Abstract
The Saccharomyces cerevisiae inhibitor of apoptosis (IAP) repeat protein Bir1 localizes as a chromosomal passenger. A deletion analysis of Bir1 identified two regions important for function. The C-terminal region is essential for growth, binds Sli15, and is necessary and sufficient for the localization of Bir1 as a chromosomal passenger. The middle region is not essential but is required to localize the inner kinetochore protein Ndc10 to the spindle during anaphase and to the midzone at telophase. In contrast, precise deletion of the highly conserved IAP repeats conferred no phenotype and did not alter the cell cycle delay caused by loss of cohesin. Bir1 is phosphorylated in a cell cycle-dependent manner. Mutation of all nine CDK consensus sites in the middle region of Bir1 significantly decreased the level of phosphorylation and blocked localization of Ndc10 to the spindle at anaphase. Moreover, immunoprecipitation of Ndc10 with Bir1 was dependent on phosphorylation. The loss of Ndc10 from the anaphase spindle prevented elongation of the spindle beyond 7 μm. We conclude that phosphorylation of the middle region of Bir1 is required to bring Ndc10 to the spindle at anaphase, which is required for full spindle elongation.
INTRODUCTION
A successful cell cycle is completed with a single round of DNA replication, followed by sister chromatid separation, spindle elongation, and cytokinesis. These events must be coordinated such that each cell receives its exact complement of genetic material. Chromosomal passenger proteins are involved in coordinating cell cycle events that are required for proper chromosome segregation (reviewed in Lens and Medema, 2003; Vagnarelli and Earnshaw, 2004). They have a dynamic localization throughout the cell cycle as they move from kinetochores to the spindle at anaphase and concentrate at the spindle midzone before cytokinesis.
Aurora kinase Ipl1 and its binding partner Sli15 are chromosomal passengers that are involved in the spindle checkpoint. Ipl1 phosphorylates targets at the kinetochore and is required to sense the lack of tension caused by mono-oriented or detached chromosomes (Biggins and Murray, 2001; Pinsky et al., 2003). Sli15 mutants have phenotypes very similar to Ipl1 mutants, and Sli15 is required for the movement of Ipl1 to the spindle at anaphase (Kim et al., 1999; Pereira and Schiebel, 2003).
The inhibitor of apoptosis (IAP) repeat protein Bir1 has been implicated in chromosome segregation (Li et al., 1998; Uren et al., 1999; Yoon and Carbon, 1999). It copurifies with Sli15 (Cheeseman et al., 2002). Bir1 shows two-hybrid and genetic interactions with the inner kinetochore protein Ndc10 (Yoon and Carbon, 1999). Depletion of Bir1 prevents localization of Ndc10 to the anaphase spindle (Bouck and Bloom, 2005). Mutants of Bir1 in Schizosaccharomyces pombe have defects in chromosome condensation, spindle elongation, and DNA repair (Morishita et al., 2001; Rajagopalan and Balasubramanian, 2002).
Here, we show that Bir1 is essential. We mapped the essential region to a 10-kDa region at the C terminus. This region is necessary and sufficient for survival and localization of Bir1 as a chromosomal passenger. This region is also sufficient to interact with Sli15. The middle region of Bir1 is not essential, but it is required to localize Ndc10 to the spindle at anaphase and to the midzone at telophase. The middle region is not required for Ndc10's kinetochore localization. The interaction between Ndc10 and Bir1 is dependent on phosphorylation of Bir1 on CDK consensus sites and is required for full spindle elongation.
MATERIALS AND METHODS
Media
YPD medium and SD medium were made as described previously (Sherman et al., 1986). YPD cycloheximide medium is YPD supplemented with 2 μg/ml cycloheximide. SD complete is SD medium supplemented with 50 μg/ml adenine, 25 μg/ml uracil, 100 μg/ml tryptophan, and 0.1% casamino acids. SD-uracil low adenine is SD complete lacking uracil and contains 5 μg/ml adenine. NP-40 buffer was described previously (Hazbun et al., 2003).
Plasmids and Strains
The plasmids used in this study are listed in Table 1. The BIR1 open reading frame (ORF) + 560 base pairs 5′ and 548 base pairs 3′ was cloned into pRS316 to create plasmid pPW02. The plasmid used for the plasmid shuffle is plasmid pPW06, which contains ADE3 (from pLI831) and BIR1 in plasmid pTD29 (Geiser et al., 1993; Muller, 1996). The plasmids containing the deletions of BIR1 were derived from plasmid pPW02 as follows. The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to delete codons 15–239 of the BIR1 ORF in pPW02 to create pPW08. Plasmid pPW09 was made by digesting plasmid pPW02 with NheI, filling in the ends using the Klenow fragment of DNA polymerase, and ligating. To create plasmids pPW11 and pPW13-16, pPW02 was digested with the enzymes indicated in Table 1. The ends were filled in using the Klenow fragment of DNA polymerase and ligated. To create plasmid pPW17, pPW02 was digested with the enzymes indicated in Table 1, treated with mung bean nuclease, and ligated. Venus and 13xMyc tags were introduced into BIR1 deletion constructs by ligating fragments from pBS7 (gift from Yeast Resource Center, University of Washington, Seattle, WA) and pFA6a-13Myc-kanMX6 (Longtine et al., 1998) containing these tags at the 3′ end of the BIR1 ORF in frame. Plasmid pPW129 was created by inserting a synthetic duplex oligonucleotide containing two copies of the SV40 nuclear localization sequence (NLS) in frame between the BIR1 ORF and the Venus tag. All point mutations in BIR1 were generated using the QuikChange or QuikChange Multi site-directed mutagenesis kits (Stratagene).
Table 1.
