/JNEUROSCI.4660-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrylenko S, Kyrylenko O, Suuronen T, Salminen A. Differential regulation of the Sir2 histone deacetylase gene family by inhibitors of class I and II histone deacetylases. Cell Mol Life Sci. 2003;60:1990–1997. doi: 10.1007/s00018-003-3090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Kitamura A, Sasaki T. Rabring7, a novel Rab7 target protein with a RING finger motif. Mol Biol Cell. 2003;14:3741–3752. doi: 10.1091/mbc.E02-08-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afar DE, Vivanco I, Hubert RS, Kuo J, Chen E, Saffran DC, Raitano AB, Jakobovits A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61:1686–1692. [PubMed] [Google Scholar]
- Vaarala MH, Porvari KS, Kellokumpu S, Kyllonen AP, Vihko PT. Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J Pathol. 2001;193:134–140. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH743>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Wilson S, Greer B, Hooper J, Zijlstra A, Walker B, Quigley J, Hawthorne S. The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem J. 2005;388:967–972. doi: 10.1042/BJ20041066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Lee E, Chung YH, Mun GH, Park J, Lomax MI, Oh SH. Subcellular localization of WD40 repeat 1 protein in PC12 rat pheochromocytoma cells. Neurosci Lett. 2004;367:399–403. doi: 10.1016/j.neulet.2004.06.053. [DOI] [PubMed] [Google Scholar]
- Fujibuchi T, Abe Y, Takeuchi T, Imai Y, Kamei Y, Murase R, Ueda N, Shigemoto K, Yamamoto H, Kito K. AIP1/WDR1 supports mitotic cell rounding. Biochem Biophys Res Commun. 2005;327:268–275. doi: 10.1016/j.bbrc.2004.11.156. [DOI] [PubMed] [Google Scholar]
- Verma R, Ramnath J, Clemens F, Kaspin LC, Landolph JR. Molecular biology of nickel carcinogenesis: identification of differentially expressed genes in morphologically transformed C3H10T1/2 Cl 8 mouse embryo fibroblast cell lines induced by specific insoluble nickel compounds. Mol Cell Biochem. 2004;255:203–216. doi: 10.1023/B:MCBI.0000007276.94488.3d. [DOI] [PubMed] [Google Scholar]
- Landolph JR, Verma A, Ramnath J, Clemens F. Molecular biology of deregulated gene expression in transformed C3H/10T1/2 mouse embryo cell lines induced by specific insoluble carcinogenic nickel compounds. Environ Health Perspect. 2002;110:845–850. doi: 10.1289/ehp.02110s5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MS, Kitchen JR, Wang X, Threat TA, Wang X, Hasegawa A, Sun T, Grahovac MJ, Kargul GJ, Lim MK, et al. Large-scale cDNA analysis reveals phased gene expression patterns during preimplantation mouse development. Development. 2000;127:1737–1749. doi: 10.1242/dev.127.8.1737. [DOI] [PubMed] [Google Scholar]
- Lee JE, Beck TW, Brennscheidt U, DeGennaro LJ, Rapp UR. The complete sequence and promoter activity of the human A-raf-1 gene (ARAF1) Genomics. 1994;20:43–55. doi: 10.1006/geno.1994.1125. [DOI] [PubMed] [Google Scholar]
- Derry JM, Barnard PJ. Physical linkage of the A-raf-1, properdin, synapsin I, and TIMP genes on the human and mouse X chromosomes. Genomics. 1992;12:632–638. doi: 10.1016/0888-7543(92)90286-2. [DOI] [PubMed] [Google Scholar]
- Fortin A, MacLaurin JG, Arbour N, Cregan SP, Kushwaha N, Callaghan SM, Park DS, Albert PR, Slack RS. The proapoptotic gene SIVA is a direct transcriptional target for the tumor suppressors p53 and E2F1. J Biol Chem. 2004;279:28706–28714. doi: 10.1074/jbc.M400376200. [DOI] [PubMed] [Google Scholar]
