Abstract
The Ras–Raf–mitogen-activated protein kinase cascade is a key growth-signaling pathway, which uncontrolled activation results in transformation. Although the exact mechanisms underlying Raf-1 regulation remain incompletely understood, phosphorylation has been proposed to play a critical role in this regulation. We report here three novel epidermal growth factor-induced in vivo Raf-1 phosphorylation sites that mediate positive feedback Raf-1 regulation. Using mass spectrometry, we identified Raf-1 phosphorylation on three SP motif sites: S289/S296/S301 and confirmed their identity using two-dimensional-phosphopeptide mapping and phosphospecific antibodies. These sites were phosphorylated by extracellular signal-regulated kinase (ERK)-1 in vitro, and their phosphorylation in vivo was dependent on endogenous ERK activity. Functionally, ERK-1 expression sustains Raf-1 activation in a manner dependent on Raf-1 phosphorylation on the identified sites, and S289/296/301A substitution markedly decreases the in vivo activity of Raf-1 S259A. Importantly, the ERK-phosphorylated Raf-1 pool has 4 times higher specific kinase activity than total Raf-1, and its phosphopeptide composition is similar to that of the general Raf-1 population, suggesting that the preexisting, phosphorylated Raf-1, representing the activatable Raf-1 pool, is the Raf-1 subpopulation targeted by ERK. Our study describes the identification of new in vivo Raf-1 phosphorylation sites targeted by ERK and provides a novel mechanism for a positive feedback Raf-1 regulation.
INTRODUCTION
Raf-1 is part of the Ras–Raf–mitogen-activated protein kinase kinase (MEK)–mitogen-activated protein kinase (MAPK) pathway, among the first mammalian signaling cascades to be elucidated (Avruch et al., 2001; Chang and Karin, 2001). The Ras–Raf–MAPK pathway is responsible for transmitting signals from membrane-bound receptors to intracellular and to nuclear targets, coordinating cellular response to a variety of environmental factors (Liebmann, 2001; Pearson et al., 2001). Aberrations at the receptor level and along the Ras–Raf–MAPK pathway are associated with a variety of diseases, especially cancer, with mutations in Ras detected in >30% of all human cancers and reaching frequencies of up to 50–90% in specific carcinomas (Porter and Vaillancourt, 1998; Lyons et al., 2001; Herrera and Sebolt-Leopold, 2002). Recently, also activating mutations in the B-Raf gene have been identified in various cancers, most predominantly melanomas (Davies et al., 2002; Mercer and Pritchard, 2003). In addition, both Ras and Raf are protooncogenes that occur naturally as viral-transmitted oncoproteins (Rapp et al., 1983). The initial elucidation of the Ras–Raf–MAPK pathway resulted from complimentary molecular and biochemical studies in mammalian cells and genetic studies in yeast, Drosophila and Caenorhabditis elegans (Avruch et al., 2001). Although the general features of the pathway and its activation mode are known, the exact regulatory mechanisms, at the molecular level, remain incompletely understood (Kerkhoff and Rapp, 2001; Dhillon and Kolch, 2002). Initially, a simple linear activation process was proposed, i.e., Ras activates Raf, Raf activates MEK, and MEK activates extracellular signal-regulated kinase (ERK). However, it is clear now that this pathway is more complex than initially thought, and other processes and accessory proteins are involved in its regulation (Avruch et al., 1994; Luo et al., 1996; Kolch, 2000).
The mammalian Raf family of serine/threonine kinases consists of three highly conserved members: A-Raf, B-Raf, and Raf-1 (or c-Raf-1). Whereas Raf-1 is ubiquitously expressed, A- and B-Raf display a more tissue-specific expression. Most of the Raf studies have focused on Raf-1, the first Raf member to be identified (Hagemann and Rapp, 1999).
Treatment of cells with factors that activate Ras initiates a complex series of events leading to Raf-1 activation. These events involve changes in Raf-1 localization, phosphorylation and protein–protein interactions (Weinstein-Oppenheimer et al., 2000; Avruch et al., 2001; Wellbrock et al., 2004a). Studies focusing on Raf-1 phosphorylation document both positive and negative effects of Raf-1 phosphorylation (Avruch et al., 2001; Chong et al., 2001). For example, p21 activated kinase (PAK) and Src mediate Raf-1 phosphorylation on positive regulatory sites and augment Raf-1 activity, whereas cAMP-dependent protein kinase (PKA) and AKT mediate Raf-1 phosphorylation on negative regulatory sites and inhibit Raf-1 activity. Conversely, protein phosphatase 2A can positively regulate Raf-1 by dephosphorylating a negative regulatory site (Dhillon et al., 2002).
Four basal and several mitogen-induced in vivo Raf-1 phosphorylation sites have been reported (Figure 1A) (Morrison et al., 1993; Avruch et al., 2001). Of the four basal sites, three were identified as S43, S259, and S621, and the fourth is yet to be identified (Morrison et al., 1993). Ser 43 is presumed to be an inhibitory phosphorylation site reported to be targeted by protein kinase A; however, its functional significance is still controversial (Sidovar et al., 2000; Zhang and Guan, 2001). The kinases responsible for basal Raf-1 S259 and S621 phosphorylation have not been defined, although several candidate kinases, including PKA, AKT, and AMP-activated kinase, have been proposed. Raf-1 phosphorylation at S259 serves as a binding point for the regulatory adapter protein 14-3-3 and stabilizes the basal inactive Raf-1 conformation, serving as a negative regulatory site (Avruch et al., 2001). Accordingly, phosphorylation of S259 by AKT or PKA was shown to negatively regulate Raf-1, whereas its dephosphorylation by protein phosphatase 2A has been reported to be part of the Raf-1 activation mechanism (Rommel et al., 1999; Zimmermann and Moelling, 1999; Dhillon et al., 2002). Raf-1 phosphorylation at the S621, on the other hand, promotes Raf-1 kinase activity by providing a second, positive binding point for 14-3-3, which binding at the S621 site is critical for Raf-1 kinase activity (Tzivion et al., 1998; Yip-Schneider et al., 2000). It is not known yet whether phosphorylation at S259 and S621 occur simultaneously on the same Raf-1 protein or whether they represent two separate Raf-1 populations (Tzivion et al., 2001; Tzivion and Avruch, 2002).
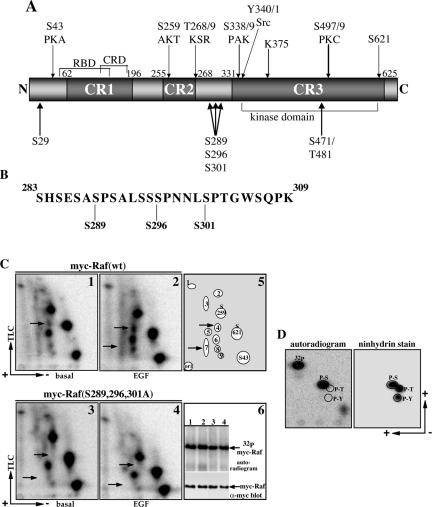
Figure 1.
Identification of Raf-1 S289, 296, and 301 as novel in vivo phosphorylation sites. (A) A diagram depicting known and newly identified Raf-1 phosphorylation sites and potential kinases reported to phosphorylate these sites. Indicated are RBD, Ras binding domain; CRD, cysteine-rich domain, CR1–3, conserved region 1–3; and K375, the ATP binding site. (B) A diagram showing Raf-1 sequence corresponding to amino acids 283–309. Indicated are the three phosphorylation sites identified by mass spectrometry analysis of myc-Raf-1 purified from COS-7 cells. (C) Serum-deprived COS-7 cells expressing wild type (1, 2 and 6, lanes 1 and 2) or S289/296/301A (3, 4 and 6, lanes 3 and 4) myc-Raf-1 were metabolically labeled with 32P and were left untreated (basal) or stimulated with 100 ng/ml EGF for 30 min. myc-Raf-1 proteins were immunoprecipitated with a myc-epitope tag antibody, and 90% of the sample was subjected to phosphopeptide map analysis as described in Materials and Methods. The recovery of phospho-myc-Raf-1 (6, autoradiogram, top) and myc-Raf-1 protein (6, anti-myc blot, bottom) was determined by separating the remaining 10% sample on a separate gel. Representative autoradiograms of Raf-1 phosphopeptide maps (1–4) and a schematic representation of the phosphopeptide spots (5) are presented. The electrophoresis direction and the TLC chromatography orientation are indicated by arrows. ori in 5 represents the origin/spotting point. The location of phospho S43, S621, and S259 peptides is indicated. The locations of spots 4 and 7 are indicated by arrows. (D) Spot 4 from 2 was excised and analyzed for phosphoamino acid composition by two-dimensional electrophoresis as described in Materials and Methods. Presented are a ninhydrin staining of the TLC plate (right) showing the migration points of standard phospho-serine (P-S), phospho-threonine (P-T), and phospho-tyrosine (P-Y) and an autoradiogramshowing the radiolabeled phosphoamino acids (indicated are the corresponding migration positions of P-S, P-T, P-Y, and the free phosphate).
Treatment of cells with growth and mitogenic factors induces Raf-1 phosphorylation at additional several minor sites. These sites include T268/269, S338/339, Y340/341, and S497/499 (Chong et al., 2003). T268 is a proposed Raf-1 autophosphorylation site (Morrison et al., 1993), and T269 was reported to be phosphorylated by kinase suppressor of Ras (KSR) (Zhang et al., 1997). Although the role of T268 and T269 phosphorylation in the regulation of Raf-1 kinase activity remains unresolved, it seems that phosphorylation at these sites does not notably affect Raf-1 activation either by oncogenic Ras or by most growth factors. Phosphorylation of Raf-1 S338/339 can be mediated by the Rac/CDC42-activated protein kinase family members PAK-2 and PAK-3 (King et al., 1998) and potentially protein kinase C (PKC) (Hamilton et al., 2001). Phosphorylation at these sites, especially S338, seems to be a crucial step in the activation of Raf-1 by oncogenic Ras and growth factors. However, phosphorylation at these sites does not activate Raf-1 per se, suggesting that other modifications may be required to achieve a stable Raf-1 activation (Mason et al., 1999). Similarly, phosphorylation of Raf-1 Y340/341, which can be mediated by the tyrosine kinase Src, results in marginal Raf-1 activation. Finally, S497 and S499 can be phosphorylated by PKC; however, these phosphorylations do not result in Raf-1 activation, and phosphorylation at these sites is not required for Raf-1 activation by PMA, growth factors or oncogenic Ras (Marais et al., 1998).
