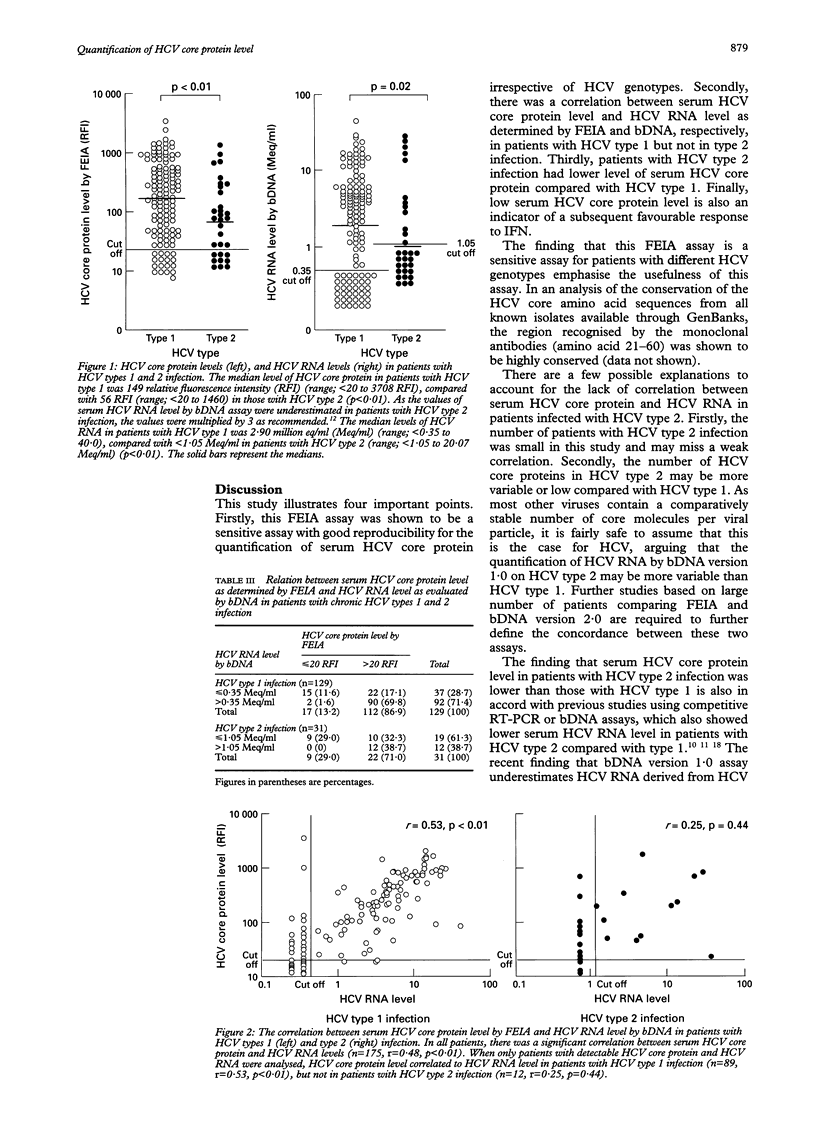
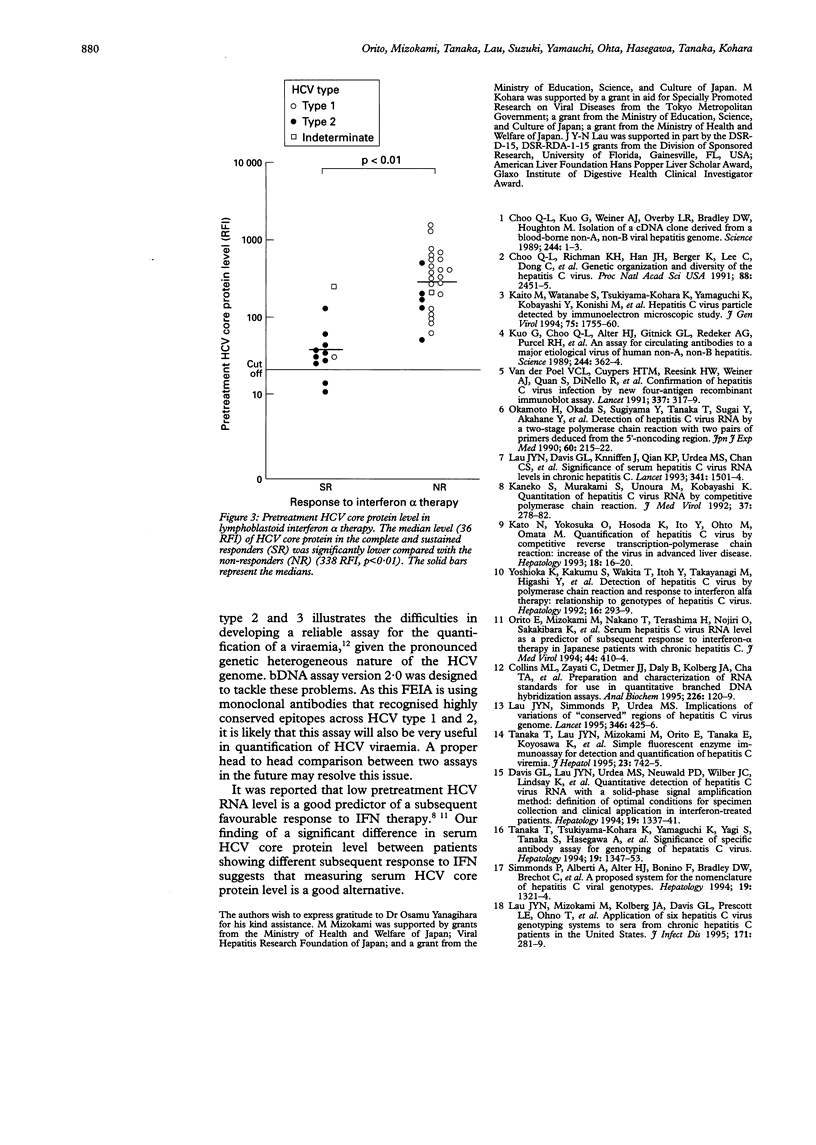
Abstract
BACKGROUND/AIM: A novel fluorescent enzyme immunoassay (FEIA) for the detection and quantification of serum hepatitis C virus (HCV) core protein was developed. The aim of this study was to evaluate the relation among serum HCV core protein level, HCV RNA level, and HCV genotype in patients with chronic HCV infection. PATIENTS AND METHODS: Serum HCV core protein, HCV RNA, HCV genotype were determined in 175 patients using the FEIA, branched DNA assay (Quantiplex HCV RNA ver 1.0), and serologically defined genotyping assay, respectively. For the specificity, all 13 patients seronegative for anti-HCV were negative for serum core antigen and HCV RNA by FEIA and bDNA, respectively. RESULTS: FEIA assay seems to be more sensitive than bDNA for patients with HCV type 2 infection (detection: 83.4% v 63.4%, p < 0.01). There was a good overall correlation between the FEIA and bDNA results. However, when the patients were stratified into their HCV types, a correlation was observed in HCV type 1 but not in type 2 infection. Patients with HCV type 2 infection had a lower serum HCV core protein level (median 56 RFI) compared with type 1 infection (median 149 RFI, p < 0.01). Thirty seven patients subsequently received interferon alpha therapy, patients who showed a complete and sustained response had a lower pretreatment serum HCV core protein level compared with patients who had a relapse and nonresponders (36 v 338 RFI, p < 0.01). CONCLUSIONS: This study showed that FEIA (1) is a good assay for the detection and quantification of serum HCV core protein level, (2) is also very sensitive in detecting HCV core protein in patients with HCV type 2 infection, and (3) may have a role as a predictor of subsequent response to interferon therapy.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Choo Q. L., Richman K. H., Han J. H., Berger K., Lee C., Dong C., Gallegos C., Coit D., Medina-Selby R., Barr P. J. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M. L., Zayati C., Detmer J. J., Daly B., Kolberg J. A., Cha T. A., Irvine B. D., Tucker J., Urdea M. S. Preparation and characterization of RNA standards for use in quantitative branched DNA hybridization assays. Anal Biochem. 1995 Mar 20;226(1):120–129. doi: 10.1006/abio.1995.1199. [DOI] [PubMed] [Google Scholar]
