Abstract
Actin assembly nucleated by Arp2/3 complex has been implicated in the formation and movement of endocytic vesicles. The dendritic nucleation model has been proposed to account for Arp2/3-mediated actin assembly and movement. Here, we explored the model by examining the role of capping protein in vivo, with quantitative tracking analysis of fluorescence markers for different stages of endocytosis in yeast. Capping protein was most important for the initial movement of endocytic vesicles away from the plasma membrane, which presumably corresponds to vesicle scission and release. The next phase of endosome movement away from the plasma membrane was also affected, but less so. The results are consistent with the dendritic nucleation model's prediction of capping protein as important for efficient actin assembly and force production. In contrast, the movement of late-stage endocytic vesicles, traveling through the cytoplasm en route to the vacuole, did not depend on capping protein. The movement of these vesicles was found previously to depend on Lsb6, a WASp interactor, whereas Lsb6 was found here to be dispensable for early endosome movement. Thus, the molecular requirements for Arp2/3-based actin assembly differ in early versus later stages of endocytosis. Finally, acute loss of actin cables led to increased patch motility.
INTRODUCTION
Understanding the molecular mechanisms for how actin filaments assemble and how that assembly generates movement in vivo are important goals in cell biology. Biochemical studies in vitro have produced a number of specific hypotheses regarding these molecular mechanisms. One important hypothesis is the dendritic nucleation model, which describes how activated Arp2/3 complex nucleates the assembly of branched networks of actin filaments and how the added activities of capping protein and ADF/cofilin can modify actin assembly to produce force and move an object from which filaments are nucleated (Loisel et al., 1999; Pollard and Borisy, 2003). In this model, the important roles proposed for the capping of barbed ends, by capping protein, are to keep the actin filaments of the branched network short, which makes the network strong, and to confine the growth of filaments to newly created free barbed ends, which are near the object to be moved. Recent studies with synthetic systems show that capping protein is crucial for actin filaments that assemble around beads to break symmetry and produce movement (Akin and Mullins, personal communication). The importance of ADF/cofilin has been proposed to be promotion of actin filament disassembly, which produces actin subunits that recycle into the system by adding to new free barbed ends. The system thus operates at a steady state far from equilibrium, with a flux of actin subunits generating force and movement.
In cells, networks of actin filaments with the branching pattern characteristic of Arp2/3 complex can be found, and Arp2/3 activity is important for actin assembly and motility in certain situations (Pollard and Borisy, 2003), supporting the relevance of the dendritic nucleation model in vivo. Our goal is to explore and test the model in living cells, and budding yeast provide a good system for this task. Yeast cells have foci of filamentous actin at the cortex, called patches, whose formation and movement depend on Arp2/3 complex and actin polymerization (Winter et al., 1997; Carlsson et al., 2002). Actin patch formation and movement is associated with the formation of endocytic vesicles at the plasma membrane and their movement into the cell (Huckaba et al., 2004; Kaksonen et al., 2003, 2005; Duncan and Payne, 2005). Patches contain actin filaments with 70° end-to-side branches, which are characteristic of Arp2/3 complex (Young et al., 2004; Rodal et al., 2005). Capping protein (Cap1/Cap2) and cofilin (Cof1) are present in patches (Engqvist-Goldstein and Drubin, 2003). Thus, the dendritic nucleation model may apply to actin patch formation and movement in yeast.
Endocytic vesicles that carry membrane proteins destined for degradation move through the cytoplasm to the vacuole. Electron microscopy (EM) morphology studies found that the process of endocytosis begins with small round vesicles, proceeds to a vesicular/tubular compartment, followed by multivesicular bodies (MVBs), and, ultimately, the vacuole (Prescianotto-Baschong and Riezman, 1998). The membrane receptor Ste2 displays ubiquitin-dependent internalization and degradation, with trafficking via multivesicular bodies to the vacuole (Hicke et al., 1997). In live cell imaging, Ste2-green fluorescent protein (GFP) is seen diffusely at the plasma membrane, as small vesicles moving rapidly through the cytoplasm, as brighter vesicles on the surface of the vacuole (presumably docked MVBs about to fuse), and in the vacuole (Chang et al., 2003). The movement of Ste2 vesicles through the cytoplasm depends on actin polymerization and WASp/Las17, which binds to Arp2/3 complex (Chang et al., 2003, 2005). Also, the speeds of Ste2 vesicles are comparable to those of actin patches (Chang et al., 2003). In contrast, Ste2 vesicles do not have detectable actin or Arp2/3 complex, by methods that detect them readily on actin patches.
To investigate the mechanism by which actin polymerization drives the assembly and movement of cortical actin patches and the movement of endocytic vesicles, we analyzed their assembly and movement using computer-assisted tracking of GFP-labeled components in movies of living cells. The method allows one to track hundreds of patches or vesicles in many cells, outputting data for their position and fluorescence intensity over time. Analyzing many patches and vesicles in a quantitative manner was important because of the high amount of variability in the behavior of the individual entities. We found differences in the results for actin patches compared with Ste2 vesicles, suggesting differences in their molecular mechanisms for actin-based assembly and motility. The results for actin patches were consistent with predictions of the dendritic nucleation model.
MATERIALS AND METHODS
Cell Growth and Light Microscopy
Cells were grown overnight at 30°C in YPD to an A600 of 0.1–0.5. Cells were harvested at low speed, resuspended in SD-complete media, and placed on 2% agarose pads made with SD-complete media. For latrunculin A (LatA) treatment, cells were treated with 200 μM LatA with 1% dimethyl sulfoxide (DMSO) in SD-complete media for 20 min at 30°C with shaking. Cells were harvested, resuspended in a smaller volume of the same media, and placed on agarose pads containing the same concentration of LatA and DMSO. For temperature shift experiments, slides were placed at the restrictive temperature for 5 min before imaging, and observations were completed within 30 min of the temperature shift.
GFP fluorescence movies were made with a spinning disk confocal system that included an upright Olympus BX52 microscope, a Yokogawa CSU10 spinning disk head, Ar laser light sources, a 100× numerical aperture (NA) 1.4 PlanApo oil objective, and an intensified charge-coupled device (CCD) videocamera (Stanford XR Mega10 S30). The temperature of the specimen and stage was maintained at 30°C, except for temperature shift experiments. The image was focused at an equatorial plane of the cells, near the mother/bud neck. Movies were collected with a frame rate of 5/s, and their duration was 40–50 s. QED software (Media Cybernetics, Silver Spring, MD) controlled the system. Dual imaging of yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) was performed with a wide-field fluorescence inverted microscope (Olympus IX 70), a 100× oil NA 1.4 objective lens, YFP and CFP filter cubes, and a cooled CCD video camera (Photometrics CoolSNAP HQ) controlled by QED software.
Computer-assisted Tracking of Movement
We tracked the position of actin patches or membrane vesicles from fluorescence microscopy movies, as described previously (Carlsson et al., 2002). Movie 1 shows an example of tracking. The experimenter viewed each movie and track to confirm the validity of the computer tracking. We often adjusted the start or end of the track by a frame or two, and we removed erroneous tracks. Zero time was the time when the patch occurred and was detected, as defined by that marker. Patches in existence at the start of the movie were not included in the analysis. A track ends when its patch disappears, and this time can differ quite a bit between individual patches.
The computer program produced values for position and intensity of the particle over time, which were imported into spreadsheets in Excel (Microsoft, Redmond, WA) for motion analysis. Mean squared displacement (MSD) versus time was often plotted to analyze the character of the motion and to quantitate the degree of motion. The method calculates the distance a patch has moved from its point of origin, at each time point. That distance is squared, and those values are then averaged for many patches to give a mean. The plots for individual patches were generally aligned with each other at their zero time points, the beginning of each curve. The plots shown are thus averages. We generally truncated them at the time when half the patches had disappeared; at longer times, the plots became too noisy to be informative. Thus, the length of the curve is an indication of the median lifetime. In certain cases, we lined up the individual curves, for the tracking of one patch, on the right hand side, at the end of their lifetime, instead of the left as described above. This method is helpful if a patch is stationary for a variable amount of time and then moves, because all the potential movements occur at the end. In addition, we collected the value for the lifetime of each patch at the membrane directly in certain cases.