Plasmids used in this study
| Plasmid | Relevant markers | Source or reference |
|---|---|---|
| pPW02 | BIR1 in pRS316 | This study |
| pPW06 | BIR1 and ADE3 in 2μm vector | This study |
| pPW08 | bir1-ΔIAP in pRS316 | This study |
| pPW09 | bir1-876stop in pRS316 | This study |
| pPW11 | bir1-ΔBSa in pRS316 | This study |
| pPW13 | bir1-ΔBclBa in pRS316 | This study |
| pPW14 | bir1-ΔSNa in pRS316 | This study |
| pPW15 | bir1-ΔMBa in pRS316 | This study |
| pPW16 | bir1-ΔMSa in pRS316 | This study |
| pPW17 | bir1-ΔMNa in pRS316 | This study |
| pPW25 | bir1 in pRS306 | This study |
| pPW33 | bir1-Venus in pRS306 | This study |
| pPW34 | bir1-ΔIAP-Venus in pRS306 | This study |
| pPW35 | bir1-876stop-Venus in pRS306 | This study |
| pPW36 | bir1-ΔBclBa-Venus in pRS306 | This study |
| pPW37 | bir1-ΔSNa-Venus in pRS306 | This study |
| pPW38 | bir1-ΔMBa-Venus in pRS306 | This study |
| pPW39 | bir1-ΔMSa-Venus in pRS306 | This study |
| pPW40 | bir1-ΔMNa-Venus in pRS306 | This study |
| pPW46 | bir1-ΔBSa-Venus in pRS306 | This study |
| pPW75 | bir1 T735A, T747A in pRS316 (bir1-2xA) | This study |
| pPW118 | BIR1-13xmyc in pRS306 | This study |
| pPW122 | bir1 T735A, T747A-13xmyc in pRS306 (bir1-2xA-13xmyc) | This study |
| pPW124 | bir1 S383A, S395A, S552A, S587A, S667A, T684A, S688A, T735A, T747A in pRS316 (bir1-9xA) | This study |
| pPW126 | bir1 S383A, S395A, S552A, S587A, S667A, T684A, S688A, T735A, T747A-13xmyc in pRS306 (bir1-9xA-13xmyc) | This study |
| pPW129 | bir1-ΔMNa-Venus +2NLS in pRS316 | This study |
| pPW131 | bir1-ΔIAP-13xmyc in pRS306 | This study |
| pPW132 | bir1-ΔBclBa-13xmyc in pRS306 | This study |
| pPW133 | bir1-ΔSNa-13xmyc in pRS306 | This study |
| pPW134 | bir1-ΔMBa-13xmyc in pRS306 | This study |
| pPW135 | bir1-ΔMSa-13xmyc in pRS306 | This study |
| pPW136 | bir1-ΔBSa-13xmyc in pRS306 | This study |
| pJL13 | DBD-BIR1 | This study |
| pJL16 | AD-SLI15 | This study |
| pJL19 | DBD-bir1C80 | This study |
| pJL20 | DBD-bir1C296 | This study |
| pACTII | Gal4 DBD LEU2 | Clontech (Mountain View, CA) |
| pGBKT7 | Gal4 AD TRP1 | Clontech |
| pRS306 | URA3 f1 origin | Sikorski and Hieter (1989) |
| pRS316 | CEN6 ARSH4 URA3 f1 origin | Sikorski and Hieter (1989) |
| pTD29 | SPC110 LYS2 ADE3 2 μm origin | Geiser et al. (1993) |
| pLI831 | ADE3 | Muller (1996) |
| pBS4 | CFP hphMX3 | Yeast Resource Center |
| pBS7 | Venus kanMX6 | Yeast Resource Center |
| pFA6a-13Myc-kanMX6 | 13xmyc kanMX6 | Longtine et al. (1998) |
| pDH6 | YFP kanMX6 | Yeast Resource Center |
| pDH3 | CFP kanMX6 | Yeast Resource Center |
| pFA6a-CTAP-MX6-2xPA | TAP kanMX6 | Tasto et al. (2001) |
Plasmids were digested with the plasmids indicated (M, MscI; Bcl, BclI; B, BglII; S, SphI; N, NheI), treated with Klenow fragment of DNA polymerase or mung bean nuclease, and ligated in frame.
The yeast strains used in this study are listed in Table 2. All strains are derived from W303. C-terminal cyan fluorescent protein (CFP), yellow fluorescent protein (YFP), Venus, 3xHA, 13xMyc, and TAP fusions were created by amplifying the CFP-hphMX3, CFP-kanMX6, YFP-kanMX6, and VenuskanMX6 cassettes from plasmids pBS4, pDH3, pDH6, and pBS7 (all gifts from Yeast Resource Center) and plasmids pFA6a-13Myc-kanMX6 and pFA6a-3HA-kanMX6 (Longtine et al., 1998) and plasmid pFA6a-CTAP-MX6-2XPA (Tasto et al., 2001). Cassettes were integrated in frame at the 3′ end of the target ORF. The BIR1 plasmid shuffle strain, PWY16-4D was constructed by replacing the BIR1 ORF with the hph gene in a diploid using a PCR cassette generated from plasmid pBS4, followed by transformation of plasmid pPW06 and subsequent sporulation. PWY91-10C is a modified BIR1 plasmid shuffle strain containing Ndc10-CFP. All functional BIR1 deletions, truncations, and point mutants were introduced as the sole copy at the BIR1 locus in PWY16-4D or PWY91-10C using the gene replacement method as described previously (Widlund and Davis, 2005).
Table 2.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| AH109 | MATa, trp1-901, leu2-3 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, | Clontech |
| URA3:: MEL1UAS-MEL1 TATA-lacZ | ||
| BESY49-6C | MATacyh2r NDC10-YFP::kanMX6 | Yeast Resource Center |
| BESY46-1D | MATα cyh2r NDC10-CFP::hphMX3 | Yeast Resource Center |
| BSY9 | MATa/MATα ade2-1oc/ade2-1oc ADE3/ade3Δ can1-100/can1-100 CYH2s/cyh2r; his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 | Hazun et al. (2003) |
| PWY8 | MATa/MATα ade3Δ/ade3Δ BIR1-Venus::kanMX6/BIR1-Venus::kanMX6 CYH2s/cyh2r NDC10-CFP:kanMX6/NDC10-CFP:kanMX6 | This study |
| PWY16-4D | MATaade3Δ bir1Δ::hphMX3 lys2Δ::HIS3 with autonomous plasmid pPW06 | This study |
| PWY19 | MATa/MATα ade3Δ/ade3Δ BIR1-Venus::kanMX6/BIR1-Venus::kanMX6 CYH2s/cyh2r SLI15-CFP::kanMX6/SLI15-CFP::kanMX6 | This study |
| PWY56 | MATaBIR1::bir1-876stop-Venus::kanMX6::URA3 | This study |
| PWY57 | MATaBIR1::bir1-ΔMN-Venus::kanMX6::URA3 | This study |
| PWY60 | MATaBIR1::BIR1-Venus::kanMX6::URA3 | This study |
| PWY61-4D | MATaade3Δ BIR1-TAP::kanMX6 cyh2r | This study |
| PWY75-6C | MATaade3Δ BIR1-Venus::kanMX6 NDC10-CFP::hphMX3 | This study |
| PWY76-1C | MATaade3Δ bir1-ΔIAP-Venus::kanMX6 NDC10-CFP::hphMX3 | This study |
| PWY77-2B | MATaade3Δ bir1-ΔBclB-Venus::kanMX6 NDC10-CFP::hphMX3 | This study |
| PWY78-6C | MATaade3Δ bir1-ΔSN-Venus::kanMX6 NDC10-CFP::hphMX3 | This study |
| PWY79-5A | MATaade3Δ bir1-ΔMB-Venus::kanMX6 NDC10-CFP::hphMX3 | This study |