- Tee AR, Proud CG. Caspase cleavage of initiation factor 4E-binding protein 1 yields a dominant inhibitor of cap-dependent translation and reveals a novel regulatory motif. Mol Cell Biol. 2002;22:1674–1683. doi: 10.1128/MCB.22.6.1674-1683.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilling MB, Germain GS, Dudkin L, Jayaraman AL, Zhang X, Harwood FC, Houghton PJ. 4E-binding proteins, the suppressors of eukaryotic initiation factor 4E, are down-regulated in cells with acquired or intrinsic resistance to rapamycin. J Biol Chem. 2002;277:13907–13917. doi: 10.1074/jbc.M110782200. [DOI] [PubMed] [Google Scholar]
- Clemens MJ. Translational regulation in cell stress and apoptosis. Roles of the eIF4E binding proteins. J Cell Mol Med. 2001;5:221–239. doi: 10.1111/j.1582-4934.2001.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFatta RJ, Turbat-Herrera EA, Li BD, Anderson W, De Benedetti A. Elevated expression of eIF4E in confined early breast cancer lesions: possible role of hypoxia. Int J Cancer. 1999;80:516–522. doi: 10.1002/(SICI)1097-0215(19990209)80:4<516::AID-IJC6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Sano Y, Harada J, Tashiro S, Gotoh-Mandeville R, Maekawa T, Ishii S. ATF-2 is a common nuclear target of Smad and TAK1 pathways in transforming growth factor-beta signaling. J Biol Chem. 1999;274:8949–8957. doi: 10.1074/jbc.274.13.8949. [DOI] [PubMed] [Google Scholar]
- Suliman HB, Welty-Wolf KE, Carraway M, Tatro L, Piantadosi CA. Lipopolysaccharide induces oxidative cardiac mitochondrial damage and biogenesis. Cardiovasc Res. 2004;64:279–288. doi: 10.1016/j.cardiores.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Jonsson H, Peng SL. Forkhead transcription factors in immunology. Cell Mol Life Sci. 2005;62:397–409. doi: 10.1007/s00018-004-4365-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Wang Y, Mahajan D, Qin X, Wang Y, Wang Y, Alexander SI, Harris DC. The role of tubulointerstitial inflammation. Kidney Int Suppl. 2005:S96–100. doi: 10.1111/j.1523-1755.2005.09423.x. [DOI] [PubMed] [Google Scholar]
- Jaeckel E, von Boehmer H, Manns MP. Antigen-specific FoxP3-transduced T-cells can control established type 1 diabetes. Diabetes. 2005;54:306–310. doi: 10.2337/diabetes.54.2.306. [DOI] [PubMed] [Google Scholar]
- Li H, Malbon CC, Wang HY. Gene profiling of Frizzled-1 and Frizzled-2 signaling: expression of G-protein-coupled receptor chimeras in mouse F9 teratocarcinoma embryonal cells. Mol Pharmacol. 2004;65:45–55. doi: 10.1124/mol.65.1.45. [DOI] [PubMed] [Google Scholar]
- Keay SK, Szekely Z, Conrads TP, Veenstra TD, Barchi JJ, Jr, Zhang CO, Koch KR, Michejda CJ. An antiproliferative factor from interstitial cystitis patients is a frizzled 8 protein-related sialoglycopeptide. Proc Natl Acad Sci U S A. 2004;101:11803–11808. doi: 10.1073/pnas.0404509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagnino L, Fry CJ, Bartley SM, Farnham P, Gallie BL, Phillips RA. Expression patterns of the E2F family of transcription factors during mouse nervous system development. Mech Dev. 1997;66:13–25. doi: 10.1016/S0925-4773(97)00083-X. [DOI] [PubMed] [Google Scholar]
- Shan B, Durfee T, Lee WH. Disruption of RB/E2F-1 interaction by single point mutations in E2F-1 enhances S-phase entry and apoptosis. Proc Natl Acad Sci U S A. 1996;93:679–684. doi: 10.1073/pnas.93.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharatos P, Kotsinas A, Evangelou K, Karakaidos P, Vassiliou LV, Rezaei N, Kyroudi A, Kittas C, Patsouris E, Papavassiliou AG, et al. Distinct expression patterns of the transcription factor E2F-1 in relation to tumour growth parameters in common human carcinomas. J Pathol. 2004;203:744–753. doi: 10.1002/path.1582. [DOI] [PubMed] [Google Scholar]
- Isacsson A, Kanje M, Dahlin LB. Induction of activating transcription factor 3 (ATF3) by peripheral nerve compression. Scand J Plast Reconstr Surg Hand Surg. 2005;39:65–72. doi: 10.1080/02844310410004892. [DOI] [PubMed] [Google Scholar]
- Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, Yonenobu K, Ochi T, Noguchi K. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: A novel neuronal marker of nerve injury. Mol Cell Neurosci. 2000;15:170–182. doi: 10.1006/mcne.1999.0814. [DOI] [PubMed] [Google Scholar]
- Keeton AB, Bortoff KD, Franklin JL, Messina JL. Blockade of rapid versus prolonged extracellularly regulated kinase 1/2 activation has differential effects on insulin-induced gene expression. Endocrinology. 2005;146:2716–2725. doi: 10.1210/en.2004-1662. [DOI] [PubMed] [Google Scholar]
- Yan C, Lu D, Hai T, Boyd DD. Activating transcription factor 3, a stress sensor, activates p53 by blocking its ubiquitination. Embo J. 2005;24:2425–2435. doi: 10.1038/sj.emboj.7600712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottone FG, Jr, Moon Y, Kim JS, Alston-Mills B, Ishibashi M, Eling TE. The anti-invasive activity of cyclooxygenase inhibitors is regulated by the transcription factor ATF3 (activating transcription factor 3) Mol Cancer Ther. 2005;4:693–703. doi: 10.1158/1535-7163.MCT-04-0337. [DOI] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci USA. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawadros T, Martin D, Abderrahmani A, Leisinger HJ, Waeber G, Haefliger JA. IB1/JIP-1 controls JNK activation and increased during prostatic LNCaP cells neuroendocrine differentiation. Cell Signal. 2005;17:929–939. doi: 10.1016/j.cellsig.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Hersh LB, Shimojo M. Regulation of cholinergic gene expression by the neuron restrictive silencer factor/repressor element-1 silencing transcription factor. Life Sci. 2003;72:2021–2028. doi: 10.1016/S0024-3205(03)00065-1. [DOI] [PubMed] [Google Scholar]
- Oda Y, Muroishi Y, Misawa H, Suzuki S. Comparative study of gene expression of cholinergic system-related molecules in the human spinal cord and term placenta. Neuroscience. 2004;128:39–49. doi: 10.1016/j.neuroscience.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Kameoka S, Gopalakrishnan V, Aldape KD, Pan ZZ, Lang FF, Majumder S. Conversion of myoblasts to physiologically active neuronal phenotype. Genes Dev. 2004;18:889–900. doi: 10.1101/gad.1179004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman RA, Tomasek JJ, McRae J, Haaksma CJ, Schwartz RJ, Lin HK, Cowan RL, Jones AN, Kropp BP. Decreased expression of smooth muscle alpha-actin results in decreased contractile function of the mouse bladder. J Urol. 2004;172:1667–1672. doi: 10.1097/01.ju.0000139874.48574.1b. [DOI] [PubMed] [Google Scholar]
- Abdelhaleem M. RNA helicases: regulators of differentiation. Clin Biochem. 2005;38:499–503. doi: 10.1016/j.clinbiochem.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Ohmori Y, Schreiber RD, Hamilton TA. Synergy between interferon-gamma and tumor necrosis factor-alpha in transcriptional activation is mediated by cooperation between signal transducer and activator of transcription 1 and nuclear factor kappaB. J Biol Chem. 1997;272:14899–14907. doi: 10.1074/jbc.272.23.14899. [DOI] [PubMed] [Google Scholar]
- Saban MR, Saban R, Hammond TG, Haak-Frendscho M, Steinberg H, Tengowski MW, Bjorling DE. LPS-sensory peptide communication in experimental cystitis. Am J Physiol Renal Physiol. 2002;282:F202–210. doi: 10.1152/ajprenal.0163.2001. [DOI] [PubMed] [Google Scholar]
- http://www.dbi.tju.edu/dbi/tools/paint
- http://www.informatics.jax.org/
- http://www.biobase.de
- http://www.dbi.tju.edu/dbi/tools/paint/index.php?op=FnetBuilder
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Separation of bladder layers (mucosa and detrusor).
Detailed methodology for SSH and analysis of the experimental procedures.
Analysis of ligation.
Analysis of PCR products.
The reduced message of a known housekeeping gene between the subtracted and un-subtracted cDNA over the same number of PCR cycles.