Abstract
Animal genomes each encode multiple highly conserved actin isoforms that polymerize to form the microfilament cytoskeleton. Previous studies of vertebrates and invertebrates have shown that many actin isoforms are restricted to either nonmuscle (cytoplasmic) functions, or to myofibril force generation in muscle cells. We have identified two temperature-sensitive and semidominant embryonic-lethal Caenorhabditis elegans mutants, each with a single mis-sense mutation in act-2, one of five C. elegans genes that encode actin isoforms. These mutations alter conserved and adjacent amino acids predicted to form part of the ATP binding pocket of actin. At the restrictive temperature, both mutations resulted in aberrant distributions of cortical microfilaments associated with abnormal and striking membrane ingressions and protrusions. In contrast to the defects caused by these dominant mis-sense mutations, an act-2 deletion did not result in early embryonic cell division defects, suggesting that additional and redundant actin isoforms are involved. Accordingly, we found that two additional actin isoforms, act-1 and act-3, were required redundantly with act-2 for cytoplasmic function in early embryonic cells. The act-1 and -3 genes also have been implicated previously in muscle function. We found that an ACT-2::GFP reporter was expressed cytoplasmically in embryonic cells and also was incorporated into contractile filaments in adult muscle cells. Furthermore, one of the dominant act-2 mutations resulted in uncoordinated adult movement. We conclude that redundant C. elegans actin isoforms function in both muscle and nonmuscle contractile processes.
INTRODUCTION
Actin is a highly conserved ATPase that assembles into cytoskeletal polymers called microfilaments. Both the hydrolysis of ATP to ADP, and the activities of actin-associated proteins, regulate microfilament assembly and disassembly, influencing microfilament dynamics and contractility (Sablin et al., 2002). Microfilaments are required for many cellular processes including cell adhesion, cell migration, cell polarity, cytokinesis, endocytosis, muscle contraction, synapse function, and transcription. The regulated dynamic properties of microfilaments are important for many if not all of these roles.
The nonmuscle roles for microfilaments are often referred to as cytoplasmic, to distinguish them from the better understood contractile processes that occur in muscle cell myofibrils. These different functions appear to depend in part on the differential expression of closely related but distinct actin isoforms (Herman, 1993; Khaitlina, 2001). For example, vertebrate genomes encode several different actin isoforms, some specifically expressed in muscle and others in non-muscle cell types. These different expression patterns correlate with limited differences in primary amino acid sequence. For example, the length and charge of the N-terminus varies among different isoform classes, and other amino acid positions also exhibit isoform-specific variations (Herman, 1993; Khaitlina, 2001). Functional studies indicate that the sequence differences used to classify actin isoforms are important. For example, the lethality associated with loss of a cardiac isoform in mice could not be rescued by cardiac expression of a smooth muscle isoform (Kumar et al., 1997). In addition, overexpression of different human cytoplasmic actin isoforms in cell culture resulted in distinct cellular phenotypes (Schevzov et al., 1992).
Invertebrate actin isoforms are less distinctive in amino acid sequence and in general more closely resemble vertebrate cytoplasmic actins (Khaitlina, 2001). Nevertheless, invertebrate isoforms also have been shown to have specific cytoplasmic or muscle functions. For example, the genome of the fruit fly Drosophila melanogaster encodes six actin isoforms. Two appear to function specifically as cytoplasmic actins, whereas two others are specifically expressed in larval muscle, and still two others in adult muscle (Fyrberg et al., 1980, 1981). One adult-specific muscle isoform, Act-88F, is essential for flight. Flight was restored in mutants lacking Act-88F, when the Act-88F promoter was used to drive expression of the other adult muscle isoform. However, neither of the two larval muscle isoforms, nor one of the cytoplasmic isoforms, could restore flight when expressed using the Act-88F promoter (Fyrberg et al., 1998). Similarly, expression of the beta form of human cytoplasmic actin, driven by the Act-88F promoter, did not restore flight, even though it was incorporated into microfilaments (Brault et al., 1999). Finally, one of the two cytoplasmic isoforms in Drosophila, Act5C, is essential, but viability was restored in mutants lacking Act5c when the other cytoplasmic isoform, Act42A, was expressed from the Act5C promoter (Wagner et al., 2002). Thus the two cytoplasmic isoforms appear interchangeable, but proper transcriptional control by the promoter is required for viability. These studies indicate that the cytoplasmic and muscle isoforms have at least some distinct functions, whereas isoforms within each class are more interchangeable.
The five different actin isoforms in the nematode Caenorhabditis elegans have not been studied as extensively as those in Drosophila. Three actin genes, act-1, -2, and -3, reside in a cluster on one chromosome, whereas act-4 and act-5 are each on different chromosomes (Files et al., 1983; Macqueen et al., 2005). As in other organisms, these actin isoforms are highly conserved in amino acid sequence. ACT-1 and -3 are 100% identical. ACT-2 and -4 are both 99%, and ACT-5 93% identical to ACT-1 and -3. Dominant mutations identified in act-1 and -3 result in disorganized contractile filaments in body-wall muscle cells and an uncoordinated mutant phenotype (Landel et al., 1984), and a GFP::act-1 fusion is expressed in body wall muscle and incorporated into contractile filaments (Dixon and Roy, 2005). A semidominant mutant with pharyngeal muscle pumping defects, but no body-wall muscle defects, was shown to have mis-sense mutations in both act-2 and -3 (Avery, 1993). Thus, either act-2 or -3, or both, function in pharyngeal muscle cells. Although no mutations have been identified in act-4, an act-4 reporter gene was expressed in body-wall and other muscle cell types (Stone and Shaw, 1993). Although studies in C. elegans have identified actin genes that function in muscle, less is known about which isoforms are required for cytoplasmic functions. A recent study has shown that act-5 encodes a cytoplasmic actin required for the assembly of intestinal microvilli (Macqueen et al., 2005). However, it remains unclear which C. elegans actin isoforms are required for other cytoplasmic processes.
Here we report our identification of two conditionally semidominant and embryonic-lethal mutations in the C. elegans act-2 gene. These mutations alter conserved amino acids in the predicted ATP binding pocket of actin and promote contractile instabilities and ectopic furrowing in early embryonic cells, implicating ACT-2 as a cytoplasmic actin. However, a recessive act-2 deletion mutant is homozygous viable, and we present evidence that both act-1 and -3 function redundantly with act-2 in early embryonic cells. Finally, we show that a GFP::act-2 fusion is expressed cytoplasmically throughout early embryos and, later in development, in epidermal cells and body wall muscle. Moreover, one of the dominant act-2 mutants exhibits uncoordinated adult movement. These results, together with previous studies implicating act-1, -2, and -3 in muscle function, show that in C. elegans three actin isoforms function redundantly as both cytoplasmic and muscle actins.
MATERIALS AND METHODS
C. elegans Strains and Culture
All strains were cultured by standard methods (Brenner, 1974). Bristol (N2) was the standard wild-type strain used in this study. The following alleles were used: LGI: dpy-5(e61); LGII: rol-6(e187); LGIII: unc-32(e189); LGIV: unc-5(e53); him-8(e1489); LGV: dpy-11(e224), act-2(or295SD,ts), act-2(or621SD,ts), act-2(ok1229), act-3(st22d), unc-42(e270), sma-1(e30), egl-3(n150ts), stDf4; and LGX: lin-2(e1309), lon-2(e678). Conditional mutants were maintained by growth at the permissive temperature of 15°C. To obtain mutant embryos for phenotypic analysis, mutant L4 larvae were shifted to 26°C and grown to adulthood overnight, or young adults were grown at 26°C for at least 8 h. To assay the motility of wild-type and act-2 mutant worms, L1 larvae produced at the permissive temperature of 15°C were shifted to the restrictive temperature until adulthood and then transferred to fresh plates for 6 h before recording images.
To determine if the or295 and or621 mutations were dominant or recessive, or295/+ and or621/+ L4 hermaphrodites raised at 15°C were shifted to 26°C and matured to adulthood overnight. Adults were transferred to fresh plates, allowed to lay eggs for several hours, and then removed. The fraction of embryos that hatched was scored 24 h after removal of the adults. In addition, unc-42(e270) or295/+ hermaphrodites maintained at 15°C were shifted to 26°C as L4s and matured to adulthood overnight to produce broods of embryos. Twenty-two percent of the hatched embryos (n = 215) matured into Unc adults that were fertile and made dead embryos, indicating that the embryonic lethality observed in the broods of the unc-42 or295/+ hermaphrodites was due to a maternal-effect dominance.
Positional Cloning of act-2(or295) and act-2(or621)
The or295 and or621 alleles were identified in screens for temperature-sensitive, embryonic-lethal mutations, using EMS and ENU mutagenesis, respectively, as described previously (Encalada et al., 2000). The or295 and or621 mutations were backcrossed to the wild-type N2 strain eight and four times, respectively, in all strains used for phenotypic analysis.
Linkage group and 3-factor mapping were done as described previously (Brenner, 1974). Linkage group mapping was done using the strains MT3751 [dpy-5(e61) I; rol-6(e187) II; unc-32(e189) III], MT464 [unc-5(e53) IV; dpy-11(e224) V; lon-2(e678) X], EU1073 [him-8(e1489) IV; act-2(e295) V], and EU1296 [him-8(e1489); act-2(or621) V]. Both or295 and or621 mapped to LGV. Three-factor mapping with act-2(or295) was done using dpy-11(e224) unc-76(e911) V (map positions 0 and + 7.12), unc-42(e270) sma-1(e30) V (map positions +2.19 and +3.46), and egl-3(n150ts) emo-1(oz1) sma-1(e30) (map positions + 2.32, +2.83 and +3.46). Homozygous emo-1(oz1) worms are sterile. Using the egl-3 emo-1 sma-1 chromosome in trans to or295, we found that in 53/53 Egl-non-Ste recombinants egl-3 became linked to or295, whereas in 12/21 Sma-non-Ste recombinants, sma-1 became linked to or295, placing or295 at 3.0 map units. Three-factor mapping for or621 was done using dpy-11(e224) unc-76(e911) in trans to or621. In 7/15 Dpy-non-Unc recombinants, dpy-11 became linked to or621, whereas in 7/10 Unc-non-Dpy recombinants, unc-76 became linked to or621, placing or621 at roughly +2.85 map units on LGV. The cluster of act-1, -2, and -3 is located to the right of emo-1, at about +3.0 map units, and act-2 mRNA has been shown to be enriched in a germline cDNA library, suggesting it is maternally expressed (Piano et al., 2000). We therefore sequenced the act-1, -2, and -3 genes in genomic DNA from homozygous or295 and homozygous or621 worms. DNA fragments spanning each gene (from 100 base pairs 5′ of the predicted translational start site to 100 base pairs 3′ of the predicted stop codon) were amplified by PCR, and the products were isolated using a QIAquick Gel Extraction Kit (Qiagen, Chatsworth, CA). For each fragment, three pools of PCR products were independently isolated and combined for DNA sequencing reactions. DNA sequencing was done using the Beckman Quickstart Dye Terminator Kit and a Beckman CEQ8000 instrument, at the University of Oregon DNA Sequencing Facility. In or295 genomic DNA, we identified a GGA to AGA mis-sense mutation at nucleotide number 46 in act-2 that predicts a G15R amino acid change. In or621 genomic DNA, we identified a TCC to GCC mis-sense mutation at nucleotide number 43 in act-2 that predicts an S14A amino acid change. The amino acid numbers we use are based on the processed forms of actin, after the expected posttranslational removal of N-terminal residues. No nucleotide changes relative to wild type were detected in the act-1 and -3 genes in or295 and or621 genomic DNA. No nucleotide changes relative to wild type were detected in act-2 after amplification from genomic DNA of the lin-2(e1309) X strain used as the background for the original isolation of or295 and or621.
