Abstract
Rck2 is a mitogen-activated protein kinase-activated protein kinase in yeast implicated in translational regulation. rck2Δ mutants are mildly sensitive to oxidative stress, a condition that causes dissociation of actively translating ribosomes (polysomes). In rck2Δ cells, polysomes are lost to an even higher degree than in the wild-type upon stress. Cells overexpressing the catalytically inactive rck2-kd allele are highly sensitive to oxidative stress. In such cells, dissociation of polysomes upon stress was instead greatly delayed. The protein synthesis rate decreased to a similar degree as in wild-type cells, however, indicating that in rck2-kd cells, the polysome complexes were inactive. Array analyses of total and polysome-associated mRNAs revealed major deregulation of the translational machinery in rck2 mutant cells. This involves transcripts for cytosolic ribosomal proteins and for processing and assembly of ribosomes. In rck2Δ cells, weakly transcribed mRNAs associate more avidly with polysomes than in wild-type cells, whereas the opposite holds true for rck2-kd cells. This is consistent with perturbed regulation of translation elongation, which is predicted to alter the ratio between mRNAs with and without strong entry sites at ribosomes. We infer that imbalances in the translational apparatus are a major reason for the inability of these cells to respond to stress.
INTRODUCTION
To adapt to changes in the environment and optimize its expression program, the cell mounts a range of responses on different levels. Through posttranslational reactions, signaling cascades receive and transmit the initiating signals. The best-investigated aspect of these responses is on the level of transcriptional initiation. Recent investigations using array technology have unraveled groups of genes that are activated by environmental stress and specific signaling pathways and transcription factors that control them (Gasch et al., 2000; Causton et al., 2001).
In addition to the long-term responses orchestrated at the promoter level, the expression program is rapidly changed at the level of preexisting mRNAs. This allows production of critical proteins to increase quickly and efficient overall optimization of energy expenditure by rapid down-regulation of noncritical proteins. Among posttranscriptional regulation mechanisms, changes in mRNA stability and mRNA loading onto ribosomes are the more prominent processes. These two processes are mechanistically interconnected; there is competition between translation initiation factors and ribonucleases for binding to mRNA. Association between the polyA tail and the 5′ end of actively translated mRNAs protects them against degradation. Quantitative assessments verify their importance for adaptation to environmental changes. Thus, a large fraction of overall regulation of protein production upon exposure to mating pheromone in yeast occurs at the translational level (MacKay et al., 2004). In mammalian cells, a major part of gene regulation after heat stress occurs at the level of mRNA stability (Fan et al., 2002). It has also been shown that Ras signaling confers more pronounced effects through mRNA recruitment to ribosomes than through transcriptional regulation (Rajasekhar et al., 2003).
There are indications that the stress-activated mitogen-activated protein (MAP) kinase (SAPK) pathways are involved in regulating both the stability and the loading onto ribosomes of mRNAs. In the fission yeast Schizosaccharomyces pombe, the SAPK Sty1 is required for stabilization of a set of mRNAs after oxidative stress; this effect also requires the RNA-binding protein Csx1 (Rodriguez-Gabriel et al., 2003). In mammalian cells, the SAPK p38, a homologue of Sty1, is required for stabilization of the tumor necrosis factor-α mRNA, and this effect is mediated through the downstream protein kinase MAP kinase-activated protein kinase (MAPKAPK)-2 (Mahtani et al., 2001). Furthermore, p38 signaling through MAPKAPK-3 leads to phosphorylation of translation eukaryotic elongation factor 2 kinase (Knebel et al., 2002). In yeast, homologues of MAPKAP kinases have been identified. The Saccharomyces cerevisiae MAPKAP kinase Rck2 binds to and is phosphorylated by the SAPK Hog1 upon hyperosmotic shock (Bilsland-Marchesan et al., 2000; Teige et al., 2001). Rck2 has been shown to phosphorylate translation elongation factor 2 (EF-2) in vitro (Melcher and Thorner, 1996) and in vivo (Teige et al., 2001). Osmotic shock causes a substantial down-regulation of the translation rate in yeast (Teige et al., 2001; Uesono and Toh, 2002), and Rck2 is implicated in the regulation of this phenomenon (Teige et al., 2001). This implies a role in regulation of translation for this kinase. Rck2 and its paralog Rck1 as well as the Hog pathway also contribute to oxidative stress resistance in budding yeast (Bilsland et al., 2004; Haghnazari and Heyer, 2004; Staleva et al., 2004). In S. pombe, the related protein Mkp2/Cmk2 contributes to resistance to oxidative stress caused by arsenite (Sanchez-Piris et al., 2002). The paralog Mkp1/Srk1 binds to Sty1 and is phosphorylated in a Sty1-dependent manner (Smith et al., 2002; Asp and Sunnerhagen, 2003) and is activated by oxidative stress (Smith et al., 2002).
There are thus reasons to think that MAPKAP kinases in yeast have a role in posttranscriptional regulation. In the present study, we investigate how mRNA association with actively translating ribosomes (polysomes) is affected by oxidative stress in wild-type and rck2 mutant cells. We do this by analyzing profiles of total and polysome-associated mRNA by array hybridization. Based on the changes in mRNA abundance in the different fractions, we find several aspects of the translational and mRNA processing machinery to be altered in rck2 mutants. This includes ribosomal proteins (RPs) and nucleolar proteins responsible for rRNA modification, ribosome assembly, and export. We conclude that mutation of RCK2 causes extensive effects on the state of the translation and mRNA processing machinery and that these effects are likely to be a major cause of cell death by oxidative stress in rck2 mutants.
MATERIALS AND METHODS
Yeast Strains and Culture, Plasmids, and Application of Oxidative Stress
S. cerevisiae strains were wild-type (wt) W303-1A or its rck2Δ derivative WΔRCK2-T (Dahlkvist and Sunnerhagen, 1994). Plasmids used were vector Yep13 (2μ, LEU2), Yep13-RCK2 (Dahlkvist and Sunnerhagen, 1994), vector pCM262 (Rodríguez-Navarro et al., 2002), and pCM262rck2-kd (mutated kinase-dead allele of RCK2 cloned into pCM262, putting the mutated gene under control of a doxycycline-repressible tet promoter; Bilsland-Marchesan et al., 2000). All experiments were performed in synthetic complete (SC) medium (Sherman, 1991). In experiments involving expression of the kinase-dead allele of RCK2, rck2-kd, cells were pregrown in medium containing 2 μg/μl doxycycline, to repress transcription of rck2-kd from the tet promoter. To permit induction of rck2-kd, cells were washed free of doxycycline and allowed to grow in doxycycline-free medium for at least 4 h. Unless stated otherwise, tert-butyl hydroperoxide (tBOOH) was used at a concentration of 0.8 mM.
Preparation of Total RNA
Logarithmically growing yeast (A595 = 0.6) cultures were harvested by centrifuging and resuspended in extraction buffer (1 mM EDTA, 0.1 M LiCl, 0.1 M Tris-HCl, pH 7.5, and 1% SDS). Glass beads and a phenol/CHCl3/isoamylalcohol (25:24:1) (PCI) mixture were added, and the cells were disrupted in a FastPrep 120 apparatus (Bio 101, Vista, CA) for 20 s at speed 5. After centrifugation at 4°C, 0.1 volume of 40% potassium acetate, pH 5.5, and PCI were added to the supernatant, and the samples were again vortexed as described above. The samples were centrifuged a second time, RNA was isolated from the supernatant by ethanol precipitation and was finally resuspended in 50 μl of RNase free water.
Separation of Polysomal RNA by Sedimentation Centrifugation
Cells were grown logarithmically to A595 = 0.6, cycloheximide was added to a final concentration of 0.1 mg/ml, and the culture was chilled for 10 min on ice. Cells were pelleted by centrifugation and resuspended in 0.04 culture volumes of 20 mM Tris-HCl, pH 7.5, 140 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 1% Triton X-100, 0.1 mg/ml cycloheximide, and 0.2 mg/ml heparin. After washing and resuspension in 0.01 culture volumes of the same buffer, glass beads were added, and cells were broken in a Bio 101 FastPrep for 20 s at speed 5. The lysates was cleared by centrifuging twice and stored at–70°C. Polysomes were separated by loading lysates onto 7–50% sucrose gradients followed by sedimentation ultracentrifugation in a Beckman SW41 rotor for 3 h at 35,000 rpm at 4°C. In the case of large-scale preparative isolations, a Beckman SW28 rotor was used instead, and centrifugation conditions were modified to 5 h at 28,000 rpm. Gradients were fractionated using isotonic pumping of 60% sucrose from the bottom, followed by recording of polysomal profiles by online UV detection. RNA was isolated from polysomal fractions through precipitation with 1 volume of 6 M guanidine thiocyanate and two volumes of ethanol, followed by storage overnight at –20°C. After centrifugation and washing with ethanol, the samples were extracted with acid phenol, followed by CHCl3 extraction. Heparin was eliminated by precipitating with LiCl to a final concentration of 1.5 M, storing overnight at –20°C, and centrifuging at 4°C. The pellet was washed twice with ethanol and dissolved in RNAse-free water. After addition of sodium acetate, pH 5.2, to a final concentration of 0.3 M, RNA was again ethanol precipitated and finally redissolved in RNAse-free water.
