Abstract
Integration of high-risk human papillomavirus (HRHPV) into the host genome is a key event in cervical neoplastic progression. Integration is associated with deregulated expression of the viral oncogenes E6 and E7 and acquisition of a selective growth advantage for cells containing integrants. Overexpression of the viral transcriptional regulator E2 from heterologous promoters has an inhibitory effect on transcription from integrated HRHPV. Therefore, we hypothesized that loss of E2-expressing episomes from cells in which integration had previously occurred would be required for such cells to gain a growth advantage. Using the unique W12 model of cervical squamous carcinogenesis, we show that cells containing integrated HPV16 reproducibly emerged during long-term culture when there had been a rapid fall in episome numbers. During the period of emergence, it is possible to isolate single-cell clones containing an intracellular mixture of the integrant being selected and episomes at reduced load. The lower level of E2 expression seen in such cells is associated with partial inhibition of transcription from the HPV16 integrant. Full deregulation is not observed until complete loss of E2-expressing episomes occurs. Microarray analysis showed that episome loss was closely associated with endogenous activation of antiviral response genes that are also inducible by the type I IFN pathway. Taken together, our results indicate that episome loss, associated with induction of antiviral response genes, is a key event in the spontaneous selection of cervical keratinocytes containing integrated HPV16. We conclude that cervical carcinogenesis requires not only HRHPV integration, but also loss of inhibitory episomes.
Keywords: human papillomavirus, cervix, integration, interferon, progression
Integration of high-risk human papillomavirus (HRHPV) into the host genome is an important step in cervical neoplastic progression (1, 2). Integrated viral genomes from which HRHPV early genes are transcribed have been detected in ≈87.5% of cervical malignancies (3). Integration usually causes deletion or disruption of the viral regulatory E2 gene, while retaining a variable segment including the E6 and E7 oncogenes and the upstream regulatory region (4, 5). Overexpression of E2 from heterologous promoters in cells harboring integrated HRHPV can repress the early promoter of the integrated virus, causing a sharp reduction in E6 and E7 expression (6). Thus, HRHPV integration and disruption/deletion of E2 leads to increased expression of the viral oncogenes (7, 8). Cells containing integrated HRHPV acquire a growth advantage over cells harboring episomal HRHPV (the natural viral state in productive infections) and show increased genomic instability (9–11).
Cervical keratinocyte cell lines established from precursor low-grade squamous intraepithelial lesions have indicated that episomal HRHPV genomes are maintained at ≈100 copies per cell in the basal region of an infected epithelium (12, 13). Viral integration is therefore most likely to occur in cells containing this number of episomes. It has recently been suggested that, whereas overexpression of E2 can inhibit the early promoter of integrated HRHPV, it has little or no effect on the transcription from episomal HRHPV (14). It follows that physiological levels of E2 expressed in episomally infected keratinocytes could inhibit transcription from coexistent integrants, preventing deregulated viral oncogene expression, cell selection, and clonal outgrowth. We therefore hypothesize that loss of E2-expressing episomes plays a key role in the emergence of cervical keratinocytes containing “selectable” HRHPV integrants (i.e., those that retain the upstream regulatory region, E6, and E7 and have disrupted/deleted E2 genes). This model represents a departure from prevailing views of HRHPV-related oncogenesis, because progression is generally assumed to be due simply to cells containing exclusively integrated HRHPV outgrowing cells with only episomal genomes (7).
We have tested our hypothesis by using the unique HPV16-containing cervical keratinocyte cell line W12 in monolayer culture (12); this represents a useful system to investigate the effects of HPV16 infection in basal cervical squamous cells, the key site of deregulation of HRHPV viral oncogenes in cervical neoplasia (10). W12 was derived from a low-grade squamous intraepithelial lesion (LG-SIL), which resulted from “natural” infection in vivo with HPV16, the HRHPV type most commonly detected in cervical carcinomas (15). At early passages, W12 retains ≈100 HPV16 episomes per cell and recapitulates a LG-SIL in organotypic culture (8, 12). We have previously demonstrated that W12 accurately models cervical neoplastic progression during long-term culture, with spontaneous transition from cells containing only episomal HPV16 to a population containing only integrated HPV16 (8, 10). Therefore, W12 represents a valuable system for studying events associated with spontaneous selection of cervical keratinocytes containing only integrated HRHPV.