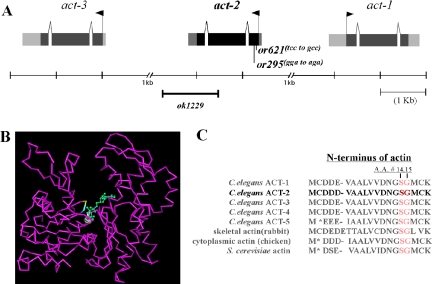
The C. elegans actin structure shown in Figure 3 was viewed using Cn3D version 4.1 from NCBI (http://www.ncbi.nlm.nih.gov). Files were exported as .png files and viewed by image J.
Figure 3.
or295 and or621 are act-2 alleles. (A) The actin gene cluster on linkage group V, where three of the five C. elegans actin genes reside, and the approximate location of the act-2 (ok1229) deletion and the act-2(or295) and act-2(or621) molecular lesions. (B) The act-2(or295) and act-2(or621) mutations alter conserved amino acids in a predicted loop that forms part of the ATP binding pocket of subdomain 1 of actin. The affected residues are highlighted in blue (S14A) and yellow (G15R) and are near the ATP molecule (green), which resides close to the center of the actin structure (purple). The structure, PDB name 1D4X, is from C. elegans Mg-ATP actin, purified from extracts and, thus, not isoform-specific (Vorobiev et al., 2003; see Materials and Methods). (C) The N-terminal amino acid sequences (amino acids 1 through 19 or 20) of different actin isoforms in C. elegans and other eukaryotes. The residues in ACT-2 altered by the or295 (G15R) and or621 (S14A) mutations are highlighted in red. As is conventional for actin, numbers are assigned to amino acid residues after removal of the first two residues (methionine and cysteine), which occurs during posttranslational processing (adapted from Ono, 1999).
Single Oocyte RT-PCR and GFP::ACT-2 Translational Fusion Construction
Preparation of single oocytes for nested RT-PCR was performed as described (Rutledge et al., 2001), but with the following changes. Isolated gonads were sheared through the glass needle of a mouth pipette to dislodge mature oocytes from the somatic gonad. Mature oocytes were transferred to watch glasses with fresh egg buffer twice before final transfer to a 0.65-ml Eppendorf tube containing 2 μl egg buffer. For RT-PCR we used the Invitrogen (Carlsbad, CA) SuperScript One-Step RT-PCR and followed the manufacturers protocol. The cDNA synthesis was carried out at 55°C for 30 min after DNase treatment. The first PCR reaction went for 15 cycles and 2 μl of this product was used for a second PCR reaction for an additional 25 cycles. Five microliters of each reaction was loaded per well. To determine that the PCR product was not amplified from genomic DNA, we used gene-specific PCR primers that flanked the last intron (at least 45 bp in length) Samples were run on a 1.5% agarose gel and a 50- and a 100-base pair DNA ladder from New England Biolabs (Ipswich, MA) were used to determine size. Using these conditions, we could clearly distinguish between PCR products that differed by 45 base pairs. Furthermore, the primer pairs were designed to amplify differently sized RT-PCR products for each isoform, to distinguish each actin isoform. The conditions used did not allow us to compare isoform mRNA expression levels.
For construction of the GFP::act-2 genomic transgene, we used PCR to amplify the act-2 promoter region (starting 2.9 kb 5′ of the first codon) from the cosmid T04C12. The primers included a 5′ Not1 site and a 3′ Spe1 site for subcloning into pBluescript II KS+, using standard methods (Ausubel et al., 1991). Subsequently, both a PCR-amplified GFP fragment (with primer-encoded 5′ and 3′ Spe1 sites), and the act-2 exons and introns, plus 1 kb of 3′ genomic sequence (with primer-encoded 5′ Spe1 and a 3′ Kpn1 sites) were subcloned into the promoter vector. A linker encoding seven amino acids (G, S, S, T, R, G, A) was used between the 3′ end of the GFP and the start of ACT-2. Proper insert orientation and sequence fidelity were determined by DNA sequence analysis. To obtain a transgenic line that expresses the GFP::ACT-2 translational fusion, we injected N2 animals using standard methods (Mello et al., 1991). The concentration of the injected GFP::act-2 plasmid was 5 ng/μl, and the concentration of the dominant rol-6 plasmid (pRF4) used as a transformation marker was 50 ng/μl. Genomic yeast DNA was used as carrier at a concentration of 50 ng/μl. One heritable extrachromosomal array was obtained in a transgenic strain that expressed GFP. We found that 33% of the progeny (n = 350) produced by transgenic animals inherited the array. Imaging of GFP expression in live embryos and anesthetized animals was done with a Bio-Rad 2100 Radiance confocal microscope (Richmond, CA), using a 60× oil immersion lens (use courtesy of Chris Doe, HHMI and the University of Oregon).
RNA Interference
Double-stranded RNA was prepared by amplifying the unique 3′UTR sequences of each actin gene with the following oligonucleotide pairs, using the indicated cosmids as templates: For act-1 (T04C12): 5′ATGCACAACTTCGTCAACTTGCAC3′ and 5′TATCAATTTTTAAATTTTTATTCACAC3′; act-2 (T04C12): 5′ACGTTTTAACAATTTATGTAATAT3′ and 5′TAGAAATATTATTAAAATAAA3′; act-3 (T04C12): 5′GCTCTTCGCCTTACCATTTTC3′ and 5′ATTCTGAAATTATTTATTGACTT3′; act-4 (MO3F4): 5′ATTTTTTGCCCCTTCCACCC3′ and 5′GTCTTATTAAAAGCTTTATT3′; act-5 (T25C8): 5′GCTGATTTTTTTTCAAATTTT3′ and 5′AAGTGGTCCTAACAAGTTT3′. PCR products were subcloned using the pGEM-t vector system (Promega, Madison, WI) and amplified with T7 and SP6 primers, with the SP6 primer including T7 sequences 5′ to the SP6 sequence. The amplified inserts were purified by phenol/chloroform extraction and ethanol precipitation (Ausubel et al., 1991) and then used as templates for bidirectional transcription using T7 Polymerase (Promega). Double-stranded RNAs were microinjected into both arms of the gonad of young adult hermaphrodites using a Narishige micro-manipulator. Injected act-2(ok1229) worms were incubated at 26°C; embryos were dissected from injected worms 12–16 h after injection. When we injected the same 3′UTR-specific dsRNAs into N2 animals, we observed fully penetrant embryonic/L1 larval lethality for the act-1, -4, and -5 actin genes, whereas the only phenotype observed in the broods of act-3 3′UTR-injected animals was adult sterility (33%; n = 12). We observed no early embryonic cell division defects except after microinjection of act-2 3′UTR dsRNA into N2. However, as the act-2(ok1229) deletion allele is homozygous viable (Table 1), and microinjection of act-2 3′UTR dsRNA into act-2(ok1229) worms does not produce any lethality (Table 2), we suspect that the embryonic cell division defects observed after microinjection of act-2 3′UTR dsRNA into N2 worms is due either to short regions of high conservation with other actin genes or to spreading of the dsRNA into more highly conserved 5′ coding sequences (Alder et al., 2003; May and Plasterk, 2005). Such spreading, if it occurred, appears to occur infrequently, as we did not observe any such effect for the other four 3′UTR dsRNA injections into N2 worms, and 3′UTR dsRNAs have also been used successfully to specifically deplete four different embryonic tubulin isoforms (Wright and Hunter, 2003; Phillips et al., 2004). For depletion of pfn-1, cyk-1, and mlc-4 gene products, exon fragments of at least 500 base pairs were amplified using PCR to generate DNA templates for the production of dsRNAs using T7 RNA polymerase (as described above).
Table 1.
Embryonic lethality of act-2 alleles and allelic combinations
| Maternal genotype | % Emb. lethality (15°C) | % Emb. lethality (26°C) |
|---|---|---|
| N2 | 0 (385) | 3 (478) |
| act-2(or295)/act-2(or295) | 62 (240) | 98 (467) |
| act-2(or295)/+ | 0 (210) | 12 (360) |
| act-2(or621)/act-2(or621) | 60 (226) | 100 (300) |
| act-2(or621)/+ | 7 (415) | 85 (325) |
| act-2(ok1229)/act-2(ok1229) | 0 (345) | 12 (515) |
| act-2(ok1229))/+ | 0 (196) | 0 (288) |
| act-2(or295)/act-2(or621) | 100 (230) | 100 (270) |
| act-2(or295)/act-2(ok1229) | na | 88 (420) |
| act-2(or295)/stDf4 | na | 100 (140) |
| act-2(or621)/act-2(ok1229) | na | 97 (762) |
| act-2(or621)/stDf4) | na | 100 (472) |
Individually plated L4 animals were shifted to the indicated temperature for 24 h at which time the adult was removed. Twenty-four to 36 h later the plates were scored for embryonic lethality. stDf4 is a small deficiency that has been shown to remove act-2 and -3, but not act-1(Wormbase). na, not available.
Table 2.
