Abstract
Background
Genital infection by herpes simplex virus (HSV)–2 offers a unique model for study of the effects of a remitting and exacerbating infection on the survival and persistence of antigen-specific T cells.
Methods
We used complementarity-determining region 3 (CDR3) length analysis to examine the complete T cell receptor (TCR) β-chain repertoire in skin-lesion biopsy samples from subjects with genital herpes.
Results
We found that herpetic skin lesions consistently demonstrated oligoclonal CDR3 DNA length distribution, indicating the presence of T cell expansions. Sequence analysis of representative HSV-specific lesional CD4+ cell clones and TCR β-variable (TCRBV) sequencing confirmed that the oligoclonal expansions were largely related to HSV-specific T cell proliferation. To assess the persistence of HSV-specific CD4+ cells that localize to genital lesions, we developed a sensitive and highly specific clonal tracking technique using a combination of TCRBV-specific polymerase chain reaction, followed by liquid hybridization with clonotype-specific probes.
Conclusion
Two different patterns of clonal persistence were observed. Some long-lasting clones appear to home to different epithelia, such as skin and genital mucosa, and to circulate in the peripheral blood, whereas others detected in lesions were absent or very rare in the peripheral blood.
Herpes simplex virus (HSV)–2, the major cause of genital herpes, causes a chronic, intermittent disease characterized by both clinical and subclinical episodes of the viral infection of skin and mucosal surfaces [1]. Herpetic skin lesions are characterized by an early dermal mononuclear cell infiltrate. This infiltrate has been shown to contain predominantly CD4+ cells during the first few days, and these are joined subsequently by CD8+ cells [2]. HSV-specific CD4+ and CD8+ cell clones can be detected in cells isolated from skin-lesion biopsy samples or lesion vesicle fluid [3]. We have previously reported that the frequency of HSV-specific T lymphocytes in lesions is ~100–1000-fold more than that found in the peripheral blood mononuclear cells (PBMCs), which suggests that T cell infiltration is an active process that is driven by the presence of viral antigens [3].
Analysis of T cell repertoires has become an important tool for understanding the immune response to infectious pathogens. In humans, T cells that express the α or β T cell receptor (TCR) represent the majority of lesional skin-infiltrating lymphocytes [4]. In the TCR β-chain, the third hypervariable complementarity-determining region (CDR3) is known to interact with peptide bound to the major histocompatibility complex [5]. CDR3 is generated by somatic recombination of the TCR variable (V), diversity (D), and joining (J) gene segments and further N-region diversification. As a result of this somatic recombination process, the length of the TCR β-chain can vary between TCRs by up to 8 amino acids. Furthermore, the CDR3 sequence de-fines a unique TCR clonotype that acts as a molecular fingerprint. We used spectratyping to evaluate the T cell repertoire by determining the distribution of lengths of CDR3 [6]. Because spectratyping provides no information about the antigen specificity of the expanded clones, we developed a liquid hybridization technique to assess the persistence of HSV-specific lesion-infiltrating T cell clones with a clonotypic probe covering the CDR3 region.
SUBJECTS, MATERIALS, AND METHODS
Subjects and specimens
Subjects with recurrent genital HSV-2 infection and lesions were recruited for a study protocol that had been approved by the University of Washington institutional review board. Informed consent was obtained from subjects who were either HIV-negative or who were at low clinical risk for HIV infection. We studied 4 female subjects and 1 male subject who had culture- and serologically proven recurrent HSV-2 infection. Subjects were selected because the HSV-2 lesion occurred on the buttocks or thigh, and these sites are suitable for multiple biopsies. Lesions were sampled on day 2 and then every 3 days during the vesicular-ulcerative stage. To obtain tissue, the area was anesthetized, and 4-mm punch biopsies were performed in erythematous skin adjacent to vesicles, pustules, ulcers, or crusts. Serial biopsies were performed at slightly different locations within the recurrence. Cervical and blood specimens were processed as described elsewhere [7]. Human experimentation guidelines of the US Department of Health and Human Services and the University of Washington were followed.
Lymphocyte culture
Biopsies were processed as described elsewhere [3]. In brief, tissue was minced into 8–10 pieces and placed into 2 48-well plates, each with 1 mL of T cell medium supplemented with 0.8 μg/mL phytohemagglutinin (PHA)–P (Murex Diagnostics), 1 ×106 allogeneic irradiated (5000 rad γ-irradiation) PBMCs, 50 U/mL human natural interleukin-2 (Hemagen), and 50 μmol/L acyclovir. The resultant first-passage bulk cultures and matching PBMCs were further stimulated with either PHA or with UV-inactivated HSV-2. T cell clones were generated, screened, and propagated as described elsewhere [3].
Lymphocyte functional assays
Proliferation assays were performed as described for clones or for bulk cell lines as responders [3]. Washed cells (1 ×104 cells/well), autologous irradiated PBMCs (1 ×105 cells/well), and antigen were incubated in triplicate in 200 μL of T cell medium in 96-well U-bottom plates for 3 days, followed by overnight incubation with 0.5 μCi/well of [3H]-thymidine (Amersham). Antigens included UV-inactivated HSV-2 and purified recombinant glycoproteins gB2 and gD2 [3]. DNA synthesis was measured by liquid scintillation.
