Summary
Phytochrome A (phyA) plays a primary role in initiating seedling de-etiolation and is the only plant photoreceptor known to be activated by far-red light (FR). The signaling intermediate FHY1 appears to either participate directly in relaying the phyA signal or to positively regulate a critical signaling event(s) downstream of phyA activation. Here we identify a homolog of FHY1 named FHL (FHY1-like) as a novel signaling factor essential for complete responsiveness to phyA. FHL possesses functional nuclear localization and nuclear export signals. Lines in which FHL function was abolished by insertional mutagenesis or attenuated by RNAi-mediated suppression displayed a weaker hyposensitivity to continuous FR than fhy1 null mutants and most reported phyA signaling mutants. However, hypocotyl elongation assays indicated that suppression of FHL expression in fhy1-3 caused an insensitivity of hypocotyl elongation to FR and blue light (B) indistinguishable from that seen in phyA. Real-time PCR indicates that in FR, FHY1 transcripts are approximately 15-fold more abundant than FHL transcripts. Although both FHY1 and FHL are capable of homo- and hetero-interaction via their C-termini, the ability of FHL overexpression to restore wild-type (WT) morphological and molecular phenotypes to fhy1-3 seedlings suggests that the extreme insensitivity to FR associated with suppression of FHL expression in fhy1-3 cannot be accounted for by a critical role for FHY1-FHL heterodimers in phyA signal transmission. Rather, we suggest that the relative abundances of FHY1 and FHL in WT plants account for the differences in the severity of fhy1 and fhl mutations. As for FHY1, FHL transcript accumulation is dependent on FHY3 and is decreased after exposure to FR, R or B light. These findings reiterate the prevalence of partial degeneracy in plant signaling networks that regulate responses crucial to survival.
Keywords: Arabidopsis, photomorphogenesis, phytochrome, signaling mutant, FHY1, phytochrome A
Introduction
Our current insight into plant signal transduction strongly indicates that plant growth and development are influenced by complex networks of interacting signaling cascades, each triggered by different stimuli. Light makes a critical contribution to the web of interactions that optimize plant adaptation to environmental fluctuations. Of the known plant photoreceptors, phytochrome A (phyA) is the only one activated by high fluence far-red light (FR). Owing to its abundance in dark-grown seedlings, phyA is also the most important factor that monitors the availability of light when an etiolated seedling grows toward the soil surface. After sensing light, the shift from skotomorphogenesis to photomorphogenic development enables seedlings to divert resources that might otherwise be needed for continued hypocotyl elongation to cotyledon opening and the development of a competent photosynthetic apparatus.
The ease with which mutant seedlings can be screened for increased and reduced sensitivity of hypocotyl elongation to FR has facilitated characterization of several phyA-specific signaling intermediates. Thus far, approximately 15 phyA-specific intermediates have been reported (Hare et al., 2003; Wang and Deng, 2003). At least a further 10 gene products or distinct regulatory protein complexes are not specific to phyA signaling, but are nonetheless essential for transmission or repression of the phyA signal. Presently, these intermediates provide an incomplete picture of the sequence of events and underlying mechanisms that link photo-activation of phyA with the cellular and molecular events that elicit changes in light-regulated gene expression. They have been identified by classical forward and reverse genetic screens in wild-type (WT) and mutant backgrounds, as well as by their ability to interact physically with light signaling components. Gene products required for phyA signaling are distributed in various subcellular locations including the nucleus, cytoplasm, chloroplast and peroxisome (Wang and Deng, 2003). Although the mechanistic details of the action of many of the intermediates have yet to be uncovered, some have been directly implicated in processes as diverse as transcriptional regulation, protein degradation and protein phosphorylation. Based on the morphological and molecular phenotypes of plants lacking FHY1 (Desnos et al., 2001; Whitelam et al., 1993; Zeidler et al., 2001), PAT1 (Bolle et al., 2000) and FHY3 (Wang and Deng, 2002; Whitelam et al., 1993), these three gene products are often thought to be the most central intermediates that act closest to phyA, prior to convergence of the phyA signal with events also activated by other photoreceptors. The dependence of FHY1 transcript accumulation on FHY3 (Desnos et al., 2001; Hudson et al., 2003; Wang and Deng, 2002) suggests a serial relationship between the two gene products.
So far, no known mutant defective in phyA signaling has an FR phenotype as severe as that found in phyA null mutants. This observation suggests either that activated phyA immediately transduces the signal to at least two separate transducers or that redundancy between the factors that act closest to phyA prevents their identification by forward genetics. Complete or partial redundancy is often invoked as an explanation why large portions of many plant signal transduction chains remain uncharacterized. Partial redundancy exists between certain members of the Arabidopsis phytochrome gene family (Franklin et al., 2003) as well as in two classes of blue light (B) receptors (Lin et al., 1998). Functional overlaps between homologous components in light signaling cascades have also been reported. The phyA-specific intermediates FHY3 and FAR1 are closely related nuclear proteins that are capable of homo- and hetero-dimerization (Wang and Deng, 2002). When overexpressed, both can compensate partially for the absence of the other (Wang and Deng, 2002). Further downstream in the phyA cascade, the transcription factors HY5 and HYH exemplify homologous homo- or hetero-interacting bZIP transcription factors that have overlapping functions in seedling responses to B (Holm et al., 2002). Dimerization of the phyA-signaling component HFR1 with another bHLH-type transcription factor PIF3 was suggested to modulate phyA signaling (Fairchild et al., 2000). In all of these cases, one member of the pair (FHY3, HY5 or HFR1) makes a greater contribution to the response to the particular wavelength of light, whereas loss of the other member alone (FAR1, HYH or PIF3) causes a more subtle phenotype. It seems reasonable to propose that the apparent prevalence of functional overlaps between plant light receptors and signaling intermediates ensures more complex regulation of light signaling and its integration with pathways activated by other signals. This permits flexibility in responses that require assessment of multiple environmental factors. Complete or partial redundancy also ensures informational homeostasis when one homolog is lost through mutation.
Prompted by the likely significance of genetic redundancy in light signaling, we investigated whether the only homolog of FHY1 in the Arabidopsis genome (named FHL for FHY1-like) might also participate in phyA signal transduction. Both FHL null mutations and RNAi-mediated suppression of FHL increase the insensitivity of fhy1 mutants to FR and B. By contrast, overexpression of FHL can rescue the fhy1 mutant phenotype and confer hypersensitivity to the inhibitory effects of FR and B on hypocotyl elongation. These findings suggest a functional overlap between FHY1 and FHL and provide further evidence of the importance of partial redundancy between members of the same gene family in ensuring optimal developmental responses to the availability and quality of light. The abilities of both FHL and FHY1 to form homocomplexes with themselves and heterocomplexes with each other suggest a potential mechanism that may account for the roles these proteins play in phyA signal transduction.
Results
FHL is a homolog of FHY1 with functional nuclear import and nuclear export signals
The existence of a single homolog of FHY1 in the Arabidopsis genome was reported previously (Desnos et al., 2001; Zeidler et al., 2001). 5′-RACE and RT-PCR analysis using mRNA extracted from 4 day-old etiolated seedlings confirmed that the gene encoding FHL (At5g02200) comprises two exons encoding 181 amino acids, as annotated in the database. Much of the difference in length between FHY1 (202 amino acids) and FHL arises from an approximately 30 amino acid N-terminal extension found in FHY1 but not in FHL (Figure 1a). A search of the EST database indicated several ESTs with homology to Arabidopsis FHY1 and FHL. The potato genome contains at least three members of the FHY1/FHL gene family, two of which (represented in Figure 1a by ESTs CK260880 and CK264717) encode proteins with slightly greater similarity to Arabidopsis FHL than to Arabidopsis FHY1. Likewise, an incomplete cDNA from tomato encodes a protein displaying 33.7% identity to FHL, but only 24.6% identity to FHY1 (Figure 1a). The expression of FHY1/FHL homologs in rice (Figure 1a) and Sorghum bicolor (Zeidler et al., 2004) indicates that the gene family is also represented in monocot species. The most conserved regions in the alignment shown in Figure 1(a) correspond to the nuclear localization sequence (NLS) and a C-terminal motif found in septins, both of which are functional in FHY1 (Zeidler et al., 2004). A nuclear export sequence (NES) recently shown to be functional in FHY1 (Zeidler et al., 2004) is absolutely conserved in FHL (Figure 1a), but not in its putative homologs in potato, tomato and rice (Figure 1a). A region in Arabidopsis FHY1 (amino acids 132–152) proposed to bear significant homology to part of the PAS-A domain of phyA (Desnos et al., 2001) is not conserved in FHL or any of the proteins aligned in Figure 1(a).
