Abstract
Recent studies of meiotic recombination in the budding yeast and the model plant Arabidopsis thaliana indicate that meiotic crossovers (COs) occur through two genetic pathways: the interference-sensitive pathway and the interference-insensitive pathway. However, few genes have been identified in either pathway. Here, we describe the identification of the PARTING DANCERS (PTD) gene, as a gene with an elevated expression level in meiocytes. Analysis of two independently generated transferred DNA insertional lines in PTD showed that the mutants had reduced fertility. Further cytological analysis of male meiosis in the ptd mutants revealed defects in meiosis, including reduced formation of chiasmata, the cytological appearance of COs. The residual chiasmata in the mutants were distributed randomly, indicating that the ptd mutants are defective for CO formation in the interference-sensitive pathway. In addition, transmission electron microscopic analysis of the mutants detected no obvious abnormality of synaptonemal complexes and apparently normal late recombination nodules at the pachytene stage, suggesting that the mutant's defects in bivalent formation were postsynaptic. Comparison to other genes with limited sequence similarity raises the possibility that PTD may present a previously unknown function conserved in divergent eukaryotic organisms.
INTRODUCTION
During meiotic prophase I, homologous chromosomes (homologues) interact through pairing, synapsis, and recombination, which are required for the proper segregation of homologues during subsequent stages of meiosis. Such interactions begin with the pairing of two homologues, followed by the polymerization of the synaptonemal complex (synapsis) between the homologues (Page and Hawley, 2004). Under a transmission electron microscope (TEM), the synaptonemal complex (SC) can be observed as a structure with three parallel filaments: two outer lateral elements and one central element (Page and Hawley, 2004). In addition, electron-dense ovoid structures (late recombination nodule, or LN) can be seen associated with the SC (Page and Hawley, 2004). LNs correlate with sites of meiotic crossovers (COs) (Page and Hawley, 2004). During late stages of the meiotic prophase I, SCs disassemble (desynapsis). By this time, homologue recombination has been completed, and the resulting COs can be seen cytologically as chiasmata. The attached homologues known as bivalents align at the metaphase plate, ensuring the correct segregation of homologues during anaphase I. The COs, along with cohesion between sister chromatids, are important for the maintenance of homologue association until the transition from metaphase I to anaphase I is complete (Page and Hawley, 2003).
Although the molecular process of recombination is not yet fully understood, studies in yeast and other organisms have led to the double-strand break repair (DSBR) model (Szostak et al., 1983). According the DSBR model, recombination is initiated by a double-strand break (DSB) generated by the SPO11 protein, originally discovered in the budding yeast Saccharomyces cerevisiae (Esposito and Esposito, 1969; Keeney, 2001; Lichten, 2001). Homologues of SPO11 have been identified in a wide range of other organisms, including other fungi, plants, and animals, suggesting that the mechanism of recombination initiation is highly conserved (Dernburg et al., 1998; McKim and Hayashi-Hagihara, 1998; Keeney et al., 1999; Romanienko and Camerini-Otero, 1999; Celerin et al., 2000; Grelon et al., 2001). After DSBs are formed, they are processed by the MRE11–RAD50–XRS2 protein complex to form 3′OH single-stranded ends (Symington, 2002). RAD51, a homologue to the bacterial recombinase RecA, and its meiotic paralog DMC1 then are localized to the 3′ single-stranded-tails to facilitate single strand invasion of intact duplexes (Petukhova et al., 2000; Sung et al., 2003).
According to the current understanding, the early interaction between the invading strand and intact duplex is transient and can have three different fates (Bishop and Zickler, 2004). The interactions can be processed to form noncrossovers (NCOs), or they can be processed through one of two genetically separate pathways for CO formation (Bishop and Zickler, 2004; Börner et al., 2004). One of the CO pathways is dependent on the two eukaryotic homologues of the bacterial MutS protein, MSH4 and MSH5, whereas the other CO pathway is dependent on the heterodimeric endonuclease with the MUS81 and MMS4/EME1 subunits (de los Santos et al., 2003; Hollingsworth and Brill, 2004). In many organisms, the presence of a CO inhibits the formation of a second CO nearby between the same pair of homologues, a phenomenon known as interference (Hillers, 2004). The MSH4–MSH5-dependent pathway forms COs that show interference, whereas the MUS81–MMS4/EME1-dependent COs do not show interference (de los Santos et al., 2003; Bishop and Zickler, 2004; Hollingsworth and Brill, 2004). In addition, the numbers of COs formed through these two pathways vary in different organisms (Hollingsworth and Brill, 2004). For example, in budding yeast and Arabidopsis the MSH4–MSH5-dependent pathway accounts for the majority of COs, whereas the MUS81–MMS4/EME1 pathway accounts for most, if not all, of the COs generated in fission yeast (Copenhaver et al., 2002; de los Santos et al., 2003; Bishop and Zickler, 2004; Hollingsworth and Brill, 2004).
There is considerable information on the interference-sensitive pathway. In yeast, the initial interaction between the invading stand and the intact duplex proceeds to form a stable intermediate, known as the single strand invasion intermediate (SEI) (Bishop and Zickler, 2004). Further DNA synthesis and ligation, through the second end capture (SEC) intermediates, leads to the formation of the recombinant intermediate, known as the double Holliday junction (dHJ). In principle, resolution of dHJ can give rise to either COs or NCOs, but recent results in S. cerevisiae indicate that dHJs tend to resolve only to form COs (Bishop and Zickler, 2004). The MSH4 and MSH5 proteins form a heterodimer and seem to function in stabilizing and preserving CO intermediates (Pochart et al., 1997; Bocker et al., 1999; Colaiacovo et al., 2003; Snowden et al., 2004). The msh4 and msh5 mutants in S. cerevisiae show a reduction in COs without affecting NCOs (Ross-Macdonald and Roeder, 1994; Hollingsworth et al., 1995). More specifically, the budding yeast msh5 mutant shows defects in the conversion of DSBs into SEI and in the formation of dHJ (Börner et al., 2004). Similarly, a mutant in the Arabidopsis homologue of MSH4, atmsh4, shows a dramatic reduction of interference-sensitive COs, whereas the formation of interference-insensitive COs occurs at the expected levels (Higgins et al., 2004), indicating that the interference-sensitive pathway in Arabidiopsis also requires MSH4. MSH4 and MSH5 homologues in mammals are also important for meiosis (de Vries et al., 1999; Kneitz et al., 2000), although analysis of CO formation in mutant mice have not yet been described. In addition, statistical modeling suggests that humans may also have two CO pathways with only one forming interference-sensitive COs (Housworth and Stahl, 2003).
In addition, the S. cerevisiae mutant mer3 and the Arabidopsis mutant (atmer3/rck) in the MER3 homologue are impaired in the formations of COs via the interference-sensitive pathway (Nakagawa and Ogawa, 1999; Chen et al., 2005; Mercier et al., 2005). Furthermore, a recent study has shown that MER3, which is a DNA helicase, is involved in DNA heteroduplex extension in the 3′–5′ direction in SEC intermediates (Mazina et al., 2004). Therefore, the interference-sensitive CO pathway seems to be conserved in yeast, plants, and probably mammals, involving at least the MSH4/MSH5 heterodimer and likely the MER3 DNA helicase.
However, relatively little is known about the protein(s) involved in dHJ resolution in eukaryotes. During dHJ resolution, DNA strands need to be cleaved and ligated in a specific orientation to form COs (Heyer et al., 2003). In prokaryotes, two classes of proteins, represented by the Escherichia coli RuvC resolvase and the archeal Hjc nuclease serve as Holliday junction resolvases (Kvaratskhelia et al., 2001; Lilley and White, 2001; Heyer et al., 2003; Parker and White, 2005). In Drosophila melanogaster, mei-9 and ercc1 mutations lead to a reduction in CO formation (Baker and Carpenter, 1972; Carpenter, 1979, 1982; Sekelsky et al., 1995; Radford et al., 2005). MEI-9 encodes a homologue of the mammalian XPF structure-specific DNA endonuclease that is critical for DNA excision repair (Sekelsky et al., 1995; Sijbers et al., 1996; de Laat et al., 1998; Yildiz et al., 2004). Furthermore, mei-9 meiocytes exhibit unaltered distribution, frequency, and morphology of LNs (Carpenter, 1979), suggesting that MEI-9 acts at a stage after LN formation, possibly at the dHJ resolution step. The Drosophila ERCC1 is a homologue of mammalian ERCC1, which forms a complex with XPF (Sijbers et al., 1996; de Laat et al., 1998; Sekelsky et al., 2000). In addition, recent studies revealed that RAD51 paralogs might also be involved at dHJ resolution (Yokoyama et al., 2003, 2004; Liu et al., 2004). However, it is expected that more genes need to be identified to fully understand the molecular process of homologous recombination during meiosis.
