Abstract
PURPOSE. To determine the subcellular distribution of components of the ubiquitin-proteasome pathway (UPP) in lens epithelium and differentiating fibers and to evaluate potential roles of the UPP in eliminating nuclei and other organelles during maturation of lens fibers.
METHODS. Adult bovine lens cryosections were stained for immunofluorescence and analyzed by confocal microscopy. The specificities of the antibodies used in this study were determined by Western blot.
RESULTS Cryosections of bovine lenses show that E1 and Ubc1 were present in both the cytoplasm and the nucleus in epithelial cells, whereas Ubc3 and ubiquitin conjugates were mostly confined to the nucleus, and Ubc4/5 was preferentially localized in clusters in the vicinity of the nuclear membrane. The 19S and 20S proteasome complexes were preferentially localized in the cytoplasm. When the epithelial cells differentiated into fiber cells at the transition zone, all components of the UPP were primarily present in the nucleus, with the exception of Ubc4/5, which was associated with the nuclear membrane.
CONCLUSIONS The results show that during lens fiber differentiation and maturation, components of the UPP are redistributed at subcellular levels. Subcellular localization of an enzyme indicates where the reaction takes place. The primary nuclear localization of the UPP components in the differentiating fibers supports the hypothesis that the UPP may play a role in elimination of nuclei and other organelles during differentiation and maturation of lens fibers.
The ubiquitin proteasome pathway (UPP) is an intracellular nonlysosomal protein-degradation system. Substrates for this pathway are recognized by covalent attachment of multiple ubiquitin molecules and subsequently degraded by the 26S proteasome. Substrates for the UPP include transcription factors, cyclins, proteins with destabilizing N termini, and misfolded or damaged proteins.1-4 Conjugation of ubiquitin to the substrate protein proceeds through a three-step mechanism. Initially, ubiquitin is activated by the ubiquitin-activating enzyme (E1). After activation, one of several ubiquitin conjugating enzymes (Ubc or E2) transfers ubiquitin from E1 to a member of the ubiquitin-protein ligase family (E3), which interacts with substrates specifically.5 The core of the 26S proteasome (also called the 20S proteasome) in eukaryotes consists of 14 different subunits arranged into four stacked, seven-member rings forming a hollow cylinder. The two inner rings are made up of β-subunits and the two outer rings are made up of α-subunits. Another 700-kDa regulatory complex (also called the 19S regulatory complex) can bind to each end of the cylinder in an adenosine triphosphate (ATP)-dependent fashion, forming a fully assembled 26S proteasome.6,7 The six related subunits of the 19S regulatory complex have adenosine triphosphatase (ATPase) activity and are involved in the recruitment and translocation of polyubiquitinated proteins to the 20S catalytic core.8
The eye lens is composed of an anterior monolayer of epithelial cells and the underlying fiber cells that form the bulk of the organ. Epithelial cells differentiate continuously in the equatorial region into lens fibers through a process that involves the synthesis of unique gene products and the degeneration of nuclei and other organelles.9-11 The differentiation-associated elimination of organelles and other cellular proteins suggests the involvement of proteolytic events during lens differentiation. Different proteolytic systems have been identified in the lens, including calpains,12 trypsin-like protease,13 proteasomes,14,15 aminopeptidases,16 and the UPP.17-23 The UPP is the most likely to be involved in differentiation of fiber cells because it appears to be the most substrate-specific proteolytic system. Consistent with the hypothesized roles of the UPP, we previously demonstrated that lens cells undergoing differentiation have a higher concentration of ubiquitin conjugates than nondifferentiated cells.17
Considering the dramatic biochemical, physiological, and morphologic changes involved in differentiation of lens epithelial cells into fiber cells, it is likely that the UPP has different functions at different stages of lens cell differentiation. Although the presence of ubiquitin, E1, some E2s, and the 20S and 19S proteasome subunits has been unequivocally demonstrated in lens epithelial cells17-23and differentiating fiber cells,17,24 the subcellular distribution of these components in lens cells has not been addressed before, nor have changes in the subcellular distribution of components of the UPP during differentiation been evaluated. The current work documented the subcellular distributions of components of the UPP in lens cells and further evaluated the changes of subcellular localizations of these components during differentiation and maturation of lens fibers. The differential subcellular localization of components of the UPP during different stages of differentiation supports the hypothesis that the UPP may have a role in the process of lens differentiation.
MATERIALS AND METHODS
Unless otherwise stated, all chemicals were obtained from Sigma-Aldrich (St. Louis, MO). Rabbit polyclonal antibodies directed against ubiquitin, E1, and Ubc1 (E225k), were produced in synthetic peptides as antigens.19 Antibodies to Ubc3 (Cdc34) and Ubc4 were generous gifts from Mark Goebl (Indiana University School of Medicine, Indianapolis, IN) and Simon Wing (McGill University, Montreal, Quebec, Canada), respectively. Mouse monoclonal antibody (clone LN43) to lamin B was a generous gift from Angus Lamond (Wellcome Trust Biocenter, University of Dundee, Dundee, UK). All other primary antibodies used in this work were purchased from Affiniti (Exeter, UK). Cy2- and tetramethylrhodamine isothiocyanate (TRITC)-conjugated goat anti-rabbit and anti-mouse IgG were obtained from Jackson ImmunoResearch Laboratories (Stratech Scientific Ltd., Luton, UK).
Cell Culture
For primary cultures of lens epithelial cells, eyeballs were removed from adult bovines and the anterior capsule of the lens with the attached epithelium was cut along the equator and cultured in a 24-well plate containing Dulbecco′s modified Eagle′s medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen-Gibco, Inchinnan, UK), 100 IU/mL penicillin, and 100 μg/mL streptomycin, at 37°C in 5% CO2. The epithelial cells were then allowed to spread out from the capsule into the plate.