Metabolic Labeling of Protein
Cells were grown to A595 = 0.6 in SC with the appropriate supplements. One hour before labeling, the cells were washed in SC lacking sulfate and then cultured in SC lacking sulfate. Five microcuries of 35S-labeled methionine (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) was added to the culture, and samples were withdrawn at various time intervals thereafter, lysed by boiling in 10% trichloroacetic acid, precipitated by chilling on ice for 10 min, and finally passed through GF/C filters. Precipitable 35S radioactivity was quantitated by liquid scintillation counting.
Northern Blot
RNA was separated on formaldehyde-containing agarose gels, blotted to GeneScreen membranes (PerkinElmer Life and Analytical Sciences, Boston, MA), and detected with 32P-labeled DNA probes representing the respective open reading frame (ORF).
DNA Array Analysis
From RNA samples, cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) using a 1:1 mixture of oligo(dT) and random hexamer primers in the presence of 33P-labeled dCTP (100 μCi/reaction). The labeled cDNAs were hybridized to Yeast GeneFilters containing 6144 S. cerevisiae ORFs (Research Genetics, Huntsville, AL) in Microhyb solution (Research Genetics) at 42°C overnight and washed twice with 2× SSC, 1% SDS at 50°C and once with 0.5× SSC, 1% SDS at room temperature. Images were recorded in a PhosphorImager by exposure for 1 to 3 d. Signals representing DNA spots were identified using the Pathways 4 software package (Research Genetics); subtraction of local background and normalization of intensities for all arrays were done with the same software using default settings, such that the sum of all intensities for each array was equalized (set to 6144; the mean value for one spot was thus 1). Polysomal association factor (PAF) for an individual gene was defined as the ratio between the normalized intensities for the polysomal and the total RNA samples for any particular mRNA species. PAF values are thus >1 for mRNAs with an higher than average association with polysomes and <1 for those mRNAs with an association weaker than the average. Hierarchical clustering was done using Cluster 3.0 (de Hoon et al., 2004) and visualized with Mapletree (http://rana.lbl.gov/EisenSoftware.htm) freewares, respectively.
Assignment of gene products to functional or compartmental categories was done mainly based on Gene Ontology (GO) information in the Saccharomyces Genome Database (http://genome-www.stanford.edu/Saccharomyces/) or the Martinsried Institute for Protein Science (http://mips.gsf.de/genre/proj/yeast/index.jsp) for the respective proteins. Lists of genes assigned to the various categories are found in Supplemental Material 6.
RESULTS
The Kinase-Dead rck2-kd Allele Is Dominant for Sensitivity to Oxidative Stress
We have shown previously that rck2Δ mutants are moderately sensitive to oxidative stress (Bilsland et al., 2004); however, they are not sensitive to hyperosmotic shock (Dahlkvist and Sunnerhagen, 1994; Bilsland-Marchesan et al., 2000). By contrast, overexpression of the catalytically inactive rck2-kd allele does cause considerable sensitivity to hyperosmotic shock (Bilsland-Marchesan et al., 2000). We wanted to investigate the behavior of the rck2-kd allele under oxidative stress. In Figure 1, right, it is seen that, similar to hyperosmolarity, oxidative stress caused by tBOOH leads to impaired survival in wild-type W303-1A overexpressing rck2-kd (second row). This effect is more severe than in rck2Δ mutants (fifth row). Even in unstressed cells, overexpression of rck2-kd causes a moderate decrease of viability (Figure 1, top). It is noteworthy that in cells lacking the wild-type RCK2 allele, overexpression of rck2-kd has a less severe effect on viability (sixth row). In line with this observation, overexpression of the wild-type RCK2 allele further aggravates the adverse effects of rck2-kd (fourth row).
Figure 1.
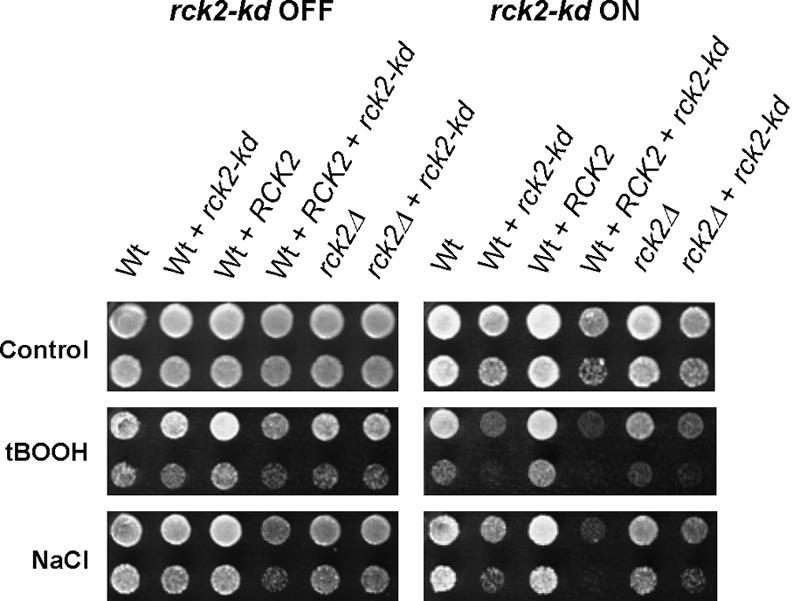
Sensitivity to oxidative stress of cells overexpressing rck2-kd. Wild-type W303-1A (wt) or WΔRCK2-T (Dahlkvist and Sunnerhagen, 1994; rck2Δ) were transformed with either the high-copy construct Yep13-RCK2 (Dahlkvist and Sunnerhagen, 1994) or with empty vector Yep13, and with either rck2-kd (mutated kinase-dead allele of RCK2 cloned into vector pCM262; Rodríguez-Navarro et al., 2002), putting the mutated gene under control of a doxycycline-repressible tet promoter (Bilsland-Marchesan et al., 2000) or with empty plasmid pCM262. Cells were grown to mid-log phase in liquid medium lacking leucine and uracil to select for both plasmids, and containing 2 μg/μl doxycycline to repress expression of rck2-kd. They were then washed in medium lacking doxycycline, serially diluted 1:5 and spotted on minimal medium with (rck2-kd OFF) or without (rck2-kd ON) doxycycline. Plates with no stress agent added and plates containing 0.4 M NaCl were incubated at 30°C; plates with tBOOH (0.05 mM) were incubated at 25°C.
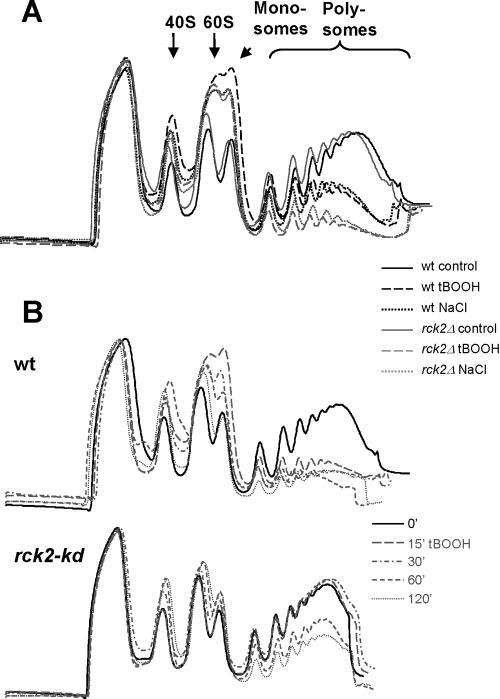
Dissociation of Polysomes upon Oxidative and Hyperosmotic Stress Is Accentuated in rck2Δ Cells and Prevented in Cells Overexpressing rck2-kd
We wanted to explore the reasons for the lethality of cells lacking RCK2 or overexpressing rck2-kd when exposed to oxidative stress. Because of the evidence linking Rck2 to translation, we followed the degree of association between mRNAs and ribosomes in wild-type and rck2 mutant cells, using sedimentation centrifugation to separate polysomes from free ribosomes (Figure 2). In line with expectations, polysome levels dropped distinctly within 15 min of exposure to tBOOH in wild-type cells and continued to stay low throughout the 2 h that they were monitored (Figure 2, A and B). For rck2Δ cells, the size of the polysomal fraction was similar to the wild-type in undisturbed cells. On addition of tBOOH, however, the polysome levels in rck2Δ cells dropped even lower than in wild-type cells (Figure 2A). A similar difference between rck2Δ and wild-type cells was also found at 15 min after hyperosmotic shock (Figure 2A).