For cytotoxicity assays [3], target autologous Epstein-Barr virus (EBV)–lymphoblastoid cell line (LCL) were infected for 1 h with HSV-2 or mock virus at an MOI of 5 in serum-free RPMI 1640 at 37°C, followed by overnight incubation in T cell medium that included 100 μCi of 51Cr. LCLs were seeded in triplicate into 96-well U-bottom plates with bulk or cloned effector cells. Effector:target ratios were 20:1. After 4 h at 37°C, 30 μL of supernatant was counted by liquid scintillation. The percentage of specific release (in counts per minute [cpm]) was determined by the equation [(mean cpm experimental − mean cpm spontaneous)/(mean cpm total − mean cpm spontaneous)× 100].
Lymphocyte phenotype analysis
Bulk lesion-derived T cells were analyzed by flow cytometry [3], by use of monoclonal antibodies specific for CD4 and CD8 antigens (BD Pharmingen). Isotype controls were also included.
Viruses and cell lines
HSV-2 strain 333, which was used throughout, was raised in Vero cells, and cell-associated virus was released by sonication and titered on Vero cells, as described elsewhere [3]. EBV-transformed LCLs were established and maintained as described elsewhere [3].
RNA extraction and CDR3 size spectratyping
Total RNA from 5–10 ×106 cells was extracted as described elsewhere [8]. cDNA was synthesized by use of random hexamer primers, Maloney murine leukemia virus (MMLV) reverse transcriptase (Gibco Invitrogen), and dNTPs. An aliquot of cDNA was used for TCR βV (TCRBV)–specific polymerase chain reaction (PCR) amplification by use of a constant (as opposed to variable) β-chain–specific reverse primer, of which 4 pmol was labeled with 32P by use of T-4 kinase (Invitrogen), and an upstream βV–specific forward primer. Primer sequences have been published elsewhere [9]. Conditions for the cycle sequencer were denaturation for 30 s at 94°C, annealing for 30 s at 55°C, and extension for 1 min at 72°C, for 35 cycles. PCR buffer conditions were 10 μmol/L Tris-HCl (pH 8.3), 50 mmol/L KCl, 2 mmol/L Mg Cl2, and 20 pmol of each primer in a 25-μ L reaction volume. Amplified products were loaded on a standard sequencing gel. Bands were visualized after overnight exposure to Kodak AR film.
Liquid hybridization
Product (7 μL) obtained after PCR with individual family- or subfamily-specific TCRBV primer was mixed with 200 mmol/L NaCl, 100 μmol/L dNTP, formamide, and 0.56 ×106 cpm of 32P-labeled probe in a total volume of 14 μL. The formamide concentration was optimized for each probe by use of the formula %formamide = [34.95 +(0.41×% GC content probe) − (675/probe length)/0.65] (where G is guanine and C is cytosine). The addition of dNTP was empirically derived in our laboratory, to reduce nonspecific hybridization. The mixture was heated to 97°C for 5 min and then cooled to room temperature over the course of 15 min. Then, 10 μL of each hybridized reaction mixture was electrophoresed in a nondenaturing 6% acrylamide gel. Gels were dried and autoradiographed for up to 18 h.
Sequencing
Direct sequencing of PCR products was performed by use of the DyeDeoxy Terminator Sequencing kit (Applied Biosystem). In some cases, TCRBV-specific amplicons were first cloned into bacteria by use of a TA cloning kit (In-vitrogen). TCRBV-specific primers were used as sequencing primers. The TCRBV, TCRBD, and TCRBJ sequence of each clone was aligned relative to other TCRB sequences published in GenBank by use of nBlast software (version 2.2.10; National Center for Biotechnology Information). On the basis of this alignment, specific oligonucleotide probes were designed to span each unique N-D-N region.
RESULTS
Oligoclonal T cell repertoire in herpetic skin lesions
The complexity of TCRBV usage of herpetic lesion–derived T cell lines was initially determined by analysis of the distribution of CDR3 lengths. The CDR3 size pattern has been shown to display 6–8 bands spaced by 3 nt, corresponding to in-frame transcripts and distributed in a Gaussian fashion. A Gaussian pattern signifies polyclonality. A spectratype that displays an increased band intensity relative to such a normal distribution is an indication of the oligoclonal expansion of ≥1 T cell lineage within that size class. The lesion and PBMC lines used were obtained after PHA stimulation. Spectratypes from 5 subjects were analyzed on the basis of band number and band intensity. The TCRBV amplification products derived from all of the skin lesions analyzed exhibited a markedly skewed pattern of TCR β-chain usage, compared with their respective PBMC patterns. A representative spectratype for a PBMC-derived cell line (figure 1A) showed that most TCRBV families detected are composed of clusters of 6–8 bands whose intensities followed a Gaussian distribution. Few families (βV 3, 6, 7, 19, and 20) displayed the bands of increased intensity that suggest clonal expansion. This is in marked contrast to the pattern of a T cell line obtained in a similar manner from the same subject’s herpetic skin-lesion biopsy sample (figure 1B). In the latter case, a majority of the TCRBV families were represented by few scattered bands with a high intensity, which suggests oligoclonal T cell expansion. These data suggest that T cell infiltrates in herpetic skin lesion constitute a restricted repertoire of expanded T cell clones.