Protein Electrophoresis and Immunoblot Analysis
The outer cortex of bovine lens was homogenized in Death buffer (50 mM Tris-HCl [pH 7.6], 5 mM EDTA, 1% NP-40, 0.1% SDS, 10 mM NEM [N-ethylmaleimide], and 2 mM AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride]). The homogenate was centrifuged at 12,000g at 4°C for 10 minutes, and the supernatant was mixed with an equal volume of 2× Laemmli buffer, boiled for 5 minutes, loaded on a 12% polyacryl- amide gel and separated by SDS-PAGE. After electrophoretic separation, the proteins were transferred to nitrocellulose with a sandwich transfer system (Bio-Rad, Herts, UK). The blots were probed with the appropriate primary antibodies, as described in the legends and shown by labels on the section of antibodies in the figures. Specific bound antibodies were detected by proper secondary antibodies and visualized by chemiluminescence (SuperSignal; Pierce, Rockford, IL).
Immunostaining of Lens Cryosections or Primary Cultured Lens Epithelial Cells
Adult bovine lenses were dissected from the eye and immediately plunged into liquid nitrogen. The lenses were subsequently sectioned (15 μm thick) in a Shandon cryostat at -20°C. The lens cryosections or primary cultures of lens epithelial cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS). The samples were then washed with PBS and permeabilized with 1% (vol/vol) Triton X-100 in PBS, and blocked with goat serum (1:10) for 20 minutes before incubation with primary antibodies. Incubation with primary antibodies proceeded for 1 hour at room temperature. The samples were then washed three times with PBS before incubation with the secondary antibody for 1 hour at room temperature, after which the specimens were rinsed in PBS and mounted (Glycergel; Dako, Carpinteria, CA). For selected sections, nuclei were stained with 4′,6′-diamino-2-phenylindole (DAPI), which was included in the secondary antibody solution to a final concentration of 1 μg/mL. All antibodies were diluted in PBS containing 0.2% (wt/vol) BSA and 0.02% sodium azide. For negative control, primary antibodies were replaced with preimmune serum (rabbit) or control IgG (mouse). The fluorescence images were collected by a confocal microscope (MRC600; Bio-Rad) equipped with a Kr-Ar laser.
RESULTS
The UPP is involved in many cellular processes, including degradation of obsolete, abnormal, and damaged proteins; the cell cycle25,26; signal transduction27-30; and differentiation.17,24,31-33 As illustrated in Figure 1, lens epithelial cells start to differentiate into fiber cells at the equatorial region of the lens. The differentiating fibers at the transition zone still have nuclei, whereas mature fibers at the center of the lens are without nuclei or other cellular organelles. To study the potential function of the UPP during differentiation and maturation of lens fiber cells, we immunostained sections containing cells at different stages of differentiation with antibodies against components of the UPP and examined the subcellular localization of these components.
FIGURE 1.
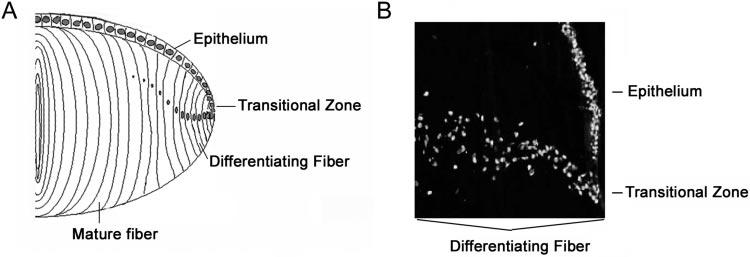
Structure of the lens. A monolayer of epithelial cells covers the anterior part of the lens. The lens epithelial cells start to differentiate into fiber cells at the equatorial region of the lens. Whereas the differentiating fibers at the transition zone retain the nuclei, the mature fibers eliminate nuclei and other organelles. (A) Schematic representation of the sections of the bovine lens. (B) DAPI staining of the equatorial region of the bovine lens to emphasize the transition zone of the lens.
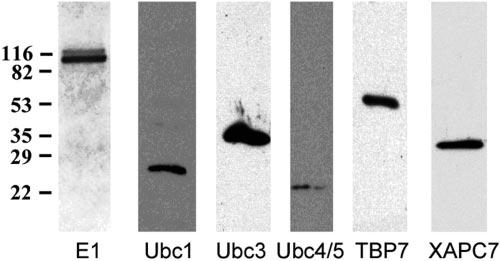
Specificities of the antibodies used in this study were determined by homogenizing the outer cortex of bovine lens, resolving by SDS-PAGE, transferring to nitrocellulose membrane, and probing with each of the antibodies. As shown in Figure 2, the antibody to E1 recognized only the two bands corresponding to E1A and E1B.19 The antibodies to Ubc1, Ubc3, Ubc4/5, and proteasome subunits TBP7 and XAPC, reacted with a single band corresponding to masses of the respective antigen protein (Fig. 2). Minor nonspecific reactions were only detectable when the films were overexposed (data not shown). These data show that the antibodies used in this study are highly specific and are suitable for immunostaining.
FIGURE 2.
Specificity of the antibodies to components of the UPP. The outer cortex of bovine lens was homogenized, resolved by SDS-PAGE, transferred to nitrocellulose membrane, and probed with each of the antibodies indicated. These antibodies reacted primarily with their corresponding antigens, and nonspecific reactions were negligible.