Figure 2.
Dissociation of mRNAs from ribosomes upon stress. Cell lysates were prepared as described in Materials and Methods, loaded on a sucrose gradient (7–50%) in a SW41 ultracentrifuge tube. Contents were separated by centrifugation at 35 krpm for 3 h at 4°C and fractionated by isotonic pumping with online reading of A280. The bottom of the gradients, representing high-molecular-weight fractions, is oriented to the right. (A) Response to tBOOH or NaCl in rck2Δ cells. Cultures were harvested after growth for 30 min in the presence of 0.8 mM tBOOH or for 15 min after addition of NaCl to 0.4 M. Solid black line, wild-type cells, no stress; dashed black line, wild-type cells with tBOOH; dotted black line, wild-type cells after 0.4 M NaCl; solid gray line, rck2Δ cells, no stress; dashed gray line, rck2Δ cells with tBOOH; and dotted gray line, rck2Δ cells after 0.4 M NaCl. (B) Time course after exposure to tBOOH in cells expressing rck2-kd. Cells were exposed to 0.8 mM tBOOH for various times. Top, wild-type cells; bottom, cells expressing rck2-kd. Solid line, before addition of tBOOH; large-dashed line, 15 min after addition of tBOOH; dashed and dotted line, 30 min; small-dashed line, 60 min; and dotted line, 120 min.
However, the behavior of polysomes in cells overexpressing rck2-kd was radically different. When such cells were undisturbed, polysomal levels were again similar to the wild-type situation. However, after addition of tBOOH, polysomal level persisted unchanged for 30 min after exposure. Not until after 60 min was a drop in polysomal levels observable (Figure 2B). The same qualitative difference between the behavior of cells carrying the null allele or the rck2-kd allele was seen after hyperosmotic shock (our unpublished data).
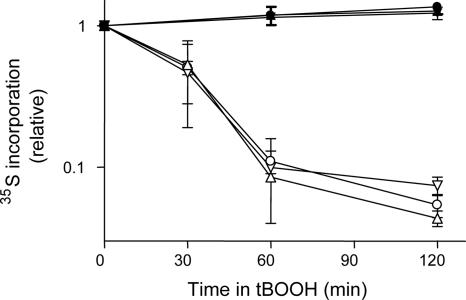
The Drop in Protein Synthesis Caused by Oxidative Stress Is Similar in Wild-Type and rck2 Mutants
The persistent association of mRNAs and ribosomes during oxidative stress in cells overexpressing rck2-kd could mean that protein synthesis continued at unaffected levels. Alternatively, ribosomes could be stuck in an unproductive association with mRNAs. To distinguish between these two possibilities, we measured total protein synthesis by metabolic labeling in wild-type and mutant cells upon exposure to tBOOH. As seen in Figure 3, protein synthesis continuously decreases throughout the 120-min period. The kinetics and degree of decrease were similar for all three strains. We conclude that the decrease of protein synthesis during oxidative stress is not significantly changed in cells expressing rck2-kd.
Figure 3.
Rate of incorporation of amino acid into protein upon oxidative stress. Cells were grown in the presence or absence of 0.8 mM tBOOH. Filled symbols, without tBOOH; empty symbols, in the presence of tBOOH. Circles, wild-type W303-1A (•/○); triangles, rck2Δ mutants (▴/▵); and inverted triangles, wild type expressing rck2-kd (▾/▿). Incorporation is expressed as fractions of the incorporation at time = 0. Data points are the average of two independent experiments with samples in triplicate. Error bars are SEM.
Oxidative Stress Caused by tBOOH Up-Regulates mRNAs for Synthesis of Sulfur-containing Amino Acids, and Rck2 Is Not Required for This Response
To appreciate the nature of the dissociation of polysomes in cells exposed to oxidative stress, and the apparent aberrations in this regard in rck2 mutant cells, we performed array analyses of total and polysomal mRNA levels. Total mRNA hybridization yields steady-state levels of individual mRNAs and mainly reflects changes at the transcriptional level. Hybridization with polysomal RNA fractions in addition reflects the degree of association with actively translating ribosomes. By comparing the abundance of an individual mRNA in the total and polysomal fractions, it is possible to estimate its degree of association with actively translating ribosomes (Kuhn et al., 2001; Arava et al., 2003; Preiss et al., 2003).
Oxidative stress causes increased expression of many gene products required for cellular defenses against its effects such as reactive oxygen species, glutathione depletion, and damaged proteins. The 30 genes most up-regulated in total mRNA by tBOOH stress in wt cells were analyzed by their GO annotation. The term most significantly associated with these genes was “sulfur metabolism,” and 23% (7/30) were annotated to this term (Table 1A) versus 0.7% in the entire genome (p = 1.9 × 10–10). We investigated the expression levels in this set (42 genes; required for biosynthesis of the amino acids cysteine and methionine as well as of glutathione, a major protector against oxidative stress) and in another set expected to be induced by tBOOH stress, those associated with the GO terms “oxidative stress response” (43 genes). Using geometric means, transcript levels increased on average 1.8-fold in the first group as a whole but only 1.3-fold in the second group (Supplemental Material 1).
Table 1.
Genes up-regulated by tBOOH stress in wild-type cells
| ORF | Gene | Total RNA stressed/unstresseda | PAF stressed/unstressedb | Polysomal RNA stressed/unstressedc |
|---|---|---|---|---|
| A. Genes most up-regulated on the transcriptional level by tBOOH stress in wild-type cells | ||||
| YML128C | MSC1 | 12.0 | 0.4 | 4.9 |
| YJR010W | MET3 | 11.0 | 0.8 | 8.4 |
| YOR120W | GCY1 | 8.3 | 0.6 | 5.8 |
| YOR382W | FIT2 | 8.3 | 1.1 | 9.1 |
| YFR053C | HXK1 | 8.2 | 0.8 | 6.3 |
| YLR303W | MET25 | 7.8 | 0.7 | 5.7 |
| YMR173W-A | 7.5 | 0.8 | 5.7 | |
| YDL124W | 7.5 | 1.0 | 7.8 | |
| YHL021C | FMP12 | 7.2 | 1.1 | 7.7 |
| YLR109W | AHP1 | 6.6 | 0.6 | 3.9 |
| YDR171W | HSP42 | 6.6 | 0.2 | 1.2 |
| YGL037C | PNC1 | 6.5 | 0.8 | 5.1 |
| YDL023C | SRF4 | 6.0 | 0.8 | 5.1 |
| YHR087W | 5.8 | 0.6 | 3.4 | |
| YMR173W | DDR48 | 5.6 | 0.9 | 5.3 |
| YBR126C | TPS1 | 5.3 | 0.7 | 3.5 |
| YNL134C | 5.1 | 1.0 | 5.3 | |
| YDR074W | TPS2 | 5.0 | 0.8 | 3.9 |
| YLR327C | 4.8 | 0.5 | 2.3 | |
| YJL101C | GSH1 | 4.6 | 0.5 | 2.2 |
| YKL001C | MET14 | 4.5 | 1.2 | 5.3 |
| YHR104W | GRE3 | 4.4 | 1.4 | 6.0 |
| YMR169C | ALD3 | 4.1 | 1.1 | 4.4 |
| YFR030W | MET10 | 4.1 | 1.4 | 5.6 |
| YML100W | TSL1 | 4.1 | 1.4 | 5.9 |
| YJL159W | HSP150 | 4.0 | 0.5 | 1.9 |
| YLR205C | HMX1 | 3.9 | 0.6 | 2.5 |
| YDR253C | MET32 | 3.9 | 0.4 | 1.6 |
| YGR209C | TRX2 | 3.9 | 1.3 | 4.9 |