Figure 1. FHL has homologs in several species and possesses functional nuclear import and export sequences.
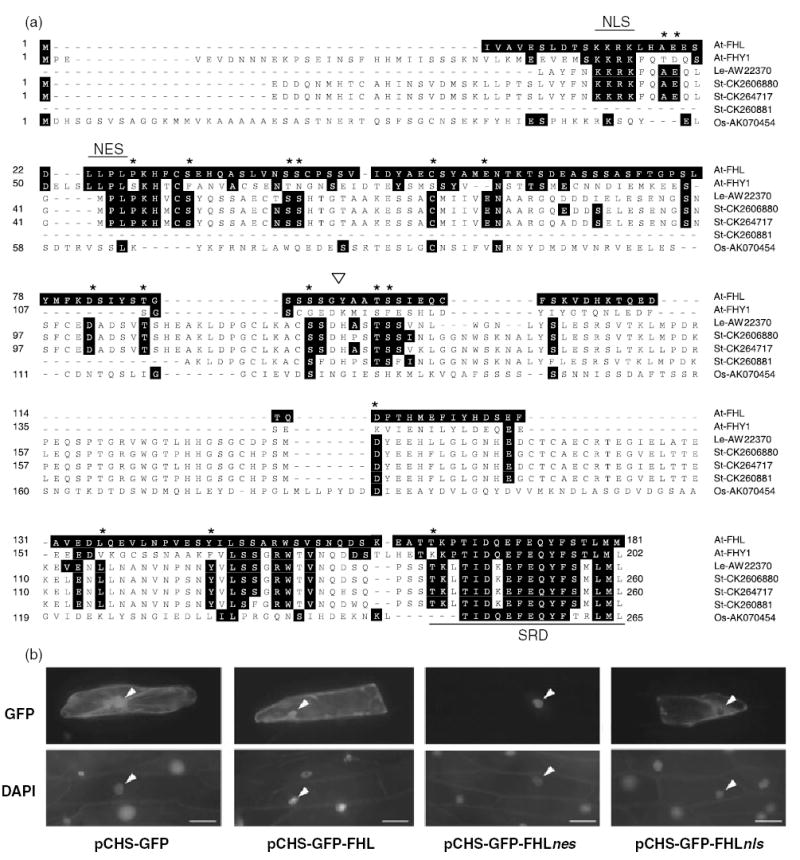
(a) Predicted amino acid sequences encoded by three classes of potato cDNAs (represented by St-CK260880, St-CK264717 and St-260881), a tomato cDNA (represented by Le-AW223570) and a rice cDNA (Os-AK070454) indicate the likely existence of FHY1 gene families in other species besides Arabidopsis. Sequences were aligned using the ClustalV algorithm. Amino acids identical to the FHL residue at that position are highlighted. Residues that are absolutely conserved in tomato and potato sequences, but not in At-FHY1, are indicated with an asterisk. Note that sequences encoded by Le-AW223570 and St-CK260881 are not complete at their N-termini. The arrow indicates the codon disrupted by T-DNA insertion in the fhl-1 mutant (SALK_024320). NLS, nuclear localization signal; NES, nuclear export signal; SRD, septin-related domain.
(b) Mutation of the putative NLS and NES motifs in FHL indicates that they mediate nuclear import and export, respectively. Arrowheads indicate nuclei. Scale bar represents 50 μm.
A caveat of investigating the function of NLS and NES motifs in small proteins is that, especially when they are overexpressed, their diffusion through nuclear pores may confound interpretation of the localization pattern. This can be overcome by fusion to a larger fluorescent protein that increases the molecular mass to a size closer to the exclusion limit of the nuclear pore. To investigate the function of the NLS and NES motifs of FHL, the WT protein was fused to the C-terminus of a 73-kDa fusion of GFP to parsley chalcone synthase (pCHS-GFP). The resultant fusion protein pCHS-GFP-FHL is approximately 93 kDa. Mutants of FHL in which the NLS and NES motifs were substituted with alanine residues (AAAA) were also fused to the C-terminus of pCHS-GFP to generate the constructs pCHS-GFP-FHLnls and pCHS-GFP-FHLnes, respectively.
As reported previously, transient expression of GFP (Zeidler et al., 2001) and pCHS-GFP (Haasen et al., 1999) results in their distribution throughout the cytoplasm and nucleus (Figure 1b). Fusion of FHL to pCHS-GFP did not significantly affect the distribution of pCHS-GFP (Figure 1b). However, mutation of the NES motif in FHL caused apparently exclusively nuclear accumulation of the pCHS-GFP-FHLnes protein, whereas mutation of the NLS in FHL appeared to result in accumulation of pCHS-GFP-FHLnls exclusively in the cytosol (Figure 1b). This indicates that both of the NLS and NES motifs are functional in the context of the FHL protein. Therefore, pCHS-GFP-FHL, and thus by implication FHL, probably cycles between the nucleus and cytoplasm when expressed in onion epidermal cells.
FHL acts redundantly with FHY1 in relaying phyA signals
To evaluate the contribution of FHL to hypocotyl elongation, we screened a plant population (SALK_024320) that segregated a T-DNA insertion in the first exon of the FHL gene. PCR was used to identify lines homozygous for the mutation. The truncated protein expressed in the fhl-1 mutant comprises only 50% of full-length FHL. Under relatively high fluence FR (4.5 μmol m−2 sec−1), fhl-1 mutants possessed slightly, but significantly, longer hypocotyls than WT and phyB mutants (Figure 2a). Significantly reduced inhibition of hypocotyl elongation was also evident at 1.4 and 3.4 μmol m−2 sec−1 (Figure 2c). Unlike fhy1 null mutants (Barnes et al., 1996), fhl-1 nulls do not display significantly decreased sensitivity to the phyA-dependent block of greening in white light after de-etiolation in high intensity FR (data not shown).
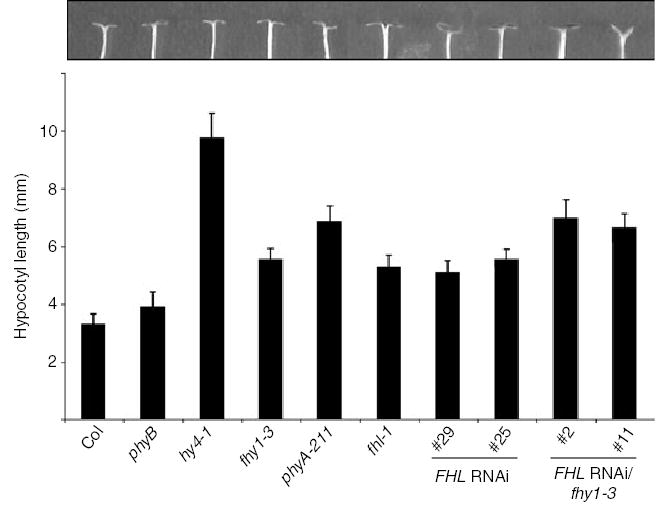
Figure 2. Loss of FHL expression attenuates responsiveness to far-red (FR) light in wild type (WT) and fhy1-3 seedlings.
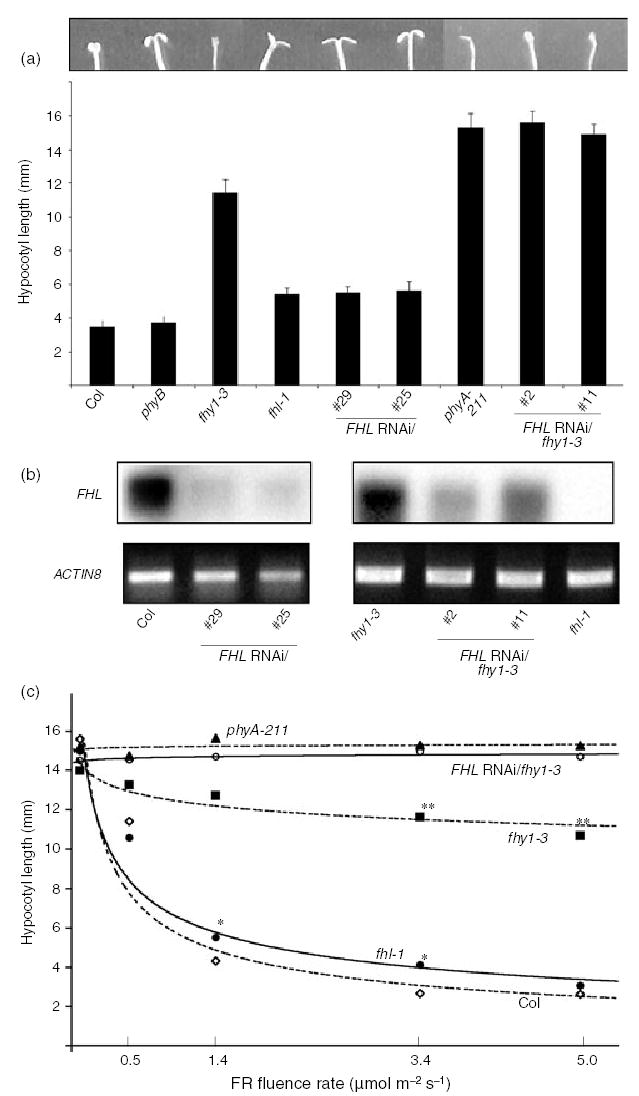
(a) Hypocotyl lengths were measured after germination and 4 day growth under continuous FR (4.5 μmol m−2 sec−1). Error bars indicate SD (n = 20). Cotyledons of representative seedlings are shown. In darkness, hypocotyl lengths of both FHL RNAi/fhy1-3 lines as well as phyA-211 mutants were indistinguishable from those of FR-irradiated seedlings.