Embryonic lethality and embryonic cell division defects after depletion of different actin isoforms by 3′UTR-specific RNAi
| Maternal genotype | % Embryonic lethality (15°C) | Early embryonic cell division defects |
|---|---|---|
| N2 | 0 (395) | No (n = 30) |
| act-2(ok1229) | 0 (310) | No (n = 12) |
| act-1(3′UTR RNAi) | 0 (175) | No (n = 10) |
| act-3(3′UTR RNAi) | 0 (185) | No (n = 12) |
| act-1(3′UTR RNAi); act-2(ok1229) | 100 (415) | Yes (n = 11) |
| act-3(3′UTR RNAi); act-2(ok1229) | 100 (345) | Yes (n = 13) |
| act-1(3′UTR RNAi); act-3(3′UTR RNAi) | 0 (196) | No (n = 8) |
| act-1(3′UTR RNAi); act-3(3′UTR RNAi);act-2(ok1229) | 100 (230) | Yes (n = 14) |
| act-4(3′UTR RNAi); act-5(3′UTR RNAi);act2(ok1229) | 0 (178) | No (n = 8) |
Individually plated L4 animals were shifted to the indicated temperature for 24 h at which time the adult was removed. Twenty-four to 36 h later the plates were scored for embryonic lethality.
Microscopy and Immunofluorescence
For time-lapse video microscopy, gravid hermaphrodites grown at the restrictive temperature of 26°C from the L4 stage were dissected in a watch glass filled with M9 buffer. Dissected embryos were transferred with a mouth-pipette to a 3% agarose pad and covered with a 22 × 22-mm coverslip (Fisher Scientific, Pittsburgh, PA). DIC images were captured every 5 s using a Dage MT1 VE1000 digital camera and Scion Image software. For immunofluorescence staining of actin, tubulin, PGL-1, and PAR-2, we permeabilized embryos using a standard freeze-crack procedure (Bowerman et al., 1992), plunging slides with coverslips into liquid nitrogen to freeze the samples, followed by removal of the coverslip and immersion of the slide for 15 min in methanol at –20°C. All rinses and antibody dilutions used phosphate-buffered saline (PBS). We blocked specimens before antibody staining with 1% BSA (PAR-2 and PGL-1 antibodies) or 3% BSA (actin and microtubules) for 30 min. We used the following antibodies: rabbit anti-PGL-1; 1:1000 (kindly provided by Dr. Susan Strome, Indiana University), rabbit anti-PAR-2; 1:15 (kindly provided by Dr. K. Kemphues, Cornell University), anti-TBB1 (kindly provided by Dr. P. Mains, University Calgary), and anti-actin (ICN Biochemicals, Costa Mesa, CA; 1 × 500 dilution in block). Primary antibodies were incubated at 4°C overnight in a humidity chamber. FITC-conjugated goat anti-mouse or anti-rabbit secondary antibodies (Jackson ImmunoResearch Laboratory, West Grove, PA) were used at 1:200 dilutions in PBS and incubated at room temperature for at least 1 h. DNA was labeled with 0.2 μM TOTO3 iodide (Molecular Probes, Eugene, OR). Specimens were sealed under coverslips using Slow Fade (Molecular Probes) and fingernail polish (Target, Eugene, OR). Fluorescent images were obtained using a Bio-Rad MRC 1024 laser scanning confocal microscope (Richmond, CA). For simultaneous staining of actin and tubulin we used identical laser settings and exposure times for all images (see Figure 7B).
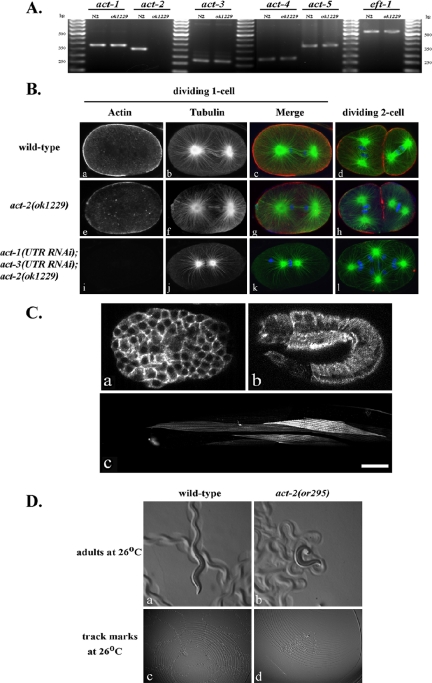
Figure 7.
Actin gene expression in oocytes, early embryos, and adult muscle; motility defects in act-2(or295) adults. (A) Amplification of actin gene transcripts from isolated oocytes. Nested RT-PCR was performed using gene-specific 3′UTR primer sequences (see Materials and Methods). (B) Immunofluorescence photomicrographs of fixed wild-type and mutant embryos stained with fluorescently labeled antibodies that recognize actin (red) or tubulin (green); DNA (blue) was labeled with TOTO3. Confocal laser strength and antibody concentration were identical in all cases (see Materials and Methods). Little or no actin was detected in act-1(3′UTR RNAi); act-3(3′UTR RNAi); act-2(ok1229) embryos (i, k, and l). Extra chromosomes in panel k presumably are due to a failure in polar body extrusion during meiosis. (C) Cytoplasmic GFP::ACT-2 expression in live embryos was detected throughout the cytoplasm and at the cortex of all early embryonic cells (a), beginning at about the 20-cell stage (unpublished data), and in epidermal cells during embryonic elongation (b). GFP::ACT-2 also was detected in contractile filaments of adult striated muscle cells (c). Scale bar, 25 μm. (D) Motility defects in act-2(or295) adult animals at the restrictive temperature. Animals were placed on individual plates for 6 h before recording images, and track marks were recorded at 400× and 125× magnification, respectively. Also see Supplementary Movies 14 and 15, in which adult worm movements were recorded at 1 frame/s. Motility defects are apparent for act-2(or295), whereas act-2(or621) mutants resemble wild-type adults in their motility.
For phalloidin staining of WT, or295, and or621 embryos (see Figure 4), we raised embryos from each strain to L4 stage at 15°C and then transferred the worms to 26°C for 12–24 h. We then cut open the gravid adults in a drop of water to release eggs directly onto 1 × 5-inch glass slides with Teflon spacers (Cell line/Erie Scientific, Portsmouth, NH), coated with 1% poly-lysine, and exposed the eggs for 1 min to 1.5% sodium hypochlorite (Sigma, St. Louis, MO), and 250 mM KOH. We then rinsed the eggs briefly three times in egg salts (118 mM NaCl, 40 mM KCl, 3.4 mM CaCl, 3.4 mM MgCl, 5 mM HEPES, pH 7.2) and allowed them to recover in egg salts for 1 min. We then fixed embryos in 3% formaldehyde and 0.2% glutaraldehyde (Electron Microscopy Sciences, Hatsfield, PA), 0.1 mg/ml lysolecithin (Sigma), 60 mM Pipes, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, 100 mM dextrose for 15 min at room temperature (RT), rinsed them briefly three times in PBS with 0.1% Triton-X (PBT), and then stained them for 1 h at RT with Bodipy fl-phallicidin (Molecular Probes), at 1 U/200 μl in PBT. We then rinsed embryos three times for 5 min in PBS; we included DAPI at 0.1 μg/ml in the final rinse to stain DNA. We mounted embryos in 15 μl Fluoromount G (Southern Biotechnology, Birmingham, AL) under No. 1.5 glass coverslips and allowed the mountant to cure for >24 h before imaging the embryos. We collected all microfilaments images using a Bio-Rad Radiance 2000 confocal scanhead mounted on a Nikon E-800 upright scope. We used identical settings for all images. We then visualized DNA in the same embryo using a Delta Vision microscope (Applied Precision, Issaquah, WA).
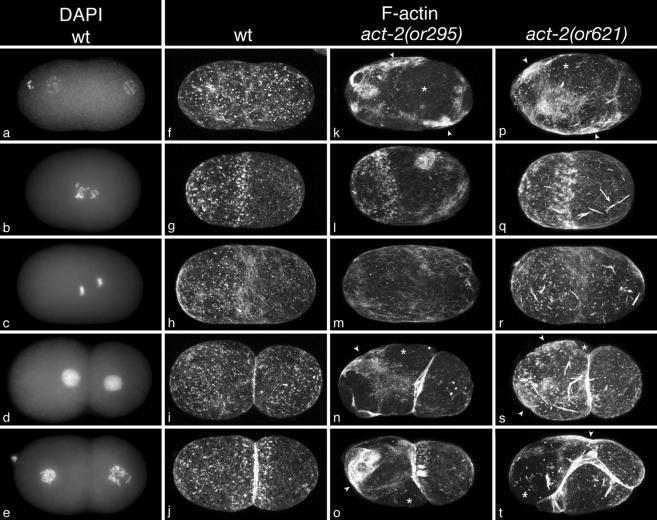
Figure 4.
Cortical microfilaments during the first two embryonic cell cycles in fixed wild-type and dominant act-2 mutant embryos. Wild-type embryos (a–j) costained with DAPI (a–e) to label DNA and Bodipy-fl-phallicidin (f–j) to label microfilaments. act-2(or295) embryos (k–o) and act-2(or621) embryos (p–t) also were stained with Bodipy-fl-phallicidin and DAPI. Embryos in each row are at the same point in the cell cycle, based on DAPI staining of chromosomes (not presented here for the mutant embryos). Each phallicidin-stained image is a maximum projection of 25 confocal sections taken at 0.25-μm intervals. All embryos were fixed and stained using the same procedures, imaged using the same confocal settings, and digitally processed identically. DAPI-stained images are maximum projections of 10–15 widefield epifluorescence images taken at 0.5-μm intervals. Both or295 and or621 embryos contain abnormally dense accumulations of cortical microfilaments (white arrowheads in k, n, o, p, s, and t). These are most pronounced within, but not restricted to, interphase embryos. Regions of dense accumulation often lie adjacent to regions that appear to be depleted of filaments relative to the wild type (white asterisks in k, n, o, p, s, and t), suggesting that microfilaments are abnormally distributed within the cortex. act-2(or621) embryos also accumulate dense microfilaments-containing structures (arrow in q), reminiscent of the barlike structures observed in yeast cells bearing the same point mutation (Chen and Rubenstein, 1995). These structures were mostly present deeper in the cytoplasm (unpublished data), but were also detected at or near the cortical surface. Late in anaphase, cortical microfilaments are reduced in density throughout the cortex outside the cleavage furrow, in both wild-type and mutant embryos (h, m, and r). The accumulation of microfilaments into a contractile ring late in anaphase was not obviously affected in the mutant embryos, although it is not apparent in this example of an or295 mutant embryo (m).