| YJL060W | BNA3 | 3.8 | 1.6 | 6.0 |
| B. Genes most up-regulated at polysomes by tBOOH stress in wild-type cells | ||||
| YDL222C | FMP45 | 3.4 | 2.9 | 9.6 |
| YOR382W | FIT2 | 8.3 | 1.1 | 9.1 |
| YJR010W | MET3 | 11.0 | 0.8 | 8.4 |
| YDL124W | 7.5 | 1.0 | 7.8 | |
| YHL021C | FMP12 | 7.2 | 1.1 | 7.7 |
| YLR178C | TFS1 | 2.9 | 2.3 | 6.6 |
| YPL162C | 0.9 | 7.2 | 6.4 | |
| YCL040W | GLK1 | 2.5 | 2.6 | 6.4 |
| YFR053C | HXK1 | 8.2 | 0.8 | 6.3 |
| YBR213W | MET8 | 3.4 | 1.8 | 6.1 |
| YHR104W | GRE3 | 4.9 | 1.4 | 6.0 |
| YJL060W | BNA3 | 3.8 | 1.6 | 6.0 |
| YML100W | TSL1 | 4.1 | 1.4 | 5.9 |
| YOL151W | GRE2 | 2.6 | 2.2 | 5.8 |
| YOR120W | GCY1 | 8.3 | 0.7 | 5.8 |
| YMR173W-A | 7.5 | 0.8 | 5.7 | |
| YLR303W | MET25 | 7.8 | 0.7 | 5.7 |
| YCL042W | 1.2 | 4.9 | 5.7 | |
| YFR030W | MET10 | 4.1 | 1.4 | 5.6 |
| YIL074C | SER33 | 2.2 | 2.5 | 5.4 |
| YLR302C | 3.1 | 1.8 | 5.4 | |
| YNL134C | 5.1 | 1.0 | 5.3 | |
| YMR173W | DDR48 | 5.6 | 0.9 | 5.3 |
| YKL001C | MET14 | 4.5 | 1.2 | 5.2 |
| YGL037C | PNC1 | 6.5 | 0.8 | 5.1 |
| YDL023C | SRF4 | 6.0 | 0.8 | 5.1 |
| YGR209C | TRX2 | 3.9 | 1.3 | 4.9 |
| YML128C | MSC1 | 12.0 | 0.4 | 4.9 |
| YBR149W | ARA1 | 3.2 | 1.5 | 4.7 |
| YOL053C-A | DDR2 | 3.2 | 1.5 | 4.7 |
Genes annotated to the term `synthesis of sulfur-containing amino acids' are bold, and genes annotated to `stress respons' are italicized. Relative changes upon oxidative stress are indicated in lettered footnotes.
Changes in total mRNA pool.
Changes in polysomal association factor (=column c/column a)
Changes in polysomal mRNA pool.
The PAF values (see Materials and Methods; DNA array analysis) for the genes involved in sulfur metabolism on average increased by a factor 1.2 upon tBOOH stress, for a total increase in the polysomal fraction of 2.1 (Supplemental Material 1A). A corresponding list of the mRNAs with the highest increase in PAF value after stress (Table 1B) gives a similar picture as that for total mRNAs (Table 1A). The effects on the transcriptional and translational levels thus reinforce each other for this gene group, with transcriptional regulation as the major factor.
Both in rck2Δ and rck2-kd cells, the degree of transcriptional induction and PAF value upon stress was similar to the situation in wt cells with respect to genes involved in metabolism of sulfur-containing amino acids or oxidative stress response. Hence, in general Rck2 is not required for those responses. This pattern is exemplified by MET3, where induction is at least as high in the mutants (Figure 4). However, in some cases, such as CYS3, the transcriptional induction was depressed in rck2-kd cells (Figure 4).
Figure 4.
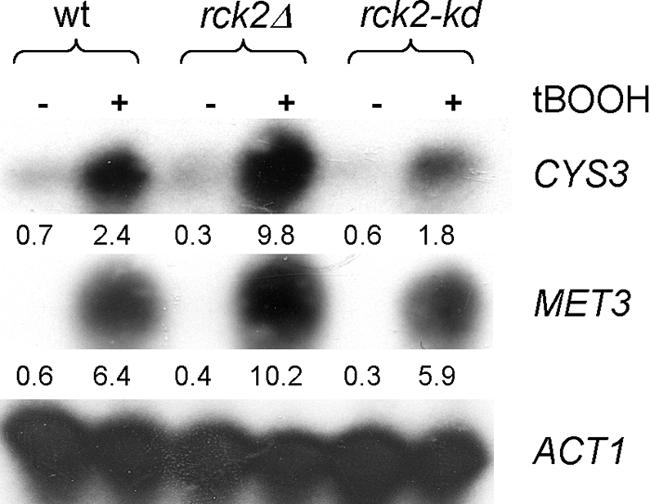
Transcriptional induction of genes required for sulfur-containing amino acid synthesis on oxidative stress. Northern blot analysis of total RNA from wild-type and mutant cells with and without exposure to tBOOH, using CYS3 and MET3 probes as indicated. Bottom, ACT1 loading control. Numbers below CYS3 and MET3 rows are the signal intensities for the corresponding experiments obtained from array hybridizations.
Mutation of RCK2 Affects Expression of Cytoplasmic but Not Mitochondrial RPs
We further wanted to compare the transcript pattern in rck2 mutants with wild-type cells in undisturbed cells. Inspection of the GO terms for genes whose transcription levels were most up-regulated in rck2Δ mutants compared with wild-type cells revealed that cytoplasmic RPs were prominent in this group. Among the 50 most up-regulated genes in rck2Δ cells compared with the wild-type, 29 encode cytoplasmic RPs (p = 2.4 × 10–34). This prompted us to explore the entire set of RPs in rck2 mutants. Genes encoding cytoplasmic or mitochondrial RPs, including translation factors, clearly fell into distinct groups. Using geometric mean, we found that the total mRNA levels for cytoplasmic RPs (181 genes) under unstressed conditions is elevated on average 1.5-fold in rck2Δ and 1.4-fold in rck2-kd cells (Figure 5A; our unpublished data). This increase in the amount of total mRNAs encoding RPs is not fully reflected at the polysomal level, because the PAF values for these mRNAs are lower than in wild-type cells.
Figure 5.
Changes in transcription of genes for ribosomal proteins. Logarithmic x-axis shows signal intensity from array analysis of total mRNA samples relative to the respective control. Arrows indicate the position for genes with no change. Open bars, cytosolic ribosomal proteins; filled bars, mitochondrial ribosomal proteins. (A) unstressed rck2Δ relative to unstressed wild-type cells. (B) stressed wild-type relative to unstressed wild-type cells.
By contrast to cytoplasmic RPs, the levels of mRNAs for mitochondrial RPs (86 genes) are not changed in rck2Δ or rck2-kd cells. A corresponding separation between the patterns for cytoplasmic and mitochondrial RPs is observed for the response to tBOOH-induced stress. A substantial down-regulation of total transcript levels for cytosolic RPs was observed to stress, on average 2.5-fold, whereas transcripts for mitochondrial RPs were unchanged or slightly increased (Figure 5B). The observed separate control of the cytoplasmic and mitochondrial RPs was confirmed by hierarchical cluster analysis, which efficiently sorts the RP genes into two distinct sets (Supplemental Material 2).
Deregulation on the Translational Level of Transcripts for Structural and Modifying Components of Cytosolic Ribosomes in rck2 Cells
We analyzed how the association with polysomes shifts upon tBOOH stress for different classes of mRNAs. In wild-type cells, the 30 genes that increase their PAF values the most upon tBOOH stress represent a variety of functions, including several hexose and vitamin transporters (Table 2). By contrast, in cells expressing rck2-kd, among the genes with the highest increase in PAF value in rck2-kd cells, 33% (11/30) are annotated to the term “ribosome biogenesis and assembly” versus 3% in the entire genome (p = 1.5 × 10–9). These encode proteins mainly localized in the nucleolus and include rRNA-modifying enzymes (Table 2). The PAF value changes upon stress in all strains for all of these genes are presented in Supplemental Material 3, where it can be seen that both genes encoding nucleolar proteins and cytosolic RP genes are differently affected in rck2 mutants.
Table 2.
