Abstract
Ubiquitin plays essential roles in various cellular processes; therefore, it is of keen interest to study the structure-function relationship of ubiquitin itself. We investigated the modification of Lys6 of ubiquitin and its physiological consequences. Mass spectrometry-based peptide mapping and N-terminal sequencing demonstrated that, of the 7 Lys residues in ubiquitin, Lys6 was the most readily labeled with sulfosuccinimidobiotin. Lys6-biotinylated ubiquitin was incorporated into high molecular mass ubiquitin conjugates as efficiently as unmodified ubiquitin. However, Lys6-biotinylated ubiquitin inhibited ubiquitin-dependent proteolysis, as conjugates formed with Lys6-biotinylated ubiquitin were resistant to proteasomal degradation. Ubiquitins with a mutation of Lys6 had similar phenotypes as Lys6-biotinylated ubiquitin. Lys6 mutant ubiquitins (K6A, K6R, and K6W) also inhibited ATP-dependent proteolysis and caused accumulation of ubiquitin conjugates. Conjugates formed with K6W mutant ubiquitin were also resistant to proteasomal degradation. The dominant-negative effect of Lys6-modified ubiquitin was further demonstrated in intact cells. Overexpression of K6W mutant ubiquitin resulted in accumulation of intra-cellular ubiquitin conjugates, stabilization of typical substrates for ubiquitin-dependent proteolysis, and enhanced susceptibility to oxidative stress. Taken together, these results show that Lys6-modified ubiquitin is a potent and specific inhibitor of ubiquitin-mediated protein degradation.
The ubiquitin/proteasome pathway (UPP)1 is involved in regulating the cell cycle, signal transduction, differentiation, stress response, and DNA repair. Most of these functions are mediated by the conditional turnover of regulatory and abnormal proteins in eukaryotic cells (1-4). In this essential pathway, the conserved ubiquitin acts as a degradation signal upon covalent linkage of multiple ubiquitins to cellular substrates. This is accomplished through the formation of an isopeptide bond between the C terminus of ubiquitin (Gly76) and an internal lysine residue of the substrate. Formation of the ubiquitin-substrate conjugates is usually followed by additional rounds of conjugation of more ubiquitins to the initial adduct, also via isopeptide bonds, leading to the assembly of long polyubiquitin chains. Although polyubiquitin chains formed via Lys6 (5), Lys11 (6), Lys29 (7), and Lys63 (7-10) are detected in cell-free systems and/or in whole cells and are competent for 26 S proteasome-dependent degradation (6, 11), Lys48-linked polyubiquitin chains represent the predominant signal for targeting substrates to the 26 S proteasome (12-14). Polyubiquitin chains linked through lysine residues other than Lys48 have functions principally independent of targeting proteins for degradation, such as DNA repair and signal transduction (8-11).
The 26 S proteasome specifically degrades ubiquitin-conjugated proteins, and it is assembled from a 20 S catalytic core and two 19 S regulatory complexes (also called PA700) that cap the entry to the cylindrical catalytic core (15-18). It is the 19 S regulatory complex that recruits ubiquitin-protein conjugates to the 26 S proteasome and translocates the ubiquitinated proteins into the catalytic cavity. It has been demonstrated that S5a (also called MBP1 and Mcb1p), the ATPase S6[H11032] subunit of the 19 S regulatory complex, and Rad23, a proteasome-associated protein, can each bind polyubiquitin chains (6, 19-23). The 19 S regulatory complex also has deubiquitinating activity, which removes ubiquitin from the substrate prior to or coordinated with degradation of the substrate proteins. Such deubiquitination may serve both in degradation and in proofreading or editing functions (24-26).
Biotinylated ubiquitin has been used as a probe to study the function of the UPP (27). However, biotinylated ubiquitin has not been characterized in detail, and the potential kinetic artifacts of biotinylated ubiquitin have not been assessed. In most cases, Lys residues are modified by biotinylating reagents such as sulfosuccinimidobiotin (sulfo-NHS-biotin). Some of the 7 Lys residues in ubiquitin are essential for formation of polyubiquitin chains. Modification of 1 or more of the critical Lys residues may abolish the ability of ubiquitin to form polyubiquitin chains. Modifications of some Lys residues may also alter the surface property of ubiquitin chains and may interfere with the interaction between ubiquitin conjugates and the proteasome. Previous studies indicated that Lys6 is the most susceptible residue to modification by various chemicals, such as p-nitrophenyl acetate (28), aspirin (29), acetic anhydride (30), and Oregon green succinimidyl ester (31). This preference for modification could be due to the differential amine surface accessibility (29) or to the catalytic microenvironment. The latter is most likely with respect to Lys6 because acetylation of Lys6 depends upon its interaction with His68 (30). In the three-dimensional structure of ubiquitin, His68 is located in the vicinity of Lys6 (32, 33). The interaction between Lys6 and His68 provides a catalytic microenvironment for the modification of Lys6 (30). Given the similarity between biotinylation and the chemical modifications mentioned above, Lys6 could also be the dominant site of biotinylation.
Individual Lys-to-Arg mutations are often used to test the role of particular Lys residues in ubiquitin chain formation. However, the physiological consequences of Lys-to-Arg mutations may be due to factors unrelated to whether that particular Lys is used for chain synthesis. Disruption of a face on ubiquitin necessary for recognition by enzymes involved in ubiquitin conjugation or degradation could be one of the factors. A detailed characterization of biotinylated ubiquitin will allow us to determine the role of a particular Lys residue in ubiquitin chain formation and in maintaining the surface property for proper interaction with enzymes involved in ubiquitination, deubiquitination, and degradation of ubiquitin conjugates.
In this study, we ascertained which Lys residues are modified by sulfo-NHS-biotin. Subsequently, we determined the ability of various biotinylated ubiquitins to form ubiquitin conjugates and to support ubiquitin-dependent degradation. The results show that 1) Lys6 was the most readily labeled with sulfo-NHS-biotin; 2) Lys6-biotinylated ubiquitin was used as efficiently as unmodified ubiquitin to form high molecular mass ubiquitin conjugates; 3) Lys6-biotinylated ubiquitin inhibited ubiquitin-dependent degradation because conjugates formed with Lys6-biotinylated ubiquitin were less susceptible to degradation by the 26 S proteasome; 4) K6W mutant ubiquitin had effects similar to those of Lys6-biotinylated ubiquitin; and 5) intracellular expression of K6W mutant ubiquitin resulted in stabilization of substrates for the UPP and enhanced susceptibility to oxidative stress. The data indicate that Lys6-modified ubiquitin is a specific inhibitor of the UPP and a valuable reagent for studying functions of ubiquitin-dependent cellular processes.
EXPERIMENTAL PROCEDURES
Materials—Tris, acrylamide, N,N′-methylenebisacrylamide, N,N,N′,N′-tetramethylenediamine, 2-mercaptoethanol, SDS, glycine, and protein molecular mass standards were purchased from Bio-Rad. Sulfo-NHS-biotin and chemiluminescence detection SuperSignal kits were purchased from Pierce. Ubiquitin, bovine α-lactalbumin, dithiothreitol, MgCl2,Me2SO, ATP, creatine phosphate, and creatine phosphokinase were purchased from Sigma. 125I-Protein A and Na125I were obtained from PerkinElmer Life Sciences. Anti-ubiquitin antibody was produced in New Zealand White rabbits. Antibodies to p21WAF1 and green fluorescent protein (GFP) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Ubiquitin aldehyde was purchased from Boston Biochem (Cambridge, MA). Sequencing grade trypsin and Lys-C were purchased from Roche Diagnostics. Cell culture medium and Lipofectamine 2000 were purchased from Invitrogen.
Labeling of Ubiquitin with Biotin and Purification of Biotin-labeled Ubiquitin—Ubiquitin was dissolved in phosphate-buffered saline at a concentration of 10 mg/ml at pH 7.4. 100 mM sulfo-NHS-biotin stock solution was made in Me2SO. To obtain ubiquitins with various levels of biotin attached, an appropriate volume of sulfo-NHS-biotin stock solution was mixed with the ubiquitin solution to achieve molar ratios of sulfo-NHS-biotin to ubiquitin of 2, 3.5, and 7. The reaction was carried out on ice for 2 h with occasional shaking and was terminated by addition of lysine at a final concentration of 20 mM. The biotinylated ubiquitins were purified by reversed-phase HPLC (HP 1100) on a Vydac C18 column (4.6 × 250 mm, 5-μm particle size, 90-Å pore size) using water/acetonitrile linear gradients containing 0.05% trifluoroacetic acid in both mobile phases. The elution gradient started at 20% acetonitrile and increased to 70% acetonitrile in a 40-min period. Elution was monitored at 254 and 280 nm. Peaks were collected individually. The masses of ubiquitin in each fraction were determined by a matrix-assisted laser desorption ionization time-of-flight mass spectrometer (Voyager-DE Biospectrometry workstation, PerSeptive Biosystems) or by an in-line electrospray ionization ion-trap mass spectrometer (Esquire-LC, Bruker Daltonik GmbH).
