Abstract
Survivin is a member of the chromosomal passenger complex implicated in kinetochore attachment, bipolar spindle formation, and cytokinesis. However, the mechanism by which survivin modulates these processes is unknown. Here, we show by time-lapse imaging of cells expressing either green fluorescent protein (GFP)-α-tubulin or the microtubule plus-end binding protein GFP-EB1 that depletion of survivin by small interfering RNAs (siRNAs) increased both the number of microtubules nucleated by centrosomes and the incidence of microtubule catastrophe, the transition from microtubule growth to shrinking. In contrast, survivin overexpression reduced centrosomal microtubule nucleation and suppressed both microtubule dynamics in mitotic spindles and bidirectional growth of microtubules in midbodies during cytokinesis. siRNA depletion or pharmacologic inhibition of another chromosomal passenger protein Aurora B, had no effect on microtubule dynamics or nucleation in interphase or mitotic cells even though mitosis was impaired. We propose a model in which survivin modulates several mitotic events, including spindle and interphase microtubule organization, the spindle assembly checkpoint and cytokinesis through its ability to modulate microtubule nucleation and dynamics. This pathway may affect the microtubule-dependent generation of aneuploidy and defects in cell polarity in cancer cells, where survivin is commonly up-regulated.
INTRODUCTION
Survivin is a member of the inhibitor of apoptosis (IAP) gene family (Salvesen and Duckett, 2002), which is overexpressed in nearly every human tumor and frequently associated with resistance to therapy, and unfavorable outcome (Altieri, 2003). Experimental work carried out in vitro (Beltrami et al., 2004) and in transgenic animals (Grossman et al., 2001; Okada et al., 2004) has assigned a dual function to survivin: protection from apoptosis and regulation of cell division. Although the cytoprotective function of survivin has recently come into better focus (Blanc-Brude et al., 2003; Marusawa et al., 2003) and has been linked to the upstream initiation of mitochondrial apoptosis (Dohi et al., 2004), the mechanism by which survivin participates in cell division is still unclear. Although survivin-like IAP molecules in model organisms seem to participate predominantly or exclusively in cytokinesis (Uren et al., 1999; Speliotes et al., 2000), reduction or loss of survivin in mammalian cells has been associated with a panoply of cell division defects that include supernumerary centrosomes (Li et al., 1999), aberrant spindle assembly (Giodini et al., 2002), mislocalization of mitotic kinases (Wheatley et al., 2001), loss of mitotic checkpoint(s) (Lens et al., 2003), and cytokinesis failure with appearance of multinucleated cells (Li et al., 1999). Adding further complexity to its potential role in mitosis, survivin localizes to multiple sites on the mitotic apparatus, including centrosomes, microtubules of the metaphase and central spindle, kinetochores, and midbodies (Fortugno et al., 2002).
Previous experiments of antibody microinjection suggested a potential role of survivin in spindle microtubule assembly, reflected in a phenotype of flattened mitotic spindles depleted of microtubules (Giodini et al., 2002). Similar observations were reported in knockout studies, and homozygous deletion of survivin resulted in nearly complete absence of mitotic spindles (Okada et al., 2004), and appearance of disorganized tubulin bundles (Uren et al., 2000). This model may fit well with the observation that survivin forms a complex with some of the chromosomal passenger proteins, notably, Aurora B (Adams et al., 2001) and the more recently described Borealin/hDasra B (Gassmann et al., 2004; Sampath et al., 2004). It has been proposed that the chromosomal passenger complex is a regulator of kinetochore attachment and cytokinesis (Adams et al., 2001) and is important for bipolar spindle formation in a pathway independent of Ran-GTP involving Aurora B-dependent phosphorylation of the microtubule-destabilizing Kin I kinesin MCAK (Gass-mann et al., 2004; Sampath et al., 2004). Survivin can enhance the activity of Aurora B (Bolton et al., 2002), suggesting a model whereby survivin regulates spindle formation through Aurora B.
In this study, we examined the role of survivin in microtubule dynamics and its potential dependence on the chromosomal passenger complex. Using time-lapse live imaging of two independent microtubule markers, the microtubule plus-end protein EB1 and α-tubulin, we found that survivin functions as a novel regulator of microtubule dynamics and nucleation in interphase and throughout mitosis and that this pathway is independent of Aurora B activity.
MATERIALS AND METHODS
Cells and Cell Cultures
Cervical carcinoma HeLa cells and monkey COS-7 cells were obtained from the American Type Culture Collection (Manassas, VA) and were maintained in culture according to the manufacturer's specifications. Diploid, telomerase-immortalized human RPE-1 cells (hTERT-RPE-1) were obtained from Clontech (Palo Alto, CA) (Morales et al., 1999).
Antibodies
Antibodies used were α-tubulin (#DM1a; Sigma-Aldrich, St. Louis, MO), acetylated-tubulin (#6-11B-1; Sigma-Aldrich), β-galactosidase (#1083 104; Roche Diagnostics, Indianapolis, IN), tyrosinated-tubulin (rabbit W2) (Gurland and Gundersen, 1995), survivin (Fortugno et al., 2002), anti-EB1(catalog no. 610534; BD Biosciences, Franklin Lakes, NJ), anti-γ-tubulin (HM2569, polyclonal peptide antibody raised against amino acids AATR; Covance, Princeton, NJ) and hemagglutinin (HA) (#3F10; Roche Diagnostics). As secondary antibodies, we used anti-mouse cy3 (Molecular Probes, Eugene, OR), anti-rabbit cy5 or fluorescein isothiocyanate (FITC) (Jackson ImmunoResearch Laboratories, West Grove, PA), or anti-rat cy3 (Molecular Probes).
Microinjection and Live Cell Imaging
COS-7 cells were synchronized by double-thymidine block (Quintyne and Schroer, 2002), and upon release from the S phase block, they were microinjected into the nucleus with plasmids containing HA-survivin or β-galactosidase (200 ng/ml) together with 25 ng/ml an EB1-GFP plasmid (a gift from L. Cassimeris, Lehigh University, Bethlehem, PA) using an Eppendorf transjector 5246 and Micromanipulator (Brinkmann, Westbury, NY). Cells were returned to the incubator for 8–10 h and then used for live imaging or fixed and stained for other antigens as indicated using methods described previously (Gromley et al., 2003).
Adenoviral Transduction
The replication-deficient adenoviruses encoding survivin (pAd-survivin) or GFP (pAd-GFP) were described previously (Mesri et al., 2001). Cells (2.5 × 107) were transduced at a multiplicity of infection of 50 for 24 h at 37°C, washed, and replenished with fresh growth medium for further analysis.
Microtubule Quantification in Fixed Cells
COS-7 cells plated at comparable density were transduced with pAd-GFP or pAd-survivin, treated with 10 μM of the microtubule-depolymerizing agent nocodazole (Sigma-Aldrich), and harvested at increasing time intervals between 5 and 60 min (Hergovich et al., 2003). Cells were fixed in ice-cold methanol, washed three times in phosphate-buffered saline with 1% bovine serum albumin and 0.5% Triton X-100 (PBSAT), and stained with antibody to acetylated tubulin (Gromley et al., 2003) followed by a secondary anti-IgG conjugated to cy3 and mounting on glass slides (Prolong Antifade; Molecular Probes). Microtubule fluorescence was quantified by acquiring 13 optical sections (333 ± 50 nm) using wide-field fluorescence microscopy (Olympus, Tokyo, Japan) and a 63× objective from six random fields. For individual cell measurements, we obtained fluorescence values of areas outside those containing microtubules and subtracted this background from the total value (typically 5–10% of total cellular fluorescence). The fluorescence intensity (integrated optical density) (Dictenberg et al., 1998) for every optical section in every full cell profile or within an entire population was calculated using MetaMorph software (Universal Imaging, Downingtown, PA) (see above) or IP Lab software version 3.5.4 (Scanalytics, Fairfax, VA) as described previously (Purohit et al., 1999; Gromley et al., 2003). These values were then averaged to calculate the fluorescence of the total population.