Strain Construction
Yeast strains are listed in Table 1. Wild-type haploid strains expressing Abp1-GFP, Cap1-GFP, Las17-GFP, or Sla2-GFP were constructed by integrating GFP at the 3′ end of the ABP1, CAP1, LAS17, SAC6, or SLA2 coding region, as described previously (Karpova et al., 1998), in a Yeast Gene Deletion Collection MATa wild-type haploid (BY4741, MATa his3Δ1 ura3Δ leu2Δ met15Δ), resulting in strains YJC 2719, 2718, 3420, 3396, and 3398, respectively. The polarization of the actin cytoskeleton was normal in strains expressing Cap1-GFP (YJC 2718), Sac6-GFP (YJC 3396), Sla1-GFP (YJC 4001), or Sla2-GFP (YJC 3398), indicating that the fusion proteins were functional. The movement of actin patches was similar in strains expressing either Cap1-GFP, Abp1-GFP, or Sac6-GFP, also consistent with normal function for the fusion proteins.
Table 1.
Yeast strains used in this study
| YJC | Name | Source | Genotype |
|---|---|---|---|
| 0093 | Wt, W303 | R. Rothstein | MATarho+ ade2-1his3-11,15leu2-3,112 trp1-1 ura3-1 |
| 0171 | cap2Δ | This laboratory | MATarho+ ade2-1 his3-11,15leu2-3,112 trp1-1 ura3-1 cap2-Δ1::HIS3 |
| 2538 | cap1Δ | Invitrogen | MATahis3Δ1 leu2Δ met15Δ ura3Δ cap1Δ::KanR |
| 2588 | Wt, Yeast Gene Collections | Invitrogen | MATahis3Δ1 leu2Δ met15Δ ura3Δ |
| 2718 | CAP1-GFP | This study | MATaCAP1-GFP-HIS3 his3Δ1 leu2Δ met15Δ ura3Δ |
| 2719 | ABP1-GFP | This study | MATaABP1-GFP-HIS3 his3Δ1 leu2Δ met15Δ ura3Δ |
| 3396 | SAC6-GFP | This study | MATaSAC6-GFP-HIS3 his3Δ1 leu2Δ met15Δ ura3Δ |
| 3398 | SLA2-GFP | This study | MATaSLA2-GFP-HIS3 his3Δ1 leu2Δ met15Δ ura3Δ |
| 3420 | LAS17-GFP | This study | MATaSLA2-GFP-HIS3 his3Δ1 leu2Δ met15Δ ura3Δ |
| 3481 | cap1Δ SAC6-GFP | This study | MATaSAC6-GFP-HIS3 cap1Δ::kanMX4 his3Δ1 leu2Δ ura3Δ lys2Δ met15Δ |
| 3482 | cap1Δ SAC6-GFP | This study | MATaSAC6-GFP-HIS3 cap1Δ::kanMX4 his3Δ1 leu2Δ ura3Δ lys2Δ met15Δ |
| 3919 | cap2Δ SAC6-GFP | This study | MATα ABP1-GFP-HIS3 cap2Δ::kanMX4 his3Δ1 leu2Δ met15Δ ura3Δ lys2Δ |
| 3920 | cap1Δ ABP1-GFP | This study | MATaABP1-GFP-HIS3 cap1Δ::kanMX4 his3Δ1 leu2Δ met15Δ ura3Δ |
| 3921 | cap1Δ ABP1-GFP | This study | MATα ABP1-GFP-HIS3 cap1Δ::kanMX4 his3Δ1 leu2Δ ura3Δ lys2Δ |
| 3922 | cap2Δ ABP1-GFP | This study | MATaABP1-GFP-HIS3 cap2Δ::kanMX4 his3Δ1 leu2Δ met15Δ ura3Δ lys2Δ |
| 3923 | cap2Δ ABP1-GFP | This study | MATα ABP1-GFP-HIS3 cap2Δ::kanMX4 his3Δ1 leu2Δ ura3Δ lys2Δ |
| 3945 | cap1Δ LAS17-GFP | This study | MATα LAS17-GFP-HIS3 cap1Δ::kanMX4 his3Δ1 leu2Δ met15Δ ura3Δ |
| 3946 | cap1Δ LAS17-GFP | This study | MATaLAS17-GFP-HIS3 cap1Δ::kanMX4 his3Δ1 leu2Δ met15Δ ura3Δ |
| 3947 | cap2Δ LAS17-GFP | This study | MATα LAS17-GFP-HIS3 cap2Δ::kanMX4 his3Δ1 leu2Δ met15Δ ura3Δ |
| 3948 | cap2Δ LAS17-GFP | This study | MATaLAS17-GFP-HIS3 cap2Δ::kanMX4 his3Δ1 leu2Δ ura3Δ |
| 3949 | cap2Δ LAS17-GFP | This study | MATα LAS17-GFP-HIS3 cap2Δ::kanMX4 his3Δ1 leu2Δ ura3Δ lys2Δ met15Δ |
| 3950 | cap1Δ SLA1-GFP | This study | MATα SLA1-GFP-HIS3 cap1Δ::kanMX4 his3Δ1 leu2Δ ura3Δ lys2Δ met15Δ |
| 3951 | cap1Δ SLA1-GFP | This study | MATaSLA1-GFP-HIS3 cap1Δ::kanMX4 his3Δ1 leu2Δ ura3Δ |
| 3952 | cap1Δ SLA1-GFP | This study | MATaSLA1-GFP-HIS3 cap1Δ::kanMX4 his3Δ1 leu2Δ ura3Δ met15Δ |
| 3953 | cap2Δ SLA1-GFP | This study | MATaSLA1-GFP-HIS3 cap2Δ::kanMX4 his3Δ1 leu2Δ ura3Δ lys2Δ |
| 3954 | cap2Δ SLA1-GFP | This study | MATaSLA1-GFP-HIS3 cap2Δ::kanMX4 his3Δ1 leu2Δ ura3Δ met15Δ |
| 3955 | cap2Δ SLA1-GFP | This study | MATα SLA1-GFP-HIS3 cap2Δ::kanMX4 his3Δ1 leu2Δ ura3Δ |
| 3995 | ABP1-GFP | This study | MATaABP1-GFP-HIS3 his3Δ1 leu2Δ ura3Δ met15Δ |
| 3996 | ABP1-GFP | This study | MATaABP1-GFP-HIS3 his3Δ1 leu2Δ ura3Δ met15Δ lys2Δ |
| 3997 | ABP1-GFP | This study | MATα ABP1-GFP-HIS3 his3Δ1 leu2Δ ura3Δ |
| 3998 | SAC6-GFP | This study | MATα SAC6-GFP-HIS3 his3Δ1 leu2Δ ura3Δ met15Δ lys2Δ |
| 3999 | SAC6-GFP | This study | MATα SAC6-GFP-HIS3 his3Δ1 leu2Δ ura3Δ lys2Δ |
| 4000 | SAC6-GFP | This study | MATα SAC6-GFP-HIS3 his3Δ1 leu2Δ ura3Δ |
| 4001 | SLA1-GFP | This study | MATaSLA1-GFP-HIS3 his3Δ1 leu2Δ ura3Δ lys2Δ |
| 4002 | SLA1-GFP | This study | MATα SLA1-GFP-HIS3 his3Δ1 leu2Δ ura3Δ met15Δ |
| 4088 | LAS17-GFP | This study | MATaLAS17-GFP-HIS his3Δ1 leu2Δ ura3Δ met15Δ |
| 4089 | LAS17-GFP | This study | MATα LAS17-GFP-HIS3 his3Δ1 leu2Δ ura3Δ lys2Δ |
| 4090 | LAS17-GFP | This study | MATaLAS17-GFP-HIS3 his3Δ1 leu2Δ ura3Δ lys2Δ |
| 4091 | CP Overexpression, ABP1-GFP | This study | MATaABP1-GFP-HIS3 his3Δ1 leu2Δ met15Δ ura3Δ [GAL1/10-CAP1&CAP2 LEU2 2μ] |
| 4244 | bnr1Δ | A. Bretscher | MATα bnr1Δ::KanR his3 leu2 lys2 ura3 |
| 4245 | bnr1Δ bni1-11 | A. Bretscher | MATα bnr1Δ::kanR bni1-11::URA3 his3 leu2 lys2 ura3 |
| 4246 | bnr1Δ bni1-12 | A. Bretscher | MATα bnr1Δ::kanR bni1-12::URA3 his3 leu2 lys2 ura3 |
| 4247 | bnr1Δ ABP1-GFP | This study | MATα bnr1Δ::KanR ABP1-GFP-HIS3MX6 his3 leu2 lys2 ura3 |