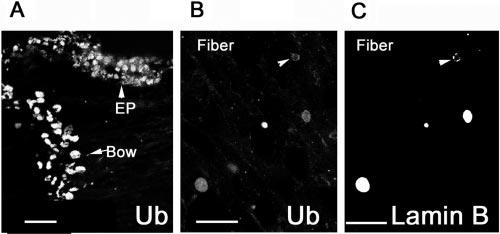
Immunostaining using monoclonal antibodies against ubiquitin-protein conjugates34 showed that most of the ubiquitin-protein conjugates were localized in the nucleus of both epithelial cells and fibers (Fig. 3A). Only a faint staining of ubiquitin conjugates was observed in the cytoplasm of epithelial cells, but not in the cytoplasm of fibers (Fig. 3A). Double staining of cryosections with polyclonal antibodies against ubiquitin and monoclonal antibodies against lamin B showed that ubiquitin was present in all nuclei of the tissue, even in the terminal stages of the lens fiber denucleation (Figs. 3B, 3C), when the nuclei became rounded, and lamin B was extensively fragmented (Figs. 3B, 3C, arrowheads). The primary nuclear staining of ubiquitin conjugates was a result of nonspecific staining of the nuclei, since both monoclonal (Fig. 3A) and polyclonal (Fig. 3B) antibodies to ubiquitin conjugates showed the same pattern of staining, whereas preimmune serum produced only background fluorescence (data not shown). Furthermore, antibody to vimentin showed only cytoplasmic staining and antibody to connexin 43 stained only the cytoplasmic membrane (data not shown). The immunostaining patterns of these molecules are consistent with their cellular localizations.
FIGURE 3.
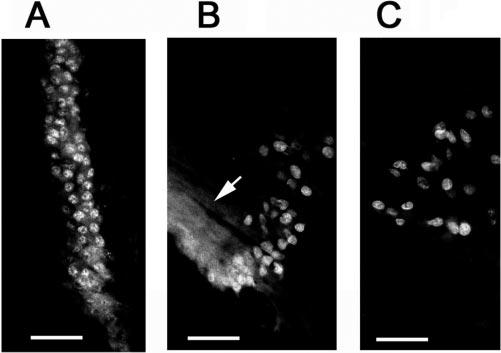
Distribution of ubiquitin in the lens. The epithelium and differentiating fibers were stained with monoclonal antibodies against ubiquitin conjugates (A), showing primarily a nuclear staining. Double staining of the nuclei on fiber cells with polyclonal antibodies to ubiquitin (B) and monoclonal antibodies to lamin B (C) show that ubiquitin was present in nuclei in all stages of denucleation, even when the nuclear membrane was fragmented, as shown by the immunostaining of lamin B. For the negative control, primary antibodies were replaced with preimmune IgG (D). Scale bars, 50 μm.
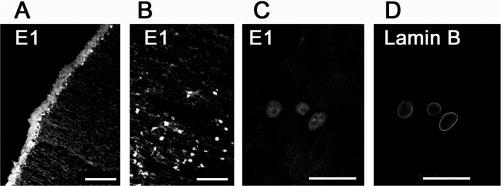
The first step in the UPP is performed by E1, which forms a high-energy thiol-ester bond with the C terminus of ubiquitin and transfers the activated ubiquitin to one of a family of E2s by transthiolation. The immunostaining of cryosections with antibodies to E1 showed that E1 was distributed evenly within the whole cell volume of epithelial cells, including the nuclear and cytosolic compartments (Fig. 4A). In the bow region, where epithelial cells differentiate into fiber cells, the cytoplasmic staining of E1 became weaker and the nuclear staining became stronger (Fig. 4B). As demonstrated by double staining of cryosections with antibodies against E1 (Fig. 4C) and lamin B (Fig. 4D), E1 was barely stained in fibers at later stage of differentiation, when the nuclei became rounded and there were signs of nuclear membrane fragmentation (compare Fig. 4D with 4C).
FIGURE 4.
Distribution of ubiquitin-activating enzyme (E1) in the lens. The epithelial cells (A) and differentiating cells in the bow region (B) were labeled with polyclonal antibodies against E1. (C, D) Double staining of differentiating fiber cells with antibodies to E1 (C) and lamin B (D). These data show that E1 was present in nuclei of lens cells at different stages of differentiation. Scale bars, 50 μm.
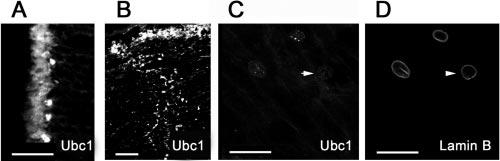
E2s are a family of enzymes that shuttle ubiquitin between E1 and protein substrates, or E3s. More than a dozen E2s have been identified, some of which are involved in specific cellular processes. All the E2s have a highly conserved core domain that contains the active-site cysteine. However, each E2 has a unique C- or N-terminal extension that is thought to contribute to the selectivity of E2 by interacting either with substrates or with a specific E3. In this study, we used antibodies against three different E2s, namely Ubc1 (E225k), Ubc3 (Cdc34), and Ubc4, to study the cellular distributions of these enzymes. Immunostaining of lens cryosections with antibodies to Ubc1 showed that, in the epithelial cell layer, Ubc1 was present in both the cytoplasm and the nucleus (Fig. 5A). Similar to the immunostaining of E1, Ubc1 was primarily localized in the nucleus of the fibers in the bow region (Fig. 5B). In inner layers of the lens, there was virtually no specific staining for Ubc1 (Fig. 5C, arrow), although the lamin B staining was still visible (Fig. 5D, arrowhead). These data indicate that Ubc1 was eliminated from nuclei before the nuclear membrane breakdown. Unlike Ubc1, which showed both cytoplasmic and nuclear staining in the epithelium, Ubc3 was located primarily in the nucleus of lens epithelial cells (Fig. 6A). Cytoplasmic staining of Ubc3 was observed only in the epithelial cells in the equatorial region, where epithelial cells exit from the cell cycle and start to differentiate (Fig. 6B, arrow). The cytoplasmic staining of this enzyme in the equatorial region may be related to the nuclear membrane breakdown during mitosis of the dividing epithelial cells. As for E1 and Ubc1, Ubc3 staining was confined to the nuclei of differentiating fibers (Figs. 6B, 6C), and it was not observable in the denucleated lens fibers.