RNA Interference (RNAi)
Double-stranded (ds)RNA oligonucleotides targeting survivin (S4), Aurora B, or a control unrelated sequence (VIII) were described previously (Altieri, 2003; Beltrami et al., 2004). Cells were transfected with 50 nM of the various dsRNA oligonucleotides using Oligofectamine (3 ml/well) reagent (Invitrogen, Carlsbad, CA) and replenished with growth medium after 4 h. After 24–48 h, in the absence of any detectable apoptosis, cells were harvested and analyzed for reduction of survivin levels by immunoblotting, or used for live analysis of EB1-GFP or GFP–α-tubulin dynamics.
Microtubule Regrow Assay (Nucleation)
Cells grown on coverslips were either transfected with the indicated dsRNA oligonucleotides or transduced with the indicated adenovirus as described above. Cells were than incubated for 4 h in 8 μM nocodazole at 37°C and then for an additional 30 min on ice before washing. Coverslips were than washed twice in 50 ml of ice-cold phosphate-buffered saline and incubated at 37°C for 2 min to allow microtubule growth and subsequently fixed in –20°C methanol, washed three times in PBSAT, and stained with antibodies to EB1 and γ-tubulin followed by cy3 and FITC-conjugated secondaries, respectively. EB1 foci were quantified by acquiring 16 optical sections (200 ± 50 nm) using wide-field fluorescence microscopy (Olympus) and a 100× objective from random fields. EB1 foci were individually counted from the entire z-series, and final images are presented as maximum projection of all planes used. Centrosomes containing two γ-tubulin foci were used. Graphs represent data taken from 25 to 30 cells in two experiments.
Live Microscopy of GFP-EB1
Cells (COS-7 or RPE) were plated on coverslips (25 mm in diameter) and were placed in a chamber (PDMI-2; Harvard Apparatus, Holliston, MA) in complete medium with CO2 exchange (0.5 l/min) at 37°C. Cells were imaged every 3 s for two or more minutes using a 100× objective on an inverted microscope (Olympus IX-70). Images were captured on a CoolSNAP HQ charge-coupled device camera (Roper Scientific, Trenton, NJ). Time-lapse movies of EB1 movements were obtained. In some cases, individual images were concatenated to produce linear elements representing the total distance traveled and providing a measure of the total amount of microtubule growth during the imaging period (MetaMorph software 4.6). Using similar visualization techniques, we quantified the number of growing microtubules in cells. Where indicated, 10 μM taxol (Sigma-Aldrich) was added to live microscopy media. Resulting movies are shown at a rate of 15 frames per second.
Time-Lapse Analysis of Cells Expressing GFP-α-Tubulin
RPE cells stably expressing GFP-α-tubulin were transfected with survivin-specific S4 or control VIII dsRNA oligonucleotides and imaged by confocal laser scanning microscopy. Microtubule growth, catastrophe, shrinking, and rescue as well as growth/catastrophe transition rates for individual microtubules were calculated from images collected as time-lapse movies from several random areas of the cytoplasm that in most cases comprised the leading edge of the cell. Average transition values were obtained from five microtubules. Similar results were obtained from five individual cells in two different experiments. For targeting of Aurora B kinase, RPE GFP-α tubulin cells were first transfected with control (VIII) or Aurora B-directed dsRNA oligonucleotide for 36 h and analyzed by Western blotting. In independent experiments, cells were treated with the Aurora B kinase inhibitor hesperadin (100 nM for 6 h) characterized in previous studies (Hauf et al., 2003; Sessa et al., 2005). For analysis of microtubule dynamics, cells prepared as described above were observed using an inverted Zeiss microscope equipped with a 100×, numerical aperture 1.4 objective lens, a spinning-disk confocal scan head (PerkinElmer Life and Analytical Sciences, Boston, MA), and a MicroMAX interline transfer cooled charge-coupled device camera (Roper Scientific). All images (16-bit) were acquired using a single-wavelength (488-nm) filter cube. Image acquisition was controlled by Ultraview RS software (PerkinElmer Life and Analytical Sciences). Time-lapse sequences were acquired at 3-s intervals by using an exposure time of 0.2 s at four optical planes per interval with a Z-step of 0.3 μm. Resulting movies are shown at a rate of 15 frames per second.
Quantification of Microtubule Dynamics
Individual microtubules were analyzed as described previously (Rusan et al., 2001). Briefly, time-lapse images were exported from the proprietary Ultraview software and imported into MetaMorph software (Universal Imaging) for further analysis. A stack of four optical planes was used to make a z-projection at each time point, and a time-lapse movie was reconstructed. The position of the microtubule end was tracked using the “track points” function in MetaMorph that was linked to Excel to produce a history plot of each microtubule. Growth and shortening phases were identified based on the history plots. The frequency of catastrophe was calculated by dividing the sum of the number of transitions from growth to shortening and pause to shortening by the sum of the duration of growth and pause. The frequency of rescue was calculated by dividing the sum of the number of transitions from shortening to growth and shortening to pause by the duration of shortening. Microtubule dynamicity was calculated as the total number of tubulin dimers exchanged at the microtubule end (using 1624 dimers/μm), considering the lifetime of the microtubule (Waterman-Storer et al., 2000; Toso et al., 1993). The value from each microtubule was used to calculate an average for each experiment and was used in Table 1. The time spent in each phase (shrink, growth, and pause) was recorded, and the percentage of time spent in each phase was calculated for each microtubule. The percentage of time was averaged individually and used in Table 1.
Table 1.
Microtubule dynamics in survivin-depleted cells
| A | ||||
| Avg. frequency rescue (s-1) | Avg. frequency catastrophe (s-1) | |||
| Control siRNA | 0.070 ± 0.0148 | 0.025 ± 0.0047 | ||
| Survivin siRNA | 0.094 ± 0.0207 | 0.045 ± 0.0044* | ||
| Aurora B siRNA | 0.064 ± 0.0464 | 0.023 ± 0.0103 | ||
| Hesperadin analogue | 0.064 ± 0.0270 | 0.030 ± 0.0082 | ||
| Hesperadin | 0.069 ± 0.0168 | 0.027 ± 0.0120 | ||
| B | ||||
| Avg. time (%) | Avg. growth rate (μm/s) | Avg. shrink rate (μm/s) | Dynamicity (dimer/s) | |
| Control siRNA | ||||
| Shrink | 24.67 | 0.383 ± 0.1167 | 0.920 ± 0.2752 | 686.38 |
| Pause | 20.42 | |||
| Growth | 54.56 | |||
| Survivin siRNA | ||||
| Shrink | 27.94 | 0.398 ± 0.1258 | 0.852 ± 0.2503 | 811.89* |
| Pause | 19.80 | |||
| Growth | 52.43 | |||
| Aurora B siRNA | ||||
| Shrink | 27.94 | 0.326 ± 0.0598 | 0.817 ± 0.2480 | 587.11 |
| Pause | 19.85 | |||
| Growth | 52.21 | |||
| Hesperadin analogue | ||||
| Shrink | 30.38 | 0.338 ± 0.1215 | 0.897 ± 0.1715 | 634.55 |
| Pause | 21.94 | |||
| Growth | 47.68 | |||
| Hesperadin | ||||
| Shrink | 22.77 | 0.391 ± 0.0592 | 0.897 ± 0.4081 | 618.12 |
| Pause | 19.80 | |||
| Growth | 57.42 |
(A) Catastrophe is increased in survivin-depleted cells compared with cells treated with control siRNAs or siRNAs targeting Aurora B (1.66-to 1.99-fold increase, respectively; value indicated by asterisk). (B) The average time microtubules spent shrinking, pausing, or growing is not significantly perturbed. There is no significant difference in growth or shrinkage rates in survivin-depleted cells compared with controls. Dynamicity is increased in cells with depleted survivin. Here, dynamicity represents the exchange of dimer at the microtubule tip over time (seconds); see Materials and Methods. Pauses represent periods between growth and shrinking or vice versa (<0.5-μm change). All data shown were acquired from 25 microtubules in five cells per experimental condition. All differences described are statistically significant (t test; p < 0.005).