| PWY80-2C | MATaade3Δ bir1-ΔMS-Venus::kanMX6 NDC10-CFP::hphMX3 | This study |
| PWY81-1B | MATaade3Δ bir1-ΔBS-Venus::kanMX6 lys2Δ::HIS3 NDC10-CFP::hphMX3 | This study |
| PWY82-2B | MATaade3Δ bir1-Venus::kanMX6 SLI15-CFP::hphMX3 | This study |
| PWY83-2A | MATaade3Δ bir1-ΔIAP-Venus::kanMX6 SLI15-CFP::hphMX3 | This study |
| PWY84-4D | MATaade3Δ bir1-ΔBclB-Venus::kanMX6 SLI15-CFP::hphMX3 | This study |
| PWY85-4D | MATaade3Δ bir1-ΔSN-Venus::kanMX6 SLI15-CFP::hphMX3 | This study |
| PWY86-1D | MATα ade3Δ bir1-ΔMB-Venus::kanMX6 SLI15-CFP::hphMX3 | This study |
| PWY87-5C | MATaade3Δ bir1-ΔMS-Venus::kanMX6 cyh2r SLI15-CFP::hphMX3 | This study |
| PWY88-10A | MATaade3Δ bir1ΔBS-Venus::kanMX6 cyh2r lys2Δ::HIS3 SLI15-CFP::hphMX3 | This study |
| PWY91-10C | MATaade3Δ bir1Δ::hphMX3 lys2Δ::HIS3 NDC10-YFP::kanMX6 with autonomous plasmid pPW06 | This study |
| PWY93-3B | MATaade3Δ BIR1—13xMyc::kanMX6 | This study |
| PWY160-1B | MATα ade3Δ NDC10-3xHA::kanMX6 | This study |
| PWY192 | MATaade3Δ bir1Δ::hphMX3::bir1-2xA-13xMyc::kanMX6::URA3 lys2Δ::HIS3 NDC10-YFP::kanMX6 | This study |
| PWY194 | MATaade3Δ bir1Δ::hphMX3:: bir1-9xA-13xMyc::kanMX6::URA3 lys2Δ::HIS3 NDC10-YFP::kanMX6 | This study |
| PWY199-5B | MATaade3Δ BIR1-13xMyc::kanMX6 cdc15-2 SLI15-3xHA::kanMX6 | This study |
| PWY200-14C | MATabir1-ΔIAP::hphMX3 his3-11::PCUP1-GFP12-lacI12::HIS3 mcd1-1 PDS1-3xHA:URA3 trp1-1::lacO::TRP1 | This study |
| PWY200-17A | MATahis3-11::PCUP1-GFP12-lacI12::HIS3 mcd1-1 PDS1-3xHA::URA3 trp1-1::lacO::TRP1 | This study |
| PWY202-11B | MATaBIR1-13xMyc::kanMX6 cdc15-2 NDC10-3xHA::kanMX6 | This study |
| PWY204 | MATaade3Δ bir1Δ::hphMX3::bir1-ΔIAP-13xMyc::kanMX6:URA3 lys2Δ:HIS3 | This study |
| PWY205 | MATaade3Δ bir1Δ:hphMX3:bir1-ΔBclB-13xMyc:kanMX6:URA3 lys2Δ:HIS3 | This study |
| PWY206 | MATaade3Δ bir1Δ:hphMX3:bir1-ΔSN-13xMyc:kanMX6:URA3 lys2Δ:HIS3 | This study |
| PWY207 | MATaade3Δ bir1Δ:hphMX3:bir1-ΔMB-13xMyc:kanMX6:URA3 lys2Δ:HIS3 | This study |
| PWY208 | MATaade3Δ bir1Δ:hphMX3:bir1-ΔMS-13xMyc:kanMX6:URA3 lys2Δ:HIS3 | This study |
| PWY209 | MATaade3Δ bir1Δ:hphMX3:bir1-ΔBS-13xMyc:kanMX6:URA3 lys2Δ:HIS3 | This study |
| JLY15 | MATa/MATα ADE3/ade3Δ BIR1-Venus:kanMX6/BIR1-Venus:kanMX6 cyh2r/cyh2r; his3-11/HIS3 HIS4/his4:CYH2s | This study |
| JLY16 | MATa/MATα ADE3/ade3Δ bir1-ΔIAP-Venus:kanMX6/bir1-ΔIAP-Venus:kanMX6 cyh2r/cyh2r; his3-11/HIS3 HIS4/his4:CYH2s | This study |
| JLY17 | MATa/MATα ADE3/ade3Δ bir1-ΔBclB-Venus:kanMX6/bir1-ΔBclB-Venus:kanMX6 cyh2r/cyh2r HIS3/HIS3 HIS4/his4:CYH2s lys2Δ:HIS3/LYS2 | This study |
| JLY18 | MATa/MATα; ADE3/ADE3 bir1-ΔSN-Venus:kanMX6/bir1-ΔSN-Venus:kanMX6 cyh2r/cyh2r HIS3/his3-11,15 his4:CYH2s/HIS4 | This study |
| JLY19 | MATa/MATα ADE3/ADE3 bir1-ΔMB-Venus:kanMX6/bir1-ΔMB-Venus:kanMX6 cyh2r/cyh2r his3-11,15/HIS3 HIS4/his4:CYH2s | This study |
| JLY20 | MATa/MATα ade3Δ/ADE3 bir1-ΔMS-Venus:kanMX6/bir1-ΔMS-Venus:kanMX6 cyh2r/cyh2r HIS3/his3-11,15 his4:CYH2s/HIS4 | This study |
| JLY21 | MATa/MATα ade3Δ/ADE3 bir1-ΔBS-Venus:kanMX6/bir1-ΔBS-Venus:kanMX6 cyh2r/cyh2r HIS3/HIS3 his4:CYH2s/HIS4 LYS2/lys2Δ:HIS3 | This study |
| TDY93 | MATa/MATα ADE3/ade3Δ cyh2r/cyh2r his3-11,15/HIS3 HIS4/his4:CYH2r | Davis (1992) |
| W303 | MATaade2—1oc can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 |
All strains, except strain AH109, have the same markers as W303 except as shown.
Fluorescence Microscopy
Live cell imaging was performed with a DeltaVision microscopy system from Applied Precision (Issaquah, WA). The system incorporates an Olympus IL-70 microscope, a u-plan-apo 100× oil objective (1.35 numerical aperture), a CoolSnap HQ digital camera from Roper Scientific (Tucson, AZ), and optical filter sets from Omega Optical (Battleboro, VT). Cells were mounted on a 1% agarose pad containing SD-complete medium (Muller et al., 2005). Single sections were taken. The images were analyzed using DeltaVision softWoRx software. Images were converted to TIFF format in 24-bit RGB.
Chromosome Loss Assays
The rates of loss of a tagged chromosome III was assayed as described previously except that cycloheximide was used at a final concentration of 2 μg/ml (Runge et al., 1991; Davis, 1992). Nineteen small colonies were picked after 2 d of growth at room temperature on YPD media and suspended in 1 ml of distilled H2O. Suspensions were then sonicated, and 100 μl of each suspension was plated onto each of 19 YPD-cycloheximide plates. Plates were incubated at 30°C for 4 d. To determine the average number of colony-forming units per colony, aliquots (50 μl) of each suspension were pooled and titered on YPD medium. Calculation of the rate of chromosome loss was performed by the method of the median (Lea and Coulson, 1949). The number of chromosome loss events per colony was determined from the median number of cycloheximide resistant cells per colony. This number was then divided by the average number of cell divisions per colony to give the rates of chromosome loss per cell division.
Protein Purification
Tandem-affinity purifications of Bir1 and mass spectrometry analysis were performed as described previously (Hazbun et al., 2003) except in the case of the low-stringency purification, where NaCl concentration was kept at 150 mM (Hazbun et al., 2003). Sites of phosphorylation were identified by mass spectrometry as described previously (MacCoss et al., 2002). All mass spectrometry data generated for this study are available at www.yeastrc.org/pdr.