Mapping the Sites of Biotinylation—Each fraction of biotinylated ubiquitin was lyophilized and redissolved in 100 mM Tris-HCl (pH 8.0) containing 1 mM EDTA at a concentration of 1 mg/ml. Twenty μg of biotinylated ubiquitin was digested with 2 μg of sequencing grade Lys-C or trypsin at 32 °C for 16 h. The resulting peptides were separated by HPLC (HP 1100) on a Vydac C18 column (1 × 50 mm, 5-μm particle size, 300-Å pore size) with gradients from 0 to 60% acetonitrile over 40 min. The masses of the separated peptides were determined with the Esquire-LC in-line electrospray ionization ion-trap mass spectrometer. The sites of biotinylation were determined by comparing the observed peptide masses with the calculated masses of all peptides that could be derived from ubiquitin after digestion with Lys-C or trypsin. The sites of biotinylation were also confirmed by N-terminal sequencing of biotinylated ubiquitins. To do this, each of the purified biotinylated ubiquitins was sequenced for 34 cycles using a protein sequencer (Ap-plied Biosystems).
Generation of Ubiquitin Mutants—K6A and K6R mutant ubiquitins were generated by PCR-based site-directed mutagenesis and expressed in Escherichia coli as described previously (34). K6W mutant ubiquitin was generated by PCR amplification of mouse polyubiquitin cDNA (35) with forward primer 5′-CGCCACCATGCAGATCTTCGTGTGGACC-3′ and reverse primer 5′-TTAGCCACCTCTCAGGCAAGGACC-3′. The PCR products were purified and inserted into the pAdTrack-CMV vector (36) for expression in mammalian cells or subcloned into the pET15b vector (Novagen) for expression in E. coli. The sequence of the mutant ubiquitin gene was confirmed by sequencing the entire coding region. Recombinant adenoviruses were generated using the AdEasy adenoviral vector system (36). To study the effect of K6W mutant ubiquitin in HEK293 cells, wild-type and K6W mutant ubiquitin constructs were introduced into 293 cells using Lipofectamine according to the manufacturer's instructions. To study the effect of K6W mutant ubiquitin in human lens epithelial cells, the cells were infected with control recombinant adenovirus (no ubiquitin) and with recombinant adenovirus encoding K6W mutant ubiquitin.
Detection of Ubiquitin Conjugates—Levels of endogenous ubiquitin conjugates were determined by Western blotting as described previously (37). Briefly, proteins were resolved by SDS-PAGE on 12% gels and transferred to nitrocellulose. The blots were probed with antibody to ubiquitin. The specifically bound antibody was detected with 125I-protein A, followed by autoradiography or by chemiluminescence detection using the SuperSignal kit.
De Novo Ubiquitin Conjugation—The ability of biotinylated ubiqui-tins to support ubiquitination was determined in fraction II of rabbit reticulocytes (38) or in the supernatant (S100) of retinal pigment epithelial (RPE) cells. Fraction II of reticulocytes and RPE cell lysate were prepared as described previously (38, 39). The conjugation activity was determined using endogenous enzymes and exogenous 125I-labeled bovine α-lactalbumin or bovine transducin-α as substrate. Briefly, the assay was conducted in a final volume of 25 μl containing (final concentrations) 12 mg/ml RPE cell supernatant or 10 mg/ml fraction II, 50 mM Tris (pH 7.6), 2 mM ATP, 1 mM dithiothreitol, 5 mM MgCl2, 4 μM 125I-labeled substrate, and 8 μM wild-type or mutant ubiquitin. The mixture was incubated at 37 °C for 20 min, and the reaction was stopped by addition of 25 μl of 2× SDS-PAGE loading buffer. After boiling at 100 °C for 3 min, aliquots of the mixture were resolved by SDS-PAGE. The ubiquitin conjugates formed de novo were visualized by autoradiography.
Ubiquitin-dependent Degradation of Model Substrates—The ability of differentially modified ubiquitins to support ATP-dependent degradation was determined in RPE cell lysates using 125I-labeled α-crystallin or transducin-α as substrate. Briefly, the assay was conducted in a final volume of 25 μl containing (final concentrations) 12 mg/ml RPE cell supernatant, 8 μM wild-type or mutant ubiquitin, 50 mM Tris (pH 7.6), 2 mM ATP, 1 mM dithiothreitol, 5 mM MgCl2, 4 mM creatine phosphate, 2 μg/ml creatine phosphokinase, and 2—4 μM 125I-labeled substrate. The same conditions were used to measure ATP-independent degradation except that ATP, creatine phosphate, and creatine phosphokinase were replaced with 30 mM 2-deoxyglucose. The reaction was carried out at 37 °C for 2 h and then stopped by addition of 200 μl of cold 1% (w/v) bovine serum albumin solution and 50 μl of 100% (w/v) trichloroacetic acid. The mixture was centrifuged at 14,000 × g for 10 min after standing on ice for 10 min, and the radioactivity in the supernatants and pellets was counted. The degradation rates are expressed as percentages of the 125I-labeled substrates that became trichloroacetic acid-soluble (40).
Proteasome Binding Assay—Bovine α-lactalbumin was first labeled with 125I and used as substrate for ubiquitin conjugation. Ubiquitin conjugates of 125I-labeled α-lactalbumin were formed in proteasome-free fraction II of rabbit reticulocytes upon addition of wild-type, Lys6-biotinylated, K6W, or L8A mutant ubiquitin. To obtain proteasome-free fraction II, rabbit reticulocyte fraction II was prepared as described (41), and the proteasome was removed from fraction II by centrifugation at 100,000 × g for 5 h (42). The ubiquitin conjugates of 125I-labeled α-lactalbumin were isolated chromatographically using a DE52 column. Proteasome from rabbit reticulocyte was resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes. After denaturation and renaturation procedures (19, 20), the membranes were probed with the respective ubiquitin conjugates of 125I-labeled α-lactalbumin. To quantitatively test the capability of binding to S5a, glutathione S-transferase (GST)-S5a fusion protein was immobilized on glutathione-agarose as described (43) and incubated overnight with ubiquitinated 125I-labeled α-lactalbumin at 4 °C. After five washes with 50 mM Tris, 150 mM NaCl, and 0.2% Tween 20, bound ubiquitin conjugates, together with GST-S5a, were eluted with 10 mM glutathione and quantified using a γ-counter. GST-glutathione-agarose was used as negative control to subtract nonspecific binding.
Sensitivity to Oxidative Stress Assays—Human lens epithelial cells at ∼50% confluence were infected with recombinant adenovirus encoding K6W mutant ubiquitin or with control adenovirus for 48 h. The cells were then exposed to ∼20 μM H2O2 constantly generated by addition of glucose oxidase to the medium (0.1 unit/dish) as described (4). After exposure to H2O2 for 8 h, cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay using re-agents from Promega. Cells detached from the plate were collected and stained with trypan blue, and the stained and unstained cells were then counted using a hemocytometer. Cells adhering to the dishes were collected with trypsin/EDTA and counted to determine the total number of cells in each dish. (>99% of the adherent cells were viable.) Susceptibility to oxidative stress was indicated by a decrease in cell viability and by an increase in the number of cells stained with trypan blue (dead cells) after exposure to H2O2.
Sensitivity to Canavanine—K6R mutant ubiquitin was constructed and expressed in yeast as described (8). In brief, wild-type or K6R mutant ubiquitin under the control of the CUP1 promoter (16) was transformed into a yeast strain (SUB328) in which the genes encoding ubiquitin-ribosomal protein fusions (UBI1 to UBI3) and polyubiquitin (UBI4) had been deleted (44). The yeast cells were cultured in the presence or absence of 1 μM canavanine, an amino acid analog.
RESULTS
Lys6 of Ubiquitin Is the Most Readily Labeled with Sulfo-NHS-biotin—To characterize relations between the stoichiometry of sulfo-NHS-biotin and the extent and location of ubiquitin modification, we isolated biotinylated ubiquitins that were formed using different amounts of sulfo-NHS-biotin (Fig. 1A). There are 7 Lys residues in a ubiquitin molecule (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63). Together with the N-terminal amine group, there are eight possible positions that can be labeled with sulfo-NHS-biotin. When the sulfo-NHSbiotin/ubiquitin ratio was 2, >95% of the ubiquitin was modified, and the majority of the ubiquitin was monobiotinylated (Fig. 1B). When the sulfo-NHS-biotin/ubiquitin ratio was increased to 3.5, 100% of the ubiquitin was modified by at least one biotin moiety (Fig. 1C). The modified ubiquitins were separated by HPLC, and their masses were determined using an in-line mass spectrometer. The peak that eluted at ∼22.5 min had a mass of 8565.0 Da, which corresponds to unmodified ubiquitin. The peak that eluted at ∼23.5 min had a mass of 8790.3 Da, which corresponds to monobiotinylated ubiquitin. The peaks that eluted at ∼24.5 and 26 min, respectively, had an identical mass of 9016.3 Da, corresponding to dibiotinylated ubiquitin. The peak that eluted at ∼27 min had a mass of 9242.8 Da, corresponding to tribiotinylated ubiquitin.
FIG. 1.
