Abstract
Barrier-to-autointegration factor (BAF) is a conserved 10-kDa chromatin protein essential in proliferating cells. BAF dimers bind double-stranded DNA, histone H3, histone H1.1, lamin A, and transcription regulators, plus emerin and other LEM-domain nuclear proteins. Two-dimensional gel analysis showed that endogenous human and Xenopus BAF are posttranslationally modified by phosphorylation and potentially other modifications and that they are hyperphosphorylated during mitosis. The invariant Ser-4 residue on BAF is a major site of phosphorylation during both interphase and mitosis. In HeLa cells that overexpressed the phosphomimetic BAF missense mutant S4E, but not S4A, emerin mislocalized from the nuclear envelope, suggesting Ser-4-nonphosphorylated BAF normally promotes emerin localization at the nuclear envelope. Supporting this model, wild-type BAF but not mutant S4E enhanced emerin binding to lamin A in vitro. Thus, Ser-4-unphosphorylated BAF has a positive role in localizing emerin; this role may be disease relevant because loss or mislocalization of emerin causes Emery–Dreifuss muscular dystrophy. Our findings further suggest Ser-4 phosphorylation inhibits BAF binding to emerin and lamin A, and thereby weakens emerin–lamin interactions during both mitosis and interphase.
INTRODUCTION
The nuclear envelope includes two membranes, outer (OM) and inner (IM), which merge periodically to form nuclear pores. Integral membrane proteins destined for the IM are thought to diffuse from their sites of synthesis in the endoplasmic reticulum (ER) and OM to the IM, where they are retained by binding to nuclear filament proteins named lamins and other stable elements (Ellenberg et al., 1997; Soullam and Worman, 1995). Lamin filaments are essential architectural components of the nucleus. Lamins are thought to provide scaffolds for other proteins required for DNA replication, gene expression, chromosome attachment, signaling, and nuclear position (Gruenbaum et al., 2005). Mutations in A-type lamins and three lamin-binding proteins (emerin, MAN1, and LBR) cause many human diseases for which the underlying pathological mechanisms are unclear (Bengtsson and Wilson, 2004; Broers et al., 2004; Gruenbaum et al., 2005).
The LEM-domain family of lamin-binding proteins in the nucleus includes LAP2β, emerin and MAN1 at the IM, and LAP2α in the nuclear interior (Foisner, 2003). Their defining feature is the “LEM-domain” (Holmer and Worman, 2001), which binds barrier-to-autointegration factor (BAF), a conserved chromatin protein (Segura-Totten and Wilson, 2004).
BAF is conserved in metazoans and is essential during embryogenesis (Zheng et al., 2000; Furukawa et al., 2003; Margalit et al., 2005). The atomic structure of the BAF dimer is known (Cai et al., 1998; Umland et al., 2000). Each BAF dimer has two binding sites for double-stranded DNA and binds nonspecifically in terms of DNA sequence (Umland et al., 2000; Zheng et al., 2000) by contacting the phosphate backbone of DNA in the minor groove face (Bradley et al., 2005). Interestingly, DNA causes BAF dimers to oligomerize (Zheng et al., 2000) and can also trigger conformational changes in BAF (Forne et al., 2003), although BAF dimer structure is unaltered in a cocrystal with seven-base pair DNA (Bradley et al., 2005). When added to cell-free Xenopus nuclear assembly reactions, BAF potently regulates higher order chromatin structure (Segura-Totten et al., 2002). Other findings also suggest roles for BAF in chromatin structure and gene expression. First, BAF binds and represses paired-like homeodomain transcription activators in vivo (Wang et al., 2002). Second, BAF blocks binding of a transcriptional repressor (GCL) to emerin in vitro (Holaska et al., 2003). Third, BAF binds core histone H3 and some (but not all) linker histones (Montes de Oca et al., 2005). Loss of BAF function in Drosophila somatic cells causes many phenotypes, including loss of cyclin gene expression and failure to proliferate (Furukawa et al., 2003). BAF also has structural roles in recruiting emerin, other LEM-domain proteins and lamins to chromosomes during nuclear assembly in mammalian cells (Dechat et al., 2004; Haraguchi et al., 2001) and Caenorhabditis elegans embryos (Margalit et al., 2005). BAF localization seems to be regulated, because BAF is predominantly chromatin-associated in Drosophila embryos (Furukawa et al., 2003) but not in mammalian somatic cells, where there are both nuclear and cytoplasmic pools of BAF (Segura-Totten et al., 2002; Shimi et al., 2004). Finally, cytoplasmic BAF is recruited by retroviruses, including human immunodeficiency virus type 1 (Lee and Craigie, 1998; Harris and Engelman, 2000; Lin and Engelman, 2003). BAF binds directly to the human immunodeficiency virus (HIV)-1-encoded matrix protein (Mansharamani et al., 2003). Both BAF and LAP2α, a soluble LEM-domain protein, are present in DNA-containing retroviral preintegration complexes (Mansharamani et al., 2003; Suzuki et al., 2004), and BAF is required for these complexes to integrate into target DNA in vitro (Suzuki et al., 2004, and references therein).
Several findings suggested BAF might be posttranslationally modified in vivo (Segura-Totten and Wilson, 2004), which might explain how BAF can localize differentially and interact meaningfully with so many different partners. However, direct evidence was lacking. We therefore tested the hypothesis that BAF is posttranslationally modified. Several of our experiments used fractionated Xenopus egg extracts as a source of endogenous kinases and other enzymes. When reconstituted, the membrane and soluble fractions of S-phase Xenopus extracts can assemble chromatin into replication-competent nuclei (Lohka and Masui, 1984; Newmeyer and Wilson, 1991). Furthermore, S-phase extracts can be converted to M phase by adding the nondegradable (Δ90) form of cyclin, which activates p34cdc2 (Glotzer et al., 1991). Previous studies showed that recombinant Xenopus and human BAF are functionally equivalent when added to Xenopus nuclear assembly reactions (Segura-Totten et al., 2002). In the present study, both endogenous and exogenous BAF gave evidence of extensive posttranslational modification. We focused on a subset of phosphorylated forms of BAF. Our results suggest that phosphorylation at Ser-4 regulates the binding activity of BAF both in vitro and in vivo.
MATERIALS AND METHODS
Bioinformatics
BAF amino acid sequences from http://www.ncbi.nlm.nih.gov/BLAST were aligned using the ClustalW program (Higgins et al., 1994) and edited using ESPript (Gouet et al., 1999). Secondary structure and residue accessibility were predicted by ESPript based on the crystal structure of BAF using the 1CI4.pdb file. The physical properties of BAF and BAF peptides were calculated using the ProtParam tool (Gasteiger et al., 2005). All programs were found at http://us.expasy.org.
Cell Culture and Indirect Immunofluorescence Microscopy
HeLa and 293T cells were cultured in DMEM supplemented with 10% fetal calf serum and 5% penicillin/streptomycin. Cells were transiently transfected using TransIT-LT1 (Mirus, Madison, WI) per the manufacturer's instructions. For indirect immunofluorescence, HeLa cells were grown on 13-mm cover-slips, fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) ∼19 h after transfection, permeabilized with 0.5% Triton X-100 in PBS (10 min; 22–25°C), blocked in PBS containing 3% bovine serum albumin (BSA) (30 min; 22–25°C), and probed for 1 h with antibodies against either the Xpress-tag (Invitrogen, Carlsbad, CA), emerin (serum 2999; Holaska et al., 2003) or A-type lamins (rabbit polyclonal against a nuclear localization sequence-containing fragment, kindly provided by Dr. Nilabh Chaudhary, Ridgeway Biosystems, Cleveland, OH), all at 1:500 dilution. Secondary antibodies, either tetramethylrhodamine B isothiocyanate- or Cy5-conjugated (The Jackson Laboratory, Bar Harbor, ME), were diluted 1:300 and incubated 1 h at 22–25°C. Cells were imaged using a fluorescence microscope (Nikon Eclipse E600; Nikon, Tokyo, Japan).
