Summary
Class III homeodomain-leucine zipper proteins regulate critical aspects of plant development, including lateral organ polarity, apical and lateral meristem formation, and vascular development. ATHB15, a member of this transcription factor family, is exclusively expressed in vascular tissues. Recently, a microRNA (miRNA) binding sequence has been identified in ATHB15 mRNA, suggesting that a molecular mechanism governed by miRNA binding may direct vascular development through ATHB15. Here, we show that miR166-mediated ATHB15 mRNA cleavage is a principal mechanism for the regulation of vascular development. In a gain-of-function MIR166a mutant, the decreased transcript level of ATHB15 was accompanied by an altered vascular system with expanded xylem tissue and interfascicular region, indicative of accelerated vascular cell differentiation from cambial/procambial cells. A similar phenotype was observed in Arabidopsis plants with reduced ATHB15 expression but reversed in transgenic plants overexpressing an miR166-resistant ATHB15. ATHB15 mRNA cleavage occurred in standard wheat germ extracts and in Arabidopsis and was mediated by miR166 in Nicotiana benthamiana cells. miR166-assisted ATHB15 repression is likely to be a conserved mechanism that regulates vascular development in all vascular plants.
Keywords: Arabidopsis, ATHB15, HD-ZIP, microRNA, mRNA cleavage, vascular development
Introduction
Vascular system is an elaborate network of conducting tissues that interconnects all plant organs and transports water, minerals, organic compounds, and signaling molecules throughout the plant body. It consists of two conducting tissues, xylem and phloem, and procambial/cambial cells.
Vascular development is initiated by the formation of provascular cells that subsequently develop into procambium, from which both conducting tissues are eventually differentiated (Steeves and Sussex, 1989). In older plant parts, vascular tissues can develop through the activity of a secondary meristem, called vascular cambium (Esau, 1965). Plant growth hormones, such as auxins and brassinosteroids, also play regulatory roles in vascular tissue differentiation (Carland et al., 2002; Jang et al., 2000; Sachs, 2000). It is now generally accepted that a unified molecular mechanism modulates temporal and spatial development of vascular tissues in different plant species, although vascular patterns and organizations are quite diverse (Baima et al., 2001). However, the molecular components and schemes that regulate vascular development are poorly understood.
Recent application of molecular genetic tools, mainly established in Arabidopsis, greatly accelerated the identification of genes involved in vascular development and the elucidation of regulatory mechanisms at the molecular level. A subset of class III homeodomain-leucine zipper (HD-ZIP III) transcription factors has been implicated in vascular development. In Arabidopsis, the HD-ZIP III gene family includes five members; ATHB15, ATHB8, PHAVOLUTA (PHV), PHABULOSA (PHB), and REVOLUTA (REV). PHV, PHB, and REV are expressed in various plant tissues, including vascular tissues, apical and floral meristems, and the adaxial domain of lateral organs (Emery et al., 2003; McConnell et al., 2001; Otsuga et al., 2001). By contrast, ATHB15 and ATHB8 are predominantly expressed in vascular tissue (Baima et al., 1995; Ohashi-Ito and Fukuda, 2003), suggesting that they may have some role in vascular development.
ATHB8 is induced by auxin and promotes procambial/ cambial cell differentiation into xylem tissues (Baima et al., 2001). However, a loss-of-function ATHB8 mutant does not show any defects in vascular patterning and development, indicating that ATHB8 is not essential for vascular development. Although ATHB15 has not yet been examined molecular genetically, the predominant expression of ATHB15 and ZEHB13, an ATHB15 gene homolog from Zinnia elegans, in vascular tissue suggests a role for ATHB15 in vascular development (Ohashi-Ito and Fukuda, 2003). Interestingly, a microRNA (miRNA) binding sequence has been recently predicted in ATHB15 mRNA (Bartel and Bartel, 2003; Reinhart et al., 2002; Rhoades et al., 2002). It is therefore envisioned that a mechanism governed by miRNA binding might direct the regulatory role of ATHB15 during vascular development.
miRNAs are small non-coding RNA molecules that regulate target genes by either mRNA cleavage or translational repression (Bartel, 2004; Kidner and Martienssen, 2003). They exert their regulatory role through complementary base pairing to target mRNAs. A handful of miRNAs and their target genes have been identified and demonstrated to regulate various plant developmental processes in recent years, including flowering time control, floral development, leaf polarity, and leaf morphogenesis (Aukerman and Sakai, 2003; Chen, 2004; Emery et al., 2003; Juarez et al., 2004; Kidner and Martienssen, 2004; Laufs et al., 2004; Mallory et al., 2004a,b; McHale and Koning, 2004; Palatnik et al., 2003; Vaucheret et al., 2004). In some cases, the regulatory mechanisms and biochemical actions of miRNAs have been extensively studied using mutants of miRNAs and their target genes. In other cases, only the putative target genes have been studied without functional characterization of the relevant miRNA mutants.
In this work, we isolated an Arabidopsis mutant in which an MIR166a gene is activated by the insertion of the CaMV 35S enhancer and demonstrated that miR166-mediated ATHB15 mRNA cleavage is a principal mechanism for the regulation of vascular development in inflorescence stems. In the gain-of-function MIR166a mutant, the ATHB15 transcript level was drastically reduced, and the vascular system was severely altered with expanded xylem tissue and interfascicular region, indicating that vascular cell differentiation is greatly promoted. ATHB15 mRNA is cleaved in standard wheat germ extracts and in Arabidopsis, and its cleavage is mediated by miR166 in Nicotiana benthamiana cells. The miR166/165 complementary sequence is highly conserved in mRNAs of ATHB15 and its gene homologs from various plant species (Floyd and Bowman, 2004). It is therefore likely that miR166-mediated HD-ZIP III gene repression is conserved in all vascular plants.
Results
Isolation of the men1 mutation
To explore the molecular genetic components and regulatory mechanisms that govern plant growth and development, we generated an Arabidopsis mutant pool by randomly integrating the CaMV 35S enhancer into the genome of Columbia accession (Col-0) (Weigel et al., 2000). We then searched for morphogenetic mutants with altered leaf and inflorescence stem morphologies. Among the isolated mutants, we chose a morphogenetic mutant with severe growth retardation.