FIGURE 5.
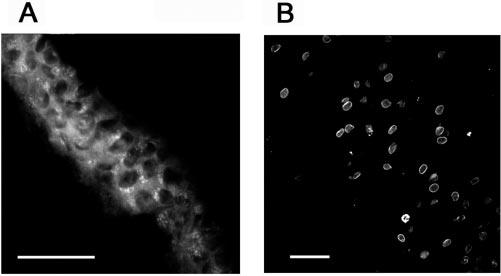
Distribution of Ubc1 (E225k) in the lens. The epithelium (A) and differentiating fiber cells (B) were stained with polyclonal antibodies to Ubc1. Double staining of fiber cells with antibodies to Ubc1 (C) and lamin B (D) showed that Ubc1 was eliminated from nuclei of fibers at later stages of differentiation (arrowheads). Scale bars, 50 μm.
FIGURE 6.
Subcellular localization of Ubc3 (Cdc34) in the lens. Bovine lens cryosections were stained with polyclonal antibodies against Ubc3. (A) Central epithelium, (B) equatorial region epithelium, and (C) differentiating fibers. Scale bars, 50 μm.
Ubc4 and -5 are a subclass of closely related E2s that are involved in selective degradation of damaged or obsolete proteins.5 Antibodies raised against Ubc4 react with both Ubc4 and -5. To indicate reactivity to both of these closely related proteins they are referred to herein as Ubc4/5. Consistent with its putative function, strong Ubc4/5 immunostaining was observed in the cytosol of epithelial cells (Fig. 7A). High-magnification examination of the cytosolic immunostaining of Ubc4/5 in the epithelium revealed that this enzyme localized preferentially in clusters in the vicinity of the nuclei or as scattered dots in the cytosol (Fig. 7A). As lens fibers differentiated in the bow region, the cytoplasmic staining became weaker and the immunostaining pattern for Ubc4/5 at this stage of differentiation was similar to that for lamin B, suggesting that Ubc4/5 is associated with the nuclear membrane (Fig. 7B).
FIGURE 7.
Immunolocalization of Ubc4/5 in the lens. Bovine lens cryosections were immunostained with polyclonal antibodies to Ubc4. (A) Epithelium; (B) differentiating fibers. Scale bars, 50 μm.
To further study the potential role of the UPP during lens cells differentiation, we determined subcellular localization of subunits of the 26S proteasome. The 26S proteasome is composed of two 19S regulatory complexes and one 20S catalytic core. Immunostaining using antibodies raised against different subunits of the 20S proteasome showed that it was localized in both the nuclear and the cytoplasmic compartments of the epithelial cells. In the cytoplasm, the 20S proteasome appeared to be evenly distributed (Figs. 8A, 8B). However, the 20S proteasome appeared to form aggregate-like structures in the nucleus of epithelial cells (Figs. 8A-D). Double staining with two different antibodies to different subunits of the 20S proteasome (XAPC7 and core) showed a complete colocalization, indicating that all these subunits are assembled into the 20S proteasome in lens epithelial cells (Figs. 8C, 8D). In contrast to observations in epithelial cells, subunits of the 20S proteasome in differentiating fibers were exclusively localized in the nucleus (Fig. 8E). Double staining of the sections with antibodies to lamin B and the core of the proteasome showed that these proteasome subunits were present in nuclei until later stages of denucleation (Fig. 8F, arrow), when lamin B was fragmented (Fig. 8G, arrow)
FIGURE 8.
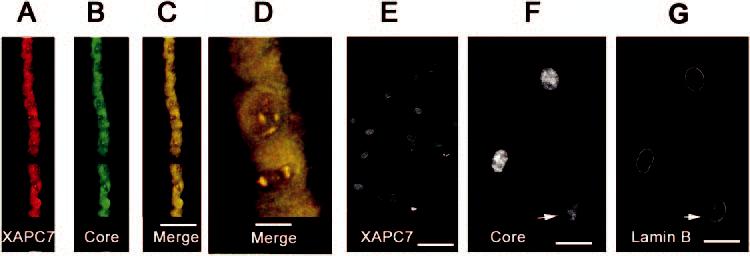
Immunolocalization of different subunits of the 20S proteasome. Lens cryosections were probed with monoclonal antibodies to the XAPC7 subunit (subunit α4) (A) and polyclonal antibodies to the “core” (B). Merged images (C, D) of lens epithelial layer show that these subunits colocalized in the cytoplasm and nucleus. In fiber cells, the 20S subunit XAPC7 localized exclusively to the nuclei (E). Double staining of nuclei with 20S core (F) and lamin B (G) antibodies shows that the proteasome remained in the nuclei when the nuclear membrane started to be dismantled. Scale bars: (C, E)50 μm; (D) 10 μm; (F, G) 20 μm.
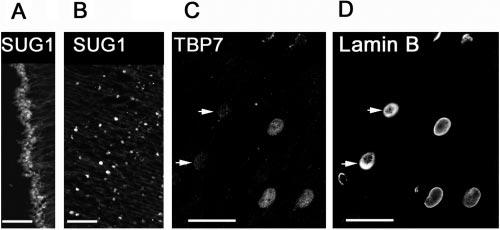
We then examined the distribution of three different subunits of the regulatory 19S particle: MSS1, TBP7, and SUG1. These subunits showed different cellular distribution in the lens. Although TBP7 (Fig. 9A) and MSS1 (Fig. 9D) showed immunostaining in the cytoplasm, SUG1 was localized predominantly in the nucleus of epithelial cells (Fig. 10A). In differentiating fiber cells at the bow region, all three subunits were restricted to the nuclei (Figs. 9A, 9E, 10B). However, the subunit TBP7 appeared to be eliminated from the nucleus before denucleation, as demonstrated by the absence of TBP7 staining in the nuclei (Fig. 10C, arrows), which still showed lamin B staining (Fig. 10D). The same result was observed for the subunit SUG1 (data not shown).