RESULTS
Abnormal Membrane Ingressions and Protrusions in or295 and or621 Mutant Embryos
To identify genes required for microfilament-dependent processes during early embryogenesis in C. elegans, we screened chemically mutagenized nematodes for temperature-sensitive (ts) embryonic-lethal mutants. From a total of ∼2300 such mutants, we found two (or295 and or621) that exhibited abnormal membrane ingressions and protrusions in early embryonic cells (Figure 1A). Both or295 and or621 are partially conditional and semidominant. At the permissive temperature of 15°C, 62% (n = 240) of embryos produced by homozygous or295 hermaphrodites (hereafter referred to as or295 mutant embryos) failed to hatch. Similarly, 60% (n = 226) of or621 mutant embryos produced at 15°C failed to hatch. At the restrictive temperature of 26°C, 98% (n = 467) of or295 mutant embryos and 100% (n = 300) of or621 mutant embryos failed to hatch (Table 1; see Materials and Methods). Furthermore, 12% (n = 300) of the embryos produced by heterozygous or295/+ mothers at 26°C and 85% (n = 325) produced by or621/+ mothers at 26°C failed to hatch (Table 1). Of the embryos from or295/+ mothers that did hatch, 22% were or295/or295 in genotype (see Materials and Methods). Thus the lethality observed in broods of heterozygous hermaphrodites was not restricted to homozygous mutant progeny but was caused by maternal expression of a dominant mutation, with or621 being substantially more dominant than or295.
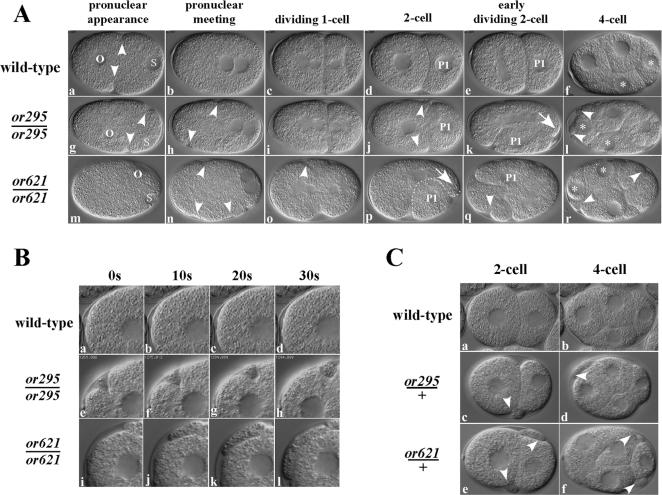
Figure 1.
Ectopic membrane ingressions and protrusions in early or295 and or621 mutant embryos. (A) DIC micrographs from time-lapse images of live wild-type (a–f), or295 (g–l) and or621 (m–r) embryos. Arrowheads indicate the pseudocleavage furrow in a wild-type embryo, and abnormal membrane ingressions and protrusions in mutant embryos (a, g, h, j, l, n, o, q, and r). Oocyte (o) and sperm (s) pronuclei in 1-cell embryos are labeled; the posterior daughter of the first mitotic division (P1) is labeled, and the two daughters of P1 are indicated with asterisks. Arrows indicate the direction of cytoplasm protrusion in AB cells of or295 and or621 mutant embryos (k and p). Beginning at the 2-cell stage, these protrusions result in the mis-positioning of embryonic cells in or295 and or621 embryos. In some or621 panels, not all nuclei are present in the focal plane shown (m). In this and all subsequent figures, anterior is to the left and posterior to the right. C. elegans embryos are ∼50 μm in length. (B) Higher magnification DIC photomicrographs of the anterior cortex of the embryonic cell AB in live 2-cell stage wild-type, or295, and or621 embryos. The membrane ingressions in mutant embryos often move along the surface of embryonic cells (or295; e–h), and the protrusions often grow in size over time (or621; i–l). Images were taken every 10 s from time-lapse video micrographs, beginning roughly at the completion of cytokinesis after the first mitotic division of each embryo. (C) Two- and 4-cell stage embryos produced by heterozygous or295/+ and or621/+ mothers raised at the restrictive temperature of 26°C often exhibit abnormal membrane ingressions and protrusions (arrowheads), due to the dominant nature of these mutations (see text for details; c-d and e-f).
To examine early embryonic cell divisions, we used differential interference contrast (DIC) microscopy to make time-lapse video micrographs, after obtaining both mutant and wild-type embryos from worms raised at the restrictive temperature of 26°C (see Materials and Methods). The time-lapse sequences we made began shortly after the completion of meiosis, when the oocyte and sperm pronuclei become visible (Figure 1). In wild-type embryos, the initially diploid oocyte chromosome content is reduced to a haploid number by the extrusion of extra chromosomes into two polar bodies, followed by appearance of the oocyte pronucleus (Albertson, 1984). The first detectable defect we observed in mutant embryos was the presence of multiple maternal pronuclei before pronuclear migration and the first mitotic division, in 2 of 24 or295 mutant embryos (8%) and in 4 of 21 or621 mutant embryos (19%). We never observed this phenotype in wild-type embryos (n = 20). These results suggest that meiosis failed in some or295 and or621 mutant embryos, presumably due to defects in meiotic cytokinesis.
The most highly penetrant defects we observed in both mutants were abnormal ingressions and protrusions of the cell surface during interphase. In wild-type one-cell embryos, a prominent pseudocleavage furrow formed during pronuclear migration and then regressed after congression of the two pronuclei (Figure 1A, a and b). Anterior to the pseudocleavage furrow, the cell surface exhibited numerous shallow membrane ingressions, whereas posterior to the pseudocleavage furrow the surface remained largely quiescent (Figure 1A; Supplementary Movie 1). Before the first mitotic division in both or295 and or621 mutant embryos, we observed abnormal membrane ingressions and, in some cases, protrusions of the cell surface anterior to the pseudocleavage furrow during pronuclear migration in one-cell stage embryos (interphase and prophase of the first mitotic cell cycle). These events were more dramatic and dynamic during interphase and prophase in 2- and 4-cell stage embryos (Figure 1, A and B; Supplementary Movies 1–5). Abnormal membrane ingressions were observed in all or295 (n = 24) and or621 (n = 21) mutant embryos. In contrast to the transient and shallow contractions observed in the anterior daughter of 2-cell stage wild-type embryos during early interphase, the deeper membrane ingressions in or295 and or621 mutant embryos persisted and often moved across the surface of embryonic cells, and the protrusions often grew in size (Figure 1B, e–h and i–l). These deeper membrane ingressions and the protrusions did not occur in wild-type embryos (Figure 1B, a–d). The abnormal membrane ingressions were more frequent in the anterior (AB) cell than in the posterior (P1) cell in 2-cell stage mutant embryos. In or295 mutants, we observed an average of 9.4 membrane ingressions during interphase in AB, whereas in P1 we observed an average of 0.4 membrane ingressions (n = 10). In or621 mutants, we observed an average of 3.8 ingressions in AB and 0.4 in P1 (n = 10). This polarized distribution suggests that the anterior enrichment of actomyosin observed in wild-type embryos (Strome, 1986; Munro et al., 2004) also occurred in these mutants. Finally, at the 2- and 4-cell stages in some or295 mutant embryos, and more often in or621 mutants, some membrane ingressions were associated with dramatic protrusions and cell migrations that disrupted the normal positions of early embryonic cells (Figure 1A, l and r; Supplementary Movies 3 and 5). We also observed abnormal membrane ingressions in embryos from heterozygous or295/+ and or621/+ mothers (Figure 1C, c-d, e-f; Supplementary Movies 6 and 7), consistent with the dominant nature of these two mutations (Table 1).
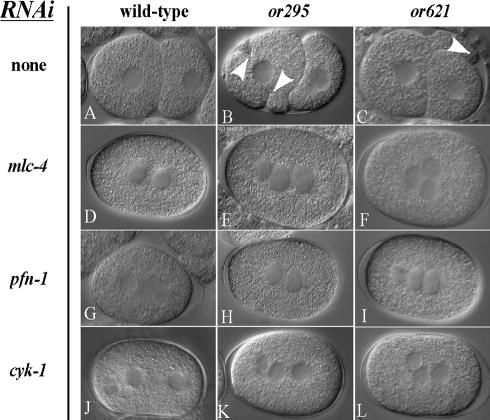
Because membrane ingressions in wild-type embryonic cells require the actomyosin cytoskeleton, we next asked if components and regulators of the cortical actomyosin cytoskeleton are required for the abnormal ingressions and protrusions observed in or295 and or621 mutant embryos. Previous work has shown that the cortical assembly of microfilaments in early embryonic cells requires a profilin called PFN-1 and a formin called CYK-1 (Severson et al., 2002). Homologues of these two proteins have recently been shown to promote the barbed-end assembly of microfilaments in other organisms (Kovar et al., 2005). Depletion of the myosin light chain MLC-4 or the nonmuscle myosin heavy chain NMY-2 does not impede the assembly or cortical enrichment of microfilaments, but these proteins are required for contractile force production in early embryonic cells (Guo and Kemphues, 1996; Shelton et al., 1999). We used RNA interference (RNAi) to deplete, in or295 and in or621 mutant embryos, PFN-1 (n = 5 for or295, n = 4 for or621), CYK-1 (n = 4 for or295; n = 5 for or621), or MLC-4 (n = 6 for or295, n = 7 for or621; see Materials and Methods). In each case, as previously shown for the normal membrane ingressions in wild-type embryos (Shelton et al., 1999; Severson et al., 2002), the abnormal ingressions and protrusions were nearly or completely eliminated, as were other microfilaments-dependent processes, including cytokinesis (Figure 2; see Supplementary Movies 8–13). We conclude that the membrane ingressions observed in or295 and or621 mutant embryos result from and require actomyosin-dependent contractile forces.
Figure 2.
Ectopic membrane ingressions and protrusions in dominant act-2 mutant embryos require a functional microfilament cytoskeleton. DIC photomicrographs of 2-cell stage wild-type, or295, and or621 embryos (A–C) and equivalent stage wild-type, or295 and or621 embryos after RNAi-mediated depletion of actomyosin function (D–L). Arrowheads indicate ectopic ingressions (B) and a protrusion (C) in the mutant embryos. Depletion of actomyosin function prevents cytokinesis in all embryos and eliminates, or nearly eliminates, abnormal membrane ingressions and protrusions in the mutant embryos (C–F). The presence of more than two nuclei in many of the RNAi-depleted embryos is due to failures of meiotic cytokinesis and consequent aneuploidy (unpublished data). Also see Supplementary Movies 8–13.
Although the abnormal membrane ingressions in or295 and or621 mutant embryos appear to involve abnormal cortical actomyosin function, we did not detect any failure of cytokinesis in time-lapse videomicrographs of the first mitotic division or the divisions of AB and P1 at the two-cell stage (n = 18 for or295; n = 15 for or621). As noted above, most of the abnormal membrane ingressions and protrusions occurred during interphase and early mitosis. During cytokinesis, ectopic furrows and protrusions were reduced in number and size (Supplementary Movies 2–5). These observations suggest that cortical actomyosin function in these mutant embryos was relatively normal during mitotic cytokinesis.