With the rapid advancements of Arabidopsis molecular and cell biological studies because of readily available genetic and cytological tools, analysis in Arabidopsis has led to novel findings in the meiotic process (Ma, 2005). For example, the isolation and analysis of the solo dancers (sds) mutant indicates that a novel cyclin protein, SDS, is required for normal recombination and bivalent formation (Azumi et al., 2002; Wang et al., 2004). In addition, analysis of the Arabidopsis mutant meiotic prophase aminopeptidase1 (mpa1) indicates that a puromycin-sensitive aminopeptidase is involved in meiotic recombination (Sanchez-Moran et al., 2004). Furthermore, plant-specific meiotic genes, such as SWI1, have been identified that have no known functional homologues in other organisms (Mercier et al., 2001, 2003). Reverse genetic studies have also demonstrated the importance of homologues of known meiotic recombination genes in Arabidopsis meiosis (Grelon et al., 2001; Bleuyard and White, 2004; Li et al., 2004, 2005; Mercier et al., 2005). These studies indicate that meiosis uses both conserved and divergent mechanisms.
Here, we report the identification of an Arabidopsis gene, PARTING DANCERS (PTD), which is involved specifically in meiotic prophase I. We present data from cytological and quantitative cytogenetic studies showing that ptd meiocytes exhibit reduced levels of crossover formation. The distribution of remaining crossovers suggests that the PTD gene is important for the promotion of interference-sensitive crossovers. The PTD gene seems to function at a postsynaptic step, indicating a role in late stages of crossover pathway. Furthermore, PTD encodes a protein with limited sequence similarity to ERCC1 proteins and other proteins, suggesting possible mechanisms for PTD function.
MATERIALS AND METHODS
Plant and Growth Conditions
Both the wild-type and the ptd mutant plants carrying a transferred DNA (T-DNA) insertion, SALK_127477 (ptd-1) and SAIL_567_D09 (ptd-2), were of the Columbia ecotype unless otherwise indicated. All plants were grown under long-day conditions (16-h day and 8-h night) in a growth room at an average of 18–22°C or in a greenhouse.
Phenotypic Analysis
Plants were photographed using Sony digital camera DSC-F828 (Sony, Tokyo, Japan). Dissected tetrads were stained with 0.01% toluidine blue. Pollen grains were stained with Alexander's solution (Alexander, 1969) to test for pollen viability. The images were obtained using a Nikon dissecting microscope (Nikon, Tokyo, Japan) with an Optronics digital camera (Optronics, Goleta, CA). Wild-type and mutant inflorescences were collected, and chromosome spreads were prepared as described by Ross et al. (1996) and stained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Images of chromosome spreads were obtained using a Nikon Eclipse E800 microscope (Nikon) and a Hamamatsu digital camera (Hamamatsu Photonics, Hamamatsu, Japan). The images were edited using Photoshop 7.0 (Adobe Systems, Mountain View, CA). Number of chiasmata was counted from diakinesis images in a way similar to that described previously (Sanchez Moran et al., 2001). Transmission electron microscopy was carried out as described previously (Li et al., 2004). All statistical analyses were carried out as described previously (Zar, 1974).
PCR and Sequencing of T-DNA Insertion Sites
All primer sequences used here are listed in Table 1. Genomic DNAs were extracted from rosette leaves using 2× CTAB buffer [2% (wt/vol) cetyltrimethyl-ammonium bromide, 1.4 M NaCl, 100 mM Tris-HCl, pH 8.0, and 20 mM EDTA]. PCR was used to identify plants that were homozygous and heterozygous for one of the two T-DNA insertions, SALK_127477 (ptd-1) and SAIL_567_D09 (ptd-2). For the SALK_127447 line, the wild-type allele was amplified using primers oMC1607 and oMC1608, and the mutant allele was amplified using oMC1607 and oMC1863, a primer specific for the T-DNA left border. For the SAIL_567_D09 line, the wild-type allele was amplified using oMC1911 and oMC1912, and the mutant allele was amplified using a gene-specific primer oMC1911 and the T-DNA left border primer oMC2009. The products from oMC1607/oMC1863 or oMC1911/oMC2009 were purified and sequenced to confirm their identity.
Table 1.
Sequence information for oligonucleotides used
| Primer name | Sequence |
|---|---|
| oMC571 | TCCCAGAATCGCTAAGATTGCC |
| oMC572 | CCTTTCCCTTAAGCTCTG |
| oMC1607 | TCAAAACACATATCGCCTA |
| oMC1608 | GTAATGTGGAGCGTATGGA |
| oMC1735 | ATCATCAAGTTTCGTCACCTCTC |
| oMC1736 | TCTGTGTTTGCTAAAATGTCTTC |
| oMC1863 | TAGACGGTTTTTCGCCCTTTGACG |
| oMC1996 | TCCGCCAAGATTTTCGAATTTCT |
| oMC1998 | AAACAAGTGTGGTCATCTATTGTG |
| oMC1887 | ACAAAAGCAATAGAAACACCTCCAC |
| oMC1911 | GACAAATCTGATGAAGATGTTTG |
| oMC1912 | TGCTTATCTGTGTTGTCGGAAAT |
| oMC2009 | TAGCATCTGAATTTCATAACCAATCTCGATACAC |
Reverse Transcription (RT)-PCR
RT–PCR was performed using ∼1 μg of total RNA from wild-type (Landsberg erecta ecotype) roots, stems, leaves, stage 12 flowers, and young inflorescences. The RNAs were treated with RNase-free DNase I, followed by the inactivation of the DNase I. The RNAs were then used to synthesize the cDNA using Superscript II (Invitrogen, Carlsbad, CA) and oligo(dT) according to manufacturer's instructions. These cDNAs were later used as template for RT-PCR using PTD-specific primers oMC1735 and oMC1887. As a control the same cDNAs were used to amplify the APT1 gene (oMC571 and oMC572) encoding the adenine phosphoribosyltransferase (Moffatt et al., 1994). PCR was carried out under standard conditions using 10 pmol of each primer and 30 cycles (for either APT1 or PTD) of 94°C for 30 s, 55°C for 45 s, and 72°C for 45 s.
To estimate the RNA expression level in mutant plants, RNA was extracted from inflorescences (containing buds younger than stage 11 flowers) of mutant plants homozygous for one of the T-DNA insertions and wild-type plants as a control. cDNA was synthesized as described above, and RT-PCR was performed with primer combinations of oMC1996 and oMC1998, oMC1735 and oMC1736, or oMC1735 and oMC1887. Positions of these PCR primers are shown in Figure 2. To confirm the PTD coding region, a cDNA fragment was obtained by RT-PCR using RNA from wild-type young inflorescence with oMC1996 and oMC1998 and was cloned into the pGEM-T vector (Promega, Madison, WI) (plasmid pMC2974) and sequenced.
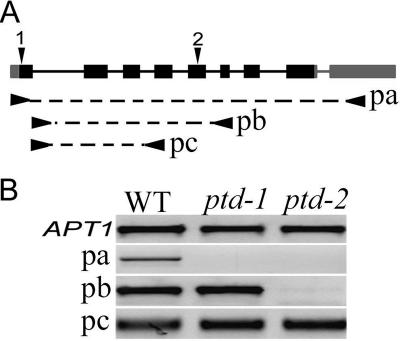
Figure 2.
The PTD gene structure and mRNA expression in the wild type and the two mutants. (A) A diagram showing PTD (At1g12790) locus with the exon/intron organization of PTD. Solid boxes indicate exons, including full coding (black) and untranslated regions (gray). The position of the T-DNA insertion sites of the ptd-1 and ptd-2 alleles are indicated by black arrowhead 1 and 2. Underneath the structure, arrow-dash bars-arrows indicate the approximate positions of different primers used for RT-PCR: pa, oMC1996/oMC1998; pb, oMC1735/oMC1736; and pc, oMC1735/oMC1887. (B) Results for RT-PCR with primer pairs of pa, pb, and pc. The APT1 control is indicated.
RNA In Situ Hybridization
Nonradioactive RNA in situ hybridization was performed essentially as described previously (Xu et al., 2002). Wild-type inflorescences were fixed in the formol-acetic-alcohol fixative for at least 2 h at room temperature and then dehydrated, cleared, and embedded in paraffin (Fisher, Hampton, NH). Sections (10 μm in thickness) were made using a Shandon Finesse (Thermo Electron, Pittsburgh, PA) microtome and mounted onto slides. All slides were dewaxed with Histoclear, treated with protein kinase for 30 min, and then dehydrated and baked at 42°C for at least 2 h. A PTD cDNA fragment was amplified with gene-specific primers oMC1735 and oMC1736 and cloned into the pGEM-T vector (Promega) to yield plasmid pMC2913. The PTD antisense and sense RNAs were labeled with digoxigenin (DIG) through in vitro transcription of linearized pMC2913, using enzymes SpeI and NcoI, respectively. The sections were hybridized with the RNA probes, and hybridization signals were detected using anti-DIG antibodies conjugated with alkaline phosphatase and nitro blue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate. Images were taken using a SPOT II RT camera (Diagnostic Instruments, Sterling Heights, MI) and were edited using Photoshop 7.0 (Adobe Systems).