The mutant, designated men1 for meristem enlargement 1, exhibits pleiotropic alterations in the inflorescence stem and leaf morphologies and floral structure in addition to repressed growth (Figure 1). Homozygous men1 lines stopped growing at the very early seedling stage (<1 cm in height) and eventually died without further development in most lines. Although one or two flowers were occasionally produced in a few lines, they were sterile, possibly due to extremely short carpels (Figure 2c). When homozygous men1 plants were pollinated with wild-type pollens, no seeds were produced. However, seeds were successfully produced from wild-type plants pollinated with homozygous men1 pollens (data not shown). Therefore, all experiments were carried out using heterozygous men1/+ plants.
Figure 1. men1/+ plants with fasciated stem, enlarged meristem, and short carpel.

Adult plants are shown. Inset shows a magnified image of the upper part of a fasciated inflorescence stem.
Figure 2. Inflorescence stem morphology and floral structure.
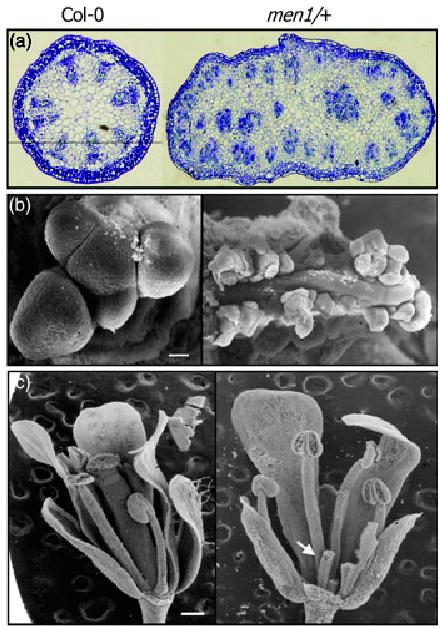
(a) Transverse inflorescence stem sections stained with toluidine blue. Note that vascular patterning is disrupted in men1/+.
(b) Scanning electron microscopic (SEM) images of shoot apical meristems. Scale bar is 20 μm.
(c) SEM images of flowers. Some sepals and petals were removed to expose carpels. Carpel is extremely short in men1/+ (arrow). Scale bar is 200 μm.
One prominent feature of men1/+ is the fasciated inflorescence stem with disrupted radial vascular patterning (Figures 1 and 2a), which may be developmentally related to apical meristem enlargement (Figure 2b). Stem fasciation may be due to disrupted cell partitioning from the central and peripheral zones to the rib zone during shoot apical meristem development (Clark et al., 1996; Laufs et al., 1998). Floral development was also affected by the men1 mutation. More flowers were produced in men1/+ than in wild-type plants. In addition, although the numbers of floral organs are unchanged, the carpels are very short and sterile (Figure 2c).
men1 is a gain-of-function MIR166a mutant
Mapping of the T-DNA insertion site by the three-step thermal asymmetric interlaced (TAIL)-PCR (Liu et al., 1995) revealed that the 35S enhancer was inserted into a region between two previously annotated genes At2g46680 and At2g46690 (Figure 3a). However, the expression of the two genes was not altered in the mutant, and transgenic plants that express either one of the two genes under the control of the CaMV 35S promoter did not show any phenotypic changes (data not shown), indicating that the phenotypic alterations observed in the mutant are not caused by the misexpression of the two genes.
Figure 3. Activation tagging of an MIR166a gene in men1/+.
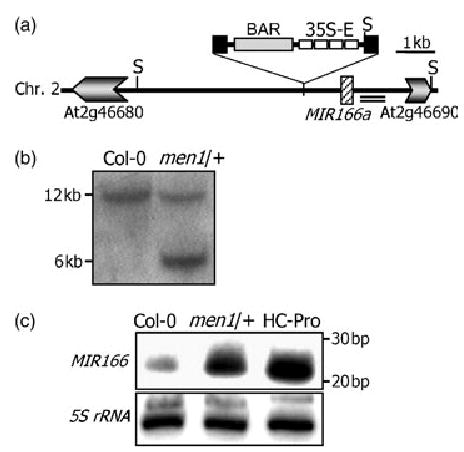
(a) The 35S enhancer insertion site relative to the MIR166a locus. The double underlining indicates a 500-bp genomic DNA fragment used as a probe for genomic Southern blot analysis. S, SacI site.
(b) Genomic Southern blot analysis. SacI-digested genomic DNA was probed with a genomic DNA fragment.
(c) miR166 expression in men1/+ and wild-type plants. Plants overexpressing tobacco etch virus (TEV) helper component proteinase (HC-Pro) were included as a comparison. Fifteen micrograms of total RNA was loaded onto each lane. 5S rRNA gene was used as a loading control.
More careful examination of the intergenic sequence around the T-DNA insertion site resulted in identification of the MIR166a gene located about 1.4 kb from the insertion site (Figure 3a). Genomic Southern blot analysis using a genomic DNA fragment as a probe confirmed a single insertion event in the mutant and placed the insertion near the MIR166a gene (Figure 3b). Northern blot analysis using an antisense MIR166a sequence as a probe revealed that miR166 greatly increased in men1/+ as well as in HC-Pro transgenic plants (Figure 3c), in which the tobacco etch virus helper component proteinase (HC-Pro) is overexpressed (data not shown). This result suggests that miR166 overexpression may be the molecular basis for the men1 phenotype. We could not recover any viable transgenic plants that overexpress MIR166a from four independent transformation attempts. The MIR166a gene sequences we tried include the miR166 sequence of 21 bp, a sequence of about 170 bp that contains the flanking genomic DNA sequences of about 75 bp on both sides of the MIR166a locus, and genomic DNA fragments of about 3 and 5 kb that include the MIR166a locus in the middle of each fragment. Their expression was driven by the CaMV 35S promoter. A few kanamycin-resistant seeds were obtained, but they died at the very early seedling stage just after germination. This could be because the CaMV 35S promoter-driven miR166 expression is more ubiquitous or more robust than that of the activation tagged lines. However, biochemical actions of miR166 on mRNAs of ATHB15 and other HD-ZIP III genes and recapitulation of the men1 phenotype in transgenic plants with reduced ATHB15 expression unequivocally demonstrate that the men1 phenotype is caused by miR166 overexpression (see below).