To characterize mitotic cytokinesis in more detail, we examined the timing of furrow initiation and the time required for completion of furrowing, during the first cell division in or295, or621 and wild-type embryos, using DIC time-lapse videomicroscopy. We first measured the duration of mitosis, using nuclear envelope breakdown (NEB) and nuclear envelope reformation (NER) as rough markers for the beginning and end of mitosis. In dividing one-cell stage embryos, the duration of mitosis for or295 and or621 mutants was on average 1.3 and 1.5 times longer than normal: wild type = 5.8 min (n = 8); or295 = 7.3 min (n = 10); or621 = 8.6 min (n = 10). We next asked if this delay was restricted to either the time between NEB and cytokinesis furrow initiation or the time from cytokinesis furrow initiation to NER. We defined the initiation of cytokinesis as the first appearance of a detectable cleavage furrow that progressed without regression toward the central spindle. We found that the average time between NEB and cytokinesis furrow initiation in or295 and or621 mutant embryos was, respectively, 1.5- and 1.6-fold longer than normal: wild-type = 2.8 min (n = 8); or295 = 4.1 min (n = 10); or621 = 4.5 min (n = 10). In contrast, the average time from the beginning of cytokinesis to NER for or295 embryos was relatively normal: wild-type = 3.0 min (n = 8); or295 = 3.2 min (n = 10). However, in or621 mutant embryos this latter interval was also longer than normal (4.1 min, or 1.4-fold longer; n = 10). Modest spindle checkpoint-dependent delays in mitosis have been reported in C. elegans mutants with defective mitotic spindles in early embryonic cells (Encalada et al., 2005). However, we have not detected obvious defects in mitotic chromosome segregation in or295 and or621 mutants (Figure 1, and unpublished data), and the timing of these events was determined using only those embryos in which meiosis appeared to occur normally (with only a single maternal pronucleus present). Thus the increased time required for mitosis may reflect delays in the initiation and duration of cytokinesis.
or295 and or621 Are Alleles of act-2
To determine the identity of the gene(s) mutated in or295 and or621 mutants, we first used genetic mapping to determine the chromosomal location of these mutations. We found that both or295 and or621 are positioned at about + 3.0 map units on chromosome V (see Materials and Methods). Three of the five C. elegans actin genes reside in a cluster at roughly this location: act-1/T04C12.4, act-2/T04C12.5, and act-3/T04C12.6 (Figure 3A). We therefore sequenced the act-1, -2, and -3 genes from genomic DNA isolated from homozygous or295 and or621 mutant worms and amplified using PCR (Materials and Methods). We did not find any mutations in the coding sequences of either act-1 or -3, but in both mutants we identified mis-sense mutations near the beginning of the act-2 open reading frame: a glycine to arginine mutation at codon 15 in or295, and a serine to alanine mutation at codon 14 in or621 (Figure 3, B and C). Consistent with both mutations affecting act-2 function, we also found that or295 and or621 failed to complement each other at the permissive temperature: none of the embryos produced at 15°C by or295/or621 mothers hatched, whereas nearly all embryos produced at 15°C by or295/+ and or621/+ mothers hatched (Table 1). We conclude that or295 and or621 are mutant alleles of the C. elegans act-2 gene. Both mutations change amino acids in the ATP-binding pocket of actin, as extrapolated from the structure of C. elegans actin (Figure 3 legend; Figure 3, B and C; Vorobiev et al., 2003). Identical actin mutations have been identified in other organisms (Chen and Rubenstein, 1995; Costa et al., 2004).
Altered Distributions of Cytoplasmic Microfilaments in act-2(or295) and act-2(or621) Mutant Embryos
To investigate the effects of the dominant act-2(or295) and act-2(or621) mutations on microfilaments in early embryonic cells, we fixed wild-type and mutant embryos and examined the distribution and organization of microfilaments using fluorescently labeled phalloidin (Materials and Methods). As documented previously (Strome, 1986; Munro et al., 2004), we observed focal accumulations of microfilaments in the cortex of early one-cell wild-type embryos (Figure 4F). These microfilament foci are associated with myosin-dependent contractions that drive cortical flow and membrane ingressions during polarization of the anterior-posterior body-axis in one-cell zygotes (Munro et al., 2004). We also observed the previously documented anterior enrichment of cortical microfilaments that occurs before the first mitotic division in wild-type embryos (Figure 4G) and the subsequent enrichment of microfilaments in the contractile ring that occurs during cytokinesis (Strome, 1986; Munro et al., 2004). In act-2(or295) and act-2(or621) mutant embryos, the general distribution of microfilaments throughout the first two mitotic cell cycles appeared relatively normal. Microfilaments were distributed throughout the cortex early in the one-cell stage (Figure 4, k and p), subsequently became enriched anteriorly as the zygote entered mitosis (Figure 4, l and q), and then concentrated in the cleavage furrow during cytokinesis (Figure 4, m–o and r–t). However, in both mutants we observed relatively large and dense patches of cortical microfilaments, particularly during interphase of the first and second cell cycles (Figure 4, k, n–p, and s), when actomyosin contractility is distributed evenly in small foci throughout the cortex in wild-type embryos (Munro et al., 2004). Early in the first mitosis in the mutant embryos, we detected these more intensely staining patches within the microfilament-enriched anterior portion of the cortex and also within the posterior cortex (Figure 4, l and q), which is largely depleted of cortical microfilaments at this time in wild-type embryos (Figure 4g). We also observed increased levels of microfilaments between the cell boundaries in 2-cell stage mutant embryos (Figure 4, n, o, s, and t). Dense patches of microfilaments in 2-cell stage mutant embryos were more pronounced in the anterior AB cell than in the posterior P1 cell, consistent with the greater number of cortical ingressions observed in AB in act-2(or295) and act-2(or621) mutant embryos (see above). Although the deep cortical ingressions observed in live mutant embryos were lost during the fixation process, we often observed shallow ingressions associated with dense patches of cortical microfilaments (arrowheads in Figure 4, n, o, s, and t).
Intriguingly, we also observed areas of the cortex with a pronounced depletion of microfilaments in act-2(or295) and act-2(or621) mutant embryos (Figure 4, k and p), in addition to the more restricted dense accumulations of microfilaments. Thus the denser accumulations may represent a redistribution of microfilaments within the plane of the cortex. As has been documented for wild-type embryos (Munro et al., 2004), these increased microfilament densities likely correspond to foci of contractile NMY-2 associated with the ectopic membrane ingressions that we observed in live mutant embryos (Figures 1 and 2). Intriguingly, we also observed prominent, intensely staining barlike microfilaments structures in act-2(or621) embryos (Figure 4, q–t). Although we observed some barlike structures at the cortex (Figure 4, q–t), most were detected away from the cortex in the deeper cytoplasm (unpublished data).
Although we observed abnormal cortical microfilament distributions throughout most of the cell cycles in or295 and or621 mutant embryos, one exception occurred during anaphase. Ectopic accumulations of cortical microfilaments were nearly absent outside the cleavage furrow during anaphase in the mutant embryos (Figure 4, m and r). The absence of abnormal microfilament aggregations late in mitosis may explain why, in spite of the abnormal membrane ingressions that occurred throughout most of the cell cycle, we never observed cytokinesis failures during mitosis in act-2(or295) and act-2(or621) mutant embryos (see above).
Cell Polarity Appears Normal in act-2(or295) and act-2(or621) Mutant Embryos
Both time-lapse DIC videomicroscopy of live embryos (Figure 1) and the distribution of microfilaments in fixed embryos (Figure 4) suggest that the actomyosin-dependent establishment of anterior-posterior polarity at the one-cell stage (reviewed in Schneider and Bowerman, 2003) occurs normally in act-2(or295) and act-2(or621) mutant embryos. In time-lapse videomicrographs of mutant embryos (n = 18 for or295, n = 16 for or621), as in wild-type embryos, the pronuclei met near the posterior pole and moved to the center of the zygote before assembly of the first mitotic spindle (Figure 1; Supplementary Movies 1–5). By late anaphase, the first mitotic spindle was displaced posteriorly in all mutant embryos and the posterior daughter divided after the anterior daughter. Moreover, microfilaments in both mutants became enriched in the anterior portion of the embryo before the first mitotic division (Figure 4). These asymmetries also occur in wild-type embryos and indicate that anterior-posterior cell polarity in the one-cell zygote was not substantially affected by these dominant act-2 mutations.
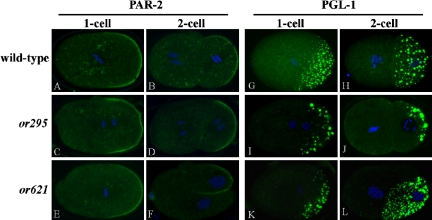
To test further for any defects in the establishment of an anterior-posterior body-axis, we examined the distribution of the cell polarity regulator PAR-2, which localizes to the posterior cortex in both the one-cell stage zygote and in P1, the posterior daughter produced by the first mitotic division (Boyd et al., 1996). We also examined the distribution of germline P granules, which are enriched in the posterior cytoplasm of the one-cell zygote and in P1 (Kawasaki et al., 1998). We stained fixed wild-type and mutant embryos with antibodies that recognize PAR-2 or antibodies that recognize the P granule component PGL-l (Boyd et al., 1996; Kawasaki et al., 1998). Both PAR-2 and P granules appeared to localize normally in act-2(or295) and act-2(or621) mutant embryos (Figure 5). We conclude that although the distribution and contractile properties of microfilaments are abnormal in act-2(or295) and act-2(or621) mutants, they nevertheless are competent for the establishment of anterior-posterior polarity. However, we have not examined the rate of polarization, and it remains possible that, as for cytokinesis, the dynamics of polarization were affected.
Figure 5.
Anterior-posterior polarity appears normal in act-2 mutant embryos. Fixed wild-type, act-2(or295), and act-2(or621) embryos stained with fluorescently labeled antibodies that recognize the polarity regulatory protein PAR-2 (A–F) and the P granule component PGL-1 (G–L). Both proteins are localized normally in 1- and 2-cell stage act-2(or295) and act-2(or621) mutant embryos. Note that in F, even though the P1 cell is becoming abnormally positioned, PAR-2 maintains a posterior cortical localization.