By growing W12 in long-term culture and undertaking single-cell cloning we demonstrate here that selection of cells containing integrated HPV16 is indeed associated with episome loss from cells also containing the integrant. Moreover, episome loss is itself associated with endogenous activation of antiviral genes. Our data suggest that models of HRHPV-related carcinogenesis must include not only viral integration but also the steps leading to loss of episome-mediated inhibition of selectable integrants.
Results
Physical State of HPV16 in Multiple Long-Term Passage Series of W12.
We previously showed that in a long-term passage series of W12, referred to here as W12.Series1, there was loss of episomal HPV16 and selection of cells containing only integrated virus (8, 10). We aimed to analyze the period of selective outgrowth more closely and to determine the characteristics of episome loss in additional long-term series (W12.Series2 and W12.Series3) originating from the same starting polyclonal population at passage 10 (W12p10EPI, which contains ≈100 episomes per cell; refs. 8 and 12).
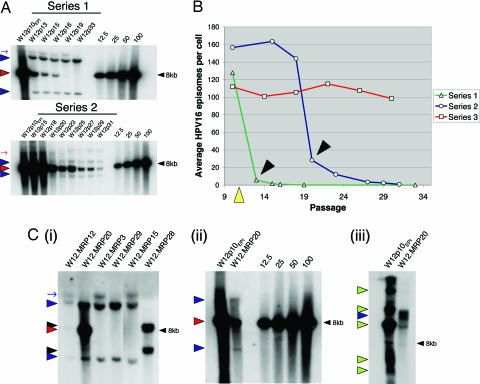
We performed Southern blot analysis using the HPV16 single-cutter BamHI (Fig. 1A, for W12.Series3, see Fig. 5, which is published as supporting information on the PNAS web site) and quantified episomes based on the intensity of the 7.9-kb episomal band (Fig. 1B). W12.Series1 demonstrated an early rapid reduction in episome numbers. By passage 13 (p13), the average episome content was reduced by more than 95%, and this was followed by a more gradual reduction in residual episomes until none was detectable by p19. The selected HPV16 integrant (indicated by the presence of two additional BamHI bands) emerged when episome numbers had rapidly fallen and was subsequently retained.
Fig. 1.
Southern blot analysis of HPV16 physical state in multiple long-term passage series of polyclonal W12. (A) Genomic DNA from independent long-term passage series of W12, each starting from episome-only cells (W12p10EPI), were digested with the HPV16 single-cutter BamHI and subjected to Southern blot analysis using 32P-labeled full-length HPV16 as probe. The images depict autoradiographs of the hybridization pattern from W12 long-term passage series 1 and 2 (for series 3, see Fig. 5), together with copy number controls (right lanes). Red and blue arrowheads indicate bands generated by unit length HPV16 and virus–host junctions, respectively. Red and blue arrows indicate partially digested HPV16 episomes and HPV16-containing fragments, respectively. (B) Line graphs depicting PhosphorImager quantification of the average HPV16 episome copy number per cell in each of the long-term passage series. Black arrowheads indicate the passage at which integrated HPV16 was first detected in W12.Series1 and Series2. Limiting dilution cloning of cells in W12.Series1 was performed at p11 (yellow arrowhead). (C) Southern blot analysis of selected single-cell clones generated from W12.Series1. (i) Selected clones digested with BamHI. (ii and iii) Comparison of W12p10EPI and clone W12.MRP20, digested with BamHI (ii) or the HPV16 noncutter HindIII (iii). Copy number controls are also shown in ii. The red and blue arrowheads and blue arrow are as in A. Black arrowheads in i indicate bands generated by nonselected virus–host junctions in W12.MRP28. Green arrowheads in iii indicate various forms of uncut HPV16 episomes.