| 4248 | bnr1Δ bni1-11 ABP1-GFP | This study | MATα bnr1Δ::kanR bni1-11::URA3 ABP1-GFP-HIS3MX6 his3 leu2 lys2 ura3 |
| 4249 | bnr1Δ bni1-12 ABP1-GFP | This study | MATα bnr1Δ::kanR bni1-12::URA3 ABP1-GFP-HIS3MX6 his3 leu2 lys2 ura3 |
| 4267 | ABP1-CFP CAP1-YFP | This study | MATaABP1-CFP-KanMX6 CAP1-YFP-HIS3 his3Δ1 leu2Δ met15Δ ura3Δ |
| 4273 | CAP1-GFP | This study | MATaCAP1-GFP-HIS3 his3Δ1 leu2Δ ura3Δ |
| 4274 | CAP1-GFP | This study | MATaCAP1-GFP-HIS3 his3Δ1 leu2Δ ura3Δ met15Δ lys2Δ |
| 4275 | CAP1-GFP | This study | MATaCAP1-GFP-HIS3 his3Δ1 leu2Δ ura3Δ met15Δ |
| 4294 | SLA2-GFP | This study | MATaSLA2-GFP-HIS3 his3Δ1 leu2Δ ura3Δ lys2Δ |
| 4295 | SLA2-GFP | This study | MATaSLA2-GFP-HIS3 his3Δ1 leu2Δ ura3Δ met15Δ |
| 4296 | SLA2-GFP | This study | MATaSLA2-GFP-HIS3 his3Δ1 leu2Δ ura3Δ met15Δ lys2Δ |
| 4297 | ABP1-GFP | This study | MATα ABP1-GFP-HIS3 his3Δ1 leu2Δ ura3Δ |
| 4298 | ABP1-GFP | This study | MATaABP1-GFP-HIS3 his3Δ1 leu2Δ ura3Δ |
| 4299 | ABP1-GFP | This study | MATaABP1-GFP-HIS3 his3Δ1 leu2Δ ura3Δ met15Δ |
| 4300 | lsb6Δ ABP1-GFP | This study | MATaABP1-GFP-HIS3 lsb6Δ::kanMX4 his3Δ1 leu2Δ met15Δ ura3Δ |
| 4301 | lsb6Δ ABP1-GFP | This study | MATaABP1-GFP-HIS3 lsb6Δ::kanMX4 his3Δ1 leu2Δ met15Δ ura3Δ lys2Δ |
| 4302 | lsb6Δ ABP1-GFP | This study | MATaABP1-GFP-HIS3 lsb6Δ::kanMX4 his3Δ1 leu2Δ met15Δ ura3Δ lys2Δ |
| 4303 | ABP1-GFP | This study | MATaABP1-GFP-HIS3 his3Δ1 leu2Δ met15Δ ura3Δ lys2Δ |
| 4381 | cap1Δ STE2-GFP | This study | MATaSTE2-GFP-HIS cap1::kanMX ura3Δ met15Δ |
| 4384 | STE2-GFP | This study | MATaSTE2-GFP-HIS ura3Δ met15Δ lys2Δ |
| 4579 | LAS17-3XGFP | This study | MATaLAS17-3GFP::TRP1 ste2::leu2 sst1-D5 leu2-3,112 ura3-52 his3-Δ1 trp1 |
Haploid wild-type, cap1Δ, and cap2Δ strains expressing various GFP fusions were prepared by crossing YJC 2719, 3420, 3396, and 3398 with the cap1Δ strain YJC 3538 and the cap2Δ strain YJC 0171. Tetrad dissection produced the strains YJC 3481–4090 listed in Table 1.
Additional independent wild-type strains for analysis were generated by crossing YJC 2719 and 3398 with the Yeast Gene Deletion Collection MATα aip1Δ strain (record number 16227). Sporulation and tetrad dissection produced the Abp1-GFP wild-type strains YJC 4297, 4298, and 4299, and the Sla2-GFP wild-type strains YJC 4294, 4295, and 4296.
A wild-type strain expressing a Las17–3XGFP fusion from the endogenous locus was prepared using a pBS-3xGFP-TRP1 plasmid as described previously (Lee et al., 2003) to produce YJC 4579, which was used in experiments for Figure 1, C and E.
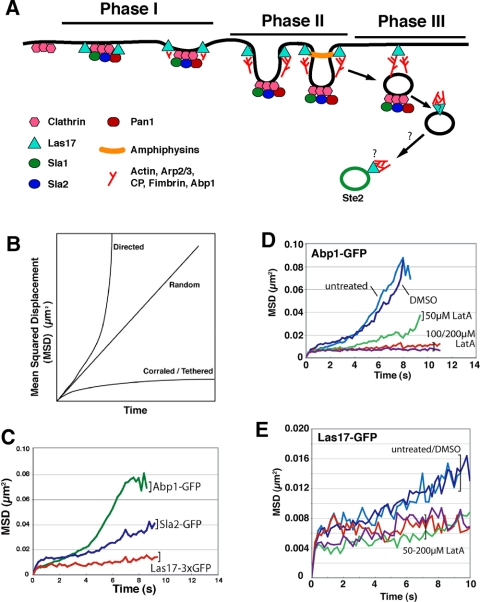
Figure 1.
(A) Diagram of actin patch formation and movement accompanying endocytosis at the plasma membrane. The diagram illustrates results and conclusions drawn from the work of Kaksonen et al. (2003, 2005) and Chang et al. (2003, 2005. (B) Illustration of how plots of MSD versus time reveal the character of particle motion. A random walk is a straight line, a directed path is concave up, and a tethered or corralled motion is concave down with a horizontal asymptote. (C) Tracking of actin patches labeled with GFP fusions of Abp1, Sla2, and Las17, in wild-type cells. For Abp1-GFP, the curve is the mean from 548 patches from five strains: YJC 2719, 4297, 4298, 4299, and 4303. For Sla2-GFP, there were 266 patches from three strains: YJC 4294, 4295, and 4296. Las17-GFP was a 3XGFP fusion, and the curve is based on 63 patches from strain YJC 4579. (D) Effect of latrunculin A on movement of Abp1-GFP patches. MSD is plotted versus time. LatA causes decreased movement and increased lifetime. Cells were treated with 200 μM LatA in SD-complete medium with 1% DMSO for 20 min. Strain YJC 2719 was used, and the number of patches averaged for each curve is as follows: untreated, 115; DMSO, 78; 50 μM, 125; 100 μM, 92; and 200 μM, 64. (E) Effect of latrunculin A on movement of Las17–3XGFP patches. MSD is plotted versus time, with an expanded y-axis. LatA causes decreased motion. The number of patches averaged for each curve is as follows: untreated, 63; DMSO, 94; 50 μM, 104; 100 μM, 78; and 200 μM, 64. The strain was YJC 4579. As a control, formaldehyde fixation caused a complete loss of patch movement in MSD plots (our unpublished data), confirming that the level of noise in the detection and tracking systems was sufficiently low that the level of phase I movement seen here was significant and above background.
lsb6Δ mutants expressing Abp1-GFP were prepared by crossing YJC 2719 with the Yeast Gene Deletion Collection MATα lsb6Δ strain (record number 11323). Sporulation and tetrad dissection produced the ABP1-GFP lsb6Δ haploids YJC 4300, 4301, and 4302 and the ABP1-GFP wild-type haploid YJC 4303. The lsb6 deletion was confirmed by PCR analysis.