Figure 1.

Complementarity-determining region 3 length analysis of peripheral blood mononuclear cells (PBMCs) and a lesion-derived T cell line. Spectratypes from PBMCs (A) and a matched lesion-derived T cell line (B) were generated by stimulation with phytohemagglutinin, as described in Materials and Methods. The autoradiographs were exposed overnight with an intensifying screen. The different T cell receptor β-variable families are indicated above the autoradiographs.
Temporal stability of the T cell repertoire of herpetic skin lesions
We evaluated 2 separate biopsy samples from lesions that occurred 9 months apart in the same subject. In between, the original lesion had healed completely and had recurred only a few days prior to the second biopsy. Representative spectra-typing profiles after PHA stimulation indicated a clonal expansion of T cells with each separate lesion (figure 2). The pattern of bands seen in one subject for any βV family varied with time either during the course of an outbreak or between each individual recurrence, which suggests that certain clonotypes expand and others decline or that the spatial distribution of T cell clones within a lesion may not be homogeneous.
Figure 2.
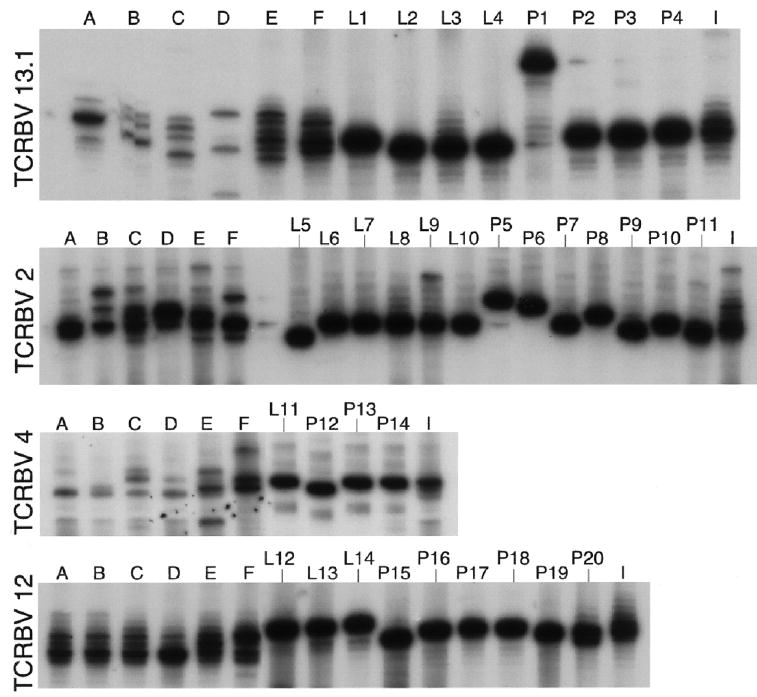
Longitudinal skin lesion repertoire analysis for different T cell receptor β-variable (TCRBV) families. Spectratypes were generated from sequential herpetic skin-lesion biopsies obtained from 1 subject during 2 different recurrences. The peripheral blood mononuclear cell (PBMC)– and lesion-derived cell lines were obtained after stimulation with phytohemagglutinin: lane A, nonlesional skin biopsy; lanes B, C, and D, 1996 skin-lesion biopsies (obtained on 20, 22, and 27 August 1996); lanes E and F, 1995 skin-lesion biopsies (obtained on 28 and 30 November 1995); lane I, autologous PBMC (obtained on 25 September 1996). Herpes simplex virus–specific CD4+ cell clones expressing matched TCRBVs were run side by side with the skin-lesion biopsy samples. Clones were established from either the 28 November 1995 skin-lesion biopsy (lane L) or the 25 September 1996 blood draw (lane P).
We sought evidence for clonal expansion by establishing several CD4+ cell clones from a skin-lesion biopsy sample and paired PBMC sample. HSV-specific clones were selected by proliferation assay and their TCRBV chain usage was determined by sequencing. Identical TCRBV sequences were observed repeatedly in clones established from the lesional skin biopsies. In contrast, many unique TCRB sequences were observed among HSV-specific clones isolated from blood. Moreover, when TCRBV PCR products were run side by side on a sequencing gel (figure 2), some bands coresolved, which indicates that dominant lesional clonotypes and lesion-derived HSV-specific T cell clones have a similar CDR3 length. These data suggest that the clonal expansion of HSV-specific clones, at the level of either cell migration or expansion in situ, contributes at least partially to the oligoclonality observed among lesional cell lines.
HSV-specific CD4+ cell clonotype tracking by liquid hybridization
To further investigate possible clonal persistence, we developed a sensitive PCR assay to track T cell clonotypes in skin-lesion biopsy and PBMC samples over time by use of liquid hybridization with specific oligonucleotide probes that span the unique N-D-N region of each clone. In preliminary experiments, cells from a selected CD4+ cell clone were diluted in a background of allogeneic PBMCs. The total RNA from each dilution was extracted and used for first-strand cDNA synthesis. As is shown in figure 3A, we could detect 100 cells of a given T cell clone mixed with 1 ×106 PBMCs. In a separate experiment, tubes that contained 10 clonal cells were positive at a detection rate of 50%. These data indicate that our methods were able to systematically detect a clonotype with a precursor frequency of 1 in 10,000 cells.