Sequence Analysis
Unless mentioned otherwise all homologous protein sequences were obtained using National Center for Biotechnology Information HOMOLOGENE function (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?DB=homologene). All sequence alignment was done using the MUSCLE sequence alignment program and further modified using GeneDoc (http://www.psc.edu/biomed/genedoc) (Nicholas et al., 1997; Edgar, 2004). Phylogenetic analyses of protein sequences were performed using MEGA version 3.0 software (Kumar et al., 2004) with default settings except for the following parameters: 1) for parsimony analysis, 500 bootstrap replicates, seed = 33,000; and 2) for Neighbor-Joining analysis, 10,000 bootstrap replicates, seed = 63,695 and pairwise deletion.
RESULTS
PTD Is Expressed in Male Meiocytes and Other Cells in Vegetative and Floral Organs
To identify genes that potentially function in male meiosis, microarray expression profiles of Arabidopsis meiotic-stage anthers were generated (Zhang, unpublished data). One gene, At1g12790, which was originally annotated as being weakly similar to bacterial DNA ligases with nonspecific DNA binding, was expressed in stage 4–6 anthers (Zhang, unpublished data). According to our published microarray data, At1g12790 expression was also detected in all organs tested, including young inflorescences (Zhang et al., 2005). This locus was named PTD for the meiotic defects of T-DNA insertion mutants in this gene (see below).
To verify the microarray results, we performed semiquantitative RT-PCR with PTD-specific primers and found that PTD was expressed at similar levels in both vegetative and reproductive organs, including anthers near the stage of meiosis (Figure 1A). To determine the PTD spatial expression pattern in reproductive tissues, nonradioactive in situ hybridization with inflorescence sections was carried out using an antisense PTD probe. Signals could be observed in early stages of floral meristem (Figure 1B). In addition, in the mature flower, the PTD signals were restricted to the reproductive organs stamens and carpels (Figure 1C). In the stamen, strong signals were observed in the male meiocytes and in tapetal cells, which surround the male meiocytes (Figure 1C). Moreover, signals were detected in mature pollen grains (Figure 1E). In the carpel, the PTD signals were present in developing ovules (Figure 1D) and in developing embryos (our unpublished data). These results indicate that PTD is expressed in the meiocytes and other dividing cells. In addition, we detected PTD expression in vegetative organs including leaves, stem, and roots.
Figure 1.
The expression patterns of PTD. (A) RT-PCR analyses of the PTD mRNA expression in wild-type root, stem, cauline leaf, silique, stage-12 flower, inflorescence, and anthers at stages 4–6. (B) PTD was expressed in the wild-type meristem and young flower buds. (C) A possible stage 5/6 (flower stage 9) wild-type anther showing PTD signal in the male meiocytes and tapetal layer. (D) A cross-section of a flower at stages 8–9 with developing ovules and anthers. (E) Anthers with mature pollen grains. Strong signals are associated some pollen grains. im, inflorescence meristem; fm, floral meristem; fb, a stage 4 flower bud; pmc, pollen mother cells; and pg, pollen grains. Bar, 20 μm.
Identification and Analysis of T-DNA Insertions in PTD
To characterize the function of PTD, we obtained two lines with a T-DNA insertion in the gene from the SIGnAL and SAIL collections (Sessions et al., 2002; Alonso et al., 2003), SALK_127447 and SAIL_567_D09 and were confirmed to have insertions in PTD. These lines showed reduced fertility (see below). The mutant alleles in these two lines were named ptd-1 (SALK_127447) and ptd-2 (SAIL_567_D09). Plants that showed the reduced fertility were genotyped using PCR and were homozygous for the T-DNA insertion (hereafter, plants that are homozygous for either one of the T-DNA insertion will be referred as ptd mutant unless mentioned otherwise).
Plants heterozygous for the T-DNA insertion were phenotypically normal, indicating that the mutations are recessive. The progenies of heterozygous plants for either mutant alleles segregated for mutant to normal phenotypes in a 1:3 ratio as expected for single Mendelian recessive mutations (ptd-1, 187: 637 mutant:normal; ptd-2, 37:114 mutant:normal). To test for linkage between the T-DNA insertions and mutant phenotypes, the genotypes of these segregating populations were determined using gene-specific and T-DNA specific primers (see Materials and Methods). For ptd-1, 60 mutant plants were all ptd-1/ptd-1; among 48 normal plants tested, 12 were PTD/PTD and 36 were PTD/ptd-1. For ptd-2, 35 mutant plants were ptd-2/ptd-2, whereas 12 of the 22 normal plants were PTD/PTD with the remaining 10 being PTD/ptd-2. These results indicate that the ptd-1 and ptd-2 insertions were tightly linked to the mutant phenotype. Furthermore, ptd-1/ptd-2 heterozygous plants were generated by crosses between PTD/ptd-1 and PTD/ptd-2 plants and showed similar phenotypes to those of ptd-1 and ptd-2 plants (including the meiotic chromosomal morphology discussed below). Because the ptd-1 and ptd-2 alleles were identified from independently generated T-DNA collections, these results strongly support the conclusion that the insertions in the PTD gene caused the mutant phenotypes.
Using gene-specific and T-DNA left border primers, we confirmed that the SALK_127447 insertion was within the first exon, 40 base pairs downstream of the start codon, whereas the SAIL_567_D09 insertion was in the fifth exon, 430 bp downstream of start codon and in both T-DNA insertions, the left border was oriented toward the 3′ end (Figure 2A). RT-PCR of the entire coding region indicated that the PTD transcripts were detectable only in the wild-type young inflorescence but were not in inflorescences of either ptd mutants even after 40 PCR cycles. However, RT-PCR with primers that amplify a region downstream of the SALK_127447 insertion but upstream of the SAIL_567_D09 insertion showed comparable levels of PTD mRNA in wild-type, ptd-1 and ptd-2 young inflorescences (Figure 2B). These results suggest that, although T-DNA insertions might have disrupted the synthesis of the full-length PTD mRNA, there can be truncated versions of mRNA in both mutant alleles.
The ptd Mutants Have Reduced Fertility and Are Defective in Meiosis
We found that the vegetative growth in the ptd mutant plants was normal under standard growth conditions. In particular, the number of rosette leaves, cauline leaves, and biomass were similar to those of wild-type plants grown under the same growth conditions (Figure 3, A and B). In addition, the mutant and wild-type flowers had similar sepals and petals. However, the mutant plants (Figure 3B) produced shorter siliques with fewer seeds than the wild type (Figure 3A). The average number of seeds was approximately three per seedpod in ptd-1 (n = 42) and ptd-2 (n = 83), compared with 42 seeds per seedpod (n = 29) in the wild type.
Figure 3.
Phenotypes of the wild type and the ptd-1 mutant. (A) A wild-type plant showing normal vegetative growth and normal siliques. (B) A mutant plant showing similar shape of the wild type with no detectable defects on vegetative growing but with shorter siliques. (C) A portion of a wild-type flower. (D) A portion of mutant flower with floral organs similar to the wild type except a fewer pollen grains on anthers. (E) Tetrads from a wild-type anther with four microspores. (F) Tetrads from the mutant anther with varied numbers of microspores. (G) A wild-type anther with functional pollen grains stained in red. (H) A mutant anther with fewer functional pollen grains stained with red and nonfunctional pollen grains stained with dark green. Bars, 1 cm (A and B), 200 μm (C and D), 10 μm (E and F), and 50 μm (G and H).
Consistent with the reduced fertility, ptd anthers showed reduced pollen production compared with wild-type anthers (Figure 3, C and D). Furthermore, the anthers of mutant plants stained with Alexander's staining showed a large number of dead pollen grains, whereas wild-type anthers stained with the same stain showed little, if any, dead pollen (Figure 3, G and H). To ascertain the cause of the defect in pollen production in the ptd mutant, we stained the tetrads from both wild type and the mutant with toluidine blue. Completion of meiosis in wild-type plants produces tetrads with four spores, whereas the mutant plants produced polyads with two to eight spores, indicating a meiotic defect and possibly chromosome missegregation during male meiosis (Figure 3, E and F). An examination of the surface of the stigma in ptd flowers did not reveal any abnormality. However, the number of seeds produced by the ptd-1 flowers whose stigma were pollinated by wild-type pollen grains was not significantly different from the number of seeds produced by self-pollinated mutant plants (24 crosses, average seed set two seeds per silique). Therefore, the ptd mutations seem to be defective in female reproduction as well.
ptd Meiocytes Contained a Reduced Number of Interference-sensitive Chiasmata
To obtain further insight into the meiosis of the ptd mutant, male meiosis in the mutant and wild type was analyzed by staining meiotic chromosome spreads with DAPI (Figure 4). The wild-type cells showed the characteristic chromosome behaviors of meiosis as described previously (Figure 4, A–D and M–P) (Ross et al., 1996). In the wild type, at leptotene, chromosomes started to condense and can be seen as thin treads (Figure 4A). During zygotene, chromosomes continue to condense, began to synapse, and could be seen to concentrate to one side of the nucleus (Figure 4B). During pachytene, completely synapsed chromosomes look like thick threads (Figure 4C). At diplotene, chromosomes have desynapsed, and homologues remain attached to each other only at the chiasmata (Figure 4D). During diakinesis, chromosomes contract length wise to produce highly condensed bivalents (Figure 4M).