(b) RT-PCR indicates that FHL transcript abundance can be decreased by RNAi-mediated silencing in four independent transgenic lines (no. 29 and no. 25 in WT Col, no. 2 and no. 11 in fhy1-3). Total RNA extracted from 4-day-old etiolated seedlings was used. RT-PCR using oligos specific to ACTIN8 transcript was used as a control.
(c) Fluence response curves indicate that FHL RNAi/fhy1-3 line no. 11 displays a hypocotyl elongation response that is indistinguishable from phyA-211 under a range of intensities of FR. The same effect was observed for FHL RNAi/fhy1-3 line no. 2 (data not shown). The lowest fluence indicates hypocotyl lengths in darkness. *, fhl-1 is significantly longer than Col at the fluence indicated (P < 0.05, Student’s t-test; n > 80); **, fhy1-3 is significantly shorter than both phyA-211 and FHL RNAi/fhy1-3 line no. 11 at the fluence indicated (P < 0.01, Student’s t-test; n > 80). The standard deviation at any point was never more than 8.3% of the mean value.
The fhy1-3 mutant lacks functional FHY1 protein and was previously referred to as pat3 (Zeidler et al., 2004). Although hypocotyls of fhy1-3 seedlings grown in FR were longer than those of fhl-1, the absence of FHY1 does not render a hypocotyl phenotype as severe as that of a phyA null (Figure 2a; Desnos et al., 2001; Whitelam et al., 1993; Zeidler et al., 2001). The cotyledons of fhl-1 seedlings were more open than those of fhy1-3, but never completely folded downward as observed in seedlings with an intact phyA response (Figure 2a). This intermediate hypocotyl and cotyledon phenotype observed for fhl-1 mutants was evident in two WT lines transformed to express an RNAi transgene designed to specifically silence expression of FHL but not FHY1. Both Northern analysis (data not shown) and RT-PCR (Figure 2b) confirmed that in the four FHL RNAi lines for which data are presented, FHL expression was silenced. No FHL transcript was detected in the fhl-1 mutant (Figure 2b).
Remarkably, silencing of FHL expression in the fhy1-3 background increased the severity of the FR hypocotyl elongation phenotype of fhy1-3 to the extent that hypocotyl lengths of two FHL-RNAi/fhy1-3 lines were indistinguishable from those of a phyA null mutant (Figure 2a). This was observed for both lines over a range of FR fluences (Figure 2c). This suggests that the incomplete insensitivities of fhy1-3 and fhl-1 to FR might arise at least in part from the abilities of either intermediate to compensate for the loss of the other. Although neither of the FHL-RNAi/fhy1-3 lines had open cotyledons, their cotyledons were more expanded than those of phyA seedlings (Figure 2a).
All of the above genotypes displayed indistinguishable hypocotyl elongation when germinated and grown in darkness (Figure 2c), 35 μmol m−2 sec−1 continuous red light (R) or 20 μmol m−2 sec−1 continuous white light (data not shown). Therefore, as observed for all other bona fide phyA signaling intermediates, the fhl-1 phenotype is dependent on phyA activation and is not apparent under light wavelengths that activate other phytochromes sharing the same downstream targets as phyA.
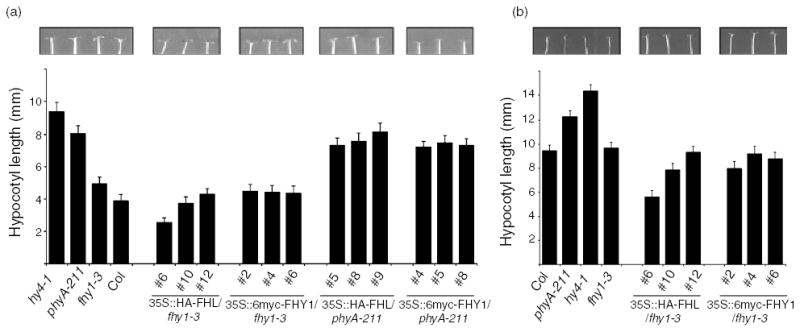
Overexpression of FHL can compensate completely for loss of FHY1
To further evaluate the partial redundancy between FHL and FHY1, epitope-tagged FHL and FHY1 proteins were expressed in fhy1-3 under the control of the CaMV 35S promoter. An antibody raised against His-FHY1 also recognizes FHL and was used to correlate the abundance of FHL or FHY1 in fhy1-3 with the ability of the transgene to complement the mutation. Under the conditions used to detect tagged FHL and FHY1 proteins under 35S control, the antibody raised against His-FHY1 was not able to detect endogenous FHL or FHY1. At relatively high fluences of continuous FR (4.5 μmol m−2 sec−1), overexpression of 6myc-FHY1 (approximately 55 kDa) was associated with complementation of fhy1-3 (Figure 3a). Expression of HA-FHL (approximately 28 kDa) was at least as effective as 6myc-FHY1 in restoring the WT hypocotyl elongation phenotype to fhy1-3 (Figure 3a). At a lower fluence of FR (0.45 μmol m−2 sec−1), hypocotyl elongation was significantly reduced in two of the three HA-FHL/fhy1-3 lines in comparison with WT and 6myc-FHY1/fhy1-3 lines (Figure 3b) and in this case, there was a clear inverse relationship between HA-FHL abundance and hypocotyl length. This indicates that HA-FHL overexpression confers hypersensitivity to low-fluence FR. Cotyledon expansion and opening were also hypersensitive to FR in both of the lines that displayed significantly less hypocotyl elongation than the WT (Figure 3b). Therefore, FHL can substitute for FHY1 when expressed at adequate levels. Moreover, in the absence of FHY1, FHL normally limits the response capacity of the cascade that relays the phyA signal. The observation that overexpression of either HA-FHL or 6myc-FHY1 did not alter photoresponses in a phyA null background (Figure 3a) confirms that these FHL overexpression effects are dependent on phyA.
Figure 3. Overexpression of FHL can compensate for the absence of FHY1.
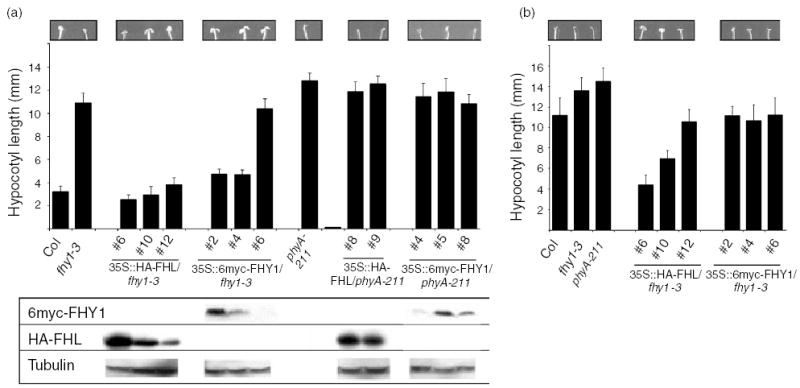
(a) Hypocotyl lengths were measured after germination and 4 day growth under high fluence continuous FR (4.5 μmol m−2 sec−1). Error bars indicate SD (n = 20). Cotyledons of representative seedlings are shown. Relative levels of HA-FHL and 6myc-FHY1 were determined by Western assays using an anti-FHY1 antibody that recognizes both overexpressed FHL and FHY1, but not the endogenous proteins under the conditions used. Tubulin levels were determined as loading controls.
(b) Hypocotyl lengths were measured after germination and 4 day growth under low fluence continuous FR (0.45 μmol m−2 sec−1). Error bars indicate SD (n = 20). Cotyledons of representative seedlings are shown.