The mechanism(s) responsible for Raf-1 inactivation is much less understood, although the common view is that Raf-1 dephosphorylation should play a key role in this process (Avruch et al., 2001).
In the present study, we report the identification of three novel epidermal growth factor (EGF)-induced in vivo Raf-1 phosphorylation sites. We show that these sites are targeted by ERK both in vivo and in vitro. In addition, our data suggests that phosphorylation at these sites provides a positive feedback input, possibly by stabilizing the active form of Raf-1 or attenuating the rate of its inactivation.
MATERIALS AND METHODS
cDNA Constructs, Antibodies and Kinase Inhibitors
The mammalian expression vector pMT2 containing wild-type myc-Raf-1, myc-Raf-1 S259A, or HA-ERK-1; the mammalian expression vector pExchange 5A containing FLAG-MEK DD; and the bacterial expression vector pGEX containing glutathione S-transferase (GST)-MEK-1 or GST-ERK-1 were as described previously (Tzivion et al., 1998; Shen et al., 2003a,b; Zhu et al., 2005). The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to introduce the S289/296/301A and S471A mutations in a pMT2-myc-Raf-1 or pMT2-myc Raf-1 S259A templates. Phosphospecific antibodies for active forms of MEK and ERK were from Cell Signaling Technology (Beverly, MA), and antibodies for hemag-glutinin (HA) and myc epitope tags were produced from the 12CA5 and 9E10 hybridoma cell lines, respectively. The MEK inhibitor U0126 was from Promega (Madison, WI). Phosphospecific antibodies for Raf-1 pS289, pS296, and pS301 were produced as described previously (Shen et al., 2003a) and screened for specificity as described in the text.
Cell Culture and Transfection
COS-7 cells were maintained in DMEM supplemented with 10% fetal calf serum. For transient expression of proteins, cells were transfected using LipofectAMINE (Invitrogen, Carlsbad, CA) as detailed in the figure legends according to the manufacturer's instructions. For myc-Raf-1 expression, 4 μg of pMT2-myc-Raf-1 DNA was used and for Raf-1/ERK coexpression experiments, 2 μg each, pMT2-myc-Raf-1 and pMT2-HA-ERK-1 were used. For ERK-1/2 knockdown experiments, 4 μg of pMT2-myc Raf-1 DNA was coexpressed with the following Stealth ERK-1/2 siRNA duplexes (Invitrogen): 5′-GGAAGCCAUGAGAGAUGUCUACAUU-3′, 5′-CCUGCGACCUUAAGAUUUGUGAUUU-3′, 5′-GCCAUGGAGCUGGAUGACCUACCUA-3′, 5′-CCUCAGCAAUGACCAUAUCUGCUAU-3′, 5′-GGCUGUUCCCAAAUGCUGACUCCAA-3′, at 400 pmol each, using 30 μl of LipofectAMINE 2000 and 10-cm culture dish. For cell stimulation, 24 h after transfection, cells were deprived of serum for 18 h before adding the agonist. Details for cell stimulation and treatment with the MEK inhibitor are provided below and in the figure legends.
32P Metabolic Labeling
Serum-deprived COS-7 cells transfected as indicated in figure legends were washed once with medium lacking phosphate, followed by 30-min incubation in the same media for depleting intracellular phosphate. Cells were radiolabeled by incubation in the presence of 0.5 mCi/ml 32P for 2 h.
Cell Extraction and Protein Purification
Cells were lysed for 30 min using ice-cold extraction buffer containing 50 mM Tris-Cl, pH 7.5, 100 mM NaCl, 1% Triton X 100, 1 mM dithiothreitol (DTT), 1 mM EDTA, 1 mM EGTA, 2 mM Na3VO4, 50 mM β-glycerophosphate, and a protease inhibitor cocktail (Amersham Biosciences, Piscataway, NJ). For immunoprecipitation, cleared cell lysates were incubated at 4°C for 3 h with the appropriate antibody precoupled to protein A/G agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA). The beads were washed twice with extraction buffer, twice with extraction buffer containing 0.5 M LiCl, and twice with kinase assay buffer (40 mM Tris-Cl, pH 7.5, 0.1 mM EDTA, 5 mM MgCl2, and 2 mM DTT). The purified proteins were used further as specified in the figure legends.
Mass Spectrometry Analysis
myc-Raf-1 protein (20 plates/sample) from variably treated cells was purified as described above and eluted in SDS sample buffer. The protein samples were alkylated by incubation with 2% iodoacetamide for 30 min at room temperature and separated on 7.5% SDS-PAGE. The proteins were visualized by Coomassie blue staining and the Raf-1 protein band was excised from the gel (4–5 μg of Raf-1 protein was used per analysis). In-gel trypsin digestion and mass spectrometry analysis of the eluted Raf-1 peptides was performed as described previously (Qin and Zhang, 2002).
Raf-1 Phosphopeptide Mapping and Phosphoamino Acid Analysis
Two-dimensional (2D) phosphopeptide mapping and subsequent phosphoamino acid analysis of individual phosphopeptide spots was performed according to previously described protocols (Boyle et al., 1991; Luo et al., 1991). Briefly, immunopurified 32P-labeled myc-Raf-1 proteins were resolved using 7.5% SDS-PAGE, transferred to a polyvinylidene membrane, and excised, and 32P incorporation in myc-Raf-1 was determined by Cherenkov counting. After incubation with 0.5% polyvinylpyrrolidone in 100 mM acetic acid for 30 min at 37°C and extensive washes, Raf-1 protein samples were digested with 10 μg of sequencing grade modified trypsin (Promega) in 50 mM ammonium bicarbonate buffer for 2 h at 37°C and with additional 10 μg of trypsin for overnight (this method routinely allowed recovery of 90–95% of the initial radioactivity in myc-Raf-1). The eluted peptides were washed twice with 50 mM ammonium bicarbonate buffer and once with pH 1.9 thin layer chromatography (TLC)-electrophoresis buffer (2.2% formic acid and 7.8% acetic acid in water). Samples were spotted on cellulose TLC plates (Merck, Darmstadt, Germany) and separated using the Hunter TLC system (CBS Scientific, Del Mar, CA) in pH 1.9 buffer for 25 min at 1000 V. The plates were dried overnight and subjected to second dimension chromatographic separation in a phospho-chromatography buffer (37.5% n-butanol, 25% pyridine, and 7.5% acetic acid). The plates were dried, and the phosphopeptide spots were visualized by autoradiography and phosphorimaging. For phosphoamino acid analysis, individual spots were scraped from the plates and eluted in 200 μl of pH 1.9 buffer and hydrolyzed in 6 N HCl for 60 min at 111°C. The samples were resolved on TLC plates by two-dimensional electrophoresis, and the labeled amino acids were identified by reference to ninhydrin stained phosphoamino standards.
Raf-1 Kinase Assay
Raf-1 kinase activity was determined using a slight modification of our previously described protocol (Tzivion et al., 1998). Briefly, after myc immunoprecipitation, myc-Raf-1-containing beads were incubated in kinase assay buffer (100 μl final volume) supplemented with 100 μM ATP, and 0.4 μg of prokaryotic recombinant GST-MEK-1 for 30 min at 30°C. Samples were separated on 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. The phosphorylation of MEK-1 was determined by phospho-MEK-immunoblotting and densitometry quantification. myc-Raf-1 recovery was determined by myc-immunoblotting. To maintain homogeneity of the kinase and blotting conditions, and because of technical challenges, the kinase activity was not measured in triplicates, instead, each experiment has been repeated at least three times, and only experiments that produced similar results in repeating experiments are presented.
ERK-1 Activation In Vitro
Fifty micrograms of recombinant GST-ERK-1 produced in bacteria was incubated with 10 μg of recombinant M2-FLAG-MEK-1 S218/222D (a constitutively active form of MEK-1; Huang and Erikson, 1994) produced in COS-7 cells and precoupled to protein A/G beads, in a 100-μl kinase assay buffer containing 100 μM ATP for 60 min at 30°C. After the incubation, the beads were spun down, and the supernatant containing activated ERK-1 was collected. ERK-1 incubated with beads alone served as a control for basal ERK-1 activity. ERK-1 activation was determined in a kinase assay using myelin basic protein as a substrate as described previously (Shen et al., 2003b).
In Vitro Raf-1 Phosphorylation for 2D Phosphopeptide Mapping and Assessment of the Phosphorylation Effect on Raf-1 Kinase Activity
Protein A/G beads containing ∼500 ng of myc-Raf-1 protein purified as described above were incubated with in vitro-activated ERK-1 or control proteins as specified in the figure legends in a 100-μl kinase assay buffer containing 100 μM ATP and 25 μCi of [γ-32P]ATP (for Raf-1 peptide mapping) or in kinase buffer omitting ATP for time periods as indicated in figure legends (for Raf-1 kinase activity assays) at 30°C. After the incubation, myc-Raf-1 containing beads were washed twice with extraction buffer and twice with kinase assay buffer and myc-Raf-1 protein was either directly eluted in sample buffer for 2D phosphopeptide analysis or was assayed for Raf-1 kinase activity as described above.
RESULTS
Identification of Raf-1 S289, S296, and S301 as Novel In Vivo Raf-1 Phosphorylation Sites
Phosphorylation has been proposed as the main modification contributing to stable Raf-1 activation (Avruch et al., 2001; Chong et al., 2003). Supporting this notion, we recently reported that specific Raf-1 dephosphorylation at sites induced by mitogenic stimulation completely deactivates Raf-1 kinase activity (Zhu et al., 2005). To comprehensively identify basal and mitogen-induced Raf-1 phosphorylation sites, Raf-1 was purified from serum-deprived or EGF-stimulated COS-7 cells, and its phosphorylation was analyzed by mass spectrometry. This analysis revealed both previously reported Raf-1 phosphorylation sites and several novel sites (Figure 1A; Zhu et al., 2005). Among the novel sites, we observed phosphorylation of Raf-1 on a tryptic peptide corresponding to Raf-1 283-309 peptide (Figure 1B). The mass spectrometry analysis identified three phosphorylations in this peptide at Ser 289, Ser 296, and Ser 301 and also detected the presence of this peptide as a doubly phosphorylated form. Phosphorylation at these sites was observed in both, Raf-1 samples purified from serum-deprived cells and Raf-1 samples purified from EGF-treated cells.