Xenopus Egg Extract Preparation
Xenopus egg S-phase extracts were prepared as described and the 200,000g soluble fraction was used (Segura-Totten et al., 2002). Where indicated, S-phase extracts were biochemically converted to M-phase by adding purified recombinant Δ90cyclin (final concentration 30 μg/ml) and incubating 1 h at 22–25°C before use (Glotzer et al., 1991).
Two-dimensional (2-D) Gel Electrophoresis and BAF Detection
Human (HeLa or 293T) cultured cells were solubilized 20 min by shaking at 4°C in 20 mM HEPES, pH 8.0, 0.1% SDS, 1:1000 (vol/vol) benzonase (Merck, Whitehouse Station, NJ), protease inhibitors [protease inhibitor cocktail, 1:100 (vol/vol); Roche Diagnostics, Indianapolis, IN], and phosphatase inhibitors [phosphatase inhibitors I and II, 1:100 (vol/vol); Sigma-Aldrich, St. Louis, MO]. Lysates were clarified by centrifugation (4°C, 15 min, 13,000 rpm; Eppendorf 5415C). Supernatant proteins (20 μg/reaction) were supplemented with IEF buffer [8 M urea, 4% (wt/vol) 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate, and 1.5% ampholytes] containing 1.5% hydroxyethyldisulfide and 1% dithiothreitol (DTT) in a total volume of 115 μl. Xenopus egg extracts (20 μg of protein) were supplemented with IEF buffer containing 2% DTT. Samples were applied to pH 3–10, 7-cm IPG Strips (linear gradient for HeLa and Xenopus samples, nonlinear for 293T samples; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) and then rehydrated and focused on IPGphor (GE Healthcare) per the manufacturer's recommendations. For the second dimension, strips were first equilibrated 15 min (22–25°C) in equilibration buffer [50 mM MES, 50 mM Tris, 1 mM EDTA, 0.1% SDS, 6 M urea, and 30% (vol/vol) glycerol] containing 1 mM DTT, followed by 15 min in equilibration buffer containing 4% (wt/vol) iodoacetamide and then resolved on NuPage Bis-Tris 4–12% gels (Invitrogen). The gels were transferred to nitrocellulose, blocked 1 h in Blotto (5% nonfat milk in PBS/0.1% Tween 20), and probed with rabbit serum 3273 (bleed 7) against human BAF. This serum was described previously (Haraguchi et al., 2001). For peptide inhibition controls, immune BAF serum 3273 was first diluted 1:100 in PBS either with or without the unconjugated antigenic peptide (50 μg) in a final volume of 200 μl and then incubated 3 h at 37°C and clarified by centrifugation (15 min, 13,000 rpm; Eppendorf 5415C). The supernatants were used to probe nitrocellulose membranes (final antibody dilution 1:5000) in Blotto at 4°C overnight. Membranes were then washed and incubated 1 h at 22–25°C with horseradish peroxidase-conjugated goat anti-rabbit antibodies (Pierce Chemical, Rockford, IL). Bands were visualized by enhanced chemiluminescence (GE Healthcare).
Plasmid Preparation and Protein Purification
Wild-type and mutant BAF cDNAs were cloned into vectors pET15b (Novagen, Madison, WI), pcDNA3.1/His C (Invitrogen), and pEYFP-C1 (Clontech, Mountain View, CA) using the XhoI and BamHI restriction sites. DNA primers used for subcloning and mutagenesis are summarized in Supplemental Table 1. All constructs were confirmed by DNA sequencing (our unpublished data).
cDNAs encoding emerin residues 1–222 (in pET11c) and prelamin A tail residues 394–664 (in pET23a) were expressed in bacteria and recombinant proteins purified as described previously (Holaska et al., 2003; Zastrow et al., 2006).
Wild-type and mutant BAF proteins were purified from bacteria as described previously (Segura-Totten et al., 2002). Where indicated, the N-terminal His6-tag on BAF was removed by digestion with thrombin (20 U/μg protein) overnight at 4°C; untagged BAF dimers were then recovered and purified by size-exclusion chromatography on a Superdex-75 fast-performance liquid chromatography column (GE Healthcare) in 20 mM Tris, pH 7.4, 150 mM NaCl, 5 mM β-mercaptoethanol, and 1 mM EDTA. When necessary, BAF dimers were concentrated using Microcon centrifugal filter devices YM3 (Millipore, Billerica, MA).
In Vitro Phosphorylation Using Xenopus Egg Extracts
Xenopus egg extracts lacking an ATP regeneration system were used as a source of endogenous S- or M-phase kinases. Typical reactions contained 30 μg (∼1 μl) of Xenopus extract soluble protein and 1–10 μg of recombinant substrate protein in phosphorylation buffer (20 mM HEPES/NaOH, pH 7.5, 0.03% Triton X-100, and 100 μM CaCl2) plus protease inhibitors [protease inhibitor cocktail, 1:100 (vol/vol); Roche Diagnostics] and phosphatase inhibitors (phosphatase inhibitors I and II, 1:100 (vol/vol); Sigma-Aldrich] in a total volume of 40 μl. This reaction was further diluted, and phosphorylation was initiated by adding 10 μl of ATP mix (75 mM MgCl2, 0.5 mM ATP, and 1 μCi/μl [γ-32P]ATP) and incubating for 30 min at 22–25°C. Reactions were stopped by adding 17 μl of 4× SDS-PAGE sample buffer and incubating 10 min at 65°C. Where indicated, reactions were incubated 1 h at 37°C with 50 U of calf intestine phosphatase (CIP) (New England Biolabs, Beverly, MA) before adding SDS-PAGE buffer. Proteins were resolved on NuPage gels, stained with Coomassie G-250, and exposed to film. Typically, good signals were obtained after 1-h exposure (PhosphorImager; Bio-Rad, Hercules, CA).
To purify recombinant His-tagged 32P-labeled BAF after modification by endogenous Xenopus kinases (Figure 6C), the phosphorylation reactions were supplemented with 300 μl of binding buffer (PBS containing 300 mM NaCl) and 100 μl Ni2+-NTA beads (QIAGEN, Valencia, CA) pre-equilibrated in binding buffer, and rotated 1 h at 4°C. Beads were then pelleted, washed three times in binding buffer, and bound proteins were eluted using 150 μl of 0.9 M imidazole in PBS. For blot overlay, 20 μl of eluate was used.
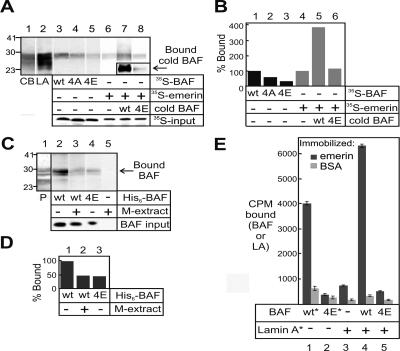
Figure 6.
Effect of BAF proteins (wild type and mutant) on the emerin–lamin A interaction. (A) Recombinant prelamin A tails immobilized on nitrocellulose membrane were probed with 35S-BAF, 35S-emerin, or 35S-emerin plus recombinant radioactive (cold) BAF. CB, strip stained with Coomassie blue. LA, strip probed with antibodies against lamin A. Bound nonradioactive recombinant BAF was detected using BAF serum 3273 (inset, lanes 7 and 8). Lowest panel (35S-input) shows 1% of each input 35S-labeled protein. (B) Densitometric quantification of results in A showing percentage of each probe bound, relative to the binding of 35S-emerin. (C) Nitro-cellulose-immobilized prelamin A tails were probed with His6-tagged bacterially expressed BAF that was preincubated with or without M-phase Xenopus extracts and then repurified on Ni2+-beads. Bound BAF was detected using BAF serum 3273. P, Ponceau S-stained lane to show prelamin A tail protein. (D) Densitometric quantification of results in C. Values are normalized to correct for different BAF input amounts. Binding of unmodified BAF was set as 100%. Results shown are typical of three repeats. (E) Recombinant emerin or BSA (control) were immobilized in microtiter wells and probed with 35S-BAF, 35S-prelamin A (full-length), or 35S-prelamin A (full length) plus recombinant radioactive (cold) BAF. Asterisks indicate 35S-labeled proteins.