The act-2 Gene Is Not Required for Early Embryonic Cell Division
Although previous studies have shown that microfilaments are required for multiple processes during early embryogenesis (Hill and Strome, 1988, 1990), our identification of dominant mutations in act-2 provides the first conclusive evidence for a specific C. elegans actin isoform being involved in cytoplasmic microfilament function in early embryonic cells. However, our analysis of these two mutants does not address whether act-2 is required for early embryonic processes or whether other actin genes also contribute to embryonic actin pools. To examine the requirements for act-2, we obtained and analyzed a deletion allele, act-2(ok1229), generated by the C. elegans knockout consortium (http://www.celeganskoconsortium.omrf.org). This deletion begins in the second exon, extends 705 base pairs beyond the act-2 3′UTR and removes 489/1131 base pairs of coding sequence (Figure 3A). Although ok1229 appears to be a null allele, we did not detect any defects during the first mitotic division of the zygote in mutant embryos produced by homozygous act-2(ok1229) animals (n = 16). Indeed, all embryos produced at 15°C hatched, although 12% of the embryos produced by mothers raised at 26°C failed to hatch (Table 1). We conclude that act-2 is not required for early embryonic cell divisions and probably functions redundantly with other actin genes at this stage.
act-1, -2, and -3 Function Redundantly in Early Embryonic Cells
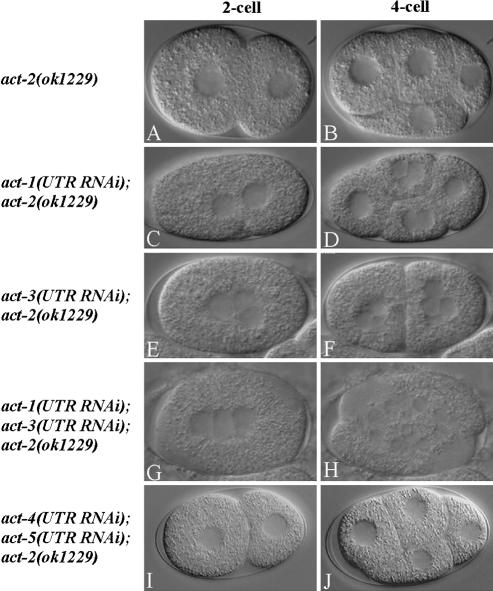
To determine if other actin genes are required redundantly with act-2 in the early embryo, we used RNAi to reduce the function of act-1, -2, -3, -4, and -5 in act-2(ok1229) mutant worms. Because the coding sequences of the different actin genes are highly conserved and thus might cross-react if used for RNAi, we used the 3′UTR sequences, which range from 26 to 29% in nucleotide identity, to make dsRNA for microinjection into the ovaries of act-2(ok1229) worms (Materials and Methods). We then determined the percent embryonic lethality and made DIC time-lapse videomicrographs of individual embryos, 20–24 h after microinjection of dsRNAs. Microinjection of act-2 3′UTR dsRNA had no effect on embryonic viability in the act-2(ok1229) background (Table 2), which was expected as the deletion removes the 3′UTR. Furthermore, although microinjection of either act-1 or -3 3′UTR dsRNAs into wild-type worms did not result in any early embryonic defects (unpublished data), microinjection of either act-1 or -3 3′UTR dsRNA into act-2(ok1229) worms resulted in 100% embryonic lethality (Table 2) and in cytokinesis defects during early embryonic cell divisions (Figure 6, C and E). We observed more severe cytokinesis defects when act-1 and -3 3′UTR dsRNAs were coinjected into act-2(ok1229) animals: cytokinesis was never successful, even at the second attempt (Table 2; Figure 6, D, F, and H). We did not detect any cytokinesis defects when act-4 and -5 3′UTR dsRNAs were microinjected, separately or together, into act-2(ok1229) worms (Table 2, Figure 6, I and J). Nevertheless, microinjection of act-4 and -5 3′UTR dsRNAs did result in highly penetrant embryonic or early larval lethality (Table 2), indicating that RNAi reduced gene function and presumably affected other processes later in development. We conclude that act-1, -2, and -3 are required redundantly for microfilament-dependent processes in early embryonic cells, whereas act-4 and -5 probably make at most minor contributions to cytoplasmic microfilament function in early embryos.
Figure 6.
act-1, -2, and -3 are redundantly required for early embryonic cell divisions. DIC photomicrographs of live 2- and 4-cell stage embryos from mothers homozygous for the act-2(ok1229) deletion (A and B) and equivalent stage embryos from act-2(ok1229) mothers after RNAi-mediated depletion of other actin isoforms, using 3′UTR dsRNA sequences specific for each isoform (see text and Materials and Methods for details). Early cell divisions appeared normal in embryos from act-2(ok1229) worms, although some embryonic lethality occurred in embryos produced at 26°C (Table 1). Depletion of either ACT-1 or -3 in the act-2(ok1229) background resulted in 100% embryonic lethality (Table 2) and in cytokinesis defects and thus multinucleate embryonic cells (C–F). Depletion of both ACT-1 and -3 in act-2(ok1229) embryos resulted in more severe cytokinesis defects (H). Depletion of both ACT-4 and -5 in act-2(ok1229) worms did not result in early cell division defects (I and J). See Table 2 for the number of embryonic cell divisions scored for cytokinesis defects. Also see Supplementary Movies 14–16.
To further examine the contributions of different actin genes, we used reverse transcriptase and PCR (RT-PCR) to detect specific actin isoform mRNAs, and immunolocalization to detect total actin protein. First, we isolated oocytes from wild-type and act-2(ok1229) mutant adults and then performed nested RT-PCR, using a 3′ primer complementary to the 3′UTR sequences specific for each actin gene and a 5′ primer corresponding to exon sequences 5′ of the last intron in each gene (Materials and Methods). We detected mRNA for all five actin genes in wild-type oocytes and mRNAs for each actin gene, except act-2, in oocytes from act-2(ok1229) worms (Figure 7A). In addition, we detected act-2 transcripts in both act-2(or295) and act-2(or621) mutant embryos (Supplementary Figure 1), confirming that act-2 transcripts are present in these dominant mutants. Finally, we used an anti-actin antibody to stain fixed embryos from wild-type and act-2(ok1229) worms and embryos from act-2(ok1229) worms 20–24 h after using microinjection of 3′UTR-specific dsRNAs to deplete both ACT-1 and -3. In wild-type and act-2(ok1229) embryos, we detected actin throughout the cortex of early embryonic cells (Figure 7B, a, c–e, g, and h). In contrast, we detected little or no cortical actin in embryos from act-2(ok1229) worms after microinjection of both act-1 and -3 dsRNAs (n = 15 embryos; Figure 7B, i, k, and l). These results are consistent with our functional analysis suggesting that act-1, -2, and -3 are required redundantly for cytoplasmic microfilament function in the early C. elegans embryo.
ACT-2 Is Expressed and Required in Both Nonmuscle and Muscle Cell Types
Dominant mutations in act-1 and -3 have been shown previously to result in severe locomotion defects and abnormal muscle morphology, and the data we report here indicate that they also function cytoplasmically in early embryos. We therefore asked if act-2 might also function in other cell types, including muscle cells, which previous reports have suggested (Avery, 1993). To address this possibility, we first constructed a GFP::ACT-2 translational fusion, using the endogenous act-2 promoter, coding sequences, and 3′ region (Materials and Methods). We used this construct to generate a transgenic line that expresses GFP::ACT-2 from an extrachromosomal array (Materials and Methods). In embryos produced by transgenic hermaphrodites, we first detected GFP expression in embryos beginning at roughly the 20-cell stage (unpublished data), at the cortex, and throughout the cytoplasm of all embryonic cells (Figure 7C, panel A). Our failure to detect maternal expression in earlier stage embryos presumably was due to germline silencing of the extrachromosomal array, which often occurs with transgenes in C. elegans (Kelly and Fire, 1998). In addition to the cytoplasmic expression we observed throughout early embryonic cells, we also detected prominent GFP expression in the epidermis during elongation of the round embryo into a long, thin larva (Figure 7C, b). Actomyosin contractile forces are known to drive epidermal cell shape changes and elongation of the embryo in C. elegans (Priess and Hirsh, 1986). Thus it appears likely ACT-2 participates in this morphogenetic process, although our data do not conclusively demonstrate that GFP::ACT-2 colocalizes with endogenous actin in the circumferential actin bundles that mediate embryonic elongation. In adult animals, we reproducibly observed GFP expression in body wall muscle cells, incorporated into contractile filaments (Figure 7C, c), and in numerous neurons (unpublished data). To ask if ACT-2 functions in body wall muscle, we examined the motility of adult worms that were shifted as L1 larvae from the permissive temperature of 15°C to the restrictive temperature of 26°C until reaching adulthood (Materials and Methods). We observed locomotive defects in act-2(or295) mutant adults, but not in either wild-type worms or act-2(or621) mutants (Figure 7D; Supplementary Movies 14 and 15; unpublished data). We conclude that, like ACT-1 and -3, ACT-2 functions in both muscle and nonmuscle cell types.
DISCUSSION
The two conditional and dominant mutations we have identified in C. elegans ACT-2 alter conserved and adjacent amino acids that are predicted to form part of the ATP binding pocket. At the restrictive temperature of 26°C, these mutations result in similar but distinguishable defects, and in highly penetrant embryonic lethality. Although these dominant mutations implicate ACT-2 in early embryonic processes, a recessive deletion that removes a large portion of the act-2 gene does not result in early embryonic cell division defects and causes only a weakly penetrant embryonic lethality. Consistent with this weak requirement for act-2, we have shown that act-1 and -3 are required redundantly with act-2 for cytoplasmic microfilament function in early embryonic cells. As all three of these actin genes have been implicated in muscle function, our results indicate that multiple actin isoforms in C. elegans are required for actomyosin contractility in both nonmuscle and muscle cell types.
Actin Gene Redundancy in the Early C. elegans Embryo
Although the genomes of budding and fission yeast each have only a single, essential actin gene, multicellular eukaryotic organisms have multiple actin genes that encode identical or nearly identical isoforms. The C. elegans genome includes five actin genes. Two, act-1 and -3, have identical open reading frame nucleotide sequences and reside in a cluster with act-2, which encodes an isoform 99% identical in amino acid sequence compared with the actin isoform encoded by its two neighbors. Similarly, ACT-4 exhibits 99% identity, whereas the ACT-5 amino acid sequence is the most divergent, with 93% identity compared with ACT-1 and -3 (Krause et al., 1989; Macqueen et al., 2005). Our identification of dominant mutations in act-2 implicates this isoform in early embryonic cytoplasmic processes. Our use of 3′UTR-specific dsRNAs, to knock down by RNAi actin gene expression in a strain deleted for act-2, indicates that act-1, -2, and -3 are required redundantly during early embryonic cell divisions. Consistent with this conclusion, we detected transcripts for the act-1, -2, and -3 genes after using RT-PCR to amplify cDNAs from isolated oocytes. Although we also detected both act-4 and -5 transcripts in isolated oocytes using RT-PCR, we did not detect any requirements for these two genes during early embryonic cell divisions. Although we cannot rule out early embryonic functions for act-4 and -5, the use of 3′UTR-specific RNA to deplete these isoforms did result in highly penetrant embryonic and larval lethality, suggesting that RNAi effectively reduced the function of each gene. Furthermore, a recent study has shown that act-5 is required only in cells with microvilli, and expression of ACT-1 driven by the act-5 promoter cannot replace ACT-5. Moreover, Macqueen et al. (2005) used an ACT-5-specific antibody to show that this isoform is expressed specifically only within cells that produce microvilli; no early embryonic expression was reported. We conclude that act-1, -2, and -3 are required redundantly for cytoplasmic processes in the early embryo, whereas act-4 and -5 are less likely to be involved in these processes in spite of their maternal expression in oocytes.