To confirm the identity of the sites by an independent method, we generated a mutant in which all three serines were substituted with alanines, Raf-1 S289/296/301A, and compared its two-dimensional tryptic phosphopeptide maps with that of wild-type Raf-1 (Figure 1C). The tryptic 2D phosphopeptide map of wild-type Raf-1 purified from serum-deprived COS-7 cells has four major spots (Figure 1C, 1). Three of these spots were previously identified as pS43, pS259, and pS621 (Morrison et al., 1993) and Figure 1C, 5). The identity of the fourth spot, marked as spot 6 (Figure 1C, 5), has not yet been determined. Beside these four major spots, there are several minor spots, marked as spots 1–5 and 7–9 (Figure 1C, 5). Their identity also remains to be determined. Treatment of cells with EGF increases the phosphorylation of some of these spots, notably spots 4, 7, and 8 (Figure 1C, compare 1 and 2). A comparison between the maps of wild-type Raf-1 with that of the triple Raf-1 mutant S289/296/301A shows that the mutant Raf-1 maintains the four major spots but lacks specifically the minor spots 4 and 7 (Figure 1C, compare 1, 2 and 3, 4, the position of the spots is indicated by arrows). The rest of the minor spots seem to be intact in the mutant. Spot 4 contains only phosphoserine (Figure 1D), and its position on the 2D map correlates with the predicted position of a singly phosphorylated Raf-1 283-309 tryptic peptide. Spot 7 may represent a doubly phosphorylated form of the same peptide [these predictions are based on calculations using parameters published by Boyle et al. (1991)). The level of 32P incorporation and Raf-1 protein recovery in the samples used for the phosphopeptide mapping are shown in Figure 1C, 6. Collectively, the phosphopeptide mapping results confirmed the mass spectrometry data and also pointed on the possibility that phosphorylation at these sites may be regulated by EGF treatment.
Raf-1 S289, 296, and 301 Are In Vivo ERK Target Sites
Raf-1 S289, 296, and 301 sites contain an MAPK/proline-directed kinase phosphorylation site signature, thus raising the possibility that these sites may serve as regulatory feedback phosphorylation sites targeted by ERK. To address this possibility, the effect of ERK-1 overexpression on Raf-1 phosphorylation was examined (Figure 2). Coexpression of Raf-1 with ERK-1 results in a substantial increase in Raf-1 phosphorylation. This increase is apparent both at the level of 32P incorporation in Raf-1 and an up-shift in the Raf-1 mobility in the gel (Figure 2A, compare lanes 1, 2, 5, 6 and 9, 10, no ERK-1, with lanes 3, 4, 7, 8 and 11, 12, plus ERK-1). To examine the effect of ERK-1 overexpression on individual Raf-1 phosphorylation sites, the 32P-Raf-1 bands were excised from the gel and equal counts of trypsin-digested 32P-Raf-1 were analyzed by 2D phosphopeptide mapping (Figure 2B). As shown in Figure 1, EGF treatment results in increased phosphorylation of several sites, namely spots 4, 7, and 8 (Figure 2B, compare 1 and 2). ERK-1 overexpression results in an increase in spots 7/11 and 12 and also in spots 1, 5, and 10, but a decrease in spot 4, especially in the EGF-treated samples (Figure 1B, compare 1, 2 with 3, 4). Comparing the maps of wild type and the triple Raf-1 mutant, S289/296/301A showed that spots 4, 7/11, and 12 are missing in the triple mutant (Figure 1B, compare 1–4 with 5–8). Notably, the effect of ERK-1 overexpression on other spots in the Raf-1 triple mutant is similar to its effect on wild-type Raf-1, i.e., increase in spots 1, 5, and 10. These data suggest that spots 4, 7/11, and 12 represent differentially phosphorylated forms of Raf-1 283-309 peptide with spot 4 being the singly phosphorylated form, spots 7/11 the doubly phosphorylated form, and spot 12 the triply phosphorylated form. It is also notable that the triply phosphorylated form occurs mainly in the ERK-1-overexpressing cells. In support of the identification of spots 7/11 and 12 as the doubly and triply phosphorylated forms of Raf-1 283-309 peptide, respectively, the results show that spot 4, the singly phosphorylated form, diminishes with the correlative appearance of spots 7/11 and 12. Furthermore, the migration position of spots 7/11 and 12 are in accordance with the predicted migration position of the doubly and triply phosphorylated forms of Raf 283-309 peptide, respectively.
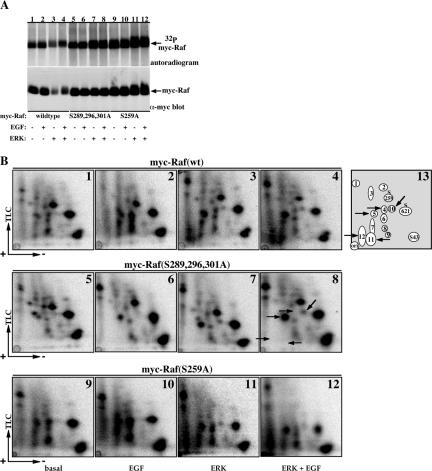
Figure 2.
ERK-1 overexpression induces Raf-1 phosphorylation on several sites, including S289, 296, and 301. (A and B) COS-7 cells expressing wild-type myc-Raf-1 (lanes 1–4), myc-Raf-1 S289/296/301A (lanes 5–8), or myc-Raf-1 S259A (lanes 9–12) alone (lanes 1, 2, 5, 6, 9, and 10) or coexpressing HA-ERK-1 (lanes 3, 4, 7, 8, 11, and 12) were metabolically labeled with 32P and were left untreated or were stimulated with 100 ng/ml EGF for 30 min as indicated. myc-Raf-1 proteins were immunoprecipitated as in Figure 1, and 10% of the sample (A) was separated using 7.5% SDS-PAGE. Shown is an autoradiogram representing 32P incorporation in myc-Raf-1 (top) and a myc immunoblot representing myc-Raf-1 protein recovery (bottom). The remaining 90% of the samples were subjected to phosphopeptide mapping (B) as described in the legends of Figure 1 and in Materials and Methods. Representative autoradiograms of Raf-1 phosphopeptide maps (1–12) and a schematic representation of the phosphopeptide spots (13) are presented. The positions of phospho S43, S621, and S259 peptides are indicated (13). The positions of spots 4, 5, 10, 11, and 12 are indicated by arrows (13 and 8). Note that the maps of the Raf-1 S259A mutant lack the spot corresponding to the pS259 peptide (9–12, indicated by arrow).
To demonstrate that the changes in the composition of the phosphopeptide spots is specific to the Raf-1 S289/296/301A mutant, we analyzed the phosphopeptide maps of another Raf-1 phosphorylation site mutant, Raf-1 S259A, which lacks the constitutive S259 phosphorylation site (Figure 2B, 9–12). The maps of this mutant are similar to those of wild-type Raf-1 with the exception of the lack of the spot corresponding to the migration position of phospho-S259 peptide (Figure 2B, 9–12). Collectively, these data show that phosphorylation of Raf-1 on S289/296/301 sites is increased by EGF treatment and to a larger extent, by ERK-1 overexpression. The data also implicate ERK-1 and/or ERK downstream target(s) as a candidate kinase(s) for phosphorylating these sites.
To further demonstrate the role of ERK in the phosphorylation of these sites in vivo, both under basal conditions and after EGF stimulation, we examined the effect of the MEK inhibitor U0126 on the basal and EGF-induced phosphorylation levels of these sites (Figure 3). Pretreatment of cells with the MEK inhibitor for 30 min before adding 32P significantly decreased 32P incorporation in Raf-1 (Figure 3, 10, compare lanes 1–4 with 5–8). To examine the effect of U0126 treatment on individual Raf-1 phosphorylation sites, equal counts of trypsin-digested 32P-Raf-1 were analyzed by 2D-phosphopeptide mapping (Figure 3, 1–8). U0126 treatment resulted in the reduction of spots 1, 4, and 7/11 under basal conditions (Figure 3, compare 1 and 5) and blocked the ability of EGF to induce their phosphorylation (Figure 3, compare 2 and 6). Similarly, U0126 treatment abolished the ability of ERK-1 overexpression to induce phosphorylation of spots 1, 4, 5, 7/11, and 12 (Figure 3, compare 3, 4 with 7, 8). These data indicate that ERK is necessary both for the basal and the EGF-induced phosphorylation of these sites in vivo. Furthermore, the ability of the MEK inhibitor to block the overexpressed ERK-1-induced Raf-1 phosphorylation suggests that ERK-1/Raf-1 coexpression activates ERK in a MEK-dependent manner, involving the endogenous MEK.
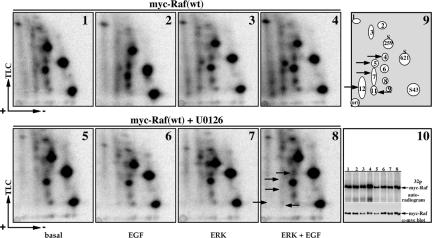
Figure 3.
MEK inhibition blocks Raf-1 phosphorylation on EGF- and ERK-1-induced sites. COS-7 cells expressing wild-type myc-Raf-1 alone (1, 2, 5, 6, and 10, lanes 1, 2, 5, and 6) or coexpressing HA-ERK-1 (3, 4, 7, 8, and 10, lanes 3, 4, 7, and 8) were treated with vehicle (1–4 and 10, lanes 1–4) or were incubated in the presence of 20 μM U0126 for 30 min before 32P metabolic labeling. After the labeling, cells were left untreated (1, 3, 5, 7, and 10, lanes 1, 3, 5, and 7) or were stimulated with 100 ng/ml EGF for 30 min (2, 4, 6, 8, and 10, lanes 2, 4, 6, and 8). myc-Raf-1 phosphopeptide mapping was performed as in Figure 1C. Presented are representative autoradiograms of Raf-1 phosphopeptide maps (1–8), a schematic representation of the phosphopeptide spots (9), and a gel showing 32P incorporation in myc-Raf-1 and myc-Raf-1 protein recovery (10). The positions of spots 4, 5, 7, 11, and 12 are indicated by arrows (8 and 9). Note the reduction in the intensity of the indicated spots, most notably in 6 and 8 compared with 2 and 4, respectively.