Immunofluorescence of Phosphorylated Histone H3
RPE-GFP-α-tubulin cells were grown on glass coverslips, treated with control or 100 nM hesperadin for 6 h, and fixed in –20°C methanol for 30 min. Coverslips were stained with 4,6-diamidino-2-phenylindole (DAPI) and an antibody to phosphorylated H3 (catalog no. 6-570; Upstate Biotechnology, Lake Placid NY) followed by cy5 secondary reagents. Images were acquired using the MetaMorph software as described above.
Quantification of Phosphorylated H3 Fluorescence
The method used was similar to the quantification EB1-GFP fluorescence (supplemental material). Briefly, 0.2 μm optical sections were taken of each mitotic cell for DAPI, GFP, and cy5. Because phosphorylated H3 specifically labels the chromatin, a region of the cytoplasm was used as background to subtract from the cy5 fluorescence. The DAPI labeling for each corresponding phosphorylated H3 image was used to define a region of interest based on the “threshold image” function in MetaMorph. The region was transferred to the appropriate phosphorylated H3 image, and the fluorescence in the defined region (occupied by the chromatin) was quantified.
Statistical Analysis
Data were analyzed using the unpaired t test on a GraphPad software package for Windows (Prism; GraphPad Software, San Diego, CA). Phosphate-buffered saline values of 0.05 were considered statistically significant.
RESULTS
Depletion of Survivin by RNA Interference Increases Microtubule Dynamics
Because of its reported localization to microtubules (Fortugno et al., 2002) and its ability to alter microtubule organization during mitosis (Giodini et al., 2002), we asked whether survivin modulated microtubule dynamics in living cells. We first examined microtubule dynamics in cells depleted of survivin by small interfering RNAs (siRNA). Transfection of RPE cells with a previously characterized dsRNA oligonucleotide targeting survivin (S4) (Beltrami et al., 2004) resulted in >90% reduction in survivin levels by immunoblotting 48 h later, whereas a control dsRNA oligonucleotide (VIII) had no effect on survivin levels (Figure 1A). We next examined several parameters of microtubule dynamics, including microtubule growth, catastrophe (transitions from growth to shrinking), shrinking, rescue (transitions from shrinking to growth) and pause (periods between growth and shrinking) in interphase cells stably expressing GFP-labeled α-tubulin (Desai and Mitchison, 1997). We found that the average frequency of catastrophe (number per second) and dynamicity (dimers exchanged at the tip/lifetime of microtubule; see Materials and Methods) (Waterman-Storer et al., 2000; Toso et al., 1993) were significantly higher in survivin-depleted cells (Figure 1, C and E) compared with control cells (Figure 1, B and D; n > 25 microtubules from 5 cells, 2 experiments/condition); other parameters were not significantly different from controls. Each parameter of microtubule dynamics is independently presented in Table 1. Histories of growth and shrinking events of five microtubules per condition are shown in Figure 1, B–E, and in Movies 1 and 2. In addition, we analyzed cultures by fluorescence microscopy with an antibody to acetylated tubulin, a posttranslationally modified tubulin found in stabilized microtubules (Bulinski et al., 1988). The acetylated tubulin signal was diminished compared with controls (our unpublished data; see below). The decrease in acetylated tubulin staining and increased frequency of microtubule catastrophe demonstrate that survivin depletion increases microtubule dynamics.
Figure 1.
Survivin silencing increases microtubule dynamics. (A) RPE cells were transfected with survivin-specific S4 or control VIII dsRNA oligonucleotides (siRNAs), harvested after 48 h, and analyzed by immunoblotting. (B and C) Analysis of microtubule growth and shrinking. RPE cells stably expressing GFP-α tubulin were transfected with control VIII (B) or survivin-specific S4 dsRNA oligonucleotide (C), and imaged by time-lapse videomicroscopy (see Movies 1 and 2). The extent of growth and shrinking (red arrows) was measured for individual microtubules in interphase. (D and E) Quantification of microtubule dynamics observed in B and C. The length of microtubule polymer growth or shrinking (distance in micrometers) was examined over time in control cells (D) or survivin-depleted cells (E). Increasing values represent microtubule growth; decreasing values represent shrinking. Transitions from growth to shrinking (catastrophe), shrinking to growth (rescue), periods of no net growth (pause), and other parameters of microtubule dynamics are quantified in Table 1. Symbols represent five individual microtubules from two cells for both D and E.
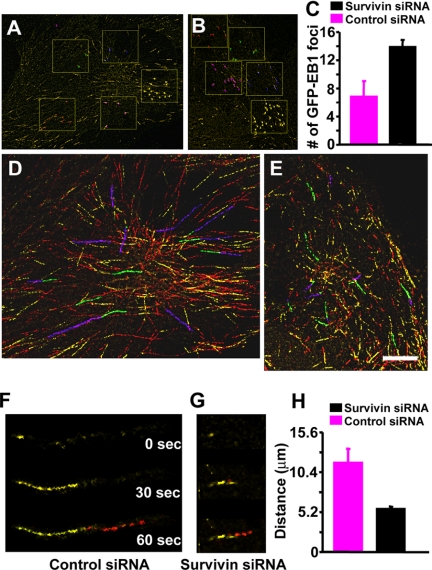
To independently validate results obtained with GFP-α tubulin-expressing cells, we used GFP-tagged EB1 as a marker for the plus ends of growing microtubules. Recent studies have shown that GFP-EB1 accurately reflects microtubule growth rates and the number of growing microtubules, including those nucleated from centrosomes (Piehl et al., 2004; Tirnauer et al., 2004). Stable expression of GFP-EB1 in control cells (VIII) revealed GFP-EB1 foci moving outward from the centrosome as previously described (Movie 3) (Piehl et al., 2004). The number of GFP-EB1 foci in survivin-depleted cells was increased compared with control cells (Figure 2, A–C). When EB1 foci were collectively displayed as a single projected image in control cells, long tracks representing extended periods of microtubule growth were observed (Figure 2, D, F, and H; Movie 3). Survivin-depleted cells expressing similar levels of GFP-EB1 (see below) had shorter EB1 tracks (Figure 2, E, G, and H; Movie 4). These results are consistent with an increase in the number of growing microtubules and a higher rate of catastrophe (Gliksman et al., 1993). The changes in microtubule parameters observed in survivin-depleted cells occurred in the absence of changes in total cellular α-, β-tubulin levels (our unpublished data). These results confirm data from GFP-α-tubulin-expressing cells and verify that GFP-EB1 is a reliable marker for microtubule dynamics.