- Davis G. L., Lau J. Y., Urdea M. S., Neuwald P. D., Wilber J. C., Lindsay K., Perrillo R. P., Albrecht J. Quantitative detection of hepatitis C virus RNA with a solid-phase signal amplification method: definition of optimal conditions for specimen collection and clinical application in interferon-treated patients. Hepatology. 1994 Jun;19(6):1337–1341. [PubMed] [Google Scholar]
- Kaito M., Watanabe S., Tsukiyama-Kohara K., Yamaguchi K., Kobayashi Y., Konishi M., Yokoi M., Ishida S., Suzuki S., Kohara M. Hepatitis C virus particle detected by immunoelectron microscopic study. J Gen Virol. 1994 Jul;75(Pt 7):1755–1760. doi: 10.1099/0022-1317-75-7-1755. [DOI] [PubMed] [Google Scholar]
- Kaneko S., Murakami S., Unoura M., Kobayashi K. Quantitation of hepatitis C virus RNA by competitive polymerase chain reaction. J Med Virol. 1992 Aug;37(4):278–282. doi: 10.1002/jmv.1890370408. [DOI] [PubMed] [Google Scholar]
- Kato N., Yokosuka O., Hosoda K., Ito Y., Ohto M., Omata M. Quantification of hepatitis C virus by competitive reverse transcription-polymerase chain reaction: increase of the virus in advanced liver disease. Hepatology. 1993 Jul;18(1):16–20. [PubMed] [Google Scholar]
- Kuo G., Choo Q. L., Alter H. J., Gitnick G. L., Redeker A. G., Purcell R. H., Miyamura T., Dienstag J. L., Alter M. J., Stevens C. E. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989 Apr 21;244(4902):362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- Lau J. Y., Davis G. L., Kniffen J., Qian K. P., Urdea M. S., Chan C. S., Mizokami M., Neuwald P. D., Wilber J. C. Significance of serum hepatitis C virus RNA levels in chronic hepatitis C. Lancet. 1993 Jun 12;341(8859):1501–1504. doi: 10.1016/0140-6736(93)90635-t. [DOI] [PubMed] [Google Scholar]
- Lau J. Y., Mizokami M., Kolberg J. A., Davis G. L., Prescott L. E., Ohno T., Perrillo R. P., Lindsay K. L., Gish R. G., Qian K. P. Application of six hepatitis C virus genotyping systems to sera from chronic hepatitis C patients in the United States. J Infect Dis. 1995 Feb;171(2):281–289. doi: 10.1093/infdis/171.2.281. [DOI] [PubMed] [Google Scholar]