ERK-1 Directly Phosphorylates Raf-1 In Vitro on Sites That Include Raf-1 S289, 296, and 301
To examine whether ERK, besides being necessary for the in vivo phosphorylation of Raf-1 on S289, 296, and 301, is also capable of directly phosphorylating these sites, we tested the ability of a bacterially expressed, in vitro-activated recombinant ERK-1 to phosphorylate Raf-1 (Figure 4). In vitro, Raf-1 has a low autophosphorylating activity (Figure 4A, lane 4 and Figure 4B, lane 3). It is important to note that this “autophosphorylating activity” results probably from a combination of true autophosphorylation and phosphorylation by copurifying kinases. Incubation of Raf-1 with active ERK-1 induces a strong Raf-1 phosphorylation (Figure 4A, lane 3 and Figure 4B, lane 1). This induction requires ERK-1 activation, because ERK-1 preparations that were not activated in vitro by incubation with active MEK did not induce Raf-1 phosphorylation (Figure 4A, lane 4). It is also notable that ERK-1 is able to phosphorylate both wild-type Raf-1 and the triple Raf-1 mutant in vitro; however, the level of 32P incorporation in the triple Raf-1 mutant was lower than that in wild-type Raf-1 (compare Figure 4A, lanes 3 and 5 and Figure 4B, lanes 1 and 2). To exclude the possibility that the observed Raf-1 phosphorylation was mediated by the kinases used to produce the active ERK-1, i.e., recombinant MEK and Raf-1, we tested the ability of these proteins to phosphorylate the myc-Raf-1 samples (Figure 4B, lanes 4–6). This control experiment showed that from the kinases used in the assay, only ERK-1 is a potent kinase for Raf-1.
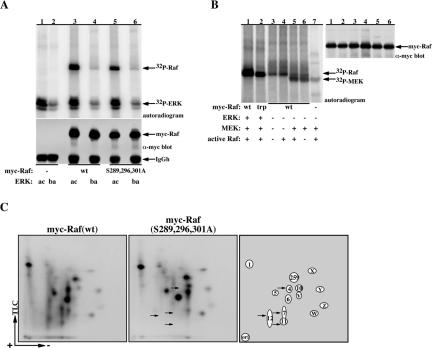
Figure 4.
ERK-1 can directly phosphorylate Raf-1 in vitro. (A and C) Approximately 500 ng of wild-type (lanes 3 and 4) or S289/296/301A myc-Raf-1 protein (lanes 5 and 6) or a mock sample (lanes 1 and 2) were purified by myc immunoprecipitation from serum-deprived COS-7 cells and incubated with recombinant bacterial ERK-1 (ba, lanes 2, 4, and 6) or with same ERK-1 activated in vitro by incubation with active MEK (ac, lanes 1, 3, and 5) as detailed in Materials and Methods. Ten percent of the sample was resolved using 7.5% SDS-PAGE (A, bottom) and assayed for Raf-1 recovery by myc-immunoblotting, and the remaining 90% (A, top) was used for Raf-1 phosphopeptide mapping (C, 1 and 2). Note that 1 contains a map of Raf-1 excised from lane 3, and 2 contains a map of Raf-1 excised from lane 5. Panel 3 is a schematic representation of the phosphopeptide spots in 1 and 2. (B) In a similar experiment as described in A, wild-type myc-Raf-1 (lanes 1 and 3–6) or myc-Raf-1 S289/296/301A (trp, lane 2) was incubated with recombinant ERK-1 activated in vitro by active MEK (lanes 1 and 2) or with the indicated combinations of recombinant active Raf-1 (produced from sf9 cells coinfected with vRas and vSrc) and recombinant bacterial MEK-1 (lanes 3–6). Lane 7 is a control lane containing only the recombinant MEK and active Raf-1. Presented are an autoradiogram showing 32P incorporation in Raf-1 (left) and a myc immunoblot showing the recovery of myc-Raf-1 (right). Indicated are the migration positions of 32P myc-Raf-1 and MEK (left) and of myc-Raf-1 protein (right).
Tryptic 2D phosphopeptide maps of Raf-1 phosphorylated in vitro by ERK-1 showed Raf-1 phosphorylation on multiple sites (Figure 4C). A comprehensive comparison of these maps with that of the in vivo phosphopeptide Raf-1 maps, including mixing experiments of in vitro and in vivo phosphorylated samples (our unpublished data) allowed us to align the in vitro and in vivo maps (Figure 4C, 3). This analysis indicates that spot 4 correlates with the singly phosphorylated Raf-1 283-309 peptide, whereas spots 7/11 and 12 correlate with the doubly and triply phosphorylated Raf-1 283-309 peptides, respectively. It is important to note that these spots are missing in the in vitro phosphopeptide map of the Raf-1 triple mutant (Figure 4C, 2, indicated by arrows). Beside these three spots, ERK-1 induces phosphorylation of Raf-1 on several other sites, namely, spots 1, 5, 6, and 10, the identity of which remains to be determined. It is also important to note that some of these sites, namely, spots 259, W, X, Y, and Z occur also on Raf-1 incubated without ERK-1 (our unpublished data), suggesting they represent the Raf “autophosphorylating” activity discussed above. Spot 6 (Figure 4C, 3) represents the major ERK-1 phosphorylation site in vitro; however, determining its identity and its in vivo counterpart needs further investigation. Also, it is possible that this site is only exposed under in vitro conditions and that in vivo it serves only as a minor target site or is highly sensitive to dephosphorylation. Collectively, the data presented show that ERK-1 can directly phosphorylate Raf-1 on the tryptic peptide containing Ser 289, 296, and 301. Combined with the in vivo data, this suggests that ERK is the main in vivo kinase responsible for the phosphorylation of these sites, both under basal and mitogen-induced conditions.
Raf-1 S296 Phosphospecific Antibodies Demonstrate EGF-regulated Raf-1 Phosphorylation
To further validate our identification of Raf S289, 296, and 301 as EGF-induced phosphorylation sites and to help in studying phosphorylation of endogenous Raf-1 on these sites, we developed phosphospecific antibodies for all the three identified sites. One of these antibodies, directed against Raf-1 S296, showed the highest specificity for phosphorylated Raf-1 in initial screens and was selected for the studies described below. To test the specificity of the antibody for Raf-1 phosphorylation on the S289/296/301 sites, wild type and the triple myc-Raf-1 mutant were expressed in COS-7 cells alone or with active MEK and examined for reactivity with the antibody (Figure 5A). These results show that the antibody has a low reactivity with wild-type Raf-1 from serum-deprived cells but binds strongly to Raf-1 from EGF-treated cells coexpressing active MEK (Figure 5A, compare lanes 1 and 2). Importantly, no reactivity of the antibody was observed with the triple Raf-1 mutant under any of the tested conditions (Figure 5A, lanes 3 and 4). The equal expression of wild-type and mutant Raf-1 is demonstrated using myc-immunoblotting (Figure 5A, bottom). This experiment demonstrates the specificity of the antibody to Raf-1 phosphorylated at one of the three ERK-induced sites. In addition, it shows the ability of the antibody to recognize endogenous Raf-1 in EGF-treated cells (Figure 5A, lanes 4 and 6). The weakly reactive band seen in the myc-immunoprecipitate lane of the triple Raf-1 mutant (Figure 5A, myc-IP part, lane 4) may be attributed to endogenous Raf-1 dimerizing and copurifying with the myc-Raf-1 mutant (we have previously reported the ability of Raf-1 proteins to oligomerize (Luo et al., 1996).
Figure 5.
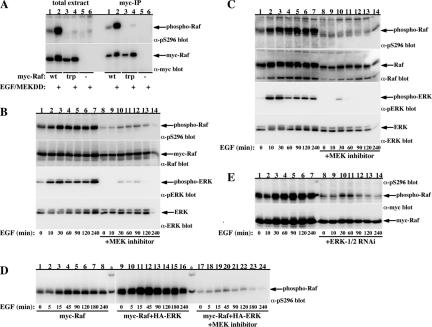
Phospho-S296 Raf-1 antibody demonstrates EGF-induced, ERK-dependent endogenous Raf-1 phosphorylation. (A) Serum-deprived COS-7 cells expressing wild-type myc-Raf-1 (lanes 1 and 2), S289/296/301A myc-Raf-1 (lanes 3 and 4), or control vector (lanes 5 and 6) alone (lanes 1, 3, and 5) or coexpressing M2-FLAG-MEKDD (lanes 2, 4, and 6) were treated with vehicle (lanes 1, 3, and 5) or 100 ng/ml EGF for 30 min (lanes 2, 4, and 6). Then, 80 μg of total cell extract or an equivalent amount of myc-immunoprecipitates was separated on 7.5% SDS-PAGE and immunoblotted using anti-Raf-1pS296 antibody (top) or myc antibody (bottom). (B and C) Serum-deprived COS-7 cells (C) or myc-Raf-1 expressing COS-7 cells (B) pretreated with vehicle (lanes 1–7) or 10 μM U0126 for 18 h followed by a fresh dose 30 min before EGF stimulation (lanes 8–14) were treated with 100 ng/ml EGF for the indicated times. Total cell extracts were immunoblotted using the indicated antibodies. Indicated are the migration positions of myc-Raf-1 (B) and endogenous Raf-1 (C). (D) Serum-deprived COS-7 cells expressing myc-Raf-1 (lanes 1–8) or coexpressing myc-Raf-1 with HA-ERK-1 (lanes 9–24) pretreated with vehicle (lanes 1–16) or 10 μM U0126 for 18 h followed by a fresh dose 30 min before EGF stimulation (lanes 17–24) were treated with 100 ng/ml EGF for the indicated times. Total cell extracts were immunoblotted with anti-pS296 Raf-1 antibody. Indicated is the migration position of phospho-myc-Raf-1. The asterisks denote protein standard markers used for signal equalization between the two gels (lanes 1–8 and 9–24). (E) Serum-deprived COS-7 cells expressing myc-Raf-1 (lanes 1–7) or coexpressing myc-Raf-1 with ERK-1/2 RNAi (lanes 8–14) were treated with 100 ng/ml EGF for the indicated time points. Total cell extracts were immunoblotted with anti-pS296 Raf-1 antibody (top) or myc (bottom). Indicated are the migration positions of pS296-myc-Raf-1 (top) and myc-Raf-1 (bottom).