Multiple C. elegans Actin Isoforms Are Required for Both Muscle and Nonmuscle Actomyosin Contractility
Our analysis of the redundant cytoplasmic requirements for three C. elegans actin genes is surprising because the same genes that we have shown are required for cell division in the early embryo also are known to be involved in muscle contractile function. A previous analysis of act-1, -2, and -3 suggested that these three isoforms are required redundantly for body wall muscle function (Landel et al., 1984), and a mutant with mis-sense mutations in both act-2 and -3 has pharyngeal muscle pumping defects (Waterston et al., 1984; Avery, 1993; Stone and Shaw, 1993). In addition, we have shown that a GFP::ACT-2 fusion protein driven by the act-2 promoter is expressed cytoplasmically in multiple cell types and is incorporated into the contractile apparatus of adult muscle cells. Furthermore, adult act-2(or295) mutants shifted to the restrictive temperature exhibit defects in locomotion, consistent with ACT-2 functioning in body wall muscle cells. We conclude that in C. elegans act-1, -2, and -3 are involved both in nonmuscle and muscle functions.
Our analysis of C. elegans actin isoform requirements contrasts sharply with several studies of actin isoforms in vertebrates and in Drosophila, which have shown that many actin isoforms are restricted in function to either muscle or nonmuscle roles (see Introduction). These restrictions have been suggested to reflect possible differences in microfilaments dynamics that depend on isoform composition, with muscle cell microfilaments being more stable than cytoplasmic microfilaments (Khaitlina, 2001). Our findings in C. elegans provide conclusive documentation that some actin isoforms can function in both cytoplasmic and muscle contractile processes. Consistent with these findings, act-1, -2, and -3 encode amino acids at several positions that are conserved in cytoplasmic isoforms, and amino acids at other positions that are conserved in vertebrate muscle actin isoforms (unpublished data).
Although our results indicate that act-1, -2, and -3 are required redundantly in the early embryo, the extent to which these three genes function in cytoplasmic processes later in development remains to be determined. Our findings that a GFP::ACT-2 translational fusion is expressed cytoplasmically throughout early stage embryos and in epidermal cells during morphogenetic processes later in development suggest that at least ACT-2 has additional cytoplasmic roles. However, the production of microvilli specifically requires ACT-5, which functions only in intestinal cells and a few other microvilli-containing cell types (Macqueen et al., 2005). This requirement appears specific for ACT-5, as the engineered expression of ACT-1 within intestinal cells could not compensate for the loss of ACT-5 (Macqueen et al., 2005). Thus at least this cytoplasmic role in C. elegans requires a distinct actin isoform. Similarly, a smooth muscle actin isoform cannot compensate for loss of a cardiac actin during mouse development (Kumar et al., 1997), and a larval muscle actin isoform in Drosophila cannot compensate for loss of an adult muscle isoform (Fyrberg et al., 1998). These results indicate that in some cases both cytoplasmic and muscle processes exhibit highly specific actin isoform requirements.
Dominant Mutations in act-2 Alter Conserved Amino Acids Previously Implicated in ATP Hydrolysis and Microfilament Dynamics
Both of the amino acids altered in the dominant act-2 mutants that we identified have been implicated in ATP binding and hydrolysis, which play critical roles in actin structure, function, and dynamics (Sablin et al., 2002; Vorobiev et al., 2003). Crystallographic data, based on studies of actin from other organisms, suggest that six amino acids, including the two affected by the dominant mutations in C. elegans act-2, form hydrogen bonds with the γ- and β-phosphates of ATP, stabilizing the nucleotide-protein complex (Kabsch et al., 1990; Kabsch and Vandekerckhove, 1992). These six amino acids are present in the actin subdomains 1 (Ser14, Gly15, Met16, Leu16) and 3 (Asp157, Gly158, Val159). Five of these amino acids, including Ser14 and Gly15, are conserved in all animal and plant actin isoforms and are thought to play critical roles in mediating the conformational changes associated with the hydrolysis of ATP to ADP (Sablin et al., 2002).
An extensive mutational analysis of the sole actin in S. cerevisiae has shown that Ser14 in subdomain 1 is important for actin function both in vivo and in vitro (Chen et al., 1995; Chen and Rubenstein, 1995; Schuler et al., 1999). Budding yeast cells expressing S14A mutant actin displayed a temperature-sensitive lethality that is preceded by the disappearance of detectable actin cables and patches and the appearance of abnormal barlike structures (Chen and Rubenstein, 1995). In addition, in vitro studies of this mutant actin documented a 40–60-fold decrease in the affinity of actin for ATP and a decreased rate of ATP hydrolysis, in addition to an increased rate of polymerization and an altered susceptibility of microfilaments to protease treatment (Chen et al., 1995; Chen and Rubenstein, 1995). Our analysis of the semidominant S14A act-2(or621) mutation in C. elegans revealed similarities to the corresponding mutation in yeast actin. These include a temperature-sensitive lethal phenotype and the appearance of abnormal barlike actin structures at the cortex and in the cytoplasm. The altered distribution of cortical microfilaments in act-2(or621) mutant embryos may reflect changes in actin dynamics consistent with the in vitro observations of a decreased rate of ATP hydrolysis and an increased rate of polymerization for yeast actin. In both the yeast and worm mutants, the accumulation of abnormal barlike microfilaments structures might reflect an alteration of microfilaments dynamics, with more stable microfilaments accumulating in dense aggregates and failing to recycle or redistribute more uniformly throughout the cortex.
A mutation identical to that in act-2(or295), G15R, has been reported in a human muscle actin called α-skeletal-muscle actin, or ACTA1. The corresponding ACTA1 mutation causes a dominant and lethal congenital myopathy in affected infants (Costa et al., 2004). Expression of this mutant actin in cell culture produced aggregated microfilaments structures, suggesting it is incorporated into filaments but interferes with normal filament structure or dynamics (Costa et al., 2004). Interestingly, we observed motility defects only in act-2(or295) animals, even though act-2(or621) results in a more highly penetrant and a more strongly dominant embryonic lethality. Moreover, act-2(or621) mutants exhibit more penetrant defects in meiosis, require more time to initiate and complete cytokinesis during mitosis, and undergo more prominent membrane protrusions. Thus the act-2(or621) S14A mutation more severely affects cytoplasmic processes, whereas only the act-2(or295) G15R mutation disrupts muscle function. Intriguingly, barlike MF aggregates in early embryonic cells are observed only in act-2(or621) mutants. Perhaps the act-2(or621) S14A mutation results in a more substantial stabilization of microfilaments and thereby disrupts cytoplasmic processes that require more dynamic microfilaments, without affecting the contractile properties in muscle cells that are thought to depend on more stable microfilaments (Khaitlina, 2001). To our knowledge, the effects of the act-2(or295) G15R mutation on actin properties and microfilaments dynamics in vitro have not been reported, making it more difficult to speculate on the consequences of this mutation. Nevertheless, one possibility is that the substitution of glycine with the much larger arginine in act-2(or295) disrupts the proper assembly or conformation of muscle actin myofibrils, independent of any effects on microfilament dynamics.
The dominant mutations we have identified in C. elegans act-2 both lead to abnormal contractility and altered cortical microfilament distribution in early embryonic cells. As we did not detect an increase in actin levels in mutant embryo extracts (unpublished data), we speculate that these mutations either increase the rate of microfilament polymerization, or increase microfilament stability, and thereby disrupt the normal turnover and distribution of cortical microfilaments. Localized actomyosin-based contractions associated with anterior-posterior body-axis polarization in wild-type embryos appear to involve a limited form of contractile instability (Munro et al., 2004). During axis polarization, focal contractions appear to be limited in size and duration by rapid actomyosin turnover within the foci (E. Munro, unpublished data). We speculate that the abnormal ingressions and protrusions we observe in act-2(or295) and act-2(or621) mutant embryos could involve an exaggerated form of the same contractile process, augmented by decreased microfilament turnover. In both mutants, the abnormal ingressions are associated with the accumulation of abnormally large and dense cortical microfilaments patches. Thus an important mechanism for the regulation of actomyosin contractility, particularly for cortical cytoplasmic function in nonmuscle cells, may be the modulation of microfilament polymerization or stability. Finally, it is interesting to note that the reduction in cortical microfilaments outside the cleavage furrow that occurs during late anaphase in wild-type embryos also occurs in act-2(or295) and act-2(or621) mutant embryos, despite their earlier accumulation of abnormally large and dense microfilament foci during interphase and early mitosis. This result suggests that a robust mechanism operates to down-regulate cortical microfilament stability outside the cleavage furrow during cytokinesis. The dominant mutations we have identified in C. elegans act-2 may therefore provide useful genetic tools for identifying factors that modulate microfilaments dynamics during cell polarization and cell division.
Supplementary Material
Acknowledgments
We thank Yuji Kohara for providing cDNA clones, the C. elegans Genetics Center, funded by the National Institutes of Health (NIH), and the C. elegans Gene Knockout Consortium, for providing strains. We are grateful to Chris Doe for use of the Bio-Rad Confocal microscope and Ken Kemphues and Susan Strome for the anti-PAR-2 and anti-PGL-1 antibodies. We thank Aaron Severson for initial characterization and genetic mapping of or295. We thank Julie Canman, Chris Doe, Morgan Goulding, Peter Rubenstein, and an anonymous reviewer for helpful comments on this manuscript. This work was supported by a NIH training grant (J.W.), by NIH Grant GM66050 (E.M.), and by NIH Grant R01GM58017 (B.B.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–09–0886) on January 11, 2006.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Albertson, D. G. (1984). Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 101, 61–72. [DOI] [PubMed] [Google Scholar]
- Alder, M. N., Dames, S., Gaudet, J., and Mango, S. E. (2003). Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA 9, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (1991). Current Protocols in Molecular Biology, New York: Greene Publishing Associates and Wiley-Interscience.