Figure 3.

Herpes simplex virus (HSV)–specific CD4+ cell clone tracking by liquid hybridization. Serial 10-fold dilutions of lymphocytes from an HSV-specific CD4+ clone were made in a background of allogeneic peripheral blood mononuclear cells (PBMCs, A). A sample without cDNA was used as negative control (C−). The 10-cell dilution was repeated 8 times. RNA extractions were performed independently for each sample. The probe was tested by liquid hybridization against all the clones expressing T cell receptor β-variable 2 (B). Hybridization on the corresponding T cell clone is shown as positive control (C+).
To assess the specificity of liquid hybridization, a clonotype-specific probe was tested on a panel of HSV-specific CD4+ clones expressing TCRBV2 (table 1). Probe hybridization was detected only with PCR products from the index clone or from clones with an identical sequence (figure 3B). No hybridization was observed on PCR products from clones with a different TCRBV sequence, including the HSV-specific CD4+ clone 18 from autologous PBMCs that had the same V and J segments, including 6 of 18 consecutive nucleotides matching in the N-D-N region.
Table 1.
Complete T cell receptor β-variable (TCRBV) gene usage, determined for herpes simplex virus–specific CD4+ cell clones expressing TCRBV2.
| Clone type and no. | Sequence |
|---|---|
| Lesion | |
| 8 | agagaccgccttcgggacagggttggggg |
| 9 | tcccgactcctcaac |
| 13 | tcccgactcctcaac |
| 22 | tcccgactcctcaac |
| 33 | tcccgactcctcaac |
| 39 | tcccgactcctcaac |
| PBMC | |
| 9 | catcctcctcggggctat |
| 10 | acgtccagc |
| 15 | atccggg |
| 18 | agagatcgagttcgga |
| 33 | gggggccgggtagcgacaggggcgtgg |
| 22 | cctaaactagcggcc |
| 39 | aggccggtcgggggacagggggta |
NOTE. After cDNA synthesis, TCRBV transcripts were amplified, and liquid hybridization was performed as described in Materials and Methods. The complete sequences are available from GenBank under the accession nos. (from top to bottom) AY751325, AY751309, AY751326, AY751327, AY751328, AY751329, AY751330, AY751331, and AY751332. A clonotype-specific probe was design on basis of the complementarity-determining region 3 (CDR3) sequence from lesion clone. The probe shared 6 of 18 consecutive nucleotides with the CDR3 sequence from peripheral blood mononuclear cell (PBMC) clone 9 (underlined nucleotides).
We then used this procedure to assay 4 biopsy samples collected simultaneously in a quadrant-like fashion within an HSV-2 lesion on a subject’s buttock on day 2 after lesion onset. Each sample was obtained from a separate quadrant at 12, 3, 6, and 9 o’clock. The biopsy samples were processed, and cells were expanded separately by use of PHA stimulation, as described elsewhere [10]. HSV-specific reactivity among the lesional T cell lines was studied by proliferation assay against a panel of viral antigens. Each T cell line displayed strong activity against HSV-1 and HSV-2 (figure 4 and table 2). Specific proliferation against the viral capsid glycoprotein gB2 was detected only in the 12 o’clock biopsy sample. A majority of CD4+ cell clones obtained from the 12 o’clock biopsy sample were found to be specific for gB2. Sequence analysis revealed that all (n = 64) clones were identical. A clonotype-specific probe based on the corresponding CDR3 region sequence was used to test each of the PHA-derived cell lines in the 4 biopsy quadrants for the presence of the gB2-specific clone. Although signal intensity varied, the clone could be detected in all 4 samples. This observation supports the hypothesis that, after tissue infiltration, HSV-specific CD4+ cells undergo local clonal expansion within the lesion area.
Figure 4.

Spatial distribution of herpes simplex virus–specific reactivity within a lesion. The complete T cell receptor β-variable (TCRBV) gene usage for the glycoprotein gB2–specific dominant CD4+ cell clone (TCRBV12S4J2S3C2; GenBank accession no. AY751304) was shown to be gccagcact at the 3′ end, ctacagagacccgga at the nucleotide sequence covering the clonotypic probe, and acgcagt at the 5′ end. Distribution of the gB2-specific clonotype among the 4 quadrant biopsies after stimulation with phytohemagglutinin was assessed by liquid hybridization, as described in Materials and Methods. MW, molecular weight; PBMCs, peripheral blood mononuclear cells.
Table 2.
Biopsy results from different quadrants, by assay type.
| Flow, % positive cells
|
CTL assay, %specific lysis
|
Proliferation assay, SI
|
||||||
|---|---|---|---|---|---|---|---|---|
| Biopsy quadrant | CD4+ | CD8+ | Mock | HSV-2 | HSV-1 | HSV-2 | gB2 | gD2 |
| 12 o’clock | 30 | 52 | 13 | 15 | 23 | 552 | 538 | 2 |
| 3 o’clock | 30 | 49 | 11 | 22 | 26 | 140 | 0 | 0 |
| 6 o’clock | 34 | 48 | 7 | 38 | 44 | 189 | 1 | 4 |
| 9 o’clock | 20 | 66 | 5 | 45 | 73 | 249 | 6 | 1 |
NOTE. CTL, cytotoxic T lymphocyte; HSV, herpes simplex virus; SI, stimulation index.