Phosphoamino Acid and Phosphopeptide Analysis
Purified recombinant BAF (1 μg) was phosphorylated in Xenopus extracts containing [γ-32P]ATP, and the whole reaction was resolved by SDS-PAGE, transferred to polyvinylidene diflouride (PVDF) membranes, Ponceau S stained, and exposed on PhosphorImager (Bio-Rad). PVDF membrane containing the radioactive BAF band was cut out, hydrolyzed in boiling HCl, and resolved as described previously (Casaday et al., 2004).
For phosphopeptide analysis, phosphorylated BAF (resolved by SDS-PAGE) was transferred to nitrocellulose membranes, and the radioactive BAF band was cut out and incubated 2 h at 22–24°C in the dark with 100 mg/ml CNBr in 70% formic acid. Supernatants were lyophilized, reconstituted in 500 μl of H2O, and lyophilized again. Dried peptides were resuspended in 10 μl of H2O, spotted on tin layer cellulose (TLC) plates, and separated either in both dimensions as described above using buffer 3.5 (pyridine/acetic acid/butanol/H2O at 1:1:2:36) for electrophoresis or separated by chromatography only. Plates were exposed overnight on PhosphorImager and aligned with the resulting autoradiograms. Resolved phosphopeptides were scraped from plates, eluted by 10-min sonication in 10 μl of 0.1% trifluoroacetic acid, 25% acetonitrile, and then 0.5 μl was used directly for matrix-assisted laser desorption ionization/mass spectrometry (MALDI-MS) analysis (Voyager DESTR MALDI/time of flight, AME Bioscience, Toroed, Norway) on 2,5-dihydroxybenzoic acid matrix. Spectra were acquired in reflectron mode using an accelerating voltage of +20 kV.
In Vitro Binding Assays
BAF, emerin, and lamin A were [35S]Met/Cys-labeled in vitro using rabbit transcription/translation extract (Promega, Madison, WI) and used to probe recombinant lamin A or emerin immobilized either on nitrocellulose membranes (“blot overlay assay”) or in microtiter wells. Blot overlays were done as described previously (Lee et al., 2001). Each lane contained ∼2 μg of recombinant lamin A tail protein and was probed with 800,000 cpm of 35S-labeled probe protein plus, where indicated, equimolar amounts of recombinant BAF. Data were quantified using VersaDoc and QuantityOne software (Bio-Rad). Microtiter well binding assays were done as described previously (Holaska et al., 2003), with ∼2 ng of protein immobilized per well and probed with the 35S-labeled protein indicated. Each assay was done in triplicate and repeated at least three times.
RESULTS
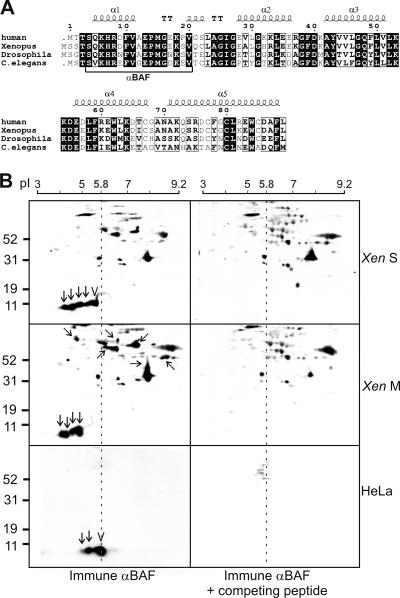
To detect potential posttranslationally modified forms of BAF, we resolved human (HeLa) cell protein lysates or the soluble fraction of S-phase or M-phase Xenopus egg extracts by 2D gel electrophoresis and detected BAF by immunoblotting with rabbit serum 3273 against human BAF residues 4–20 (plus an N-terminal Cys to allow conjugation; Haraguchi et al., 2001). This peptide is identical in 16 of 17 residues to Xenopus BAF (Figure 1A), and serum 3273 recognized Xenopus BAF (Figure 1B, top and middle). To control for specific antibody recognition, we probed duplicate immunoblots in the presence of the competing antigenic peptide (Figure 1B, right). Several spots were specifically recognized in both HeLa and Xenopus samples (Figure 1B, left, arrows and chevrons). The calculated mass of BAF monomers is 10.1 kDa, and the calculated isoelectric points of unmodified human and Xenopus BAF are 5.8 and 5.65, respectively. The strongest signal for ∼10 kDa BAF spots in HeLa cells resolved at pH 5.8 (Figure 1B, bottom left, chevron), suggesting most BAF in unsynchronized HeLa cells was unmodified. Two additional spots migrated at more acidic pH (Figure 1B, bottom left, arrows), suggesting endogenous BAF is phosphorylated or acetylated (or both) in HeLa cells. Three spots were also detected in the nucleoplasmic fraction of HeLa cells (our unpublished data). No other modified forms of BAF were detected by serum 3273 in HeLa lysates (see Discussion).
Figure 1.
Two-dimensional gel analysis of endogenous BAF. (A) Amino acid sequences of BAF from different species. The five α-helices (α1–5) and α-turn (“TT”) in human BAF are indicated. Serum 3273 antibodies were raised against residues 4–20 of human BAF (underlined). (B) Two-dimensionally (isoelectric focusing and SDS-PAGE) resolved HeLa cell and Xenopus egg extract proteins were immunoblotted with immune BAF serum 3273 (left) or immune serum plus competing peptide (right). M, M phase; S, S phase. Arrows and chevron indicate spots specifically recognized by serum 3273; chevron indicates unmodified BAF. Tilted and horizontal arrows indicate spots enriched in mitosis. The linear pI gradient is indicated above each set of panels. The vertical dotted line indicates pI 5.8, the calculated pI for unmodified human BAF.
A more complicated pattern was seen in Xenopus extracts, which differ significantly from HeLa cells: Xenopus extracts are embryonic (not somatic), and each extract represents one phase of the cell cycle (S or M phase). Thus Xenopus extracts are enriched for S- or M-phase BAF modifications. In addition to the unmodified ∼10-kDa BAF spot at pI 5.65 (Figure 1B, top left, chevron), S-phase extracts contained four additional acidic-shifted ∼10-kDa spots of approximately equal abundance (Figure 1B, top left, arrows). Mitotic extracts lacked unmodified BAF but had four BAF spots shifted toward acidic pH, suggesting BAF is hyperphosphorylated and/or hyperacetylated during mitosis (Figure 1B, middle left, arrows).
BAF serum 3273 also specifically recognized at least six slow-migrating spots with apparent masses of 30–100 kDa in Xenopus extracts, many of which were enriched in mitosis (Figure 1B, left middle, slanted and horizontal arrows) and many of which were also strongly shifted toward basic (relative to unmodified ∼10-kDa BAF). These BAF spots, which were not studied further, are speculated to carry additional bulky modifications (see Discussion). For this study, we focused on the putatively phosphorylated 10-kDa forms of BAF.