Immunological Techniques
Lysate was prepared from strains carrying Myc-tagged Bir1 or hemagglutinin (HA)-tagged Ndc10 according to the tandem-affinity purification protocol except that the clarification step was at 15,000 rpm for 10 min. Aliquots (1 ml) were prepared from the 50 ml of total cleared lysate and frozen at –80°C. Bir1-Myc was immunoprecipitated from 1 ml of lysate with 25 μl of Sepharose beads conjugated with 9E11 monoclonal α-Myc antibodies (Covance, Princeton, NJ). Beads were washed with NP-40 buffer and then buffer 3 (New England Biolabs, Ipswich, MA). The beads were incubated at 37°C for 1 h in buffer 3 in the presence or absence of 100 U/ml calf intestinal phosphatase (CIP). Lysate from cells carrying Ndc10-3xHA was precleared by incubating with 100-μl bed volume of Sepharose 6B (GE Healthcare, Buckinghamshire, United Kingdom) for 30 min. The precleared lysate was added to α-Myc-conjugated Sepharose beads (control) and to the Bir1-bound beads and incubated for 2 h. Beads were then washed three times in NP-40 buffer, and proteins were eluted by boiling in denaturing SDS-PAGE gel loading buffer. All samples were subjected to electrophoresis on SDS-polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked with Odyssey block (Li-Cor, Lincoln, NE). The anti-HA antibody was mouse monoclonal 12CA5 (Roche Diagnostics, Indianapolis, IN). The anti-Myc antibody used was mouse monoclonal 9E10 (Santa Cruz Biotechnology, Santa Cruz, CA). The secondary antibody was goat α-mouse Alexa Fluor 680 (Molecular Probes, Eugene, OR). Membranes were scanned and quantified using the Odyssey system (Li-Cor).
RESULTS
S. cerevisiae Bir1 Localizes as a Chromosomal Passenger
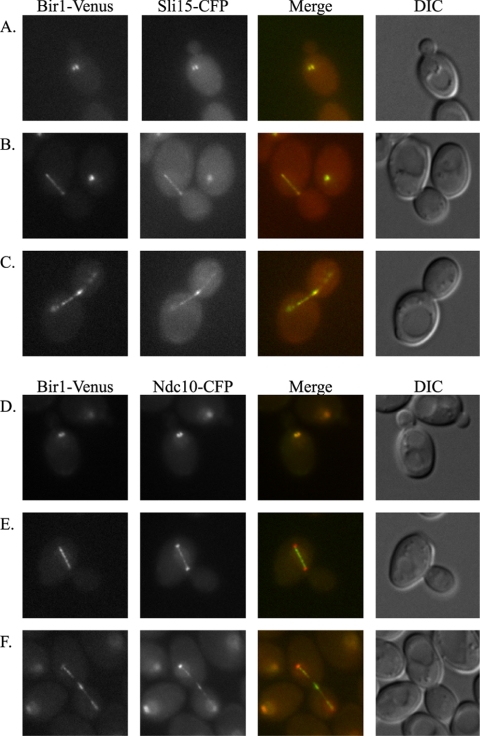
Localization of full-length Bir1 has proven difficult, although it seems to associate with microtubules (Uren et al., 1999; Yoon and Carbon, 1999; Huh et al., 2003). We successfully tagged and localized endogenous full-length Bir1 with the YFP variant Venus. Bir1-Venus shows a dynamic localization through the cell cycle that is similar to chromosomal passenger proteins in vertebrates (Figure 1). Bir1 colocalizes with Sli15 at all stages of the cell cycle. Bir1 seems to move from kinetochores to the spindle at anaphase and concentrates at the midzone late in the cell cycle.
Figure 1.
Bir1 localizes as a chromosomal passenger. (A–C) Strain PWY19, expressing Bir1 with a C-terminal Venus tag and Sli15 with a C-terminal CFP tag expressed under control of their endogenous promoters, was imaged as described in Materials and Methods. (D–F) Strain PWY8, expressing Bir1 with a C-terminal Venus tag and Ndc10 with a C-terminal CFP tag, expressed under control of their endogenous promoters, was imaged as described in Materials and Methods. (A) Bir1-Venus and Sli15-CFP colocalize on kinetochores before anaphase. (B) At anaphase, Bir1 and Sli15 colocalize on the mitotic spindle. (C) Bir1-Venus and Sli15-CFP colocalize on the spindle in late anaphase and begin to concentrate at the spindle midzone as cells approach cytokinesis. (D) Bir1-Venus colocalizes with Ndc10-CFP before anaphase. (E) Bir1-Venus and Ndc10-CFP localization overlaps on the spindle but not on kinetochores during anaphase. (F) Bir1-Venus and Ndc10-CFP localization overlaps on the spindle and at the midzone as cells approach cytokinesis.
We compared the localization of Bir1-Venus and Ndc10-CFP. Bir1 and Ndc10 colocalize at kinetochores before anaphase. During anaphase, Bir1 leaves the kinetochore, whereas the majority of Ndc10 remains. Bir1 and a pool of Ndc10 colocalize to the spindle during anaphase and at the midzone during telophase.
BIR1 Is Essential
There has been some dispute as to whether BIR1 is essential for viability. Two groups have reported BIR1 is not essential (Uren et al., 1999; Yoon and Carbon, 1999), and one group reported that BIR1 is essential (Li et al., 2000). We found that BIR1 is essential. We replaced one copy of the BIR1 ORF with the hygromycin B resistance gene, hph, in a diploid. Colony PCR using primers flanking the ORF confirmed the presence of a deleted copy and a wild-type copy in the diploid. Sporulation and tetrad dissection of the diploid isolates gave two live and two dead spores. The two live spores in each of 24 tetrads were hygromycin B sensitive. Li and coworkers proposed that any viable bir1Δ haploids isolated from a heterozygous diploid contain a wild-type copy of BIR1. We tested this possibility directly. Our BIR1/bir1Δ::hph heterozygous diploid is also heterozygous for the recessive cyhr allele. We sporulated this strain and selected for haploids on medium containing cycloheximide. Forty-eight of 276 cycloheximide-resistant spores isolated were resistant to hygromycin B indicating the presence of a disrupted copy of BIR1. Colony PCR of six of these viable cells containing bir1Δ::hph confirmed the presence of both a wild-type and a disrupted copy of BIR1.
Finally, we constructed a plasmid shuffle strain for BIR1, where a haploid has the endogenous copy of BIR1 disrupted and carries a multicopy plasmid carrying BIR1 and ADE3. If BIR1 is not essential, then the strain should form colonies with white sectors, the white sectors being cells that have lost the only copy of BIR1, which is on the ADE3 plasmid. However, this strain forms solid red colonies, indicating that BIR1 is essential for viability.
