Abstract
Slow Wallerian degeneration (WldS) mutant mice express a chimeric nuclear protein that protects sick or injured axons from degeneration. The C-terminal region, derived from NAD+ synthesizing enzyme Nmnat1, is reported to confer neuroprotection in vitro. However, an additional role for the N-terminal 70 amino acids (N70), derived from multiubiquitination factor Ube4b, has not been excluded. In wild-type Ube4b, N70 is part of a sequence essential for ubiquitination activity but its role is not understood. We report direct binding of N70 to valosin-containing protein (VCP; p97/Cdc48), a protein with diverse cellular roles including a pivotal role in the ubiquitin proteasome system. Interaction with WldS targets VCP to discrete intranuclear foci where ubiquitin epitopes can also accumulate. WldS lacking its N-terminal 16 amino acids (N16) neither binds nor redistributes VCP, but continues to accumulate in intranuclear foci, targeting its intrinsic NAD+ synthesis activity to these same foci. Wild-type Ube4b also requires N16 to bind VCP, despite a more C-terminal binding site in invertebrate orthologues. We conclude that N-terminal sequences of WldS protein influence the intranuclear location of both ubiquitin proteasome and NAD+ synthesis machinery and that an evolutionary recent sequence mediates binding of mammalian Ube4b to VCP.
INTRODUCTION
The E4 ubiquitination factor Ube4b (or Ufd2a) has a 123-amino acid N-terminal region that is essential for ubiquitination activity (Mahoney et al., 2002). It is unclear why this region is essential because it does not contain the U box, and it appears to be absent in invertebrate orthologues that ubiquitinate effectively (Koegl et al., 1999; Hatakeyama et al., 2001; Mahoney et al., 2002; Hoppe et al., 2004; Richly et al., 2005). It is important to understand the molecular mechanism of Ube4b because it has a key role in the ubiquitin proteasome system (UPS; Hoppe, 2005), it is neuroprotective in polyglutamine disorders (Matsumoto et al., 2004) and an important candidate gene for neuroblastoma (Krona et al., 2003). Information on the substrates of Ube4b is beginning to emerge (Hoppe et al., 2004; Okumura et al., 2004; Spinette et al., 2004; Richly et al., 2005) but there is much still to learn about its regulation.
In the slow Wallerian degeneration mutant mouse (WldS), 70 amino acids of this essential domain of Ube4b form the N-terminus of a chimeric protein that delays Wallerian degeneration of injured axons in mice and rats by 10-fold (see Figure 1A; Lunn et al., 1989; Mack et al., 2001; Adalbert et al., 2005). The chimeric protein is absent in wild-type mice. This sequence (N70) is fused in WldS protein to the full coding sequence of nicotinamide mononucleotide adenylyltransferase (Nmnat1; Conforti et al., 2000; Emanuelli et al., 2001; Mack et al., 2001), implicating the UPS or NAD+ metabolism in regulating axon degeneration. WldS also delays axon degeneration in a wide range of neurodegenerative disorders and acute retrograde axonal degeneration after spinal injury, indicating that axon degeneration mechanisms are more closely related than previously thought (Wang et al., 2002; Ferri et al., 2003; Samsam et al., 2003; Coleman, 2005; Kerschensteiner et al., 2005; Mi et al., 2005). Surprisingly, WldS in vivo has only been found in nuclei, suggesting that downstream axonal effector(s) mediate its remarkable effect on axon degeneration (Mack et al., 2001; Samsam et al., 2003; Sajadi et al., 2004).
Figure 1.
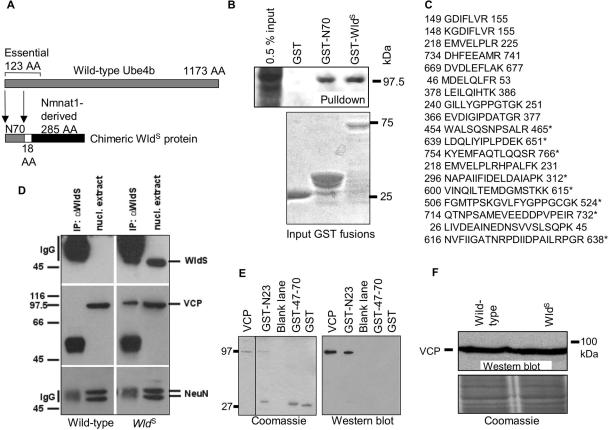
Direct binding of WldS/N70 sequence to VCP. (A) The N-terminal 70 amino acids of murine Ube4b (N70) are incorporated into the neuroprotective, chimeric WldS protein, fused in-frame to the entire coding sequence (285 amino acids) of Nmnat1 and an 18-amino acid linking region. In wild-type Ube4b, N70 forms part of a 123-amino acid sequence that is essential for ubiquitination activity. (B) A protein of ca. 97 kDa was pulled down from wild-type mouse brain by GST-WldS and by GST-N70. Bottom panel, a Coomassie-stained gel of the amounts of each protein used for pulldown, except that the GST sample has been diluted 1:10. The larger N70 and WldS proteins were obtained in lower quantities but were still able to precipitate significant amounts of the 97-kDa protein. (C) Tryptic peptides of VCP were identified by MALDI-TOF. Peptides marked with an asterisk (*), plus three additional VCP peptides (unpublished results), were present both in the protein pulled down by N70 and in the one pulled down by WldS. (D) Coimmunoprecipitation of VCP with WldS protein. Antibody Wld-18 was able to immunoprecipitate WldS protein (top) from nuclear extracts of mutant mouse brain (right) but not from wild-type mouse brain (left). Coimmunoprecipitation of VCP showed the same pattern (middle), whereas control protein NeuN is not coimmunoprecipitated (bottom) showing that the VCP result is specific. (E) Evidence that VCP binds directly to the N-terminus of WldS and Ube4b. Left, Coomassie-stained gel showing purified VCP and its precipitation by WldS peptides; right, Western blot confirming identity of the precipitated VCP and its complete absence in control lanes. The N-terminal 23 amino acids (N23) fused to GST (see also Figure 3) are able to precipitate purified His-tagged VCP. Neither GST alone, nor amino acids 47–70 possess this specific binding activity. (F) VCP Western blot of wild-type and WldS mouse brain homogenates showing that steady state levels of VCP are not altered by the presence of WldS. Below: Coomassie stain showing equal loading. D–F are representative of at least two experiments.
Nmnat1 activity was reported to preserve injured axons for 3 d in vitro (Araki et al., 2004; Wang et al., 2005). However, axons in vivo are far longer, have a profoundly different environment, and are protected by full-length WldS for up to 3 wk (Crawford et al., 1995; Adalbert et al., 2005). A contribution of N70 to strengthening the neuroprotective phenotype in vivo is suggested by the fact that transgenic mice overexpressing Nmnat1 show a normal rate of Wallerian degeneration (Coleman and Perry, 2002; Conforti and Coleman, unpublished data). N70 could contribute to neuroprotection by perturbing the UPS, which is important for regulating axon degeneration (Zhai et al., 2003; Macinnis and Campenot, 2005). Alternatively, it could influence intranuclear targeting of Nmnat1, with its intrinsic NAD+ synthesis activity, as WldS protein clusters to discrete intranuclear foci in skeletal muscle (Mack et al., 2001) and in many neuronal subtypes in vivo (Haley et al., unpublished results). The inability of successive investigators to detect any increase in NAD+ level when Nmnat1 is manipulated (Mack et al., 2001; Anderson et al., 2002; Araki et al., 2004) suggests that NAD+ is synthesized at highly localized intranuclear sites or immediately is passed to downstream components within the same complex. Either way, intranuclear targeting should be important.