To overexpress capping protein, we expressed Cap1 and Cap2 from a 2μ plasmid containing the bidirectional inducible promoter GAL 1/10. A wild-type Abp1-GFP strain, YJC2719, was transformed with the plasmid pBJ114 to produce YJC 4091. Capping protein (CP) overexpression was induced by overnight growth in 2% raffinose, followed by 2% galactose overnight.
To track patches in formin mutants, haploid bnr1Δ, bnr1Δ bni1-11 and bnr1Δ bni1-12 strains, kindly provided by Dr. Anthony Bretscher (Cornell University, Ithaca, NY) as strain numbers Y2304, Y3084, and Y3089 (Evangelista et al., 2002), were transfected with a PCR product prepared from a Longtine vector (Longtine et al., 1998), which integrated GFP at the 3′ end of ABP1. The parent strains were numbered YJC 4244, 4245, and 4246, and the resulting Abp1-GFP strains were YJC 4247, 4248, and 4249.
FM4-64 Pulse Labeling
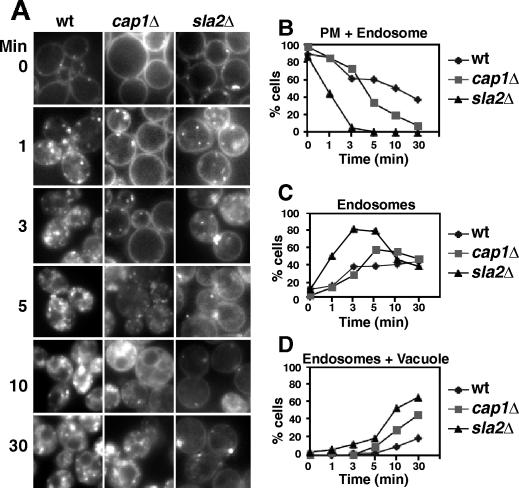
Cells were incubated with FM4-64 as described previously (Chang et al., 2003), with some modifications. Cells were incubated with 10 μM FM4-64 on ice for 2 min and then washed into ice-cold nonfluorescent media. Next, the cells were incubated with fresh nonfluorescent (NF) medium at 30°C. Aliquots were removed at various times and fixed with 3.7% formaldehyde. The cells were examined with a wide-field fluorescence inverted microscope (Olympus IX 70), a 100× oil NA 1.4 objective lens, a rhodamine filter cube, and a cooled CCD videocamera (Photometrics CoolSNAP HQ) controlled by QED software. Individual cells were scored into one of the following categories: plasma membrane + endosome, fluorescence on the plasma membrane and endocytic vesicles near or on the plasma membrane; endosome alone, fluorescence exclusively on endocytic vesicles in the cytoplasm; or endosome and vacuole, fluorescence on vacuoles and cytoplasmic vesicles. Cells (100–200) were scored per time point. Wt (YJC 2588), cap1Δ (YJC 2538), and sla2Δ haploid MATa strains were from the Yeast Gene Deletion Collection (Invitrogen, Carlsbad, CA) (Winzeler et al., 1999).
Tracking of FM4-64 and Ste2-GFP Cytoplasmic Vesicles
For FM4-64 labeling, cells were incubated on ice with 9 μM FM4-64 for 1–2 min and then washed into nonfluorescent medium at room temperature (RT). Wild-type cells were kept at RT for 2 min and cap1Δ cells for 5 min because membrane is internalized more slowly in cap1Δ cells. Time-lapse fluorescence images were acquired at a rate of 1 frame/s, with an integration time of 80 ms/frame for a total of 90–100 frames. Images were collected on a wide-field inverted fluorescence microscope as described above. To generate yeast strains expressing Ste2-GFP, strains YJC 2588 (wild-type) and 2538 (cap1Δ) were transformed with a multicopy Ste2-GFP plasmid based on pRS426 (Chang et al., 2003, 2005). GFP imaging was performed on a spinning disk system, as described above. FM4-64 and Ste2-GFP fluorescence were both observed at the plasma membrane, in freely moving particles, in the vacuole, and in particles adherent to the vacuole. Only freely moving particles in cytoplasm were tracked, and we required that they be visible for >6 s. Single-Copy Expression of Ste2-GFP. We repeated the Ste2-GFP experiments in strains expressing Ste2-GFP from the endogenous STE2locus. A haploid MATa strain expressing Ste2-GFP from the endogenous locus was taken from the Yeast GFP Clone Collection (Invitrogen) (Huh et al., 2003) and mated with a MATα cap1Δ haploid from the Yeast Gene Deletion Collection (Invitrogen) (Winzeler et al., 1999). Two resulting diploids were sporulated, and tetrads were dissected to produce haploid CAP1 (YJC 4384) and cap1Δ (YJC 4381) strains expressing Ste2-GFP. Cells were grown overnight at 30°C. Time-lapse movies at 1 frame/s were collected on an upright Olympus fluorescence microscope, as described above, with an exposure time of 860 ms. One hundred images were collected per movie. One or two freely moving Ste2-GFP vesicles could be seen per cell, and their motion was tracked. Thirty and 26 vesicles were tracked in the wild-type and cap1Δ strain, respectively. The MSD plots for wild-type and cap1Δ strains were similar to each other, as observed with plasmid-based higher expression of Ste2-GFP. Ste2-GFP vesicles are entirely of endocytic origin because they are absent in cells expressing a form of Ste2-GFP that is completely blocked for endocytic internalization but is otherwise well expressed and functional at the plasma membrane (Stefan and Blumer, 1999).
RESULTS
Defining the Phases of Actin Patch Assembly and Movement
For actin patches, the phases in their life have been defined by the presence of different components and by the character of their movement (Figure 1A). Phase I is characterized by the presence of WASp/Las17, a protein that targets active Arp2/3 complex (Wen and Rubenstein, 2005). Phase I patches show a limited amount of motion, which is random in direction and restricted in range, as though the patch were in a corral or anchored by a tether on the plasma membrane.
To quantitatively analyze the motion of many patches under different conditions, we developed a computer-assisted method to track the position of patches over time in movies of living cells expressing GFP fusion proteins (Carlsson et al., 2002). Movie 1 helps to illustrate the method. One useful way to analyze these data is to calculate the square of the distance of the patch from its starting point. The average of these values, the mean squared displacement (MSD), is then plotted versus time. Such plots reveal the character of the motion as well as quantitating its magnitude. Tethered random motion, characteristic of phase I, produces an MSD plot that is concave down, approaching a horizontal asymptote (corralled/tethered in Figure 1B; Las17–3XGFP in Figure 1C). Actin patches accumulate endocytic adaptors and actin-binding proteins over time. The tethered movement characteristic of phase I is also observed at the start of MSD plots with GFP labeling of the endocytic adaptors Sla1 and Sla2, and the actin-binding proteins Abp1, fimbrin (Sac6), and capping protein (Cap1/Cap2), which bind to actin filaments in different ways.