Ser-4 Is a Major Phosphorylation Site on Human BAF
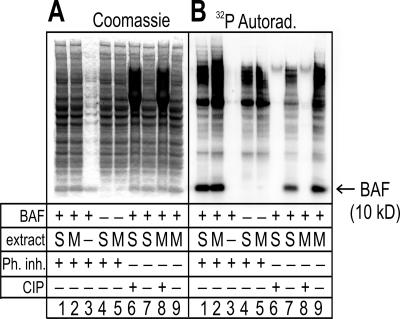
Our BAF antibody did not immunoprecipitate endogenous modified BAF efficiently enough to obtain amounts sufficient for phosphosite analysis. To identify phosphorylation sites on BAF, we therefore used an in vitro phosphorylation reaction that allowed endogenous Xenopus kinases and other enzymes to modify recombinant human BAF dimers. Bacterially expressed, purified, untagged BAF dimers (see Materials and Methods) were incubated at final concentrations of 0.25–0.5 μg/μl for 30 min at 22–24°C in the 50-fold-diluted soluble fraction of S- or M-phase Xenopus egg extracts containing [32P]ATP plus or minus phosphatase inhibitors (Figure 2). Control samples were further treated with CIP (Figure 2, CIP+). Each entire sample was then resolved by SDS-PAGE and either Coomassie stained to view total protein, including recombinant BAF (Figure 2A), or immunoblotted with BAF serum 3273 (our unpublished data), or exposed to detect incorporated 32P (Figure 2B). Exogenous BAF was detectable by Coomassie staining as a 10-kDa band (Figure 2A, compare lanes 1–3 with lane 4). BAF was 32P-labeled in both S-phase and M-phase Xenopus extracts (Figure 2B, lanes 1 and 2), even in reactions that lacked phosphatase inhibitors (Figure 2B, lanes 7 and 9). The 32P-signals were removed by treatment with CIP (Figure 2B, lanes 6 and 8). Under these SDS-PAGE conditions the mobility of 10-kDa BAF was unaffected by phosphorylation (Figure 2, A and B, compare lanes 2 and 3). In 2D gels, the S-phase-modified BAF further resolved as one major and three minor 32P-labeled acidic-shifted spots (Supplemental Figure 1). These results demonstrated phosphorylation of at least one residue in BAF.
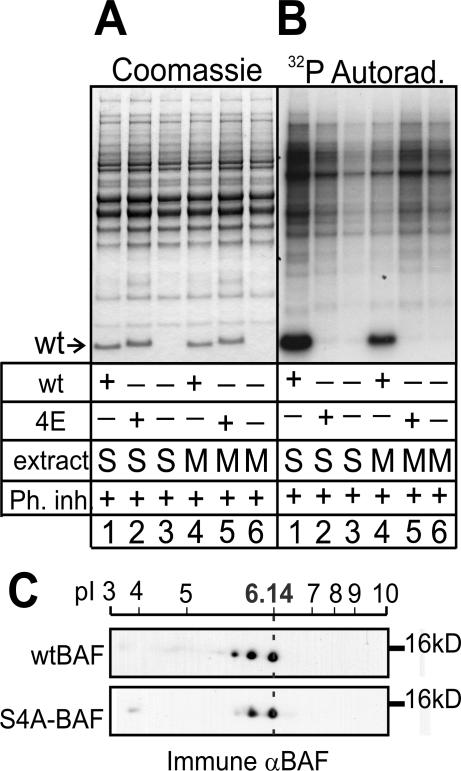
Figure 2.
Recombinant BAF is phosphorylated in both S- and M-phase Xenopus extracts. BAF dimers were incubated in S- or M-phase Xenopus extracts, and each entire reaction was resolved by SDS-PAGE and then Coomassie stained (A) and autoradiographed to detect incorporated 32P (B). M, M phase; Ph inh, phosphatase inhibitors; S, S phase.
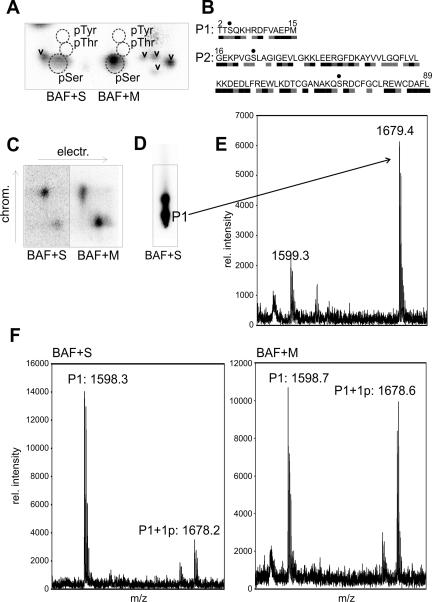
To identify the phosphorylated residue(s), we first did a phosphoamino acid analysis. PVDF membrane-immobilized 32P-BAF from parallel experiments (as in Figure 2) was hydrolyzed in boiling HCl and analyzed by electrophoresis and chromatography on TLC plates. Each plate was stained to detect phosphoamino acid standards and then overlaid with the corresponding autoradiography image (Figure 3A). This analysis demonstrated unequivocally that human BAF was phosphorylated on serine residue(s) in both M- and S-phase extracts (Figure 3A); other free phosphoamino acids were not detected. We concluded that serine was the predominantly phosphorylated residue in the 10-kDa form of BAF. The phosphoserine signal was stronger in M- than S-phase samples (Figure 3A), suggesting more efficient phosphorylation during mitosis.
Figure 3.
Phosphoamino acid and mass spectrometric analysis of 10-kDa forms of BAF. (A) Phosphoamino acid analysis of phosphorylated BAF on TLC plates. Recombinant 10-kDa BAF incubated in Xenopus S- or M-phase extract was transferred to PVDF membrane, recovered from bands as in Figure 2, hydrolyzed, and resolved on TLC plates. Incorporated 32P was visualized by autoradiography, and phosphoamino acid standards (dotted circles) were visualized by ninhydrin staining. Chevrons indicate incomplete BAF hydrolysis products. (B) CNBr cleavage of BAF yields two peptides, P1 and P2. Serines are dotted. The predicted solvent accessibility of each residue, based on the crystal structure of the BAF dimer (Umland et al., 2000) is indicated by black (accessible), gray (intermediate), or white (buried). (C) The 32P-signal after thin layer electrophoresis and chromatography of CNBr-cleaved BAF. (D) The same fragments as in C but separated by chromatography only to maximize subsequent peptide recovery. (E) MALDI-MS spectrum of the peptide eluted from the P1 spot in D. (F) MALDI-MS spectra for P1 (mono- and unphosphorylated) in samples incubated in S-phase (BAF+S) or M-phase (BAF+M) Xenopus extracts.
To compare the sites phosphorylated in M- versus S-phase extracts, we cleaved nitrocellulose-immobilized 32P-BAF with CNBr and analyzed the products by electrophoresis and chromatography on TLC plates. CNBr cleaves BAF after residue 15, yielding two peptides: P1 and P2 (Figure 3B). Peptide P1 (theoretical pI 6.51) is more basic than P2 (theoretical pI 5.38) and significantly less hydrophobic, as predicted by their grand average of hydropathy values of –1.086 (P1) and –0.261 (P2; calculated by ProtParam; Gasteiger et al., 2005). We therefore resolved P1 and P2 by combined electrophoretic and chromatographic separation and detected phosphorylated products by autoradiography (Figure 3C). Both S- and M-phase-phosphorylated BAF samples yielded two major radioactive spots that separated in the chromatographic dimension. By resolving samples in this dimension only, we recovered enough of the lower, less hydrophobic spot (Figure 3D) to determine its mass by MALDI-MS. We detected two peaks of mass 1599.3 and 1679.4 Da, consistent with un- and monophosphorylated P1, respectively (Figure 3E). The identity of these peptides was confirmed by nanoelectrospray MS (our unpublished data). Because there is only one serine in P1 (Figure 3B), this demonstrated BAF was phosphorylated at Ser-4, which is both absolutely conserved (Figure 1A) and solvent accessible in the crystal structure (Figure 3B, gray bar; Umland et al., 2000).
The second 32P-labeled CNBr spot (Figure 3, C and D) could not be positively identified, but logically represents either 1) P2 phosphorylated on serine(s), 2) P1 Ser-4-phosphorylated plus additional modification(s), or 3) uncleaved Ser-4-phosphorylated BAF. Besides Ser-4, human BAF has only two other serines, both in peptide P2 at positions 22 and 74 (Figure 3B, dots), which are less conserved (see Discussion). Attempts to identify potential phosphorylated site(s) in P2 were unsuccessful. We therefore focused further studies on the invariant Ser-4.