Numbers refer to independent transgenic lines. Hypocotyl lengths of etiolated seedlings of all genotypes indicated were not significantly different (P < 0.01, Student’s t-test).
FHL and FHY1 act together to control phyA-regulated gene expression
FHY1 is known to be essential for appropriate expression of a subset of phyA-regulated genes during photomorphogenesis (Zeidler et al., 2004). To ascertain whether the relationships between FHL and FHY1 observed through characterization of morphological phenotypes are also manifested at the molecular level through the expression of genes unrelated to hypocotyl elongation and cotyledon expansion, we compared levels of transcripts encoding chalcone synthase (CHS), chlorophyll a/b-binding protein (CAB), ferredoxin NADP-oxidoreductase (PET H), plastocyanin (PET E), a xyloglucan endotransglycosylase (XTR7) and an asparagine synthetase (ASN1) in various genotypes after exposure of etiolated seedlings to FR for 3 and 18 h (Figure 4). Whereas CHS, CAB, PET H and PET E are upregulated by FR, XTR7 and ASN1 are negatively regulated by phyA.
Figure 4.
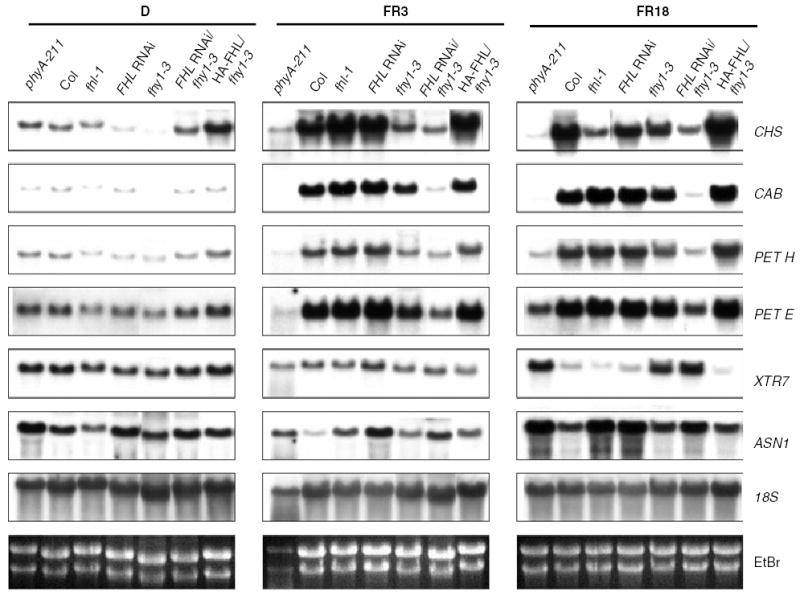
Loss of both FHY1 and FHL has a synergistic effect on phyA-regulated gene expression whereas FHL overexpression compensates for the effects of the fhy1-3 mutation on phyA-regulated gene expression. Three-day-old etiolated seedlings were either grown in darkness for 18 h (D), or exposed to continuous FR (4.5 μmol m−2 sec−1) for 3 h (FR3) or 18 h (FR18). Total RNA (10 μg) was loaded per lane and hybridized to gene-specific probes. Transgenic lines used: FHL RNAi line no. 29, FHL RNAi/fhy1-3 line no. 11, HA-FHL/fhy1-3 line no. 6.
The same relationships between FHY1 and FHL observed through assessing the effects of loss of either or both intermediates on seedling morphology (Figure 2) were evident at the level of phyA-regulated gene expression. Whereas the effects of fhl-1 or RNAi-mediated suppression of FHL on phyA-regulated gene expression are subtle and usually less severe than those observed in fhy1-3, suppression of FHL appeared to act synergistically with the fhy1-3 mutation in attenuating phyA signaling (Figure 4).
Furthermore, consistent with the ability of FHL overexpression to compensate for the absence of FHY1 in morphological assays (Figure 3), expression of HA-FHL in the fhy1-3 background restored phyA-responsive gene expression to levels resembling those seen in WT seedlings (Figure 4). This molecular evidence substantiates the functional overlap between FHL and FHY1 in relaying the phyA signal that is suggested by morphological analyses of HA-FHL/fhy1-3 and FHL RNAi/fhy1-3 seedlings.
FHL and FHY1 contribute to inhibition of hypocotyl elongation in continuous B
In addition to activating responses to FR and R, loss of phyA and at least certain phyA-specific signaling intermediates (Whitelam et al., 1993) confers hyposensitivity to B. As reported previously (Whitelam et al., 1993), fhy1 mutants display reduced hypocotyl elongation in continuous B when compared with WT seedlings. This hyposensitivity to B is not as severe as that observed following mutation of CRY1, the primary B receptor in high-intensity B (Figure 5). Under high-fluence B (12 μmol m−2 sec−1), fhl-1 mutant seedlings and lines with RNAi-mediated suppression of FHL expression had hypocotyl lengths comparable to those of fhy1-3. Loss of FHL or FHY1 results in a phenotype intermediate between those observed in the WT and phyA backgrounds (Figure 5). Expression of FHL RNAi in the fhy1-3 background decreased the level of inhibition of hypocotyl elongation observed in either of the monogenic mutants so that hypocotyl lengths of FHL-RNAi/fhy1-3 seedlings were indistinguishable from phyA mutants (Figure 5). This suggests that both gene products may normally be essential for phyA-mediated inhibition of hypocotyl elongation in this intensity of B, although definitive evidence for this proposal requires analysis of the effects of fhl-1, fhy1-3 and FHL RNAi/fhy1-3 in a cry1/cry2 background.
Figure 5.
Loss of FHL expression compromises responsiveness to blue light. Hypocotyl lengths were measured after germination and 4-day growth under continuous B (12.0 μmol m−2 sec−1). Error bars indicate SD (n = 20). Cotyledon opening and expansion of representative seedlings of each genotype are shown. The same lines were used to assess sensitivity to FR (Figure 2).
Overexpression of FHL causes hypersensitivity of B-mediated inhibition of hypocotyl elongation
The same lines used to show that FHL overexpression can complement the long hypocotyl phenotype of fhy1-3 in FR were used to examine the effects of ectopic FHL expression on fhy1-3 hypocotyl elongation in B (Figure 6a,b). Under relatively high fluence continuous B (12 μmol m−2 sec−1), HA-FHL expression was at least as effective as 6myc-FHY1 in restoring the WT hypocotyl elongation phenotype to fhy1-3 (Figure 6a). The possibility that HA-FHL overexpression might even confer hypersensitivity to B is supported by the observation that at a lower fluence of B (1.2 μmol m−2 sec−1), hypocotyl elongation was significantly less in two of the three HA-FHL/fhy1-3 lines than in comparable WT and 6myc-FHY1/fhy1-3 lines (Figure 6b). Overexpression of FHY1 did not appear to be associated with hypersensitivity of hypocotyl elongation to B, although definitive evidence of this proposal requires demonstration that our antibody has the same affinity for FHY1 and FHL. Nonetheless, it is clear that in both B and FR, FHL can substitute for FHY1 when expressed at adequate levels. Furthermore, in the absence of FHY1, FHL normally limits the response capacity of the cascade that relays the phyA signal. The observation that overexpression of either HA-FHL or 6myc-FHY1 did not confer hyposensitivity in a phyA null background (Figure 6a) confirms that these FHL overexpression effects in B are phyA-dependent and not reliant on cryptochrome activation.
Figure 6. Overexpression of FHL, but not FHY1, in an fhy1-3 background confers phyA-dependent hypersensitivity to blue light.
(a) Hypocotyl lengths were measured after germination and 4 day growth under continuous B (12 μmol m−2 sec−1). Error bars indicate SD (n = 20). Cotyledon opening and expansion of representative seedlings of each genotype are shown.
(b) Hypocotyl lengths were measured after germination and 4-day growth under continuous B (1.2 μmol m−2 sec−1). Error bars indicate SD (n = 20). Cotyledon opening and expansion of representative seedlings of each genotype are shown. The same lines were used to assess sensitivity to assess sensitivity to FR (Figure 3).
FHL and FHY1 self-associate and interact physically via their C-terminal domains
The sequence similarity and genetic interactions between FHY1 and FHL prompted us to examine whether the two proteins might interact physically. Unfortunately, yeast two-hybrid assays indicated that both proteins activate transcription when fused to the yeast GAL4 DNA binding domain. This transcriptional activity is decreased by physical association of the proteins as bait and prey (data not shown).