Phase II is the initial movement of the patch away from the membrane. This movement is slow and short, compared with subsequent movement in phase III. Phase II movement is observed with GFP labeling of the endocytic adaptors and the actin-binding proteins, but not with WASp/Las17 (Figure 1C). In MSD plots, phase II movement occurs as a relatively straight segment of low slope. Phase III consists of longer and faster movement directed away from the membrane, which shows in MSD plots as a concave-up segment of the curve after phase II. Phase III movement is only seen when tracking actin-binding proteins (Figure 1C).
Dependence of Movement on Actin Polymerization
In the current model relating actin assembly to endocytosis (Kaksonen et al., 2005), the movements associated with phase II and phase III depend on actin polymerization, and we found this to be the case by using Abp1-GFP to track the patches. LatA at 50 μM partially inhibited patch movement, and 100–200 μM LatA provided complete inhibition, as seen in MSD plots (Figure 1D). The lifetime of the patches was increased in LatA, manifested in MSD plots by the curves extending to longer times. These plots were cut off when half the patches disappeared, indicating the median lifetime of the patches. The Abp1-GFP fluorescence intensity per patch decreased over time upon addition of LatA (our unpublished data), consistent with LatA inhibiting the progress of actin polymerization.
Patches in phase I do move, albeit relatively little. We asked whether this movement depended on actin polymerization by adding LatA to Las17-GFP cells. Patch tracking revealed that the movement was inhibited (Figure 1E). The lifetime of Las17-GFP patches was greatly prolonged by LatA, and the fluorescence intensity was not decreased (our unpublished data). The inhibition of movement may seem contradictory to the notion that actin polymerization is “downstream” of Las17 recruiting active Arp2/3 complex to the membrane. However, a locus of accumulated Las17 should be subject to forces coming from the actin filaments that polymerize around the site.
The Role of Capping Protein in Actin-based Motility
CP is proposed to play a central role in the dendritic nucleation model (Pollard and Borisy, 2003), and CP is necessary to reconstitute the actin-based motility of Listeria with pure proteins (Loisel et al., 1999). We asked whether CP was important for actin-based motility in yeast actin patches.
First, we examined the assembly of CP on the actin patch with respect to other components. A Cap1-GFP fusion protein functioned normally, measured by rescue of polarization of the actin cytoskeleton. We compared Cap1-GFP with fimbrin/Sac6-GFP and Abp1-GFP by tracking actin patches in wild-type cells. The results, as MSD plots, were essentially the same (our unpublished data). We examined the order of assembly by dual-label imaging of Cap1-YFP and Abp1-CFP (Figure 2). Following individual patches in wild-type cells, we found a nearly complete coincidence, in space and time, between the two labels, as shown in frames from a movie (Figure 2A) and a graph of the fluorescence intensity over time (Figure 2B). Thus, the components of the actin network assemble in a concerted manner, suggesting that the assembly process is cooperative and rapid.
Figure 2.

(A) Images of a single patch labeled with Cap1-YFP and Abp1-CFP, in YJC 4267. The time interval between frames is 1 s. The YFP and CFP images were collected for 0.5 s, consecutively, not simultaneously, so that the interval between columns is 1 s. The width of each panel is 0.4 μm. (B) Mean fluorescence intensity of Cap1-YFP and Abp1-CFP patches over time. Results for five patches were averaged and normalized. The values for fluorescence intensity of YFP and CFP cannot be compared with each other.
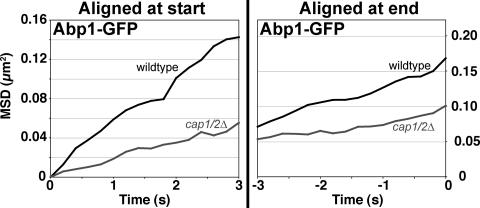
Next, we examined the effect of the loss of CP on actin patch assembly and early endocytosis by imaging and tracking actin patches in cells expressing Las17-GFP, Sla1-GFP, Abp1-GFP, or Sac6-GFP. Actin patches did form in CP mutants, but their movement away from the membrane was less, as seen in time-lapse images from movies (Figure 3 and Movies 1 and 2). To analyze the data quantitatively, we tracked patches in movies of several independent isolates of each genotype. To examine the behavior of all the patches within a sample, we first produced plots of MSD versus time in which we aligned the data for individual patches on the left, at the start of the patch lifetime, and then averaged the results for all the patches from one isolate (Figure 4, left). The results for the different isolates were similar to each other, and we show examples here to illustrate the degree of variation. We also aligned the data for individual patches on the right, at the end of the patch lifetime, to focus on motion that might have occurred at the end of the track.
Figure 3.
Time-lapse microscopy of wild-type or cap1Δ cells expressing Abp1-GFP or Sla1-GFP. Note the different time intervals between frames in each column. In wild-type cells, the arrowhead indicates the starting position of a patch that moves, and the arrow follows the patch. In cap1Δ cells, the arrowhead indicates a patch that remains tethered. Some patches in wild-type cells also do not move, corresponding to phase I behavior. Strains are as follows: wild-type Abp1-GFP, YJC 3996; cap1Δ Abp1-GFP, YJC 3920; wild-type Sla1-GFP, YJC 4002; and cap1Δ Sla1-GFP, YJC 3950. Bar, 1 μm.
Figure 4.
Quantitative analysis of patch motility in the presence and absence of capping protein. Patches labeled with GFP fusions of Abp1, Sac6, Sla1, or Las17, were tracked in wild-type, cap1Δ or cap2Δ cells. In the left panels, each curve is from one independent strain and represents the average of plots from tracks of 25–100 patches from four to seven cells. Results from independent strains are plotted to illustrate the level of variability in the results, which is usually low but can be significant. For the left panels, tracks of individual patches were aligned on the left side, at the start of their lifetime, before averaging. Plots end at the median lifetime of the patches, i.e., when half the patches have disappeared, or at 14 s, whichever is less. In the panels on the right, tracks were aligned on their right edges, at the end of their life, to focus on movement away from the membrane. Here, results from independent strains were averaged together. LatA treatment caused a greater inhibition of movement than did the loss of CP in each case, as shown in Figure 1D. The results with Sac6-GFP were similar to those with Abp1-GFP and are not shown. Strain numbers were YJC 3481–4090.
To examine phase I, we tracked Las17-GFP patches. There was no effect from the loss of CP, with patch tracks aligned at the start or the end of their lifetime before averaging to create MSD plots (Figure 4). Next, we measured the time that patches spent at the membrane, from the time of their initial appearance until they either moved away from the membrane or disappeared, which can result from disassembly or z-axis movement (Figure 5). In CP null mutants, Las17-GFP patches did spend slightly more time at the membrane, compared with wild-type (wt) cells, by an amount that was statistically significant (Figure 5).
Figure 5.

Mean time spent by patches in the tethered phase on the membrane. The time measured was the interval from the appearance of a patch until its movement away from the membrane or disappearance. The error bar is 1 s. Dimethyl sulfoxide. n = 24–50 patches for each strain. Wild-type is significantly different from cap1Δ in each case, with p < 0.001. Strains are as follows: wild-type Abp1-GFP, YJC 3995; cap1Δ Abp1-GFP, YJC 3918; wild-type Sac6-GFP, YJC 3998; cap1Δ Sac6-GFP, YJC 3481; wild-type Sla1-GFP, YJC 4001; cap1Δ Sla1-GFP, YJC 3950; wild-type Las17-GFP, YJC 4088; cap1Δ Las17-GFP, YJC 3945; wild-type Cap1-GFP, YJC 2718; cap2Δ Abp1-GFP, YJC 3922 and 3923; cap2Δ Sac6-GFP, YJC 3919; cap2Δ Sla1-GFP, YJC 3953–3955; and cap2Δ Las17-GFP, YJC 3947–3949.