The radioactive peptides from S- and M-phase samples migrated identically on TLC plates (Figure 3C), confirming that no additional sites on human BAF were phosphorylated during mitosis. MALDI-MS analysis of the raw (not separated by TLC) CNBr-cleavage products suggested more efficient Ser-4 phosphorylation during mitosis; the 1678-Da peak (P1 + one phosphate) was enriched in M-phase samples relative to the 1598-Da peak (nonphosphorylated P1; Figure 3F). We conclude that BAF is efficiently phosphorylated on Ser-4 during mitosis but that it is also phosphorylated at Ser-4 during interphase.
We used site-directed mutagenesis to change Ser-4 to glutamate (S4E). BAF S4E protein was readily expressed in bacteria and purified as dimers (our unpublished data), indicating proper folding, and migrated slightly above ∼10 kDa in SDS-PAGE (Figure 4A, lanes 2 and 5). The corresponding alanine mutant (S4A) was difficult to recover from bacteria; only a minor fraction purified as dimers. This mutant was therefore included only in later studies (see below) using protein expressed in eukaryotic extracts or living cells. The purified recombinant human wild-type BAF and BAF S4E proteins were incubated in either S- or M-phase Xenopus extracts (Figure 4). Wild-type exogenous BAF was phosphorylated in both S- and M-phase extracts, as expected (Figure 4B, lanes 1 and 4), whereas S4E was not detectably phosphorylated in either S- or M-phase extracts (Figure 4B, lanes 2 and 5). Thus, Ser-4 seemed to be a major site of phosphorylation on BAF (see Discussion).
Figure 4.
Mutations at Ser-4 block phosphorylation of BAF in vitro and in vivo. Purified His-tagged BAF (wild type or S4E) were each incubated in S- or M-phase Xenopus egg extracts in the presence of [32P]ATP. Each whole reaction was then resolved by SDS-PAGE, Coomassie stained (A), and autoradiographed to detect incorporated 32P (B). M, M-phase; Ph inh, phosphatase inhibitors; S, S-phase. (C) Western blots of human kidney (293T) cells that were transiently transfected to overexpress either wild-type (WT) or S4A-mutant His/Xpress-tagged BAF and then separated on 2D gels, blotted and probed with BAF serum 3273. Dotted line indicates pI of unmodified His/Xpress-tagged BAF, which migrated at ∼16 kDa; endogenous BAF (10 kDa) is not shown.
Mutation at Ser-4 Reduces BAF Phosphorylation In Vivo
To determine whether Ser-4 was phosphorylated in vivo, we transiently overexpressed human BAF (S4A mutant or wild type) with N-terminal Xpressand His6-tags in human kidney (293T) cells for 19 h. Cells were then lysed, and proteins were resolved on 2D gels using a nonlinear pI gradient for maximum resolution of BAF spots. The tagged overexpressed BAF migrated at ∼16 kDa (Figure 4C), clearly distinct from endogenous BAF (our unpublished data), and was detected using BAF serum 3273 (Figure 4C). Overexpressed wild-type BAF resolved into three spots (Figure 4C), as seen for endogenous BAF in HeLa cells (Figure 1B). In contrast, BAF S4A resolved as only two spots corresponding to unmodified and monophosphorylated (or monoacetylated) BAF, consistent with loss of one phosphorylation site. This result suggested BAF Ser-4 was phosphorylated in vivo.
Overexpression of Wild-Type BAF or Mutant S4E Causes Emerin to Mislocalize
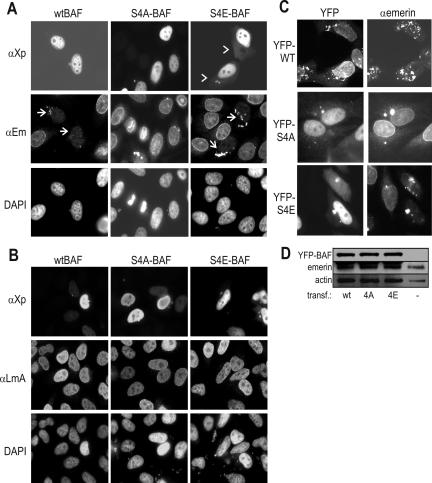
To test the physiological relevance of Ser-4 in human (HeLa) cells, we transiently overexpressed either wild-type human BAF or mutants S4A or S4E, each bearing N-terminal Xpress- and His6-tags. Each mutation was expected to block phosphorylation on Ser-4 in vivo; however S4E had the potential to mimic Ser-4 phosphorylation in vivo. HeLa cells were double labeled by indirect immunofluorescence 19–24 h after transfection using antibodies against the Xpress-tag (Figure 5A, αXp) plus antibodies against endogenous emerin (Figure 5A, αEm) or endogenous A-type lamins (Figure 5B, αLmA). Wild-type BAF and both mutants localized predominantly to the nucleus (Figure 5, A and B, αXp). Interestingly, 72% of cells expressing S4E (n = 350) also had consistently low but detectable mutant BAF in the cytoplasm (Figure 5A, αXp, arrowheads). This result suggested the S4E mutation either mildly inhibited nuclear import of BAF or mildly promoted its cytoplasmic localization, perhaps by mimicking phosphorylation at Ser-4.
Figure 5.
Localization of endogenous emerin and A-type lamins in cells that overexpress wild-type or mutant BAF. (A and B) HeLa cells transiently transfected with cDNAs encoding either wild-type or mutant BAF were fixed and double stained by indirect immunofluorescence 19 h after transfection, using monoclonal antibodies against the N-terminal Xpress-tag (αXp) to visualize transfected BAF, and rabbit antibodies against emerin (A, αEm) or A-type lamins (B, αLmA). DNA was stained with 4,6-diamidino-2-phenylindole (DAPI). Chevrons in (A, top row) indicate cytoplasmic localization of BAF S4E protein. Arrows in (A, middle row) indicate emerin aggregates in cytoplasm. (C) HeLa cells expressing YFP-fused BAF (wild type, S4A, or S4E) stained by indirect immunofluorescence using antibodies against endogenous emerin (αEm). YFP-BAF was localized by YFP-autofluorescence. (D) Cells from C and control untransfected cells (–) were lysed and immunoblotted using antibodies against BAF, emerin, or actin.
In 73% of cells (n = 106) that overexpressed wild-type BAF and 95% of cells that overexpressed S4E-BAF (n = 107), emerin failed to localize at the nuclear envelope (Figure 5A, αEm, arrows); instead, the emerin signal localized as punctate aggregates in the cytoplasm, presumably in association with ER membranes. Loss of the envelope-localized emerin signal was not because of masking of the emerin epitope by overexpressed BAF, because green fluorescent protein (GFP)-tagged emerin also mislocalized when the experiment was repeated in cells that stably expressed GFP-emerin (our unpublished data). In striking contrast to cells overexpressing mutant S4E, emerin localized normally at the nuclear envelope in 85% of cells (n = 123) that overexpressed BAF mutant S4A (Figure 5A, αEm, middle).
To ask whether emerin was unstable or degraded, we transfected HeLa cells with each BAF protein tagged on the N terminus with yellow fluorescent protein (YFP) (∼90% transfection efficiency; our unpublished data). Overexpressed YFP-tagged BAF gave phenotypes similar to Xpress-tagged BAF (emerin mislocalized in cells expressing YFP-fused wild-type or S4E, but not S4A; Figure 5C). YFP-BAF-transfected cells were lysed and immunoblotted using antibodies against emerin, BAF, or actin as a loading control (Figure 5D). Exogenous BAF had no detectable effect on emerin protein levels, relative to actin (Figure 5D). Thus, emerin was mislocalized, not degraded, in cells overexpressing wild-type or S4E BAF.