The C Terminus of Bir1 Is Essential
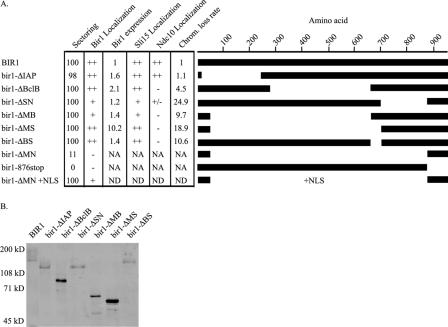
Because BIR1 is essential, we performed a deletion analysis to define the essential regions of BIR1. Deletions and truncations of Bir1 were expressed from the BIR1 promoter in an ARS/CEN vector (Figure 2A). These constructs were transformed into the plasmid shuffle strain PWY16-4D to assess their ability to support the growth of yeast cells. Only the C-terminal truncation (bir1-876stop) and the largest deletion (bir1-ΔMN) were unable to support growth (Figure 2A).
Figure 2.
Bir1 deletions and truncations. (A) Internally deleted and truncated versions of BIR1 were constructed as described in Materials and Methods. Sectoring is shown as the percentage of colonies that formed white sectors in red colonies when transformed with the version of BIR1 shown, expressed from an autonomously replicating plasmid (plasmids are given in Table 1). For all other analyses, the deletions and truncations were integrated as the only copy of bir1 at the endogenous locus as described previously (Widlund and Davis, 2005). Strains are listed in Table 2. Bir1, Sli15, and Ndc10 localizations are scored as absent (–), present at a reduced level in some cells (+/–), present at a reduced level in all cells (+), or present at a normal level in all cells (++). Expression levels are given relative to wild type as quantified from the Western blot shown in B. Chromosome loss rate was measured as described in Materials and Methods. The rate of loss of chromosome III in a wild-type strain was 1.05 × 10–5 per cell division. NA, not applicable; ND, not determined. (B) Western blot analysis of the protein levels of Bir1 wild-type and Bir1 deletions. 13xMyc tagged versions of all deletions under control of the BIR1 promoter were used to replace BIR1 at the endogenous locus. Strains are listed in Table 2. Strains were grown to mid-log phase, and extracts were prepared by precipitation with trichloroacetic acid as described previously (Wright et al., 1989). Equal amounts of total protein were loaded in each lane as determined from a Coomassie blue-stained gel. Protein was detected by antibodies against the Myc epitope (Santa Cruz Biotechnology).
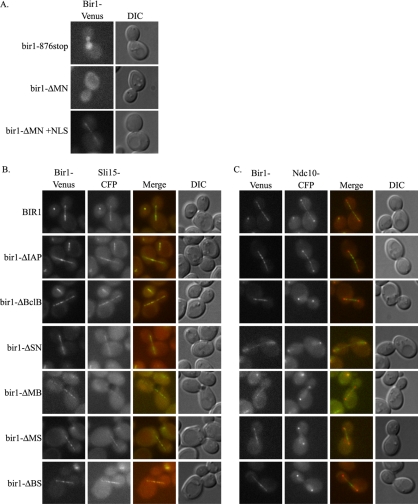
We localized the two inactive deletions. Bir1-876stop is present in the nucleus but does not localize to the kinetochore or spindle (Figure 3A). Bir1-ΔMN seems uniformly spread throughout the cell and is not found in the nucleus (Figure 3A). Addition of a nuclear localization sequence to Bir1-ΔMN partially restored proper localization (Figure 3A). The NLS-Bir1-ΔMN is also able to support growth, indicating that Bir1-ΔMN lacks an NLS but is otherwise functional. Therefore, the C terminus is the essential region of Bir1 and is required for kinetochore and spindle localization.
Figure 3.
Localization of Bir1, Sli15, and Ndc10 in bir1 deletion mutants. (A) Two bir1 mutants that could not independently support growth, Bir1-876stop and Bir1-ΔMN, were tagged with Venus and integrated into a wild-type haploid and imaged as described in Materials and Methods. Bir1-876stop-Venus shows diffuse nuclear localization and is absent from kinetochores and the spindle. Bir1-ΔMN-Venus is spread throughout the cell and is unable to efficiently localize to the nucleus. Bir1-ΔMN +NLS on an ARS/CEN vector was transformed into PWY16]hyphen]4D. Addition of an SV40 NLS partially restores localization of Bir1-ΔMN to the spindle. (B) Functional deletion mutants of BIR1 were tagged with Venus and were used to replace wild-type BIR1 as the only copy as described previously (Widlund and Davis, 2005). The localizations of the tagged mutant proteins were compared with the localization of Sli15-CFP. First column, the deletion mutants of Bir1 localize normally except the bir1-ΔSN and bir1-ΔMB mutants, which display fainter spindle localizations. Second column, Sli15 localizes in all the mutants with reduced levels in bir1-ΔSN and bir1-ΔMB. Third column, merged image with Bir1-Venus in green and Sli15-CFP in red. Fourth column, differential interference contrast (DIC) image. (C) Localization of Ndc10-CFP in the bir1 deletion mutants. First column, deletion mutants of Bir1. Second column, Ndc10 is mislocalized in all but the bir1-ΔIAP mutant strain. Third column, merged image with Bir1-Venus in green and Ndc10-CFP in red. Fourth column, DIC image.
The Essential C Terminus of Bir1 Interacts with Sli15
We purified TAP-tagged Bir1 and identified the copurifying proteins by mass spectrometry. In six of six purifications, Sli15 copurified with Bir1. Ipl1 was not detected even when Bir1 was purified under low stringency. These results are consistent with a model in which Bir1 forms a complex with Sli15 that does not contain Ipl1.
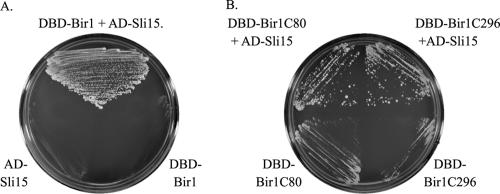
The region of Bir1 responsible for interaction with Sli15 was mapped by directed two-hybrid experiments. Full-length Bir1 showed a strong interaction with full-length Sli15 (Figure 4A). The C-terminal 297 amino acids (C terminus of bir1-ΔMB) and the C-terminal 80 amino acids (C terminus of bir1-ΔMN) also showed interaction with full-length Sli15 (Figure 4B).
Figure 4.
The C terminus of Bir1 interacts with Sli15. Full-length Sli15 was fused to the Gal4 DNA binding domain (DBD). Full-length and fragments of Bir1 were fused to the Gal4 activation domain. Combinations of Sli15 and Bir1 fragments were cotransformed into strain AH-109 and plated on SD-Leu-Trp medium to select for both plasmids. Transformants were then tested for growth on SD-Leu-Trp-His plates containing 3 mM 3-aminotriazole (3AT). (A) Strain AH109 containing Bir1 tagged with the Gal4 DNA binding domain and Sli15 tagged with the Gal4 activation domain was assayed for growth on SD-Leu-Trp-His plates containing 3 mM 3AT. AH109 cotransformed with AD-Sli15 and the DBD alone or DBD-Bir1 and the AD alone were used as controls. (B) Strain AH109 containing AD-SLI15 and one of two DBD tagged C-terminal fragments of Bir1 were assayed for growth as described in A. AH109 cotransformed with the AD alone and either DBD tagged C-terminal fragment of Bir1 were used as controls.