Genes with the highest increase in PAF upon stress
| Wild type
|
rck2Δ
|
rck2-kd
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF | Gene | Description | Fold PAF increase | ORF | Gene | Description | Fold PAF increase | ORF | Gene | Description | Fold PAF increase |
| YPL162C | 7.2 | YIL052C | RPL34B | Ribosome (cytosolic) | 10.8 | YNR040W | 6.2 | ||||
| YDL229W | SSB1 | Heat-shock protein | 5.5 | YGL147C | RPL9A | Ribosome (cytosolic) | 8.0 | YLR343W | 5.8 | ||
| YCL042W | 4.9 | YLR325C | RPL38 | Ribosome (cytosolic) | 7.6 | YCL042W | 5.1 | ||||
| YMR011W | HXT2 | Hexose transporter | 3.7 | YDL191W | RPL35A | Ribosome (cytosolic) | 7.6 | YHR081W | LRP1 | rRNA processing | 4.8 |
| YDR101C | ARX1 | Ribosomal biogenesis | 3.6 | YBL027W | RPL19A | Ribosome (cytosolic) | 7.5 | YOR181W | LAS17 | Actin assembly | 4.3 |
| YDR411C | 3.5 | YOR096W | RPS7A | Ribosome (cytosolic) | 7.5 | YKR002W | PAP1 | Polyadenylation | 4.2 | ||
| YAL040C | CLN3 | Cyclin | 3.5 | YER056C-A | RPL34A | Ribosome (cytosolic) | 7.2 | YNL009W | IDP3 | Isocitrate dehydrogenase | 4.0 |
| YOR161C | PNS1 | 3.4 | YLR062C | Overlaps with RPL22A | 6.9 | YMR011W | HXT2 | Hexose transporter | 4.0 | ||
| YDL213C | NOP6 | rRNA processing | 3.3 | YNL174W | 6.4 | YKL009W | MRT4 | rRNA processing | 3.9 | ||
| YDR133C | 3.2 | YKL009W | MRT4 | rRNA processing | 6.4 | YGR103W | NOP7 | rRNA processing | 3.8 | ||
| YER091C | MET6 | Methionine biosynthesis | 3.2 | YJL177W | RPL17B | Ribosome (cytosolic) | 6.3 | YLR198C | 3.7 | ||
| YLR269C | 3.0 | YKL180W | RPL17A | Ribosome (cytosolic) | 6.3 | YDL222C | FMP45 | 3.7 | |||
| GL209W | MIG2 | Transcription factor | 2.9 | YDL208W | NHP2 | rRNA processing | 6.1 | YML093W | UTP14 | rRNA processing | 3.6 |
| YDL222C | FMP45 | Cell wall | 2.9 | YKL156W | RPS27A | Ribosome (cytosolic) | 5.9 | YLR302C | 3.4 | ||
| YLL061W | MMP1 | Amino acid permease | 2.8 | YDR449C | UTP6 | rRNA processing | 5.8 | YOL077C | BRX1 | rRNA processing | 3.4 |
| YKL043W | PHD1 | Transcription factor | 2.7 | YPL079W | RPL21B | Ribosome (cytosolic) | 5.7 | YLR042C | 3.4 | ||
| YKL060C | FBA1 | Glycolysis, gluconeogenesis | 2.7 | YJL188C | Overlaps with RPL39 | 5.6 | YLR197W | SIK1 | rRNA processing | 3.4 | |
| YDR476C | 2.7 | YKL172W | EBP2 | rRNA processing | 5.5 | YKR081C | RPF2 | rRNA processing | 3.3 | ||
| YGL215W | CLG1 | Cyclin-like | 2.7 | YPL090C | RPS6A | Ribosome (cytosolic) | 5.5 | YGL209W | MIG2 | Transcription factor | 3.3 |
| YGR162W | TIF4631 | eIF-4γ | 2.6 | YDR184C | BAT1 | Nuclear | 5.4 | YOR004W | rRNA processing | 3.2 | |
| YCL040W | GLK1 | Glucokinase | 2.6 | YHR049W | FSH1 | rRNA processing | 5.3 | YDR365C | ESF1 | rRNA processing | 3.2 |
| YDR324C | UTP4 | rRNA processing | 2.6 | YOR310C | NOP58 | rRNA processing | 5.3 | YHR196W | UTP9 | rRNA processing | 3.2 |
| YJR027W | Ty element | 2.6 | YHR152W | SPO12 | nucleolar | 5.2 | YKL097C | 3.2 | |||
| YLR197W | SIK1 | rRNA processing | 2.6 | YHR099W | TRA1 | HAT complex | 5.1 | YMR321C | 3.1 | ||
| YBR092C | PHO3 | Phosphatase | 2.6 | YGR085C | RPL11B | Ribosome (cytosolic) | 5.1 | YDR324C | UTP4 | rRNA processing | 3.1 |
| YDR483W | KRE2 | Protein glycosylation | 2.6 | YLR197W | SIK1 | rRNA processing | 5.1 | YJL163C | 3.1 | ||
| YDR072C | IPT1 | Sphingolipid metabolism | 2.6 | YLR293C | GSP1 | rRNA processing | 4.9 | YNL175C | NOP13 | rRNA processing? | 3.1 |
| YDL134C | PPH21 | Protein phosphatase | 2.6 | YJL189W | RPL39 | Ribosome (cytosolic) | 4.8 | YGR192C | TDH3 | Glycolysis, gluconeogenesis | 3.1 |
| YNL110C | NOP15 | Ribosome biogenesis | 2.6 | YGR148C | RPL24B | Ribosome (cytosolic) | 4.7 | YDR184C | BAT1 | 3.0 | |
| YIL074C | SER33 | Serine biosynthesis | 2.5 | YGL078C | DBP3 | rRNA processing | 4.7 | YKL043W | PHD1 | 3.0 | |
Genes for proteins involved in ribosomal biogenesis and rRNA processing are bold; cytosolic ribosomal proteins are italicized.
A corresponding analysis of the mRNAs exhibiting the greatest shift away from polysomes after stress (i.e., with the largest decrease in PAF value) reveals that in wild-type cells, 27% (8/30) of mRNAs are annotated to the term “cytosolic large ribosomal subunit” versus 1.2% in the total genome (p = 8.7 × 10–8). By contrast, in rck2Δ or rck2-kd cells, no genes related to RPs are found in the corresponding position. Examination of the entire set of mRNAs encoding cytosolic RPs reveals that on average, no prominent change of PAF value occurs upon stress in wild-type cells. However, in rck2 mutants, particularly rck2Δ mutants, PAF values increase strongly upon stress for the entire class of mRNAs encoding cytosolic RPs. Thus, it seems that down-regulation of RPs under tBOOH stress occurs chiefly at the transcriptional level in wild-type cells, and that polysomal association of the corresponding mRNAs is perturbed in rck2 mutants.
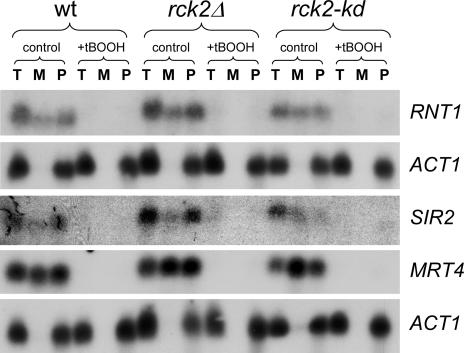
We performed Northern analysis of some of the mRNAs with products involved in rRNA modifications and ribosome assembly, with localization in the nucleolus, which were predicted by array results to be enriched at polysomes in rck2 mutants (Figure 6). For all nucleolar genes shown (RNT1, SIR2, and MRT4), full-length mRNAs were almost completely absent after tBOOH treatment, indicating extensive mRNA degradation. This is not surprising given that mRNAs coupled to functions required for rapid cell growth, such as protein synthesis, are known to be destabilized under severe stress conditions. However, this was not fully reflected in the array data, and we interpret this to mean that these mRNAs are degraded to fragments still long enough to hybridize efficiently on arrays. We thus had to restrict the Northern analysis to unstressed cells. It should be noted that other mRNAs not belonging to this category, such as ACT1, MET3, and CYS3, were not degraded (Figures 4 and 6). Both RNT1 and SIR2 are predicted by array results to be more abundant in the polysomal fraction in rck2Δ cells than in the wild type and less abundant in rck2-kd cells. This was confirmed by Northern analysis (Figure 6, rows 1 and 3).
Figure 6.
Northern blot analysis of total RNA, monosomal, and polysomal fractions for genes encoding nucleolar proteins. RNA was isolated from untreated or tBOOH-treated wild-type or mutant cells and probed with ORFs representing genes for nucleolar proteins as indicated. T, total RNA; M, monosomal RNA fraction; P, polysomal RNA fraction. The relevant ACT1 loading controls are shown directly under the respective rows. Note the consistent absence of ACT1 in the monosomal fractions.