To understand better the function of N70 in both WldS and Ube4b proteins, we sought N70 binding partners. We identified valosin containing protein (VCP) as a direct binding partner of N70 that becomes targeted to discrete intranuclear foci when WldS protein is present. VCP, one of the AAA family of ATPases associated with a variety of activities, has critical roles in the UPS (Dai and Li, 2001; Jarosch et al., 2002; Wang et al., 2004) and many other cellular roles dictated by interacting proteins (Meyer et al., 2000; Mogk et al., 2004; Wang et al., 2004). It accumulates in neuronal nuclei in a range of neurodegenerative diseases and can influence neurodegeneration both positively and negatively (Hirabayashi et al., 2001; Higashiyama et al., 2002; Mizuno et al., 2003; Watts et al., 2004; Schroder et al., 2005). Its intranuclear roles include interaction with Werner protein to influence the DNA damage response pathway (Indig et al., 2004) and the nuclear import of the T-cell-specific adaptor protein (Marti and King, 2005). Our data suggest the possibility of further intranuclear roles for VCP and an important function for the N70 domain in both wild-type Ube4b and WldS proteins.
MATERIALS AND METHODS
Constructs
VCP full-length cDNA was PCR-amplified with the Expand High Fidelity PCR System (Roche, Mannheim, Germany) from a mouse fibroblast cDNA library (kindly provided by R. Lange) using the 5′ BamHI- and 3′ SalI-tagged primers (restriction sites in bold): 5′-ATATATGGATCCCCATGGCCTCTGGAGCCGATTC-3′; 5′-AATATTGTCGACTTAGCCATACAGGTCATCGTC-3′.
The products were cloned with BamHI/SalI into pGEX5X-1 (Amersham Biosciences, Freiburg, Germany), and with NcoI/SalI into the pGBKT7 vector (BD Biosciences Clontech, Heidelberg, Germany). The WldS protein, N70 and further truncation products were similarly PCR-amplified and cloned using the WldS transgene construct (Mack et al., 2001) as template and appropriate primers based on the WldS cDNA (GenBank AF260924). Human Ube4b was amplified from a construct kindly provided by Professor James Mahoney using primers 5′-ATCCCGGAATTCATGGAGGAGCTGAGCGCTGAT-3′ and 5′-CCGCCTCGAGTTAGTGATCGCTGTTCTGTTT-3′ (EcoRI and XhoI sites for cloning into pGEX5X-1 in bold). cDNA sequence encoding full-length WldS protein, Nmnat-1 or Ube4b amino acids 1–70 (N70) was PCR-amplified from the WldS transgene template (Mack et al., 2001) using the high-fidelity enzyme Pfu (Stratagene, Heidelberg, Germany) and appropriate combinations of the following primers (1 + 2, 4 + 2, and 1 + 3, respectively). 5′ restriction enzyme tags, added for cloning purposes, are shown in bold and the first or last three bases of sequence derived from the WldS gene are underlined. A single base change to repress the stop codon of WldS Rev and allow read-through of the C-terminal EGFP is double-underlined: 1) 5′-TAGATCCCAAGCTTAACCTTTCACCATTAAGAGGAAAGCGATG-3′; 2) 5′-GCGGGATCCCGTCCCAGAGTGGAATGGTTGTG-3′; 3)5′-TCCTCCCCGCGGGTCTGCTGCACCTATGGGGGA-3′; and 4) 5′-GACTAGCTAGCATGGACTCATCCAAGAAGACAG-3′.
After cloning of pEGFP-N1 (BD Biosciences), all sequences were verified using the Taq FS BigDye-terminator cycle sequencing method on a ABI 377 prism sequencer and the corresponding ABI software. The sequences were analyzed using the GCG program Wisconsin Package Version 10.2 (Accelrys, San Diego, CA).
Full-length WldS DsRed construct was generated by cloning the HindIII/BamHI insert from pEGFP-N1 (above) into pDsRed2-N1 (BD Biosciences). WldS lacking N16 was generated using the following HindIII and BamHI-tagged primers: 5′-TAGCCCAAGCTTTAGGCCGCCACCATGCTTGCTGGTGGACAGACCT-3′; 5′-GCGGGATCCCGTCCCAGAGTGGAATGGTTG-3′.
Pulldown Assays
C57BL/6J mouse brain homogenates were used in this experiment to avoid competition from endogenous WldS protein. Brains were flash-frozen and homogenized in 50 volumes of 50 mM Tris, pH 9.0, and 1% deoxycholate containing protease inhibitor mix (Sigma, Taufkirchen, Germany). After incubating for 30 min at 37°C, insoluble material was removed by centrifugation (10,000 × g, 30 min) and the supernatant was dialyzed against binding buffer (50 mM Tris, pH 7.4, and 0.1% Triton X-100) overnight at 4°C. Homogenates were incubated overnight at 4°C with GST fusion proteins bound to glutathione Sepharose 4B. The beads were washed four times with phosphate-buffered saline (PBS)/0.1% Triton X-100 and resuspended in 3× standard Laemmli sample buffer. Proteins were separated by SDS-PAGE on 12% gels and analyzed by Western blotting.
MALDI-TOF Mass Spectrometry
Proteins fished by a pulldown assay were eluted with double-concentrated SDS-PAGE sample buffer and separated by SDS-PAGE. From the gel individual Coomassie blue-stained protein bands were excised with a scalpel and destained by washing with 25 mM NH4HCO3/50% acetonitrile. For MALDI-TOF mass spectrometry analysis, the samples were dissolved in 5 μl 0.1% aqueous trifluoroacetic acid. MALDI-MS was carried out in linear mode on a Bruker Reflex IV equipped with a video system (Rheinstetten, Germany), a nitrogen UV laser (Omax = 337 nm), and a HiMass detector. One microliter of the sample solution was placed on the target and 1 μl of a freshly prepared saturated solution of sinapinic acid in acetonitrile/H2O (2:1) with 0.1% trifluoroacetic acid was added. The spot was then recrystallized by addition of another 1 μl acetonitrile/H2O (2:1), which resulted in a fine crystalline matrix. For recording of the spectra an acceleration voltage of 20 kV was used, and the detector voltage was adjusted to 1.9 kV. Approx. 500 single laser shots were summed into an accumulated spectrum. Calibration was carried out using the single and doubly protonated ion signal of bovine serum albumin for external calibration. Identification of the mass fingerprint spectra was performed using the Mascot program available from Matrix Science on the World Wide Web (http://www.matrixscience.com/home.html).
Isolation and Immunoprecipitation of Nuclear Proteins
Six mouse brains (ca. 2.5 g) were each homogenized using a Dounce homogenizer in 40 ml precooled nuclear isolation medium (NIM; 0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 10 mM Tris/HCl, pH 7.4) supplemented with protease inhibitor cocktail (Sigma). Unbroken cells and connective tissue were removed by filtration. The filtered homogenate was then diluted with an equal volume of ice-cold NIM, centrifuged (10 min, 800 × g) and the pellet, including lipids, was resuspended in 40 ml NIM supplemented with protease inhibitor cocktail. After repeating the centrifugation, the new pellet was resuspended in 8 ml NIM, 2 ml sucrose density barrier (SDB; 2.3 M sucrose, 25 mM KCl, 5 mM MgCl2, 10 mM Tris/HCl, pH 7.4) solution was added and mixed thoroughly. 10 ml of the suspension was underlayered with 2.3 M sucrose and centrifuged for 1 h at 100,000 × g in a Beckman SW41 Ti rotor (Krefeld, Germany). The pellet, together with remaining lipids, was resuspended in PBS containing 1% Triton X-100 and incubated for 2 h at 4°C with continuous agitation. Samples were then centrifuged at 16,000 × g to generate a supernatant containing solubilized nuclear proteins.
For the subsequent immunoprecipitation, protein lysates were preincubated with 30 μl of protein G agarose (Roche) for 2 h at 4°C under constant agitation. Samples were centrifuged (5 min, 500g) to remove proteins unspecifically bound to the protein G agarose and 3 μl of anti-WldS rabbit serum and another 30 μl of protein G agarose were then added and incubated overnight at 4°C with constant agitation. The precipitated proteins bound to protein G agarose were then repeatedly washed by centrifugation at 500 × g at 4°C for 5 min. The final pellet was resuspended in 1 ml PBS, 1% Triton X-100. Samples were then analyzed by SDS-PAGE and immunoblotting.