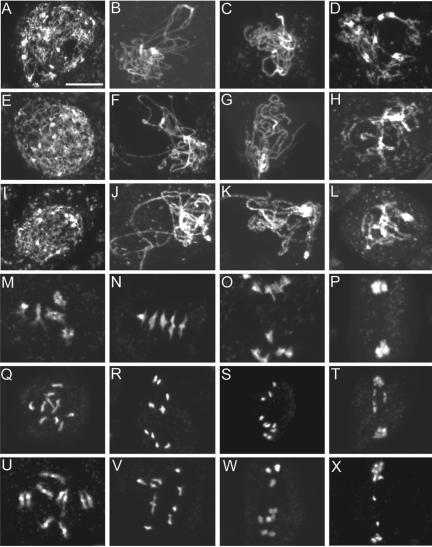
Figure 4.
Male meiosis I in the wild type and the ptd mutants. Shown are images of DAPI-stained chromosomes. (A–D) Wild-type prophase I at leptotene, zygotene, pachytene, and diplotene, respectively. (E–H) ptd-1 and (I-L) ptd-2 prophase I at similar stages showing no obvious difference to the wild type. (M–P) Wild-type diakinesis of prophase I, early metaphase I, anaphase I, and telophase I, respectively; note that M shows five brightly stained entities, representing five attached pairs of condensed homologues. (N) Chromosomes align at the equator. (O) Homologues are separating. (P) Homologues separate and form two clusters. (Q–T) Similar stages of meiosis from ptd-1. (U-X) ptd-2. Note that in Q, 10 stained bodies can be seen, indicating that the condensed homologues formed univalents. (R and V) Chromosomes condense further, similarly to normal metaphase I chromosomes, but there were no five perfectly aligned bivalents at the equator and contained univalents as in N. Chromosomes are starting to separate similar to (S and W), suggesting that they might be pulled by the spindle as normal homologues are at anaphase I. In T and X, however, the chromosome distribution seems abnormal. Bar, 10 μm.
In both mutant alleles, under the light microscope, no significant abnormality of the chromosome behaviors from leptotene to pachytene was observed (Figure 4, E–G and I–K). It was also difficult to see any defect at diplotene (Figure 4, H and L). However, at diakinesis or prometaphase I, instead of five bivalents as in the wild type, the mutant contained fewer than five bivalents with the remainder being univalents (Figure 4, Q and U). Among 139 ptd-1 mutant meiocytes at diakinesis, 128 cells (∼92%) had less than five bivalents with the following distributions: four bivalents (21 cells; 15%), three bivalents (50 cells; 36%), two bivalents (32 cells; 23%), one bivalent (16 cells; 12%) and zero bivalents (9 cells; 6%). Among 88 cells counted for ptd-2, 87 cells (99%) contained fewer than five bivalents with the following distribution: four bivalents (6 cells; 7%), three bivalents (15 cells; 17%), two bivalents (31 cells; 35%), one bivalent (25 cells; 28%), and zero bivalents (10 cell; 11%). Thus, both ptd-1 (average bivalents per cell 2.7) and ptd-2 (average bivalents per cell 1.8) showed a reduced number of bivalents compared with the wild type (average bivalents per cell 5). As mentioned in Azumi et al. (2002), the wild-type chromosome interactions and condensation at prophase I has a resemblance to a highly choreographed duet dance in which homologues “dance” in pairs. Because the ptd mutant showed the ability to form normal pachytene chromosomes, but then separate at diakinesis, we named the mutants “parting dancers,” meaning that the chromosomes start the dance as pairs but separate subsequently.
Furthermore, comparison of wild-type and mutant meiocytes at late stages of meiosis I revealed additional defects, which may be a result of the defective prophase I (Figure 4, N–P, R–T, and V–X). In the wild type at metaphase I, the five bivalents are aligned at the equatorial plane of the cell (Figure 4N). At anaphase I, homologous chromosomes separated and migrated toward the two opposite poles (Figure 4O). In contrast, mutant cells with fewer than five bivalents were observed at metaphase I, and cells with five perfectly aligned bivalents were not observed (Figure 4, R and V). In ptd-1, among 49 cells at metaphase I, 47 cells (∼95%) had fewer than five bivalents with the following distributions: four bivalents (6 cells; 12.2%), three bivalents (12 cells; 24.5%), two bivalents (14 cells; 28.6%), one bivalent (10 cells; 20.4%), and zero bivalents (5 cells; 10.2%). Similarly in ptd-2, 47 cells of 47 cells (100%) at metaphase I had fewer than five bivalents with the following distributions: four bivalents (1 cell; 2%), three bivalents (10 cells; 21%), two bivalents (11 cells; 23%), one bivalent (22 cells; 47%), and zero bivalents (3 cells; 6%). Some of the univalents were far from the equator, suggesting that they were not aligned because of the lack of opposing forces from the spindle. The abnormal metaphase I was followed by uneven chromosomal distribution at anaphase I (Figure 4, S and W). In addition, chromosome bridges were observed frequently in the mutant during anaphase I, suggesting possible incomplete recombination.
During wild-type meiosis II, two groups of chromosomes could be observed with a characteristic organelle band in between (Supplemental Figure S1, A–E). At metaphase II the groups of chromosomes (5 in each) assembled at the two equators perpendicular to the organelle band (Supplemental Figure S1, B). The sister chromatids separated and moved to the opposite poles at anaphase II (Supplemental Figure S1, C). Then, the newly formed chromosomes decondensed to form four haploid nuclei (Supplemental Figure S1, D and E). In the mutant meiocytes, chromosome distribution was abnormal, most likely because of the defective prophase I (Supplemental Figure S1, F–O).
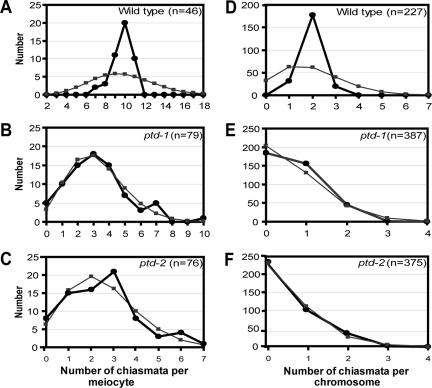
The bivalent formation is highly reduced in both ptd mutant alleles. Because the majority of the chiasmata in Arabidopsis are interference sensitive, it is possible that the ptd mutations block the formation of crossovers through the interference-sensitive CO pathway. To test this hypothesis, we counted the distribution of residual chiasmata present in ptd-1 and ptd-2 and compared them with that of the wild type (Figure 5). The chiasmata distribution among wild-type cells deviated significantly from the Poisson prediction (Figure 5A; χ2(18) = 57.90, p < 0.001), whereas the distribution of the residual chiasmata among cells in both mutant alleles (Figure 5, B and C; ptd-1: χ2(8) = 4.16, p > 0.7; ptd-2: χ2(5) = 4.22, p > 0.5) did not deviate significantly from the predicted Poisson distribution. In addition, distribution of chiasmata per chromosome in the wild type deviated significantly from the predicted distribution (Figure 5D; wild type χ2(8) = 308, p < 0.001). In contrast, in the ptd mutants this distribution was dramatically different from the distribution in the wild type (Figure 5, E and F; ptd-1: χ2(4) = 15.472, p < 0.05; ptd-2: χ2(4) = 7.895, p > 0.05). This suggested that the majority of the residual chiasmata present in the mutants were randomly distributed and were not sensitive to interference.
Figure 5.
Chiasma distribution in wild type, ptd-1 and ptd-2. (A–C) Chiasma distribution per meiocytes. (D–F) Chiasma distribution per chromosome. Black lines and closed circles indicate observed distributions, whereas gray line and closed squares show predicted Poisson distribution. A and D indicate that the observed distributions of the chiasmata deviate from predicted distributions in the cells and on the chromosome of the wild type, respectively. B, C, E, and F show the observed distributions of the chiasmata do not deviate significantly from the predicted Poisson distributions in mutant cells and on mutant chromosomes.