FIGURE 9.
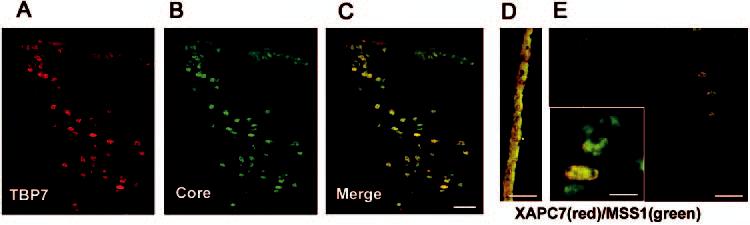
Colocalization of the 19S regulatory complex and the 20S catalytic core. The lens cryosections were double stained with antibodies against the 19S subunit TBP7 and the 20S core and (A, B, C) or double stained with antibodies to the 20S subunit XAPC7 and the 19S subunit MSS1 (D, E). (A, B, C) Staining of the epithelial layer and bow regions is shown. (D) Staining of central epithelium; (E) staining of inner fibers. (D, E) Merged images showing the colocalization of 20S and 19S subunits. In all cases the 20S subunits were stained with TRITC (red) and 19S subunits were stained with Cy2 (green). Colocalization of the subunits of the 20S proteasome and the 19S regulatory complex appeared as yellow spots. (E, inset) Staining showed that the 20S and 19S complex of the 26S proteasome did not always colocalize. Scale bars: (C-E) 50 μm; (E, inset) 10 μm.
FIGURE 10.
Immunolocalization of different subunits of the 19S proteasome. Lens sections stained with antibodies to the 19S subunit SUG1 show both nuclear and cytoplasmic staining in the epithelium (A) and exclusively nuclear staining in the differentiating fibers (B. Double staining with antibodies to the 19S subunit TBP7 (C) and lamin B (D) shows that TBP7 is eliminated from some nuclei at late stages of fiber differentiation. Scale bars, 50 μm.
Double staining for subunits of the 20S proteasome and the 19S regulatory complex showed that although both of these complexes were present in the same compartments, they did not always colocalize (Fig. 9C). These data indicate that the 20S proteasome is not always assembled with the 19S regulatory complex to form a functional 26S proteasome. This finding is further manifested in the nuclei of differentiating fibers at the bow region, where only a small fraction of the 20S proteasome colocalized with the 19S regulatory complex (Fig. 9E). Similar results were observed in cultured lens epithelial cells, in which only ∼50% of the 20S proteasome colocalized with the 19S regulatory complex (data not shown). Taken together, these data indicate that the assembly of the 19S with 20S subunit is a dynamic process.
DISCUSSION
Maintenance of lens transparency involves elimination of large organelles that scatter light in most tissues. These include degradation of nuclei, mitochondria, lysosomes, and endoplasmic reticulum. This, together with expression of fiber-specific proteins, results in dramatic changes in cellular protein composition. We previously showed that cells undergoing differentiation have higher concentrations of high mass ubiquitin conjugates than nondifferentiated cells.17 In a recent study, we demonstrated that different components of the ubiquitin conjugating system were differentially regulated in the in vitro lens differentiating model.24 Taken together, these data suggest that the UPP is involved in removal of nuclei and other organelles during maturation of lens fibers.
The work reported here describes the immunohistochemical distribution of different components of the UPP in the lens. As summarized in Table 1, components of the UPP redistributed at subcellular levels during lens differentiation and maturation. The subcellular localizations of these components support the hypothesis that the UPP plays a role in elimination of nuclei and other organelles during the differentiation process. However, this does not exclude other functions of the UPP during differentiation. For example, caspase-3 is involved in lens fiber differentiation,35,36 and the proteasome is involved in control caspase-3 activity and apoptosis.37 Thus, it is conceivable that in the lens, the sequestering of the proteasome in the nuclei may be part of a general strategy that regulates the precise sequence of proteolytic events for proper differentiation of lens epithelial cells into fibers.
TABLE 1.
Summary of Subcellular Localizations of Components of the UPP in the Lens
| Epithelial Cells | Differentiating Fibers | |
|---|---|---|
| Ubiquitin conjugates | Mainly in nuclei | Nuclei |
| E1 | Cytoplasm and nuclei | Nuclei |
| Ubc1 | Cytoplasm and nuclei | Nuclei |
| Ubc3 | Nuclei | Nuclei |
| Ubc4/5 | Cytoplasm and perinuclear | Nuclear membrane |
| 20S core complex of proteasome | Cytoplasm and nuclei | Nuclei |
| 19S regulatory complex of proteasome | Cytoplasm and nuclei | Nuclei |
The data obtained by immunostaining of cryosections with antibodies raised against ubiquitin showed that ubiquitin conjugates in lens cells were primarily present in nuclei, and cytoplasm levels of ubiquitin conjugates were very low, although active ubiquitin conjugation is present in cytoplasm.An explanation of these data is that ubiquitin conjugates in the cytoplasm were rapidly degraded by the 26S proteasome, whereas those in the nuclei were relatively stable, probablybecause of the absence of assembled 26S proteasome. The presence of ubiquitin conjugates in all nuclei at different stages of lens fiber maturation indicates that the UPP may be involved in the elimination of nuclei during terminal differentiation oflens fiber cells.