A subset of HD-ZIP III genes is affected by the men1 mutation
The five HD-ZIP III genes are regulated by miR166/165 in Arabidopsis (Floyd and Bowman, 2004; Mallory et al., 2004b). miR166 directs mRNA cleavage of PHV and PHB in wheat germ extracts (Tang et al., 2003). REV mRNA is cleaved within the miR166/165 binding sequence in planta as demonstrated by 5′-RACE experiments (Emery et al., 2003; Zhong and Ye, 2004), although it is currently unclear whether it is cleaved by miR166 or miR165. A similar biochemical mechanism has been proposed in the cleavage of the ATHB8 and ATHB15 mRNAs, based on the conservation of the miR166/165 complementary sequences in mRNAs of ATHB8 and ATHB15 (Juarez et al., 2004; Reinhart et al., 2002; Rhoades et al., 2002; Zhong and Ye, 2004) (Figure 4a). It has been later demonstrated that their mRNAs are also cleaved by miR165/166 in Arabidopsis through the 5′-RACE method (Mallory et al., 2004b).
Figure 4. Transcript levels of HD-ZIP III genes in men1/+.
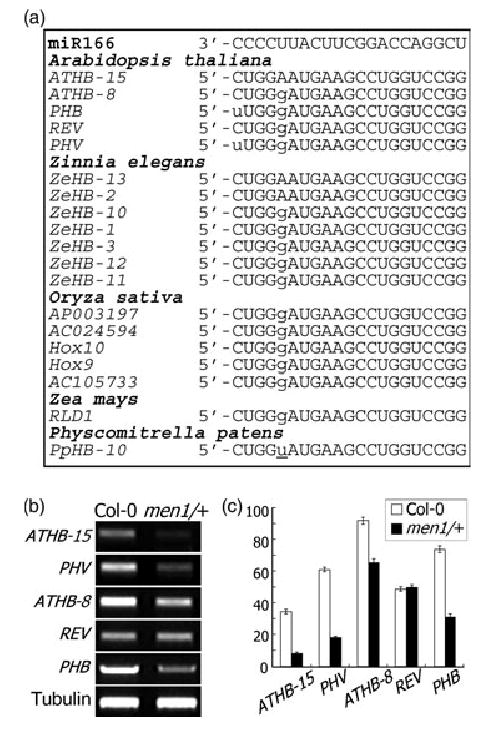
(a) Sequence comparison of miR166 and its putative target sequences. The sequences of ATHB15 gene homologs from plants and moss were aligned.
(b) Transcript levels of HD-ZIP III genes in men1/+.
(c) Graphic view of the transcript levels as measured in (b).
To examine the biochemical relationship between miR166 and its target genes, the transcript levels of the target genes were analyzed in men1/+. Semiquantitative RT-PCR analyses showed that the mRNA levels of ATHB15 and PHV drastically decreased, whereas those of ATHB8 and PHB moderately decreased in men1/+ (Figure 4b,c). It is therefore likely that ATHB15 and ATHB8 are regulated by miR166 through mRNA cleavage like PHV and PHB. In particular, ATHB15 mRNA was barely detectable in men1/+, suggesting that at least some features of the men1 phenotype could be attributed to ATHB15 functions. The mRNA level of REV was identical in men1/+ and wild-type plants. REV mRNA is cleaved within the miR166/165 target sequence as shown by 5′-RACE experiments, and a semidominant mutation of REV, amphivasal vascular bundle 1 (avb1), inhibits the mRNA cleavage (Emery et al., 2003; Zhong and Ye, 2004). It seems that REV mRNA is cleaved by miR166/165 in specific plant tissues such as vascular tissues. These observations indicate that the pleiotropic alterations caused by the men1 mutation are due to multiple effects conferred by ATHB8, PHB, and PHV as well as by ATHB15.
ATHB15 is regulated by miR166 through mRNA cleavage
Inflorescence stems are severely fasciated, and vascular differentiation and radial patterning are greatly affected by the men1 mutation (Figure 2a). Whereas PHV, PHB, and REV are expressed in a broad range of plant tissues (Baima et al., 1995; Emery et al., 2003; McConnell et al., 2001), ATHB15 and ATHB8 are expressed more specifically in vascular tissues (Baima et al., 2001; Ohashi-Ito and Fukuda, 2003). Therefore, it is expected that ATHB15 and ATHB8 may be more directly related to vascular development than other HD-ZIP III genes. However, it is evident that ATHB8 is not essential for vascular cell differentiation, although it promotes procambial/cambial cell differentiation into xylem tissues (Baima et al., 2001). These results strongly suggest that miR166-mediated ATHB15 repression may play a major role in vascular development. We therefore decided to focus on the putative role of ATHB15 in the regulation of vascular development.
The ATHB15 transcript level drastically decreased in men1/+ (Figure 4b), suggesting that ATHB15 is regulated by miR166 through mRNA cleavage. We further examined the biochemical relationship between ATHB15 and miR166 in vitro and in planta. To more directly investigate ATHB15 mRNA cleavage by miR166 in planta, an MIR166a gene sequence of about 170 bp or the full-size ATHB15 gene sequences were subcloned under the control of the CaMV 35S promoter in plant expression vector constructs, and the constructs were transformed into Agrobacterium cells. The Agrobacterium cells were then injected independently or in various combinations into the leaves of N. benthamiana for transient expression. Total RNAs were extracted from the leaf tissues at 4 days after injection and analyzed by Northern blot hybridization. ATHB15 mRNA was cleaved to some extent in the leaf tissues of N. benthamiana even in the absence of Arabidopsis miR166, possibly due to intrinsic miR166/165 activity in this plant species. However, it was almost completely cleaved in the presence of Arabidopsis miR166 (Figure 5a, lane 5). By contrast, an ATHB15 mutant mRNA (mATHB15) that has base substitutions within the miR166/165 target sequence but without any amino acid changes (Figure 5c, lane 7) was not cleaved at all by miR166. These results clearly show that ATHB15 mRNA is cleaved by miR166 through near-perfect base complementarity. ATHB15 mRNA was also cleaved in standard wheat germ extracts (Figure 5b), indicating that wheat also possesses miR166/165 activity (Tang et al., 2003). It is unclear whether ATHB15 is also cleaved by miR165.
Figure 5. ATHB15 mRNA cleavage by miR166 in Nicotiana benthamiana, wheat germ extract, and in Arabidopsis.