Figure 2.
Survivin silencing increases the number of growing microtubules and decreases the duration of microtubule growth. (A and B) Number of EB1 foci. GFP-EB1-expressing RPE cells were transfected with control dsRNA (A) or survivin-specific S4 dsRNA (B), and EB1 foci were examined in composite images made from three consecutive frames of each movie (see Movies 3 and 4). (C) GFP-EB1 foci, representing individual microtubules, are quantified from fields covering 50–70% of a cell's area (boxes in A and B) from five cells in two separate experiments (C; each bar represents the average number of EB1 tracks). (D and E) GFP-EB1 movements. GFP-EB1-expressing RPE cells were transfected with control dsRNA (D) or survivin-specific S4 dsRNA (E), and EB1 movements were examined over 1-min (see Movies 3 and 4). All GFP-EB1 movements, representing microtubule growth, are displayed as linear tracings in D and E. The first 15 s of microtubule growth is represented in yellow, and the final 45 s is in red. Bar (E), 5 μm for D and E. Examples of microtubules used for analysis are displayed as green to blue instead of yellow to red. (F and G) Higher magnification images of individual growing microtubules in D (control; VIII transfectants) and E (survivin; S4 transfectants) at times indicated. (H) Quantification of GFP-EB1 tracking distances after transfection of dsRNA VIII (control) or survivin-directed S4 (survivin) oligonucleotide. Length in micrometers. Data represent 10 measurements from each of 10 cells from two separate experiments. Examples of microtubules analyzed for H are shown as green (first 15 s) and blue (next 45 s) in D and E. Bars (C and H) represent the mean ± SD.
The increase in the number of EB1 foci demonstrated that more microtubules were present in survivin-depleted cells and suggested an increase in the number of microtubules nucleated from centrosomes. To test this directly, cells were treated with nocodazole to depolymerize microtubules and then washed to remove the drug and to allow regrowth of centrosomal microtubules. By counting the number of EB1 foci stained with anti-EB1 emanating from centrosomes, an accurate determination of microtubule nucleation could be determined as described previously (Piehl et al., 2004; Tirnauer et al., 2004). We found a significant increase in the number of EB1 foci after siRNA-mediated depletion of survivin compared with cells treated with a control siRNA (Figure 3A), demonstrating an increase in the number of centrosome-nucleated microtubules.
Figure 3.
Survivin modulates the number of growing microtubules emanating from the centrosome. (A) Maximum projection of z-series taken of individual centrosomes in interphase RPE cells treated with nocodazole and then allowed to regrow microtubules for 2 min. Cells had been treated with either survivin-specific S4 dsRNA (A; left) or control dsRNA (A; right). Cells were stained by immunofluorescence using an antibody to EB1 (red) and γ-tubulin (green) (A; left and right), and foci were quantified in each z-plane and compared with adjacent planes to ensure that individual foci were not counted multiple times (graph; A, far right). (B) Maximum projection as in A where cells had been transduced either by pAd-GFP-survivin (B; left) or pAd-GFP-survivin (B; right). Cells were stained by immunofluorescence and quantified as in A (B; far right). (C) Maximum projection as in A and B where cells had been treated with either survivin-specific S4 dsRNA (C; left) or control dsRNA (C; right) in the presence of hesperadin. Cells were stained by immunofluorescence and quantified as in A and B (C; far right).
Expression of Survivin Suppresses Microtubule Dynamics and Nucleation at Multiple Cell Cycle Phases
Based on the increase in microtubule dynamics and nucleation observed in survivin-depleted cells, we reasoned that elevated survivin levels would suppress these parameters. To test this prediction, we first examined GFP-EB1 movements in living COS-7 cells microinjected with a plasmid encoding GFP-EB1 together with a plasmid encoding HA-survivin or a control protein, β-galactosidase. As expected, multiple GFP-EB1 foci emanated from the centrosome in control β-galactosidase-expressing cells (Figure 4, A and B, and Movie 5) and long GFP-EB1 tracks marking EB1 movements over time were observed (Figure 4, B and D). When the same cell was subsequently treated with taxol to suppress microtubule dynamics, GFP-EB1 movements were abolished, and no GFP-EB1 foci or tracks of EB1 movements were detected (Figure 4C and Movie 6). Survivin-expressing cells revealed a phenotype similar to that of taxol-treated cells. In many cells (∼80%), no detectable GFP-EB1 foci were observed and GFP-EB1 track projections yielded little to no linear dimension (Figure 4E and Movie 7). GFP-EB1 levels achieved in these experiments were roughly similar in all cells examined (±11%; see below) and never approached levels known to induce microtubule changes (Ligon et al., 2003). Consistent with the decrease in the number of GFP-EB1 foci, the number of centrosomal EB1 foci and hence the number of microtubules nucleated from centrosomes was decreased (Figure 3B).
Figure 4.
Increased levels of survivin suppress microtubule growth in interphase and mitotic cells. (A and B) Individual frames from a movie of GFP-EB1 in an interphase COS-7 cell coexpressing β-galactosidase (control) showing multiple foci at time 0 (A) that became more visible after growth (displayed as tracks that extend for long distances over 1 min; B; see Movie 5). First 15 s of microtubule growth are in yellow, and last 45 s are in red. (C) Taxol treatment. The same cell as in B treated with 10 μM taxol for 31 min; no detectable GFP-EB1 foci are seen (see Movie 6). (D) Higher magnification of a growing microtubule in B at 0, 30, and 60 s of filming. (E) Suppression of microtubule dynamics by survivin. Cell coexpressing survivin and GFP-EB1 showing no detectable GFP-EB1 foci (see Movie 7). (F) Quantification of spindles with detectable GFP-EB1 foci (or short tracks) in living cells expressing β-galactosidase or survivin. Data are the average of two separate experiments. n > 20 cells per bar. (G and H) Individual frames from movies of GFP-EB1 in mitotic cells expressing either survivin (G; see Movie 11) or β galactosidase (H; see Movie 10). Bar (C), 5 μm for A–H. (I–N) Cells coexpressing GFP-EB1 together with β-galactosidase (I–K) or survivin (L–N) were analyzed for GFP-EB1 foci (I and L), β-galactosidase and HA-survivin expression (J and M), or microtubules (K and N). Insets, DNA labeled with DAPI. For all injection studies, an average of 91% of cells survived injections, and 96% of those expressed both GFP-EB1 and either survivin or β-galactosidase at levels that do not affect microtubule dynamics or organization (survivin). Bar (N), 10 μm for I–N.
We next examined the effect of elevated survivin levels in mitotic cells, where microtubules are more dynamic than interphase cells (Rusan et al., 2001). We used living COS-7 cells overexpressing either survivin or β-galactosidase (control), and expressing similar levels of GFP-EB1. We found that survivin-expressing cells had significantly less spindle-associated GFP-EB1 and more cytoplasmic GFP-EB1, which was reversed in control cells (Supplemental Figure 1). Moreover, survivin-expressing cells showed little to no detectable microtubule growth, because most spindles (85%; n = 20) contained a negligible number of organized GFP-EB1 foci (Figure 4, F and G, and Movie 11). Although EB1 foci were present in some spindles (∼15%; n = 20), their number never exceeded 10% of control levels (our unpublished data). As expected, control cells showed multiple GFP-EB1 foci moving away from both spindle poles (Figure 4, F and H, and Movie 10). Results from living cells were also confirmed in fixed cells. In control experiments, fixed β-galactosidase-expressing cells contained organized bipolar spindles with numerous EB1 foci (Figure 4, I–K). Conversely, fixed cells expressing survivin had little to no spindle-associated GFP-EB1 foci (Figure 4, L–N) and revealed small or disorganized mitotic spindles as reported previously (Giodini et al., 2002).