A comparable pattern of events was observed in W12.Series2. Rapid episome loss was observed between p18 and p20, where average episome content was reduced by >80%. A different selected integrant (producing two virus–host junction bands of different sizes to those in Series1) emerged when episome numbers had fallen rapidly. There was a more gradual reduction of residual episomes during subsequent passages. In contrast, in W12.Series3 neither loss of episomes nor selection of integrated HPV16 were observed after 20 passages and ≈100 population doublings (Fig. 5). These observations indicate that spontaneous selection of integrated HPV16 is associated reproducibly with rapid loss of episomes, although these events are not inevitable and their timing varies.
Clonal Analysis of W12 Long-Term Passage Series1 During the Period of Rapid Episome Loss.
Although Southern blot analysis was consistent with our hypothesis, we considered the alternative explanation that the kinetics of episome loss may simply have reflected integrant-only cells outgrowing variably less “fit” episome-only populations. Therefore, we generated single-cell clones by limiting dilution of W12.Series1 at p11; i.e., during the period of rapid episome loss and selection of integrated HPV16 (Fig. 1B). Thirty clones were generated, designated W12.MRP1–30.
We have shown that the upstream virus–host junction of the selected HPV16 integrant in W12.Series1 comprises a disrupted L2 gene and sequences at chromosome 5p15.1 (10). Therefore, we were able to design a PCR protocol for screening the physical state of HPV16 in the single-cell clones (see Fig. 6 and Supporting Text, which are published as supporting information on the PNAS web site). Based on the PCR results, Southern blot analysis was performed on six selected clones; W12.MRP12 and -20 (suggested by PCR to contain episomes and the integrant being selected), W12.MRP3 and -29 (containing the integrant only), and W12.MRP15 and -28 (E7-positive but negative for episomes and the integrant) (Fig. 1C).
BamHI digestion of DNA from W12.MRP12 and -20 confirmed the presence of an intracellular mixture of episomes and the integrant being selected, as indicated by both the 7.9-kb band and the virus–host junction fragments (Fig. 1Ci). The episome content in W12.MRP12 was lower than in W12.MRP20. Quantitative PhosphorImager analysis demonstrated an average of five episomes per cell in W12.MRP20, ≈5% of the load in W12p10EPI (Fig. 1Cii). Southern blot analysis of W12.MRP20 with the HPV16 noncutter HindIII confirmed that the 7.9-kb band represented episomal HPV16 and not unit-length head-to-tail integrated concatamers (Fig. 1Ciii). Clones W12.MRP3 and -29 contained only the HPV16 integrant. W12.MRP15 also showed virus–host junction bands of the selected HPV16 integrant, indicating failure of PCR amplification of the virus–host junction in this clone. Interestingly, W12.MRP28 showed the presence of two previously undetected virus–host junction bands, demonstrating the presence of at least one additional integrant within W12.Series1 that was out-competed during evolution of the polyclonal population.
Isolation of cells containing an intracellular mixture of episomes and the integrant being selected argues against a process of selection in which integrant-only cells outgrew episome-only cells. Given that all cells containing a selected integrant must ultimately be derived from the same cell of origin, the observation that the integrant in the “mixed” clones W12.MRP12 and -20 was identical to that in the fully selected integrant-only cells indicated that all of the integrant-bearing cells must have contained episomal HPV16 at some point during their evolution. Because retention of HPV episomes requires ≈100 copies per cell (12, 13), we conclude that the residual episomes in clones W12.MRP12 and -20 were in the process of being lost rapidly. Consistent with this, a parallel cloning experiment where the number of population doublings was higher before analysis of HPV16 physical state yielded 35 clones containing only the selected integrant. Thus, the mixed clones W12.MRP12 and -20 represented valuable isolates from a highly transient intermediate step in the selection process. The higher episome content in W12.MRP20 indicated that this clone represented an earlier stage of selection than W12.MRP12.
Viral Gene Expression in Selected W12.MRP Clones.