- Lau J. Y., Simmonds P., Urdea M. S. Implications of variations of "conserved" regions of hepatitis C virus genome. Lancet. 1995 Aug 12;346(8972):425–426. doi: 10.1016/s0140-6736(95)92786-7. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Okada S., Sugiyama Y., Tanaka T., Sugai Y., Akahane Y., Machida A., Mishiro S., Yoshizawa H., Miyakawa Y. Detection of hepatitis C virus RNA by a two-stage polymerase chain reaction with two pairs of primers deduced from the 5'-noncoding region. Jpn J Exp Med. 1990 Aug;60(4):215–222. [PubMed] [Google Scholar]
- Orito E., Mizokami M., Nakano T., Terashima H., Nojiri O., Sakakibara K., Mizuno M., Ogino M., Nakamura M., Matsumoto Y. Serum hepatitis C virus RNA level as a predictor of subsequent response to interferon-alpha therapy in Japanese patients with chronic hepatitis C. J Med Virol. 1994 Dec;44(4):410–414. doi: 10.1002/jmv.1890440418. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Alberti A., Alter H. J., Bonino F., Bradley D. W., Brechot C., Brouwer J. T., Chan S. W., Chayama K., Chen D. S. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology. 1994 May;19(5):1321–1324. [PubMed] [Google Scholar]
- Tanaka T., Lau J. Y., Mizokami M., Orito E., Tanaka E., Kiyosawa K., Yasui K., Ohta Y., Hasegawa A., Tanaka S. Simple fluorescent enzyme immunoassay for detection and quantification of hepatitis C viremia. J Hepatol. 1995 Dec;23(6):742–745. doi: 10.1016/0168-8278(95)80043-3. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Tsukiyama-Kohara K., Yamaguchi K., Yagi S., Tanaka S., Hasegawa A., Ohta Y., Hattori N., Kohara M. Significance of specific antibody assay for genotyping of hepatitis C virus. Hepatology. 1994 Jun;19(6):1347–1353. [PubMed] [Google Scholar]
- Van der Poel C. L., Cuypers H. T., Reesink H. W., Weiner A. J., Quan S., Di Nello R., Van Boven J. J., Winkel I., Mulder-Folkerts D., Exel-Oehlers P. J. Confirmation of hepatitis C virus infection by new four-antigen recombinant immunoblot assay. Lancet. 1991 Feb 9;337(8737):317–319. doi: 10.1016/0140-6736(91)90942-i. [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Kakumu S., Wakita T., Ishikawa T., Itoh Y., Takayanagi M., Higashi Y., Shibata M., Morishima T. Detection of hepatitis C virus by polymerase chain reaction and response to interferon-alpha therapy: relationship to genotypes of hepatitis C virus. Hepatology. 1992 Aug;16(2):293–299. doi: 10.1002/hep.1840160203. [DOI] [PubMed] [Google Scholar]