Tracking the endocytic adaptor Sla1-GFP in CP null mutants, compared with wt cells, revealed substantial changes in the dynamics of patches in phase II. With MSD plots aligned on the left before averaging, cap1/2 cells showed much less motion for Sla1-GFP than did wt cells (Figure 4, left). We considered two reasons for this difference. We thought that Sla1-GFP patches might persist at the membrane for longer and variable times or that they might move less after leaving the membrane. To test these possibilities, we first measured the lifetime of Sla1-GFP patches at the membrane and found it increased by a substantial amount, more than twofold (Figure 5). Second, we aligned the MSD plots on the right-hand side before averaging them, i.e., at the end of their lifetime, to focus on the phase where inward movement normally exists. This analysis showed that the average movement away from the membrane was greatly inhibited by the loss of CP, albeit not completely (Figure 4, right).
To examine phase III, we tracked patches using the actin-binding proteins Abp1-GFP and Sac6-GFP. Both markers gave similar results; only the Abp1-GFP results are shown here. With MSD plots aligned on the left, CP null mutant strains showed a substantial loss of the concave-up portion of the normal curve (Figure 4, left). The lifetime of the patches at the membrane was increased, by approximately threefold (Figure 5). With MSD plots aligned on the right-hand side, to focus on motion at the end of the track, CP null mutant cells showed a substantial decrease in average motion compared with wt cells (Figure 4, right). The effect of the loss of CP here was less than the effect on patch lifetime or on MSD plots aligned on the left. In addition, the effect of the loss of CP on Abp1-GFP, with MSD plots aligned on the right, was less than what was seen for Sla1-GFP. To investigate further the movement of phase III, we tracked Abp1-GFP patches after they had moved away from the membrane, defined by a distance of 200 nm. The average movement was still decreased by the loss of CP, based on MSD plots aligned either on the left or the right before averaging (Figure 6).
Figure 6.
Average movement of Abp1-GFP patches after leaving the plasma membrane in wild-type, cap1Δ, or cap2Δ cells. Patches were tracked after they had moved 0.2 μm away from the plasma membrane. MSD plots on the left and right were aligned at the start and end before averaging, respectively. Conditions and strains as in Figure 4. The wild-type curve is the average of MSD plots of 205 patches from 17 cells from three strains, YJC 3995–3997, and the cap1/2 curve is the average of 218 patches from 18 cells from four strains, YJC 3920–3923.
When actin-based motility of Listeria was reconstituted from pure proteins in vitro, the curve describing how motility depends on the concentration of CP was bell-shaped, with motility falling off at high and low concentrations (Loisel et al., 1999). This was also the case for ADF/cofilin and Arp2/3 complex. To test this prediction of the dendritic nucleation model in vivo, we overexpressed the two subunits of CP, Cap1 and Cap2, from a bidirectional promoter. We observed strong inhibition of movement, similar to the effect of the loss of CP, based on tracking of Abp1-GFP patches (Figure 7). Thus, the concentration of CP in vivo is optimal for actin patch assembly and motility.
Figure 7.
Movement of Abp1-GFP patches in cells overexpressing CP, compared with wild-type and cap1Δ cells. MSD plots were aligned on the left for averaging. Strain numbers are as follows: overexpression, YJC 4091; wild-type, YJC 3995; and cap1Δ, YJC 3920.
Patch Motility in the Absence of Actin Cables
Cells without CP are known to have a moderate defect in actin cable assembly and polarization (Karpova et al., 1998), so we asked whether their cable defect could account for their decrease in patch movement. Actin cables are highly dynamic bundles of filaments, polymerizing at their ends near the bud tip on a rapid time scale comparable with that of actin patch assembly (Yang and Pon, 2002). Actin patches are sometimes seen at the ends of cables and vice-versa (Karpova et al., 1998). Some actin patches have been observed to move along actin cables, seeming to attach to the cable and move rearward as the cable treadmilled (Huckaba et al., 2004). We reasoned that the decrease in the number of cables in CP mutant cells might be a contributing factor in the decrease in patch movement.
To test this hypothesis, we tracked actin patch assembly and movement with Abp1-GFP in conditional mutants known to lose actin cables completely and rapidly upon shift to restrictive temperature. We used two formin double mutants, bnr1Δ bni1-12 and bnr1Δ bni1-11 (Evangelista et al., 2002). Formins nucleate and sustain the polymerization of the actin filaments in the cable, so that loss of formin function leads to rapid disappearance of cables. We confirmed that these mutants did lose cables rapidly at the restrictive temperature under our conditions, by rhodamine-phalloidin staining (our unpublished data). Surprisingly, tracking actin patches revealed a modest increase of patch motility upon loss of cables, not a decrease (Figure 8). Thus, a decrease in cables cannot account for the decrease in patch movement in capping protein mutants.
Figure 8.
Acute loss of actin cables increases patch motility. The motion of Abp1-GFP patches was tracked in wild-type, bnr1Δ, bnr1Δ bni1-11, and bnr1Δ bni1[hyphen]12 cells, at permissive (30°C) and restrictive temperatures (35°C). Each curve represents the average from >100 patches from eight cells. MSD plots were aligned on the left for averaging. Cells on slides were placed at the restrictive temperature on the microscope stage for 5 min before observation. Observations were completed within the ensuing 25 min. Strains and number of patches per curve are as follows: wild-type, YJC 2719, 115; bnr1Δ, YJC 4247, 92; bnr1Δ bni1-11, YJC 4248, 154; and bnr1Δ bni1-12, YJC 4249, 96.
Endocytosis with Membrane Markers
Because loss of CP inhibited the movement of patches away from the membrane, and patches have been associated with endocytic vesicles as defined by FM4-64 labeling (Huckaba et al., 2004), we asked whether the loss of CP delayed the transit of FM4-64 through the endosomal system in a pulse experiment (Figure 9A). The initial internalization step was delayed in the cap1Δ mutant, to a degree similar to that seen for sla2Δ, a positive control endocytosis mutant (Wesp et al., 1997), based on the fraction of cells that persist in the initial pattern of fluorescence distribution (Figure 9B). This result confirmed our results with actin patch tracking in the capping protein mutants.
Figure 9.
Endocytosis traffic assessed with FM4-64 pulse labeling. Cells were incubated with FM4-64 on ice, washed, and incubated at 30°. (A) Representative fluorescence images at different time points for wild-type, cap1Δ, and sla2Δ cells. (B–D) Quantification of the types of membrane staining at each time point. (B) Plasma membrane and endosome. (C) Endosome only. (D) Endosome and vacuole.
In contrast, when examining the timing for the appearance of subsequent distribution patterns for the FM4-64 fluorescence, the cap1Δ curve was closer to the wild-type curve than it was to the sla2Δ curve (Figure 9, C and D). This result suggested that movement of vesicles to the vacuole might be less affected by the loss of capping protein. We tested this hypothesis directly with time-lapse movies of living cells in which we examined the movement of FM4-64 vesicles in the cytoplasm at intermediate times during a pulse label experiment. In wild-type cells, vesicles that were free in the cytoplasm moved rapidly, with frequent sharp turns. In cap1Δ cells, the vesicle behavior was similar to that of wild-type cells. Tracking analysis and MSD plots showed that cap1Δ and wild-type vesicles were indistinguishable (Figure 10A and Movies 3 and 4), supporting the hypothesis. The high frequency of turning caused the MSD plots to be linear, characteristic of random paths, as opposed to concave up, characteristic of directed motion.
Figure 10.
Movement of FM4-64 and Ste2-GFP vesicles in the cytoplasm. Single vesicles, not adherent to a larger membrane, were tracked. MSD plots were aligned on the left for averaging. (A) FM4-64. n = 133 and 144 vesicles for wild-type (YJC 2588) and cap1Δ (YJC 3920), respectively. (B) Ste2-GFP. n = 55 and 45 vesicles for wild-type (YJC 2588) and cap1Δ (YJC 2538) strains, respectively, carrying a multicopy Ste2-GFP plasmid (Chang et al., 2003). Similar results were obtained when Ste2-GFP was expressed from the STE2 endogenous locus in the CAP1 and cap1Δ haploid strains YJC4384 and YJC4381, respectively.