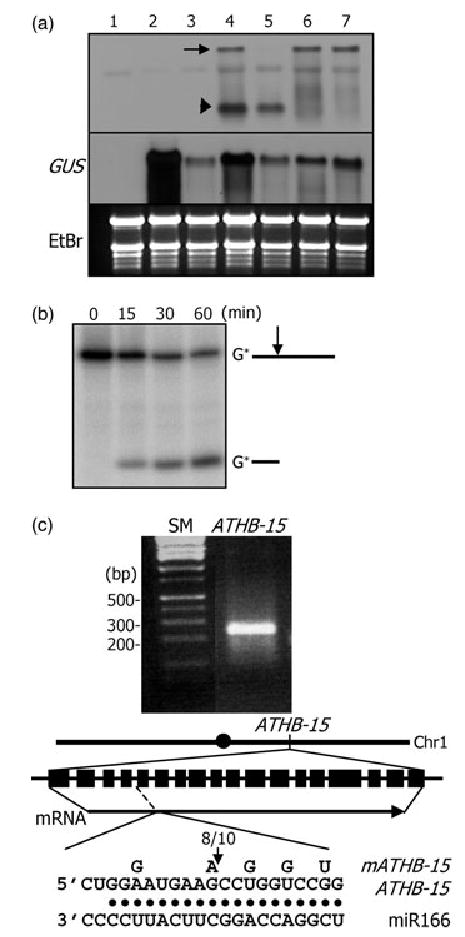
(a) ATHB15 mRNA cleavage in N. benthamiana. Total RNAs were extracted from plants without any vector constructs (1), with pBI121 control alone (2), with pBI121 + miR166 vector (3), with pBI121 + ATHB15 vector (4), with pBI121 + ATHB15 vector + miR166 vector (5), with pBI121 + mATHB15 vector (6), and with pBI121 + mATHB15 vector + miR166 vector (7). RNA gel blots were probed with the 5′-end cleavage fragment of ATHB15. The arrow marks the full-size mRNA, and the arrowhead the 5′-end cleavage product.
(b) ATHB15 mRNA cleavage in wheat germ extract. A 5′-end labeled ATHB15- specific RNA was used in the assay.
(c) 5′-RACE to determine the cleavage site in Arabidopsis. The arrow in the diagram marks the cleavage site. The number refers to that of independent clones with the 5′-end as determined.
To confirm the ATHB15 mRNA cleavage site, the 3′-end cleavage product of ATHB15 mRNA was isolated from Arabidopsis floral organs, and RNA ligase-mediated 5′-RACE experiments were carried out (Kasschau et al., 2003; Llave et al., 2002). As expected, ATHB15 mRNA was cleaved within the miR166/165 target sequence that encodes the putative sterol/lipid-binding START domain (Ponting and Aravind, 1999) (Figure 5c). Interestingly, the cleavage site is identical to those confirmed with PHV and REV mRNAs (Emery et al., 2003; Tang et al., 2003; Zhong and Ye, 2004).
ATHB15 is essential for vascular development
The fasciated inflorescence stems have altered vascular system and radial patterning in men1/+ (Figure 2). ATHB15 is primarily expressed in vascular tissues (Baima et al., 1995; Ohashi-Ito and Fukuda, 2003) and regulated by miR166 via mRNA cleavage (Figure 5). Consistent with this, the transcript level of ATHB15 significantly decreased in men1/+ (Figure 4b). It is therefore predicted that miR166 directs vascular development by regulating ATHB15 mRNA.
To investigate the physiological role of ATHB15 in plant development, especially in vascular development, we generated transgenic Arabidopsis plants that overexpress sense or antisense ATHB15. The miR166-resistant mATHB15 was also included as a control. Overexpression of transgenes and suppression of intrinsic ATHB15 gene by antisense expression were verified by Northern blot hybridization (Figure 6c). By contrast, it is evident that expression of other HD-ZIP III genes was unaffected in the transgenic plants. The Northern probe was a 483-bp-containing DNA fragment encoding the N-terminal region of ATHB15. The observed size difference between the endogenous and the transgenic ATHB15 transcripts would be due to different sizes of the 5′- and 3′-untranslated sequences.
Figure 6. ATHB15 transgenic plants and transverse sections of inflorescence stems.
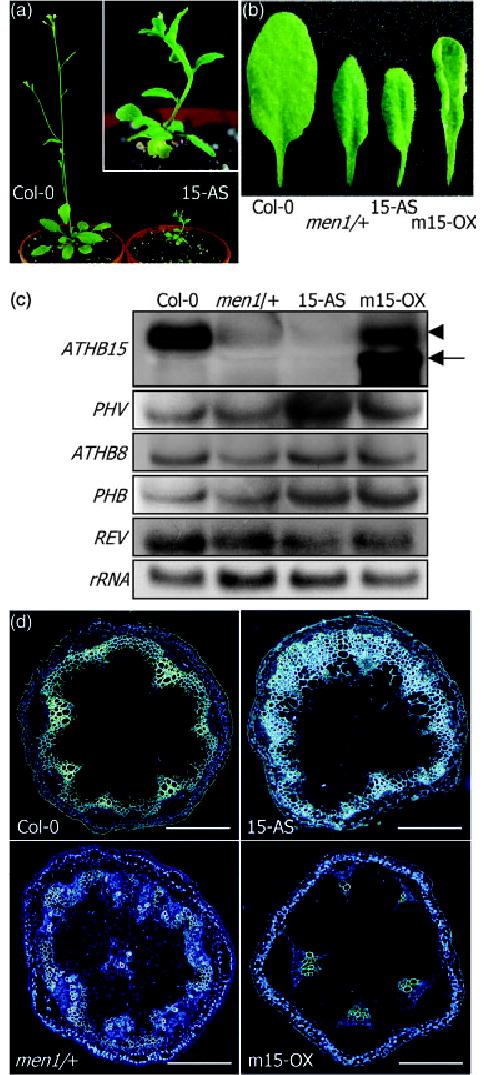
(a) Antisense ATHB15 transgenic plant. Note the fasciated inflorescence stem in antisense ATHB15 transgenic plant, although not as prominent as that in men1/+.
(b) Leaf morphology. Upward leaf curling is observed in mATHB15 overexpressors, whereas a slight downward curling is evident in men1/+ and antisense ATHB15 transgenic plants.
(c) Transgene expressions. Expression of ATHB15 transgenes was confirmed by Northern blot hybridization. Note that the sizes of transgene transcripts (arrow) are smaller than that of intrinsic ATHB15 transcript (arrowhead). Expression of endogenous HD-ZIP III genes was also analyzed by Northern blot hybridizations.