Next, we used the antibody to examine the kinetics of Raf-1 phosphorylation at these sites after EGF stimulation and the role of ERK in mediating this phosphorylation (Figure 5, B–E). Both, recombinant Raf-1 and the endogenous Raf-1 undergo robust phosphorylation at these sites after EGF treatment (Figure 5, B and C, respectively). In terms of time kinetics, Raf-1 phosphorylation was induced at 10 min, peaked at 30 min, and stayed stable for up to 4 h. Pretreatment of the cells with a MEK inhibitor abolished both the basal and the EGF-induced Raf-1 phosphorylation at these sites, demonstrating that the activity of endogenous MEK/ERK is required for these phosphorylations. Similar results were obtained using ERK-1/2 RNA interference (RNAi) (Figure 5E) demonstrating that ERK-1/2 are needed for the EGF-induced Raf-1 phosphorylation at these sites. Conversely, Raf-1 coexpression with ERK-1 markedly enhanced the basal and the EGF-induced Raf-1 phosphorylation at these sites (Figure 5D, compare lanes 1–8 with 9–16). This enhancement was completely blocked in cells pretreated with the MEK inhibitor (Figure 5D, lanes 17–24). These experiments validate the results obtained using phosphopeptide mapping and further strengthen the role of ERK in mediating phosphorylation of these sites, both under basal and EGF-stimulated conditions. In addition, the results establish that endogenous Raf-1 phosphorylation at these sites holds the same pattern as the recombinant Raf-1, demonstrating a physiological relevance of these phosphorylations in Raf-1 regulation.
ERK-induced Raf-1 Phosphorylation Sustains Raf-1 Kinase Activity
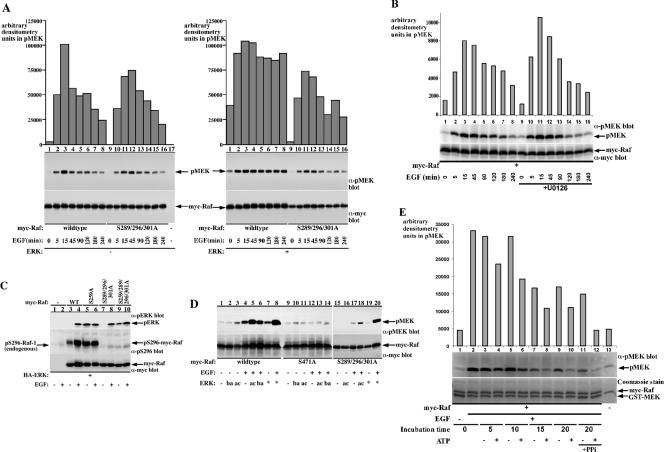
To determine the functional significance of the ERK-induced Raf-1 phosphorylation, we examined the effect of ERK-1 overexpression and mutation of the ERK-induced sites on Raf-1 kinase activity (Figure 6A). Treatment of COS-7 cells with EGF induces Raf-1 kinase activation, peaking at 15 min after EGF addition and returning to near basal activity at 240 min (Figure 6A, left, lanes 1–8). The triple Raf-1 mutant shows similar time kinetics (Figure 6A, left, compare lanes 1–8 with 9–16). Coexpression of ERK-1 with Raf-1 sustained Raf-1 kinase activity at almost peak levels for up to 240 min (Figure 6A, right, lanes 1–8). This effect of ERK-1 was abolished in the Raf-1 triple mutant (Figure 6A, right, compare lanes 1–8 with 9–16), suggesting that the ability of ERK-1 overexpression to sustain Raf-1 kinase activity is mediated by the ability of ERK-1 to phosphorylate the Raf-1 S289/296/301 sites. Notably, ERK-1 overexpression sustained an up shift in Raf-1 mobility in the gel (Figure 6A, right, lower part, lanes 1–8), suggesting that ERK-1 phosphorylation may stabilize the phosphorylated, active form of Raf-1. Also, the basal activity of Raf-1 from cells coexpressing ERK-1 was significantly higher than Raf-1 activity from cells expressing Raf-1 alone (Figure 6A, compare lane 1, right, with lane 1, left), correlating with the increased Raf-1 basal phosphorylation seen in Figure 5D.
Figure 6.
ERK-1 phosphorylation stabilizes the active form of Raf-1 by attenuating its inactivation rate. (A) Serum-deprived COS-7 cells expressing wild-type myc-Raf-1 (left, lanes 1–8) or S289/296/301A myc-Raf-1 (left, lanes 9–16) or coexpressing HA-ERK-1 (right) were treated with 100 ng/ml EGF for the indicated time points. After stimulation, Raf-1 kinase activity in the samples was assayed as described in Materials and Methods. Presented are a phospho-MEK immunoblot showing the level of MEK phosphorylation (middle), a myc-immunoblot showing Raf-1 recovery (bottom), and a bar graph showing a densitometric quantification of the phospho-MEK band (top). The migration positions of phospho-MEK and myc-Raf-1 are indicated. (B) Serum-deprived COS-7 cells expressing myc-Raf-1 were treated with vehicle (lanes 1–8) or 20 μM U0126 for 30 min before EGF treatment. The samples were analyzed for Raf-1 kinase activity as described in A. (C) Serum-deprived COS-7 cells expressing HA-ERK-1 (lanes 1 and 2) or coexpressing HA-ERK-1 and either wild-type myc-Raf-1 (lanes 3 and 4), myc-Raf-1 S259A (lanes 5 and 6), myc-Raf-1 S289/296/301A (lanes 7 and 8), or myc-Raf-1 S259/289/296/301A (lanes 9 and 10) were stimulated with EGF as indicated for 20 min, and total cell extracts were immunoblotted for phospho-ERK, pS296 Raf-1 and myc-Raf-1 as indicated. (D) Wild-type myc-Raf-1 (lanes 1–8), S471A myc-Raf-1 mutant (a kinase inactive Raf mutant, lanes 9–14), and S289/296/301A myc-Raf-1 (lanes 15–20) were immunopurified from serum-deprived COS-7 cells (lanes 1–3, 7, 9–11, 15, 16, and 19) or from cells stimulated with EGF for 15 min (lanes 4–6, 8, 12–14, 17, 18, and 20). Raf kinase activity in the samples was assayed directly (lanes 7, 8, 19, and 20, representing a 0-time point) or after incubation with vehicle (lanes 1, 4, 9, 12, 15, and 17), recombinant ERK-1 (ba, lanes 2, 6, 10, and 14) or recombinant ERK-1 activated in vitro with MEK-1 (ac, lanes 3, 5, 11, 13, 16, and 18) under conditions provided in the experimental procedures. Presented are a phospho-MEK immunoblot showing the level of MEK phosphorylation (top) and a myc-immunoblot showing myc-Raf-1 recovery (bottom). The migration positions of phospho-MEK and myc-Raf-1 are indicated. (E) Wild-type myc-Raf-1 purified from serum-deprived (lane 1) or EGF-stimulated cells (lanes 2–12) was incubated for the indicated time points in a complete kinase reaction buffer (lanes 4, 6, 8, 10, and 12) or in a kinase reaction buffer lacking ATP (lanes 3, 5, 7, 9, and 11) at 30°C and assayed for Raf-1 kinase activity as described in A. Bottom, a Coomassie blue staining presenting myc-Raf-1 recovery and GST-MEK amounts. Lane 13 is a control lane containing GST-MEK alone.
Next, we wanted to determine to what extent endogenous ERK plays a role in sustaining the kinase activity of Raf-1 (Figure 6, B and C). Initially, we used an MEK inhibitor to block ERK activation and tested its effect on the time kinetics of Raf-1 activation (Figure 6B). MEK inhibition resulted in a slightly faster inactivation rate of Raf-1, without significantly affecting the maximal activation rate (Figure 6B, compare lanes 1–8 with 9–16), similarly to the results observed with the triple Raf-1 mutant (Figure 6A). However, because it has been reported that ERK can negatively regulate several upstream Raf effectors, e.g., Sos and EGF receptor, the effect of ERK on the time kinetics of Raf-1 activation involves both direct effects on Raf-1 through phosphorylation and effects on recurring Raf-1 activation through the upstream signals. Thus, to focus more on the direct effect of ERK on Raf-1 activity, we introduced the triple S289/296/301A mutation in the constitutively active S259A Raf-1 form and tested its effect on Raf-1 activity in vivo (Figure 6C). The Raf-1 S259A mutant has a slightly increased basal kinase activity than wild-type Raf-1 when measured in vitro (Tzivion et al., 1998); however, this elevated basal activity is sufficient to induce full activation of ERK in vivo (Figure 6C, compare lanes 3 and 5). Introduction of the triple mutation resulted in a marked decrease in this basal activity without affecting the activity level seen after EGF stimulation (Figure 6C, compare lanes 5, 6 and 9, 10). In addition, a pS296 immunoblot shows that Raf-1 S259A is constitutively phosphorylated on the S296 site (Figure 6C, compare lanes 3, 4 and 5, 6), indicating that the constitutive activation of ERK in these cells results in a feedback phosphorylation of Raf-1 by ERK. Together, these results suggest that the ERK-mediated feedback phosphorylation of Raf-1 S259A contributes to its constitutive activity.
Several early studies, in an effort to establish the hierarchy of signal transmission downstream of Ras, reported Raf-1 phosphorylation by ERK in vitro (Anderson et al., 1991; Kyriakis et al., 1993). One of these studies pointed to a negative effect of ERK on Raf-1 activity (Kyriakis et al., 1993). Our results showing a positive effect of ERK on Raf-1 activity in vivo, prompted us to reexamine the effect of ERK on Raf-1 activity in vitro (Figure 6, D and E). To our surprise, incubation of immunopurified Raf-1 from EGF-treated cells either alone or with ERK resulted in a decreased Raf-1 kinase activity (Figure 6D, compare lane 8 with lanes 4–6). This inactivation, however, was markedly attenuated when Raf-1 was incubated with active ERK compared with kinase buffer alone or inactive ERK (Figure 6D, compare lane 5 with 4 and 6). As a negative control in these experiments we used inactive Raf-1 mutant S471A (Zhu et al., 2005), which showed no detectable kinase activity at any of the treatment conditions (Figure 6D, lanes 9–14). Thus, the in vitro results support to some extent the in vivo data by demonstrating that although ERK phosphorylation may not be activating Raf-1 per se, it can attenuate Raf-1 inactivation. In vitro, ERK was able to also inhibit the inactivation rate of the triple Raf-1 mutant to a certain degree (Figure 6D, lanes 15–20); however, this could be the result of different mechanisms behind the in vivo and in vitro ERK activity.
To further validate the finding that Raf-1 preincubation in kinase reaction conditions results in the inactivation of the kinase, we examined the effect of varying incubation times and conditions on this inactivation (Figure 6E). These experiments showed that Raf-1 is rapidly inactivated when incubated in kinase reaction buffer at 30°C and that the presence of ATP accelerates the inactivation rate (Figure 6E, compare lanes 3, 5, 7, 9, 11, no ATP, with lanes 4, 6, 8, 10, 12, with ATP). This inactivation was not affected by including a mixture of phosphatase inhibitors in the reaction buffer (Figure 6E, lanes 11 and 12) and was not accompanied by a detectable reduction in Raf-1 protein amounts or the appearance of Raf-1 degradation products, suggesting that Raf-1 dephosphorylation or degradation are not the cause for Raf-1 inactivation. The mechanisms of this inactivation and ERK ability to attenuate it remain to be determined, with one suitable mode being Raf-1 autophosphorylation or its phosphorylation by coassociated kinases.