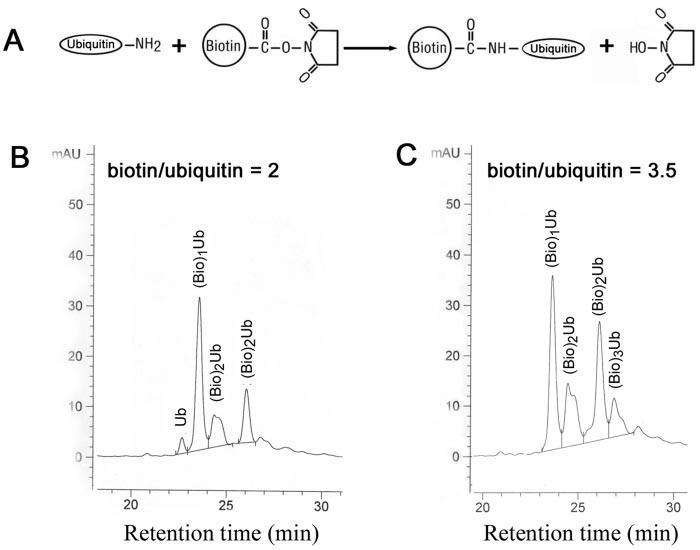
Characterization of biotin-labeled ubiquitin. Sulfo-NHS-biotin modifies primary amine groups on proteins to form amide bonds (A). Ubiquitin was labeled in PBS (pH 7.5) with sulfoNHS-biotin at molar ratios of 2 (B) and 3.5 (C). The labeled ubiquitin was separated by HPLC, and elution was monitored at 280 nm (B and C). Peaks were collected, and the molecular mass of each peak was determined by mass spectrometry. The peak that eluted at ∼22.6 min had a mass of 8565.0 Da, which corresponds to unmodified ubiquitin (Ub). The peak that eluted at ∼23.5 min had a mass of 8790.3 Da, which corresponds to monobiotinylated ubiquitin ((Bio)1Ub). The peaks that eluted at ∼24.5 and 26 min had an identical mass of 9016.3 Da, which corresponds to dibiotinylated ubiquitin ((Bio)2Ub). The peak that eluted at ∼27 min had a mass of 9242.8 Da, which corresponds to tribiotinylated ubiquitin ((Bio)3Ub). mAU, milli-absorbance units.
There are eight positions where a single biotin might attach and 28 or 56 combinations of possible di- or tribiotinylated ubiquitins, respectively. To determine which Lys residues were labeled with biotin, the differentially labeled ubiquitins were digested with endopeptidase Lys-C, which cleaves peptide bonds after Lys residues. The resulting peptides were separated by HPLC, and the masses were determined as described above. Except for the Ala28—Lys29 and Ile30—Lys33 peptides, all peptides were recovered stoichiometrically during peptide mapping. Comparison of the masses of peptides from monobiotinylated ubiquitin and from unmodified ubiquitin showed that, upon biotinylation, only the mass of the Met1—Lys11 peptide increased by the mass of the biotin adduct (227 atomic mass units) (Table I). Thus, the biotin in monobiotinylated ubiquitin is exclusively located between Met1 and Lys11. One of the biotins in both dibiotinylated and tribiotinylated ubiquitins is also located within this fragment. Edman degradation analysis of this fragment revealed that only Lys6 was labeled and that Met1 and Lys11 were free of modification. The other biotin in dibiotinylated ubiquitin that eluted at 26 min was located in the Gln49—Lys63 fragment (Table I). Thus, this dibiotinylated ubiquitin was labeled at Lys6 and Lys63.
TABLE I.
Biotinylation sites of biotin-labeled ubiquitin as determined by peptide mass mapping after endopeptidase Lys-C digestion. Biotin-labeled ubiquitin was purified by reversed-phase HPLC and digested by endopeptidase Lys-C. The resulting peptides were separated by reversed-phase HPLC, and their masses were determined using an in-line coupled mass spectrometer. In monobiotinylated ubiquitin ((Bio)1Ub), the biotin was exclusively attached to residues between Met1 and Lys11. In dibiotinylated ubiquitin ((Bio)2Ub), one biotin was located between Met1 and Lys11, and the other biotin was located between Glu34 and Lys63 (peak eluting at 24.5 min) or Gln49 and Lys63 (peak eluting at 26 min). In tribiotinylated ubiquitin ((Bio)3Ub), one biotin was located between Met1 and Lys11, and the other two biotins were located between Glu34 and Lys63. The mass for the biotin moiety is 227 atomic mass units. WT Ub, wild-type ubiquitin.
| WT Ub |
Peak at 23.5 min, (Bio)1Ub |
Peak at 24.5 min, (Bio)2Ub |
Peak at 26 min, (Bio)2Ub |
Peak at 27 min, (Bio)3Ub |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Masses | Positions | Masses | Positions | Masses | Positions | Masses | Positions | Masses | Positions |
| Da | Da | Da | Da | Da | |||||
| 764.4 | Met1-Lys6 | 1490.0 | Met1-Lys11 biotin | 1490.2 | Met1-Lys11 biotin | 1490.2 | Met1-Lys11 biotin | 1490.0 | Met1-Lys11 biotin |
| 518.3 | Thr7-Lys11 | ||||||||
| 1786.7 | Thr12-Lys27 | 1786.5 | Thr12-Lys27 | 1786.2 | Thr12-Lys27 | 1786.7 | Thr12-Lys27 | 1786.2 | Thr12-Lys27 |
| 1667.9 | Glu34-Lys48 | 1668.2 | Glu34-Lys48 | 3773.2 | Glu34-Lys63 biotin | 1668.0 | Glu34-Lys48 | 3999.5 | Glu34-Lys63 dibiotin |
| 1778.9 | Gln49-Lys63 | 1779.2 | Gln49-Lys63 | 2006.3 | Gln49-Lys63 biotin | ||||
| 1449.8 | Glu64-Gly76 | 1450.0 | Glu64-Gly76 | 1449.5 | Glu64-Gly76 | 1450.0 | Glu64-Gly76 | 1449.5 | Glu64-Gly76 |
One of the biotins in dibiotinylated ubiquitin that eluted at ∼24.5 min was located at Glu34—Lys63 (Table I). This dibiotinylated ubiquitin had a different retention time compared with Lys6/Lys63-dibiotinylated ubiquitin, suggesting that it was labeled at Lys6 and Lys48. To definitively determine this labeling site, this dibiotinylated ubiquitin was digested with trypsin, which cleaves peptide bonds after Lys and Arg residues. Peptide mapping showed that one biotin was located in the Met1—Lys11 fragment and that the other was located at the Leu43—Arg54 fragment (Table II), confirming that this ubiquitin was labeled at Lys6 and Lys48.
TABLE II.
Biotinylation sites of dibiotin-labeled ubiquitin determined by peptide mass mapping after trypsin digestion. Unmodified and dibiotinylated ((Bio)2Ub; peak eluting at 24.5 min) ubiquitins were digested by trypsin. The resulting peptides were separated by reversed-phase HPLC, and their masses were determined using an in-line coupled mass spectrometer. One biotin was located between Met1 and Lys11, and the other biotin of this dibiotinylated ubiquitin was located between Leu43 and Arg54. WT Ub, wild-type ubiquitin.
| WT Ub |
Peak at 24.5 min, (Bio)2Ub |
||
|---|---|---|---|
| Masses | Positions | Masses | Positions |
| Da | Da | ||
| 764.2 | Met1-Lys6 | 1490.0 | Met1-Lys11 biotin |
| 518.2 | Thr7-Lys11 | ||
| 1786.7 | Thr12-Lys27 | 1786.5 | Thr12-Lys27 |
| 1038 | Glu34-Arg42 | 1038.2 | Glu34-Arg42 |
| 643.4 | Leu43-Lys48 | 1572.2 | Leu43-Arg54 biotin |
| 716.8 | Gln49-Arg54 | ||
| 1080.3 | Thr55-Lys63 | 1080.5 | Thr55-Lys63 |
| 1066.4 | Glu64-Arg72 | 1066.7 | Glu64-Arg72 |
Peptides containing Lys29 and Lys33 were not detected by peptide mapping because of poor retention of these small fragments on the HPLC column. To determine whether these residues were labeled with biotin, biotinylated ubiquitins were sequenced by Edman degradation for 34 cycles, which revealed that ∼10% of the dibiotinylated ubiquitin in the peak that eluted at ∼24.5 min was labeled at Lys6 and Lys33. No modification at Lys11, Lys27, or Lys29 was observed unless the Sulfo-NHS-biotin/ubiquitin ratio was >7 (data not shown). Peptide mapping and Edman degradation analysis showed that biotins were at Lys6, Lys48, and Lys63 in tribiotinylated ubiquitin (Table I). Taken together, these data indicate that the order of susceptibility of amino groups in ubiquitin to modification by sulfo-NHS-biotin is Lys6 >> Lys63 > Lys48 >> Lys33 >> Lys11, Lys27, and Lys29.