In Vitro Binding Assays
GST fusion proteins were purified and coupled to glutathione-Sepharose 4B according to the protocol of the manufacturer (Amersham Biosciences). The pGBKT7 constructs containing the T7 promoter were in vitro transcribed and translated incorporating 35S-methionine using the TNT T7 Reticulocyte Lysate Coupled Transcription/Translation kit from Promega (Promega GmbH, Mannheim, Germany). The obtained proteins were mixed with equal amounts of GST fusion proteins and the binding assay was performed as previously described (Dai et al., 1998). Reactions were analyzed by SDS-PAGE, and the gel was fixed with 10% methanol/10% acetic acid before autoradiography. Purified His-tagged VCP was a kind gift from Dr. Sarah Spinette (Johns Hopkins).
Cell Culture and Transfection
Plasmid DNA was isolated using the endonuclease free plasmid kit (QIAGEN, Hilden, Germany). DNA was transfected using LipofectAMINE 2000 (Invitrogen) into COS-7, PC12, or HeLa cells immediately before differentiation by culturing in 100 ng/μl NGF on a type IV collagen substrate (Sigma). The “TV” PC12 subline, stably transfected with a tet-off inducible C-terminal EGFP-tagged VCP construct (Kobayashi et al., 2002) was grown in 1.0 μg/ml doxycycline (Sigma), which was removed to induce VCP/EGFP expression. Protein location was observed 1–5 d after transfection. Spinal motor neurons from embryonic day 14 (E14) embryonic rats were cultured and electroporated in suspension as previously described (Henderson et al., 1995; Raoul et al., 2002).
Immunocytochemistry
Slices of 100 μm were cut from the cerebellum of WldS mice and fixed in 4% paraformaldehyde (Fisher, Schwerte, Germany). Lumbar spinal cord dorsal root ganglia (DRG) were removed from WldS mice or rats and fixed in 4% paraformaldehyde, and then 20-μm sections cut on a cryostat. Slices were incubated for 2 h at room temperature in serum blocker consisting of 4% bovine serum albumin (Sigma) and 0.5% Triton X-100 (Sigma) in PBS. Wld-18 antibody (Samsam et al., 2003; 1:500 dilution) plus VCP antibody (against amino acids 9–130; BD Biosciences; 1:200 dilution; or mouse monoclonal to VCP (ab11433); AbCam, Cambridge, United Kingdom, 1:500 dilution) were applied simultaneously in serum blocking solution overnight at 4°C. After washing with PBS, slices were incubated overnight at 4°C in a solution containing TRITC-conjugated anti-rabbit (DAKO, Hamburg, Germany; 1:20 dilution in PBS) plus Alexa488-conjugated anti-mouse (Molecular Probes, Leiden, Netherlands; 1:200) secondary antibodies. Slices were then washed in PBS and incubated in To-pro3 (Molecular Probes) for 10 min before mounting in Mowiol/DABCO preparation. Staining was visualized on a laser scanning confocal microscope (Bio-Rad Radiance 2000, Hemel Hempsted, United Kingdom) and Z-series were merged using Lasersharp (Bio-Rad) software.
Cultured cells were fixed for 30 min in 4% paraformaldehyde, permeabilized with Triton-X-100 (0.1%, 5 min), blocked with horse serum (5%, 1 h), and incubated with primary (1 h) and secondary (45 min) antibodies with multiple washes in PBS between each stage.
VCP antibody was used as described above. Anti-ubiquitin polyclonal antibody (DAKO) was used at 1:100 dilution and anti-neurofilament medium chain polyclonal antibody (Chemicon, Hofheim, Germany; Ab1987) was used as 1:1000. Images were taken on a Zeiss LSM 510 META confocal system (Oberkochen, Germany; LSM Software Release 3.2) coupled to a Zeiss Axiovert 200 microscope.
Western Blotting
WldS protein expression was analyzed in mouse cerebella homogenized in five volumes of RIPA buffer, respectively, plus 1× Complete protease inhibitor cocktail (Roche) or in PC12 cells scraped from the dish in a minimal volume of the same buffer. Proteins were separated by SDS-PAGE and semidry blotted onto nitrocellulose (Bio-Rad). Blocking and incubation with primary antibodies and horseradish peroxidase-conjugated secondary antibodies (1:5000; Serotec, Heidelberg, Germany) were performed in PBS plus 0.02% Tween-20 and 5% low fat milk. Proteins were visualized using the ECL detection kit (Amersham Biosciences) according to the manufacturer's instructions.
RESULTS
WldS Protein Binds Directly to VCP through its N-terminal Domain
A screen for WldS binding partners revealed that a protein of ca. 97 kDa can be pulled down in large quantities from mouse brain homogenate by GST-WldS (Figure 1B). N70 is sufficient for this activity. Isolation of this protein from an SDS-PAGE gel followed by MALDI-TOF mass spectrometry protein identification revealed several tryptic peptides exactly matching sequences from murine VCP (Figure 1C). To confirm that binding of WldS to VCP is biologically relevant, we coimmunoprecipitated VCP specifically from nuclear extracts of WldS mouse brain using antibody Wld-18 and confirmed the identity of VCP by Western blotting (Figure 1D). VCP could not be coimmunoprecipitated from wild-type mouse brain, which lacks WldS protein. The reciprocal coimmunoprecipitation was inconclusive because VCP itself was not immunoprecipitated by the BD Biosciences anti-VCP antibody, but these data strongly suggest that the WldS/VCP complex is biologically relevant. To determine whether the binding was direct or mediated by other factors, we precipitated purified His-tagged VCP (a kind gift from Dr. Sarah Spinette and Prof. Antony Rosen, Johns Hopkins) using GST-tagged N-terminal fragments of WldS (Figure 1E; see also Figure 3). The ability of WldS-derived peptides to precipitate VCP was retained in the absence of other relevant proteins, showing that VCP binds directly to the shared N-terminus of WldS and Ube4b. Because WldS protein has been suggested to alter the UPS (Zhai et al., 2003; Coleman and Ribchester, 2004), we then tested by Western blotting of WldS and wild-type brain homogenates whether WldS alters turnover of VCP, and thus its steady-state level, and found that it does not (Figure 1F). Thus, VCP directly binds to WldS protein through its N70 domain, and this binding does not significantly alter steady-state level of VCP in brain.
Figure 3.
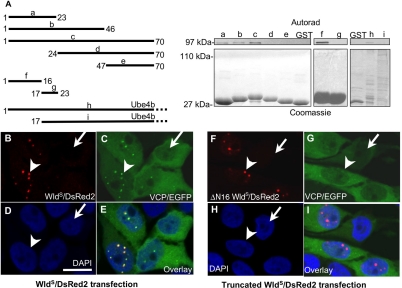
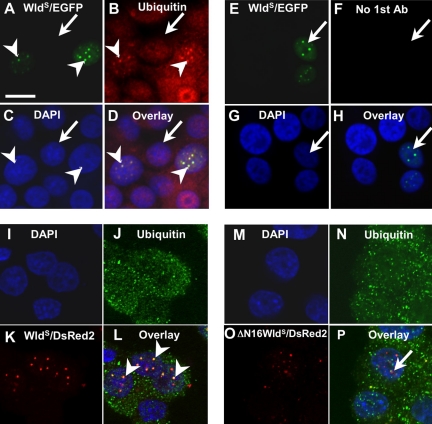
VCP binds within the first 16 residues of WldS. (A) Left: truncated GST fusion constructs of WldS and Ube4b. Right top, autoradiogram of 35S-labeled, IVTT expressed VCP pulled down with these constructs; right bottom, corresponding Coomassie-stained gel. Constructs a, b, and c, containing the first 23 residues, were able to pull down VCP, but the more C-terminal constructs d and e were not. The region necessary and sufficient for binding was subsequently narrowed to amino acids 1–16 using constructs f and g. Ube4b also pulled down VCP but failed to bind when this sequence was removed. Note that very little soluble full-length GST-Ube4b could be recovered by this method. However, this was sufficient to pull down VCP (lane h), whereas a greater amount of truncated GST-Ube4b (lane i) was not. (B–E) Full-length WldS/DsRed2 fusion protein partially redistributes VCP/EGFP fusion protein in the PC12 subline TV. (B) Transiently transfected WldS/DsRed2 fusion protein. (C) Stably transfected VCP/EGFP fusion protein. (D) DAPI. (E) Overlay. In cells not transfected by WldS/DsRed2, VCP/EGFP (arrow in C), like endogenous VCP (Supplementary Figure 6), is expressed throughout the cytoplasm and most of the nucleus as previously reported (Hirabayashi et al., 2001; Kobayashi et al., 2002). In cells transfected by WldS/DsRed2, VCP/EGFP signal colocalizes with the punctate intranuclear pattern of WldS/DsRed2 (arrowheads in B and C), while retaining the cytoplasmic and nuclear VCP signal. (F–I) WldS/DsRed2 fusion protein truncated by 16 amino acids at the N-terminus does not alter VCP distribution. (F) Transiently transfected, N-terminally truncated WldS/DsRed2 fusion protein. (G) Stably transfected VCP/EGFP fusion protein. (H) DAPI. (I) Overlay. VCP/EGFP distribution in the absence of truncated WldS/DsRed2 is similar to that in C (arrow in G). Although truncated WldS/DsRed2 still shows a punctate intranuclear distribution (arrowhead in F), it does not alter the distribution of VCP/EGFP (arrowhead in G). Scale bar, 10 μm. Figures are representative of two (A) or three (B–I) experiments.