The IAP Repeats of Bir1 Are Not Required for the Spindle Checkpoint
The function of the IAP repeats in Bir1 is not known. Deletion of the repeats did not affect Bir1's localization or expression (Figures 2 and 3B). The IAP repeats were not required for Sli15 localization or Ndc10 localization (Figure 3, B and C). Deletion of the IAP repeats did not alter the rate of chromosome loss as measured by a fluctuation analysis (Figure 2A). Because Bir1 has been linked to Ipl1 function and Ipl1 homologues can phosphorylate IAP repeats in vitro (Leverson et al., 2002; Wheatley et al., 2004), we examined whether the IAP repeats were required for the tension checkpoint. Cells carrying a temperature-sensitive mutation in cohesin, mcd1-1, delay in the cell cycle, and this delay is dependent on Ipl1 (Biggins and Murray, 2001). We found that deletion of the IAP repeats did not alter the cell cycle delay caused by the mcd1-1 mutation (Figure 5).
Figure 5.
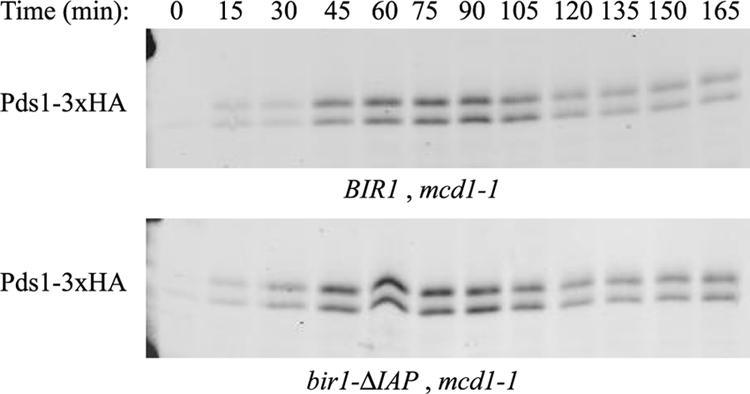
The IAP repeats of Bir1 are not required for the tension checkpoint. Cells carrying the bir1-ΔIAP allele and a temperature-sensitive cohesin mutation, mcd1-1, were arrested in G1 with α-factor and released to the restrictive temperature of 37°C as described previously (Yoder et al., 2005). HA-tagged securin (Pds1) levels were monitored by Western blot analysis to determine if cells arrested in anaphase. Pds1 persists at the restrictive temperature in both the IAP mutant and the controls, indicating that the IAP repeats are dispensable for the tension checkpoint.
Deletions of the Middle Region of Bir1 Alter Ndc10 Localization and Increase the Rate of Chromosome Loss
Deletions in the middle region of Bir1 could support growth. These deletions localized properly except the bir1-ΔSN and the bir1-ΔMB mutants showed weaker spindle localization (Figure 3). All deletions also expressed well (Figure 2). The deletions of the middle region did not alter localization of Sli15 (Figure 3B). Localization of Ndc10 to the kinetochore was not altered. However, localization of Ndc10 to the anaphase spindle and the midzone was prevented (Figure 3C). Moreover, the middle region deletions caused a moderate-to-severe increase in the rate of chromosome loss as analyzed by a fluctuation analysis (Figure 2A) (Runge et al., 1991; Davis, 1992).
Bir1 Is Cell Cycle Regulated
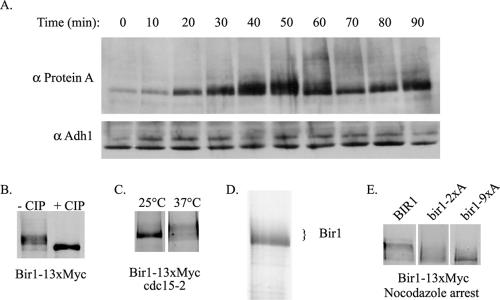
Bir1 expression levels fluctuate through the cell cycle (Figure 6A). Bir1 is barely expressed in cells arrested with α-factor. Protein levels increase after release from the arrest. Bir1 is also phosphorylated in a cell cycle-dependent manner. Phosphorylation occurs as the cells enter S phase and is removed late in the cell cycle (Figure 6A). Immunoprecipitated Bir1 migrates on SDS-PAGE as multiple bands, which can be condensed to one band by treatment with phosphatase, demonstrating that the mobility shift is because of phosphorylation (Figure 6B). Bir1 is heavily modified at a cdc15 cell cycle arrest, indicating phosphorylation is maintained into telophase (Figure 6C).
Figure 6.
Bir1 is phosphorylated in a cell cycle-dependent manner. (A) Haploid cells expressing TAP-tagged Bir1 were synchronized by treatment with α-factor and released as described previously (Yoder et al., 2005). At each time point, extracts were prepared by precipitation with trichloroacetic acid (Wright et al., 1989). Bir1 protein levels were detected by probing for the protein A motif in the TAP tag using a peroxidase α-peroxidase antibody (Sigma, St. Louis, MO). Adh1 was used as a control for equal protein loading in all lanes. (B) Cells containing Myc-tagged Bir1 and the temperature-sensitive cdc15-2 allele were arrested at telophase by shifting to the restrictive temperature and lysed. Immunoprecipitated Bir1 was incubated at 37°C for 1 h in the absence or presence of CIP. (C) Cells containing the temperature-sensitive cdc15-2 allele and Myc-tagged Bir1 were grown at the permissive temperature (25°C) or arrested at the restrictive temperature (37°C). Extracts were prepared by precipitation with trichloroacetic acid (Wright et al., 1989). Precipitated protein was analyzed by Western blot using antibody to the Myc epitope. (D) Bir1-TAP was purified by tandem affinity purification in the presence of phosphatase inhibitors from 4 liters of culture grown to 150 Klett units as described in Materials and Methods. One-fourth of the purified protein was subjected to electrophoresis on a 10% polyacrylamide gel. The gel was silver stained (Bio-Rad, Hercules, CA). The remaining purified Bir1 was analyzed by mass spectrometry to identify phosphorylated peptides. (E) Strains expressing Myc-tagged versions of the indicated phospho-mutants of Bir1 were grown to mid-log phase, and extracts were prepared by precipitation with trichloroacetic acid and analyzed by Western blot analysis using antibody against the Myc epitope.
Phosphorylation of Bir1 Is Required for Interaction with Ndc10 and Efficient Spindle Elongation
We identified which sites were phosphorylated in vivo by analyzing purified Bir1 by mass spectrometry (Figure 6D). The two sites, T735 and T747, were minimal consensus sites for Cdc28 phosphorylation.