We next examined the effect of survivin on microtubule dynamics during cytokinesis. In control cells expressing β-galactosidase (Figure 5, A–C), GFP-EB1 foci in midbodies were numerous (Figure 5A). They moved away from the center of the midbody (the zone that does not stain for microtubules at asterisk; Figure 5D, arrow; Gromley et al., 2005) as well as toward the midbody center (Figure 5E, arrow), showing that microtubules were growing in both directions at this site (better visualized in Movies 8 and 9). These data suggest that midbody microtubules are highly dynamic, of dual polarity, and undergo bidirectional growth both toward and away from the midbody center. In comparison, GFP-EB1 foci in survivin-expressing cells (Figure 5, F and H) were either undetectable or diminished in number. The expressed survivin (Figure 5G) primarily accumulated with the endogenous protein at the midbody (Li et al., 1999), suggesting that this was the site of action of the ectopically expressed protein. Consistent with previous observations (Uren et al., 1999; Speliotes et al., 2000), survivin-expressing cells with reduced microtubule dynamics often failed cytokinesis and generated multinucleated cells (our unpublished data).
Figure 5.
Increased levels of survivin suppress microtubule growth in midbodies during cytokinesis. (A–C) GFP-EB1 staining at midbodies in telophase cells expressing control protein (β-galactosidase) or survivin (F–H). Midbody GFP-EB1 labeling is significant in control cells (A–C), with movements (representing microtubule growth) both away from the central midbody region (D; arrow) and toward the central midbody (E; arrow) (see Movies 8 and 9). Schematic in D shows midbody region examined in this figure. In survivin-expressing cells (F–H), little GFP-EB1 labeling is observed at the midbody (F) although the cell is at a similar stage of cytokinesis to that in A. Insets (A–C and F–H), DNA labeled with DAPI. Images in A and F represent enlargements of midbodies seen in B and G, respectively. Bar (H), 10 μm for B, C, G, and H and 5 μm for A and F. Bar (E), 5 μm for D and E.
Expression of Survivin Stabilizes Microtubules in Interphase and Mitosis
Because of its role in modulating microtubule dynamics, we next asked whether survivin influenced microtubule stability in fixed cell preparations. We expressed GFP-tagged survivin in COS-7 cells using a replication-deficient adenovirus (GFP-survivin) (Mesri et al., 2001) and analyzed cultures by fluorescence microscopy for acetylated tubulin (see above). Interphase cells expressing GFP-survivin showed an increase in the amount of acetylated tubulin compared with GFP-expressing control cells (Figure 6, A–D). In addition, cells in cytokinesis showed an increase in the amount of actylated tubulin at midbodies when survivin levels were increased (Figure 6, E–H). Moreover, survivin-expressing interphase cells treated with the microtubule-depolymerizing agent nocodazole showed increased resistance to microtubule depolymerization compared with controls (Figure 6K). Twenty minutes after nocodazole treatment, most microtubules were depolymerized in control cells (Figure 6I, GFP), whereas microtubules persisted in survivin-expressing cells at this time (Figure 6J) and for an additional 40 min.
Figure 6.
Increased levels of survivin increase microtubule stability. (A and B) Survivin increases acetylated tubulin content. COS-7 cells transduced by pAd-GFP (A) or pAd-GFP-survivin (B) were stained by immunofluorescence using an antibody to a stabilized acetylated form of α-tubulin. Bar (B), 5 μm for A and B. Individual cells outlined. (C and D) Quantification of acetylated tubulin signal in individual cells (C; total integrated fluorescence of a single representative experiment) and in all cells (D; expressed as an average, n > 2 × 103 measurements from optical sections taken from >200 cells/bar). All data for A–D were acquired from interphase cells). (E and F) Survivin increases acetylated microtubules at midbodies. Cells were transduced with pAd-GFP (E) or pAd-GFP-survivin (F) and analyzed with an antibody to acetylated tubulin by fluorescence microscopy. Bar (F), 5 μm for E and F. (G and H) Quantification of acetylated tubulin signal at midbodies in individual cells (G) or whole cell population (H). (I and J) Nocodazole resistance. Cells were transduced with pAd-GFP or pAd-GFP-survivin, exposed to nocodazole for 20 min (I and J), and analyzed for α-tubulin staining by fluorescence microscopy. I and J represent high-magnification images of cells acquired from random fields for analysis. Bar (J), 5 μm for I and J. (K) Quantification of nocodazole resistance of microtubules (α-tubulin staining) in cells expressing GFP or survivin at 20 min. Fluorescence intensity is in arbitrary units.
Depletion or Pharmacologic Inhibition of Aurora B Kinase Does Not Affect Microtubule Dynamics or Nucleation
To investigate the mechanism by which survivin modulates microtubule dynamics and nucleation, we first tested whether Aurora B perturbed microtubule dynamics when depleted by RNAi. Aurora B depletion was achieved using siRNAs previously used in studies to deplete Aurora B (Hauf et al., 2003). These effectively reduced Aurora B levels in RPE cells by 60–80% (Figure 7A). Reduction of Aurora B expression by siRNA was associated with formation of binucleated cells presumably because of cytokinesis failure (Figure 7, B and C), in agreement with published results. However, analysis of microtubule stability using acetylated tubulin antibodies under these experimental conditions revealed no significant differences between control and Aurora B siRNA-treated cultures (Figure 7, D–F). To formally test whether Aurora B suppression by siRNA affected microtubule dynamics, we used time-lapse imaging of stably transfected cells expressing GFP-labeled α-tubulin. In these experiments, the frequency of microtubule rescue and catastrophe, the duration of microtubule pause, growth and shrinking, and the rate of growth and shrinking were indistinguishable from cultures treated with control (VIII) or Aurora B-directed siRNA (Figure 8, A and B, and Table 1). To independently validate these results, we used time-lapse imaging of microtubule growth in living cells expressing GFP-EB1. In these experiments, Aurora B suppression by siRNA did not significantly alter microtubule growth distances and the number of GFP-EB1 foci, compared with control (VIII)-transfected cells (Figure 8, C and D). We also showed that immunoprecipitation of survivin from logarithmically growing or mitotic HeLa cells did not pull down detectable Aurora B, although the survivin-binding protein heat-shock protein of 90 kDa (Hsp90) effectively coimmunoprecipitated with survivin (Figure 8E, cells overexpressing survivin did not pull down detectable levels of Aurora B; our unpublished data). These biochemical experiments suggest that at least some survivin was not bound to Aurora B kinase in HeLa cells under these conditions, a result different from studies done in Xenopus extracts or cells ectopically expressing the proteins (see Discussion) (Bolton et al., 2002; Beardmore et al., 2004, Temme et al., 2005).
Figure 7.