Formation of Synaptonemal Complexes and Late Recombination Nodules in ptd
To investigate synapsis in the mutant, we performed TEM analysis on wild-type, ptd-1 and ptd-2 meiocytes (Figure 6, A–O). These TEM images revealed no detectable morphological abnormalities in SCs of mutant cells, with properly developed central elements and dimensions that are identical to the wild-type SC (compare Figure 6, E and F, with D and Figure 6, K and L, with J). Pachytene nuclei with fully synapsed chromosomes were observed in both mutant meiocytes. However, delay in formation of SCs in some meiocytes of the mutant was observed. We used the morphology of meiocytes and surrounding tapetal cells to determine the ptd-1 and ptd-2 meiocyte stages that corresponded to early or late pachytene in the wild type. All axial elements were assembled into SCs in the wild type at early pachytene (Figure 6A). In ptd-1, most axial elements were synapsed, but a few unsynapsed axial elements were also present in the “early pachytene” nuclei (Figure 6B), whereas only short SC stretches were detected in ptd-2 (Figure 6C). At this stage, most axial elements remained unsynapsed in ptd-2, resembling the zygotene stage. In addition, early nodules (ENs) were observed on the central element of SC in both mutant alleles (Figure 6, D–F). During zygotene, 34 ENs were found after examining 44 wild-type serial sections and 22 early nodules from 37 ptd-2 serial sections. The difference was not statistically significant (χ2(1) = 3.1316, p > 0.05). LNs were also observed on the central elements of SC in ptd-1 and ptd-2 as meiosis progressed (Figure 6, G and J, H and K, and I and L). At mid-pachytene, there were 16 LNs on 62 wild-type serial sections and 12 LNs on 67 ptd serial sections (not significantly different: χ2(1) = 1.1826, p > 0.25). At late pachytene, we found nine LNs on 45 wild-type sections and eight LNs on 45 ptd-2 sections (not significantly different: χ2(1) = 0.071, p > 0.75). Furthermore, we did not observe any precocious desynaptic abnormality in the mutant at pachytene. Synaptonemal complexes surrounded with condensed chromatin were regularly seen in late pachyteneearly diplotene nuclei in both mutant alleles (compare Figure 6, N and O, with M).
Figure 6.
Transmission electron micrographs of wild type (A, D, G, J, and M), the ptd-1 (B, E, H, K, and N), and the ptd-2 (C, F, I, L, and O) meiocytes (white arrowheads point to the synaptonemal complexes, black arrows point to unsynapsed elements, and white arrows point to the recombination nodules). (A) Fully synapsed chromosomes in wild type at early pachytene. Almost normal level of synaptonemal complexes in ptd-1 (B) and reduced level of synaptonemal complexes in ptd-2 at the same stage (C). Apparently normal level of early recombination nodules in the ptd-1 and ptd-2 mutants compared with wild type (A and D, B and E, and C and F). At mid-pachytene in the wild type (G) and ptd-1 (H), morphology of synaptonemal complex seems identical, but in ptd-2 development of synaptonemal complexes was delayed (I) (white arrowheads point to synaptonemal complexes). Late recombination nodule on mid-pachytene SC in wild type (J, arrow), in ptd-1 (K, arrow), and ptd-2 (L, arrows) at the same stage. Normal level of synaptonemal complexes in both mutant alleles (N, ptd-1; O, ptd-2) compared with wild type (M) at late pachytene–early diplotene stages. D, E, F, J, K, and L are magnified images from A, B, C, G, H, and I, respectively. Bars, 1000 nm (A–C, G–I, and M–O) and 250 nm (D–F and J–L).
PTD is a Founding Member of Plant Gene Family with Limited Sequence Similarity to ERCC1
The PTD gene is predicted to be 2119 base pairs and is located on chromosome 1. PTD contains nine exons and eight introns, as supported by the sequence of both our cDNA clone and a reported cDNA (Haas et al., 2002). A 753-base pair PTD open reading frame is predicted to encode a 250-amino acid protein with a pI of 9.17 and molecular mass of ∼28 kDa. Searches of public databases using BLASTp and PHI-BLASTp revealed no significant overall homology to any other Arabidopsis protein but showed a significant sequence similarity to sequences from rice genome that seem to encode a PTD homologue (NCBI accession nos. AAT37994 and AAT44166). This putative homologue, named OsPTD, shared 63% identity and 78% similarity over a 223-aa region with PTD. In addition, there were at least two expressed sequence tag (EST) clones for this rice sequence, indicating that OsPTD is expressed (EST, CR288441 and AK109323).
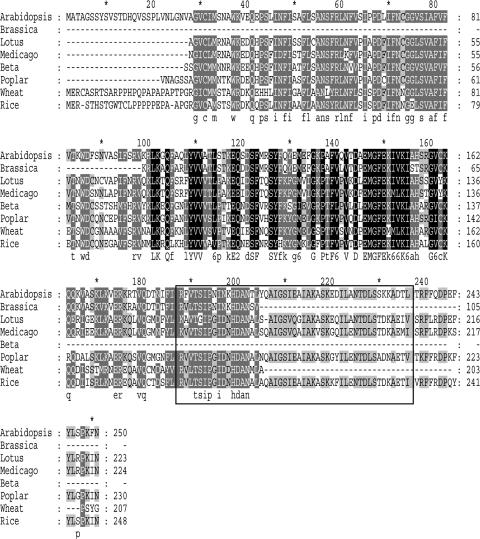
In addition, searches using tBLASTn against National Center for Biotechnology Information nonredundant database for Lotus corniculatus (In clone, LjT44D21, TM0127b from 64782 to 66205 bp) and Medicago truncatula (In clone, mth2-36b7 from 101727 to 100223) and using JGI (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html) for Populus trichocarpa (sequence, LG_I from 30981347 to 30983435) revealed similarity to several genomic sequences (BLAST expected value 2e–19 and 4e–18 for L. corniculatus and M. truncatula, respectively). These genomic sequences were manually annotated using three reading frames generated by Gene Runner software version 3.05 (Hastings Software, Hastings-on Hudson, NY) using PTD and OsPTD amino acid sequences as precursors. In addition, searches against EST databases using tBLASTn yielded several truncated cDNA sequences. One of these is a cDNA from wheat meiotic floret cDNA library that encodes a protein with 51% identity and 67% similarity to PTD over a 217-aa region. Another is a cDNA from developing roots of Beta vulgaris that encodes a protein with 65% identity and 79% similarity to PTD over a 135-aa region. A third sequence is from Brassica whose predicted protein product has 85% identity and 92% similarity to PTD over a 105-aa region. Therefore, PTD is the founding member of a novel gene family in plants. An alignment of the PTD amino acid sequence with representative protein sequences is shown in Figure 7.
Figure 7.
Sequence alignment of PTD homologues in plants. Arabidopsis, A. thaliana; Brassica, Brassica oleracea (partial EST); Lotus, Lotus corniculatus; Medicago, Medicago truncatula; Beta, B. vulgaris (partial EST); Poplar, Populus trichocarpa; Wheat, Triticum aestivum (partial EST); and Rice, Oriza sativa. Square encloses the putative one pseudo- and one classic HhH motif.
Furthermore, the C-terminal region of the PTD protein shares low levels of sequence similarity to several other proteins, including ERCC1 (and its homologues RAD10 and SWI10 from S. cerevisiae and Schizosaccharomyces pombe, respectively), the UvrC subunit of the ABC exonucleases in bacteria (∼24% identities and ∼45% similarities in an ∼80-aa region) and a bacterial NAD-dependent DNA ligase (36% identity and 58% similarity in a 58-aa region). To obtain clues about the evolutionary relationship between PTD and ERCC1 and XPF, members of the XPF superfamily, we generated a phylogenetic tree using representative sequences from the PTD, ERCC1, and XPF families (protein alignments for construction of the tree is provided in Supplemental Figure S3). This tree showed that putative PTD, ERCC1, and XPF homologues formed three distinct clades, with the ERCC1 and XPF clades represented by members from all major eukaryotic kingdoms and the PTD clade containing only plant sequences (Figure 8B). In addition, this tree suggests that ERCC1 and PTD are more closely related than they are to XPF.
Figure 8.
Protein sequence comparison of PTD, ERCC1, and XPF homologues. (A) Schematic representation of protein structure of XPF superfamily members XPF and ERCC1 and PTD. XPF contains an N-terminal helicase domain (helicase), an endonuclease domain (endo), and C-terminal DNA binding domain represented by two HhH motifs. The catalytic domains of XPF share a highly conserved signature motif, GDXnERKX3D, that is important for endonuclease activity (Heyer et al., 2003; Nishino et al., 2003). ERCC1 contains putative endonuclease domain and DNA binding domain with double HhH motifs, but it lacks functionally important residues in its putative catalytic domain (Heyer et al., 2003; Tsodikov et al., 2005). The PTD proteins possess DNA binding domain similar to ERCC1 but lacks catalytic residues important for endonuclease activity. (B) Maximum parsimony tree for PTD, ERCC1, and XPF homologues in different organisms. Bootstrap (base pairs) values in parentheses indicate base pairs for a tree generated using Neighbor-Joining method for the same sequence alignment. Protein sequences used are as follows: for PTD, A. thaliana (At PTD), B. oleracea (Bo PTD) (partial EST), L. corniculatus (Lc PTD), M. truncatula (Mt PTD), P. trichocarpa (Pt PTD), and Oriza sativa (Os PTD); for ERCC1: Homo sapiens (Hs ERCC1), Rattus norvegicus (Rn ERCC1), D. melanogaster (Dm ERCC1), Anopheles gambiae (Ag ERCC1), S. pombe (Sp SWI10), Neurospora crassa (Nc ERCC1), A. thaliana (At ERCC1), Oryza sativa (Os ERCC1); and Lilium longiflorum (Ll ERCC1); and for XPF, H. sapiens (Hs XPF), R. norvegicus (Rn XPF), D. melanogaster (Dm XPF), An. gambiae (Ag XPF), S. pombe (Sp XPF), N. crassa (Nc XPF), M. grisea (Mg XPF), and A. thaliana (At XPF).