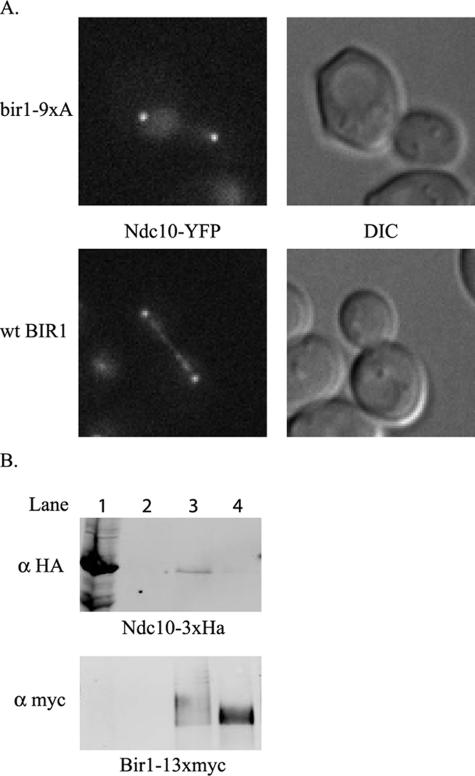
Mutation of the two identified sites diminished but did not abolish phosphorylation (Figure 6E, bir1-2A). Mutation of all nine cdc28 consensus sites in the central region (S383A, S395A, S552A, S587A, S667A, T684A, S688A, T735A, and T747A) dramatically decreased the shift (Figure 6E, bir1-9xA). Mutation of these nine sites prevents localization of Ndc10 to the spindle at anaphase (Figure 7A). Moreover, treatment of Bir1-13xMyc with phosphatase disrupts the in vitro interaction between Ndc10 and Bir1 (Figure 7B).
Figure 7.
Phosphorylation of Bir1 is required for interaction with Ndc10. (A) The bir1-9xA mutant was transformed into the modified plasmid shuffle strain PWY91-10c. Ndc10-YFP was localized in the presence of this bir1 mutant as described in Materials and Methods. Localization of Ndc10-YFP in the presence of wild-type Bir1 is shown as a control. B. The interaction between Ndc10 and Bir1 depends on phosphorylation of Bir1. Bir1-13xMyc was immunoprecipitated from whole cell extract with Myc-conjugated Sepharose beads and either dephosphorylated with CIP or left untreated. Whole cell extract carrying Ndc10-3xHA was then added to both the CIP-treated and untreated beads. The presence of Bir1 and Ndc10 were assayed by Western blot analysis using antibodies recognizing Myc and HA, respectively. Lane 1, Ndc10-3xHA extract alone. Lane 2, Ndc10-3xHA extract bound directly to Myc-conjugated Sepharose beads (control). Lane 3, Ndc10 bound to phosphorylated Bir1. Lane 4, Ndc10 bound to dephosphorylated Bir1.
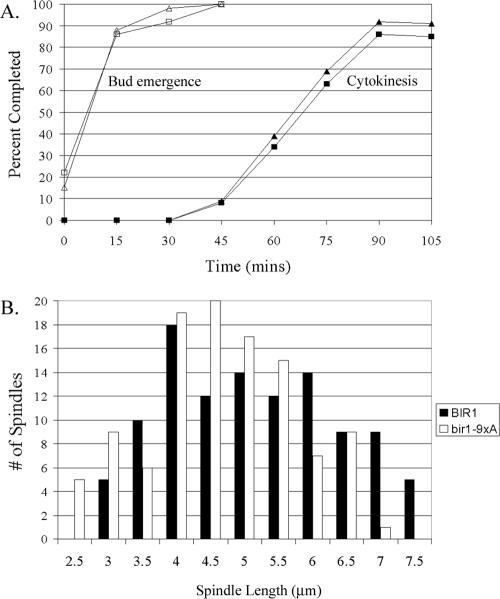
Finally, we examined the consequence of loss of Ndc10 from the anaphase spindle and midzone by characterizing the bir1-9xA mutant. Cells were synchronized by treatment with α-factor and then released and progression through the cell cycle was analyzed. Bud emergence and cytokinesis occurred at the same time in mutant and wild-type cultures (Figure 8A). Next, we measured the length of the mutant and wild-type anaphase spindles in a cycling population of cells. The mutant anaphase spindles were shorter with an average length of 4.9 μm compared with an average length of 5.4 μm for wild-type spindles. This was a statistically significant difference to a 99% confidence level with Student's t test. The main difference between wild-type and mutant was a lack of long spindles greater than 7 μm in length in the mutant cells. Less than 1% of the mutant anaphase spindles were above 7 μm in length compared with 12% of wild-type anaphase spindles (Figure 8B).
Figure 8.
Full spindle elongation is prevented in the bir1-9xA mutant. (A) Cultures of strains BESY49-6c and PWY194 were synchronized by treatment with α-factor as described previously (Yoder et al., 2005). At the given times, samples were removed and fixed in 3.7% formaldehyde. Budding and cytokinesis was determined by phase contrast microscopy. Time is in minutes after release from α-factor. Triangles denote strain BESY49-6c (control) and squares denote PWY194 (bir1-9xA). At least 100 cells were analyzed at each time point. (B) Strains BESY49-6c and PWY194 were imaged as described in Materials and Methods except that 12 0.4-μm sections were taken. Spindles were considered to be at anaphase when one spindle pole had entered the bud. Distance between spindles poles (as detected by Ndc10-Venus) three or fewer sections apart were measured in three dimensions using softWoRx software; 108 spindles were measured for each strain.
DISCUSSION
Bir1 is a chromosomal passenger protein implicated in chromosome segregation. We have shown that the C terminus is necessary and sufficient for Bir1 kinetochore and spindle localization and is therefore critical for its functions as a chromosomal passenger. In fact, when fused to a nuclear localization sequence, the C-terminal 80 amino acids are sufficient for survival. The C-terminal 80 amino acids are also sufficient to interact with Sli15.
The IAP repeats of Bir1 seem to be completely dispensable for normal growth. Precise deletion of these repeats did not affect localization of Bir1 itself, localization of Sli15 or localization of Ndc10. They were also not important for high-fidelity chromosome segregation or the spindle checkpoint because the bir1-ΔIAP mutant behaved normally in response to loss of cohesin. This suggests that in Bir1 homologues, such as survivin, the C terminus but not the IAP repeats are important in their role at the mitotic spindle.
The middle region of Bir1 is required to localize Ndc10 to the anaphase spindle and to the midzone but is not required for kinetochore localization of Ndc10. The C-terminal half of Bir1 is required for a strong two-hybrid interaction with Ndc10 (Yoon and Carbon, 1999). We find that even small deletions in this region abolish Ndc10 localization. Moreover, the interaction between Bir1 and Ndc10 is regulated by phosphorylation. Mutation of the nine CDK minimal consensus sites in the middle region of Bir1 reduced the mobility shift as seen by Western blot analysis and prevented Ndc10 localization to the anaphase spindle.
We found that Bir1-TAP copurified with Sli15. Sli15-TAP copurifies with Bir1 (Cheeseman et al., 2002). Bir1 and Sli15 colocalize throughout the cell cycle. Both Bir1 and Sli15 are heavily modified in a cdc20-based metaphase arrest (Sullivan et al., 2004). However, the modification of Sli15 is removed earlier in the cell cycle than the modification of Bir1. Sli15 is dephosphorylated at anaphase by the FEAR network (Pereira and Schiebel, 2003), whereas we find that Bir1 remains modified in a cdc15-2 arrest when the FEAR network is active but the mitotic exit network is not. Despite their colocalization throughout the cell cycle, Bir1 and Sli15 are regulated differently.