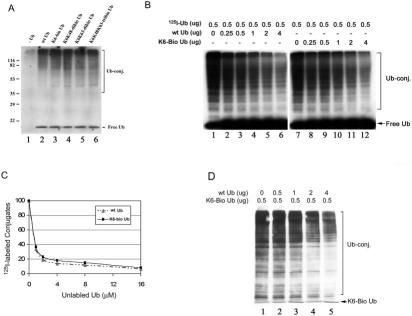
Biotinylated Ubiquitin Is Efficiently Used by the Ubiquitin Conjugation System to Form Ubiquitin Conjugates de Novo—The differentially biotinylated ubiquitins were isolated by HPLC, and their abilities to form ubiquitin conjugates were tested using reticulocyte fraction II. Fraction II is largely free of ubiquitin, but contains E1 and most E2 and E3 enzymes. Therefore, it allowed us to evaluate the utilization of biotinylated ubiquitin by the endogenous cellular ubiquitin conjugation machinery rather than by a single combination of E1, E2, and E3. To determine the relative extent of ubiquitination of endogenous substrates with biotinylated ubiquitin, 2 μg of the differentially biotinylated ubiquitins was added to each 25-μl assay, and the levels of ubiquitin conjugates were determined by Western blotting. A dramatic increase in high molecular mass ubiquitin conjugates was observed when wild-type ubiquitin was added to the assay (Fig. 2A, compare lanes 1 and 2), confirming that fraction II is largely devoid of ubiquitin. Addition of biotinylated ubiquitin to the assay resulted in a comparable increase in ubiquitin conjugates (Fig. 2A, compare lane 2 with lanes 3—6), indicating that these biotinylated ubiquitins were efficiently incorporated into high molecular mass ubiquitin conjugates. Because it is thought that Lys48 and Lys63 are usually used to form the inter-ubiquitin linkages that allow for polyubiquitin chain assembly, it was surprising to find that even when Lys6 and Lys48 or Lys6 and Lys63 were blocked (Lys6/Lys48- and Lys6/Lys63-dibiotinylated ubiquitins, respectively), these biotinylated ubiquitins were capable of forming high molecular mass ubiquitin conjugates in this system. These data indicate that, in addition to Lys48 and Lys63, other Lys residues in ubiquitin are capable of forming high molecular mass conjugates, particularly when Lys48 or Lys63 is not available. The specific linkage of polyubiquitin chains may also be substrate-dependent because both substrate specificity and polyubiquitin chain linkage are governed by combinations of specific E2 and E3 enzymes (45). As shown in Fig. 3A, Lys6/Lys48-dibiotinylated ubiquitin could not form high molecular mass conjugates with transducin-α. Under the steady-state conditions used in this assay, the levels of ubiquitin conjugates reflect the balance between conjugate formation and degradation/deconjugation. Therefore, although the levels of ubiquitin conjugates appear to be very similar for each of the modified ubiquitins, the rates of conjugation and degradation/deconjugation may vary for the differentially modified ubiquitins.
FIG. 2.
Biotin-labeled ubiquitin is efficiently used by the ubiquitin conjugation system to form ubiquitin conjugates. A, 2 μg of wild-type ubiquitin (wt Ub) or purified Lys6-biotinylated ubiquitin (K6-Bio Ub) was added to fraction II of rabbit reticulocytes in the presence of 2 mM ATP in a 25-μl assay. After incubation at 37 °C for 30 min, the reaction was terminated by addition of 25 μl of 2× SDS gel loading buffer. The mixture was resolved by SDS-PAGE and transferred to a nitrocellulose membrane. The levels of ubiquitin conjugates (Ub-conj.) were detected by Western blot analysis using anti-ubiquitin antibodies. K6K48-dibio Ub, Lys6/Lys48-dibiotinylated ubiquitin; K6K63-dibio Ub, Lys6/Lys63-dibiotinylated ubiquitin; K6K48K63-tribio Ub, Lys6/Lys48/Lys63-tribiotinylated ubiquitin. B, 0—4 μg of unmodified ubiquitin (lanes 1—6) or Lys6-biotinylated ubiquitin (lanes 7—12) was added to the conjugation assays, which contained 0.5 μgof 125I-labeled wild-type ubiquitin. Ubiquitin conjugation was performed as described for A, and ubiquitin conjugates were monitored by autoradiography. C, the results in B were quantitated by densitometry. D, 0—4 μg of wild-type ubiquitin was added to the conjugation assays, which contained 0.5 μg of Lys6-biotinylated ubiquitin. Biotinylated ubiquitin conjugates were determined with horseradish peroxidase-conjugated avidin using a chemiluminescence detection kit.
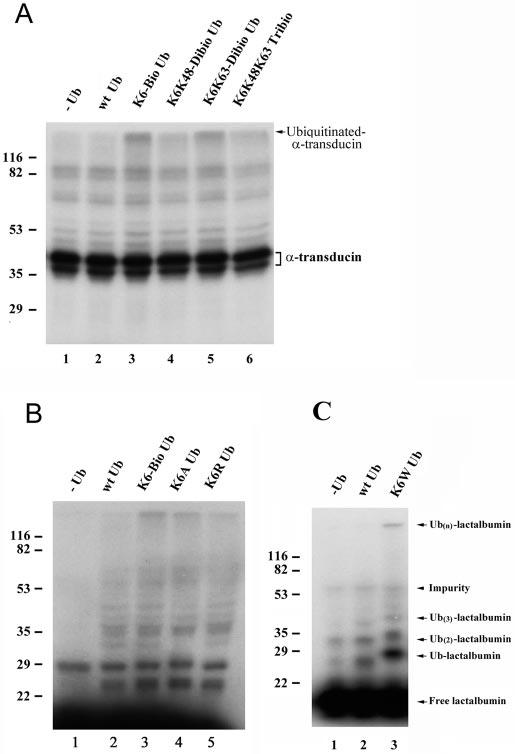
FIG. 3.
Lys6-modified ubiquitin causes accumulation of high molecular mass ubiquitin conjugates. Transducin (A) and α-lactalbumin (B and C) were labeled with 125I and used as substrates for conjugation assays in the RPE cell supernatant. Each 25-μl assay contained 2 μg of native or modified ubiquitin (Ub) as indicated. The reaction mixture was resolved by SDS-PAGE, and the levels of ubiquitinated substrates were determined by autoradiography. In C, K6W mutant ubiquitin was His6-tagged; therefore, the masses of the mono-, di, and triubiquitinated α-lactalbumin conjugates formed with this mutant ubiquitin were a little higher than those formed with wild-type ubiquitin (wt Ub). K6-Bio Ub, Lys6-biotinylated ubiquitin; K6K48-Dibio Ub, Lys6/Lys48-dibiotinylated ubiquitin; K6K63-Dibio Ub, Lys6/Lys63-dibiotinylated ubiquitin; K6K48K63 Tribio, Lys6/Lys48/Lys63-tribiotinylated ubiquitin.
To further determine whether Lys6-biotinylated ubiquitin was used as efficiently as unmodified ubiquitin by ubiquitin conjugation enzymes, we determined the abilities of Lys6-biotinylated ubiquitin to compete with 125I-labeled wild-type ubiquitin in the formation of high molecular mass ubiquitin conjugates. Fig. 2B (compare lane 1 with lanes 2—6 and lane 7 with lanes 8—12) shows that unmodified and Lys6-biotinylated ubiquitins were capable of competing with 125I-labeled ubiquitin as indicated by the progressively reduced levels of 125I-labeled ubiquitin conjugates in the presence of increasing levels of wild-type or Lys6-modified ubiquitin, respectively. Lys6-biotinylated ubiquitin competed with 125I-labeled ubiquitin almost as efficiently as unmodified ubiquitin (Fig. 2, B, compare lanes 1—6 with lanes 7—12; and C). In a reciprocal experiment, we determined the competition of wild-type ubiquitin with Lys6-biotinylated ubiquitin (Fig. 2D). As shown by the progressive decrease in the levels of biotinylated ubiquitin conjugates, which were detected by horseradish peroxidase-conjugated avidin, wild-type ubiquitin competed with Lys6-biotinylated ubiquitin in a dose-dependent manner. These data demonstrate that unmodified and Lys6-biotinylated ubiquitins are equally used by the ubiquitin conjugation system.
Lys6-modified Ubiquitin Inhibits ATP-dependent Degradation—To further characterize the ability of biotinylated ubiquitin to support ATP-dependent degradation, proteolysis of exogenous 125I-labeled substrates was tested in RPE cell supernatants. These RPE cell preparations have limited amounts of free ubiquitin (39). Endogenous ubiquitin supported modest ATP-dependent degradation (22%) (Table III). Addition of wild-type ubiquitin to the RPE cell supernatant increased the ATP-dependent degradation by ∼100% (Table III). In contrast, addition of Lys6-biotinylated ubiquitin had the opposite effect, inhibiting the ATP-dependent degradation by ∼45%. The dominant-negative effect of biotinylated ubiquitin appears not to be due to steric effects because K6A mutant ubiquitin, which has no steric bulk, inhibited degradation to a similar extent (Table III). Similarly, K6W mutant ubiquitin also inhibited ATP-dependent degradation. Lys6/Lys48- and Lys6/Lys63-dibiotinylated ubiquitins and Lys6/Lys48/Lys63-tribiotinylated ubiquitin also inhibited proteolysis in the RPE cell supernatant (Table III).