(d) Transverse sections of inflorescence stems of 5.5-week-old plants. Sections were stained with aniline blue and observed under a fluorescence microscope. Scale bar is 20 μm.
Sense transgenic plants with mATHB15 were moderately dwarfed with an upward curling of the leaf blade (Figure 6b). Leaf size was slightly smaller than that of wild-type plants. Transgenic plants overexpressing ATHB15 were indistinguishable from wild-type plants (data not shown). By contrast, antisense ATHB15 transgenic plants were severely dwarfed with small, short rosette leaves (Figure 6a,b). Among 39 homozygous transgenic lines we obtained, 14 lines exhibited such phenotypic alterations, and four lines were lethally affected and died at the very early seedling stage. Others were mildly affected or similar to wild-type plants. Northern blot analysis showed that ATHB15 is specifically suppressed but that other HD-ZIP III genes are unaffected in the antisense transgenic plants (Figure 6c). The phenotype of the antisense ATHB15 transgenic plants may be again related to the arrested growth of homozygous men1 lines. A slight downward curling of the leaf blade was also observed as in men1/+. Thismayhave been caused by partial abaxialization of leaf polarity. Consistent with this view, the expression of KANADI andYABBY genes, which are abaxially expressed in leaf organs and in phloem tissues and regulate leaf polarity (Emery et al., 2003; Eshed et al., 2001; Kerstetter et al., 2001; Sawa et al., 1999; Siegfried et al., 1999), was upregulated in men1/+ and antisense ATHB15 transgenic plants but repressed in mATHB15 transgenic plants (data not shown). This altered expressionmaybe caused by alterations in vascular organization as well as in leaf morphology.
It is apparent that ATHB15 has a partially overlapping function with other HD-ZIP III genes such as PHV and PHB (McConnell and Barton, 1998; McConnell et al., 2001) in the specification of leaf polarity, although its role is not as critical as that of PHV and PHB. Consistent with this notion, the adaxial and abaxial cell fate determination during vascular development is governed by a mechanism similar to that functioning in the formation of lateral organ polarity (Emery et al., 2003; Juarez et al., 2004).
Especially, many antisense ATHB15 transgenic plants showed fasciated inflorescence stems (Figure 6a), further supporting the view that the fasciated inflorescence stems observed in men1/+ are caused at least in part by miR166-mediated ATHB15 repression. These observations suggest that ATHB15 may have a role in apical meristem formation as well as in vascular development in inflorescence stems, which are developmentally related (Clark et al., 1996; Laufs et al., 1998; McHale and Koning, 2004).
Transverse sections of fully developed inflorescence stems at a growth stage of green siliques, which are routinely obtained 3 weeks after bolting (5.5-week-old plants) under our growth conditions, exhibit striking differences among men1/+, mATHB15 transgenic plants, and wild-type plants. Lignified tissue was greatly expanded in men1/+ and antisense ATHB15 transgenic plants but was significantly reduced in mATHB15 transgenic plants (Figure 6d). Lignified tissue formation is mainly contributed by the activity of fascicular cambium and interfascicular cambium that eventually differentiate into secondary xylem (Baima et al., 2001), indicating that cambium activity and its differentiation into xylem is markedly promoted in men1/+ and in antisense ATHB15 transgenic plants.
The number of vascular bundles was also different. Whereas it was six to eight in wild-type plants, it was 11–12 in men1/+ and antisense ATHB15 transgenic plants and four to five in mATHB15 transgenic plants (Figure 6d). This alteration may be due to the differences in stem size – fasciated stems may have more vascular bundles, and thinner stems may have fewer vascular bundles. However, the radial patterning of vascular bundles was unaltered, and the adaxial–abaxial polarity was maintained in both plants, although it was somewhat irregular. Amphivasal vascular bundles were observed in very few lines of men1/+ and antisense ATHB15 transgenic plants, unlike plants with the REV mutation (avb1) (Emery et al., 2003; Zhong and Ye, 2004). These results indicate that ATHB15 has distinct as well as overlapping functions compared with REV, ATHB8, and other HD-ZIP genes involved in vascular development.
It is also evident that ATHB15 is necessary for vascular development, which is contrary to the role of ATHB8. The alteration of vascular bundles and vascular development observed in men1/+ and ATHB15 transgenic plants is also consistent with the localized expression of ATHB15 in the procambial and cambial cells (Ohashi-Ito and Fukuda, 2003).
ATHB15 negatively regulates vascular cell differentiation
Close examination of individual vascular bundles revealed that the formation of xylem tissue and interfascicular tissue was greatly promoted in men1/+ and antisense ATHB15 transgenic plants (Figure 7), consistent with the expanded lignification pattern (Figure 6d). Protoxylem and metaxylem were also greatly expanded. However, the phloem formation was only marginally affected by the men1 mutation and by the antisense expression of ATHB15, although it was somewhat flattened compared with that in wild-type plants. Arabidopsis cambium produces more xylems than phloems (Baima et al., 2001). The increased production of primary and secondary xylems is apparently caused by the acceleration of procambial/cambial cell differentiation into xylem parenchyma cells (Baima et al., 2001).
Figure 7.
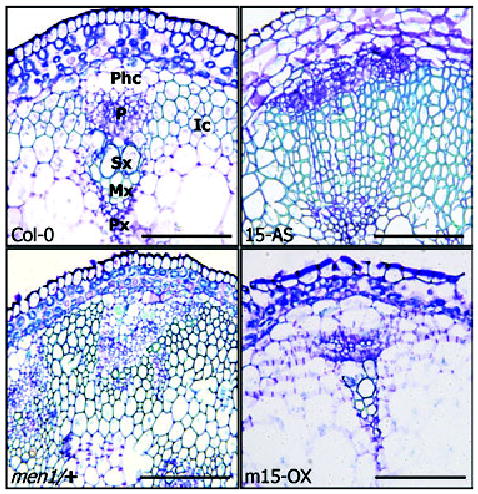
Vascular bundles in men1/+ and ATHB15 transgenic plants. Transverse sections were stained with toluidine blue and observed under a bright- field microscope. Phc, phloem cap cell; P, phloem; Sx, secondary xylem; Mx, metaxylem; Px, Protoxylem; Ic, interfascicular cells. Scale bars are 5 μm.