To gain further insight into the possible function of the PTD protein, its sequence was used to search public databases to identify similar protein domains. Searches in the Protein Families database of alignments and hidden Markov models (Bateman et al., 2004) did not identify any known protein domains. In addition, searches in the Simple Modular Architecture Research Tool (SMART) (Letunic et al., 2004) database indicated that PTD contains a region (190–234 aa) similar to the Rad51 N-terminal domain; this region in PTD possibly represents one pseudo- and one classic helix-hairpin-helix (HhH) motif. The 190–234-aa region of PTD showed 64–73% aa sequence identities and 77–84% similarities to putative PTD homologues from rice, P. trichocarpa, L. corniculatus, and M. truncatula. In addition, searches against the National Center for Biotechnology Information conserved domain database revealed that a portion of the PTD protein is similar to a domain in the MUS81 protein (37.8%) (Marchler-Bauer et al., 2005). However, the low expected value (2e–05) and the lack of a functional motif that is characteristic of members of the MUS81 endonuclease family suggest that PTD may not contain a true MUS81 domain. To investigate whether there was any putative subcellular target sequence in PTD, we searched the TargetP database (Emanuelsson et al., 2000) and found that PTD did not have any predicted organelle targeting sequence.
DISCUSSION
PTD Is Required for Normal CO Formation and Affects the Interference-sensitive Pathway
Our analysis of two independent ptd T-DNA mutants indicates that they are defective in meiotic CO formation. The ptd meiocytes have dramatically reduced numbers of chiasmata, which seem to be distributed randomly among cells. We also determined the distribution of chiasmata per chromosome, again with apparently random distribution, in contrast to the interference-sensitive distribution in the wild type. The distribution per chromosome was obtained without knowing the specific chromosome identity because the five Arabidopsis chromosomes are hardly distinguishable in DAPI-stained images. Nevertheless, because individual Arabidopsis chromosomes do not seem to have a preference for interference-sensitive or interference-insensitive crossovers (Copenhaver et al., 2002; Higgins et al., 2004; Lam et al., 2005), the observed single-chromosome chiasma distributions revealed a real difference between the wild type and the ptd mutants. Therefore, the results strongly support the idea that PTD is important for the interference-sensitive CO pathway, which requires the AtMSH4 and RCK/AtMER3 genes in Arabidopsis (Higgins et al., 2004; Chen et al., 2005; Mercier et al., 2005). In the yeast msh5 and Arabidopsis atmsh4 mutants, only 10–20% crossovers remain (Börner et al., 2004; Higgins et al., 2004); however, in ptd mutants, the residual chiasmata represent ∼30% of the wild-type number per cell (wild type, 9.7 chiasmata per cell; ptd-1, 3.2 chiasmata per cell; and ptd-2, 2.5 chiasmata per cell). A similar number of residual chiasmata were observed in the Arabidopsis rck mutant (32% of the wild-type number) (Chen et al., 2005).
The difference in the number of residual chiasmata can be explained by several possible reasons. First, because both alleles produce truncated mRNAs that might encode truncated proteins, the ptd alleles might be leaky. In the slightly weaker ptd-1, the insertion is in the first exon and the truncated mRNA can potentially encode most of the PTD protein including the C-terminal conserved domain. In ptd-2, the insertion is in the fifth exon, leading to a shorter truncated mRNA that would code for a protein lacking the conserved domain. Therefore, ptd-1 is more likely to have partial function than ptd-2. In addition, it is possible that the CO pathway bifurcates downstream of MSH4/MSH5 into two branches, only one of which requires PTD. The idea of a bifurcated pathway is also supported by recent studies of the yeast MLH1 gene (homologue of the bacterial MutL), which seems to function downstream of MSH4-MSH5 and may be necessary for only a subset of the MSH4-MSH5-dependent COs (Argueso et al., 2004). By analogy, the Arabidopsis MSH4-dependent COs may be divided into PTD-dependent and PTD-independent COs.
PTD Is Not Crucial for Synaptonemal Complex Formation
Recombination and synapsis are closely associated processes; for example, several Arabidopsis meiotic recombination mutants also show defects in synapsis (Grelon et al., 2001; Li et al., 2004; Li et al., 2005). In addition, SC formation might initiate at the sites of axial associations, which in yeast may mark future CO sites (Sym et al., 1993; Rockmill et al., 1995). Yeast mutations in genes encoding synaptonemal initiation complex (SIC) proteins (Fung et al., 2004; Page and Hawley, 2004), are defective in formation of both SC and MSH4-MSH5-dependent CO intermediates (Börner et al., 2004). Similarly, the Arabidopsis atmsh4 and rck/atmer3 mutants exhibit defects in SC formation in addition to reduced CO formation (Higgins et al., 2004; Chen et al., 2005; Mercier et al., 2005). In contrast, the presence of morphologically normal SCs in both ptd mutants strongly suggests that PTD functions in the recombination pathway after synapsis has approached completion. Nevertheless, a delay in SC formation in ptd-2 suggests PTD may have a nonessential role in promoting SC formation.
Another difference between the ptd mutants and other mutants defective in CO formation, such as rck (Chen et al., 2005) or atrad51 (Li et al., 2004), is that the ptd mutants had a nearly normal number of late recombination nodules. Recombination nodules are categorized into two types: ENs and LNs. The timing of their appearance and number suggest that EN correspond to the sites of early events where homologue pairing occurs, whereas LNs are associated with the sites of COs (Page and Hawley, 2004; Anderson and Stack, 2005). In addition, there is a close morphological resemblance between LN and SIC and thus LN may be composed of the same SIC proteins, at least in yeast (Page and Hawley, 2004; Anderson and Stack, 2005). The formation of nearly normal numbers of LNs in pachytene meiocytes of the ptd mutants suggests that PTD may not be part, nor required for the formation, of the LN complex in pachytene nuclei, unlike AtMSH4 and AtMER3/RCK.
Sequence Similarity of PTD and ERCC1 Suggests a Role for PTD in the Resolution of Meiotic dHJs
The conservation of PTD in both monocots and eudicots indicates that the PTD gene family had originated before the divergence of these two major angiosperm groups. Our findings suggest that PTD function may be conserved in flowering plants, supporting a role for PTD in plant meiosis. Whether PTD function is also conserved in other eukaryotes, including fungi and animals, is less certain. Nevertheless, an ∼80-aa PTD C-terminal region is ∼24% identical and ∼45% similar to several DNA repair proteins, including ERCC1 (Hoeijmakers, 2001; Sancar et al., 2004). It is possible that in nonplant organisms, ERCC1 and related proteins fulfill the meiotic function that in plants requires PTD, as supported by the observed CO reduction in Drosophila mei-9 (affecting an XPF homologue) and ercc1 mutants (Baker and Carpenter, 1972; Sekelsky et al., 1995; Radford et al., 2005). Conversely, the fact that PTD is also expressed widely in many nonmeiotic cells, somewhat similar to a plant ERCC1 gene (Xu et al., 1998), suggests that it may also have a general function, such as DNA repair, as does the Arabidopsis ERCC1 gene (Hefner et al., 2003; Dubest et al., 2004).
ERCC1 forms a complex with XPF, and this complex performs a single-strand 5′ incision in DNA nucleotide excision repair and participates in cleavage of in vitro structures that resemble recombinational intermediates (Habraken et al., 1994; de Laat et al., 1998; Adair et al., 2000; Prakash and Prakash, 2000; Sargent et al., 2000; Hoeijmakers, 2001; Sancar et al., 2004). ERCC1 and XPF are structure-specific endonucleases and are members of the XPF super-family (Heyer et al., 2003; Yang, 2003). In XPF but not ERCC1, a conserved signature motif is important for its endonuclease activity (Enzlin and Scharer, 2002; Nishino et al., 2003; Newman et al., 2005). Like ERCC1, PTD lacks the XPF signature motif and phylogenetic analysis suggests that PTD is more similar to ERCC1 than to XPF (Figure 8, A and B). Therefore, PTD may act like ERCC1 as a noncatalytic subunit in a complex with another endonuclease to perform a meiotic function (Nishino et al., 2003; Newman et al., 2005). The ptd phenotypes are consistent with the hypothesis that PTD acts downstream of AtMSH4 and RCK/AtMER3, possibly by cutting recombination intermediates, such as dHJ, to complete the process of CO formation. However, PTD may not interact with AtRAD1 (AtXPF) during meiosis because meiotic defects were not detected in the atrad1 mutant (Liu et al., 2000; Dubest et al., 2002). Although understanding the mechanism of PTD action needs future studies, the ptd phenotypes strongly support its role in CO formation.