To assess whether T cell clones recovered from a skin-lesion biopsy sample are representative of the infiltrating lymphocyte repertoire found in a lesion, we investigated the distribution of different CD4+ cell clones established from the 12 o’clock biopsy sample, compared with the distribution of T cell clones for each of the other 3 biopsy samples. A total of 5 different clonotypic probes were designed. The detection profile for each probe in the different PHA-stimulated cell lines is summarized in table 3. One clone, expressing TCRVB12, was detected in all skin-lesion biopsy samples and in autologous PBMCs. Two others were detected in 3 of 4 skin-lesion biopsy samples and in PBMCs. Only 1 clone was detected exclusively in the 12 o’clock biopsy sample. These data suggest that most HSV-specific CD4+ cell clones infiltrating recurrent skin lesion are broadly distributed topographically.
Table 3.
Clonotype distribution detected by liquid hybridization in a herpetic skin lesion.
| Biopsy quadrant
|
|||||
|---|---|---|---|---|---|
| Clonotype-specific probe sequence | 12 o’clock | 3 o’clock | 6 o’clock | 9 o’clock | PBMC |
| ctctacagagacccgga (gB2 specific) | + | + | + | + | + |
| cccgggacaccctt | + | + | − | + | + |
| ttccgggacacc | + | − | − | − | − |
| tctagcaaagt | + | + | − | + | + |
| cttggccccctcgtc | + | + | − | − | + |
NOTE. Four punch biopsies were collected simultaneously in a quadrant-like fashion within a lesion from a subject with a recurrence of herpes simplex virus (HSV) type 2 on the buttock. The lesion and matching peripheral blood mononuclear cell (PBMC) lines were all obtained after stimulation with phytohemagglutinin. A total of 5 different clonotypic probes were designed on the basis of the T cell receptor β-variable sequence of several HSV-specific T cell clones obtained from the 12 o’clock biopsy. The complete sequences are available from GenBank under accession nos. (from top to bottom) AY751304, AY751305, AY751306, AY751307, and AY751308. +, detectable; −, undetectable.
Persistence of HSV-specific CD4+ clonotypes in herpetic skin lesions
As a chronic, relapsing disorder, HSV disease can potentially lead to clonal exhaustion. We therefore undertook a longitudinal analysis of HSV-specific T cell persistence using liquid hybridization. For these studies, sequential tissue samples were collected from subjects 1 and 2. Subject 1 provided samples twice during an episode of HSV reactivation in November 1995 and 3 times during another episode 9 months later. Subject 2 provided samples twice during an episode in February 1996. Autologous blood samples were also collected for a period up to 25 months after the initial skin-lesion biopsy sample was obtained for subject 1 and 24 months for subject 2. HSV-specific CD4+ cell clones were isolated from a skin-lesion biopsy sample from both subjects. A panel of clonotypic probes was designed on the basis of the TCRBV sequence of each clone. Liquid hybridization was performed on the different cell lines after stimulation with either PHA- or UV-inactivated HSV-2, to maximize the chance of detection of each clonotype.
The hybridization pattern for each probe is shown in table 4. In subject 1, 4 of 9 different T cell clonotypes tested could be positively detected in another biopsy sample from the same lesion or in subsequent biopsies from a different recurrence. The detection of the other clonotypes was limited to the lesion from which they originated. Interestingly, CD4+ cell clones that were detected repetitively in herpetic skin-lesion biopsies were also found persistently in PBMC samples 25 months later. Although biopsies from only 1 lesion were available from subject 2, a similar pattern of clonal persistence was detected. In this subject, 3 of 7 clonotypes detected in both lesional biopsies were also present in blood samples for up to 24 months. These findings demonstrate the existence of a pool of long-lasting HSV-specific CD4+ cells that are capable of trafficking between lesional skin and PBMCs. Whether the inability to detect some clonotypes over time relates to sampling bias, T cell exhaustion, or the limits of detection of the assay is unclear.
Table 4.
Summary of clones detected in lesion biopsies and autologous peripheral blood mononuclear cells (PBMCs).