We propose that unphosphorylated Sli15 brings phosphorylated Bir1 to the spindle at anaphase. Phosphorylated Bir1, in turn, brings Ndc10 to the spindle and midzone. Ndc10 remains associated with Bir1 until after spindle depolymerization because it has been shown that Ndc10 tracks plus ends of depolymerizing interpolar microtubules (Bouck and Bloom, 2005). At or after mitotic exit, Bir1 is dephosphorylated, disrupting interaction with Ndc10.
Loss of Ndc10 from the anaphase spindle by mutating the phosphorylation sites in Bir1 caused both an increase in chromosome loss and a defect in spindle elongation. However, we do not see a severe cytokinesis defect as is described for the ndc10-1 mutant (Bouck and Bloom, 2005). Ndc10's major role in cytokinesis seems to be independent of its spindle and midzone localization. The inability to fully elongate the anaphase spindle may increase the possibility of missegregation of rare lagging chromosomes resulting in the increase in the rate of chromosome loss we observed in mutants in the middle region of Bir1.
Acknowledgments
We are grateful to Eric Muller for critical reading of the manuscript. We thank Susan Francis and Brian Kennedy for helpful discussions. P.O.W., J.S.L., and T.N.D. were funded by National Institutes of Health Grant R01 GM-40506. P.O.W. was funded in part by Public Health Service National Research Service Award T32 GM-07270 from National Institute of General Medical Sciences. J.R.Y., S. A., and S. N. were funded by National Institutes of Health Grant P41 RR-11823
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–07–0640) on December 28, 2005.
Abbreviations used: CDK, cyclin-dependent kinase; IAP, inhibitor of apoptosis; ORF, open reading frame.
References
- Biggins, S., and Murray, A. W. (2001). The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15, 3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck, D. C., and Bloom, K. S. (2005). The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA 102, 5408–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I. M., Anderson, S., Jwa, M., Green, E. M., Kang, J., Yates, J. R., 3rd, Chan, C. S., Drubin, D. G., and Barnes, G. (2002). Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172. [DOI] [PubMed] [Google Scholar]
- Davis, T. N. (1992). A temperature-sensitive calmodulin mutant loses viability during mitosis. J. Cell. Biol. 118, 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser, J. R., Sundberg, H. A., Chang, B. H., Muller, E. G., and Davis, T. N. (1993). The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. Mol. Cell. Biol. 13, 7913–7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazbun, T. R., et al. (2003). Assigning function to yeast proteins by integration of technologies. Mol. Cell 12, 1353–1365. [DOI] [PubMed] [Google Scholar]
- Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- Kim, J. H., Kang, J. S., and Chan, C. S. (1999). Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell. Biol. 145, 1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea, D. E., and Coulson, C. A. (1949). The distribution of the numbers of mutants in bacterial populations. J. Genet. 49, 264–284. [DOI] [PubMed] [Google Scholar]
- Lens, S. M., and Medema, R. H. (2003). The survivin/Aurora B complex: its role in coordinating tension and attachment. Cell Cycle 2, 507–510. [DOI] [PubMed] [Google Scholar]
- Leverson, J. D., Huang, H. K., Forsburg, S. L., and Hunter, T. (2002). The Schizosaccharomyces pombe aurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol. Biol. Cell 13, 1132–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Ambrosini, G., Chu, E. Y., Plescia, J., Tognin, S., Marchisio, P. C., and Altieri, D. C. (1998). Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396, 580–584. [DOI] [PubMed] [Google Scholar]
- Li, F., Flanary, P. L., Altieri, D. C., and Dohlman, H. G. (2000). Cell division regulation by BIR1, a member of the inhibitor of apoptosis family in yeast. J. Biol. Chem. 275, 6707–6711. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., 3rd, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- MacCoss, M. J., et al. (2002). Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. USA 99, 7900–7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita, J., Matsusaka, T., Goshima, G., Nakamura, T., Tatebe, H., and Yanagida, M. (2001). Bir1/Cut17 moving from chromosome to spindle upon the loss of cohesion is required for condensation, spindle elongation and repair. Genes Cells 6, 743–763. [DOI] [PubMed] [Google Scholar]
- Muller, E. G. (1996). A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol. Biol. Cell 7, 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, E. G., Snydsman, B. E., Novik, I., Hailey, D. W., Gestaut, D. R., Niemann, C. A., O'Toole, E. T., Giddings, T. H., Jr., Sundin, B. A., and Davis, T. N. (2005). The organization of the core proteins of the yeast spindle pole body. Mol. Biol. Cell 16, 3341–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., and Schiebel, E. (2003). Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 302, 2120–2124. [DOI] [PubMed] [Google Scholar]
- Pinsky, B. A., Tatsutani, S. Y., Collins, K. A., and Biggins, S. (2003). An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell 5, 735–745. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, S., and Balasubramanian, M. K. (2002). Schizosaccharomyces pombe Bir1p, a nuclear protein that localizes to kinetochores and the spindle midzone, is essential for chromosome condensation and spindle elongation during mitosis. Genetics 160, 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge, K. W., Wellinger, R. J., and Zakian, V. A. (1991). Effects of excess centromeres and excess telomeres on chromosome loss rates. Mol. Cell. Biol. 11, 2919–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink, G. R., Hicks, J. B., and Cold Spring Harbor Laboratory. (1986). Laboratory Course Manual for Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sullivan, M., Hornig, N. C., Porstmann, T., and Uhlmann, F. (2004). Studies on substrate recognition by the budding yeast separase. J. Biol. Chem. 279, 1191–1196. [DOI] [PubMed] [Google Scholar]
- Tasto, J. J., Carnahan, R. H., McDonald, W. H., and Gould, K. L. (2001). Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast 18, 657–662. [DOI] [PubMed] [Google Scholar]
- Uren, A. G., Beilharz, T., O'Connell, M. J., Bugg, S. J., van Driel, R., Vaux, D. L., and Lithgow, T. (1999). Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc. Natl. Acad. Sci. USA 96, 10170–10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli, P., and Earnshaw, W. C. (2004). Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma 113, 211–222. [DOI] [PubMed] [Google Scholar]
- Wheatley, S. P., Henzing, A. J., Dodson, H., Khaled, W., and Earnshaw, W. C. (2004). Aurora-B phosphorylation in vitro identifies a residue of survivin that is essential for its localization and binding to inner centromere protein (INCENP) in vivo. J. Biol. Chem. 279, 5655–5660. [DOI] [PubMed] [Google Scholar]
- Widlund, P. O., and Davis, T. N. (2005). A high efficiency method to replace essential genes with mutant alleles in yeast. Yeast 22, 769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, A. P., Bruns, M., and Hartley, B. S. (1989). Extraction and rapid inactivation of proteins from Saccharomyces cerevisiae by trichloroacetic acid precipitation. Yeast 5, 51–53. [DOI] [PubMed] [Google Scholar]
- Yoder, T. J., McElwain, M. A., Francis, S. E., Bagley, J., Muller, E. G., Pak, B., O'Toole, E. T., Winey, M., and Davis, T. N. (2005). Analysis of a spindle pole body mutant reveals a defect in biorientation and illuminates spindle forces. Mol. Biol. Cell 16, 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, H. J., and Carbon, J. (1999). Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl. Acad. Sci. USA 96, 13208–13213. [DOI] [PMC free article] [PubMed] [Google Scholar]