WldS Colocalizes with and Partially Redistributes VCP
To investigate further whether WldS/VCP complexes form inside living cells, we carried out colocalization studies in vitro and in vivo. We took advantage of the fact that WldS clusters into discrete intranuclear foci in some neuronal subtypes (Haley et al., unpublished results) and other cells (Mack et al., 2001; Figure 2). After transient transfection of WldS/EGFP fusion construct into PC12 cells, VCP partially shifted from the widespread nuclear and cytoplasmic distribution that was previously described (Hirabayashi et al., 2001; Kobayashi et al., 2002) and is present in untransfected cells (Figure 2B, arrow) to produce the same pattern of intranuclear foci as WldS (Figure 2B, arrowhead). VCP is an abundant protein (Dai and Li, 2001), so the wider nuclear and cytoplasmic pool remained without any obvious depletion (Figure 2, A–C). VCP was also partially redistributed by WldS in HeLa cells which, like PC12, remained healthy (Supplementary Figures 2 and 4).
Figure 2.
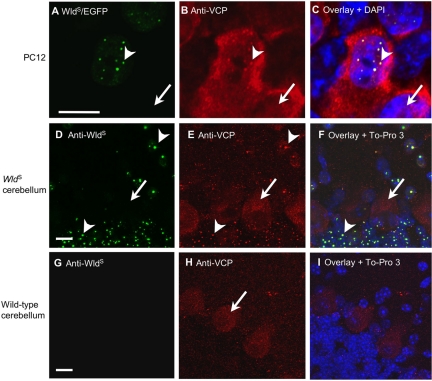
WldS partially relocalizes VCP in vitro and in vivo. (A–C) PC12 cells transiently transfected with WldS/EGFP fusion construct. (A) WldS/EGFP shows a punctate intranuclear pattern in transfected cells (arrowhead). Arrow marks nucleus of an untransfected cell. (B) VCP immunocytochemistry reveals the same pattern of intranuclear foci in transfected cells (arrowhead) together with widespread nuclear and cytoplasmic staining. Untransfected cells show only the wider nuclear and cytoplasmic signal (arrow). (C) Overlay, together with DAPI stain, confirms that the WldS and VCP puncta colocalize within nuclei. The specificity of the staining and the healthy status of the cells is demonstrated in Supplementary Figures 1 and 3. (D–I) Double immunofluorescence staining of sections of WldS and wild-type mouse cerebellum with WldS (green) and VCP (red). (D) As in PC12 cells, WldS localizes in mouse cerebellar granule cells and molecular layer neurons to intranuclear spots, although there are fewer spots per nucleus than in most PC12 cells (arrowheads). Purkinje cells (arrow) appear not to express WldS. (E) Immunostaining for VCP reveals the same pattern of spots as WldS (arrowheads), as well as staining both cytoplasm and nuclei of the large Purkinje cells (arrow). (F) Overlay, together with To-pro3 nuclear stain (blue), confirms that the WldS and VCP puncta colocalize within nuclei. (G) WldS antibody showed no specific stain in wild-type cerebellum. (H) VCP intranuclear foci were not found in cerebellar granule cells or molecular layer neurons in wild-type mice. The faintly stained Purkinje cells (arrow) confirm that E and H were similarly processed and that cells were healthy. (I) Overlay, together with To-pro3 stain, indicates the position of nuclei that lack WldS protein. The nonhomogeneous staining by DAPI in C and to-pro3 in F and I reflects nonhomogeneous distribution of heterochromatin (Wu et al., 2005). Scale bars, 10 μm. Each figure is representative of four or more experiments.
To study this in vivo, we made use of the finding that cerebellar granule cells and molecular layer neurons also show intranuclear foci of WldS protein, albeit of different size and number from PC12 cells (Haley et al., unpublished results). Again WldS and VCP colocalized within intranuclear foci in these neurons from WldS mice (Figure 2, D–F), whereas in wild-type cerebellum, there were no intranuclear VCP foci (Figure 2, G–I). Some DRG neurons in both WldS mice and rats also showed good colocalization (Supplementary Figure 5), indicating that the WldS/VCP interaction occurs in at least some neurons known to express the neuroprotective WldS phenotype and that this binding is conserved between species. The situation is less clear in motor neurons, where WldS is more homogenously distributed in vivo (Mack et al., 2001; Samsam et al., 2003), and VCP colocalization with WldS spots was not evident in vitro (Supplementary Figure 5). Nevertheless, the data are sufficient to conclude that WldS and VCP bind one another under physiological conditions, including in at least some neurons that display the WldS phenotype.
VCP Binds within the First 16 Residues of WldS
We show above that WldS binds to and partially redistributes VCP. To establish that binding causes redistribution, we mapped and then deleted key residues of WldS required for VCP binding. Truncated N70 constructs were expressed as GST-fusion proteins and used to pull down IVTT-expressed VCP. First the N-terminal 23 amino acids were found to be necessary and sufficient for VCP binding, and this was subsequently narrowed to the N-terminal 16 amino acids (N16; Figure 3A). Ube4b lacking this sequence also did not bind VCP. Two WldS/DsRed2 constructs were then made: one full-length and one lacking N16, and transiently transfected into the TV PC12 stable subline that expresses VCP/EGFP fusion protein in a tet-off-inducible manner (a kind gift from Prof. Akira Kakizuka). Full-length WldS/DsRed2 was able to bring about partial redistribution of VCP/EGFP into intranuclear foci in transfected cells (arrowhead; Figure 3, B–E) but removal of N16 containing the VCP binding site prevented this (Figure 3, F–I). Interestingly, the ability of WldS protein to accumulate in intranuclear foci does not require binding to VCP (Figure 3F). Thus, VCP has to bind WldS N16 to be partially redistributed in the manner we describe.
The Sequence Targeting WldS Protein to Intranuclear Foci Is Not Nmnat1
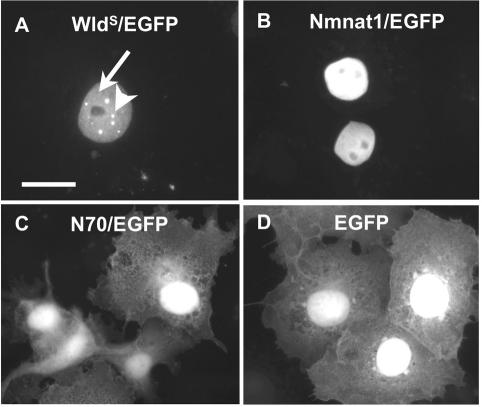
Because VCP binding is not required to target WldS to intranuclear foci, we asked whether this is a property of the Nmnat1 sequence by fusing Nmnat1 to EGFP and transiently transfecting it (Figure 4). Nmnat1, but not N70, was restricted to the nucleus, confirming previous reports that Nmnat1 is a nuclear protein and suggesting that nuclear targeting of WldS protein is due to the putative nuclear localization signal of Nmnat1 (Raffaelli et al., 2002; Araki et al., 2004; Magni et al., 2004). However, unlike WldS/EGFP, intranuclear distribution of Nmnat1/EGFP was homogeneous apart from its exclusion from nucleoli. The fraction of N70/EGFP that entered the nucleus was also homogeneously distributed, suggesting that WldS protein is targeted to intranuclear foci either by a conformational structure involving both N70 and Nmnat1 sequence or by the short unique sequence that separates these two parts of the protein. Cell and nuclear shape indicated that the cells were healthy in all cases. We conclude that Nmnat1 and WldS protein differ in their intranuclear distribution, but as shown above, their differing abilities to bind VCP do not underlie this difference.