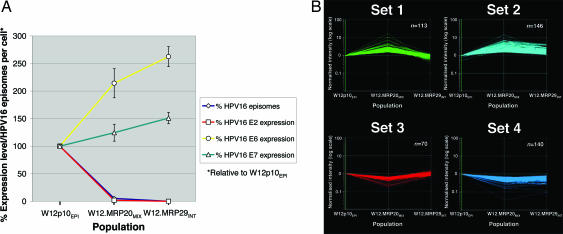
We analyzed viral gene expression in W12 populations representing different stages in the process of integrant selection. We compared W12p10EPI (episome-only) with clones W12.MRP20 (undergoing selection for the integrant) and W12.MRP29 (containing only the selected integrant), which are henceforth referred to as W12.MRP20MIX and W12.MRP29INT. E2 expression from episomes in W12.MRP20MIX was reduced by a level similar to the reduction in episome content (Fig. 2A); to ≈2% of levels in W12p10EPI. E2 was undetectable in W12.MRP29INT. Expression levels of E6 and E7 were increased in W12.MRP29INT relative to W12p10EPI. Given that HPV16 copy number in W12.MRP29INT was ≈5% of that in W12p10EPI (8, 12), this represented significant deregulation of each viral transcription center after complete selection of integrated HPV16. Interestingly, levels of E6 and E7 in W12.MRP20MIX were intermediate between those in W12p10EPI and W12.MRP29INT. The incomplete deregulation of viral oncogene levels in W12.MRP20MIX suggested that residual E2 expressed from the homologous HPV16 episomal promoter (even after a 98% reduction in levels) could still exert a partial inhibitory effect on expression from the integrant.
Fig. 2.
Patterns of viral and host gene expression associated with episome loss and selection of integrated HPV16. (A) Relationship between HPV16 episome number and viral gene expression in clones, presented as a percentage relative to W12p10EPI. W12.MRP20MIX demonstrates ≈5% episome copy number and 2% E2 expression level relative to W12p10EPI, whereas W12.MRP29INT demonstrates 0% relative to W12p10EPI for both. Error bars show the standard error of mean percentage expression levels, normalized by using four host housekeeping genes. (B) Selected k-means clusters generated from microarray analysis of W12p10EPI, W12.MRP20MIX, and W12.MRP29INT. Normalized intensity (y axis) refers to a per gene normalization using the mean value for W12p10EPI as a reference, and therefore equates for each gene to expression ratio versus W12p10EPI.
Expression Microarray Analysis.
Microarray analysis was performed to determine whether the episome loss was associated with specific changes in host gene expression, and whether the expression of any such genes subsequently reverted following complete episome clearance. The populations W12p10EPI, W12.MRP20MIX, and W12.MRP29INT were analyzed as a time course. A total of 776 characterized genes showed a significant change in expression in at least one population. K means cluster analysis of these genes generated four sets (sets 1–4, Fig. 2B) for which patterns of expression change included a significant difference between W12p10EPI and W12.MRP20MIX, the period when the major reduction in episomal load occurred. Four additional sets showing significant change overall between W12p10EPI and W12.MRP29INT but not between W12p10EPI and W12.MRP20MIX were also generated (data not shown). These were not considered further because it was much less likely that they would contain genes contributing to episome loss. Sets 1 and 2 were closely related, in that they contained genes that showed increased expression in W12.MRP20MIX relative to W12p10EPI, followed by a drop in W12.MRP29INT. The drop was back to baseline (or just below) in set 1 and to above baseline in set 2. Sets 3 and 4 were also closely related to each other, showing reciprocal patterns to those seen in sets 1 and 2, respectively. Genes showing >5-fold changes in expression in W12.MRP20MIX versus W12p10EPI were: SCGB1A1 (GenBank accession no. NM_003357) in set 1; C3 (GenBank accession no. NM_000064); CEACAM1 (GenBank accession no. X16352), DUSP4 (GenBank accession no. NM_001394), GALNT5 (GenBank accession no. BF002195), GDF15 (GenBank accession no. AF003934), MSMB (GenBank accession no. NM_002443), MUC4 (GenBank accession no. AJ242547), PTGS2 (GenBank accession no. AY151286), SLC16A14 (GenBank accession no. R15072), and WFDC2 (GenBank accession no. NM_006103) in set 2; EMP3 (GenBank accession no. NM_001425) in set 3; and CXCL14 (GenBank accession no. NM_004887), ECHDC3 (GenBank accession no. NM_024693), FLJ20366 (GenBank accession no. NM_017786), and TNC (GenBank accession no. NM_002160) in set 4.