The ERK-Phosphorylated Raf-1 Pool Displays Up to Fourfold Higher Specific Kinase Activity than the General Raf-1 Population
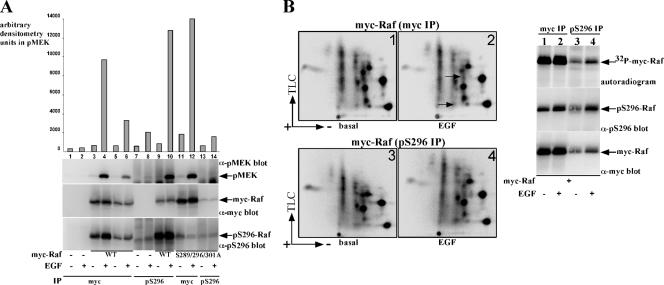
The finding that ERK sustains Raf-1 kinase activity suggested that ERK would need to phosphorylate the activated Raf-1 pool and that possibly the ERK-phosphorylated Raf-1 pool will display higher specific kinase activity than the general Raf-1 population. To examine this hypothesis, we compared the specific kinase activities and the phosphopeptide compositions of Raf-1 immunoprecipitated using a mycepitope tag antibody, representing the general Raf-1 population and Raf-1 immunoprecipitated using the pS296 antibody, representing the ERK-phosphorylated Raf-1 population (Figure 7). Raf-1 in pS296 pull-downs had up to fourfold higher specific kinase activity than Raf-1 in myc pull-downs (Figure 7A, compare lane 10 with lane 6). In these experiments, because the total amount of Raf-1 recovered by myc was higher than that recovered by pS296 antibody, the Raf-1 amounts were standardized before the kinase assay such that equal amounts of Raf-1 were used in the myc and pS296 pull-down samples (Figure 7A, middle, compare lanes 5, 6 and 9, 10). These experiments also showed the enrichment of pS296 phosphorylated Raf-1 in the pS296 pull-downs compared with the myc pull-downs (Figure 7A, bottom, compare lanes 5, 6 and 9, 10). As expected, pS296 antibody recovered only traces of myc-Raf-1 or Raf-1 kinase activity from cells expressing the S289/296/301A Raf-1 mutant (Figure 7A, lanes 13 and 14). The trace myc-Raf-1 seen in these pull-downs can be attributed to the ability of pS296 to precipitate endogenous Raf-1 (Figure 7A, compare lanes 7, 8, mock, and lanes 13, 14, S289/296/301 Raf-1), which can oligomerize with the mutant Raf-1 (Luo et al., 1996). These experiments demonstrate that the ERK phosphorylated Raf-1 is active and because this pool of Raf-1 has a higher specific activity than the general Raf-1 population, suggest that ERK preferentially phosphorylates the activated Raf-1 form. This notion is supported by the phosphopeptide map analysis of Raf-1 immunoprecipitated using the pS296 antibody (Figure 7B). In this experiment, Raf-1 from 32P metabolically labeled cells was immunoprecipitated either using the myc antibody (Figure 7B, lanes 1 and 2) or using the pS296 antibody (Figure 7B, lanes 3 and 4) and analyzed by 2D phosphopeptide mapping. The results show that the phosphopeptide composition of Raf-1 immunoprecipitated using either of the antibodies is highly similar, indicating that ERK phosphorylates the Raf-1 pool, which is already phosphorylated on the other sites. Because this pool of Raf-1 is considered to represent the active or the activatable pool of Raf-1, these results support the notion that ERK preferentially phosphorylates the activated Raf-1 pool in vivo. Similarly to the results shown in Figure 7A, the total Raf-1 recovery in the pS296 immunoprecipitates was lower than Raf-1 recovery using myc immunoprecipitation; however, it was enriched with Raf-1 phosphorylated on the S296 site.
Figure 7.
The ERK-phosphorylated Raf-1 population has increased specific kinase activity. (A) Serum-deprived COS-7 cells expressing a vector control (lanes 1, 2, 7, and 8), myc-Raf-1 (lanes 3–6, 9, and 10), or myc-Raf-1 S289/296/301A (lanes 11–14) were subjected to immunoprecipitation using myc antibodies (lanes 1–6, 11, and 12) or pS296 antibodies (lanes 7–10, 13, and 14). The immunoprecipitates were assayed for Raf-1 kinase activity as in Figure 6A. To help in evaluating Raf-1-specific kinase activity and standardizing for total Raf-1 amounts used in the kinase assay, 75% (lanes 3 and 4) or 25% (lanes 5 and 6) of the myc-Raf-1 immunoprecipitates were used in the kinase assays. Note that the total myc-Raf-1 used in the pS296 immunoprecipitates (lanes 9 and 10) is equal to the total myc-Raf-1 used in lanes 5 and 6, demonstrating up to fourfold increase in Raf-1 specific activity in the pS296 immunoprecipitated samples. The bottom panel represents the pS296-phosphorylated myc-Raf-1 recovery in the samples. (B) myc-Raf-1 from 32P metabolically labeled COS-7 cells were immunoprecipitated using myc antibodies (samples 1 and 2) or pS296 antibodies (samples 3 and 4) and analyzed by 2D-phosphopeptide mapping (panels 1–4, the position of the ERK-induces sites 4 and 11 are indicated by arrows). The right part shows myc-Raf-1 recovery (bottom), myc-Raf-1 phosphorylation on S296 (middle), and total 32P incorporation in myc-Raf-1 (top). Equal amounts of 32P counts (1500 cpm) were used to generate the phosphopeptide maps.
DISCUSSION
The Raf activation process is one of the most tightly controlled processes along the growth factor signaling cascade (Avruch et al., 2001). It involves regulation by both modulating protein–protein interactions and posttranslational modifications, such as phosphorylation. Constitutive activation of Raf, either by direct mutation in Raf or by constitutive activation of its upstream effectors, is transforming and is commonly found in a variety of human cancers (Weinstein-Oppenheimer et al., 2000; Mercer and Pritchard, 2003). Thus, a comprehensive understanding of Raf regulation and its activation/deactivation process would be highly significant. In the present study, we describe three novel in vivo Raf-1 phosphorylation sites that are positively regulated by EGF treatment. Phosphorylation of these sites in vivo can be enhanced by ERK-1 overexpression, and ERK activity is necessary both for basal Raf-1 phosphorylation and the mitogen-induced phosphorylation. Furthermore, using direct in vitro phosphorylation we show that these sites are direct targets of ERK-1 and using phosphospecific antibodies developed against one of the sites, S296, show that these sites are physiological phosphorylation sites induced in vivo after mitogen stimulation. Most importantly, we show that the ERK-phosphorylated Raf-1 is highly active and that these sites serve for positive Raf-1 regulation in vivo. In addition, our data suggest that ERK preferentially phosphorylates the active pool of Raf-1 and positively regulates its function, potentially, by contributing to the stabilization of the active form. These findings add a new layer of complexity to the Raf-1 regulation mechanism and suggest that varying ERK activation levels in the cell can modulate the output from Raf-1.