Figure 4.
Nmnat1 is not sufficient to target WldS protein to intranuclear foci. C-terminal EGFP fusion constructs transiently transfected into COS cells. (A) WldS/EGFP localizes primarily to discrete intranuclear foci (arrowhead), although some remains homogeneously distributed inside the nucleus (arrow). (B) Nmnat1/EGFP is homogeneously distributed, apart from exclusion from nucleoli. N70/EGFP (C) and EGFP (D) alone are homogeneously distributed throughout the cell but EGFP has a preference for the nucleus as previously described (Alonso et al., 2004). Neither is targeted to intranuclear foci. Scale bar, 10 μm. Figures are representative of three experiments.
WldS Also Can Partially Relocalize Ubiquitin inside the Nucleus
VCP binds long ubiquitin chains (Dai and Li, 2001) so one consequence of its partial redistribution by WldS protein could be redistribution of bound polyubiquitinated proteins. This was supported by immunocytochemistry of PC12 cells transiently transfected with WldS/EGFP fusion construct (Figure 5, A–H). As with VCP, the normal distribution of ubiquitin throughout the nucleus and cytoplasm of untransfected cells is joined by a punctate intranuclear pattern in transfected cells, where ubiquitin puncta colocalize with WldS/EGFP. Cell and nuclear shape indicated that the cells were healthy (see also Supplementary Figure 3). The association of ubiquitin epitopes with WldS also depended on the N16 sequence, suggesting it is secondary to VCP binding to WldS (Figure 5, I–P).
Figure 5.
Full-length WldS can also partially relocalize ubiquitin inside the nucleus. (A) PC12 cells transiently transfected with WldS/EGFP fusion construct show a punctate intranuclear distribution (arrowheads), whereas untransfected cells show no signal (arrow). (B) Ubiquitin immunofluorescence shows colocalizing punctate signal in transfected cells (arrowheads), together with a widespread nuclear and cytoplasmic signal. Untransfected cells show only the more homogenous distribution (arrow). (C) DAPI. (D) Overlay. (E and F) Control experiment in which the first antibody against ubiquitin was omitted. Transfected cells (arrow) show no signal in the red channel. Panel order corresponds to A–D as indicated. Scale bar, 10 μm. (I–P) A repeat of the experiment in Figure 3 but immunostaining for ubiquitin instead of VCP. Ubiquitin shows the same dependence on N16 for colocalization with WldS/DsRed2 in intranuclear spots. (I–L) Transfection with full-length WldS/DsRed2 results in partial colocalization, which is retained when the confocal image stack is rotated (arrowheads in L). (M–P) Transfection with N-terminally truncated WldS/DsRed2 does not result in colocalization. The small region of overlapping signal (arrow in P) is not retained when the confocal image stack is rotated. (I and M) DAPI staining shows position of nuclei; (J and N) ubiquitin immunostain. (K and O) DsRed2 signal fused, respectively, to full-length WldS and WldS lacking N16. (L and P) Overlays.
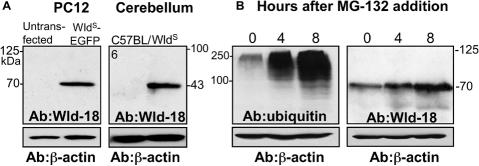
An alternative explanation for the accumulation of ubiquitin epitopes at these sites could be ubiquitination of the WldS/EGFP fusion protein, so we looked for these putative ubiquitinated species by Western blotting (Figure 6). Wld-18 antibody detected only a single band of the expected size, even on long-exposure ECLs after a generalized accumulation of ubiquitination products due to proteasome inhibition. The intensity of the WldS band did increase after proteasome inhibition, suggesting that WldS protein is degraded by the UPS but despite this, ubiquitinated WldS did not reach detectable levels on the Western blots. Although we cannot rule out the possibility that ubiquitinated WldS contributes to the ubiquitin speckles, the inability to detect this putative species on a Western blot and the probable dependence on VCP binding suggest that putative ubiquitination of WldS is unlikely to be the sole explanation for the ubiquitin speckles. Thus, we propose that polyubiquitinated proteins may accumulate in the WldS/VCP complex.
Figure 6.
Ubiquitinated WldS protein cannot be detected. (A) Western blotting with antibody to WldS protein did not detect any ubiquitinated WldS/EGFP fusion protein or ubiquitinated WldS in transfected PC12 cells (left) or in WldS mouse cerebellum (right), respectively. The 70- and 43-kDa bands correspond to the predicted molecular weights of the unmodified proteins. (B) The proteasome inhibitor MG-132 (10 μM for times shown) caused ubiquitination products to build up in PC12 cells (left), but ubiquitinated WldS/EGFP (>70 kDa) still could not be detected even on an overexposed ECL (right). Figures are representative two experiments.
VCP Binding to N16 Is an Evolutionary “Recent” Development
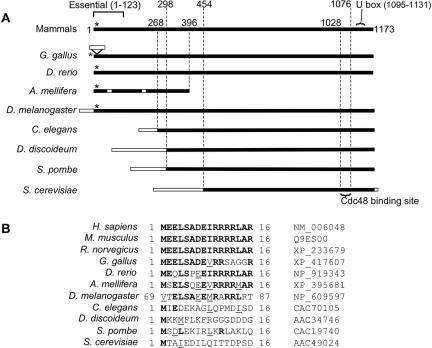
An alignment of human Ube4b with known orthologues shows that an N-terminal extension containing the N16 sequence is present in mammals, birds, fish, and insects, but this entire region is absent in nematodes, slime molds, and yeasts (Figure 7). Nevertheless, the VCP ortholog Cdc48 does bind Ufd2 in Saccharomyces cerevisiae (Koegl et al., 1999; Richly et al., 2005) and Caenorhabditis elegans (C. Kähler and T. Hoppe, personal communication) so the interaction in these species must have a different molecular basis. The Cdc48 binding site in S. cerevisiae has been mapped (Richly et al., 2005) and mammals do have a sequence homologous to it. However, this sequence appears to be insufficient for VCP binding in human, because N-terminally truncated Ube4b did not bind VCP (Figure 3A). Mammalian Ube4b was previously reported to bind VCP (Meyer et al., 2000; Kaneko et al., 2003) but the interacting sequence was not fully mapped. Our data show that VCP binds to N16, and this appears to be a recent event in evolutionary terms with distantly related higher eukaryotes employing a different mode of interaction.
Figure 7.
VCP binding to N16 is an evolutionary “recent” event. (A) Full-length alignment of Ube4b and Ufd2 sequences. (B) Amino acid alignment of N16-like or other N-terminal sequences in the same proteins. The database accession numbers are also shown. Mammals, birds, fish, and insects have sequence showing significant homology to the VCP binding site in N16 at positions denoted by the asterisk (*). Nematodes, slime molds, and yeasts lack the entire N-terminal extension containing this sequence. The Cdc48 binding site of S. cerevisiae Ufd2p (Richly et al., 2005), the U box (Hatakeyama et al., 2001), and the sequence essential for in vitro ubiquitination activity (Mahoney et al., 2002) are also marked. Black boxes, regions of significant homology to human Ube4b; open boxes, regions not homologous to human Ube4b; bold, amino acids identical to human sequence; underlined, conservative substitutions.
DISCUSSION
We report direct binding between VCP and the N-terminal 16 amino acids of WldS protein (N16) that drives partial redistribution of VCP in nuclei in vivo and in vitro when full-length WldS is present. N16 is necessary and sufficient for VCP binding and is necessary for relocalization of VCP, but is not required to target WldS to discrete intranuclear foci. Thus, sequences outside N16 cause WldS to accumulate at these sites and VCP is redistributed through binding to N16. The N-terminal domain of WldS also influences the intranuclear distribution of the covalently attached Nmnat1 sequence and hence the distribution of nuclear NAD+ synthesis machinery.