TABLE III.
Lys6-modified ubiquitin inhibits ATP-dependent degradation of transducin in the RPE cell lysate. The ability of differentially labeled ubiquitin (Ub) and K6A or K6W mutant ubiquitin to support ATP-dependent degradation was determined in supernatants of RPE cells using 125I-labeled transducin as substrate. The final concentration of the added ubiquitin was 8 μM in a 25-μl assay. The degradation rates are expressed as percentages of 125I-labeled substrate degraded into acid-soluble fragments in a 2-h period. WT Ub, wild-type ubiquitin.
| ATP-dependent degradation | Stimulation | |
|---|---|---|
| % | % | |
| Without addition of Ub | 22 ± 2.2 | 0 |
| WT Ub | 44 ± 3.4 | 100 |
| Lys6-biotinylated Ub | 12 ± 1.9 | -45 |
| K6A Ub | 13 ± 1.2 | -41 |
| K6W Ub | 10 ± 2.5 | -54 |
| Lys6/Lys48-dibiotinylated Ub | 18 ± 1.1 | -18 |
| Lys6/Lys63-dibiotinylated Ub | 19 ± 1.8 | -14 |
The dominant-negative effect of Lys6-modified ubiquitin was not limited to RPE cells. Addition of Lys6-biotinylated or K6W mutant ubiquitin to rabbit reticulocyte lysate, which has sufficient endogenous ubiquitin, also inhibited the ATP-dependent degradation of αA-crystallin by 60—70%, whereas addition of wild-type ubiquitin had little effect (Table IV).
TABLE IV.
K6W mutant ubiquitin inhibits ATP-dependent degradation of α-crystallin in reticulocyte lysate. The effects of wild-type (WT), Lys6-biotinylated, or K6W or L8A mutant ubiquitin (Ub) on ATP-dependent degradation of αA-crystallin were determined in supernatants of rabbit reticulocyte lysate. The final concentration of the added ubiquitin was 8 μM in a 25-μl assay. The degradation rates are expressed as percentages of 125I-labeled αA-crystallin degraded into acid-soluble fragments in a 90-min period.
| ATP-dependentdegradation | Stimulation | |
|---|---|---|
| % | % | |
| Without addition of Ub | 13.4 ± 2.5 | 0 |
| WT Ub | 12.9 ± 3.1 | -3 |
| Lys6-biotinylated Ub | 5.4 ± 2.1 | -60 |
| K6W Ub | 4.1 ± 1.8 | -69 |
| L8A Ub | 7.7 ± 1.4 | -42 |
Lys6-modified Ubiquitin Causes Accumulation of High Molecular Mass Ubiquitin Conjugates—To determine the mechanism of the inhibitory effect of Lys6-modified ubiquitin on ATP-dependent degradation, we studied the stability of ubiquitinated substrates. Transducin and α-lactalbumin were labeled with 125I and used as substrates for conjugation assays in the RPE cell supernatant. Fig. 3A (compare lanes 1 and 2) shows that a slight increase in the levels of ubiquitin conjugates was observed when additional wild-type ubiquitin was included in the assay. When equivalent amounts of Lys6-modified ubiquitin were used in the conjugation assays, significant amounts of high molecular mass ubiquitin conjugates were observed (Fig. 3A, compare lanes 2 and 3). Similar results were obtained with Lys6/Lys63-dibiotinylated ubiquitin as with Lys6-monobiotinylated ubiquitin (Fig. 3A, compare lane 2 with lanes 3 and 5). In contrast, the level of high molecular mass conjugates of transducin was much lower when Lys6/Lys48-dibiotinylated or Lys6/Lys48/Lys63-tribiotinylated ubiquitin was used (Fig. 3A, lanes 4 and 6), indicating that high molecular mass conjugates of transducin are linked mainly via the Lys48 isopeptide bond. The higher levels of ubiquitin conjugates observed in assays containing Lys6-monobiotinylated and Lys6/Lys63-dibiotinylated ubiquitins may result from enhanced conjugate formation or diminished degradation/deconjugation of these conjugates. However, the latter is more likely because Lys6-modified ubiquitin competed with 125I-labeled ubiquitin similarly compared with unmodified ubiquitin for conjugate formation (Fig. 2, B and C), but it inhibited proteolysis (Tables III and IV). The increased levels of high molecular mass ubiquitin conjugates in these assays suggest that Lys6 is required for degradation and/or deconjugation of ubiquitin conjugates and that blocking Lys6 by biotinylation diminishes the degradation or deconjugation process. This hypothesis is supported by the observation that an increase in high molecular mass conjugates was also detected when K6A, K6R, or K6W mutant ubiquitin was used in place of Lys6-modified ubiquitin (Fig. 3, B, compare lane 2 with lanes 3—5; and C, compare lanes 2 and 3).
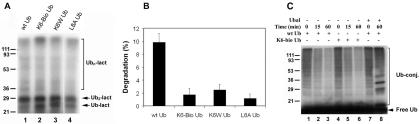
Conjugates Formed with Lys6-modified Ubiquitin Are Resistant to Degradation by the Proteasome, but Are Not Resistant to Isopeptidases—The data in Figs. 2 and 3 and Tables III and IV suggest that, although Lys6-modified ubiquitin is conjugation-competent, it does not support degradation, perhaps due to an inability to degrade conjugates formed with modified or mutant ubiquitin. To directly test the susceptibility of conjugates formed with Lys6-modified ubiquitin to proteasomal degradation, 125I-labeled α-lactalbumin was ubiquitinated in protea-some-free fraction II of rabbit reticulocyte using wild-type, Lys6-biotinylated, or mutant ubiquitin. The conjugates were then separated from non-ubiquitinated 125I-labeled α-lactalbumin by ion-exchange chromatography and subjected to degradation by the reconstituted 26 S proteasome. The profiles of the isolated conjugates formed with each modified or mutant ubiquitin are shown in Fig. 4.A Although the profiles of conjugates formed with Lys6-modified or mutant ubiquitin were not identical to those formed with wild-type ubiquitin, they were similar. The rates of proteasome-dependent degradation of conjugates formed with Lys6-modified or mutant ubiquitin were ∼80% lower than those formed with wild-type ubiquitin (Fig. 4B). It has been reported that mutation of the hydrophobic patch (Leu8, Ile44, and Val70) also results in accumulation of ubiquitin conjugates due to resistance to proteasomal degradation (46, 47). We compared the susceptibilities of conjugates formed with Lys6-modified ubiquitin with those formed with L8A mutant ubiquitin. The data show that conjugates formed with L8A mutant ubiquitin were even less susceptible to proteasomal degradation than those formed with Lys6-modified or K6W mutant ubiquitin. The proteasome-dependent degradation of conjugates formed with L8A mutant ubiquitin was only ∼10% of those formed with wild-type ubiquitin (Fig. 5B). Although conjugates formed with L8A mutant ubiquitin were more resistant to proteasomal degradation than those formed with Lys6-modified ubiquitin (Fig. 5B), L8A mutant ubiquitin was less potent than Lys6-biotinylated or K6W mutant ubiquitin in inhibiting ATP-dependent degradation of α-crystallin in rabbit reticulocyte lysate (Table IV). This may be due to reduced incorporation of L8A mutant ubiquitin into polyubiquitin conjugates in this system.
FIG. 4.
Conjugates formed with Lys6-modified ubiquitin are relatively resistant to proteasomal degradation, but are not resistant to isopeptidases. Ubiquitin conjugates were formed with wild-type, Lys6-biotinylated, or K6W or L8A mutant ubiquitin in proteasome-free fraction II of reticulocytes using 125I-labeled α-lactalbumin as substrate. The ubiquitinated 125I-labeled α-lactalbumin was separated from non-ubiquitinated 125I-labeled α-lactalbumin by ion-exchange chromatography, and its susceptibility to purified proteasome was determined. A, profiles of ubiquitinated α-lactalbumin (Ub-lact) formed with ubiquitin variants. Lane 1, wild-type ubiquitin (wt); lane 2, Lys6-biotinylated ubiquitin (K6-Bio); lane 3, K6W mutant ubiquitin; and lane 4, L8A mutant ubiquitin. B, proteasomal degradation of ubiquitinated 125I-labeled α-lactalbumin in the RPE cell supernatant. Ubiquitin conjugates of 125I-labeled α-lactalbumin were incubated with the reconstituted 26 S proteasome (composed of 0.25 μg of 20 S and 0.5 μg of 19 S) in the presence of ATP. Degradation rates were determined as the percentage of the substrates that became trichloroacetic acid-soluble after incubation with proteasome. wt Ub, wild-type ubiquitin; K6-Bio Ub, Lys6-biotinylated ubiquitin; K6W Ub, K6W mutant ubiquitin; L8A Ub, L8A mutant ubiquitin. C, deubiquitination assay. Wild-type (lanes 1—3) or Lys6-modified (lanes 4—6) ubiquitin was labeled with 125I, and ubiquitin conjugates (Ub-conj.) were formed in proteasome-free fraction II of rabbit reticulocyte. Deubiquitination by isopeptidases of the 125I-labeled ubiquitin conjugates was determined in the presence of a 20-fold excess of unlabeled wild-type ubiquitin conjugates. In lanes 7 and 8, ubiquitin aldehyde (Ubal) was added to inhibit isopeptidases.