An interesting case is MRT4, encoding a nucleolar protein required for mRNA decay (Zuk et al., 1999). In Figure 6, row 4, it can be seen that MRT4 mRNA is enriched in the monosomal fraction in undisturbed rck2Δ cells and even more in cells expressing rck2-kd.
Abundant mRNAs Associate with Polysomes in Cells Expressing rck2-kd, whereas the Opposite Trend Is Seen in rck2Δ Cells
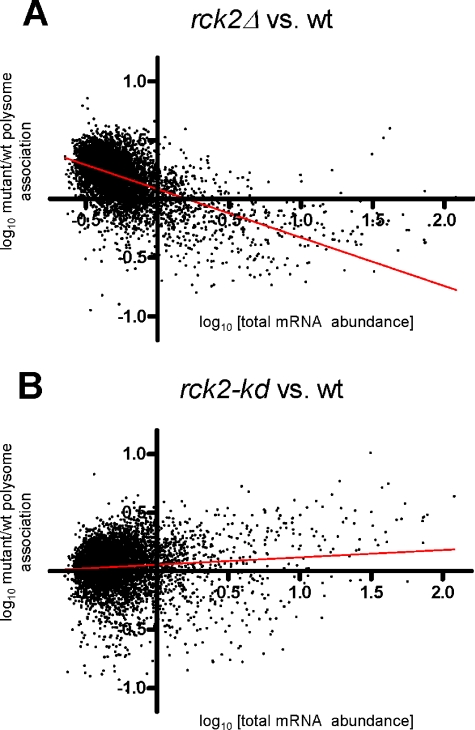
The stress response analyzed by polysomal profiles seemed opposite in rck2Δ versus rck2-kd cells. To describe main differences in the composition of polysome-associated mRNA in stressed cells of the two genetic setups, we investigated which mRNAs had the highest ratio between PAF values in rck2-kd cells (with remaining high polysomal content) compared with rck2Δ cells (with low polysomal content). It was clear that most of genes with this property are very highly expressed, including Ty elements and glycolytic enzymes (our unpublished data). We wanted to see the generality of this finding and plotted the PAF values in mutants, normalized for the corresponding value in wild-type cells, as a function of expression level. As seen in Figure 7A, there is a strong negative correlation between expression level and relative PAF value in rck2Δ mutants, i.e., the more abundant an mRNA, the more the presence of Rck2 tends to associate it with polysomes. When the gene set is subdivided into smaller intervals of expression levels and analyzed separately, the trend remains (Supplemental Material 4). For rck2-kd mutants, there is instead a weaker but statistically significant trend of the opposite (Figure 7B). A separate analysis of the Ty elements, which are generally highly transcribed, similarly shows markedly higher PAF values for strongly transcribed Ty elements in cells expressing rck2-kd and lower for the same elements in rck2Δ mutants (Supplemental Material 5).
Figure 7.
Correlation between polysomal association and expression level is altered in rck2 mutants. Scatter diagram of array data from unstressed cells. Relative PAF value (PAFmutant/PAFwild-type) on y-axis; total mRNA abundance in wild-type cells on x-axis. Note logarithmic scale on both x- and y-axes. Linear regression trend line is shown in each diagram. Regression data for rck2Δ versus wild type: slope = –0.41 ± 0.008, r = –0.55, ***p < 0.0001; for rck2-kd versus wild type: slope = –0.06 ± 0.008, r = 0.09, ***p < 0.0001.
DISCUSSION
Regulation of Gene Groups in Wild-Type and rck2 Mutant Cells
In this study, we have analyzed the responses to tBOOH-induced oxidative stress in S. cerevisiae cells on the transcriptional level, and on the level of association to actively translating ribosome complexes (polysomes). If we consider first the responses of the wild-type cell for gene products expected to have an active role in protection against oxidative stress, and that become up-regulated, we can make the following generalizations: 1) Induction of mRNAs for proteins involved in sulfur metabolism is apparent. This has been observed under Cd2+ stress (Fauchon et al., 2002) and can be ascribed to the need for increased glutathione synthesis. 2) Most of the changes occur on the level of total mRNA, whereas increased polysomal association makes a minor contribution. In a study investigating, a wide range of experimental conditions, it was likewise found that changes on the transcriptional and translational levels tend to occur in the same direction for a particular gene (Preiss et al., 2003). 3) As a rule, Rck2 is not required for induction of these gene products.
To optimize energy conservation and timing of the response to stress, the cell must rapidly down-regulate the translational apparatus, which represents a significant fraction of the total energy expenditure of the cell. Thus, hyperosmotic shock will lead to repression of many gene products directly required for translation (Varela et al., 1992; Mager and Varela, 1993; Gasch et al., 2000). Among proteins involved in translation, three regulatory patterns are clearly discernible in this study: 1) mitochondrial RPs, 2) cytosolic RPs, and 3) proteins located in the nucleolus and involved in modification of rRNA, assembly and transport of newly synthesized ribosomes. The mRNAs in the mitochondrial RP group are not affected by mutation in RCK2. This may mean that Rck2, a cytoplasmic protein, is excluded from mitochondria. Mitochondrial RP mRNAs are also not affected by tBOOH treatment, which could mean that tBOOH does not penetrate into the mitochondrial compartment. The mRNAs encoding cytosolic RPs increase in rck2Δ and rck2-kd mutants. They also decrease in abundance by oxidative stress. It is well established that these genes are under tight transcriptional control and are down-regulated by various stress conditions (Warner, 1999), although there is also evidence for regulation at the level of mRNA stability (Yin et al., 2003). In one study investigating shift from fermentable to nonfermentable carbon source (Kuhn et al., 2001), it was found that mRNAs for cytosolic RPs were selectively dissociated from polysomes; however, this was not clearly seen in our results. Likewise, other workers have reported that the main level of RP regulation in yeast is on transcription (Zaragoza et al., 1998; Powers and Walter, 1999), in contrast to mammalian cells. Down-regulation of the total transcript levels for RPs has been seen earlier in array studies for many types of stress, such as glucose deprivation (Kuhn et al., 2001), hyperosmotic shock, hydrogen peroxide, heat, acid, and stationary phase (Gasch et al., 2000; Causton et al., 2001; Segal et al., 2003; O'Rourke and Herskowitz, 2004), but the distinction between cytosolic and mitochondrial ribosomes has not always been made clear. For proteins located in the nucleolus and involved in modification of rRNA, assembly and transport of newly synthesized ribosomes, no net change of mRNA abundance takes place either upon stress or mutation of RCK2, and so transcriptional regulation is not the major factor here. Instead, these mRNAs display changes in polysomal association: For some genes, their PAF value is increased in rck2Δ mutants and decreased in rck2-kd (cf. Figure 6). For the majority in this group, PAF values increase upon stress in rck2 mutants, rather than to decrease as in the wild type.
As a first approach to understand the changes in mRNA regulation that take place in rck2 mutants, it is natural to compare with previous investigations of global transcription patterns. Because Rck2 binds to Hog1 and is downstream of Hog1 with respect to signaling upon hyperosmotic and oxidative stress (Bilsland-Marchesan et al., 2000; Bilsland et al., 2004), one could hypothesize that changes of the transcript pattern in rck2 mutants would simply be a reflection of changes in Hog1 activity. If so, transcript profiles of rck2 and hog1 mutants would be similar. This is clearly not the case, however. The most striking feature of transcript profiles in rck2 mutants, transcriptional up-regulation of genes encoding cytoplasmic RPs, does not occur in hog1 mutants (O'Rourke and Herskowitz, 2004). Many RPs are repressed after osmotic shock, including RPS27B. HOG1 is required for reactivation of these genes during the recovery phase; however, they are not deregulated in unstressed hog1 mutants as they are in unstressed rck2 mutants (Uesono and Toh, 2002; O'Rourke and Herskowitz, 2004). In this context, it is noteworthy that genes involved in synthesis of sulfur-containing amino acids are at least as strongly induced by oxidative stress in rck2 mutants as they are in the wild type. In addition to Hog1, Rck2 could be regulated by another pathway; indeed, there is some residual phosphorylation of Rck2 in pbs2 mutants upon oxidative or hyperosmotic stress (Bilsland et al., 2004). The TOR pathway has a strong influence on the activity level of the translational apparatus (Powers and Walter, 1999), and this pathway is a candidate for such additional regulation of Rck2.