Supplementary Material
Acknowledgments
We thank to A. Surcel, W. Li, W. Pawlowski, and anonymous reviewers for helpful comments; G. Ning, R. Haldeman, and M. Hazen for assistance in electron microscopy; and A. Omeis, J. Wang, and C. L. Hendrix Hord for plant care. We also thank The Ohio State University Arabidopsis Stock Center for providing the SALK and SAIL lines. A.J.W. was partially supported by the Intercollege Graduate Program in Plant Physiology and Department of Biology, The Pennsylvania State University. This work was conducted using material generated with support from the National Science Foundation under Grant 0215923 and was supported by National Institutes of Health Grant R01 GM-63871-01 and National Science Foundation Grant MCB-0092075 (to H. M.). H. M. gratefully acknowledges the generous support of the John Simon Guggenheim Memorial Foundation.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–09–0902) on January 4, 2006.
Abbreviations used: CO, crossover; dHJ, double Holliday junction; DSB, double-strand break; LN, late recombination nodule; NCO, noncrossover; RT, reverse-transcription; T-DNA, transferred DNA; TEM, transmission electron microscope.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adair, G. M., Rolig, R. L., Moore-Faver, D., Zabelshansky, M., Wilson, J. H., and Nairn, R. S. (2000). Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 19, 5552–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, M. P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122. [DOI] [PubMed] [Google Scholar]
- Alonso, J. M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Anderson, L. K., and Stack, S. M. (2005). Recombination nodules in plants. Cytogenet. Genome Res. 109, 198–204. [DOI] [PubMed] [Google Scholar]
- Argueso, J. L., Wanat, J., Gemici, Z., and Alani, E. (2004). Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168, 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azumi, Y., Liu, D., Zhao, D., Li, W., Wang, G., Hu, Y., and Ma, H. (2002). Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J. 21, 3081–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, B. S., and Carpenter, A. T. (1972). Genetic analysis of sex chromosomal meiotic mutants in Drosophilia melanogaster. Genetics 71, 255–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., et al. (2004). The Pfam protein families database. Nucleic Acids Res. 32, D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, D. K., and Zickler, D. (2004). Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117, 9–15. [DOI] [PubMed] [Google Scholar]
- Bleuyard, J. Y., and White, C. I. (2004). The Arabidopsis homologue of Xrcc3plays an essential role in meiosis. EMBO J. 23, 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocker, T., et al. (1999). hMSH 5, a human MutS homologue that forms a novel heterodimer with hMSH4 and is expressed during spermatogenesis. Cancer Res. 59, 816–822. [PubMed] [Google Scholar]
- Börner, G. V., Kleckner, N., and Hunter, N. (2004). Crossover/noncrossover differentiation, synaptonemal complex formation and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45. [DOI] [PubMed] [Google Scholar]
- Carpenter, A. T. (1979). Recombination nodules and synaptonemal complex in recombination-defective females of Drosophila melanogaster. Chromosoma 75, 259–292. [DOI] [PubMed] [Google Scholar]
- Carpenter, A. T. (1982). Mismatch repair, gene conversion, and crossing-over in two recombination-defective mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 79, 5961–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celerin, M., Merino, S. T., Stone, J. E., Menzie, A. M., and Zolan, M. E. (2000). Multiple roles of SPO11 in meiotic chromosome behavior. EMBO J. 19, 2739–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Zhang, W., Timofejeva, L., Gerardin, Y., and Ma, H. (2005). The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. Plant J. 43, 321–334. [DOI] [PubMed] [Google Scholar]
- Colaiacovo, M. P., MacQueen, A. J., Martinez-Perez, E., McDonald, K., Adamo, A., La Volpe, A., and Villeneuve, A. M. (2003). Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but not for proper completion of recombination. Dev. Cell 5, 463–474. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G. P., Housworth, E. A., and Stahl, F. W. (2002). Crossover interference in Arabidopsis. Genetics 160, 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat, W. L., Appeldoorn, E., Jaspers, N. G., and Hoeijmakers, J. H. (1998). DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 273, 7835–7842. [DOI] [PubMed] [Google Scholar]
- de los Santos, T., Hunter, N., Lee, C., Larkin, B., Loidl, J., and Hollingsworth, N. M. (2003). The Mus81/Mms81 endonuclease acts independently of double Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg, A. F., McDonald, K., Moulder, G., Barstead, R., Dresser, M., and Villeneuve, A. M. (1998). Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapses. Cell 94, 387–398. [DOI] [PubMed] [Google Scholar]
- de Vries, S. S., Baart, E. B., Dekker, M., Siezen, A., de Rooij, D. G., de Boer, P., and te Riele, H. (1999). Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13, 523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubest, S., Gallego, M. E., and White, C. I. (2002). Role of the AtRad1p endonuclease in homologous recombination in plants. EMBO Rep. 3, 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubest, S., Gallego, M. E., and White, C. I. (2004). Roles of the AtERCC1 protein in recombination. Plant J. 39, 334–342. [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and Heijne, G. V. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Enzlin, J. H., and Scharer, O. D. (2002). The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 21, 2045–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito, M. S., and Esposito, R. E. (1969). The genetic control of sporulation in Saccharomyces. I. The isolation of temperature-sensitive sporulation-deficient mutants. Genetics 61, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, J. C., Rockmill, B., Odell, M., and Roeder, G. S. (2004). Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116, 795–802. [DOI] [PubMed] [Google Scholar]
- Grelon, M., Vezon, D., Gendrot, G., and and Pelletier, G. (2001). AtSPO11–1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B. J., Volfovsky, N., Town, C. D., Troukhan, M., Alexandrov, N., Feldmann, K. A., Flavell, R. B., White, O., and Salzberg, S. L. (2002). Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol. 3, RESEARCH0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habraken, Y., Sung, P., Prakash, L., and Prakash, S. (1994). Holliday junction cleavage by yeast Rad1 protein. Nature 371, 531–534. [DOI] [PubMed] [Google Scholar]
- Hefner, E., Preuss, S. B., and Britt, A. B. (2003). Arabidopsis mutants sensitive to gamma radiation include the homologue of the human repair gene ERCC1. J. Exp. Bot. 54, 669–680. [DOI] [PubMed] [Google Scholar]
- Heyer, W. D., Ehmsen, K. T., and Solinger, J. A. (2003). Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends Biochem. Sci. 28, 548–557. [DOI] [PubMed] [Google Scholar]
- Higgins, J. D., Armstrong, S. J., Franklin, F. C., and Jones, G. H. (2004). The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18, 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers, K. J. (2004). Crossover interference. Curr. Biol. 14, R1036–R1037. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers, J. H. (2001). Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374. [DOI] [PubMed] [Google Scholar]
- Hollingsworth, N. M., and Brill, S. J. (2004). The mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18, 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, N. M., Ponte, L., and Halsey, C. (1995). MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9, 1728–1739. [DOI] [PubMed] [Google Scholar]
- Housworth, E. A., and Stahl, F. W. (2003). Crossover interference in humans. Am. J. Hum. Genet. 73, 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S. (2001). Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52, 1–53. [DOI] [PubMed] [Google Scholar]
- Keeney, S., Baudat, F., Angeles, M., Zhou, Z. H., Copeland, N. G., Jenkins, N. A., Manova, K., and Jasin, M. (1999). A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics 61, 170–182. [DOI] [PubMed] [Google Scholar]
- Kneitz, B., Cohen, P. E., Avdievich, E., Zhu, L., Kane, M. F., Hou, H., Jr., Kolodner, R. D., Kucherlapati, R., Pollard, J. W., and Edelmann, W. (2000). MutS. homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14, 1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA 3, Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5, 150–163. [DOI] [PubMed] [Google Scholar]
- Kvaratskhelia, M., Wardleworth, B. N., and White, M. F. (2001). Multiple Holliday junction resolving enzyme activities in the Crenarchaeota and Euryarchaeota. FEBS Lett. 491, 243–246. [DOI] [PubMed] [Google Scholar]
- Lam, S. Y., Horn, S. R., Radford, S. J., Housworth, E. A., Stahl, F. W., and Copenhaver, G. P. (2005). Crossover interference on nucleolus organizing region-bearing chromosomes in Arabidopsis. Genetics 170, 807–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., Ponting, C. P., and Bork, P. (2004). SMART 4. 0, towards genomic data integration. Nucleic Acids Res. 32, D142–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Chen, C., Markmann-Mulisch, U., Timofejeva, L., Schmelzer, E., Ma, H., and Reiss, B. (2004). The Arabidopsis AtRAD51gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 101, 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Yang, X., Lin, Z., Timofejeva, L., Xiao, R., Makaroff, C. A., and Ma, H. (2005). The AtRAD51Cgene is required for normal meiotic chromosome synapsis and double-stranded break repair in Arabidopsis. Plant Physiol. 138, 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten, M. (2001). Meiotic recombination: breaking the genome to save it. Curr. Biol. 11, R253–R256. [DOI] [PubMed] [Google Scholar]