Why did emerin mislocalize? One possibility was that exogenous wild-type BAF (which can be phosphorylated in vivo) and S4E-BAF (which might mimic phosphorylation) might directly inhibit emerin anchorage at the nuclear envelope. Alternatively, nonphosphorylated BAF might promote emerin localization (e.g., during interphase), and this activity might be lost or misregulated in the presence of excess phosphorylatable (wild-type) or phosphomimetic (S4E) BAF. Emerin depends on A-type lamins to localize at the nuclear envelope, as shown in lmna-null cells where emerin drifts throughout the ER network (Sullivan et al., 1999). A-type lamins localized normally in cells that overexpressed Xpress-tagged wild-type BAF, S4A-BAF, or S4E-BAF (Figure 5B, αLmA). Both models (emerin localization inhibited by phospho-BAF or enhanced by unphosphorylated BAF) are plausible, given that recombinant BAF binds lamin A with moderate (micromolar) affinity in vitro (Holaska et al., 2003), and forms three-way complexes with emerin and lamin A in vitro (Holaska et al., 2003). We therefore hypothesized that Ser-4 phosphorylation of BAF would either 1) inhibit or 2) fail to promote binding between emerin and lamin A in vivo. Both models were consistent with mitotic hyperphosphorylation at Ser-4 potentially contributing to nuclear envelope breakdown at mitosis.
Unmodified BAF Enhances Emerin Binding to Lamin A In Vitro
To test the above-mentioned models, we asked whether recombinant BAF influenced 35S-emerin binding to purified, bacterially expressed prelamin A tails (residues 394–664; residues 384–566 are sufficient to bind emerin in vitro; Sakaki et al., 2001). Recombinant prelamin A tails were resolved by SDS-PAGE, transferred to nitrocellulose, cut into strips, and probed with either 35S-BAF or 35S-emerin (Figure 6A, lanes 3–8). Control strips were either stained with Coomassie (Figure 6A, lane 1, CB) or immunoblotted with antibodies against A-type lamins (Figure 6A, lane 2, LA) to locate the recombinant prelamin A tail; smaller bands are presumed breakdown products. The signals (percentage of input probe that bound lamin A) were then compared. Binding of 35S-labeled BAF mutants S4A and S4E to lamin A was reduced by 43 and 71%, respectively, relative to wild-type 35S-BAF (Figure 6A, lanes 4 and 5; Figure 6B, lanes 1–3). We also probed the lamin-containing strips with 35S-emerin plus or minus equimolar amounts of recombinant nonradioactive (“cold”) BAF (wild-type or mutant S4E). The binding of 35S-emerin was enhanced fourfold by wild-type BAF (Figure 6A, compare lanes 6 and 7; quantified by densitometry in Figure 6B, lanes 4 and 5). In contrast, BAF S4E had no effect on emerin binding to lamin A (Figure 6A, compare lanes 6 and 8, quantified in Figure 6B, lanes 4 and 6), consistent with its weak binding to lamin A (Figure 6A, lane 5; confirmed by the weak immunoblot signal for bound BAF S4E in lane 8, inset). These results are representative of three repeats, and the same results were obtained with blot-immobilized full-length prelamin A (our unpublished data).
To investigate whether BAF mutant S4E “acted” phosphorylated, we compared its binding to nitrocellulose-immobilized prelamin A tails (Figure 6C, lane 4) with that of His6-tagged recombinant wild-type BAF that had been preincubated in M-phase extracts and ATP (to efficiently phosphorylate Ser-4) and then repurified using Ni2+-beads (Figure 6C, lane 3). As controls, we used His6-tagged wild-type BAF not exposed to Xenopus extracts (Figure 6C, lane 2) or Xenopus extracts alone (Figure 6C, lane 5). In all cases, bound BAF was detected using BAF serum 3273 (Figure 6C). Only the unmodified wild-type BAF bound the prelamin A tail efficiently (Figure 6C, lane 2); both the in vitro phosphorylated BAF and the untreated S4E mutant had at least 50% reduced binding, as estimated by densitometry (Figure 6D), normalized with respect to the amount of each input protein. The data shown are representative of three repeats. Similar results were obtained when wild-type and S4E mutant BAF were used to probe nitrocellulose-immobilized emerin (n = 2; our unpublished data). Although not definitive, these experiments suggest that in terms of its binding to lamin A and emerin, BAF S4E mimics the behavior of wild-type BAF phosphorylated by endogenous kinases.
The above-mentioned results suggested that Ser-4-nonphosphorylated BAF enhances emerin binding to lamin A. To test this hypothesis by an independent method, we used a microtiter well binding assay (Figure 6E). Either recombinant emerin or BSA as the negative control was immobilized on microtiter wells and then probed with either 35S-BAF (wild-type or mutant) or 35S-labeled full-length prelamin A, plus or minus unlabeled recombinant BAF (wild-type or S4E, Figure 6E). The 35S-labeled wild-type BAF bound emerin efficiently and specifically (Figure 6E, lanes 1), as expected. Mutant S4E had only background levels of binding to emerin (Figure 6E, lanes 2). 35S-lamin A bound detectably to emerin (lanes 3), and this binding was enhanced ∼10-fold in the presence of equimolar amounts of nonradio-labeled recombinant wild-type BAF but not BAF S4E (Figure 6E, compare lanes 3–5). We concluded that nonphosphorylated BAF enhances emerin binding to lamin A in vitro and hypothesized that phosphorylation at Ser-4 (mimicked by the S4E mutant) inhibits this activity.
DISCUSSION
BAF has many different binding partners (Segura-Totten and Wilson, 2004), and we hypothesized that its binding properties must be regulated to be meaningful. As shown here, naturally existing forms of BAF support this hypothesis. Endogenous forms of BAF from both Xenopus and human cells resolved as multiple spots on 2D gels. The endogenous BAF spots detected by our antibody ranged widely in apparent mass and pI. Thus, in addition to being phosphorylated, endogenous BAF is subject to additional modifications, potentially including bulky moieties (e.g., SUMO and ubiquitin) that remain to be characterized. The slow-migrating forms of BAF were readily detected by 2D gel analysis of Xenopus extracts but not HeLa cells. However, comparable slow-migrating BAF bands were detected on one-dimensional-gels when we immunoblotted whole HeLa cell lysates rather than the supernatant fraction required for 2D gels (Bengtsson, unpublished observations). We therefore hypothesize that slow-migrating forms of BAF also exist in HeLa cells, but they were potentially of lower abundance than in S- or M-phase-enriched Xenopus extracts. Indeed, slow-migrating forms of BAF are biologically relevant because p55Gag, encoded by HIV-1, binds preferentially to an ∼50-kDa-migrating form of BAF in HeLa cells (Mansharamani et al., 2003).
Our in vitro phosphorylation assay and subsequent phosphoamino acid analysis as well as 2D-analysis of wild-type and S4A-BAF overexpressed in cells showed definitively that BAF is phosphorylated on Ser-4 both in vitro and in vivo. Human BAF has three serines: Ser-4, Ser-22, and Ser-74. Ser-4 is both absolutely conserved across species and solvent exposed and was identified here as a major site of BAF phosphorylation in both Xenopus egg extracts and human cells. For CNBr peptide P1, which contains Ser-4, the monophosphorylated form was enriched during M phase compared with S phase. Based on this result and the phosphoamino acid analysis, we conclude that Ser-4 is phosphorylated during interphase, but it is phosphorylated much more efficiently during mitosis. Importantly, Ser-4 is also phosphorylated in vivo: when Ser-4 was switched to Ala, the resulting mutant BAF overexpressed in HeLa cells resolved as two spots on 2D gels, in contrast to three acidically shifted spots in cells that overexpressed wild-type BAF.