To test the hypothesis further, we asked whether the movement of endocytic vesicles in the cytoplasm labeled in another manner had a similar lack of dependence on CP. The membrane receptor Ste2 is constitutively endocytosed, and Ste2-GFP can be seen as vesicles moving rapidly though the cytoplasm on their way to the vacuole (Chang et al., 2003, 2005). We tracked the movement of Ste2-GFP vesicles and found no difference between cap1Δ and wild-type (Figure 10B and Movies 5 and 6). Therefore, the movement of endocytic vesicles at a later phase in the endocytic pathway seemed not to depend on capping protein, whereas the movement of early endocytic vesicles did.
To explore further the idea that actin patches and Ste2 vesicles move with different mechanisms, we examined the dependence of their movement on Lsb6, a Las17/WASp binding protein. Las17/WASp and Lsb6 were found to be important for the movement of Ste2-GFP vesicles in the cytoplasm (Chang et al., 2003, 2005). Therefore, we tested actin patch motility in an lsb6Δ null mutant, by Abp1-GFP tracking. We found no effect of the loss of Lsb6 (Figure 11), supporting the conclusion that the mechanism of actin-based motility for actin patches differs from that for Ste2 vesicles.
Figure 11.
Normal actin patch motility in the absence of Lsb6, based on Abp1-GFP patch tracking, with alignment of individual patch plots on the left before averaging. LatA treatment of the wt strain provides a positive control. Aligning the plots on the right also showed no difference between wt and mutant (our unpublished data). The wild-type strain was YJC 2719. One hundred and fifteen patches were analyzed for the wild-type control, and 64 patches were analyzed for the LatA-treated wild-type sample. For lsb6Δ, strains YJC 4300, 4301, and 4302 were analyzed, with 92, 118, and 110 patches per strain, respectively.
DISCUSSION
The Dendritic Nucleation Model and Actin Patch Motility
Our results here on the role of capping protein in the assembly and movement of actin patches support the relevance of the dendritic nucleation model in vivo. The model predicts that capping is not required for actin to assemble on an object or surface but that capping is important for efficient conversion of actin assembly into motion. Our results with actin patches agree with both predictions. First, the loss of capping protein did not affect the initial polymerization of actin at the site of nucleation, where WASp/Las17 presumably recruits active Arp2/3 complex (Wen and Rubenstein, 2005). In the absence of capping protein, actin still polymerized and components of the actin patch still assembled, as revealed by tracking of WASp/Las17, endocytic adaptors, and actin-binding proteins.
Second, the loss of capping protein did inhibit the movement of the endocytic vesicle away from the plasma membrane. The first motion of the endocytic vesicle away from the plasma membrane, which may correspond to scission, is most readily revealed by tracking endocytic adaptors, such as Sla1. Here, we saw that Sla1-GFP motion was severely compromised in capping protein mutants. A small amount of movement was seen but only after a substantial delay.
Normally, the endocytic vesicle then moves farther away from the plasma membrane, at a faster speed. Detecting this motion requires tracking actin filaments, with any of several different actin-binding proteins, such as Abp1-GFP. The loss of capping protein caused a substantial loss of this motion but not to the degree seen for the initial movement of the vesicle away from the membrane. Even focusing on the final phases of motion observed by tracking Abp1-GFP, we saw less motion in the absence of CP.
In a previous article (Kim et al., 2004), we reported an analysis of actin patch motion that revealed no difference between wild-type and capping protein null strains. Those studies were limited to the short-range movements of phase I, near the plasma membrane, because only a single z-axis plane at the top of the cell was examined, by confocal microscopy. We see here that phase I movement is not affected by the loss of CP, which is consistent with those previous results. In this study, equatorial planes were selected, which reveals motion into the cell interior more clearly.
The Molecular Mechanism of Actin-based Movement on Actin Patches versus Ste2 Vesicles
Our results reveal differences in the molecular basis of the movement of actin patches, in which endocytic vesicles form and move away from the plasma membrane, compared with the movement of Ste2 vesicles, in which endocytic vesicles travel through the cytoplasm to the vacuole. In previous studies, both processes were found to depend on actin polymerization (Carlsson et al., 2002; Chang et al., 2003). Actin patch movement was found to depend on the activity of Arp2/3 complex (Winter et al., 1997), and Ste2 vesicle movement to depend on WASp/Las17 (Chang et al., 2003), which binds Arp2/3 complex. These results suggest that both movements involve dendritic nucleation of actin assembly by Arp2/3 complex. In contrast, actin filaments and Arp2/3 complex have been observed to assemble and move with the actin patch (Kaksonen et al., 2003), but actin and Arp2/3 complex have not been visualized on Ste2 vesicles (Chang et al., 2005). This difference may simply reflect the sensitivity of detection methods, with Ste2 vesicles having less F-actin and Arp2/3 complex than actin patches do. A more telling difference may be that, in our results, the absence of capping protein inhibited movement of actin patches away from the plasma membrane to a large degree, whereas the movement of Ste2 vesicles in the cytoplasm was not affected by the loss of capping protein. In addition, Lsb6, a WASp/Las17 interactor, which is required for the movement of Ste2 vesicles (Chang et al., 2005), was not found to be required for the movement of actin patches here.
How do Ste2 vesicles move? One hypothesis is that their movement is a product of inertia resulting from the force that pushes the vesicle away from the plasma membrane. This model is consistent with the apparent absence of F-actin and Arp2/3 complex on the Ste2 vesicle, in that the vesicle would move passively, without the need for an active process to continuously drive its motion. We view this model as extremely unlikely. First, the Reynolds number for an object of this size moving through an aqueous solution, especially one with the viscosity of cytoplasm, is so low that inertia should be negligible. Second, Ste2 vesicles display sharp turns in the path of their movement through the cytoplasm, which is not predicted by inertial movement, unless one envisions the vesicles striking and bouncing off a larger object. Third, in this model, the same molecular process would power the movement of the early endosome away from the membrane and its subsequent movement through the cytoplasm. To the contrary, we found that early endosome movement depends on capping protein but not Lsb6, whereas Ste2 vesicle movement depends on Lsb6 but not capping protein. Thus, the molecular processes seems to be distinct.
We favor the hypothesis that F-actin and Arp2/3 complex are present on the Ste2 vesicle but below current detection methods. The lack of dependence on capping protein may be rationalized if a relatively low amount of force is required to move a vesicle that is free in the cytoplasm. Dendritic nucleation would occur and be less efficient than in the presence of capping protein. That the actin assembly becomes polarized to one side of the vesicle as the vesicle leaves the membrane may enable less efficient assembly to suffice for movement. To move objects in a synthetic system, Arp2/3-based dendritic nucleation needs capping protein to break symmetry and produce force (Akin and Mullins, personal communication), which seems consistent with our in vivo findings that capping protein is more important in the early stages of vesicle scission and movement.
The Nature of the Endocytic Pathway for Ste2
Relatively little is known about whether the pathway taken by a membrane receptor, such as Ste2, to the vacuole is the same as the pathways revealed by tracking membrane dyes, such as FM4-64, or actin patch components. Here, we observed similar results with Ste2-GFP and FM4-64 when we followed the movement of vesicles free in the cytoplasm in the presence and absence of capping protein, consistent with Ste2 following the common pathway. Alternatively, the prospect that a vesicle containing Ste2 would be able to exclude FM4-64 is unlikely, and so the Ste2 pathway may represent only a small part of what is observed with FM4-64. It remains possible that Ste2 vesicles form at the plasma membrane at locations distinct from actin patches, as suggested by immuno-EM results in which Ste2 at the plasma membrane was localized to foci distinct from actin patches (Mulholland et al., 1999).