A recent report indicates that Nmnat1, but not N70, is sufficient to confer a WldS-like phenotype on neurons in vitro (Araki et al., 2004). However, the relative strengths of the neuroprotective phenotypes induced by Nmnat1 and WldS were not tested and Nmnat1 was not shown to protect in vivo, where WldS robustly protects longer axons for far greater time periods (Mack et al., 2001; Adalbert et al., 2005). Our data show that N70 is not an inert addition to Nmnat1, but instead concentrates VCP, NAD+ synthesis activity and probably its associated ubiquitinated proteins into subnuclear sites, functions that each have potential to influence the strength of the WldS phenotype. Thus, WldS joins a growing list of chimeric proteins whose biological activity amounts to more than the sum of their parts (Fujimoto et al., 1996; Campbell et al., 1997; Blume-Jensen and Hunter, 2001).
Further studies are needed to test whether VCP binding is required for the neuroprotective WldS phenotype, but this possibility was upheld in two important tests. First, WldS and VCP show colocalization in at least some DRG neurons known to express the neuroprotective phenotype, as well as in cerebellum where the phenotype in vivo remains to be tested. Second, colocalization was conserved across species in DRG of WldS rats (Adalbert et al., 2005). The more homogeneous intranuclear distribution of WldS in motor neurons in vivo makes colocalization in this cell type more difficult to test (Mack et al., 2001; Samsam et al., 2003). Cultured motor neurons electroporated with WldS/EGFP show WldS puncta without obvious colocalization of VCP but this does not exclude a role for VCP in the neuroprotective phenotype in vivo (Supplementary Figure 5).
Colocalization studies cannot ultimately answer the question of whether VCP is required for the WldS phenotype. Unfortunately, VCP knockout mice are not available to test the hypothesis and a more complex strategy may be needed. Given the essential role of VCP in endoplasmic reticulum-associated protein degradation (ERAD) and several reports of damaging effects of VCP RNAi, mutation, and deletion in other species or cell lines, it is unlikely that such knockout mice would be viable (Hirabayashi et al., 2001; Kobayashi et al., 2002; Wojcik et al., 2004; Yamanaka et al., 2004). In contrast, WldS alters VCP in a way that leaves all cells and organisms healthy, with no overt harmful effect in the original mutant (Lunn et al., 1989), four lines of transgenic mice (Mack et al., 2001), three of rats (Adalbert et al., 2005) or in double homozygous mice or rats (unpublished observations). This is likely to reflect the confinement of alterations to the nucleus, leaving the essential role of VCP in ERAD unaffected.
Studies in yeast suggest VCP stimulates substrate binding but limits the subsequent growth of a polyubiquitin chain (Richly et al., 2005). However, VCP is dispensable for Ube4b activity when concentrated preparations of protein are used in vitro (Hatakeyama et al., 2001; Mahoney et al., 2002) and in these circumstances removal of the N-terminal 123 amino acids of Ube4b blocks activity for unknown reasons (Mahoney et al., 2002). Our data further highlight the importance of the N-terminus of Ube4b as a regulatory sequence. Many invertebrates lack this sequence but still bind the VCP ortholog Cdc48, so this molecular interaction appears to have changed during evolution.
VCP plays pivotal roles in ERAD, nuclear envelope reconstruction, cell cycle, postmitotic Golgi reassembly, and suppression of apoptosis (Kondo et al., 1997; Dai and Li, 2001; Hetzer et al., 2001; Rabinovich et al., 2002; Wang et al., 2004). Its nuclear functions are equally diverse. First, a nuclear transport role is suggested by its association with adapter proteins Ufd1 and Np14, and by VCP-dependent transport of T-cell-specific adaptor protein into eukaryotic nuclei (Meyer et al., 2000; Marti and King, 2005). Second, VCP controls nucleolar retention of Werner syndrome helicase and its release after DNA damage (Indig et al., 2004). Interestingly, our data fit with the proposal of these authors that other nuclear binding partners regulate VCP distribution and control other pathways. Third, the nucleus has a quality control pathway (Gardner et al., 2005) likely to involve VCP. Interaction and colocalization with expanded polyglutamine and other intranuclear inclusions suggests VCP may be trying to clear misfolded nuclear proteins (Hirabayashi et al., 2001; Doss-Pepe et al., 2003; Mizuno et al., 2003). Finally, VCP regulates the stability of the transcription factor SPT23 (Richly et al., 2005), consistent with the tight regulation of transcription by the UPS (Muratani and Tansey, 2003).
VCP is altered in several neurodegenerative disorders so its interaction with the neuroprotective WldS protein is particularly interesting. VCP missense mutations cause inclusion body myopathy with Paget disease of bone and frontotemporal dementia, a disease characterized by ubiquitin-containing nuclear inclusions and white matter pathology (Watts et al., 2004; Schroder et al., 2005). VCP is present in Lewy-like inclusions in amyotrophic lateral sclerosis, in nigral Lewy neurites in Parkinson's disease (Ishigaki et al., 2004), and in ubiquitin-positive intraneuronal inclusions in motor neuron disease with dementia, ballooned neurons in Creutzfeldt-Jakob disease, and dystrophic neurites of senile plaque in Alzheimer's disease (Mizuno et al., 2003). In polyglutamine disorders such as Huntington's disease and Machado-Joseph disease, specific binding to expanded polyglutamine targets VCP to intranuclear inclusions (Hirabayashi et al., 2001). This interaction appears to influence disease severity, as loss-of-function mutations in the Drosophila VCP ortholog, ter4, dominantly suppress neurodegeneration caused by expanded polyglutamine (Higashiyama et al., 2002).
It is now important to determine whether WldS protein, or N70 can interfere with the effect of VCP in any of these neurodegenerative disorders. Ectopic expression in Drosophila of mammalian Ube4b suppresses polyglutamine disease, and loss of function mutations in VCP have a similar effect (Higashiyama et al., 2002; Matsumoto et al., 2004). It is also now clear that WldS and expanded polyglutamine both bind VCP directly and partially relocalize it to intranuclear foci (Hirabayashi et al., 2001). Taken together, these observations raise the intriguing possibility that N70 and VCP antagonize each other in some circumstances as a result of the binding interaction we describe.
In summary, we report direct interaction between VCP and the N-terminal 16 amino acids of both WldS and Ube4b proteins. This interaction drives focal intranuclear clustering of VCP in WldS neurons, probably together with associated multiubiquitinated proteins, and helps understand the function of an important, evolutionarily recent regulatory sequence in the Ube4b protein. It is now important to determine whether the redistribution of covalently attached Nmnat1 or of bound VCP influences the strength of the WldS phenotype in vivo and to understand how the N-terminal binding of VCP to Ube4b influences this ubiquitin ligase.
Supplementary Material
Acknowledgments
We thank Prof. Akira Kakizuka (Kyoto University) for the “TV” PC12 cell line stably transfected with VCP/EGFP fusion construct; Dr. Sarah Spinette, Professor James Mahoney, and Professor Antony Rosen (Johns Hopkins) for purified VCP and human Ube4b cDNA; and Rita Lange for mouse fibroblast cDNA library. We thank Dr. Thorsten Hoppe (University of Hamburg) for helpful discussion and Dr. Jonathan Gilley (Babraham Institute) for practical assistance. This work was funded primarily by Deutsche Forschungsgemeinschaft Grant CO 276/1-1 and by the Biotechnology and Biological Sciences Research Council. Additional funding came from the Federal Ministry of Education and Research (FKZ; Grant 01 KS 9502) and Center for Molecular Medicine, University of Cologne (CMMC), the Wellcome Trust, the Medical Research Council, the Amyotrophic Lateral Sclerosis Association and Koeln Fortune.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–04–0375) on December 21, 2005.