Notably, vascular tissues, especially primary and secondary xylems, were very poorly developed in mATHB15 transgenic plants (Figure 7). The number of vascular bundles was also fewer than in wild-type plants (Figure 6c), although the collateral organization of xylem and phloem was maintained in individual vascular bundles. These observations unequivocally demonstrate that ATHB15 is important for vascular development and is responsible for its negative regulation. By contrast, ATHB8, which is exclusively expressed in xylem tissue, positively regulates vascular cell differentiation (Baima et al., 2001). Consistent with this, the expansion of the lignified tissue in men1/+ and antisense ATHB15 transgenic plants is very similar to that of transgenic plants overexpressing ATHB8 (Baima et al., 2001). ATHB15 and ATHB8 may have antagonistic roles during vascular development.
Auxin induces ATHB8 (Baima et al., 1995) and stimulates vascular cell differentiation, suggesting that ATHB8 mediates auxin signals in the promotion of vascular development (Sachs, 2000; Ye, 2002). However, auxin does not influence the expression of ATHB15 and MIR166a (data not shown) under the same experimental conditions as used with ATHB8 (Baima et al., 1995). Our results indicate that ATHB15 is essential for vascular development. Antisense overexpression of ATHB15 causes severe dwarfism, as in the men1 mutation, possibly due to the suppressed vascular system. Altogether, we propose a model for the regulation of vascular development in which miR166-mediated regulation of ATHB15 is a primary molecular device for the proper maintenance of the vascular system, which is further modulated by ATHB8 through both the posttranscriptional regulation by miR166 and the transcriptional regulation by auxin.
Discussion
miRNAs are emerging as critical post-transcriptional regulators in various plant developmental processes. Although the first plant miRNAs have been reported very recently (Llave et al., 2002; Reinhart et al., 2002), much later than those in animals, more miRNAs and their target genes have been identified and functionally characterized in plants.
One of the best characterized miRNAs in plants is miR166/ 165 and its target genes that encode the HD-ZIP III transcription factors (Baima et al., 2001; Emery et al., 2003; Kidner and Martienssen, 2004; McConnell and Barton, 1998; McConnell et al., 2001). Dominant or semidominant gain-of-function mutations in PHB, PHV, and REV all map to the START domain that has a limited sequence similarity to sterol-binding proteins in metazoans. Furthermore, the presence of the miR166/165 complementary sequences within the START domain strongly supports the view that miRNA-directed post-transcriptional regulation of the target mRNAs is related to the phenotypic alterations of the mutants. This has been experimentally demonstrated by molecular genetic and biochemical studies of miRNA-resistant target genes. Although ATHB15 has been suggested to have a role in vascular development (Ohashi-Ito and Fukuda, 2003), it has not been systematically examined yet.
In this work, we demonstrated that miR166 regulates ATHB15 through mRNA cleavage during vascular development using a morphogenetic Arabidopsis mutant that is featured by fasciated inflorescence stems with altered vascular development. An MIR166a gene is activated by the insertion of the CaMV 35S enhancer in the mutant. The transcript levels of ATHB15, PHV, PHB, and ATHB8 were reduced in the mutant with the reduction of ATHB15 mRNA most significant. Consistent with this, miR166 efficiently cleaves ATHB15 mRNA in planta as well as in vitro.
Transgenic plants with reduced ATHB15 expression are severely dwarfed with fasciated inflorescence stems as observed in men1/+. The vascular system is also altered by the men1 mutation and by the reduction or overexpression of ATHB15. The expansion of lignified tissue was greatly promoted in men1/+ and antisense ATHB15 transgenic plants but significantly reduced in mATHB15 transgenic plants (Figure 6d). The number of vascular bundles was also influenced. Altogether, it is evident that ATHB15 is critical for vascular development and negatively regulates vascular cell differentiation.
However, the mechanisms that regulate vascular development are not simple. Several growth hormones have been demonstrated or implicated in vascular tissue formation and differentiation. A HD-ZIP III gene encoding an ATHB8 homolog is mainly expressed in xylem cells and induced by brassinosteroids (BRs) in cultured Z. elegans cells (Ohashi-Ito et al., 2002). Consistent with this, xylem development is reduced in BR-deficient Arabidopsis (Choe et al., 1999). Brassinazole, a BR synthesis inhibitor, represses xylem formation (Nagata et al., 2001). ZeHB-13, an ATHB15 gene homolog in Z. elegans, is suppressed by brassinazole but reversed by the addition of BR, indicating that BR is not required for the ZeHB-13 induction but promotes its expression (Ohashi-Ito and Fukuda, 2003). However, we could not observe any effects of brassinazole and BRs on ATHB15 expression (data not shown). Auxin is the major signaling determinant in the development of vascular system in Arabidopsis. Its polar transport seems to promote the formation of vascular strands (Sachs, 1991). Some mutants with defective vascular system are caused by defects in auxin transport or auxin signaling (Berleth and Sachs, 2001; Dengler and Kang, 2001; Mahonen et al., 2000). Notably, ATHB8 that promotes cambial/procambial cell differentiation into xylem tissue is induced by auxin, indicating that ATHB8 modulates auxin signals during vascular development (Baima et al., 1995, 2001).
Both ATHB15 and ATHB8 are regulated by miR166 through mRNA cleavage (Figure 4b). However, ATHB15 is distinct from ATHB8 in several aspects. ATHB15 does not respond to auxin, unlike ATHB8. Whereas ATHB15 negatively regulates xylem tissue formation, ATHB8 has a promoting effect on it. In addition, ATHB15 is essential for vascular development, but ATHB8 is not (Baima et al., 2001). It is therefore hypothesized that miR166/165 is a modulator that balances ATHB15 and ATHB8 functions during vascular development. Auxin seems to further adjust the developmental process by transcriptionally regulating ATHB8. Analysis of double mutants of ATHB15 and ATHB8 or of transgenic plants overexpressing both ATHB15 and ATHB8, in combination with auxin and auxin transport inhibitors, would clarify this hypothesis. Other HD-ZIP III genes seem to play additional roles in the fine regulation of vascular development. PHV, PHB, and REV regulate organ polarity by specifying adaxial (xylem) and abaxial (phloem) cell fates and ascertain vascular patterning (Emery et al., 2003; Juarez et al., 2004; Kidner and Martienssen, 2004; McConnell and Barton, 1998; McConnell et al., 2001; Zhong and Ye, 2004).