| Sample type, date
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lesion, 28 Nov 1995
|
Lesion, 30 Nov 1995
|
Lesion, 20 Aug 1996
|
Lesion, 22 Aug 1996
|
Lesion, 27 Aug 1996
|
PBMC, 25 Sep 1996
|
PBMC, 2 Dec 1996
|
PBMC, 1 Feb 1998
|
|||||||||
| Clonotype-specific probe sequence | PHA | HSV | PHA | HSV | PHA | HSV | PHA | HSV | PHA | HSV | PHA | HSV | PHA | HSV | PHA | HSV |
| Subject 1 | ||||||||||||||||
| tcccgactcctcaact (gD) | + | + | − | − | − | + | − | + | + | − | − | + | − | + | − | + |
| gctcagggggcg | − | + | − | − | − | − | − | − | − | − | − | + | − | + | − | + |
| ccgggggggccatc | + | + | − | + | − | − | − | + | + | + | − | + | − | + | − | + |
| ggggaccaccgggacaggggg | − | + | + | + | + | + | + | + | + | − | + | + | + | + | − | + |
| agcgggtttgtgg | + | + | + | + | − | − | − | + | − | + | − | − | − | + | − | + |
| tggacaggactacaat | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| cccccatgggggacct | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| tcgggctgatgggggctac (VP16) | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| aacgccccggccag (gB) | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Lesion, 12 Feb 1996
|
Lesion, 16 Feb 1996
|
PBMC, 12 Feb 1996
|
PBMC, 23 Jun 1997
|
PBMC, 24 Feb 1998
|
||||||||||||
| PHA | HSV | PHA | HSV | PHA | HSV | PHA | HSV | PHA | HSV | |||||||
| Subject 2 | ||||||||||||||||
| agttcgagcggggccaag | − | + | + | + | − | + | − | + | − | + | ||||||
| agcggggaagggccttggag | − | + | + | + | + | + | + | + | − | + | ||||||
| gtgcgagggggaacctaaacggggg | + | − | − | + | + | + | − | − | + | − | ||||||
| tacagggccagccaac | + | + | − | + | − | + | − | − | − | − | ||||||
| gggaggggacagcaat | − | + | − | − | − | + | − | − | − | − | ||||||
| tcggaggccacctcaat | + | + | − | − | − | + | − | − | − | − | ||||||
| gctggtgggacaggggtcgg | − | + | − | − | − | − | − | − | − | − | ||||||
NOTE. Sequential tissue samples were collected from 2 subjects. A panel of clonotype-specific probes was designed on the basis of a collection of herpes simplex virus (HSV)–specific T cell clones isolated from a skin-lesion biopsy sample from each subject (on 28 November 1995 and 12 February 1996, respectively). Clonotype detection was performed in cell culture from each tissue specimen after stimulation with phytohemagglutinin (PHA)– or UV-inactivated HSV-2, as described in Material and Methods. The complete sequences are available from GenBank under accession nos. (from top to bottom) AY751309, AY751310, AY751311, AY751312, AY751313, AY751314, AY751315, AY751316, AY751317, AY751318, AY751319, AY751320, AY751321, AY751322, AY751323,andAY751324. When known, the clone antigenic specificity is shown in parentheses. +, detectable; −, undetectable.
Cervical CD4+ cell response
Lymphocytes from cervical secretions were also available from subjects 1 and 2. We analyzed the tissue distribution and temporal persistence of cervix-derived HSV-specific CD4+ cell clonotypes. For both subjects, we selected the most abundant clonotype found among the few HSV-specific CD4+ cells isolated from each cytobrush specimen [7]. The cervical T cell clone from subject 1 was strongly detected in the cervical cytobrush sample from which it was derived (figure 5) but could not be detected in subsequent cervical samples. Interestingly, this clone was also positively identified by liquid hybridization not only in a T cell culture from a skin-lesion biopsy sample collected simultaneously with the cyto-brush specimen but also in matched PBMC samples. Moreover, a skin-lesion biopsy cell culture from a previous outbreak that occurred 16 months before the cervical specimen was collected was also positive by liquid hybridization. The presence of an identical CD4+ cell clone in the cytobrush specimen and the lesion biopsy was confirmed by cloning and sequencing the TCRBV-specific PCR product.
Figure 5.

Tissue distribution and temporal persistence of cervix-derived herpes simplex virus (HSV)–specific CD4+ cell clonotypes. Clonotype persistence of 2 cervical CD4+ cell clones from both subject 1 (A) and subject 2 (B) was assessed by liquid hybridization in cell culture from different tissue specimens. Cell lines established from blood samples (peripheral blood mononuclear cells [PBMCs]), herpetic skin lesions (skin), or cervical cytobrush (cervix) were stimulated with phytohemagglutinin (PHA) or with HSV-2, as described in Materials and Methods. The complete T cell receptor β-variable (TCRBV) gene usage is shown for both cervical CD4+ cell clones. Clone Cy2 (TCRBV2S1J2S7C2; GenBank accession no. AY751333) was shown to be cagtgc at the 3′ end, aagaaaggggaggggcg at the nucleotide sequence covering the clonotypic probe, and cctacg at the 5′ end. Clone Cy4 (TCRBV4S1J2S1C2; GenBank accession no. AY751334) was shown to be agcgtt at the 3′ end, cgaggggacagacac at the nucleotide sequence covering the clonotypic probe, and tacgag at the 5′ end. Hybridization on the corresponding T cell clone is shown as the positive control (C+). MW, molecular weight.
A similar clonal tracking experiment was performed by use of a cervix-derived HSV-specific CD4+ cell clone from subject 2. Again, the clone could be detected easily in the cytobrush sample from which it was derived. Liquid hybridization also revealed the presence of this particular clonotype in a skin-lesion biopsy sample that was obtained 16 months before the cytobrush collection. Similarly, in the peripheral blood, the clonotype was detected at the time of the outbreak before the cytobrush sample was collected but not at a later time point.