The different subcellular localization and redistribution of the E2s during lens fiber differentiation and maturation is consistent with the notion that each E2 mediates the ubiquiti-nation and turnover of specific substrates and is involved in specific cellular processes. The nuclear localization of Ubc3 and cytoplasmic localization of Ubc4/5 in the epithelial cells and the differentiating fibers suggests that substrates for Ubc3 are primarily nuclear proteins, whereas substrates for Ubc4/5 are mainly cytoplasmic or nuclear membrane-associated proteins. The subcellular localizations of these E2s are consistent with their functions demonstrated in other types of cells. The demonstrated substrates for Ubc3 include cyclin E and various inhibitors of cyclin-dependent kinases, such as p21WAF and p27Kip.23,38-41 These proteins are located in nuclei and play important roles in the regulation of the cell cycle process. Our unpublished data also show that overexpression of mutant Ubc3 in lens epithelial cells delay the cell cycle at the G2/M phase. As mentioned earlier, Ubc4/5 are involved in the degradation of damaged or obsolete proteins. The perinuclear localization of Ubc4/5 suggests that substrates for these E2s are associated with the endoplasmic reticulum. The association of Ubc4/5 with the nuclear membrane of differentiating fibers suggests that Ubc4/5 may be involved in degradation of the nuclear membrane or membrane-associated proteins during fiber cell maturation. When Ubc4 was supplemented to the supernatant of the outer cortex of the bovine lens, there was a distinct ubiquitin conjugate formed, but this conjugate did not appear in the supernatant from the inner lens cortex.22 Thes data suggest that Ubc4/5 is probably involved in the elimination of organelles during lens differentiation. However, further evidence to support this hypothesis is needed.
It is also noteworthy that ubiquitin conjugates were still retained in the nuclei when the nuclear membrane started to break down at the late stages of differentiating fibers (Fig. 3), whereas E1, Ubc1, and Ubc3 were eliminated from nuclei before nuclear membrane breakdown (Figs. 4, 5, 6, 7). These data suggest that ubiquitin conjugates in the fiber nuclei are formed before the disassembly of the nuclear membrane. Ubiquitination of these proteins may predispose them for protea-some-dependent degradation upon nuclear membrane breakdown.
Previous studies regarding the localization of the 26S proteasome in higher eukaryotes have shown controversial results. Some reports showed that both the 20S proteasome and the 19S regulatory complexes were localized in the nucleus and the cytoplasm, and some were associated with the cytoplasmic surface of the endoplasmic reticulum.42-44 A study performed in mammalian living cells using a green fluorescent protein (GFP)-tagged 20S proteasome showed that the protea-some is equally distributed in the nucleus (although excluded from the nucleoli) and in the cytoplasm.44 Other studies found that the 20S proteasome was localized almost exclusively in the nucleus45 or cytoplasm.46,47 There are also reports that show that the 20S proteasome, the 19S regulatory complex, and ubiquitin conjugates accumulate in the centrosome.48 The association of the 26S proteasome with intermediate filaments was also reported.49,50 A recent work that investigated the subcellular distribution of the different forms of proteasomes in mammalian cells showed that both the 20S proteasome and the regulatory complexes were found in nuclear, cytosolic, and microsomal preparations isolated from rat liver.51 The apparent discrepancy regarding subcellular distribution of the subunits of the 20S proteasome and the 19S regulatory complex may result, among other factors, from the use of different types of cells or cells at different stages of the cell cycle.
The results reported herein show that there was a clear redistribution of the components of the UPP during different stages of lens cell differentiation. In the quiescent epithelial cells, most components of the UPP were present in both the cytoplasm and the nucleus, whereas in the differentiating fiber cells most of these components were restricted to the nucleus. Moreover, our results also demonstrated that different subunits of the 19S complex have different distributions and that these subunits are not always colocalized with the 20S proteasome. This suggests that the 19S complex and 20S proteasome are not always assembled to form an intact 26S proteasome, or that the assembly of functional 26S proteasome is a dynamic process. Similar data have also been reported, showing that the components of the 19S regulatory complex could exist in multiple complexes or as a free subunit that may have specific functions other than proteolysis.52,53 The different immuno-staining patterns observed for the 19S and the 20S subunits may also reflect the dynamic reorganization (assembly- disassembly) of the 26S proteasome at different stages of cell differentiation. Except for Ubc4, all components of the UPP examined in this study showed exclusive nuclear staining in the fibers. The disappearance of these components from the fiber cytoplasm may be due to nuclear importation of these components during differentiation or to antigen mask or dilution by the dramatic increase in cell volume and expression crystallins in these cells.
In summary, this is the first systematic examination of subcellular localization of components of the UPP during lens differentiation and development. The redistribution of components of the UPP observed at the bow region may also suggest that specific enzymes of this pathway are recruited to particular subcellular compartments where they may participate in the destruction of obsolete proteins during the differentiation process. One of the hypothesized functions of the UPP during differentiation is to remove nuclei of fibers. The primarily nuclear localization of most components of the UPP in differentiating fibers supports this hypothesis. Thus, this work laid a solid foundation for future studies to test mechanistically the hypothesis that the UPP plays a role in elimination of nuclei and other organelles during lens fiber maturation.
Acknowledgments
The authors thank Mark Siegal for help in preparation of the manuscript.
Footnotes
Supported by grants from the Portuguese Foundation for Science and Technology (FCT), Program POCTI, with the support of FEDER (PP, HG); National Eye Institute Grants EY11717 (FS) and EY13250 (AT); and Core Grant EY13078.