Vascular system formation is a complex developmental process that is regulated by diverse genetic components and growth hormones. Furthermore, the pattern and organization of vascular systems are quite variable in different plant species. However, the molecular mechanisms that underlie the spatial and temporal regulation of vascular systems are probably shared by all vascular plants. The conserved basic architecture of vascular systems and HD-ZIP III genes in dicots, monocots, and moss (Floyd and Bowman, 2004) suggest that the miR166-mediated regulation of vascular development via HD-ZIP III genes may be a general rule in all vascular plants.
Experimental procedures
Plant materials and growth conditions
All Arabidopsis plants used in this work were in the Columbia background (Col-0). Plants were grown in a controlled culture room at 24–25°C with a relative humidity of 60% in long-day condition (16 h light and 8 h dark). Nicotiana benthamiana was also grown in soil under the same growth conditions.
Transgenic Arabidopsis plants
Transgenic Arabidopsis plants were produced by a simplified floral dip method as described (Clough and Bent, 1995). Full-size cDNAs of sense and antisense ATHB15 and mATHB15 were expressed under the control of the CaMV 35S promoter in pBI121 (Clontech, Palo Alto, CA, USA). The P1/HC-Pro expression was also driven by the CaMV 35S promoter as described (Kasschau et al., 2003). Homozygous transgenic lines were isolated through two additional generations after primary selection for each transformation.
Isolation of men1 mutant
The men1 mutant was isolated from an Arabidopsis mutant pool generated using the activation tagging vector pSKI015 as described (Weigel et al., 2000). The T-DNA insertion site was mapped by the TAIL-PCR method (Liu et al., 1995). The single insertion event of T-DNA in men1 was confirmed by segregation analysis and by genomic Southern blot analysis using SacI-digested genomic DNAs and a genomic DNA fragment of 500 bp near the insertion site as a probe (Figure 3a).
miRNA Northern blot analysis
For miRNA Northern blot hybridization, total RNA was extracted from plant materials using the TRIAZOL reagent (Invitrogen, Carlsbad, CA, USA) according to the procedure previously described (Pfeffer et al., 2003) but with some modifications. After isopropanol precipitation, the RNA pellet was briefly centrifuged without rinsing with ethanol, and the remaining isopropanol was completely removed by pipetting. The RNA pellet was then dissolved in 95% formamide/25 mm EDTA. We found that these steps greatly improve the yield and solubility of miRNA in total RNA preparations. Northern blot hybridizations were carried out using the ULTRA-Hyb Oligo solution according to the procedure supplied by the manufacturer (Ambion, Houston, TX, USA). The oligonucleotide probes were end-labeled at the 5′-end using P32-γATP and T4 polynucleotide kinase. An oligonucleotide complementary to 5S rRNA was also processed in the same way and used as a control.
Semiquantitative RT-PCR analyses of HD-ZIP III gene expression
The transcript levels of HD-ZIP III genes were analyzed by semiquantitative RT-PCR runs. Total RNA samples were extracted from aerial parts of plants and treated extensively with RNase-free DNase I to eliminate any contaminating genomic DNA. The first-strand cDNA was synthesized using Pfu Turbo polymerase (Stratagene, La Jolla, CA, USA) from 2 μg of total RNA in a 20-μl reaction volume, and 2 μl of the reaction mixture was subject to subsequent PCR in a 50-μl reaction volume. RT-PCR runs were 15–30 cycles, depending on the linear range of PCR amplification for each gene, each cycle at 94°C for 1 min, 60°C for 1 min, and 72°C for 4 min with a final cycle at 72°C for 10 min to allow the completion of polymerization. The PCR primer pairs were designed from the sequence regions of each gene with the least sequence identities among the HD-ZIP III genes. The primer sequences are 5′-TCTTGCAAGGATGGTAAGTTGG (forward, F) and 5-CTATTAGTCTGAGTAACCTCCTGAGC (reverse, R) for ATHB15, 5′-AGGAAGCAATAATAGTCACAATATGG (F) and 5′-ATACTTGGCCCGTTTTGTGTATT (R) for ATHB8, 5′-CCATGGACGATAGAGACTCTCC (F) and 5′-ACCACTTCCAAAACTTGGAAGA (R) for PHV, 5′-GAGATATGATGAACAGAGAGTCGCC (F) and 5′-ACCAAACTTCCCAGGGGACA (R) for PHB, and 5′-AACCACCGTGAGAGAAGCAGT (F) and 5′-CCGGGAACATAGTGAAAACTTC (R) for REV.
Analysis of ATHB15 mRNA cleavage
Target mRNA cleavage by miR166 was examined in three different ways. For analysis in N. benthamiana, Agrobacterium cells containing plant expression vector constructs with either an MIR166a gene of about 170 bp or ATHB15 target genes under the control of the CaMV 35S promoter were injected directly into leaves independently or in various combinations. Total RNAs were extracted from the leaf tissues at 4 days after injection as described (Llave et al., 2000) and analyzed in a 1.2% formaldehyde-agarose gel. The Northern gel blots were probed with the putative 5′-end cleavage fragment labeled with digoxigenin-UTP by in vitro transcription with SP6 or T7 RNA polymerase using the DIG RNA Labeling Kit (Roche Applied Science, Penzberg, Germany).
For analysis in standard wheat germ extracts, a 5′-end labeled target RNA corresponding to a fragment of ATHB15 mRNA spanning the miR166/165 cleavage site was incubated in wheat germ extracts under miRNA processing conditions as described (Tang et al., 2003). Aliquots were removed at different time intervals, and products resolved in a 6% sequencing gel.
To map the ATHB15 mRNA cleavage site in vivo, total RNAs were extracted from floral organs of Arabidopsis. The 5′-end of the cleavage product was determined by a modified RNA ligasemediated 5′-RACE method (Kasschau et al., 2003; Llave et al., 2002) using the RLM-RACE kit (Ambion).
Histochemical analysis
Transverse sectioning of 5.5-week-old inflorescence stems, 3 weeks after bolting under our growth conditions, and tissue staining were carried out essentially as described (Baima et al., 2001) but using aniline blue for the analysis of vascular tissues and toluidine blue for the analysis of vascular bundles. The aniline blue-stained sections were observed under a fluorescence microscope, and the toluidine blue-stained sections were observed under a bright field microscope with appropriate magnification.