DISCUSSION
Our analysis of the T cell infiltrate in herpetic skin-lesion biopsy samples indicates an oligoclonal immune response. Although several previous studies have shown TCR expression at different tissue sites to be oligoclonal, those experiments did not link this limited repertoire to antigen specificity [11]. Our study demonstrates that the clonal expansions observed among lesion-derived T cell lines were largely related to HSV-specific CD4+ cell proliferation. To define the persistence of infiltrating T cells in genital lesions, we developed a sensitive and highly specific clonal tracking technique using a combination of TCRBV-specific PCR followed by liquid hybridization with clonotype-specific probes.
In both subjects studied, we demonstrated CD4+ cell clones that persisted over the duration of an outbreak and for several months to years afterward. Moreover, T cells with identical TCRB chains were isolated from different and nonadjacent segments of tissue, which suggests that the T cells proliferated in vivo and were not simply an artifact of in vitro T cell isolation. This suggests that the colonization of peripheral tissues by CD4+ cells results from specific recruitment of T cells directed against HSV antigens. Moreover, we have shown that these HSV-specific CD4+ clonotypes were not exclusively compartmentalized in a particular tissue but were also circulating in the peripheral blood. Our observations are in agreement with recently published data demonstrating that T cell clones traffic between the mucosal and systemic compartments [12]. These lymphocytes could represent a pool of long-lasting, memory-type T cell clones that may be rapidly recruited by the immune system and mobilized in response to a HSV recurrence. Such a circulating lymphocyte population is thought to acquire its skin or mucosa homing specificity only after antigen-specific activation in peripheral lymph nodes [13]. Expression of the mucosal lymphocyte–associated antigen (MLA) appears to be a specific and stable characteristic of lymphocytes recovered from the genital tract [14], whereas the cutaneous lymphocyte–associated antigen (CLA) is preferentially expressed on skin-in-filtrating T cells [15]. We have recently reported that CLA expression is involved in HSV-specific CD8+ and CD4+ memory cells trafficking to the skin [16, 17]. To date, there has been no report of T cells expressing both CLA and MLA homing receptors. It is thus of particular interest that cervical mucosa–derived T cells can also be detected in herpetic skin lesions. Because there is a marked difference in the nature of these 2 epithelia, this suggests that some as-yet-undefined homing receptors might be similar for these 2 sites or that T cell clonotypes can express >1 homing receptor.
Our data do not allow us to distinguish whether the detected cells persist as memory T cells or whether the immune system reselected the same clone from the naive T lymphocyte pool in response to the same antigen. It has been postulated that the survival of memory T cells implies antigen persistence [18]. It is tempting to hypothesize that, because HSV is known to produce frequent asymptomatic shedding [19], this low local viral production might be the source for antigens responsible for maintaining a long-lasting pool of HSV-specific memory T cells, even during prolonged antiviral therapy [20].
We did detect several HSV-specific T cell clones transiently (i.e., only once during a recurrence). Several possibilities for this observation exist. We may have been unable to detect them because of clonal frequencies below our detection threshold (1 in 10,000). Alternatively, these particular clones may be skin-resident T cells that are part of the skin’s immune system [21, 22]. The lack of persistence observed for some of our HSV-specific T cell clones might be explained by clonal amplification at the site of the infection after stimulation by local antigen-presenting cells (APCs), followed by cell death and a reduction in the clonotype population below our limit of detection. In this scenario, a defective activation signal delivered by HSV-infected local APCs might fail to generate long-term persistent memory T cells [23].
In summary, our study indicates that the local specific immune response against HSV involves an oligoclonal expansion of effector cells. CD4+ clones may persist for prolonged periods in vivo in the presence of a chronic, intermittently productive viral infection. It is clear that viral replication still occurs in the presence of such T cells, which suggests that ways to improve the function or number of such responses may offer potential options for improving the clinical outcome of these infections.
Acknowledgments
We thank Matthew L. Johnson (University of Washington), for technical support; Mike Remington, Gail Barnum, Peter Trethewey, Kathleen Stine, and Mary Shaughnessy (University of Washington), for specimen collection; Drs. Rae Lyn Burke and Michael A. Tigges (Chiron), for providing purified gD2 and gB2 glycoproteins; Dr. Marc Schomogyi, for providing the cervical T cell clones; and Nancy Coomer, for help in preparing the manuscript.
Footnotes
Financial support: National Institutes of Health (grants AI-30731, R37 AI-42528, and AI-50132).