Aurora B silencing has no detectable effect on microtubule dynamics. (A) RPE cells were transfected with a survivinspecific siRNA or an Aurora B-specific siRNA, harvested 48 h later, and analyzed by immunoblotting. (B) Binucleated cells were quantified 24 and 48 h after transfection of Aurora B and control siRNAs (n = 300 cells/bar). (C) Image showing two binucleated cells after treatment with Aurora B siRNA (arrows). Nuclei, blue, microtubules, red. (D) Quantification of microtubule acetylation following Aurora B or control siRNA treatment (n = 200 cells for each bar). (E and F) Images of acetylated microtubules in cells treated with Aurora B (E) or control (F) siRNAs. Nuclei, blue, acetylated microtubules, red. Bar (C), 5 μm; bar (F), 5 μm for E and F.
Figure 8.
Aurora B silencing has no detectable effect on microtubule dynamics. (A and B) RPE cells stably expressing GFP α-tubulin were transfected with Aurora B (B) or control (A; VIII) siRNAs. Images of microtubules in interphase cells were acquired every 3 s, and the resulting time-lapse movies were used to construct history plots of individual microtubules. No significant differences were observed in microtubule catastrophe or rescue frequencies (6 microtubules from 2 cells are shown), the duration of microtubule pause, growth or shrinking, or the rate of microtubule growth or shrinking (see Table 1). (C and D) Silencing of Aurora B has no detectable effect on the distance traveled for GFP-EB1 foci (C; tracking distances) or the number of GFP-EB1 foci per unit area (D) (n = 16 microtubules from 3 or more cells for each bar). See legends to Figures 1 and 2 and Table 1 for more details. (E) Survivin was immunoprecipitated from asynchronous cultures of HeLa cells using previously characterized antibodies (Giodini et al., 2002) and probed for survivin, Hsp90 and Aurora B as indicated. Although Aurora B can be detected in the cell lysates it is undetectable in the survivin immunoprecipitation. IgG is nonimmune rabbit IgG used for immunoprecipitation control. Asterisk (*) represents the IgG band.
Because recent data suggests that depletion of Aurora B can affect the cellular levels of survivin and vice versa (Honda et al., 2003), we used the pharmacologic agent hesperadin to inhibit the activity of the kinase as done previously (Hauf et al., 2003). Treatment with hesperadin dramatically reduced phosphorylation of the Aurora B target protein histone H3 compared with cells exposed to an inactive hesperadin analogue (Figure 9A). Hesperadin induced defects in spindle organization and chromosome alignment (17/17 spindles and 0/10 controls; Figure 9, B–D). Spindles were usually more narrow and sometimes longer than controls, and the two halves of the spindles were misoriented in that they were not aligned 180° from one another but crescent shaped. Chromosomes were often positioned outside the area occupied by spindle microtubules (Figure 9, C and D). The spindle defects in hesperadin-treated cells were distinct from those observed in survivin-expressing cells, where spindles were often shortened in the pole-to-pole dimension but had normally aligned chromosomes (Figure 4G) (Giodini et al., 2002). The different spindle disruption phenotypes suggested that Aurora B and survivin affected spindles by different mechanisms, providing additional support for the idea that these proteins functioned independently and not as members of a common protein complex. Cells treated with either hesperadin or the inactive analogue showed no differences in the rates of microtubule growth or shrinking, the duration of microtubule growth, shrinking or pause, the frequency of microtubule catastrophe, or rescue as collectively measured by time-lapse and fixed cell imaging of GFP-α tubulin or GFP-EB1 (Figure 9, B–F, and Table 1). Moreover, hesperadin treatment of survivin-depleted cells showed no effect on microtubule nucleation, suggesting that there was no contribution of Aurora B in the context of reduced survivin (Figure 3C). This was consistent with our data showing that survivin depletion did not significantly affect Aurora B levels and vice versa (Figure 7A; see Discussion). Together with data from the Aurora B depletion experiments, these results show that inhibition of Aurora B activity or levels has no effect on microtubule dynamics despite induction of dramatic defects in mitosis under both conditions. The microtubule dynamics and nucleation changes seen in cells with altered survivin levels seem to be independent of Aurora B and could be induced by a fraction of survivin that is not associated with the chromosomal passenger complex or by survivin within the complex.
Figure 9.
Chemical inhibition of Aurora B activity by hesperadin does not affect microtubule dynamics in interphase or mitotic cells. (A) RPE cells treated with 100 nM hesperadin or a nonfunctional analog for 6 h were used to quantify the phosphorylation of the Aurora B target, histone H3 (A; phos-H3). Graph, n = 10 randomly chosen mitotic cells/bar. Representative images are shown below bars. DAPI panel shows DNA pattern, and the phos-H3 panel shows staining with antibody specific for phosphorylated H3. DAPI staining was used to identify regions occupied by chromatin given the drastic loss of phos-H3 in hesperadin-treated cells. (B–D) Mitotic spindles in hesperidin-treated cells are narrower (C) and angled (D) compared with controls (B). Microtubule growth as measured by the distribution of GFP-EB1 foci (red) in mitotic cells treated with hesperadin (C and D) or the control analog (B) show no significant differences. Large images represent merge of microtubules (green), GFP-EB1 (red), and DNA (DAPI; blue). Smaller images below show, from left to right, α-tubulin staining (microtubules), GFP-EB1, and DNA. Bar (D), 5 μm for B–D. (E and F) Microtubule dynamics in interphase cells treated with hesperadin (E) or the analogue (F) show no significant differences (6 microtubules from 2 cells shown; also see Table 1).
DISCUSSION
In this study, we have shown that survivin functions as a novel regulator of microtubule dynamics and microtubule nucleation throughout the cell cycle and that this pathway is independent of the expression or activity of the chromosomal passenger protein Aurora B. Time-lapse imaging of living cells using two independent GFP-labeled microtubule markers, the plus-end protein EB1 and α-tubulin, combined with quantitative analysis of multiple parameters of microtubule dynamics revealed that survivin affected rates of microtubule catastrophe and the degree of centrosomal microtubule nucleation.
Our results are consistent with a role for survivin in microtubule dynamics and microtubule nucleation independent of at least one protein of the chromosomal passenger complex (Aurora B) for several reasons. First, we found no effect on microtubule dynamics or nucleation when Aurora B was pharmacologically inactivated during a short incubation periods (up to 6 h), even though inhibitory activity dramatically affected spindle function. Second, inhibition of Aurora B activity or depletion of Aurora B levels produced spindle phenotypes dramatically different from survivin-depleted or -overexpressing cells. Third, survivin affected microtubule dynamics and nucleation in interphase when Aurora B is thought to be absent or drastically reduced. Fourth, our previous studies show that survivin is in multiple separate compartments within cells (Fortugno et al., 2002), whereas Aurora B and other chromosomal passenger proteins have not been localized to these other sites. Fifth, our biochemical studies suggest that cells possess a fraction of survivin that is not physically associated with the chromosomal passenger complex and even if it was in the complex, survivin could still affect microtubule dynamics and nucleation independent of other members of the complex.
A Unifying Model for the Multiplicity of Survivin Phenotypes?
The work in this manuscript provides new mechanistic insights into the complex functions of survivin at multiple cell cycle stages. We propose a model in which survivin modulation of microtubule dynamics and nucleation contributes to microtubule-based functions at multiple cell cycle stages and at all cellular sites to which the protein is localized. In this model, survivin could modulate the organization and/or function of mitotic spindles (Li et al., 1999; Giodini et al., 2002; Okada et al., 2004), the spindle checkpoint (Carvalho et al., 2003; Lens et al., 2003), midbody activities (Adams et al., 2001), centrosome-mediated microtubule nucleation and organization (Li et al., 1999), and interphase microtubule-based processes.