Endogenous human BAF resolved as an unmodified spot plus two acidically shifted spots on 2D gels. Thus, in addition to Ser-4, Ser-22 and/or Ser-74 might be phosphorylated in vivo. Ser-22 is solvent exposed but not conserved outside human BAF (Figures 1A and 3B). In contrast, Ser-74 is conserved among vertebrates (Figure 1A), but it is predicted to be solvent inaccessible (Figure 3B; Umland et al., 2000), and its phosphorylation would require a conformational change of the BAF dimer. Indeed, it is formally possible that Ser-4 is the only phosphorylated site in human BAF, because 1) other spots might acid-shift because of acetylation and we have no positive evidence yet for phosphorylation of the P2 peptide, and 2) BAF S4E protein was not detectably phosphorylated by Xenopus extracts (Figure 4B). Further work is needed to determine whether human BAF is phosphorylated at sites other than Ser-4, or acetylated. Endogenous Xenopus BAF resolved into as many as four acidic spots, in addition to unmodified BAF. Xenopus BAF has two additional hypothetically phosphorylatable residues (Ser-2 and Tyr-78; Figure 1A) not conserved in human BAF, and we speculate that phosphorylation at these sites, or acetylation, might explain the total of four acid-shifted spots seen with endogenous Xenopus BAF during S and M phase.
Our findings suggest that Ser-4 phosphorylation is biologically relevant to BAF's interactions with emerin and potentially other LEM-domain proteins in vivo. Overexpression of either wild-type BAF (which can be phosphorylated in vivo) or the phosphomimetic S4E mutant disrupted emerin localization at the nuclear envelope of HeLa cells. At first, this result was puzzling, because purified emerin and lamin A can bind each other directly in vitro (reviewed in Bengtsson and Wilson, 2004; Zastrow et al., 2004). However, both proteins also directly bind BAF (Lee et al., 2001; Holaska et al., 2003). Thus, to explain our in vivo emerin mislocalization results, we propose that nonphosphorylated BAF normally enhances binding between emerin and lamin A. This model is supported by three findings. First, previous biochemical studies showed that emerin, BAF and lamin A can form oligomeric complexes in vitro (Holaska et al., 2003). Second, we show that unmodified BAF enhanced emerin binding to lamin A in vitro, whereas mutant S4E lacked this activity. Third, BAF mutant S4E and wild-type BAF that had been phosphorylated in M-phase extracts both had reduced binding to lamin A in vitro compared with unmodified BAF. The S4E mutant also had reduced in vitro binding to emerin (shown here) and LAP2β as well as DNA (Bengtsson, unpublished observations). Consistent with Ser-4 phosphorylation as a negative regulator of DNA binding, Ser-4 was recently shown to form a water-mediated hydrogen bond with the phosphate backbone of DNA (Bradley et al., 2005). The S4E mutant protein expressed and localized like wild-type BAF in vivo, suggesting no gross defects in protein folding or stability. Furthermore, because the BAF S4E protein disrupted emerin localization in vivo, this protein must retain an activity (e.g., binding to other partner[s]) that allows it to act dominantly. Curiously, overexpression of the S4E mutant BAF caused emerin to concentrate in ER aggregates, in contrast to lmna-null cells where emerin redistributes uniformly throughout the ER (reviewed in Bengtsson and Wilson, 2004). We speculate that the S4E protein retains an unknown activity(ies) (e.g., binding to other partner[s] or competition for a regulator) that somehow promotes (or fails to block) emerin aggregation. Interestingly, one major difference between lmna-null cells and cells that overexpress BAF S4E is that all proteins that are anchored (directly or indirectly) by A-type lamins are released into the ER network in lmna-null cells, whereas only “BAF-dependent” proteins (including emerin) would be released in cells that overexpress BAF S4E. Thus, our results suggest that the BAF S4E mutation mimics at least some aspects of negative regulation at Ser-4, and importantly also suggest that phosphorylation at Ser-4 might not inhibit all activities of BAF. Further analysis of the S4E mutant will be informative.
Our most interesting finding was that phosphorylation at Ser-4 in BAF blocks its ability to promote emerin binding to lamins in vitro, and overexpression of the S4E (phosphomimetic) BAF releases emerin from the nuclear lamina in vivo. Why would cells want to weaken emerin binding to lamin A during interphase? One possibility is that emerin might need to release lamin A to associate with other partners. Supporting this model, emerin purified from HeLa nuclear extracts is found in a variety of distinct native high-mass complexes, several of which lack lamins (Holaska and Wilson, unpublished observations). A second possibility is that emerin might need to release lamins at special times, for example, to facilitate 1) DNA replication of emerin-associated chromatin (Li et al., 1998), 2) changes in the expression status of emerin-associated genes, or 3) gross changes in chromatin position during interphase (Levi et al., 2005). Cells also need to weaken emerin-lamin binding during nuclear envelope breakdown in mitosis, when emerin redistributes into the ER network (Haraguchi et al., 2001). Subsequent dephosphorylation of BAF Ser-4 would allow BAF to serve positive roles in recruiting both emerin and lamins to the reforming nuclear envelope, as shown in C. elegans embryos (Margalit et al., 2005) and human cells (Haraguchi et al., 2001; Dechat et al., 2004). Many of BAF's known binding partners at the nuclear envelope are also mitotically phosphorylated (Ellis et al., 1998; Foisner, 2003; Gajewski et al., 2004; Martins et al., 2003). Thus, our results collectively support models in which phosphorylation and dephosphorylation at Ser-4 regulates BAF activity during both interphase and mitosis. Additional regulatory mechanisms are suggested by the existence of other uncharacterized modification(s) on BAF, which will be important to pursue in the future, along with the identification of the kinase(s) and pathways that regulate BAF.
Supplementary Material
Acknowledgments
We are indebted to Rhonda Stolle for purified BAF proteins, Henrik Molina for peptide sequencing, Brenda Lee for Xenopus egg extracts, and Roberto Diez and Wigbert Bengtsson in the Mass Spectrometry/Proteomics facility for isoelectric focusing. We gratefully acknowledge Wade Gibson for advice and access to equipment for phosphoamino acid and phosphopeptide analysis, Petra Knaus for support in the last months of this project, and Wilson laboratory members for advice and comments on the manuscript. MALDI- and nanoelectrospray-MS were done using equipment in the Mass Spectrometry/Proteomics Facility at Johns Hopkins School of Medicine, which is supported by National Center for Research Resources shared instrumentation Grant 1S10-RR14702, the Johns Hopkins Fund for Medical Discovery and the Institute for Cell Engineering. This work was supported by the Deutsche Forschungsgemeinschaft (to L. B.), National Institutes of Health Grant R01 GM-48646 (to K.L.W.), and the Ray Mills fund (to K.L.W.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–04–0356) on December 21, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Bengtsson, L., and Wilson, K. L. (2004). Multiple and surprising new functions for emerin, a nuclear membrane protein. Curr. Opin. Cell Biol. 16, 73–79. [DOI] [PubMed] [Google Scholar]
- Bradley, C. M., Ronning, D. R., Ghirlando, R., Craigie, R., and Dyda, F. (2005). Structural basis for DNA bridging by barrier-to-autointegration factor. Nat. Struct. Mol. Biol. 12, 935–936. [DOI] [PubMed] [Google Scholar]
- Broers, J. L., Hutchison, C. J., and Ramaekers, F. C. (2004). Laminopathies. J. Pathol. 204, 478–488. [DOI] [PubMed] [Google Scholar]
- Cai, M., Huang, Y., Zheng, R., Wei, S. Q., Ghirlando, R., Lee, M. S., Craigie, R., Gronenborn, A. M., and Clore, G. M. (1998). Solution structure of the cellular factor BAF responsible for protecting retroviral DNA from autointegration. Nat. Struct. Biol. 5, 903–909. [DOI] [PubMed] [Google Scholar]
- Casaday, R. J., Bailey, J. R., Kalb, S. R., Brignole, E. J., Loveland, A. N., Cotter, R. J., and Gibson, W. (2004). Assembly protein precursor (pU.L80.5 homolog) of simian cytomegalovirus is phosphorylated at a glycogen synthase kinase 3 site and its downstream “priming” site: phosphorylation affects interactions of protein with itself and with major capsid protein. J. Virol. 78, 13501–13511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat, T., Gajewski, A., Korbei, B., Gerlich, D., Daigle, N., Haraguchi, T., Furukawa, K., Ellenberg, J., and Foisner, R. (2004). LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J. Cell Sci. 117, 6117–6128. [DOI] [PubMed] [Google Scholar]
- Ellenberg, J., Siggia, E. D., Moreira, J. E., Smith, C. L., Presley, J. F., Worman, H. J., and Lippincott-Schwartz, J. (1997). Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138, 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J. A., Craxton, M., Yates, J. R., and Kendrick-Jones, J. (1998). Aberrant intracellular targeting and cell cycle-dependent phosphorylation of emerin contribute to the Emery-Dreifuss muscular dystrophy phenotype. J. Cell Sci. 111, 781–792. [DOI] [PubMed] [Google Scholar]
- Foisner, R. (2003). Cell cycle dynamics of the nuclear envelope. Scientific-WorldJournal 3, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forne, I., Carrascal, M., Martinez-Lostao, L., Abian, J., Rodriguez-Sanchez, J. L., and Juarez, C. (2003). Identification of the autoantigen HB as the barrier-to-autointegration factor. J. Biol. Chem. 278, 50641–50644. [DOI] [PubMed] [Google Scholar]
- Furukawa, K., Sugiyama, S., Osouda, S., Goto, H., Inagaki, M., Horigome, T., Omata, S., McConnell, M., Fisher, P. A., and Nishida, Y. (2003). Barrier-to-autointegration factor plays crucial roles in cell cycle progression and nuclear organization in Drosophila. J. Cell Sci. 116, 3811–3823. [DOI] [PubMed] [Google Scholar]
- Gajewski, A., Csaszar, E., and Foisner, R. (2004). A phosphorylation cluster in the chromatin-binding region regulates chromosome association of LAP2alpha. J. Biol. Chem. 279, 35813–35821. [DOI] [PubMed] [Google Scholar]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., and Bairoch, A. (2005). Protein identification and analysis tools on the ExPASy server. In: The Proteomics Protocols Handbook, ed. John M. Walker, Totowa, NJ: Humana Press.