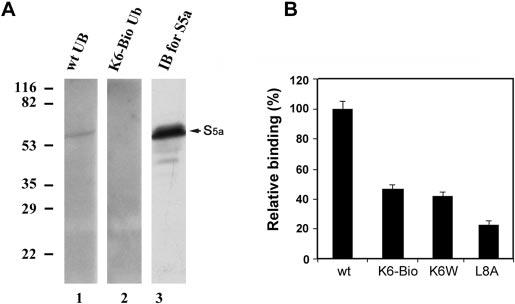
FIG. 5.
Conjugates formed with Lys6-modified ubiquitin bind S5a of the proteasome with reduced avidity. Ubiquitin conjugates of α-lactalbumin were formed with wild-type, Lys6-biotinylated, or K6W or L8A mutant ubiquitin and isolated as described in the legend to Fig. 4. A, proteasome isolated from rabbit reticulocytes was resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes. After denaturation and renaturation procedures, the membranes were incubated overnight with conjugates of 125I-labeled α-lactalbumin formed with wild-type or Lys6-biotinylated ubiquitin. After removal of nonspecifically bound radioactivity, the membranes were exposed to x-ray film, and the specific binding was visualized by autoradiography. Lane 1, conjugates formed with wild-type ubiquitin (wt UB); lane 2, conjugates formed with Lys6-biotinylated ubiquitin (K6-Bio Ub); lane 3, Western blot analysis (immunoblotting (IB)) of the 26 S proteasome using antibody to S5a. B, shown are the results from S5a binding assay. Equal amounts of ubiquitinated 125I-labeled α-lactalbumin (4 × 104 cpm) formed with wild-type, Lys6-biotinylated, or K6W or L8A mutant ubiquitin were incubated with GST-S5a, which was immobilized overnight on glutathione-agarose beads with constant shaking. After extensive washing, ubiquitin conjugates were eluted from the beads with glutathione and quantified using a γ-counter. The data presented are the relative binding capabilities, where the binding capability of conjugates formed with wild-type ubiquitin is designated as 100%.
To determine whether altered susceptibility to isopeptidases also plays a role in the accumulation of conjugates formed with Lys6-modified ubiquitin, we determined the stability of these conjugates in proteasome-free fraction II. Wild-type and Lys6-biotinylated ubiquitins were labeled with 125I, and ubiquitin conjugates were formed in proteasome-free fraction II. The stability of the 125I-labeled ubiquitin conjugates was determined in the presence of a 20-fold excess of unlabeled wild-type ubiquitin. As shown in Fig. 4C, the levels of conjugates formed with wild-type ubiquitin (lanes 1—3) or Lys6-modified ubiquitin (lanes 4—6) decreased rapidly during the chase period. There were no significant differences in the rates of deubiquitination (Fig. 4C, compare lanes 1—3 with lanes 4—6). If isopeptidases were inhibited by ubiquitin aldehyde, ubiquitin conjugates were stable during the chase period (Fig. 4C, lanes 7 and 8). In addition, several low molecular mass ubiquitin conjugates were observed in the presence of ubiquitin aldehyde (Fig. 4C, lane 8). These data indicate that conjugates formed with Lys6-modified ubiquitin are not resistant to deubiquitination by isopeptidases.
Conjugates Formed with Lys6-modified Ubiquitin Bind the 26 S Proteasome with Reduced Avidity—To determine the molecular mechanisms that underlie the resistance to proteasomal degradation of conjugates formed with Lys6-modified ubiquitin, we determined the capability of these conjugates to interact with the proteasome. To do this, ubiquitinated 125I-labeled α-lactalbumin was formed and isolated as described above. Proteasome from rabbit reticulocyte was resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes. After denaturation and renaturation procedures (19, 20), the membranes were probed with the respective ubiquitin conjugates of 125I-labeled α-lactalbumin. Conjugates formed with wild-type ubiquitin bound a proteasome subunit with an apparent molecular mass of 53 kDa on the membrane (Fig. 5A, lane 1). Binding to this or any other proteasome subunit was not observed when Lys6-biotinylated ubiquitin was used to form the conjugates (Fig. 5B, lane 2). The 53-kDa proteasome subunit that bound the ubiquitin conjugates was recognized by antibody to S5a (Fig. 5A, lane 3). Because only S5a bound ubiquitin conjugates under these conditions, we further quantitatively compared the binding to recombinant S5a. Fig. 5B shows that conjugates formed with Lys6-biotinylated ubiquitin bound immobilized S5a ∼80% less than those formed with wild-type ubiquitin. Conjugates formed with K6W mutant ubiquitin also bound S5a substantially less than those formed with wild-type ubiquitin (Fig. 5B). We also compared the proteasome-binding capabilities of conjugates formed with Lys6-modified ubiquitins with those of conjugates formed with L8A mutant ubiquitin, a ubiquitin mutant demonstrated to impair proteasome binding (46-48). As shown in Fig. 5B, conjugates formed with Lys6-modified ubiquitin bound S5a a little stronger than those formed with L8A mutant ubiquitin.
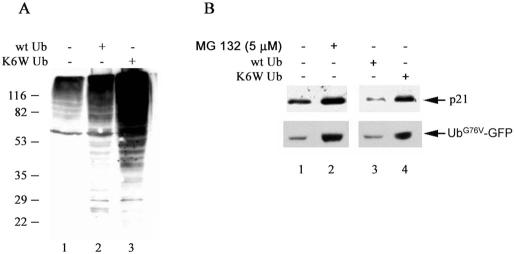
Intracellular Expression of Lys6-modified Ubiquitin Results in Accumulation of Ubiquitin Conjugates, Stabilization of Substrates for the UPP, and Enhanced Susceptibility to Oxidative Stress—As described above, Lys6-modified ubiquitin stabilized conjugates and inhibited ATP-dependent degradation in a cell-free system. To determine whether Lys6-modified ubiquitin also inhibits the UPP in intact cells, wild-type and K6W mutant ubiquitins were expressed in HEK293 cells. The levels of endogenous ubiquitin-protein conjugates (Fig. 6A) and the levels of a known UPP substrate, p21WAF1, were compared in cells transfected with wild-type or K6W mutant ubiquitin (Fig. 6B). Expression of wild-type ubiquitin increased the levels of endogenous ubiquitin conjugates by up to 2-fold (Fig. 6A, compare lanes 1 and 2). The levels of p21WAF1 decreased slightly under these conditions (Fig. 6B, compare lanes 1 and 3). In contrast, expression of K6W mutant ubiquitin increased the levels of high molecular mass ubiquitin conjugates by ∼10-fold (Fig. 6A, compare lanes 1 and 3). The difference in the levels of high molecular mass conjugates between wild-type and K6W mutant ubiquitins was not due to different levels of expression because the levels of free ubiquitin were comparable (data not shown). Expression of K6W mutant ubiquitin increased the levels of p21WAF1 by >2-fold (Fig. 6B, compare lanes 1 and 4). Similar accumulation of p21WAF1 was observed when the cells were incubated with a proteasomal inhibitor (MG132) (Fig. 6B, compare lanes 1 and 2). Likewise, overexpression of K6W mutant ubiquitin in HeLa cells resulted in stabilization of G76V mutant ubiquitin (UbG76V)-GFP (Fig. 6B, compare lanes 3 and 4), another known substrate of the UPP (49). Taken together, these data demonstrate that Lys6-modified ubiquitin is a potent and specific inhibitor for the UPP in intact cells.
FIG. 6.
Expression of Lys6-modified ubiquitin results in accumulation of intracellular high molecular mass ubiquitin conjugates and stabilization of p21WAF1. HEK293 cells were transfected with plasmids encoding wild-type or K6W mutant ubiquitin. Forty-eight h after transfection, the cells were collected. The levels of endogenous ubiquitin conjugates and p21WAF1 were determined by Western blotting. To study the effects of expression of K6W mutant ubiquitin on the stability of UbG76V-GFP, HeLa cells were cotransfected with plasmid encoding UbG76V-GFP and plasmid encoding wild-type or K6W mutant ubiquitin. The levels of UbG76V-GFP in the cells were determined at 40 h after transfection. For a positive control, HeLa cells transfected with plasmid encoding UbG76V-GFP were treated with or without MG132 for 8 h and collected to determine the levels of UbG76VGFP in the cells. A, levels of ubiquitin conjugates. Lane 1, cells transfected with empty vector; lane 2, cells transfected with plasmid encoding wild-type ubiquitin (wt Ub); lane 3, cells transfected with plasmid encoding K6W mutant ubiquitin (K6W Ub). B, levels of p21WAF1 and UbG76V-GFP. Lane 1, control cells; lane 2, cells treated with MG132; lane 3, cells transfected with wild-type ubiquitin; lane 4, cells transfected with K6W mutant ubiquitin.