- Lilley, D. M., and White, M. F. (2001). The junction-resolving enzymes. Nat. Rev. Mol. Cell. Biol. 2, 433–443. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Masson, J. Y., Shah, R., O'Regan, P., and West, S. C. (2004). RAD51C is required for Holliday junction processing in mammalian cells. Science 303, 243–246. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Hossain, G. S., Islas-Osuna, M. A., Mitchell, D. L., and Mount, D. W. (2000). Repair of UV damage in plants by nucleotide excision repair: Arabidopsis UVH1 DNA repair gene is a homolog of Saccharomyces cerevisiae Rad1. Plant J. 21, 519–528. [DOI] [PubMed] [Google Scholar]
- Ma, H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., et al. (2005). CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33, D192–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina, O. M., Mazin, A. V., Nakagawa, T., Kolodner, R. D., and Kowalczykowski, S. C. (2004). Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell 117, 47–56. [DOI] [PubMed] [Google Scholar]
- McKim, K. S., and Hayashi-Hagihara, A. (1998). mei-W68 in Drosophila melanogaster encodes a SPO11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 12, 2932–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier, R., Armstrong, S. J., Horlow, C., Jackson, N. P., Makaroff, C. A., Vezon, D., Pelletier, G., Jones, G. H., and Franklin, F. C. (2003). The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130, 3309–3318. [DOI] [PubMed] [Google Scholar]
- Mercier, R., et al. (2005). Two meiotic crossover classes cohabit in Arabidopsis: one is dependent on MER3, whereas the other one is not. Curr. Biol. 15, 692–701. [DOI] [PubMed] [Google Scholar]
- Mercier, R., Vezon, D., Bullier, E., Motamayor, J. C., Sellier, A., Lefèvre, F., Pelletier, G., and Horlow, C. (2001). Switch1 (Swi1): a novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev. 15, 1859–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt, B. A., McWhinnie, E. A., Agarwal, S. K., and Schaff, D. A. (1994). The adenine phosphoribosyl transferase-encoding gene of Arabidopsis thaliana. Gene 143, 211–216. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., and Ogawa, H. (1999). The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J. 18, 5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, M., Murray-Rust, J., Lally, J., Rudolf, J., Fadden, A., Knowles, P. P., White, M. F., and McDonald, N. Q. (2005). Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition. EMBO J. 24, 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas, K. B., Nicholas, H. B., Jr., and Deerfield, D.W.I. (1997). GeneDoc: analysis and visualization of genetic variation. EMBNEW.NEWS 4, 14. [Google Scholar]
- Nishino, T., Komori, K., Ishino, Y., and Morikawa, K. (2003). X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease: similarity between its endonuclease domain and restriction enzymes. Structure 11, 445–457. [DOI] [PubMed] [Google Scholar]
- Page, S. L., and Hawley, R. S. (2003). Chromosome choreography: the meiotic ballet. Science 301, 785–789. [DOI] [PubMed] [Google Scholar]
- Page, S. L., and Hawley, R. S. (2004). The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20, 525–558. [DOI] [PubMed] [Google Scholar]
- Parker, J. L., and White, M. F. (2005). The endonuclease Hje catalyses rapid, multiple turnover resolution of Holliday junctions. J. Mol. Biol. 350, 1–6. [DOI] [PubMed] [Google Scholar]
- Petukhova, G., Sung, P., and Klein, H. (2000). Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev. 14, 2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochart, P., Woltering, D., and Hollingsworth, N. M. (1997). Conserved properties between functionally distinct MutS homologs in yeast. J. Biol. Chem. 272, 30345–30349. [DOI] [PubMed] [Google Scholar]
- Prakash, S., and Prakash, L. (2000). Nucleotide excision repair in yeast. Mutat. Res. 451, 13–24. [DOI] [PubMed] [Google Scholar]
- Radford, S. J., Goley, E., Baxter, K., McMahan, S., and Sekelsky, J. (2005). Drosophila ERCC1 is required for a subset of MEI-9-dependent meiotic crossovers. Genetics 170, 1737–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill, B., Sym, M., Scherthan, H., and Roeder, G. S. (1995). Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 9, 2684–2695. [DOI] [PubMed] [Google Scholar]
- Romanienko, P. J., and Camerini-Otero, R. D. (1999). Cloning, characterization, and localization of mouse and human SPO11. Genomics 61, 156–169. [DOI] [PubMed] [Google Scholar]
- Ross, K. J., Fransz, P., and Jones, G. H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4, 507–516. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald, P., and Roeder, G. S. (1994). Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79, 1069–1080. [DOI] [PubMed] [Google Scholar]
- Sancar, L. A., Lindsey-Boltz, K., Unsal-Kacmaz, K., and Linn, S. (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage check-points. Annu. Rev. Biochem. 73, 39–85. [DOI] [PubMed] [Google Scholar]
- Sanchez Moran, E., Armstrong, S. J., Santos, J. L., Franklin, F. C., and Jones, G. H. (2001). Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 9, 121–128. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran, E., Jones, G. H., Franklin, F. C., and Santos, J. L. (2004). A puromycin-sensitive aminopeptidase is essential for meiosis in Arabidopsis thaliana. Plant Cell 11, 2895–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent, R. G., Meservy, J. L., Perkins, B. D., Kilburn, A. E., Intody, Z., Adair, G. M., Nairn, R. S., and Wilson, J. H. (2000). Role of the nucleotide excision repair gene ERCC1 in formation of recombination-dependent rearrangements in mammalian cells. Nucleic Acids Res. 28, 3771–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky, J. J., Hollis, K. J., Eimerl, A. I., Burtis, K. C., and Hawley, R. S. (2000). Nucleotide excision repair endonuclease genes in Drosophila melanogaster. Mutat. Res. 459, 219–228. [DOI] [PubMed] [Google Scholar]
- Sekelsky, J. J., McKim, K. S., Chin, G. M., and Hawley, R. S. (1995). The Drosophila meiotic recombination gene mei-9 encodes a homologue of the yeast excision repair protein Rad1. Genetics 141, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijbers, A. M., et al. (1996). Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86, 811–822. [DOI] [PubMed] [Google Scholar]
- Snowden, T., Acharya, S., Butz, C., Berardini, M., and Fishel, R. (2004). hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol. Cell 15, 437–451. [DOI] [PubMed] [Google Scholar]
- Sung, P., Krejci, L., Van Komen, S., and and Sehorn, M. G. (2003). Rad51 recombinase and recombination mediators. J. Biol. Chem. 278, 42729–42732. [DOI] [PubMed] [Google Scholar]
- Sym, M., Engebrecht, J. A., and Roeder, G. S. (1993). ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72, 365–378. [DOI] [PubMed] [Google Scholar]
- Symington, L. S. (2002). Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak, J. W., Orr-Weaver, T. L., Rothstein, R. J., and Stahl, F. W. (1983). The double strand-break model for recombination. Cell 33, 25–35. [DOI] [PubMed] [Google Scholar]
- Tsodikov, O. V., Enzlin, J. H., Scharer, O. D., and Ellenberger, T. (2005). Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF-ERCC1. Proc. Natl. Acad. Sci. USA 102, 11236–11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., Kong, H., Sun, Y., Zhang, X., and Zhang, W. (2004). Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol. 135, 1084–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H., Swoboda, I., Bhalla, P. L., Sijbers, A. M., Zhao, C., Ong, E. K., Hoeijmakers, J. H., and Singh, M. B. (1998). Plant homologue of human excision repair gene ERCC1 points to conservation of DNA repair mechanisms. Plant J. 13, 823–829. [DOI] [PubMed] [Google Scholar]
- Xu, Y. Y., Chong, K., Xu, Z. H., and Tan, K. H. (2002). Practical techniques of in situ hybridization with RNA probe. Chin. Bull. Bot. 19, 234–238. [Google Scholar]
- Yang, W. (2003). Pruning DNA: structure-specific endonucleases (XPF/Rad1/Mus81). Structure 11, 365–366. [DOI] [PubMed] [Google Scholar]
- Yildiz, O., Kearney, H., Kramer, B. C., and Sekelsky, J. J. (2004). Mutational analysis of the Drosophila DNA repair and recombination gene mei-9. Genetics 167, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, H., Kurumizaka, H., Ikawa, S., Yokoyama, S., and Shibata, T. (2003). Holliday junction binding activity of the human Rad51B protein. J. Biol. Chem. 278, 2767–2772. [DOI] [PubMed] [Google Scholar]
- Yokoyama, H., Sarai, N., Kagawa, W., Enomoto, R., Shibata, T., Kurumizaka, H., and Yokoyama, S. (2004). Preferential binding to branched DNA strands and strand-annealing activity of the human Rad51B, Rad51C, Rad51D and Xrcc2 protein complex. Nucleic Acids Res. 32, 2556–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar, J. H. (1974). Biostatistical Analysis, Upper Saddle River, NJ: Prentice Hall.
- Zhang, X., Feng, B., Zhang, Q., Zhang, D., Altman, N., and Ma, H. (2005). Genome-wide expression profiling and identification of gene activities during early flower development in Arabidopsis. Plant Mol. Biol. 58, 401–419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.