Role of Rck2 in Regulation of Translation
These considerations point at other mechanisms than transcriptional regulation as mainly responsible for the effects seen in rck2 mutants. Previous work in vitro (Melcher and Thorner, 1996) and in vivo (Teige et al., 2001) has implicated eEF-2 as a direct substrate of Rck2. Phosphorylation of eEF-2 at either of two conserved threonine residues is known to be inhibitory and will result in stalling of elongation (Redpath et al., 1993). Several observations in this article can be interpreted in the light of effects on the translation elongation step. First, more MRT4 messenger was found in the monosomal fraction in rck2 mutants than in wild-type cells. This would be consistent with a slowing down of translation once the mRNA has been loaded on the first ribosome. Second, inhibition of elongation can cause increased accumulation of polysomes (Hovland et al., 1999). The dissociation of polysomes in stressed rck2Δ mutants and the persistence of polysomes in oxidative stress in cells expressing rck2-kd might thus be ascribed to increased stalling of elongation when cells express rck2-kd, and decreased stalling in rck2Δ mutants. Third, stalling of translational elongation is predicted to promote translation of mRNAs with weak binding ability to initiation factors or the small ribosomal subunit at the expense of mRNAs with stronger binding properties (Walden and Thach, 1986). The reason is that, when binding to initiation factors is no longer limiting, the rate of entry of mRNAs at ribosomes is solely determined by their abundance. The observation in this article that less abundant mRNAs associate more avidly with actively translating ribosomes in rck2Δ mutants than in wild-type cells (Figure 7) clearly points in this direction. By contrast, we find no correlation between changes in polysomal association and transcript length (our unpublished data).
Given the strict control of the amounts of ribosomal components, it is perplexing that there is such a widespread deregulation of mRNAs for cytosolic RPs in rck2 mutants. Given the broad imbalances within the translational apparatus that we observe in such mutants, it is natural to interpret this as a primary effect on translation: decreased PAF values could result from an effect on the translation elongation step, and an increase of transcription of cytosolic RP genes could occur as a compensatory mechanism. The end result at the polysomal level, and by inference on the protein level, is not drastically different from the wild type.
The rck2-kd allele has unusual properties. In some respects, it behaves as a mirror image of the null allele: upon stress, polysomes persist for a long time in the former, whereas they almost vanish in the latter. Also, the trend line for polysomal association as a function of expression level is inversed in rck2-kd-expressing cells relative to rck2Δ cells (Figure 7B). An important observation is that overexpression of rck2-kd in a strain lacking wild-type Rck2 has only marginal effects on survival, whereas simultaneous overexpression of rck2-kd and wild-type RCK2 further deteriorates survival in oxidative stress (Figure 1). One model to account for this is that effects of rck2-kd are mediated through hyperactivation of the wild-type Rck2 protein. For example, in oligomeric Rck2 involving Rck2-kd subunits, negative autoregulation in trans could be blocked. Alternatively, Rck2 could have both a role as a kinase (in dissociation of polysomes) and a structural role (in loading ribosomes onto mRNAs). This could explain why highly expressed mRNAs remain polysome-associated in cells expressing rck2-kd. A third possibility is that the Rck2 protein would harbor both activating and inhibitory functions; such a situation for the MAP kinase Kss1 leads to kss1Δ mutants being totally defective for filamentous growth, whereas certain point mutated kss1 alleles are hyperactive for filamentous growth (Madhani et al., 1997).
Regulation of Polysomal Disassembly
Translational control is intimately coupled to regulation of mRNA stability. Thus, yeast mutants with deregulated mRNA degradation also have lost translational control (Holmes et al., 2004). The best-characterized cis control elements for mRNA stability are A/U-rich elements (AREs). In mammalian cells, p38-mediated signaling through the Rck2 homologue MAPKAPK-2 induces stabilization of mRNAs containing AREs, whereas a kinase-dead version of the same protein has a destabilizing effect on mRNA (Winzen et al., 1999). Under severe stress conditions, stress-induced granules in animal, plant, and fission yeast cells, occur shortly after different stress treatments, including oxidative stress, heat shock, and hyperosmotic shock. They contain mRNA and RNA-binding proteins and are thought to act as sorting points for mRNAs to be degraded or maintained for later translation after recovery from stress (Dunand-Sauthier et al., 2002). In budding yeast, inactive mRNAs are located to cytoplasmic processing bodies, where they can undergo decapping and degradation (Sheth and Parker, 2003). The exact relationship between stress granules and processing bodies is presently not clear. We observed that mRNAs encoding nucleolar proteins were hardly detectable as full-length species after stress by Northern analysis, yet in rck2 mutants they occurred as increasingly associated with polysomes. A rationalization of these findings is that they could persist as partially degraded fragments in mutants after stress. This might be related to a defect in mRNA degradation in rck2 mutants; however, we have not been able to establish the nature of such a hypothetical defect (our unpublished data). That mRNA stability is the major regulatory factor for proteins involved in ribosome biogenesis and modification proteins was also found in a previous array study (Grigull et al., 2004).
The behavior of cells expressing rck2-kd, where polysomes persist after oxidative and hyperosmotic stress even though protein synthesis is shut down, is reminiscent of mammalian cells exposed to the polysome-stabilizing drug emetine (Kedersha et al., 2000). In such cells, the formation of stress granules is inhibited. The authors interpret this as interdependency between polysomes and stress granules, because components (mRNA and proteins) can move between these two pools. Likewise, a dominant mutation in the mammalian TIA-1 protein (homologous to S. cerevisiae Pub1) inhibits formation of stress granules (Kedersha et al., 1999). The opposite is seen with the polysome-destabilizing drug puromycin, which is able to induce formation of stress granules in the absence of external stress (Kedersha et al., 2000). MAPKAPK-2 inhibits degradation of mRNAs in stress granules by phosphorylating the protein tristetraprolin (Stoecklin et al., 2004). To some extent, this mimics the situation in rck2Δ cells, where the polysomal peaks decline more drastically upon stress. The protein homologous to tristetraprolin in budding yeast is Tis11, which has a role in regulating mRNA stability in the iron metabolism regulon (Puig et al., 2005). These observations suggest that Rck2 acts along the same pathway of polysome disassembly and formation of stress granules as TIA-1 and tristetraprolin. It implies that one function of Rck2 is to counteract polysomal dissociation and by extension formation of stress granules. We speculate that Rck2 targets one or several proteins involved in the dynamic balancing of mRNA between polysomes and stress granules. The dominant allele rck2-kd, which blocks polysome disassembly, would then be predicted to also block formation of stress granules.
What could be the downstream targets responsible for the effect of RCK2 on polysomal abundance after stress? As we have seen, it is possible to rationalize many of the phenomena observed in this work through the known Rck2 target eEF-2 and its effect on translation elongation. However, in view of the apparent complex nature of regulation of stress granules (processing bodies), their interdependency with polysomes, and the involvement of mRNA degradation pathways in this regulation, we find it likely that there are additional targets, presumably with a role in mRNA metabolism.
Supplementary Material
Acknowledgments
Support for this work was provided by grants from the Swedish Cancer Fund (2163-B03-14XAB) and the Swedish Research Council for Science and Technology (2003-3189) to P. S. and by the Czech Science Foundation (204/03/1487) to M. P. S. S. was the recipient of a fellowship from the Foundation for Strategic Research, T. M. was supported in part by a Marie Curie Early stage Training grant at the Göteborg Yeast Center funded by the European Commission (QLK-CT2000-60036), and C. M. by the Swedish National Research School for Genomics and Bioinformatics.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–07–0632) on December 28, 2005.