In vitro pull-down assays indicated that full-length FHL (Figure 7b) and full-length FHY1 (Figure 7c) form homodimers in vitro and that FHL and FHY1 are capable of heterodimerization (Figure 7d). Secondary structure predictions suggested that six and seven α-helices are likely to be fairly evenly spaced along the lengths of FHL and FHY1, respectively. Five classes of deletion mutants were constructed for each protein to investigate the roles of these predicted helices in homo- and hetero-dimerization (Figure 7a). The data demonstrate a critical role for the putative helix corresponding to the septin-related domain (Zeidler et al., 2004) in homodimerization of FHL (Figure 7b) and FHY1 (Figure 7c) as well as heterodimerization of both proteins (Figure 7d). This fragment alone is incapable of interaction with either of the full-length proteins. The C-termini of both proteins (helices 3–6 of FHL and helices 4–7 of FHY1) were adequate for homo- and heterointeractions, albeit with slightly reduced affinity in comparison with full-length protein fusions (Figure 7). No determinants of dimerization appear to reside in the N-terminal halves of either FHL or FHY1.
Figure 7. FHL and FHY1 homodimerize and heterodimerize in vitro.
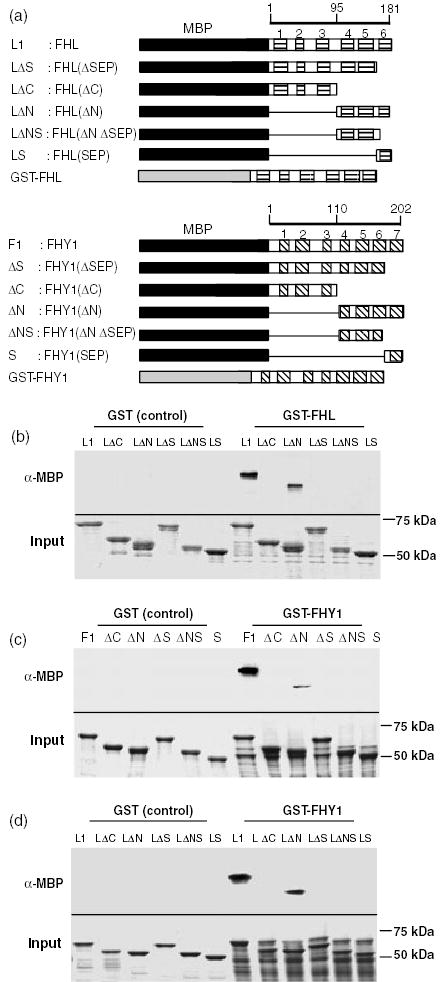
(a) Schematic representations of the full-length and deleted FHL and FHY1 proteins fused to the C-terminus of MBP or GST. Scale bars represent amino acids. Six helices in FHL and seven helices in FHY1 predicted by the SOPMA secondary structure prediction algorithm (http://npsa-pbil.ibcp.fr) are represented by horizontal and diagonal shading, respectively. Amino acids included in each deletion are indicated in Experimental procedures.
(b) In vitro pull-down using GST-FHL as the bait and fusions of full-length and deletion mutants of FHL to MBP as prey molecules.
(c) In vitro pull-down using GST-FHY1 as the bait and fusions of full-length and deletion mutants of FHY1 to MBP as prey molecules.
(d) In vitro pull-down using GST-FHY1 as the bait and fusions of full-length and deletion mutants of FHL to MBP as prey molecules.
For (b)–(d), the upper panel denotes proteins detected by anti-MBP antibody and the lower panel demonstrates Coomassie-stained total input protein.
Therefore, FHY1 and FHL are capable of forming homocomplexes with themselves as well as forming heterocomplexes with one another. The 87 residues closest to the C-terminus of FHL are sufficient for its homo- and hetero-dimerization with full-length FHY1. The 94 residues closest to the C-terminus of FHY1 are sufficient for its homodimerization. For both proteins, the helix closest to the C-terminus is critical for homo- and hetero-dimerization, but requires the adjacent two helices (in the case of FHL) or the adjacent three helices (in the case of FHY1) for its interaction with a full-length partner.
FHL transcript abundance depends on FHY3 and is negatively regulated by light
Previous studies indicated that FHY1 transcript abundance is negatively regulated by R and FR light (Desnos et al., 2001; Zeidler et al., 2001), and that FHY1 transcript levels are severely attenuated in fhy3 mutants (Desnos et al., 2001; Hudson et al., 2003; Wang and Deng, 2002). Real-time quantitative RT-PCR was used to evaluate the roles of light and the fhy3-1 mutation on FHY1 and FHL transcript levels. Levels of FHL and FHY1 were normalized relative to the abundance of ACTIN2 transcript and the levels of both transcripts in etiolated WT seedlings was arbitrarily assigned a value of 1. Monochromatic FR, R and B all decreased FHL and FHY1 transcript levels to around 60–70% of their levels in darkness (Figure 8). In etiolated fhy3-1 mutant seedlings, FHL and FHY1 transcript levels were around 43 and 27% of their levels in dark-grown WT seedlings, respectively (Figure 8). This indicates that FHY3 is required for normal accumulation of both transcripts even in the absence of light. FR, R and B all caused a further reduction of FHY1 transcript levels to <10% of the level in etiolated WT seedlings (Figure 8b), although for FHL, a slight additive effect of light on the fhy3-1 effect was only observed in R (Figure 8a).
Figure 8.
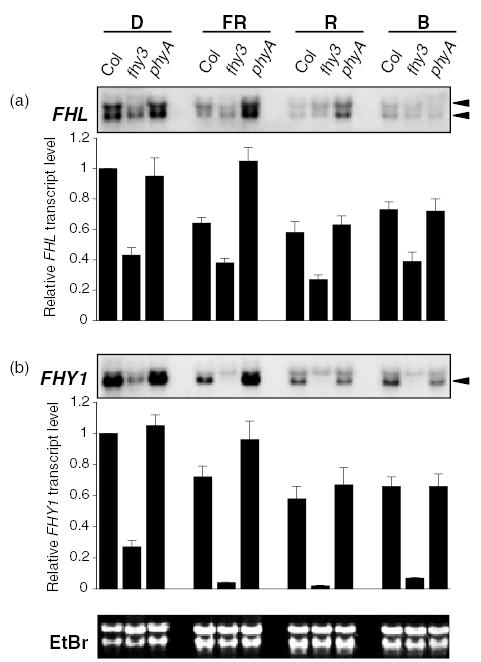
FHL transcript level is dependent on FHY3 and is decreased by exposure to FR and R. Wild-type (Col), fhy3-1 and phyA-211 seedlings were grown in darkness or continuous FR (4.5 μmol m−2 sec−1), R (12.0 μmol m−2 sec−1) or B (12.0 μmol m−2 sec−1) for 4 days and relative transcript levels were determined by real-time quantitative RT-PCR and Northern analysis. For real-time RT-PCR, relative expression levels were calculated using standard curves generated by serial dilutions of a cDNA mixture. Expression levels for FHY1 and FHL were normalized by dividing them by the abundance of the ACT2 transcript, which does not display significant regulation by light conditions (Hare et al., 2003). Data are mean ± SD of nine determinations (triplicate determinations from three independent RNA preps with a single RT reaction per prep). For Northern analysis, the same blot was probed with FHL and FHY1 cDNAs spanning the translated region of both genes. EtBr-stained rRNA on the membrane is shown as a loading control. The smaller of the two transcripts that hybridize with the FHY1 probe (denoted by the arrow) is regulated by FHY3 and light, whereas the larger fragment results from cross-hybridization. By contrast, two FHL-specific transcripts (arrows) display indistinguishable regulation by FHY3 and light.
As the PCRs for amplification of cDNA derived from the FHL, FHY1 and ACT2 transcripts worked with comparable efficiencies and all standard curves were prepared using data from FR-treated Col seedlings, we used the expression 240-Ct to broadly estimate relative differences in the abundances of the three transcripts (Czechowski et al., 2004). The threshold cycle (Ct) is the fractional PCR cycle at which a significant increase in signal above the baseline is first detected. The lower the Ct value, the greater the level of gene expression. The mean values and standard deviations of Ct values were 22.53 ± 0.45, 24.37 ± 0.46 and 28.23 ± 0.47 for the ACT2, FHY1 and FHL genes, respectively. Based on these mean Ct values, we estimate that in 4 day-old FR-grown seedlings, the FHY1 transcript is approximately 15-fold more abundant than the FHL transcript and approximately fourfold less abundant than the ACT2 transcript. In terms of absolute concentration, it seems likely that levels of the FHL transcript in irradiated fhy3-1 seedlings are probably comparable to those of FHY1 mRNA.