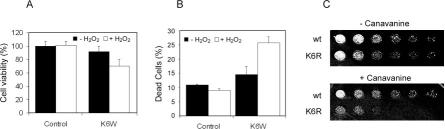
We showed previously that the UPP is involved in the response to oxidative stress and the removal of oxidized proteins (3, 4, 37, 50, 51). In this work, we determined the effect of K6W mutant ubiquitin on the ability of cells to cope with oxidative stress. Cells infected with control adenovirus showed limited toxicity as indicated by ∼10% cell death. Exposure of these cells to 20 μM H2O2 for 8 h did not significantly alter cells viability or the percentage of dead cells (Fig. 7, A and B), indicating that these cells could withstand this modest level of oxidative stress and that infection with control adenovirus had no effect on the susceptibility of these cells to oxidative stress. In contrast, cells infected with the same amount of K6W mutant ubiquitin-encoding adenovirus resulted in a slight decrease in cell viability compared with cells infected with control adenovirus (Fig. 7, A and B). The cytotoxicity of K6W mutant ubiquitin may be associated with the inhibition of ubiquitin-dependent proteolysis because proteasomal inhibition also resulted in cytotoxicity in these cells (data not shown). Furthermore, exposure of the cells expressing K6W mutant ubiquitin to 20 μM H2O2 for 8 h dramatically decreased cell viability and increased cell death by ∼80% as compared with those not treated with H2O2 (Fig. 7, B). Likewise, yeast cells expressing K6R mutant ubiquitin as the sole source of ubiquitin grew similarly to yeast cells expressing wild-type ubiquitin under normal conditions. However, in the presence of canavanine, yeast cells expressing K6R mutant ubiquitin grew significantly slower than yeast cells expressing wild-type ubiquitin (Fig. 7C), although wild-type and K6R mutant ubiquitins were expressed at similar levels (data not shown). The enhanced susceptibility of Lys6 mutant ubiquitin-expressing cells to stresses may be due to impaired proteasomal degradation of damaged proteins or canavanine-containing proteins. It has been shown that some oxidized and canavanine-containing proteins are degraded by the UPP. However, the effects of expression of Lys6 mutant ubiquitin on cell viability may also be related to the abrogation of Lys6-linked ubiquitin chains (52).
FIG. 7.
Expression of Lys6 mutant ubiquitin increases the susceptibility of cells to stress. Human lens epithelial cells at ∼50% confluence were infected with recombinant adenovirus encoding K6W mutant ubiquitin or with control adenovirus for 48 h. The cells were then exposed to ∼20 μM H2O2 constantly generated by addition of glucose oxidase to the medium for 8 h. A, cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium bromide assay. B, the percentage of dead cells that were stained by trypan blue was determined. The data represent the means ± S.D. of six measurements. C, yeast cells expressing wild-type (wt) or K6R mutant ubiquitin were cultured in the absence or presence of 1 μM canavanine.
DISCUSSION
The major observations of this work include the following. 1) Lys6 was the most readily chemically modified of the 7 Lys residues in human ubiquitin. 2) Lys6-modified ubiquitin was incorporated into ubiquitin conjugates as efficiently as wild-type ubiquitin. 3) Mutation or chemical modification of Lys6 of ubiquitin reduced its ability to stimulate ATP-dependent protein degradation. 4) Conjugates formed with Lys6-modified ubiquitin were resistant to degradation by the proteasome, as the conjugates formed with Lys6-modified ubiquitin bound to the proteasome with reduced avidity. 5) Expression of K6W mutant ubiquitin in mammalian cells also inhibited ubiquitin-dependent proteolysis. 6) Impairment of ubiquitin function by modification or mutation of Lys6 compromised cell viability and survival, particularly under stress conditions.
The results show that Lys6-modified ubiquitin is a potent and specific inhibitor of the UPP. Lys6-modified ubiquitin in cell-free assays significantly and specifically inhibited ATP-dependent proteolysis (Table III). Overexpression of K6W mutant ubiquitin in cultured cells resulted in accumulation of high molecular mass ubiquitin conjugates and stabilization of p21WAF1 and UbG76V-GFP (Fig. 6), typical substrates for the ubiquitin-proteasome pathway. In human lens epithelial cells, expression of K6W mutant ubiquitin enhanced the susceptibility of cells to oxidative stress (Fig. 7). The modest cytotoxicity of K6W mutant ubiquitin under non-stress conditions may be due to the insufficient expression of the mutant ubiquitin, which only partially inhibited the UPP. Under these experimental conditions, only ∼60% of the cells expressed the mutant ubiquitin. It is likely that only the cells expressing high levels of the mutant ubiquitin showed cytotoxicity.
The mechanism of the inhibitory effects of Lys6-modified ubiquitin appears to be related to the attenuated avidity of the interaction between ubiquitin conjugates and the 26 S protea-some (Fig. 5). It was well established that the hydrophobic patch formed by Leu8, Ile44, and Val70 on the surface of ubiquitin is critical for docking polyubiquitinated substrates to the 26 S proteasome (46-48), and conjugates formed with L8A mutant ubiquitin were resistant to proteasomal degradation (Fig. 4B). Incorporation of Lys6-modified ubiquitin into polyubiquitin chains (formed with wild-type and Lys6-modified ubiquitins) may disrupt the arrangement of signals in polyubiquitin chains that are required for proteasomal recognition. This hypothesis is consistent with previous results showing that incorporation of one L8A mutant ubiquitin into a tetramer composed of three other wild-type ubiquitins dramatically reduced the binding affinity of the heterotetramer for the proteasome (46).
It is intriguing to discover that conjugates formed with L8A mutant ubiquitin were more resistant to proteasomal degradation than those formed with K6W mutant ubiquitin (Fig. 4B), but that L8A mutant ubiquitin was not as potent as K6W mutant ubiquitin in inhibiting ATP-dependent degradation in reticulocyte lysate (Table IV). Preliminary data indicate that L8A mutant ubiquitin was not used as efficiently as wild-type ubiquitin in fraction II of reticulocytes (data not shown). The hydrophobic patch formed by Leu8, Ile44, and Val70 on the ubiquitin surface that is recognized by the 19 S complex of the proteasome may also be recognized by other ubiquitin-binding proteins such as E1. It has been reported that this hydrophobic patch is also required for monoubiquitin-mediated processes such as endocytosis (53). Lys6 of ubiquitin is important for proteasome-dependent degradation, but it may not be essential for other ubiquitin-dependent processes. This may explain why mutations of residues related to the hydrophobic patch are lethal, whereas yeast cells expressing K6A mutant ubiquitin can survive under normal conditions (53).
The 26 S proteasome (rather than the 20 S proteasome) is involved in the degradation of ubiquitinated substrates. However, none of the commercially available proteasomal inhibitors, including MG132, lactacystin, and epoxomicin, allow distinction of degradation via the 26 S proteasome versus the 20 S proteasome (54). Thus, stabilization of a protein by these proteasomal inhibitors does not prove that the protein is degraded in the UPP because the 20 S proteasome degrades proteins in a ubiquitin-independent manner (54). Our demonstration that Lys6-modified ubiquitin is a potent and specific inhibitor of the UPP provides a useful reagent for studying the function of this pathway. Together with the ability to express K6W mutant ubiquitin at high levels in intact cells, Lys6-modified ubiquitin can be used to distinguish ubiquitin-dependent from ubiquitin-independent protein degradation. The accumulation of ubiquitin conjugates by Lys6-modified ubiquitin will also facilitate the isolation and identification of ubiquitinated proteins in the cells.
Our observation that, of the 7 Lys residues, Lys6 is the most readily modified with sulfo-NHS-biotin in the ubiquitin molecule (Tables I and II) is consistent with prior observations that Lys6 is the most readily modified by p-nitrophenyl acetate (28), aspirin (29), acetic anhydride (30), and Oregon green succinimidyl ester (31). Acetylation and biotinylation of Lys residues share the same mechanism as other nonenzymatic modifications such as nonenzymatic glycosylation (glycation). This result suggests that Lys6 of ubiquitin may also be more susceptible to glycation or modification by 4-hydroxy-2-nonenal, a lipid peroxidation product. Thus, it is reasonable to expect that other types of modifications of ubiquitin Lys6 will also interfere with proteasomal degradation and result in the accumulation of ubiquitin conjugates. This might explain why ubiquitin conjugates are accumulated in response to oxidative stress and upon aging (3, 37, 50, 55-58). Accumulation of ubiquitin conjugates in the brain is a characteristic of age-related diseases such as Alzheimer and Parkinson and cataracts (59, 60). Further studies regarding relationships between Lys6 modification and accumulation of ubiquitin per se or ubiquitin conjugates will reveal new roles for ubiquitin in health and disease.
Acknowledgments
We thank Dr. Rob Layfield for providing GSTS5a and Mark Siegal and Matthew Gallagher for help in the preparation of this manuscript.
Footnotes
This work was supported in part by National Institutes of Health Grants EY11717 (to F. S.), EY13250 (to A. T.), and GM34009 (to A. L. H.) and by the United States Department of Agriculture under Agreement 58-1950-9-001. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: UPP, ubiquitin/proteasome pathway; sulfo-NHS-biotin, sulfosuccinimidobiotin; GFP, green fluorescent protein; HPLC, high pressure liquid chromatography; RPE, retinal pigment epithelial; GST, glutathione S-transferase; E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; UbG76V, G76V mutant ubiquitin.