Two Pathways to Regulate Microtubule Dynamics
The mitotic function of survivin is thought to be related to its localization to kinetochores, and in particular its association with at least some “chromosomal passenger proteins” (Wheatley et al., 2001). In addition to its potential involvement in proper kinetochore attachment, central spindle formation, and cytokinesis, the chromosomal passenger complex has been more recently implicated in a Ran-GTP-independent pathway of bipolar spindle assembly via Aurora B inhibitory phosphorylation of the microtubule-depolymerizing activity of the Kin I kinesin MCAK. It remains possible that survivin indirectly regulates MCAK activity, and therefore microtubule stability, at the kinetochore. However, our data argue for the existence of a separate, survivin-selective/specific pathway for modulating microtubule dynamics. For example, the complex could be selectively involved in modulating microtubule stability at kinetochores, whereas survivin could participate in more global mechanisms of spindle assembly/function, midbody function and interphase microtubule nucleation/organization. The differences in the ability to detect an interaction of Aurora B and survivin observed by different investigators will require further investigation. It is possible that these differences reflect an interaction that is labile or transient. In any case, we think that the ability of survivin to modulate microtubule nucleation and dynamics in an Aurora B-independent manner could be achieved whether the protein is alone or in a complex with Aurora B and other members of the chromosomal passenger complex.
Survivin as a Dual-Function Protein
The IAP gene family is comprised of two classes of molecules, one implicated in cell division and a second that suppresses apoptosis via inhibition of caspase maturation and/or activity. However, as a structurally unique IAP family member, survivin is an apparent exception to this rigid definition, and accumulating evidence indicates a role for the protein in both functions (Altieri, 2003). Survivin seems to inhibit apoptosis through a pathway centered on intermolecular cooperation with cofactors (Marusawa et al., 2003) and dynamic subcellular shuttling of a mitochondrial pool of survivin. The role of survivin in mitotic control has remained controversial. Although survivin is unanimously viewed as indispensable for cell division, given the panoply of catastrophic mitotic defects induced by functional abrogation of the protein, the mechanism of mitotic disruption and the cell cycle stages regulated by this protein are unclear. Our results suggest that the complex series of interphase and mitotic defects induced by changes in survivin levels are modulated through its ability to control microtubule nucleation and dynamics.
Survivin-dependent Changes in Microtubule Dynamics during Cytokinesis
Our data demonstrate a role for survivin in cytokinesis, which was originally proposed by analogy with ancestral IAP proteins in model organisms (Speliotes et al., 2000) and by observations linking survivin depletion to regression of cleavage furrow (Chen et al., 2000). In this study, we first show that midbody microtubules are highly dynamic in normal cells, a surprising result given the apparent static nature and high stability of this population of microtubules (high level of acetylated tubulin). Moreover, we show that microtubules grow toward the central midbody as might be expected, but also away from the midbody. This suggests the presence of several microtubule populations at this site. They could arise from overlapping microtubules of the central spindle (plus ends at center), cytoplasmic microtubules that subsequently invade the midbody, or midbody-generated microtubules possibly nucleated by γ-tubulin at the central midbody. Further studies will be required to elucidate the functions of these populations of midbody microtubules. The dynamics of these microtubule populations is severely inhibited when survivin levels are altered, and this may account for the observed cytokinesis defects.
Survivin-dependent Changes in Microtubule Dynamics in Interphase
An important conclusion of this study is that the effect of survivin depletion or overexpression on microtubule dynamics was not restricted to mitosis but occurred throughout multiple cell cycle phases, including interphase. There is already convincing evidence that survivin expression may be induced outside mitosis, and this has been experimentally validated for cytokine-stimulated hematopoietic progenitors, angiogenic endothelial cells, and tumor cells where survivin is abundantly overexpressed at all cell cycle phases (Altieri, 2003). This may reflect distinct transcriptional mechanisms of survivin gene expression at different cell cycle phases, as exemplified by the recently reported role of E2F family proteins at inducing survivin expression at the G1/S transition. Accordingly, the ability of survivin to control microtubule dynamics at multiple cell cycle phases may have dramatic repercussions for cancer cells, promoting aneuploidy at cell division but also potentially altering cell polarity through disruption of microtubule dynamics and nucleation in interphase. Moreover, the interphase effects of survivin on microtubules provide additional evidence for an Aurora B-independent activity because the kinase is thought to be mitosis specific.
In summary, our data demonstrate that survivin functions in a continuum throughout multiple cell cycle phases and its role is centered on the regulation of microtubule dynamics and nucleation. The independence of this pathway from Aurora B kinase expression and activity suggests that it may provide a novel mechanism of microtubule regulation in both mitotic and interphase cells, and a potential critical point of intervention for molecular antagonists of survivin as rational anticancer agents (Altieri, 2003). The overexpression of survivin in nearly every human tumor underlines the importance of this endeavor.
Supplementary Material
Acknowledgments
We thank Drs. Claire E. Walczak and Patricia Wadsworth for assistance with microtubule dynamics analysis. EB1-GFP was generously provided by Dr. Lynne Cassimeris. This work was supported by National Institutes of Health Grants HL-54131, CA-78810, and CA-90917 (to D.C.A.) and GM-51994 and CA-82834 (to S.J.D.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–08–0723) on January 11, 2006.