References
- 1.Wald A, Corey L, Cone R, Hobson A, Davis G, Zeh J. Frequent genital herpes simplex virus 2 shedding in immunocompetent women: effect of acyclovir treatment. J Clin Invest. 1997;99:1092–7. doi: 10.1172/JCI119237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunningham AL, Turner RR, Miller AC, Para MF, Merigan TC. Evolution of recurrent herpes simplex lesions: an immunohistologic study. J Clin Invest. 1985;75:226–33. doi: 10.1172/JCI111678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koelle DM, Abbo H, Peck A, Ziegweid K, Corey L. Direct recovery of herpes simplex virus (HSV)–specific T lymphocyte clones from recurrent genital HSV-2 lesions. J Infect Dis. 1994;169:956–61. doi: 10.1093/infdis/169.5.956. [DOI] [PubMed] [Google Scholar]
- 4.Foster CA, Yokozeki H, Rappersberger K, et al. Human epidermal T cells predominantly belong to the lineage expressing α/β T cell receptor. J Exp Med. 1990;171:997–1013. doi: 10.1084/jem.171.4.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chothia C, Boswell DR, Lesk AM. The outline structure of the T-cell αβ receptor. EMBO J. 1988;7:3745–55. doi: 10.1002/j.1460-2075.1988.tb03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorski J, Yassai M, Zhu X, et al. Circulating T cell repertoire complexity in normal individuals and bone marrow recipients analyzed by CDR3 size spectratyping: correlation with immune status. J Immunol. 1994;152:5109–19. [PubMed] [Google Scholar]
- 7.Koelle DM, Schomogyi M, Corey L. Antigen-specific T cells localize to the uterine cervix in women with genital herpes simplex virus type 2 infection. J Infect Dis. 2000;182:662–70. doi: 10.1086/315749. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Monteiro J, Hingorani R, Choi IH, Silver J, Pergolizzi R, Gregersen PK. Oligoclonality in the human CD8+ T cell repertoire in normal subjects and monozygotic twins: implications for studies of infectious and autoimmune diseases. Mol Med. 1995;1:614–24. [PMC free article] [PubMed] [Google Scholar]
- 10.Koelle DM, Posavad CM, Barnum GR, Johnson ML, Frank JM, Corey L. Clearance of HSV-2 from recurrent genital lesions correlates with infiltration of HSV-specific cytotoxic T lymphocytes. J Clin Invest. 1998;101:1500–8. doi: 10.1172/JCI1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stamenkovic I, Stegagno M, Wright KA, et al. Clonal dominance among T-lymphocyte infiltrates in arthritis. Proc Natl Acad Sci USA. 1988;85:1179–83. doi: 10.1073/pnas.85.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musey L, Ding Y, Cao J, et al. Ontogeny and specificities of mucosal and blood human immunodeficiency virus type 1-specific CD8+ cytotoxic T lymphocytes. J Virol. 2003;77:291–300. doi: 10.1128/JVI.77.1.291-300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabbri M, Bianchi E, Fumagalli L, Pardi R. Regulation of lymphocyte traffic by adhesion molecules. Inflamm Res. 1999;48:239–46. doi: 10.1007/s000110050454. [DOI] [PubMed] [Google Scholar]
- 14.Shaw SK, Brenner MB. The β 7 integrins in mucosal homing and retention. Semin Immunol. 1995;7:335–42. doi: 10.1016/1044-5323(95)90014-4. [DOI] [PubMed] [Google Scholar]
- 15.Berg EL, Yoshino T, Rott LS, et al. The cutaneous lymphocyte antigen is a skin lymphocyte homing receptor for the vascular lectin endothelial cell-leukocyte adhesion molecule 1. J Exp Med. 1991;174:1461–6. doi: 10.1084/jem.174.6.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koelle DM, Liu Z, McClurkan CM, et al. Expression of cutaneous lymphocyte-associated antigen by CD8+ T cells specific for a skin-tropic virus. J Clin Invest. 2002;110:537–48. doi: 10.1172/JCI15537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez JC, Kwok WW, Wald A, McClurkan CL, Huang J, Koelle DM. Expression of cutaneous lymphocyte-associated antigen and E-selectin ligand by circulating human memory CD4+ T lymphocytes specific for herpes simplex virus type 2. J Infect Dis. 2005;191:243–54. doi: 10.1086/426944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oehen S, Waldner H, Kundig TM, Hengartner H, Zinkernagel RM. Antivirally protective cytotoxic T cell memory to lymphocytic choriomeningitis virus is governed by persisting antigen. J Exp Med. 1992;176:1273–81. doi: 10.1084/jem.176.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rattray MC, Corey L, Reeves WC, Vontver LA, Holmes KK. Recurrent genital herpes among women: symptomatic v. asymptomatic viral shedding. Br J Vener Dis. 1978;54:262–5. doi: 10.1136/sti.54.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frenkel L, Pineda E, Garratty E, Fall H, Dillon M, Bryson Y. A prospective study of the effects of acyclovir treatment on the HSV-2 lymphoproliferative response of persons with frequently recurring HSV-2 genital infections. J Infect Dis. 1989;159:845–50. doi: 10.1093/infdis/159.5.845. [DOI] [PubMed] [Google Scholar]
- 21.Washington EA, Kimpton WG, Holder JE, Cahill RN. Role of the thymus in the generation of skin-homing αβ and γδ virgin T cells. Eur J Immunol. 1995;25:723–7. doi: 10.1002/eji.1830250314. [DOI] [PubMed] [Google Scholar]
- 22.Kimpton WG, Washington EA, Cahill RN. Virgin αβ and γδ T cells recirculate extensively through peripheral tissues and skin during normal development of the fetal immune system. Int Immunol. 1995;7:1567–77. doi: 10.1093/intimm/7.10.1567. [DOI] [PubMed] [Google Scholar]
- 23.Barcy S, Corey L. Herpes simplex inhibits the capacity of lymphoblastoid B cell lines to stimulate CD4+ T cells. J Immunol. 2001;166:6242–9. doi: 10.4049/jimmunol.166.10.6242. [DOI] [PubMed] [Google Scholar]