REFERENCES
- 1.Hershko A, Ciechanover A. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A, Orian A, Schwartz AL. J. Cell. Biochem. Suppl. 2000;34:40–51. doi: 10.1002/(sici)1097-4644(2000)77:34+<40::aid-jcb9>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 3.Shang F, Gong X, Taylor A. J. Biol. Chem. 1997;272:23086–23093. doi: 10.1074/jbc.272.37.23086. [DOI] [PubMed] [Google Scholar]
- 4.Shang F, Nowell TR, Jr., Taylor A. Exp. Eye Res. 2001;73:229–238. doi: 10.1006/exer.2001.1029. [DOI] [PubMed] [Google Scholar]
- 5.Haas A, Reback PM, Pratt G, Rechsteiner M. J. Biol. Chem. 1990;265:21664–21669. [PubMed] [Google Scholar]
- 6.Baboshina OV, Haas AL. J. Biol. Chem. 1996;271:2823–2831. doi: 10.1074/jbc.271.5.2823. [DOI] [PubMed] [Google Scholar]
- 7.Arnason T, Ellison MJ. Mol. Cell. Biol. 1994;14:7876–7883. doi: 10.1128/mcb.14.12.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spence J, Sadis S, Haas AL, Finley D. Mol. Cell. Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. Nature. 2001;412:346–351. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 10.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann RM, Pickart CM. J. Biol. Chem. 2001;276:27936–27943. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- 12.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 13.Gregori L, Poosch MS, Cousins G, Chau V. J. Biol. Chem. 1990;265:8354–8357. [PubMed] [Google Scholar]
- 14.Pickart CM. FASEB J. 1997;11:1055–1066. doi: 10.1096/fasebj.11.13.9367341. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman L, Pratt G, Rechsteiner M. J. Biol. Chem. 1992;267:22362–22368. [PubMed] [Google Scholar]
- 16.DeMartino GN, Moomaw CR, Zagnitko OP, Proske RJ, Chu-Ping M, Afendis SJ, Swaffield JC, Slaughter CA. J. Biol. Chem. 1994;269:20878–20884. [PubMed] [Google Scholar]
- 17.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 18.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 19.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. J. Biol. Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 20.van Nocker S, Deveraux Q, Rechsteiner M, Vierstra RD. Proc. Natl. Acad. Sci. U. S. A. 1996;93:856–860. doi: 10.1073/pnas.93.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Nat. Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 22.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. J. Biol. Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 23.Verma R, Oania R, Graumann J, Deshaies RJ. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Lam YA, Xu W, DeMartino GN, Cohen RE. Nature. 1997;385:737–740. doi: 10.1038/385737a0. [DOI] [PubMed] [Google Scholar]
- 25.Lam YA, DeMartino GN, Pickart CM, Cohen RE. J. Biol. Chem. 1997;272:28438–28446. doi: 10.1074/jbc.272.45.28438. [DOI] [PubMed] [Google Scholar]
- 26.Yao T, Cohen RE. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 27.Corsi D, Galluzzi L, Crinelli R, Magnani M. J. Biol. Chem. 1995;270:8928–8935. doi: 10.1074/jbc.270.15.8928. [DOI] [PubMed] [Google Scholar]
- 28.Jabusch JR, Deutsch HF. Arch. Biochem. Biophys. 1985;238:170–177. doi: 10.1016/0003-9861(85)90153-5. [DOI] [PubMed] [Google Scholar]
- 29.Macdonald JM, LeBlanc DA, Haas AL, London RE. Biochem. Pharmacol. 1999;57:1233–1244. doi: 10.1016/s0006-2952(99)00039-8. [DOI] [PubMed] [Google Scholar]
- 30.Macdonald JM, Haas AL, London RE. J. Biol. Chem. 2000;275:31908–31913. doi: 10.1074/jbc.M000684200. [DOI] [PubMed] [Google Scholar]
- 31.Wee KE, Lai Z, Auger KR, Ma J, Horiuchi KY, Dowling RL, Dougherty CS, Corman JI, Wynn R, Copeland RA. J. Protein Chem. 2000;19:489–498. doi: 10.1023/a:1026501515450. [DOI] [PubMed] [Google Scholar]
- 32.Vijay-Kumar S, Bugg CE, Cook WJ. J. Mol. Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 33.Vijay-Kumar S, Bugg CE, Wilkinson KD, Vierstra RD, Hatfield PM, Cook WJ. J. Biol. Chem. 1987;262:6396–6399. [PubMed] [Google Scholar]
- 34.Burch TJ, Haas AL. Biochemistry. 1994;33:7300–7308. doi: 10.1021/bi00189a035. [DOI] [PubMed] [Google Scholar]
- 35.Finch JS, Bonham K, Krieg P, Bowden GT. Nucleic Acids Res. 1990;18:1907–1907. doi: 10.1093/nar/18.7.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shang F, Taylor A. Biochem. J. 1995;307:297–303. doi: 10.1042/bj3070297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciechanover A, Elias S, Heller H, Hershko A. J. Biol. Chem. 1982;257:2537–2542. [PubMed] [Google Scholar]
- 39.Obin M, Nowell T, Taylor A. Curr. Eye Res. 1995;14:751–760. doi: 10.3109/02713689508995796. [DOI] [PubMed] [Google Scholar]
- 40.Shang F, Huang L, Taylor A. Curr. Eye Res. 1994;13:423–431. doi: 10.3109/02713689408999870. [DOI] [PubMed] [Google Scholar]
- 41.Huang LL, Jahngen-Hodge J, Taylor A. Biochim. Biophys. Acta. 1993;1175:181–187. doi: 10.1016/0167-4889(93)90021-g. [DOI] [PubMed] [Google Scholar]
- 42.Gaczynska M, Goldberg AL, Tanaka K, Hendil KB, Rock KR. J. Biol. Chem. 1996;271:17275–17280. doi: 10.1074/jbc.271.29.17275. [DOI] [PubMed] [Google Scholar]
- 43.Layfield R, Tooth D, Landon M, Dawson S, Mayer J, Alban A. Proteomics. 2001;1:773–777. doi: 10.1002/1615-9861(200106)1:6<773::AID-PROT773>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 44.Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. Mol. Cell. Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pickart CM, Fushman D. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Proc. Natl. Acad. Sci. U. S. A. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beal RE, Toscano-Cantaffa D, Young P, Rechsteiner M, Pickart CM. Biochemistry. 1998;37:2925–2934. doi: 10.1021/bi972514p. [DOI] [PubMed] [Google Scholar]
- 48.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 49.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Nat. Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 50.Shang F, Gong X, Palmer HJ, Nowell TR, Taylor A. Exp. Eye Res. 1997;64:21–30. doi: 10.1006/exer.1996.0176. [DOI] [PubMed] [Google Scholar]
- 51.Dudek EJ, Shang F, Taylor A. Free Radic. Biol. Med. 2001;31:651–658. doi: 10.1016/s0891-5849(01)00634-7. [DOI] [PubMed] [Google Scholar]
- 52.Morris JR, Solomon E. Hum. Mol. Genet. 2004;13:807–817. doi: 10.1093/hmg/ddh095. [DOI] [PubMed] [Google Scholar]
- 53.Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. J. Biol. Chem. 2001;276:30483–30489. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- 54.Grune T, Reinheckel T, Davies KJ. FASEB J. 1997;11:526–534. [PubMed] [Google Scholar]
- 55.Ivy GO, Kitani K, Ihara Y. Brain Res. 1989;498:360–365. doi: 10.1016/0006-8993(89)91117-7. [DOI] [PubMed] [Google Scholar]
- 56.Kudo T, Iqbal K, Ravid R, Swaab DF, Grundke-Iqbal I. Brain Res. 1994;639:1–7. doi: 10.1016/0006-8993(94)91757-4. [DOI] [PubMed] [Google Scholar]
- 57.Scrofano MM, Shang F, Nowell TR, Jr., Gong X, Smith DE, Kelliher M, Dunning J, Mura CV, Taylor A. Mech. Ageing Dev. 1998;105:273–290. doi: 10.1016/s0047-6374(98)00097-9. [DOI] [PubMed] [Google Scholar]
- 58.Scrofano MM, Shang F, Nowell TR, Jr., Gong X, Smith DE, Kelliher M, Dunning J, Mura CV, Taylor A. Mech. Ageing Dev. 1998;101:277–296. doi: 10.1016/s0047-6374(97)00178-4. [DOI] [PubMed] [Google Scholar]
- 59.Jahngen-Hodge J, Cyr D, Laxman E, Taylor A. Exp. Eye Res. 1992;55:897–902. doi: 10.1016/0014-4835(92)90016-l. [DOI] [PubMed] [Google Scholar]
- 60.Jahngen JH, Lipman RD, Eisenhauer DA, Jahngen EG, Taylor A. Arch. Biochem. Biophys. 1990;276:32–37. doi: 10.1016/0003-9861(90)90006-k. [DOI] [PubMed] [Google Scholar]