The same trends were observed when the effects of monochromatic light and the fhy3-1 mutation were determined by Northern analysis using full-length FHL and FHY1 cDNA probes (Figure 8). A greater abundance of FHY1 mRNA compared with FHL mRNA is consistent with the difficulty we experienced in reliable detection of FHL by Northern hybridization. Under our hybridization conditions, both probes detected double bands. For the FHY1 probe, the upper band appeared to result from non-specific cross-hybridization. By contrast, the levels of both fragments detected by the FHL probe are negatively regulated by R and FR light and by the fhy3-1 mutation (Figure 8a). The significance of the apparent existence of two FHL transcripts requires further investigation, but their relative levels do not appear to be regulated by FHY3 or the availability of light.
Discussion
A detailed understanding of the phyA signal transduction cascade in de-etiolating seedlings should provide a useful paradigm describing how perception of an essential environmental stimulus is translated into altered gene expression. Thus far, several recessive mutations that confer reduced sensitivity of hypocotyl growth to FR have been reported (Wang and Deng, 2003). Our findings not only identify a new participant in the chain of events necessary for a full response to phyA, but also reinforce the likely role of gene duplication in the fine-tuning of developmental responses to environmental fluctuations. The existence of an FHY1-related gene family in potato suggests that divergence of FHY1 and FHL functions is not unique to Arabidopsis. Conceivably, a slight divergence of the functions of these gene products in phyA signal transmission contributes to the complexity of the decision-making process required to initiate photomorphogenesis and ensures greater developmental plasticity during seedling establishment.
The much weaker phyA-specific loss of signaling phenotype evident in mutants and RNAi lines affected specifically in FHL expression probably accounts for the failure to recover fhl null mutants in previous screens for reduced de-etiolation in FR. Nonetheless, the apparent synergistic effects of fhl and fhy1 mutations emphasize the limitations of the classical hypocotyl elongation screen to identify the full complement of intermediates essential for phyA signaling. Mutagenesis of other known phyA-related signaling mutants will enable enhancer screens to identify novel signaling intermediates that participate in phyA signaling, but are not indispensable for morphological responses to FR.
FHL and FHY1 are present in disproportionate levels and probably define separate yet partially overlapping functions
Any model that attempts to explain the mode(s) of action of FHL and FHY1 should ideally account for (i) the synergistic consequences of decreases in FHL and FHY1 functions (Figures 2a and 4), (ii) the ability of FHL overexpression to restore a WT phenotype to fhy1-3 mutants (Figures 3a and 4), (iii) the much greater severity of fhy1 mutations in comparison with fhl mutations (Figure 2) and (iv) the ability of FHL and FHY1 to self-associate as well as interact with each other (Figure 7).
We recently reported that expression of FHY1 mutant proteins lacking the NLS and septin-related domains increased the insensitivity of fhy1-3 mutant seedlings to FR (Zeidler et al., 2004). A dominant negative phenotype of any gene product capable of homodimerization, but incapable of heterodimerization, requires the presence of the WT protein. One interpretation of the greater insensitivity of FHY1Δsep/fhy1-3 and GFP:FHY1nls/fhy1-3 mutants to FR than the insensitivity seen in fhy1-3 is that both FHY1Δsep and GFP:FHY1nls proteins interfere with activity of an FHY1 reaction partner (Zeidler et al., 2004). Although the capacity of FHL to interact physically with FHY1 through the three helices closest to its C-terminus (Figure 7) makes FHL an excellent candidate for this partner, a special role for FHY1-FHL heterodimers in phyA signaling is not consistent with the weak phenotype associated with loss of FHL alone (Figures 2 and 4). No inherent property of FHY1 that is absent from FHL is likely to account for the greater severity of fhy1 mutations in comparison with loss of FHL as overexpression of FHL can complement fhy1-3 (Figures 3 and 6). We currently favor the proposal that the extreme insensitivity of FHY1Δsep/fhy1-3 and GFP:FHY1nls/fhy1-3 mutants arises from their interaction with a phyA signal transducer other than FHL.
We suggest that both FHL and FHY1, either as homodimers or heterodimers, are essential for transmission of the phyA signal to inhibit hypocotyl elongation in FR. As our real-time PCR analysis indicates that FHY1 transcript abundance is approximately 15-fold in excess of FHL transcript level in FR-grown seedlings, we propose that fhy1 mutations have more severe phenotypic consequences simply because the much lower level of cellular FHL is incapable of supporting the flux through the signaling pathway that can be sustained by FHY1 alone. Ablation of FHL expression has weaker phenotypic consequences than loss of FHY1 because the much larger FHY1 pool can compensate better for the deficiency of FHL.
The extent of the overlap between FHY1 and FHL activities is not clear. When overexpressed, FHL can compensate for a deficiency in FHY1 (Figures 2 and 6). Given the difficulties in definitive interpretation of overexpression data, it is impossible to conclude whether this observation implies that FHY1 and FHL normally display complete redundancy. We cannot formally eliminate the possibility that ectopic expression of HA-FHL causes it to assume a novel role, although the ability of FHL to substitute for FHY1 clearly demonstrates a significant functional overlap between the two proteins. Caution should be exercised in inferring partial redundancy between signaling intermediates whenever overexpression in a mutant background is concerned, as the effects of FHL overexpression might only be apparent in the absence of FHY1. It is also not yet clear whether this apparent redundancy between FHY1 and FHL is reciprocal (that is, whether FHY1 overexpression might compensate for loss of FHL). Better insight into the relationships between the two homologs may be obtained by expressing both WT and Δsep mutants of FHY1 and FHL under their respective endogenous promoters in the fhy1 and fhy1/fhl mutant backgrounds, respectively.
We do not favor the view that FHL functions identically to FHY1. First, divergence of FHY1 family members is not restricted to Arabidopsis (Figure 1a). Secondly, complete redundancy between gene products is improbable simply because it cannot be sustained indefinitely in the absence of selective pressure. A model invoking separate but partially overlapping roles is an attractive possibility at least during seedling de-etiolation, although we cannot eliminate the possibility that FHL and FHY1 may serve unrelated roles later in the life cycle. If their functions do overlap, then it is reasonable to predict that the two genes might display similar patterns of expression. Accordingly, when compared with previously reported data for FHY1 (Desnos et al., 2001; Zeidler et al., 2001), a decrease in FHL transcript abundance under light of wavelengths capable of activating phyA (Figure 8) indicates similar negative regulation of both transcripts by light.
The relationship between FHL/FHY1 and other phyA signaling components
Evaluation of the effects of FHL and FHY1 overexpression in a phyA background on hypocotyl and cotyledon phenotypes in FR (Figure 3a) and B (Figure 6a) confirms that functioning of FHL and FHY1 requires activation of phyA. In this regard, FHL differs from the phyA signal transducer HFR1 that can elicit photomorphogenic responses when overexpressed in darkness (Jang et al., 2005). Rapid proteolytic degradation of the phyA photoreceptor is an important desensitization mechanism in phyA signaling (Seo et al., 2004). The observation that suppression of FHL expression in fhy1-3 phenocopies hypocotyl elongation in a phyA null mutant led us to investigate whether the severity of the FHL RNAi/fhy1-3 phenotype relative to isolated fhy1-3 and fhl-1 phenotypes could be attributed to decreased stability of the phyA photoreceptor in FR. Although an fhy1 null mutant was previously shown to have normal phyA levels in darkness (Whitelam et al., 1993), to our knowledge, the effect of fhy1 mutations on the photolability of phyA has never been reported. phyA levels are identical in WT and FHL RNAi/fhy1-3 seedlings in darkness as well as after exposure to R and FR for periods that promote proteolytic degradation of the photoreceptor (data not shown). This indicates that the synergistic effects of loss of both FHL and FHY1 do not arise at the level of phyA abundance and that FHL and FHY1 are bona fide signaling intermediates that transduce phyA signals. The occurrence of WT phyA levels in HA-FHL/fhy1-3 lines confirms this conclusion, as the ability of HA-FHL overexpression to restore a WT phenotype to fhy1-3 mutants cannot be attributed to stabilization of phyA in FR (data not shown).
The relationship between FHY3 and both FHL (Figure 8) and FHY1 (Desnos et al., 2001; Hudson et al., 2003; Wang and Deng, 2002) appears to be based on the requirement of FHY3 for transcriptional induction of genes encoding both FHL and FHY1. The observation that this effect is independent of light, but that fhl, fhy1 and fhy3 mutants display phyA-dependent phenotypes, can be interpreted as indicating that the role of FHY3 in phyA signaling may primarily concern the establishment in etiolated seedlings of a signaling network comprising the full complement of phyA response intermediates in anticipation of exposure to light. Nonetheless, evidence that FHY1 and FHY3 may define separate branches of the phyA signaling network has been presented elsewhere (Desnos et al., 2001; Okamoto et al., 2001; Wang and Deng, 2002). Whether FAR1, a phyA-specific homolog of FHY3, also affects FHL transcript abundance remains to be seen. However, the far1 mutation does not affect FHY1 transcript levels (Hudson et al., 2003) and causes a more severe insensitivity to FR than fhl-1 (Hudson et al., 1999). It remains to be established whether the negative effects of light and fhy3 mutations on FHL and FHY1 transcript levels occur at the level of transcriptional initiation or destabilization of transcripts.