Disclosure: H. Girẽo, None; P. Pereira, None; A. Taylor, None; F. Shang, None
References
- 1.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 2.King RW, Deshaies RJ, Peters J-M, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1658. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 3.Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varshavsky A, Turner G, Du F, Xie Y. The ubiquitin system and the N-end rule pathway. Biol Chem. 2000;381:779–789. doi: 10.1515/BC.2000.101. [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover A, Orian A, Schwartz AL. The ubiquitin-mediated proteolytic pathway: mode of action and clinical implications. J Cell Biochem Suppl. 2000;34:40–51. doi: 10.1002/(sici)1097-4644(2000)77:34+<40::aid-jcb9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 6.DeMartino GN, Slaughter CA. The proteasome, a novel protease regulated by multiple mechanisms. J Biol Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- 7.Gorbea C, Taillandier D, Rechsteiner M. Assembly of the regulatory complex of the 26S proteasome. Mol Biol Rep. 1999;26:15–19. doi: 10.1023/a:1006957802028. [DOI] [PubMed] [Google Scholar]
- 8.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. Aproteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 9.Kuwabara T. The maturation of lens cells: a morphological study. Exp Eye Res. 1975;20:427–433. doi: 10.1016/0014-4835(75)90085-8. [DOI] [PubMed] [Google Scholar]
- 10.Bassnett S, Beebe DC. Coincident loss of mitochondria and nucleiduring lens fiber cell differentiation. Devel Dyn. 1992;194:85–93. doi: 10.1002/aja.1001940202. [DOI] [PubMed] [Google Scholar]
- 11.Piatigorsky J. Lens differentiation in vertebrate: a review of cellular and molecular features. Differentiation. 1981;19:134–153. doi: 10.1111/j.1432-0436.1981.tb01141.x. [DOI] [PubMed] [Google Scholar]
- 12.Lipman RD, Cyr DE, David LL, Taylor A. Calpain in culture bovinelens epithelial cells. Curr Eye Res. 1991;10:11–17. doi: 10.3109/02713689109007606. [DOI] [PubMed] [Google Scholar]
- 13.Tse SS, Ortwerth BJ. Activation of proteases with trypsin-likespecificity from bovine and human lenses. Exp Eye Res. 1981;32:605–614. doi: 10.1016/s0014-4835(81)80009-7. [DOI] [PubMed] [Google Scholar]
- 14.Murakami K, Jahngen JH, Lin SW, Davies KJA, Taylor A. Lensproteosome shows enhanced rates of degradation of hydroxylradical modified alpha-crystallin. Free Rad Biol Med. 1990;8:217–222. doi: 10.1016/0891-5849(90)90066-r. [DOI] [PubMed] [Google Scholar]
- 15.Wagner BJ, Margolis JW. Thermal stability and activation of bovinelens multicatalytic proteinase complex (proteasome) Arch Biochem Biophys. 1993;307:146–152. doi: 10.1006/abbi.1993.1573. [DOI] [PubMed] [Google Scholar]
- 16.Talor A, Sanford D, Nowell T. Structure and function of bovinelens aminopeptidase and comparison with homologous aminopeptidases. In: Taylor A, ed. Aminopeptidases. Austin, TX: RG Landes. 1996:21–59. [Google Scholar]
- 17.Shang F, Gong X, McAvoy JW, Chamberlain C, Nowell TR, Taylor A. Ubiquitin-dependent pathway is up-regulated in differentiatinglens cells. Exp Eye Res. 1999;68:179–192. doi: 10.1006/exer.1998.0576. [DOI] [PubMed] [Google Scholar]
- 18.Shang F, Nowell TR, Jr, Taylor A. Removal of oxidatively damaged proteins from lens cells by the ubiquitin-proteasome pathway. ExpEye Res. 2001;73:229–238. doi: 10.1006/exer.2001.1029. [DOI] [PubMed] [Google Scholar]
- 19.Shang F, Deng G, Obin M, et al. Ubiquitin-activating enzyme (E1)isoforms in lens epithelial cells: origin of translation, E2 specificity and cellular localization determined with novel site-specific antibodies. Exp Eye Res. 2001;73:827–836. doi: 10.1006/exer.2001.1091. [DOI] [PubMed] [Google Scholar]
- 20.Shang F, Gong X, Taylor A. Activity of ubiquitin dependent pathway in response to oxidative stress: ubiquitin activating enzyme(E1) is transiently upregulated. J Biol Chem. 1997;272:23086–23093. doi: 10.1074/jbc.272.37.23086. [DOI] [PubMed] [Google Scholar]
- 21.Shang F, Taylor A. Oxidation and protein degradation in the lens. Mol Aspects Med. 1997;18:339–352. [Google Scholar]
- 22.Pereira P, Shang F, Girão H, Taylor A. Lens fibers have a fullyfunctional ubiquitin-proteasome pathway. Exp Eye Res. 2003;76:623–631. doi: 10.1016/s0014-4835(03)00020-4. [DOI] [PubMed] [Google Scholar]
- 23.Liu Q, Shang F, Guo W, Hobbs M, Valverde P, Reddy V, Taylor A. Regulation of the ubiquitin proteasome pathway in human lensepithelial cells during the cell cycle. Exp Eye Res. 2004;78:197–205. doi: 10.1016/j.exer.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Guo W, Shang F, Liu Q, Urim L, West-Mays J, Taylor A. Differential regulation of components of the ubiquitin-proteasome pathwayduring lens cell differentiation. Invest Ophthalmol Vis Sci. 2004;45:1194–1201. doi: 10.1167/iovs.03-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin geneis essential for resistance to high temperature, starvation, andother stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 26.Peters JM. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 27.Obin M, Lee BY, Meinke G, et al. Ubiquitylation of the transducinbetagamma subunit complex: regulation by phosducin. J BiolChem. 2002;277:44566–44575. doi: 10.1074/jbc.M205308200. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 isa ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 29.Deng L, Wang C, Spencer E, et al. Activation of the IkappaB kinasecomplex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 30.Spencer E, Jiang J, Chen ZJ. Signal-induced ubiquitination of IkappaBalpha by the F-box protein Slimb/beta-TrCP. Genes Dev. 1999;13:284–294. doi: 10.1101/gad.13.3.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shang F, Taylor A. Function of the ubiquitin proteolytic pathway inthe eye. Exp Eye Res. 2004;78:1–14. doi: 10.1016/j.exer.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Haldeman MT, Finley D, Pickart CM. Dynamics of ubiquitin conjugation during erythroid differentiation in vitro. J Biol Chem. 1995;270:9507–9516. doi: 10.1074/jbc.270.16.9507. [DOI] [PubMed] [Google Scholar]