Several early studies indicated Raf-1 as a potential ERK target (Anderson et al., 1991; Kyriakis et al., 1993); however, the functional significance of this phosphorylation in vivo was not determined. Later, Laird et al. (1999) showed that Raf-1 phosphorylation on a fragment corresponding to Raf-1 283-302 was responsible for Raf-1 mobility shift during mitosis, correlating with Raf-1 activation, and Alessandrini et al. (1996) and Zimmerman et al. (1997) showed that expression of constitutively active forms of MEK in NIH 3T3 or human embryonic kidney 293 cells promote Raf-1 activity, possibly through increased Raf-1 phosphorylation mediated by effectors downstream of MEK. Our present findings are in agreement with these previous reports offering a positive role of MEK and ERK in Raf-1 regulation. More recently, after the initial submission of this manuscript, two groups reported Raf-1 phosphorylation on similar sites as reported here (Dougherty et al., 2005; Hekman et al., 2005). Both studies proposed a negative regulatory role for the identified phosphorylations sites in Raf-1 regulation, leading to the enticing hypothesis that ERK participates in a negative feedback loop responsible for down-regulating Raf-1 activity (Dumaz and Marais, 2005). This model, however, is in strict contrast to ours and the above-mentioned previous reports. As regards this discrepancy, Hekman et al. (2005) identified Raf-1 phosphorylation on S296 and S301 and showed that alanine substitution of these sites is activating, whereas their substitution with aspartic acid has no effect. Raf-1 activity in these experiments was examined in COS-7 cells activated with EGF for 2 min. In our experiments, we were unable to detect any increase in Raf-1 kinase activity in the alanine-substituted mutants, either the S289/296/301A mutant (Figure 6) or several other combinations such as S289/296A or S296/301A (our unpublished data). As regards the time course, our data show that Raf-1 activation in COS-7 cells peaks at 15–20 min after EGF stimulation, followed by Raf-1 phosphorylation on the ERK-induced sites, which peaks at 30–45 min. Thus, the difference at 2 min after EGF stimulation between wild type and the phosphorylation site Raf-1 mutants observed by Hekman et al. (2005) may not necessarily represent the effects of ERK-induces Raf-1 phosphorylation. At an early time point, also Dougherty et al. (2005) did not observe differences in the activities of wild type and mutant Raf-1. The effect of ERK on Raf-1 activity either in vitro or in vivo has not been examined in this work. Dougherty et al. (2005) reported that Raf-1 is phosphorylated in response to platelet-derived growth factor (PDGF) treatment, resulting in an upshift in Raf-1 mobility on SDS-PAGE that coincided with decreased kinase activity of Raf-1. The authors identified five ERK-mediated Raf-1 phosphorylation sites, including S29, S289, S296, S301, and S642 and showed that their mutation together with S43, a previously identified negative Raf-1 regulatory site targeted by PKA, prevents the upshift in mobility and decreases its inactivation rate. In addition, this work showed a potential role of these sites in mediating Raf-1 binding to Pin1, a prolyl isomerase identified in the study to associate with Raf-1 and mediate its dephosphorylation. Based on these data, the authors suggested a negative role of the ERK-mediated Raf-1 phosphorylation in Raf-1 regulation. This report, however, similarly to Hekman et al. (2005) did not examine the effect of ERK on Raf-1 activity or provided evidence that indeed Pin1/PP2A plays a role in Raf-1 inactivation. Rather, Pin1 knockout cells were found to be impaired in Raf-1 activation. The authors explain this observation by suggesting that the ERK-phosphorylated Raf-1 is inactive and needs the dephosphorylation to get activated. However, they do not provide evidence to show that indeed the ERK-phosphorylated Raf-1 is inactive or whether ERK activation negatively regulates Raf-1 per se. Our results, which directly examine Raf-1 activity in the ERK-phosphorylated Raf-1 pool, show that this fraction of Raf-1 represents the active form and not the inactive form as suggested by Dougharty et al. (2005) In addition, several previous reports proposing a positive role of PP2A in Raf-1 regulation can provide a better explanation to the authors finding that Pin1 knockout cells are impaired in Raf-1 activation (Abraham et al., 2000; Jaumot and Hancock, 2001). Also, the rational behind including the S43 site when studying the effects of ERK on Raf-1 function is not clear to us. Our data, in contrast to the authors claims that this site is regulated downstream of ERK, show little or no change in S43 phosphorylation in the MEK inhibitor-treated cells, whereas resulting in a complete block of the ERK-induced sites (Figure 3). In addition, in our experiments, we did not observe an increase in the Raf-1 activation kinetics in MEK-inhibited cells (Figure 6B) as observed by Dougherty et al. (2005); however, we used COS-7 cells and EGF as stimulus, whereas Dougherty et al. (2005) used NIH 3T3 cells and PDGF stimulation. Although remotely likely, it remains a possibility that the differences in our results regarding this specific observation are cell type/mitogen dependent. In this regard, a previous work showed that treating Swiss 3T3 cells with the MEK inhibitor PD 098059 increases Raf-1 activation by PDGF but not by EGF (Alessi et al., 1995). It is important to note, however, that a main caveat in experiments using a MEK inhibitor to study ERK-mediated phosphorylation effects on Raf-1 activity is that blocking MEK/ERK has multiple indirect effects on Raf-1 activity through affecting targets upstream of Raf-1. Thus, our use of ERK overexpression (Figure 6A) and an activating Raf-1 mutant (Figure 6C), in addition to the MEK inhibitor, allows a more comprehensive analysis of the role of ERK in Raf-1 regulation. In our opinion, our study, which does not rely solely on analyzing phosphorylation site Raf-1 mutants, but also demonstrates a positive effect of ERK expression on Raf-1 activity, supported by time kinetic data establishing a positive correlation between Raf-1 phosphorylation and Raf-1 kinase activity and showing the ability of the pS296 antibody to immunoprecipitate highly active Raf-1, strongly tips the scales toward the idea that ERK is a positive Raf-1 regulator. The rationalization for ERK being a negative Raf regulator can be easily conveyed and follows many paradigms of negative feedback regulation resulting in signal termination. However, rationalizing a positive Raf-1 regulation by ERK, whereas at the same time negatively regulating various upstream Raf-1 effectors, is more challenging. One possible explanation for our finding could be that whereas ERK attenuates responses to new/recurring signals originating from the receptor, it potentiates the existing signal downstream of Raf-1. This possibility, viewed in the context of the current prevailing opinion that Raf-1 may have other effectors besides MEK (Murakami and Morrison, 2001), suggests that these so called “MEK-independent” Raf-1 functions could be affected indirectly by ERK through the positive feedback loop on Raf-1. This positive feedback loop may have significant implications in cancers with Ras or B-Raf mutations, where there is a constitutive activation of the pathway. In these cases, ERK may contribute to sustaining a continuous signal. This possibility is demonstrated to some degree in Figure 6C showing that the constitutive in vivo activity of the S259A Raf-1 mutant involves the ERK-mediated Raf-1 phosphorylation sites.
The three Raf-1 phosphorylation sites described in this work are unique to Raf-1, though there is some conservation with A-Raf in the S301 site and with B-Raf in the S296 site. The uniqueness of these sites to Raf-1 may suggest that the positive feedback regulation we identified is unique to Raf-1, thus providing a potential isoform-specific regulatory mechanism (Hagemann and Rapp, 1999). This notion, however, needs to be directly examined because it is possible that ERK can regulate A-Raf and B-Raf by phosphorylating other sites than on Raf-1.
It has been reported that B-Raf, which is a strong ERK activator, can indirectly activate Raf-1 (Wan et al., 2004; Wellbrock et al., 2004b). Our finding that ERK can be a positive Raf-1 effector may provide one mechanism to this reported B-Raf function. In addition, it is possible that some of the physiological effects of native B-Raf and the transforming effects of B-Raf mutants are a result of its indirect effect on Raf-1.
The mechanism of Raf-1 inactivation is largely unknown. The common thinking is that dephosphorylation of Raf-1 should be a major player in this process; however, to the extent that activating phosphorylation sites of Raf-1 are not yet well defined, very little is known on Raf-1 dephosphorylation (Avruch et al., 2001). Our finding that, in vitro, incubation of Raf-1 in the presence of ATP results in rapid Raf-1 kinase inactivation (Figure 6E) suggests that Raf-1 inactivation in vivo may involve both Raf-1 phosphorylation on inhibitory sites and its dephosphorylation/recycling. The ability of ERK-induced phosphorylation to stabilize the active form of Raf-1 may provide a new venue to study Raf-1 inactivation by comparing the stable active form of Raf-1 purified using pS296 immunoprecipitation and the remaining Raf-1 population. Also, it will be of interest to define the mechanism by which Raf-1 phosphorylation on the identified sites sustains its kinase activity.
We analyzed in the present study three Raf-1 phosphorylation sites that are targeted by ERK. However, the 2D phosphopeptide maps, both in vivo and in vitro, suggest the presence of several additional Raf-1 phosphorylation sites induced by growth factors and mediated by ERK. These include spots 1, 5, and 10 in vivo (Figure 2B, 13) and spots 1, 5, 6, and 10 in vitro (Figure 4C, 3). It will be important to identify these additional sites and determine their functional significance to gain a more comprehensive understanding of the feedback regulation of Raf-1 by ERK.
The Ras–Raf–MEK–ERK signaling cascade was among the first mammalian signaling pathways to be elucidated (Chang and Karin, 2001). Besides its contribution to understanding the function of growth factors, it helped in establishing a paradigm for signaling cascades and helped in unveiling other signaling cascades such as the stress-activated MAPK signaling pathways and the PI3K–AKT pathway (Avruch et al., 2001). It has long become clear that the initially defined linear cascade of Ras activates Raf, which in turn activates MEK that activates ERK is an over simplified model. Although we know now of many other factors that are involved in Raf regulation, such as accessory molecules such as heat-shock proteins, 14-3-3 and KSR, protein phosphatases, and inhibitory molecules such as RKIP, the linearity of the signal transmission along the cascade has been less challenged. We have recently demonstrated a cross-talk relationship between the stress-activated MAPK pathway and the ERK–MAPK pathway, showing that sustained MLK-3-induced c-Jun NH2-terminal kinase and c-Jun activation results in the inhibition of ERK activation by growth factors via uncoupling ERK activation from MEK (Shen et al., 2003b). Thus, demonstrating that the initially perceived linear MAPK pathways branch and cross-talk one with another. The findings of the present study demonstrate a positive feedback regulation of Raf-1 by ERK and add another level of branching to this signaling cascade. The existence of cross-talk relationships and feedback responses may provide some explanation to the ongoing challenge of understanding how cells respond differently to the same factor under varying cellular conditions.
Acknowledgments
We thank Joseph Avruch and Ajay Rana for helpful discussions. This work was supported by the National Institute of Health Grant R01 GM 067134 (to G. T.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–12–1123) on January 11, 2006.