We envisage that future attempts to dissect the unique and overlapping roles of FHL and FHY1 will reveal the mechanisms by which these partially redundant gene homologs fine-tune plant responses to subtle changes in the quantity and quality of light.
Experimental procedures
Plant materials and growth conditions
The fhl-1, fhy1-3 (Zeidler et al., 2001, 2004), fhy3-1 (Whitelam et al., 1993) and phyA-211 (Reed et al., 1994) mutants and all transgenic lines used in this study are in the Columbia background and were compared with WT Col-0 in all analyses. Although the hy4-1 (cry1-1, formerly called hy4-2.23N; Koornneef et al., 1980) mutant is in the Ler background, hypocotyl elongation of the Ler and Col ecotypes is indistinguishable under the fluences of B used (data not shown, Whitelam et al., 1993). Homozygous fhl-1 seedlings were selected from a segregating population (SALK_024320) obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH, USA). This putative null allele contains a T-DNA insertion in the first exon of the FHL gene (At5g02200). The fhy1-3 allele was previously named pat3 (Zeidler et al., 2001, 2004).
Surface-sterilized seeds were germinated on solidified Murashige and Skoog media (pH 5.7) without any supplemental carbohydrate. After 4 days of incubation in darkness at 4°C, the plates were exposed to white light (20 μmol m−2 sec−1) for 1 h and then incubated in darkness for 24 h at 22°C. To assess seedling morphological phenotypes, plates were either kept in darkness or transferred to constant monochromatic light using light sources described previously (Bolle et al., 2000). Hypocotyl elongation and cotyledon opening were assessed after 4-day growth. To assess the effects of light on transcript and protein levels, stratified seeds that had been exposed to a pulse of white light were kept in darkness at 22°C for 3 days, before transfer to the designated light condition. After light treatment, seedlings were harvested immediately, frozen in liquid nitrogen and stored at −70°C until RNA or protein extraction. Fluence rates for the various light treatments were as follows: FR high fluence treatment, 4.5 μmol m−2 sec−1; FR low fluence treatment, 0.45 μmol m−2 sec−1, R treatment, 35 μmol m−2 sec−1; B high fluence treatment, 12 μmol m−2 sec−1; B low fluence treatment, 1.2 μmol m−2 sec−1.
Molecular analyses
For RT-PCR to assess RNAi-mediated inhibition of FHL transcript accumulation, total RNA was extracted from 4 day-old etiolated seedlings using the RNeasy Plant mini kit (Qiagen, Valencia, CA, USA). Reverse transcription was performed using the Advantage RT-for-PCR kit (Clontech, Palo Alto, CA, USA), according to the manufacturer’s instructions. The oligos 5′-ATGATAGTTGCTGTGGAATCTC-3′ and 5′-CATCATGAGTGTAGAAAAGTACT-3′ were used for FHL amplification. The oligos 5′-TAAAGAGACATCGTTTCCATG-3′ and 5′-CAAGAATAGAGAATGAAGCTGAG-3′ were used for ACTIN8 amplification. PCR involved the following temperature profile: 95°C (3 min); 95°C (30 sec), 60°C (30 sec), 72°C (30 sec) – five cycles; 95°C (30 sec), 58°C (30 sec), 72°C (30 sec) – 25 cycles; 72°C (5 min). The PCR products were resolved on a 1.2% agarose gel. ACTIN8 amplification was visualized by EtBr staining. FHL amplification was assessed by southern transfer to Hybond N+ membrane (Amersham, Buckinghamshire, UK) and probing with 32P-labeled FHL cDNA probe spanning the entire coding sequence of FHL.
Northern analysis was performed as described previously (Zeidler et al., 2001). Probes used to assess the abundances of light-regulated transcripts were described elsewhere (Hare et al., 2003). Fragments encoding the complete open reading frames of FHY1 and FHL were used as probes. Hybridization was performed at 65°C over night and blots were washed once with 2X SSC, 0.1% SDS and twice times with 0.1X SSC, 0.1% SDS at 65°C.
For Western analysis involving total protein extracts, seedlings were ground under liquid nitrogen and the powder resuspended in 5X SDS-PAGE sample buffer. Immunoblotting was performed as described previously (Zeidler et al., 2001). Polyclonal anti-FHY1 antiserum was obtained from rabbits immunized with hexahistidine-tagged FHY1 expressed and purified from Escherichia coli strain BL21 (Invitrogen, Carlsbad, CA, USA). Monoclonal anti-α-tubulin (clone DM 1A) was obtained from Sigma (St Louis, MO, USA). Anti-phyA monoclonal antibody mAA1 was kindly donated by Dr A. Nagatani.
Transgenic lines
A full-length FHL cDNA was prepared by RT-PCR using RNA extracted from 4-day-old etiolated seedlings. The cDNA was amplified using the oligos 5′-ACGCTCGAGCGATGATAGTTGCTGTGGAATC-3′ and 5′-CGCGAGCTCTGTTTACATCATGAGTGTAG-3′. The product was cloned into pGEM-T (Promega, Madison, WI, USA). All transgenes were placed under the regulation of the CaMV 35S promoter. To generate the HA-FHL construct, a single copy of the hemaglutinin (HA) tag (MYPYDVPDYASL) was fused to the N-terminus of FHL by PCR and cloned into the binary transformation vector VIP96a, a derivative of a vector conferring kanamycin resistance (van der Krol and Chua, 1991) to which XbaI, BglII, AscI, PacI, SpeI, EcoRI and StuI sites had been introduced into the multicloning site between the CaMV 35S promoter and the pea RBCS E9 polyA addition sequence (S.G. Møller, University of Leicester, Leicester, UK, personal communication). For the FHL-RNAi construct, two fragments of FHL genomic DNA were amplified by PCR. The first genomic fragment, comprising the first exon and the first intron, was amplified using the oligos 5′-TGTGGAATCTCTAGACACAAGCA-3′ and 5′-TAACTCGAGTCGTGGTAGCTTCTTTGGAA3′. The second genomic fragment, comprising the first exon, was amplified using the oligos 5′-TAAGGCCTGAATCTCTAGACACAAGCAAA-3′ and 5′-AACTCGAGCTTGGTTGCTGACACTCCAC-3′. This enabled it to be cloned in the reverse orientation after the intron of the first fragment. The two PCR fragments were digested with XbaI/XhoI and XhoI/StuI, respectively, and ligated into XbaI/StuI-digested pVIP96a. A binary vector myc-pBA was constructed containing a multicloning site and six copies of the myc epitope tag ligated into the AscI and PacI sites of the vector pBA002 that confers Basta resistance (Kost et al., 1998). This introduced unique BsrGI, AatII, SmaI, BstBI, BamHI, SnaBI, AvrII and MluI sites to enable translational fusions upstream and downstream of the 6xmyc tag. Full-length FHY1 cDNA amplified using the oligos 5′-CATCCCGGGTTATGCCTGAAGTGGAAGTG-3′ and 5′-CATCCTAGGCCGGGTTACAGCATTAGCGTTGAG-3′ was cloned into the SmaI and AvrII sites of myc-pBA. The 6myc-FHY1 fusion was transferred as an AscI/PacI fragment into VIP96a to generate the 6xmyc-FHY1 construct. Binary vector constructs were transformed into WT (Col) or fhy1-3 (Basta resistant) backgrounds using the Agrobacterium tumefaciens-mediated floral dip method and transformants were selected on media supplemented with 50 mg l−1 kanamycin (Sigma). For each line, two independent transformants were selected for further analysis using the T3 generation.
Transient expression of fluorescent protein fusions in onion cells
Mutations of the NLS and NES motifs in FHL were generated using the oligos 5′-CTCCGGCCGCAGCTCTCCATGCAGAAGAATCAGAC-3′/5′-GACCGGCCGCGCTTGTGTCTAGAGATTCCAC-3′ and 5′-CTCCGGCCGCAGCTCCAAAACACTTTTGTTCTGAG-3′/5′-GACCGGCCGCGTATGATTCTTCAGCATGCAG-3′, respectively. Both 12KKRK15 to 12AAAA15 and 23LLPL26 to 23AAAA26 mutations were confirmed by sequencing. The FHL, FHLnls and FHLnes cDNAs were cloned into the vector pCHS-GFP to generate the constructs pCHS-GFP-FHL, pCHS-GFP-FHLnls and pCHS-GFP-FHLnes, respectively. This enabled fusion of WT and mutant FHL cDNAs to the C-terminus of a fusion of GFP to parsley chalcone synthase. These constructs, as well as the pCHS-GFP control, were transiently expressed in onion epidermal cells as described previously (Zeidler et al., 2001, 2004), and the subcellular localization of the transgene products visualized by fluorescence microscopy.