- 33.Wefes I, Mastrandrea LD, Haldeman M, et al. Induction of ubiquitin-conjugating enzymes during terminal erythroid differentiation. Proc Natl Acad Sci USA. 1995;92:4982–4986. doi: 10.1073/pnas.92.11.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujimuro M, Sawada H, Yokosawa H. Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains ofpolyubiquitinated proteins. FEBS Lett. 1994;349:173–180. doi: 10.1016/0014-5793(94)00647-4. [DOI] [PubMed] [Google Scholar]
- 35.Wride MA, Parker E, Sanders EJ. Members of the bcl-2 and caspasefamilies regulate nuclear degeneration during chick lens fibre differentiation. Dev Biol. 1999;213:142–156. doi: 10.1006/dbio.1999.9375. [DOI] [PubMed] [Google Scholar]
- 36.Ishizaki Y, Jacobson MD, Raff MC. A role for caspases in lens fiberdifferentiation. J Cell Biol. 1998;140:153–158. doi: 10.1083/jcb.140.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersson M, Sjostrand J, Petersen A, Honarvar AK, Karlsson JO. Caspase and proteasome activity during staurosporin-induced apoptosis in lens epithelial cells. Invest Ophthalmol Vis Sci. 2000;41:2623–2632. [PubMed] [Google Scholar]
- 38.Pagano M, Tam SW, Theodoras AM, et al. Role of the ubiquitinproteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 39.Reymond F, Wirbelauer C, Krek W. Association of human ubiquitin-conjugating enzyme CDC34 with the mitotic spindle in anaphase. J Cell Sci. 2000;113:1687–1694. doi: 10.1242/jcs.113.10.1687. [DOI] [PubMed] [Google Scholar]
- 40.Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with theSKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin Dproteins. Proc Natl Acad Sci USA. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koepp DM, Schaefer LK, Ye X, et al. Phosphorylation-dependentubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 42.Rivett AJ, Palmer A, Knecht E. Electron microscopic localization of the multicatalytic proteinase complex in rat liver and in culturedcells. J Histochem Cytochem. 1992;40:1165–1172. doi: 10.1177/40.8.1619280. [DOI] [PubMed] [Google Scholar]
- 43.Palmer A, Rivett AJ, Thomson S. Subpopulations of proteasomes inrat liver nuclei, microsomes and cytosol. Biochem J. 1996;316:401–407. doi: 10.1042/bj3160401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reits EA, Benham AM, Plougastel B, Neefjes J, Trowsdale J. Dynamics of proteasome distribution in living cells. EMBO J. 1997;16:6087–6094. doi: 10.1093/emboj/16.20.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stauber WT, Fritz VK, Maltin CA, Dahlmann B. Localization of amulti-catalytic, high-molecular mass proteinase in the nuclei ofmuscle cells. Histochem J. 1987;19:594–597. doi: 10.1007/BF01687368. [DOI] [PubMed] [Google Scholar]
- 46.Kloetzel PM, Falkenburg PE, Hossl P, Glatzer KH. The 19S ring-type particles of Drosophila: cytological and biochemical analysis oftheir intracellular association and distribution. Exp Cell Res. 1987;170:204–213. doi: 10.1016/0014-4827(87)90130-3. [DOI] [PubMed] [Google Scholar]
- 47.Beyette JR, Mykles DL. Immunocytochemical localization of the multicatalytic proteinase (proteasome) in crustacean striated muscles. Muscle Nerve. 1992;15:1023–1035. doi: 10.1002/mus.880150907. [DOI] [PubMed] [Google Scholar]
- 48.Wigley WC, Fabunmi RP, Lee MG, et al. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grossi de Sa MF, Martins de Sa C, Harper F, Olink-Coux M, Huesca M, Scherrer K. The association of prosomes with some of the intermediate filament networks of the animal cell. J Cell Biol. 1988;107:1517–1530. doi: 10.1083/jcb.107.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pal JK, Gounon P, Grossi de Sa MF, Scherrer K. Presence and distribution of specific proteasome antigens change as a function of embryonic development and tissue-type differentiation in Pleurodeles waltl. J Cell Sci. 1988;90:555–567. doi: 10.1242/jcs.90.4.555. [DOI] [PubMed] [Google Scholar]
- 51.Brooks P, Fuertes G, Murray RZ, et al. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem J. 2000;346:155–161. [PMC free article] [PubMed] [Google Scholar]
- 52.Russell SJ, Sathyanarayana UG, Johnston SA. Isolation and characterization of SUG2: a novel ATPase family component of the yeast26 S proteasome. J Biol Chem. 1996;271:32810–32817. doi: 10.1074/jbc.271.51.32810. [DOI] [PubMed] [Google Scholar]
- 53.Fraser RA, Rossignol M, Heard DJ, Egly JM, Chambon P. SUG1, aputative transcriptional mediator and subunit of the PA700 proteasome regulatory complex, is a DNA helicase. J Biol Chem. 1997;272:7122–7126. doi: 10.1074/jbc.272.11.7122. [DOI] [PubMed] [Google Scholar]