Abbreviations used: IAP, inhibitor of apoptosis.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Adams, R. R., Carmena, M., and Earnshaw, W. C. (2001). Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11, 49–54. [DOI] [PubMed] [Google Scholar]
- Altieri, D. C. (2003). Validating survivin as a cancer therapeutic target. Nat. Rev. Cancer 3, 46–54. [DOI] [PubMed] [Google Scholar]
- Beardmore, V. A., Ahonen, L. J., Gorbsky, G. J., and Kallio, M. J. (2004). Survivin dynamics increases at centromeres during G2/M phase transition and is regulated by microtubule-attachment and Aurora B kinase activity. J. Cell Sci. 117, 4033–4042. [DOI] [PubMed] [Google Scholar]
- Beltrami, E., Plescia, J., Wilkinson, J. C., Duckett, C. S., and Altieri, D. C. (2004). Acute ablation of survivin uncovers p53-dependent mitotic checkpoint functions and control of mitochondrial apoptosis. J. Biol. Chem. 279, 2077–2084. [DOI] [PubMed] [Google Scholar]
- Blanc-Brude, O. P., Mesri, M., Wall, N. R., Plescia, J., Dohi, T., and Altieri, D. C. (2003). Therapeutic targeting of the survivin pathway in cancer: initiation of mitochondrial apoptosis and suppression of tumor-associated angiogenesis. Clin. Cancer Res. 9, 2683–2692. [PubMed] [Google Scholar]
- Bolton, M. A., Lan, W., Powers, S. E., McCleland, M. L., Kuang, J., and Stukenberg, P. T. (2002). Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol. Biol. Cell 13, 3064–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulinski, J. C., Richards, J. E., and Piperno, G. (1988). Posttranslational modifications of alpha tubulin: detyrosination and acetylation differentiate populations of interphase microtubules in cultured cells. J. Cell Biol. 106, 1213–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, A., Carmena, M., Sambade, C., Earnshaw, W. C., and Wheatley, S. P. (2003). Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 116, 2987–2998. [DOI] [PubMed] [Google Scholar]
- Chen, J., et al. (2000). Down-regulation of survivin by antisense oligonucleotides increases apoptosis, inhibits cytokinesis and anchorage-independent growth. Neoplasia 2, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., and Mitchison, T. J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117. [DOI] [PubMed] [Google Scholar]
- Dictenberg, J. B., Zimmerman, W., Sparks, C. A., Young, A., Vidair, C., Zheng, Y., Carrington, W., Fay, F. S., and Doxsey, S. J. (1998). Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 141, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohi, T., Beltrami, E., Wall, N. R., Plescia, J., and Altieri, D. C. (2004). Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J. Clin. Investig. 114, 1117–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortugno, P., Wall, N. R., Giodini, A., O'Connor, D. S., Plescia, J., Padgett, K. M., Tognin, S., Marchisio, P. C., and Altieri, D. C. (2002). Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J. Cell Sci. 115, 575–585. [DOI] [PubMed] [Google Scholar]
- Gassmann, R., Carvalho, A., Henzing, A. J., Ruchaud, S., Hudson, D. F., Honda, R., Nigg, E. A., Gerloff, D. L., and Earnshaw, W. C. (2004). Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 166, 179–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giodini, A., Kallio, M., Wall, N. R., Gorbsky, G. J., Tognin, S., Marchisio, P. C., Symons, M., and Altieri, D. C. (2002). Regulation of microtubule stability and mitotic progression by survivin. Cancer Res. 62, 2462–2467. [PubMed] [Google Scholar]
- Gliksman, N. R., Skibbens, R. V., and Salmon, E. D. (1993). How the transition frequencies of microtubule dynamic instability (nucleation, catastrophe, and rescue) regulate microtubule dynamics in interphase and mitosis: analysis using a Monte Carlo computer simulation. Mol. Biol. Cell 4, 1035–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley, A., Jurczyk, A., Sillibourne, J., Halilovic, E., Mogensen, M., Groisman, I., Blomberg, M., and Doxsey, S. (2003). A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161, 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley, A., Yeaman, C., Rosa, J., Redick, S., Chen, C. T., Mirabelle, S., Guha, M., Sillibourne, J., and Doxsey, S. J. (2005). Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell 123, 75–87. [DOI] [PubMed] [Google Scholar]
- Grossman, D., Kim, P. J., Blanc-Brude, O. P., Brash, D. E., Tognin, S., Marchisio, P. C., and Altieri, D. C. (2001). Transgenic expression of survivin in keratinocytes counteracts UVB-induced apoptosis and cooperates with loss of p53. J. Clin. Investig. 108, 991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurland, G., and Gundersen, G. G. (1995). Stable, detyrosinated microtubules function to localize vimentin intermediate filaments in fibroblasts. J. Cell Biol. 131, 1275–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf, S., Cole, R. W., LaTerra, S., Zimmer, C., Schnapp, G., Walter, R., Heckel, A., van Meel, J., Rieder, C. L., and Peters, J. M. (2003). The small molecule hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich, A., Lisztwan, J., Barry, R., Ballschmieter, P., and Krek, W. (2003). Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat. Cell Biol. 5, 64–70. [DOI] [PubMed] [Google Scholar]
- Honda, R., Korner, R., and Nigg, E. A. (2003). Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell 14, 3325–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens, S. M., Wolthuis, R. M., Klompmaker, R., Kauw, J., Agami, R., Brummelkamp, T., Kops, G., and Medema, R. H. (2003). Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 22, 2934–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Ackermann, E. J., Bennett, C. F., Rothermel, A. L., Plescia, J., Tognin, S., Villa, A., Marchisio, P. C., and Altieri, D. C. (1999). Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat. Cell Biol. 1, 461–466. [DOI] [PubMed] [Google Scholar]
- Ligon, L. A., Shelly, S. S., Tokito, M., and Holzbaur, E. L. (2003). The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol. Biol. Cell 14, 1405–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusawa, H., Matsuzawa, S., Welsh, K., Zou, H., Armstrong, R., Tamm, I., and Reed, J. C. (2003). HBXIP functions as a cofactor of survivin in apoptosis suppression. EMBO J. 22, 2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesri, M., Wall, N. R., Li, J., Kim, R. W., and Altieri, D. C. (2001). Cancer gene therapy using a survivin mutant adenovirus. J. Clin. Investig. 108, 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales, C. P., Holt, S. E., Ouelette, M., Kaur, K. J., Yan, Y., Wilson, K. S., White, M. A., Wright, W. E., and Shay, J. W. (1999). Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat. Genet. 21, 115–118. [DOI] [PubMed] [Google Scholar]
- Okada, H., et al. (2004). Survivin loss in thymocytes triggers p53-mediated growth arrest and p53-independent cell death. J. Exp. Med. 199, 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehl, M., Tulu, U. S., Wadsworth, P., and Cassimeris, L. (2004). Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc. Natl. Acad. Sci. USA 101, 1584–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit, A., Tynan, S. H., Vallee, R., and Doxsey, S. J. (1999). Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 147, 481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne, N. J., and Schroer, T. A. (2002). Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J. Cell Biol. 159, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan, N. M., Fagerstrom, C. J., Yvon, A. M., and Wadsworth, P. (2001). Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol. Biol. Cell 12, 971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvesen, G. S., and Duckett, C. S. (2002). Apoptosis: IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 3, 401–410. [DOI] [PubMed] [Google Scholar]
- Sampath, S. C., Ohi, R., Leismann, O., Salic, A., Pozniakovski, A., and Funabiki, H. (2004). The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 118, 187–202. [DOI] [PubMed] [Google Scholar]
- Sessa, F., Mapelli, M., Ciferri, C., Tarricone, C., Areces, L. B., Schneider, T. R., Stukenberg, P. T., and Musacchio, A. (2005). Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell 18, 379–391. [DOI] [PubMed] [Google Scholar]
- Speliotes, E. K., Uren, A., Vaux, D., and Horvitz, H. R. (2000). The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell 6, 211–223. [DOI] [PubMed] [Google Scholar]
- Temme, A., Diestelkoetter-Bachert, P., Schmitz, M., Morgenroth, A., Weigle, B., Rieger, M. A., Kiessling, A., and Rieber, E. P. (2005). Increased p21(ras) activity in human fibroblasts transduced with survivin enhances cell proliferation. Biochem. Biophys. Res. Commun. 327, 765–773. [DOI] [PubMed] [Google Scholar]
- Tirnauer, J. S., Salmon, E. D., and Mitchison, T. J. (2004). Microtubule plus-end dynamics in Xenopus egg extract spindles. Mol. Biol. Cell 15, 1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toso, R. J., Jordan, M. A., Farrell, K. W., Matsumoto, B., and Wilson, L. (1993). Kinetic stabilization of microtubule dynamic instability in vitro by vinblastine. Biochemistry 32, 1285–1293. [DOI] [PubMed] [Google Scholar]
- Uren, A. G., Beilharz, T., O'Connell, M. J., Bugg, S. J., van Driel, R., Vaux, D. L., and Lithgow, T. (1999). Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc. Natl. Acad. Sci. USA 96, 10170–10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren, A. G., Wong, L., Pakusch, M., Fowler, K. J., Burrows, F. J., Vaux, D. L., and Choo, K. H. (2000). Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol. 10, 1319–1328. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer, C. M., Salmon, W. C., and Salmon, E. D. (2000). Feedback interactions between cell–cell adherens junctions and cytoskeletal dynamics in newt lung epithelial cells. Mol. Biol. Cell 11, 2471–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley, S. P., Carvalho, A., Vagnarelli, P., and Earnshaw, W. C. (2001). INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr. Biol. 11, 886–890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.