In vitro pull-down assays
cDNAs encoding full-length and deletion mutants of FHL and FHY1 were amplified by PCR and cloned into pMAL-C2 (New England Biolabs, Beverly, MA, USA) or pGEX-4T-1 (Amersham) for fusion to the C-termini of MBP and GST, respectively. Deletion mutant FHL fusions to the C-terminus of MBP were: FHL(Δsep): residues 1–158; FHL(ΔC): residues 1–93; FHL(ΔN): residues 94–181; FHL(ΔNΔsep): residues 94–158; and FHL(sep): residues 159–181. Deletion mutant FHY1 fusions to the C-terminus of MBP were: FHY1(Δsep): residues 1–166; FHY1(ΔC): residues 1–107; FHY1(ΔN): residues 108–202; FHY1(ΔNΔsep): residues 108–166; and FHY1(sep): residues 167–202.
All constructs were transformed into E. coli BL21 cells that were treated with isopropyl-β-d-thiogalactoside to induce fusion protein expression. Treated cells were broken by a French pressure cell press in purification buffer (20 mm Tris-HCl, pH 7.4; 200 mm NaCl; 1 mm EDTA; 1% Triton X-100; 2 mm phenylmethylsulphonyl fluoride) containing a proteinase inhibitor cocktail (Roche, Mannheim, Germany). Proteins were bound to amylose resin (New England Biolabs) for MBP-fused proteins and glutathione Sepharose 4B (Amersham) for GST-fused proteins, washed with buffer (phosphate-buffered saline; 1% Triton X-100), and eluted from the column using purification buffer containing 10 mm maltose (for MBP-fused proteins) or 10 mm glutathione (for GST-fused proteins).
For in vitro pull-down assays, 1 μg of bait protein (full-length or deletion mutants of FHL and FHY1) and 1 μg of prey protein (GST-FHL or GST-FHY1) were incubated at 25°C for 2 h in binding buffer (50 mm Tris-HCl, pH 7.5; 100 mm NaCl; 1.0% Triton X-100; 0.5 mm β-mercaptoethanol) and further incubated with glutathione sepharose 4B at 25°C for 2 h. After washing six times with buffer (50 mm Tris-HCl, pH 7.5; 100 mm NaCl; 1% Triton X-100), pull-down proteins were separated on 6% SDS-polyacrylamide gels and detected by Western blotting using anti-MBP antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Complexes were visualized by chemiluminescence with horseradish peroxidase-coupled second antibodies (Amersham).
Quantitative real-time RT-PCR
Oligonucleotides that amplify 100 bp amplicons were selected using Primer Express software (PE Applied Biosystems, Warrington, UK). ACT2: 5′-CCTTCGTCTTGATCTTGCGG-3′ and 5′-CAATTTCCCGTTCTGCGGTA-3′ (amplify nt 531–631 of the coding sequence), FHY1: 5′-ATACGGAGTATTCAATGTCTTCTTATGTG-3′ and 5′-TTCACCGCATGATCCAGATG-3′ (amplify nt 236–336 of the coding sequence), FHL: 5′-TCTGAGCATCAAGCCTCTCTTG-3′ and 5′-TCATCGCTGGTTTTTGTGTTCT-3′ (amplify nt 94–194 of the coding sequence).
Triplicate total RNA preps of each genetic background in each light condition (36 preps total) were used as templates for first-strand synthesis. Total RNA was treated with RNase-free DNase I (Invitrogen) before cDNA synthesis according to the manufacturer’s instructions. cDNA was generated from 5 μg of total RNA using oligo(dT)20 and the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Real-time qRT-PCRs (25 μl) were performed using 1 μl of cDNA, 12.5 μl of 2x SYBR® Green PCR Master Mix (Applied Biosystems, Warrington, UK) and 300 nm of each oligo. Amplification and detection of dsDNA synthesis were performed using an ABI Prism® 7900 sequence detection system (Applied Biosystems) under the following conditions: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 sec at 95°C and 1 min at 60°C.
For all three genes, template from FR-treated Col seedlings was used to construct a standard curve relating the threshold cycle (Ct) to log(ng of original RNA template). Eight twofold serial dilution points (128–1 ng) were used to construct standard curves. The remaining 72 wells in 96-well plates were used for duplicate determinations each using first-strand synthesis product from 50 ng of the original RNA template. The standard curve of each gene from was used to determine the Ct value corresponding to the mean of each duplicate determination. The abundance of ACT2 transcript, which is not affected by light treatments (Hare et al., 2003), was used to normalize template levels. FHY1/ACT2 and FHL/ACT2 ratios in dark-grown Col seedlings were arbitrarily assigned values of 1. Relative abundances in different genotypes and under the four different light conditions were determined. Data are mean of nine replicates ± SD. The median coefficient of variation (based on calculated quantities) was 11.1%.
Distribution of materials
Upon request, all novel materials described in this publication will be made available for non-commercial purposes, subject to the requisite permission from any third party owners of all or parts of the material. Obtaining any permission will be the responsibility of the requestor.
Acknowledgments
We are grateful to Thomas Merkle for the pCHS-GFP vector, to Garry Whitelam for the fhy3-1 mutant, to the ABRC for SALK_024320, phyA-211 and hy4-1 mutants, to Akira Nagatani for phyA antibody and to Wenxiang Zhang (Rockefeller University Gene Array Resource Center) for excellent technical assistance. This work was supported by NIH grant GM-44640 to N.-H.C.
References
- Barnes SA, Nishizawa NK, Quaggio RB, Whitelam GC, Chua NH. Far-red light blocks greening of Arabidopsis seedlings via a phyA-mediated change in plastid development. Plant Cell. 1996;8:601–605. doi: 10.1105/tpc.8.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua N-H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 2004;38:366–379. doi: 10.1111/j.1365-313X.2004.02051.x. [DOI] [PubMed] [Google Scholar]
- Desnos T, Puente P, Whitelam GC, Harberd NP. FHY1: a phytochrome A-specific signal transducer. Genes Dev. 2001;15:2980–2990. doi: 10.1101/gad.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transductions. Genes Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC. Phytochromes B, D and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol . 2003;131:1340–1346. doi: 10.1104/pp.102.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasen D, Köhler C, Neuhaus G, Merkle T. Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 1999;20:695–705. doi: 10.1046/j.1365-313x.1999.00644.x. [DOI] [PubMed] [Google Scholar]
- Hare PD, Møller SG, Huang LF, Chua NH. LAF3, a novel factor required for normal phytochrome A signaling. Plant Physiol. 2003;133:1592–1604. doi: 10.1104/pp.103.028480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev . 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Lisch DM, Quail PH. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 2003;34:453–471. doi: 10.1046/j.1365-313x.2003.01741.x. [DOI] [PubMed] [Google Scholar]
- Jang IC, Yang JY, Seo HS, Chua NH. HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 2005;19:593–602. doi: 10.1101/gad.1247205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- Kost B, Spielhofer P, Chua NH. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 1998;16:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- van der Krol AR, Chua NH. The basic domain of plant bZIP proteins facilitates import of a reporter protein into plant nuclei. Plant Cell. 1991;3:667–675. doi: 10.1105/tpc.3.7.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H, Matsui M, Deng XW. Overexpression of the heterotrimeric G-protein α-subunit enhances phytochrome-mediated inhibition of hypocotyl elongation in Arabidopsis. Plant Cell. 2001;13:1639–1652. doi: 10.1105/TPC.010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004;18:617–622. doi: 10.1101/gad.1187804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW. Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 2002;21:1339–1349. doi: 10.1093/emboj/21.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Deng XW. Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci. 2003;8:172–178. doi: 10.1016/S1360-1385(03)00049-9. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M, Bolle C, Chua NH. The phytochrome A specific signaling component PAT3 is a positive regulator of Arabidopsis photomorphogenesis. Plant Cell Physiol. 2001;42:1193–1200. doi: 10.1093/pcp/pce177. [DOI] [PubMed] [Google Scholar]
- Zeidler M, Zhou Q, Sarda X, Yau CP, Chua NH. The nuclear localization signal and the C-terminal region of FHY1 are required for transmission of phytochrome A signals. Plant J. 2004;40:355–365. doi: 10.1111/j.1365-313X.2004.02212.x. [DOI] [PubMed] [Google Scholar]