- Glotzer, M., Murray, A.W., and Kirschner, M. W. (1991). Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138. [DOI] [PubMed] [Google Scholar]
- Gouet, P., Courcelle, E., Stuart, D. I., and Metoz, F. (1999). ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., Margalit, A., Goldman, R. D., Shumaker, D. K., and Wilson, K. L. (2005). The nuclear lamina comes of age. Nat. Rev. Mol. Cell. Biol. 6, 21–31. [DOI] [PubMed] [Google Scholar]
- Haraguchi, T., Koujin, T., Segura-Totten, M., Lee, K. K., Matsuoka, Y., Yoneda, Y., Wilson, K. L., and Hiraoka, Y. (2001). BAF is required for emerin assembly into the reforming nuclear envelope. J. Cell Sci. 114, 4575–4585. [DOI] [PubMed] [Google Scholar]
- Harris, D., and Engelman, A. (2000). Both the structure and DNA binding function of the barrier-to-autointegration factor contribute to reconstitution of HIV type 1 integration in vitro. J. Biol. Chem. 275, 39671–39677. [DOI] [PubMed] [Google Scholar]
- Higgins, D., Thompson, J., Gibson, T., Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska, J. M., Lee, K. K., Kowalski, A. K., and Wilson, K. L. (2003). Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J. Biol. Chem. 278, 6969–6975. [DOI] [PubMed] [Google Scholar]
- Holmer, L., and Worman, H. J. (2001). Inner nuclear membrane proteins: functions and targeting. Cell Mol. Life Sci. 58, 1741–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. K., Haraguchi, T., Lee, R. S., Koujin, T., Hiraoka, Y., and Wilson, K. L. (2001). Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J. Cell Sci. 114, 4567–4573. [DOI] [PubMed] [Google Scholar]
- Lee, M. S., and Craigie, R. (1998). A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA 95, 1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi, V., Ruan, Q., Plutz, M., Belmont, A. S., and Gratton, E. (2005). Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 89, 4275–4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., Sudlow, G., and Belmont, A. S. (1998). Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J. Cell Biol. 140, 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. W., and Engelman, A. (2003). The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J. Virol. 77, 5030–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka, M. J., and Masui, Y. (1984). Roles of cytosol and cytoplasmic particles in nuclear envelope assembly and sperm pronuclear formation in cell-free preparations from amphibian eggs. J. Cell Biol. 98, 1222–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansharamani, M., Graham, D. R., Monie, D., Lee, K. K., Hildreth, J. E., Siliciano, R. F., and Wilson, K. L. (2003). Barrier-to-autointegration factor BAF binds p55 Gag and matrix and is a host component of human immunodeficiency virus type 1 virions. J. Virol. 77, 13084–13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit, A., Segura-Totten, M., Gruenbaum, Y., and Wilson, K. L. (2005). Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc. Natl. Acad. Sci. USA 102, 3290–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins, S. B., Marstad, A., and Collas, P. (2003). In vitro modulation of the interaction between HA95 and LAP2beta by cAMP signaling. Biochemistry 42, 10456–10461. [DOI] [PubMed] [Google Scholar]
- Montes de Oca, R., Lee, K. K., and Wilson, K. L. (2005). Binding of barrier-to-autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J. Biol. Chem. 280, 42252–42262. [DOI] [PubMed] [Google Scholar]
- Newmeyer, D. D. and Wilson, K. L. (1991) Xenopus eggs for nuclear import and nuclear assembly reactions. Methods Enzymol. 36, 607–634. [DOI] [PubMed] [Google Scholar]
- Sakaki, M., Koike, H., Takahashi, N., Sasagawa, N., Tomioka, S., Arahata, K., and Ishiura, S. (2001). Interaction between emerin and nuclear lamins. J. Biochem. 129, 321–327. [DOI] [PubMed] [Google Scholar]
- Segura-Totten, M., Kowalski, A. K., Craigie, R., and Wilson, K. L. (2002). Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J. Cell Biol. 158, 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Totten, M., and Wilson, K. L. (2004). BAF: roles in chromatin, nuclear structure and retrovirus integration. Trends Cell Biol. 14, 261–266. [DOI] [PubMed] [Google Scholar]
- Shimi, T., Koujin, T., Segura-Totten, M., Wilson, K. L., Haraguchi, T., and Hiraoka, Y. (2004). Dynamic interaction between BAF and emerin revealed by FRAP, FLIP, and FRET analyses in living HeLa cells. J. Struct. Biol. 147, 31–41. [DOI] [PubMed] [Google Scholar]
- Soullam, B., and Worman, H. J. (1995). Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J. Cell Biol. 130, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, T., Escalante-Alcalde, D., Bhatt, H., Anver, M., Bhat, N., Nagashima, K., Stewart, C. L., and Burke, B. (1999). Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Yang, H., and Craigie, R. (2004). LAP2alpha and BAF collaborate to organize the Moloney murine leukemia virus preintegration complex. EMBO J. 23, 4670–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umland, T. C., Wei, S. Q., Craigie, R., and Davies, D. R. (2000). Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry 39, 9130–9138. [DOI] [PubMed] [Google Scholar]
- Wang, X., et al. (2002). Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J. Biol. Chem. 277, 43288–43300. [DOI] [PubMed] [Google Scholar]
- Zastrow, M. S., Vlcek, S., and Wilson, K. L. (2004). Proteins that bind A-type lamins: integrating isolated clues. J. Cell Sci. 117, 979–987. [DOI] [PubMed] [Google Scholar]
- Zastrow, M. S., Flaherty, D. B., Benian, G. M., and Wilson, K. L. (2006). Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J. Cell Sci. (in press). [DOI] [PubMed]
- Zheng, R., Ghirlando, R., Lee, M. S., Mizuuchi, K., Krause, M., and Craigie, R. (2000). Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl. Acad. Sci. USA 97, 8997–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.