Acknowledgments
We thank Y.-H. Seo for plant culture and the members of the Molecular Signaling Laboratory for technical support and scientific discussion. This work was supported by the Brain Korea 21 program (KRF), the BioGreen21 program, grants from KOSEF (R02-2003-000-10001-0), KISTEP (M1-0219-00-0003), and the Plant Signaling Network Research Center, and NIH grant GM-44640 (to N.-H.C.). J.L.R. was supported by a Pew Latin American fellowship.
References
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baima S, Nobili F, Sessa G, Lucchetti S, Ruberti I, Morelli G. The expression of the ATHB-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development. 1995;121:4171–4182. doi: 10.1242/dev.121.12.4171. [DOI] [PubMed] [Google Scholar]
- Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G. The Arabidopsis ATHB-8 HD-Zip protein acts as a differentiation-promoting transcriptional factor of the vascular meristems. Plant Physiol. 2001;126:643–655. doi: 10.1104/pp.126.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanisms, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel B, Bartel DP. MicroRNAs: at the root of plant development. Plant Physiol. 2003;132:709–717. doi: 10.1104/pp.103.023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T, Sachs T. Plant morphogenesis: long distance coordination and local patterning. Curr Opin Plant Sci. 2001;4:57–62. doi: 10.1016/s1369-5266(00)00136-9. [DOI] [PubMed] [Google Scholar]
- Carland FM, Fujioka S, Takasuto S, Yoshida S, Nelson T. The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell. 2002;14:2045–2058. doi: 10.1105/tpc.003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022– 2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, et al. The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylene-cholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999;119:897–907. doi: 10.1104/pp.119.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1995;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dengler N, Kang J. Vascular patterning and leaf shape. Curr Opin Plant Sci. 2001;4:50–56. doi: 10.1016/s1369-5266(00)00135-7. [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Esau, K. (1965) Plant Anatomy, 2nd edn. New York, NY: John Wiley & Sons.
- Eshed Y, Baum SF, Perea JV, Bowman JL. Establishment of polarity in lateral organs of plants. Curr Biol. 2001;11:1251–1260. doi: 10.1016/s0960-9822(01)00392-x. [DOI] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL. Ancient microRNA target sequences in plants. Nature. 2004;428:485–486. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
- Jang JC, Fujioka S, Tasaka M, Seto H, Takasuto S, Ishii A, Aida M, Yoshida S, Sheen J. A critical role of sterols in embryonic patterning and meristem programming revealed by the fackel mutants of Arabidopsis thaliana. Genes Dev. 2000;14:1485– 1497. [PMC free article] [PubMed] [Google Scholar]
- Juarez MT, Kul JS, Thomas J, Heller BA, Timmermans MCP. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell. 2003;4:205–217. doi: 10.1016/s1534-5807(03)00025-x. [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Tayler RA, Bomblies K, Poethig RS. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. Macro effects of micro RNAs in plants. Trends Genet. 2003;19:13–16. doi: 10.1016/s0168-9525(02)00011-2. [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. Spatially restricted micro RNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81– 84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- Laufs P, Dockx J, Kronenberger J, Traas J. MGOUN1 and MGOUN2: two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development. 1998;125:1253–1260. doi: 10.1242/dev.125.7.1253. [DOI] [PubMed] [Google Scholar]
- Laufs P, Peaucelle A, Morin H, Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004;131:4311–4322. doi: 10.1242/dev.01320. [DOI] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457– 463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Carrington JC. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc Natl Acad Sci USA. 2000;97:13401–13406. doi: 10.1073/pnas.230334397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Mahonen AR, Bonke M, Kauppinen L, Riikonen M, Benfey PN, Helariutta Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the vascular root. Genes Dev. 2000;14:2938–2943. doi: 10.1101/gad.189200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Dugas DV, Bartel DP, Bartel B. Micro RNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol. 2004a;14:1035–1046. doi: 10.1016/j.cub.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J. 2004b;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery JF, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–712. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- McHale N, Koning E. MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell. 2004;16:1730– 1740. doi: 10.1105/tpc.021816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata N, Asami T, Yoshida S. Brassinazole, an inhibitor of brassinosteroid biosynthesis, inhibits development of secondary xylem in cress plants. Plant Cell Physiol. 2001;42:1006– 1011. doi: 10.1093/pcp/pce122. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Fukuda H. HD-zip III homeobox genes that include a novel member, ZeHB-13 (Zinnia)/ATHB-15 (Arabidopsis), are involved in procambium and xylem cell differentiation. Plant Cell Physiol. 2003;44:1350–1358. doi: 10.1093/pcp/pcg164. [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Demura T, Fukuda H. Promotion of transcript accumulation of novel Zinnia immature xylem-specific HD-Zip III homeobox genes by brassinosteroids. Plant Cell Physiol. 2002;43:1146–1153. doi: 10.1093/pcp/pcf135. [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. Plant J. 2001;25:223–236. doi: 10.1046/j.1365-313x.2001.00959.x. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S., Lagos-Quintana, M. and Tuschl, T. (2003) Cloning of small RNA molecules. In Current Protocols in Molecular Biology (Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidmann, J.G., Smith, J.A. and Struhl, K. eds). New York, NY: John Wiley & Sons. pp. 26.4.1–26.4.18. [DOI] [PubMed]
- Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signaling proteins. Trends Biochem Sci. 1999;24:130–132. doi: 10.1016/s0968-0004(99)01362-6. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Sachs T. Cell polarity and tissue patterning in plants. Dev Suppl. 1991;1:83–93. [Google Scholar]
- Sachs T. Integrating cellular and organismic aspects of vascular differentiation. Plant Cell Physiol. 2000;41:649–656. doi: 10.1093/pcp/41.6.649. [DOI] [PubMed] [Google Scholar]
- Sawa S, Watanabe K, Goto K, Liu YG, Shibata D, Kanaya E, Morita EH, Okada K. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 1999;13:1079–1088. doi: 10.1101/gad.13.9.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Ostuga D, Drews GN, Bowman JL. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- Steeves, T.A. and Sussex, I.M. (1989) Patterns in Plant Development, 2nd edn. Cambridge, UK: Cambridge University Press.
- Tang G, Reinhart B, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye ZH. Vascular tissue differentiation and pattern formation in plants. Annu Rev Plant Biol. 2002;53:183–202. doi: 10.1146/annurev.arplant.53.100301.135245. [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. amphival vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems, and carpels. Plant Cell Physiol. 2004;45:369–385. doi: 10.1093/pcp/pch051. [DOI] [PubMed] [Google Scholar]