- Avery, L. (1993). The genetics of feeding in Caenorhabditis elegans. Genetics 133, 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman, B., Eaton, B. A., and Priess, J. R. (1992). skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68, 1061–1075. [DOI] [PubMed] [Google Scholar]
- Boyd, L., Guo, S., Levitan, D., Stinchcomb, D. T., and Kemphues, K. J. (1996). PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development 122, 3075–3084. [DOI] [PubMed] [Google Scholar]
- Brault, V., Reedy, M. C., Sauder, U., Kammerer, R. A., Aebi, U., and Schoenenberger, C. (1999). Substitution of flight muscle-specific actin by human (beta)-cytoplasmic actin in the indirect flight muscle of Drosophila. J. Cell Sci. 112 (Pt 21), 3627–3639. [DOI] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Peng, J., Pedram, M., Swenson, C. A., and Rubenstein, P. A. (1995). The effect of the S14A mutation on the conformation and thermostability of Saccharomyces cerevisiae G-actin and its interaction with adenine nucleotides. J. Biol. Chem. 270, 11415–11423. [DOI] [PubMed] [Google Scholar]
- Chen, X., and Rubenstein, P. A. (1995). A mutation in an ATP-binding loop of Saccharomyces cerevisiae actin (S14A) causes a temperature-sensitive phenotype in vivo and in vitro. J. Biol. Chem. 270, 11406–11414. [DOI] [PubMed] [Google Scholar]
- Costa, C. F., Rommelaere, H., Waterschoot, D., Sethi, K. K., Nowak, K. J., Laing, N. G., Ampe, C., and Machesky, L. M. (2004). Myopathy mutations in alpha-skeletal-muscle actin cause a range of molecular defects. J. Cell Sci. 117, 3367–3377. [DOI] [PubMed] [Google Scholar]
- Dixon, S. J., and Roy, P. J. (2005). Muscle arm development in Caenorhabditis elegans. Development 132, 3079–3092. [DOI] [PubMed] [Google Scholar]
- Encalada, S. E., Martin, P. R., Phillips, J. B., Lyczak, R., Hamill, D. R., Swan, K. A., and Bowerman, B. (2000). DNA replication defects delay cell division and disrupt cell polarity in early Caenorhabditis elegans embryos. Dev. Biol. 228, 225–238. [DOI] [PubMed] [Google Scholar]
- Encalada, S. E., Willis, J., Lyczak, R., and Bowerman, B. (2005). A spindle checkpoint functions during mitosis in the early Caenorhabditis elegans embryo. Mol. Biol. Cell 16, 1056–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Files, J. G., Carr, S., and Hirsh, D. (1983). Actin gene family of Caenorhabditis elegans. J. Mol. Biol. 164, 355–375. [DOI] [PubMed] [Google Scholar]
- Fyrberg, E. A., Bond, B. J., Hershey, N. D., Mixter, K. S., and Davidson, N. (1981). The actin genes of Drosophila: protein coding regions are highly conserved but intron positions are not. Cell 24, 107–116. [DOI] [PubMed] [Google Scholar]
- Fyrberg, E. A., Fyrberg, C. C., Biggs, J. R., Saville, D., Beall, C. J., and Ketchum, A. (1998). Functional nonequivalence of Drosophila actin isoforms. Biochem. Genet. 36, 271–287. [DOI] [PubMed] [Google Scholar]
- Fyrberg, E. A., Kindle, K. L., and Davidson, N. (1980). The actin genes of Drosophila: a dispersed multigene family. Cell 19, 365–378. [DOI] [PubMed] [Google Scholar]
- Guo, S., and Kemphues, K. J. (1996). A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature 382, 455–458. [DOI] [PubMed] [Google Scholar]
- Herman, I. M. (1993). Actin isoforms. Curr. Opin. Cell Biol. 5, 48–55. [DOI] [PubMed] [Google Scholar]
- Hill, D. P., and Strome, S. (1988). An analysis of the role of microfilaments in the establishment and maintenance of asymmetry in Caenorhabditis elegans zygotes. Dev. Biol. 125, 75–84. [DOI] [PubMed] [Google Scholar]
- Hill, D. P., and Strome, S. (1990). Brief cytochalasin-induced disruption of microfilaments during a critical interval in 1-cell C. elegans embryos alters the partitioning of developmental instructions to the 2-cell embryo. Development 108, 159–172. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., Mannherz, H. G., Suck, D., Pai, E. F., and Holmes, K. C. (1990). Atomic structure of the actin:DNase I complex. Nature 347, 37–44. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., and Vandekerckhove, J. (1992). Structure and function of actin. Annu. Rev. Biophys. Biomol. Struct. 21, 49–76. [DOI] [PubMed] [Google Scholar]
- Kawasaki, I., Shim, Y. H., Kirchner, J., Kaminker, J., Wood, W. B., and Strome, S. (1998). PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635–645. [DOI] [PubMed] [Google Scholar]
- Kelly, W. G., and Fire, A. (1998). Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125, 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitlina, S. Y. (2001). Functional specificity of actin isoforms. Int. Rev. Cytol. 202, 35–98. [DOI] [PubMed] [Google Scholar]
- Kovar, D. R., Wu, J. Q., and Pollard, T. D. (2005). Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol. Biol. Cell 16, 2313–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause, M., Wild, M., Rosenzweig, B., and Hirsh, D. (1989). Wild-type and mutant actin genes in Caenorhabditis elegans. J. Mol. Biol. 208, 381–392. [DOI] [PubMed] [Google Scholar]
- Kumar, A. et al. (1997). Rescue of cardiac alpha-actin-deficient mice by enteric smooth muscle gamma-actin. Proc. Natl. Acad. Sci. USA 94, 4406–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landel, C. P., Krause, M., Waterston, R. H., and Hirsh, D. (1984). DNA rearrangements of the actin gene cluster in Caenorhabditis elegans accompany reversion of three muscle mutants. J. Mol. Biol. 180, 497–513. [DOI] [PubMed] [Google Scholar]
- Macqueen, A. J., Baggett, J. J., Perumov, N., Bauer, R. A., Januszewski, T., Schriefer, L., and Waddle, J. A. (2005). ACT-5 is an essential Caenorhabditis elegans actin required for intestinal microvilli formation. Mol. Biol. Cell 16, 3247–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, R. C., and Plasterk, R. H. (2005). RNA interference spreading in C. elegans. Methods Enzymol. 392, 308–315. [DOI] [PubMed] [Google Scholar]
- Mello, C. C., Kramer, J. M., Stinchcomb, D., and Ambros, V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, E., Nance, J., and Priess, J. R. (2004). Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell 7, 413–424. [DOI] [PubMed] [Google Scholar]
- Ono, S. (1999). Purification and biochemical characterization of actin from Caenorhabditis elegans: its difference from rabbit muscle actin in the interaction with nematode ADF/cofilin. Cell Motil. Cytoskelet. 43, 128–136. [DOI] [PubMed] [Google Scholar]
- Phillips, J. B., Lyczak, R., Ellis, G. C., and Bowerman, B. (2004). Roles for two partially redundant alpha-tubulins during mitosis in early Caenorhabditis elegans embryos. Cell Motil. Cytoskelet. 58, 112–126. [DOI] [PubMed] [Google Scholar]
- Piano, F., Schetter, A. J., Mangone, M., Stein, L., and Kemphues, K. J. (2000). RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10, 1619–1622. [DOI] [PubMed] [Google Scholar]
- Priess, J. R., and Hirsh, D. I. (1986). Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev. Biol. 117, 156–173. [DOI] [PubMed] [Google Scholar]
- Rutledge, E., Bianchi, L., Christensen, M., Boehmer, C., Morrison, R., Broslat, A., Beld, A. M., George, A. L., Greenstein, D., and Strange, K. (2001). CLH-3, a ClC-2 anion channel ortholog activated during meiotic maturation in C. elegans oocytes. Curr. Biol. 11, 161–170. [DOI] [PubMed] [Google Scholar]
- Sablin, E. P., Dawson, J. F., VanLoock, M. S., Spudich, J. A., Egelman, E. H., and Fletterick, R. J. (2002). How does ATP hydrolysis control actin's associations? Proc. Natl. Acad. Sci. USA 99, 10945–10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevzov, G., Lloyd, C., and Gunning, P. (1992). High level expression of transfected beta- and gamma-actin genes differentially impacts on myoblast cytoarchitecture. J. Cell Biol. 117, 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S. Q., and Bowerman, B. (2003). Cell polarity and the cytoskeleton in the Caenorhabditis elegans zygote. Ann. Rev. Genet. 37, 221–249. [DOI] [PubMed] [Google Scholar]
- Schuler, H., Korenbaum, E., Schutt, C. E., Lindberg, U., and Karlsson, R. (1999). Mutational analysis of Ser14 and Asp157 in the nucleotide-binding site of beta-actin. Eur. J. Biochem. 265, 210–220. [DOI] [PubMed] [Google Scholar]
- Severson, A. F., Baillie, D. L., and Bowerman, B. (2002). A formin homology protein and a profilin are required for cytokinesis and Arp2/3-independent assembly of cortical microfilaments in C. elegans. Curr. Biol. 12, 2066–2075. [DOI] [PubMed] [Google Scholar]
- Shelton, C. A., Carter, J. C., Ellis, G. C., and Bowerman, B. (1999). The nonmuscle myosin regulatory light chain gene mlc-4 is required for cytokinesis, anterior-posterior polarity, and body morphology during Caenorhabditis elegans embryogenesis. J. Cell Biol. 146, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S., and Shaw, J. E. (1993). A Caenorhabditis elegans act-4:lacZ fusion: use as a transformation marker and analysis of tissue-specific expression. Gene 131, 167–173. [DOI] [PubMed] [Google Scholar]
- Strome, S. (1986). Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J. Cell Biol. 103, 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobiev, S., Strokopytov, B., Drubin, D. G., Frieden, C., Ono, S., Condeelis, J., Rubenstein, P. A., and Almo, S. C. (2003). The structure of nonvertebrate actin: implications for the ATP hydrolytic mechanism. Proc. Natl. Acad. Sci. USA 100, 5760–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, C. R., Mahowald, A. P., and Miller, K. G. (2002). One of the two cytoplasmic actin isoforms in Drosophila is essential. Proc. Natl. Acad. Sci. USA 99, 8037–8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R. H., Hirsh, D., and Lane, T. R. (1984). Dominant mutations affecting muscle structure in Caenorhabditis elegans that map near the actin gene cluster. J. Mol. Biol. 180, 473–496. [DOI] [PubMed] [Google Scholar]
- Wright, A. J., and Hunter, C. P. (2003). Mutations in a beta-tubulin disrupt spindle orientation and microtubule dynamics in the early Caenorhabditis elegans embryo. Mol. Biol. Cell 14, 4512–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.