Abbreviations used: DRG, dorsal root ganglion; ERAD, endoplasmic reticulum-associated protein degradation; IVTT, in vitro transcription and translation; N16, N-terminal 16 amino acids of WldS and Ube4b; N70, N-terminal 70 amino acids of WldS and Ube4b; Nmnat1, nicotinamide mononucleotide adenylyltransferase; Ube4b, ubiquitination factor E4b; Ufd2, ubiquitin fusion degradation protein 2; UPS, ubiquitin proteasome system; VCP, valosin-containing protein; WldS, slow Wallerian degeneration gene or protein.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adalbert, R. et al. (2005). A rat model of slow Wallerian degeneration (WldS) with improved preservation of neuromuscular synapses. Eur. J. Neurosci. 21, 271–277. [DOI] [PubMed] [Google Scholar]
- Alonso, M. B. et al. (2004). Identification and characterization of ZFP-57, a novel zinc finger transcription factor in the mammalian peripheral nervous system. J. Biol. Chem. 279, 25653–25664. [DOI] [PubMed] [Google Scholar]
- Anderson, R. M., Bitterman, K. J., Wood, J. G., Medvedik, O., Cohen, H., Lin, S. S., Manchester, J. K., Gordon, J. I., and Sinclair, D. A. (2002). Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277, 18881–18890. [DOI] [PubMed] [Google Scholar]
- Araki, T., Sasaki, Y., and Milbrandt, J. (2004). Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305, 1010–1013. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen, P., and Hunter, T. (2001). Oncogenic kinase signalling. Nature 411, 355–365. [DOI] [PubMed] [Google Scholar]
- Campbell, R. K., Bergert, E. R., Wang, Y., Morris, J. C., and Moyle, W. R. (1997). Chimeric proteins can exceed the sum of their parts: implications for evolution and protein design. Nat. Biotechnol. 15, 439–443. [DOI] [PubMed] [Google Scholar]
- Coleman, M. P. (2005). Axon degeneration mechanisms: commonality amid diversity. Nat. Rev. Neurosci. 6, 889–898. [DOI] [PubMed] [Google Scholar]
- Coleman, M. P., and Perry, V. H. (2002). Axon pathology in neurological disease: a neglected therapeutic target. Trends Neurosci. 25, 532–537. [DOI] [PubMed] [Google Scholar]
- Coleman, M. P., and Ribchester, R. R. (2004). Programmed axon death, synaptic dysfunction and the ubiquitin proteasome system. Curr. Drug Targets CNS Neurol. Disord. 3, 227–238. [DOI] [PubMed] [Google Scholar]
- Conforti, L., Tarlton, A., Mack, T. G., Mi, W., Buckmaster, E. A., Wagner, D., Perry, V. H., and Coleman, M. P. (2000). A Ufd2/D4Cole1e chimeric protein and overexpression of rbp7 in the slow wallerian degeneration (WldS) mouse. Proc. Natl. Acad. Sci. USA 97, 11377–11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, T. O., Hsieh, S. T., Schryer, B. L., and Glass, J. D. (1995). Prolonged axonal survival in transected nerves of C57BL/Ola mice is independent of age. J. Neurocytol. 24, 333–340. [DOI] [PubMed] [Google Scholar]
- Dai, R. M., Chen, E., Longo, D. L., Gorbea, C. M., and Li, C. C. (1998). Involvement of valosin-containing protein, an ATPase Co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha. J. Biol. Chem. 273, 3562–3573. [DOI] [PubMed] [Google Scholar]
- Dai, R. M., and Li, C. C. (2001). Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 3, 740–744. [DOI] [PubMed] [Google Scholar]
- Doss-Pepe, E. W., Stenroos, E. S., Johnson, W. G., and Madura, K. (2003). Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol. Cell. Biol. 23, 6469–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelli, M., Carnevali, F., Saccucci, F., Pierella, F., Amici, A., Raffaelli, N., and Magni, G. (2001). Human NMN adenylyltransferase: molecular cloning, chromosomal localization, tissue mRNA levels, bacterial; Expression, and enzymatic properties. J. Biol. Chem. 276, 406–412. [DOI] [PubMed] [Google Scholar]
- Ferri, A., Sanes, J. R., Coleman, M. P., Cunningham, J. M., and Kato, A. C. (2003). Inhibiting axon degeneration and synapse loss attenuates apoptosis and disease progression in a mouse model of motoneuron disease. Curr. Biol. 13, 669–673. [DOI] [PubMed] [Google Scholar]
- Fujimoto, J., Shiota, M., Iwahara, T., Seki, N., Satoh, H., Mori, S., and Yamamoto, T. (1996). Characterization of the transforming activity of p80, a hyperphosphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5). Proc. Natl. Acad. Sci. USA 93, 4181–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, R. G., Nelson, Z. W., and Gottschling, D. E. (2005). Degradation-mediated protein quality control in the nucleus. Cell 120, 803–815. [DOI] [PubMed] [Google Scholar]
- Hatakeyama, S., Yada, M., Matsumoto, M., Ishida, N., and Nakayama, K. I. (2001). U-Box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276, 32111–33120. [DOI] [PubMed] [Google Scholar]
- Henderson, C. E., Bloch-Gallego, E., and Camu, W. (1995). Purified embryonic motoneurons. In: Nerve Cell Culture: A Practical Approach, ed. J. Cohen and G. Wilkin, London: Oxford University Press, 69–81.
- Hetzer, M., Meyer, H. H., Walther, T. C., Bilbao-Cortes, D., Warren, G., and Mattaj, I. W. (2001). Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat. Cell Biol. 3, 1086–1091. [DOI] [PubMed] [Google Scholar]
- Higashiyama, H., Hirose, F., Yamaguchi, M., Inoue, Y. H., Fujikake, N., Matsukage, A., and Kakizuka, A. (2002). Identification of ter94, Drosophila VCP, as a modulator of polyglutamine-induced neurodegeneration. Cell Death Differ. 9, 264–273. [DOI] [PubMed] [Google Scholar]
- Hirabayashi, M. et al. (2001). VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ. 8, 977–984. [DOI] [PubMed] [Google Scholar]
- Hoppe, T. (2005). Multiubiquitylation by E4 enzymes: `one size' doesn't fit all. Trends Biochem. Sci. 30, 183–187. [DOI] [PubMed] [Google Scholar]
- Hoppe, T., Cassata, G., Barral, J. M., Springer, W., Hutagalung, A. H., Epstein, H. F., and Baumeister, R. (2004). Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell 118, 337–349. [DOI] [PubMed] [Google Scholar]
- Indig, F. E., Partridge, J. J., Kobbe Cv, C., Aladjem, M. I., Latterich, M., and Bohr, V. A. (2004). Werner syndrome protein directly binds to the AAA ATPase p97/VCP in an ATP-dependent fashion. J. Struct. Biol. 146, 251–259. [DOI] [PubMed] [Google Scholar]
- Ishigaki, S., Hishikawa, N., Niwa, J., Iemura, S., Natsume, T., Hori, S., Kakizuka, A., Tanaka, K., and Sobue, G. (2004). Physical and functional interaction between Dorfin and Valosin-containing protein that are colocalized in ubiquitylated inclusions in neurodegenerative disorders. J. Biol. Chem. 279, 51376–51385. [DOI] [PubMed] [Google Scholar]
- Jarosch, E., Taxis, C., Volkwein, C., Bordallo, J., Finley, D., Wolf, D. H., and Sommer, T. (2002). Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4, 134–139. [DOI] [PubMed] [Google Scholar]
- Kaneko, C., Hatakeyama, S., Matsumoto, M., Yada, M., Nakayama, K., and Nakayama, K. I. (2003). Characterization of the mouse gene for the U-box-type ubiquitin ligase UFD2a. Biochem. Biophys. Res. Commun. 300, 297–304. [DOI] [PubMed] [Google Scholar]
- Kerschensteiner, M., Schwab, M. E., Lichtman, J. W., and Misgeld, T. (2005). In vivo imaging of axonal degeneration and regeneration in the injured spinal cord. Nat. Med. 11, 572–577. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., Tanaka, K., Inoue, K., and Kakizuka, A. (2002). Functional ATPase activity of p97/valosin-containing protein (VCP) is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells. J. Biol. Chem. 277, 47358–47365. [DOI] [PubMed] [Google Scholar]
- Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H. D., Mayer, T. U., and Jentsch, S. (1999). A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96, 635–644. [DOI] [PubMed] [Google Scholar]
- Kondo, H., Rabouille, C., Newman, R., Levine, T. P., Pappin, D., Freemont, P., and Warren, G. (1997). p47 is a cofactor for p97-mediated membrane fusion. Nature 388, 75–78. [DOI] [PubMed] [Google Scholar]
- Krona, C., Ejeskar, K., Abel, F., Kogner, P., Bjelke, J., Bjork, E., Sjoberg, R. M., and Martinsson, T. (2003). Screening for gene mutations in a 500 kb neuroblastoma tumor suppressor candidate region in chromosome 1p; mutation and stage-specific expression in UBE4B/UFD2. Oncogene 22, 2343–2351. [DOI] [PubMed] [Google Scholar]
- Lunn, E. R., Perry, V. H., Brown, M. C., Rosen, H., and Gordon, S. (1989). Absence of Wallerian degeneration does not hinder regeneration in peripheral nerve. Eur. J. Neurosci. 1, 27–33. [DOI] [PubMed] [Google Scholar]
- Macinnis, B. L., and Campenot, R. B. (2005). Regulation of Wallerian degeneration and nerve growth factor withdrawal-induced pruning of axons of sympathetic neurons by the proteasome and the MEK/Erk pathway. Mol. Cell Neurosci. 28, 430–439. [DOI] [PubMed] [Google Scholar]
- Mack, T. G. et al. (2001). Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat. Neurosci. 4, 1199–1206. [DOI] [PubMed] [Google Scholar]
- Magni, G., Amici, A., Emanuelli, M., Orsomando, G., Raffaelli, N., and Ruggieri, S. (2004). Structure and function of nicotinamide mononucleotide adenylyltransferase. Curr. Med. Chem. 11, 873–885. [DOI] [PubMed] [Google Scholar]
- Mahoney, J. A., Odin, J. A., White, S. M., Shaffer, D., Koff, A., Casciola-Rosen, L., and Rosen, A. (2002). The human homologue of the yeast polyubiquitination factor Ufd2p is cleaved by caspase 6 and granzyme B during apoptosis. Biochem. J. 361, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, F., and King, P. D. (2005). The p95–100 kDa ligand of the T cell-specific adaptor (TSAd) protein Src-homology-2 (SH2) domain implicated in TSAd nuclear import is p97 Valosin-containing protein (VCP). Immunol. Lett. 97, 235–243. [DOI] [PubMed] [Google Scholar]
- Matsumoto, M., Yada, M., Hatakeyama, S., Ishimoto, H., Tanimura, T., Tsuji, S., Kakizuka, A., Kitagawa, M., and Nakayama, K. I. (2004). Molecular clearance of ataxin-3 is regulated by a mammalian E4. EMBO J. 23, 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, H. H., Shorter, J. G., Seemann, J., Pappin, D., and Warren, G. (2000). A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 19, 2181–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, W., Beirowski, B., Gillingwater, T. H., Adalbert, R., Wagner, D., Grumme, D., Osaka, H., Conforti, L., Arnhold, S., Addicks, K., Wada, K., Ribchester, R. R., and Coleman, M. P. (2005). The slow Wallerian degeneration gene, WldS, inhibits axonal spheroid pathology in gracile axonal dystrophy mice. Brain 128, 405–416. [DOI] [PubMed] [Google Scholar]
- Mizuno, Y., Hori, S., Kakizuka, A., and Okamoto, K. (2003). Vacuole-creating protein in neurodegenerative diseases in humans. Neurosci. Lett. 343, 77–80. [DOI] [PubMed] [Google Scholar]
- Mogk, A., Dougan, D., Weibezahn, J., Schlieker, C., Turgay, K., and Bukau, B. (2004). Broad yet high substrate specificity: the challenge of AAA+ proteins. J. Struct. Biol. 146, 90–98. [DOI] [PubMed] [Google Scholar]
- Muratani, M., and Tansey, W. P. (2003). How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell. Biol. 4, 192–201. [DOI] [PubMed] [Google Scholar]
- Okumura, F., Hatakeyama, S., Matsumoto, M., Kamura, T., and Nakayama, K. I. (2004). Functional regulation of FEZ1 by the U-box-type ubiquitin ligase E4B contributes to neuritogenesis. J. Biol. Chem. 279, 53533–53543. [DOI] [PubMed] [Google Scholar]
- Rabinovich, E., Kerem, A., Frohlich, K. U., Diamant, N., and Bar-Nun, S. (2002). AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22, 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaelli, N., Sorci, L., Amici, A., Emanuelli, M., Mazzola, F., and Magni, G. (2002). Identification of a novel human nicotinamide mononucleotide adenylyltransferase. Biochem. Biophys. Res. Commun. 297, 835–840. [DOI] [PubMed] [Google Scholar]
- Raoul, C., Estevez, A. G., Nishimune, H., Cleveland, D. W., deLapeyriere, O., Henderson, C. E., Haase, G., and Pettmann, B. (2002). Motoneuron death triggered by a specific pathway downstream of Fas potentiation by ALS-linked SOD1 mutations. Neuron 35, 1067–1083. [DOI] [PubMed] [Google Scholar]
- Richly, H., Rape, M., Braun, S., Rumpf, S., Hoege, C., and Jentsch, S. (2005). A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120, 73–84. [DOI] [PubMed] [Google Scholar]
- Sajadi, A., Schneider, B. L., and Aebischer, P. (2004). Wld(s)-mediated protection of dopaminergic fibers in an animal model of Parkinson disease. Curr. Biol. 14, 326–330. [DOI] [PubMed] [Google Scholar]
- Samsam, M., Mi, W., Wessig, C., Zielasek, J., Toyka, K. V., Coleman, M. P., and Martini, R. (2003). The Wlds mutation delays robust loss of motor and sensory axons in a genetic model for myelin-related axonopathy. J. Neurosci. 23, 2833–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder, R., Watts, G. D., Mehta, S. G., Evert, B. O., Broich, P., Fliessbach, K., Pauls, K., Hans, V. H., Kimonis, V., and Thal, D. R. (2005). Mutant valosin-containing protein causes a novel type of frontotemporal dementia. Ann. Neurol. 57, 457–461. [DOI] [PubMed] [Google Scholar]
- Spinette, S., Lengauer, C., Mahoney, J. A., Jallepalli, P. V., Wang, Z., Casciola-Rosen, L., and Rosen, A. (2004). Ufd2, a novel autoantigen in scleroderma, regulates sister chromatid separation. Cell Cycle 3, 1638–1644. [PubMed] [Google Scholar]
- Wang, J., Zhai, Q., Chen, Y., Lin, E., Gu, W., McBurney, M. W., and He, Z. (2005). A local mechanism mediates NAD-dependent protection of axon degeneration. J. Cell Biol. 170, 349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. S., Davis, A. A., Culver, D. G., and Glass, J. D. (2002). WldS mice are resistant to paclitaxel (taxol) neuropathy. Ann. Neurol. 52, 442–447. [DOI] [PubMed] [Google Scholar]
- Wang, Q., Song, C., and Li, C. C. (2004). Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J. Struct. Biol. 146, 44–57. [DOI] [PubMed] [Google Scholar]
- Watts, G. D., Wymer, J., Kovach, M. J., Mehta, S. G., Mumm, S., Darvish, D., Pestronk, A., Whyte, M. P., and Kimonis, V. E. (2004). Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 36, 377–381. [DOI] [PubMed] [Google Scholar]
- Wojcik, C., Yano, M., and DeMartino, G. N. (2004). RNA interference of valosin-containing protein (VCP/p97) reveals multiple cellular roles linked to ubiquitin/proteasome-dependent proteolysis. J. Cell Sci. 117, 281–292. [DOI] [PubMed] [Google Scholar]
- Wu, R., Terry, A. V., Singh, P. B., and Gilbert, D. M. (2005). Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol. Biol. Cell 16, 2872–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka, K., Okubo, Y., Suzaki, T., and Ogura, T. (2004). Analysis of the two p97/VCP/Cdc48p proteins of Caenorhabditis elegans and their suppression of polyglutamine-induced protein aggregation. J. Struct. Biol. 146, 242–250. [DOI] [PubMed] [Google Scholar]
- Zhai, Q., Wang, J., Kim, A., Liu, Q., Watts, R., Hoopfer, E., Mitchison, T., Luo, L., and He, Z. (2003). Involvement of the ubiquitin-proteasome system in the early stages of wallerian degeneration. Neuron 39, 217–225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.