References
- Abraham, D., Podar, K., Pacher, M., Kubicek, M., Welzel, N., Hemmings, B. A., Dilworth, S. M., Mischak, H., Kolch, W., and Baccarini, M. (2000). Raf-1-associated protein phosphatase 2A as a positive regulator of kinase activation. J. Biol. Chem. 275, 22300–22304. [DOI] [PubMed] [Google Scholar]
- Alessandrini, A., Greulich, H., Huang, W., and Erikson, R. L. (1996). Mek1 phosphorylation site mutants activate Raf-1 in NIH 3T3 cells. J. Biol. Chem. 271, 31612–31618. [DOI] [PubMed] [Google Scholar]
- Alessi, D. R., Cuenda, A., Cohen, P., Dudley, D. T., and Saltiel, A. R. (1995). PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 270, 27489–27494. [DOI] [PubMed] [Google Scholar]
- Anderson, N. G., Li, P., Marsden, L. A., Williams, N., Roberts, T. M., and Sturgill, T. W. (1991). Raf-1 is a potential substrate for mitogen-activated protein kinase in vivo. Biochem. J. 277, 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch, J., Khokhlatchev, A., Kyriakis, J. M., Luo, Z., Tzivion, G., Vavvas, D., and Zhang, X. F. (2001). Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog. Horm. Res. 56, 127–155. [DOI] [PubMed] [Google Scholar]
- Avruch, J., Zhang, X. F., and Kyriakis, J. M. (1994). Raf meets Ras: completing the framework of a signal transduction pathway. Trends Biochem. Sci. 19, 279–283. [DOI] [PubMed] [Google Scholar]
- Boyle, W. J., van der Geer, P., and Hunter, T. (1991). Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201, 110–149. [DOI] [PubMed] [Google Scholar]
- Chang, L., and Karin, M. (2001). Mammalian MAP kinase signalling cascades. Nature 410, 37–40. [DOI] [PubMed] [Google Scholar]
- Chong, H., Lee, J., and Guan, K. L. (2001). Positive and negative regulation of Raf kinase activity and function by phosphorylation. EMBO J. 20, 3716–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, H., Vikis, H. G., and Guan, K. L. (2003). Mechanisms of regulating the Raf kinase family. Cell Signal. 15, 463–469. [DOI] [PubMed] [Google Scholar]
- Davies, H., et al. (2002). Mutations of the BRAF gene in human cancer. Nature 417, 949–954. [DOI] [PubMed] [Google Scholar]
- Dhillon, A. S., and Kolch, W. (2002). Untying the regulation of the Raf-1 kinase. Arch. Biochem. Biophys. 404, 3–9. [DOI] [PubMed] [Google Scholar]
- Dhillon, A. S., Meikle, S., Yazici, Z., Eulitz, M., and Kolch, W. (2002). Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J. 21, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty, M. K., Muller, J., Ritt, D. A., Zhou, M., Zhou, X. Z., Copeland, T. D., Conrads, T. P., Veenstra, T. D., Lu, K. P., and Morrison, D. K. (2005). Regulation of Raf-1 by direct feedback phosphorylation. Mol. Cell 17, 215–224. [DOI] [PubMed] [Google Scholar]
- Dumaz, N., and Marais, R. (2005). Raf phosphorylation: one step forward and two steps back. Mol. Cell 17, 164–166. [DOI] [PubMed] [Google Scholar]
- Hagemann, C., and Rapp, U. R. (1999). Isotype-specific functions of Raf kinases. Exp. Cell Res. 253, 34–46. [DOI] [PubMed] [Google Scholar]
- Hamilton, M., Liao, J., Cathcart, M. K., and Wolfman, A. (2001). Constitutive association of c-N-Ras with c-Raf-1 and PKC epsilon in latent signaling modules. J. Biol. Chem. 276, 29079–29090. [DOI] [PubMed] [Google Scholar]
- Hekman, M., Fischer, A., Wennogle, L. P., Wang, Y. K., Campbell, S. L., and Rapp, U. R. (2005). Novel C-Raf phosphorylation sites: serine 296 and 301 participate in Raf regulation. FEBS Lett. 579, 464–468. [DOI] [PubMed] [Google Scholar]
- Herrera, R., and Sebolt-Leopold, J. S. (2002). Unraveling the complexities of the Raf/MAP kinase pathway for pharmacological intervention. Trends Mol. Med. 8, S27–31. [DOI] [PubMed] [Google Scholar]
- Huang, W., and Erikson, R. L. (1994). Constitutive activation of Mek1 by mutation of serine phosphorylation sites. Proc. Natl. Acad. Sci. USA 91, 8960–8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaumot, M., and Hancock, J. F. (2001). Protein phosphatases 1 and 2A promote Raf-1 activation by regulating 14–3-3 interactions. Oncogene 20, 3949–3958. [DOI] [PubMed] [Google Scholar]
- Kerkhoff, E., and Rapp, U. R. (2001). The Ras-Raf relationship: an unfinished puzzle. Adv. Enzyme Regul. 41, 261–267. [DOI] [PubMed] [Google Scholar]
- King, A. J., Sun, H., Diaz, B., Barnard, D., Miao, W., Bagrodia, S., and Marshall, M. S. (1998). The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature 396, 180–183. [DOI] [PubMed] [Google Scholar]
- Kolch, W. (2000). Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 351, 289–305. [PMC free article] [PubMed] [Google Scholar]
- Kyriakis, J. M., Force, T. L., Rapp, U. R., Bonventre, J. V., and Avruch, J. (1993). Mitogen regulation of c-Raf-1 protein kinase activity toward mitogen-activated protein kinase-kinase. J. Biol. Chem. 268, 16009–16019. [PubMed] [Google Scholar]
- Laird, A. D., Morrison, D. K., and Shalloway, D. (1999). Characterization of Raf-1 activation in mitosis. J. Biol. Chem. 274, 4430–4439. [DOI] [PubMed] [Google Scholar]
- Liebmann, C. (2001). Regulation of MAP kinase activity by peptide receptor signalling pathway: paradigms of multiplicity. Cell Signal. 13, 777–785. [DOI] [PubMed] [Google Scholar]
- Luo, K. X., Hurley, T. R., and Sefton, B. M. (1991). Cyanogen bromide cleavage and proteolytic peptide mapping of proteins immobilized to membranes. Methods Enzymol. 201, 149–152. [DOI] [PubMed] [Google Scholar]
- Luo, Z., Tzivion, G., Belshaw, P. J., Vavvas, D., Marshall, M., and Avruch, J. (1996). Oligomerization activates c-Raf-1 through a Ras-dependent mechanism. Nature 383, 181–185. [DOI] [PubMed] [Google Scholar]
- Lyons, J. F., Wilhelm, S., Hibner, B., and Bollag, G. (2001). Discovery of a novel Raf kinase inhibitor. Endocr. Relat. Cancer 8, 219–225. [DOI] [PubMed] [Google Scholar]
- Marais, R., Light, Y., Mason, C., Paterson, H., Olson, M. F., and Marshall, C. J. (1998). Requirement of Ras-GTP-Raf complexes for activation of Raf-1 by PKC. Science 280, 109–112. [DOI] [PubMed] [Google Scholar]
- Mason, C. S., Springer, C. J., Cooper, R. G., Superti-Furga, G., Marshall, C. J., and Marais, R. (1999). Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. EMBO J. 18, 2137–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer, K. E., and Pritchard, C. A. (2003). Raf proteins and cancer: B-Raf is identified as a mutational target. Biochim. Biophys. Acta 1653, 25–40. [DOI] [PubMed] [Google Scholar]
- Morrison, D. K., Heidecker, G., Rapp, U. R., and Copeland, T. D. (1993). Identification of the major phosphorylation sites of the Raf-1 kinase. J. Biol. Chem. 268, 17309–17316. [PubMed] [Google Scholar]
- Murakami, M. S., and Morrison, D. K. (2001). Raf-1 without MEK? Sci. STKE 2001, E30. [DOI] [PubMed] [Google Scholar]
- Pearson, G., Robinson, F., Beers Gibson, T., Xu, B. E., Karandikar, M., Berman, K., and Cobb, M. H. (2001). Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22, 153–183. [DOI] [PubMed] [Google Scholar]
- Porter, A. C., and Vaillancourt, R. R. (1998). Tyrosine kinase receptor-activated signal transduction pathways which lead to oncogenesis. Oncogene 17, 1343–1352. [DOI] [PubMed] [Google Scholar]
- Qin, J., and Zhang, X. (2002). Identification of in vivo protein phosphorylation sites with mass spectrometry. Methods Mol. Biol. 194, 211–221. [DOI] [PubMed] [Google Scholar]
- Rapp, U. R., Goldsborough, M. D., Mark, G. E., Bonner, T. I., Groffen, J., Reynolds, F. H., Jr., and Stephenson, J. R. (1983). Structure and biological activity of v-raf, a unique oncogene transduced by a retrovirus. Proc. Natl. Acad. Sci. USA 80, 4218–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommel, C., Clarke, B. A., Zimmermann, S., Nunez, L., Rossman, R., Reid, K., Moelling, K., Yancopoulos, G. D., and Glass, D. J. (1999). Differentiation stage-specific inhibition of the Raf-MEK-ERK pathway by Akt. Science 286, 1738–1741. [DOI] [PubMed] [Google Scholar]
- Shen, Y. H., Godlewski, J., Bronisz, A., Zhu, J., Comb, M. J., Avruch, J., and Tzivion, G. (2003a). Significance of 14–3-3 self-dimerization for phosphorylation-dependent target binding. Mol. Biol. Cell 14, 4721–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y. H., Godlewski, J., Zhu, J., Sathyanarayana, P., Leaner, V., Birrer, M. J., Rana, A., and Tzivion, G. (2003b). Cross-talk between JNK/SAPK and ERK/MAPK pathways: sustained activation of JNK blocks ERK activation by mitogenic factors. J. Biol. Chem. 278, 26715–26721. [DOI] [PubMed] [Google Scholar]
- Sidovar, M. F., Kozlowski, P., Lee, J. W., Collins, M. A., He, Y., and Graves, L. M. (2000). Phosphorylation of serine 43 is not required for inhibition of c-Raf kinase by the cAMP-dependent protein kinase. J. Biol. Chem. 275, 28688–28694. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., and Avruch, J. (2002). 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 277, 3061–3064. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., Luo, Z., and Avruch, J. (1998). A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature 394, 88–92. [DOI] [PubMed] [Google Scholar]
- Tzivion, G., Shen, Y. H., and Zhu, J. (2001). 14–3-3 proteins; bringing new definitions to scaffolding. Oncogene 20, 6331–6338. [DOI] [PubMed] [Google Scholar]
- Wan, P. T., et al. (2004). Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855–867. [DOI] [PubMed] [Google Scholar]
- Weinstein-Oppenheimer, C. R., Blalock, W. L., Steelman, L. S., Chang, F., and McCubrey, J. A. (2000). The Raf signal transduction cascade as a target for chemotherapeutic intervention in growth factor-responsive tumors. Pharmacol. Ther. 88, 229–279. [DOI] [PubMed] [Google Scholar]
- Wellbrock, C., Karasarides, M., and Marais, R. (2004a). The RAF proteins take centre stage. Nat. Rev. Mol. Cell Biol. 5, 875–885. [DOI] [PubMed] [Google Scholar]
- Wellbrock, C., Ogilvie, L., Hedley, D., Karasarides, M., Martin, J., Niculescu-Duvaz, D., Springer, C. J., and Marais, R. (2004b). V599EB-RAF is an oncogene in melanocytes. Cancer Res. 64, 2338–2342. [DOI] [PubMed] [Google Scholar]
- Yip-Schneider, M. T., Miao, W., Lin, A., Barnard, D. S., Tzivion, G., and Marshall, M. S. (2000). Regulation of the Raf-1 kinase domain by phosphorylation and 14–3-3 association. Biochem. J. 351, 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. H., and Guan, K. L. (2001). Regulation of the Raf kinase by phosphorylation. Exp. Lung Res. 27, 269–295. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Yao, B., Delikat, S., Bayoumy, S., Lin, X. H., Basu, S., McGinley, M., Chan-Hui, P. Y., Lichenstein, H., and Kolesnick, R. (1997). Kinase suppressor of Ras is ceramide-activated protein kinase. Cell 89, 63–72. [DOI] [PubMed] [Google Scholar]
- Zhu, J., Balan, V., Bronisz, A., Balan, K., Sun, H., Leicht, D. T., Luo, Z., Qin, J., Avruch, J., and Tzivion, G. (2005). Identification of Raf-1 S471 as a novel phosphorylation site critical for Raf-1 and B-Raf kinase activities and for MEK binding. Mol. Biol. Cell 16, 4733–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, S., and Moelling, K. (1999). Phosphorylation and regulation of Raf by Akt (protein kinase B). Science 286, 1741–1744. [DOI] [PubMed] [Google Scholar]
- Zimmermann, S., Rommel, C., Ziogas, A., Lovric, J., Moelling, K., and Radziwill, G. (1997). MEK1 mediates a positive feedback on Raf-1 activity independently of Ras and Src. Oncogene 15, 1503–1511. [DOI] [PubMed] [Google Scholar]