One should note that our experiments involved constitutive, not ligand-induced, endocytosis of Ste2, which is consistent with events observed here with Ste2-GFP representing only a part of the entire endocytic pathway. Relatively little is known regarding the similarity of the molecular mechanisms for constitutive versus ligand-induced endocytosis. For Ste2, both types of endocytosis are known to occur and to depend on phosphorylation and ubiquitination of the same targeting motif (Hicke and Riezman, 1996; Hicke et al., 1998). In addition, the motility of Ste2 endosomes produced by constitutive as well as ligand-induced endocytosis is defective in las17ΔVCA mutants (Chang et al., 2003; Chang and Blumer, unpublished data).
The Relationship between Actin Cables and Patches
An early striking observation about the yeast actin cytoskeleton was that cables and patches are polarized in the same manner, toward sites of polarized cell growth (Adams and Pringle, 1984; Kilmartin and Adams, 1984). We now understand that cables are the tracks for myosin-powered movement of secretory vesicles to these sites (Pruyne et al., 2004), which explains why cables should be polarized in this manner. Actin patches seem to mediate endocytosis, and why endocytosis should occur at the same site as polarized growth is less clear. Perhaps retrieval and recycling of bulk membrane components is more efficient if the uptake occurs near the site of insertion.
An alternative hypothesis is that the assembly or movement of patches depends on cables. Patches have been observed to localize near the ends of cables and to move along cables (Karpova et al., 1998; Huckaba et al., 2004). This hypothesis predicts that loss of cables should impair patch movement. Capping protein mutants have a moderate loss of cables and a defect in patch movement, which seemed consistent. We tested the prediction by examining patch movement in conditional mutants that lose cables. Formins nucleate the unbranched actin filaments that compose the cable, so that a formin mutant can lose cables rapidly and completely on shift to restrictive temperature (Evangelista et al., 2002). We found that patch motility did not decrease, as predicted, but instead increased in such mutants at restrictive temperature. We suspect that the rapid disassembly of the actin cables increased the concentration of actin subunits, which increased the polymerization rate of filaments growing in the patches and thus the speed of patch movement, on average. These results are not inconsistent with a fraction of patches depending on cables for their movement (Huckaba et al., 2004), because that fraction may be small. Alternatively, association with a cable may slow patch movement, compared with movement of a free patch.
Supplementary Material
Acknowledgments
We are grateful to Jill Jeanblanc and Parker Seidel for computer programming and to Rick Heil-Chapdelaine and Scott Nelson for advice and assistance with light microscopy. The work was supported by National Institutes of Health Grants GM-47337 and GM-38542 (to J.A.C.) and Grant GM-44592 (to K.J.B.) and by an American Heart Association postdoctoral fellowship to K. K.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–10–0925) on January 4, 2006.
Abbreviations used: CP, capping protein; LatA, latrunculin A; MSD, mean squared displacement.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adams, A.E.M., and Pringle, J. R. (1984). Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98, 934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson, A. E., Shah, A. D., Elking, D., Karpova, T. S., and Cooper, J. A. (2002). Quantitative analysis of actin patch movement in yeast. Biophys. J. 82, 2333–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, F. S., Han, G. S., Carman, G. M., and Blumer, K. J. (2005). A WASp-binding type II phosphatidylinositol 4-kinase required for actin polymerization-driven endosome motility. J. Cell Biol. 171, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, F. S., Stefan, C. J., and Blumer, K. J. (2003). A WASp homolog powers actin polymerization-dependent motility of endosomes in vivo. Curr. Biol. 13, 455–463. [DOI] [PubMed] [Google Scholar]
- Duncan, M. C., and Payne, S. G. (2005). Cell biology: protein choreography. Nature 438, 571–573. [DOI] [PubMed] [Google Scholar]
- Engqvist-Goldstein, A. E., and Drubin, D. G. (2003). Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19, 287–332. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Pruyne, D., Amberg, D. C., Boone, C., and Bretscher, A. (2002). Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4, 260–269. [DOI] [PubMed] [Google Scholar]
- Hicke, L., and Riezman, H. (1996). Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell 84, 277–287. [DOI] [PubMed] [Google Scholar]
- Hicke, L., Zanolari, B., Pypaert, M., Rohrer, J., and Riezman, H. (1997). Transport through the yeast endocytic pathway occurs through morphologically distinct compartments and requires an active secretory pathway and Sec18p/N-ethylmaleimide-sensitive fusion protein. Mol. Biol. Cell 8, 13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke, L., Zanolari, B., and Riezman, H. (1998). Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 141, 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckaba, T. M., Gay, A. C., Pantalena, L. F., Yang, H. C., and Pon, L. A. (2004). Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 167, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., and O'Shea, E. K. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Sun, Y., and Drubin, D. G. (2003). A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 115, 475–487. [DOI] [PubMed] [Google Scholar]
- Kaksonen, M., Toret, C. P., and Drubin, D. G. (2005). A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123, 305–320. [DOI] [PubMed] [Google Scholar]
- Karpova, T. S., McNally, J. G., Moltz, S. L., and Cooper, J. A. (1998). Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J. Cell Biol. 142, 1501–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin, J. V., and Adams, A.E.M. (1984). Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98, 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K., Yamashita, A., Wear, M. A., Maeda, Y., and Cooper, J. A. (2004). Capping protein binding to actin in yeast: biochemical mechanism and physiological relevance. J. Cell Biol. 164, 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W. L., Oberle, J. R., and Cooper, J. A. (2003). The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 160, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel, T. P., Boujemaa, R., Pantaloni, D., and Carlier, M. F. (1999). Reconstitution of actin-based motility of Listeriaand Shigella using pure proteins. Nature 401, 613–616. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., 3rd, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Mulholland, J., Konopka, J., Singer-Kruger, B., Zerial, M., and Botstein, D. (1999). Visualization of receptor-mediated endocytosis in yeast. Mol. Biol. Cell 10, 799–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T. D., and Borisy, G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465. [DOI] [PubMed] [Google Scholar]
- Prescianotto-Baschong, C., and Riezman, H. (1998). Morphology of the yeast endocytic pathway. Mol. Biol. Cell 9, 173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D., Legesse-Miller, A., Gao, L., Dong, Y., and Bretscher, A. (2004). Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20, 559–591. [DOI] [PubMed] [Google Scholar]
- Rodal, A. A., Sokolova, O., Robins, D. B., Daugherty, K. M., Hippenmeyer, S., Riezman, H., Grigorieff, N., and Goode, B. L. (2005). Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat. Struct. Mol. Biol. 12, 26–31. [DOI] [PubMed] [Google Scholar]
- Stefan, C. J., and Blumer, K. J. (1999). A syntaxin homolog encoded by VAM3mediates down-regulation of a yeast G protein-coupled receptor. J. Biol. Chem. 274, 1835–1841. [DOI] [PubMed] [Google Scholar]
- Wen, K. K., and Rubenstein, P. A. (2005). Acceleration of yeast actin polymerization by yeast Arp2/3 complex does not require an Arp2/3-activating protein. J. Biol. Chem. 280, 24168–24174. [DOI] [PubMed] [Google Scholar]
- Wesp, A., Hicke, L., Palecek, J., Lombardi, R., Aust, T., Munn, A. L., and Riezman, H. (1997). End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 8, 2291–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, D., Podtelejnikov, A. V., Mann, M., and Li, R. (1997). The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr. Biol. 7, 519–529. [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Yang, H. C., and Pon, L. A. (2002). Actin cable dynamics in budding yeast. Proc. Natl. Acad. Sci. USA 99, 751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, M. E., Cooper, J. A., and Bridgman, P. C. (2004). Yeast actin patches are networks of branched actin filaments. J. Cell Biol. 166, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.