Abbreviations used: PAF, polysomal association factor; RP, ribosomal protein; tBOOH, tert-butyl hydroperoxide.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Arava, Y., Wang, Y., Storey, J. D., Liu, C. L., Brown, P. O., and Herschlag, D. (2003). Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100, 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp, E., and Sunnerhagen, P. (2003). Mkp1 and Mkp2, two MAPKAP-kinase homologues in Schizosaccharomyces pombe, interact with the MAP kinase Sty1. Mol. Genet. Genomics 268, 585–597. [DOI] [PubMed] [Google Scholar]
- Bilsland, E., Molin, C., Swaminathan, S., Ramne, A., and Sunnerhagen, P. (2004). Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol. Microbiol. 53, 1743–1756. [DOI] [PubMed] [Google Scholar]
- Bilsland-Marchesan, E., Ariño, J., Saito, H., Sunnerhagen, P., and Posas, F. (2000). Rck2 kinase is a substrate for the osmotic-stress activated MAP kinase Hog1. Mol. Cell. Biol. 20, 3887–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E., Jennings, E. G., Lee, T. I., True, H. L., Lander, E. S., and Young, R. A. (2001). Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell 12, 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlkvist, A., and Sunnerhagen, P. (1994). Two novel deduced serine/threonine protein kinases from Saccharomyces cerevisiae. Gene 139, 27–33. [DOI] [PubMed] [Google Scholar]
- de Hoon, M. J., Imoto, S., Nolan, J., and Miyano, S. (2004). Open source clustering software. Bioinformatics 20, 1453–1454. [DOI] [PubMed] [Google Scholar]
- Dunand-Sauthier, I., Walker, C., Wilkinson, C., Gordon, C., Crane, R., Norbury, C., and Humphrey, T. (2002). Sum1, a component of the fission yeast eIF3 translation initiation complex, is rapidly relocalized during environmental stress and interacts with components of the 26S proteasome. Mol. Biol. Cell 13, 1626–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J., Yang, X., Wang, W., Wood, W. H., 3rd, Becker, K. G., and Gorospe, M. (2002). Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl. Acad. Sci. USA 99, 10611–10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauchon, M., Lagniel, G., Aude, J. C., Lombardia, L., Soularue, P., Petat, C., Marguerie, G., Sentenac, A., Werner, M., and Labarre, J. (2002). Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell 9, 713–723. [DOI] [PubMed] [Google Scholar]
- Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D., and Brown, P. O. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigull, J., Mnaimneh, S., Pootoolal, J., Robinson, M. D., and Hughes, T. R. (2004). Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 24, 5534–5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghnazari, E., and Heyer, W. D. (2004). The Hog1 MAP kinase pathway and the Mec1 DNA damage checkpoint pathway independently control the cellular responses to hydrogen peroxide. DNA Repair 3, 769–776. [DOI] [PubMed] [Google Scholar]
- Holmes, L. E., Campbell, S. G., De Long, S. K., Sachs, A. B., and Ashe, M. P. (2004). Loss of translational control in yeast compromised for the major mRNA decay pathway. Mol. Cell. Biol. 24, 2998–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovland, R., Eikhom, T. S., Proud, C. G., Cressey, L. I., Lanotte, M., Doskeland, S. O., and Houge, G. (1999). cAMP inhibits translation by inducing Ca2+/calmodulin-independent elongation factor 2 kinase activity in IPC-81 cells. FEBS Lett. 444, 97–101. [DOI] [PubMed] [Google Scholar]
- Kedersha, N., Cho, M. R., Li, W., Yacono, P. W., Chen, S., Gilks, N., Golan, D. E., and Anderson, P. (2000). Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N. L., Gupta, M., Li, W., Miller, I., and Anderson, P. (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knebel, A., Haydon, C. E., Morrice, N., and Cohen, P. (2002). Stress-induced regulation of eukaryotic elongation factor 2 kinase by SB 203580-sensitive and -insensitive pathways. Biochem. J. 367, 525–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, K. M., DeRisi, J. L., Brown, P. O., and Sarnow, P. (2001). Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell. Biol. 21, 916–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay, V. L., et al. (2004). Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol. Cell. Proteomics 3, 478–489. [DOI] [PubMed] [Google Scholar]
- Madhani, H. D., Styles, C. A., and Fink, G. R. (1997). MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91, 673–684. [DOI] [PubMed] [Google Scholar]
- Mager, W. H., and Varela, J. C. (1993). Osmostress response of the yeast Saccharomyces. Mol. Microbiol. 10, 253–258. [PubMed] [Google Scholar]
- Mahtani, K. R., Brook, M., Dean, J. L., Sully, G., Saklatvala, J., and Clark, A. R. (2001). Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 21, 6461–6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher, M. L., and Thorner, J. (1996). Identification and characterization of the CLK1 gene product, a novel CaM kinase-like protein kinase from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 271, 29958–29968. [DOI] [PubMed] [Google Scholar]
- O'Rourke, S. M., and Herskowitz, I. (2004). Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol. Biol. Cell 15, 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, T., and Walter, P. (1999). Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10, 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss, T., Baron-Benhamou, J., Ansorge, W., and Hentze, M. W. (2003). Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat. Struct. Biol. 10, 1039–1047. [DOI] [PubMed] [Google Scholar]
- Puig, S., Askeland, E., and Thiele, D. J. (2005). Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120, 99–110. [DOI] [PubMed] [Google Scholar]
- Rajasekhar, V. K., Viale, A., Socci, N. D., Wiedmann, M., Hu, X., and Holland, E. C. (2003). Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell 12, 889–901. [DOI] [PubMed] [Google Scholar]
- Redpath, N. T., Price, N. T., Severinov, K. V., and Proud, C. G. (1993). Regulation of elongation factor-2 by multisite phosphorylation. Eur. J. Biochem. 213, 689–699. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Gabriel, M. A., Burns, G., McDonald, W. H., Martin, V., Yates, J. R., 3rd, Bähler, J., and Russell, P. (2003). RNA-binding protein Csx1 mediates global control of gene expression in response to oxidative stress. EMBO J. 22, 6256–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro, S., Llorente, B., Rodríguez-Manzaneque, M. T., Ramne, A., Uber, G., Marchesan, D., Dujon, B., Herrero, E., Sunnerhagen, P., and Pérez-Ortín, J. E. (2002). Functional analysis of yeast gene families involved in metabolism of vitamins B1 and B6. Yeast 19, 1261–1276. [DOI] [PubMed] [Google Scholar]
- Sanchez-Piris, M., Posas, F., Alemany, V., Winge, I., Hidalgo, E., Bachs, O., and Aligue, R. (2002). The serine/threonine kinase Cmk2 is required for oxidative stress response in fission yeast. J. Biol. Chem. 277, 17722–17727. [DOI] [PubMed] [Google Scholar]
- Segal, E., Shapira, M., Regev, A., Peter, D., Botstein, D., Koller, D., and Friedman, N. (2003). Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 34, 166–176. [DOI] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3–21. [DOI] [PubMed] [Google Scholar]
- Sheth, U., and Parker, R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D. A., Toone, W. M., Chen, D., Bähler, J., Jones, N., Morgan, B. A., and Quinn, J. (2002). The Srk1 protein kinase is a target for the Sty1 stress-activated MAPK in fission yeast. J. Biol. Chem. 277, 33411–33421. [DOI] [PubMed] [Google Scholar]
- Staleva, L., Hall, A., and Orlow, S. J. (2004). Oxidative stress activates FUS1 and RLM1 transcription in the yeast Saccharomyces cerevisiae in an oxidant-dependent manner. Mol. Biol. Cell 15, 5574–5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin, G., Stubbs, T., Kedersha, N., Wax, S., Rigby, W. F., Blackwell, T. K., and Anderson, P. (2004). MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23, 1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teige, M., Scheikl, E., Reiser, V., Ruis, H., and Ammerer, G. (2001). Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc. Natl. Acad. Sci. USA 98, 5625–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesono, Y., and Toh, E. A. (2002). Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 277, 13848–13855. [DOI] [PubMed] [Google Scholar]
- Walden, W. E., and Thach, R. E. (1986). Translational control of gene expression in a normal fibroblast. Characterization of a subclass of mRNAs with unusual kinetic properties. Biochemistry 25, 2033–2041. [DOI] [PubMed] [Google Scholar]
- Varela, J. C., van Beekvelt, C., Planta, R. J., and Mager, W. H. (1992). Osmostress-induced changes in yeast gene expression. Mol. Microbiol. 6, 2183–2190. [DOI] [PubMed] [Google Scholar]
- Warner, J. R. (1999). The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440. [DOI] [PubMed] [Google Scholar]
- Winzen, R., Kracht, M., Ritter, B., Wilhelm, A., Chen, C. Y., Shyu, A. B., Muller, M., Gaestel, M., Resch, K., and Holtmann, H. (1999). The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18, 4969–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Z., Wilson, S., Hauser, N. C., Tournu, H., Hoheisel, J. D., and Brown, A. J. (2003). Glucose triggers different global responses in yeast, depending on the strength of the signal, and transiently stabilizes ribosomal protein mRNAs. Mol. Microbiol. 48, 713–724. [DOI] [PubMed] [Google Scholar]
- Zaragoza, D., Ghavidel, A., Heitman, J., and Schultz, M. C. (1998). Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18, 4463–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk, D., Belk, J. P., and Jacobson, A. (1999). Temperature-sensitive mutations in the Saccharomyces cerevisiae MRT4, GRC5, SLA2 and THS1 genes result in defects in mRNA turnover. Genetics 153, 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.