Gene Ontology (GO) Analysis of Genes in k Means Cluster Sets 1–4.
Significantly enriched GO biological processes in k means cluster sets 1–4 are shown in Table 1, which is published as supporting information on the PNAS web site. The most significant representation in set 1 was from “response to virus” genes. Other GO categories in this set were closely related, for example “response to external biotic stimulus” and “immune response.” A further overrepresented biological pathway in set 1, and also in the closely related set 2, was TGF-β receptor signaling. In set 2, this was the only significantly enriched GO biological process. The GO categories in sets 3 and 4 were markedly different to those in sets 1 and 2. Many were related to cell cycle entry and progression, particularly M phase.
Changes in Relative Expression of Antiviral Genes Inducible by the Type I IFN Pathway.
The majority of the response to virus genes in set 1, such as MX1, MX2, OAS1, TRIM22, and G1P3, are induced by the type I IFN pathway. Also present in set 1 was a key regulator of IFN pathway, IRF7. Review of the array data showed that the majority of IFN-inducible genes had an essentially comparable pattern of gene expression change, with an increase in W12.MRP20MIX relative to W12p10EPI, followed by a decrease back to baseline or below in W12.MRP29INT (Fig. 7, which is published as supporting information on the PNAS web site). Array data for two IFN-inducible and two mitotic genes was validated by real-time RT-PCR (Fig. 8, which is published as supporting information on the PNAS web site). Taken together, these data indicated that an increase in expression of antiviral genes (that are inducible by type I IFN) was closely associated with episome loss during the selection of cells containing only integrated HPV16.
Expression of Antiviral Genes in W12 Long-Term Passage Series 2.
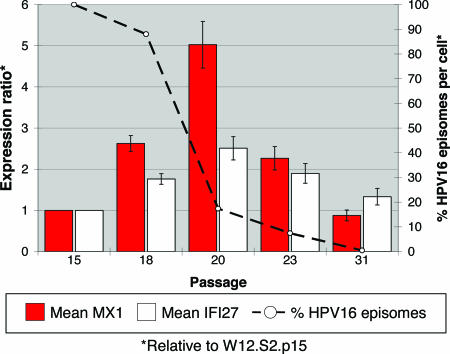
In view of the data obtained from W12.Series1, we performed quantitative PCR analysis of polyclonal long-term passage W12.Series2. We again observed a close association between increased expression of the IFN-inducible antiviral genes MX1 and IFI27 and the period of rapid episome loss. Both genes showed increased expression at p18 and a further increase at p20, with rapid episome loss occurring between these passages (Fig. 3). Expression of both genes reverted to baseline or just below by p31, when virtually all episomes had been lost. These data demonstrated that activation of antiviral response genes is associated reproducibly with loss of episomal HPV16 and selection of integrated virus in the W12 system.
Fig. 3.
Long-term passage W12.Series2: Relationship between episome copy number and antiviral gene expression in polyclonal cells. HPV16 episome copy number (dashed line) is presented as a percentage of that at P15 (W12.S2.p15), as determined by PhosphorImager analysis (right y axis). Expression levels of MX1 (red bars) and IFI27 (white bars) are ratios relative to W12.S2.p15, as determined by quantitative PCR (left y axis), with error bars representing the standard error.
Discussion
We have demonstrated that selection of cervical keratinocytes containing integrated HPV16 reproducibly occurred during long-term passage of W12 when numbers of HPV16 episomes had fallen rapidly. To our knowledge, ours is the first indication that low levels of E2 expressed from the homologous early promoter of HPV16 episomes (in cells representing an intermediate step in the selection process) can partially inhibit expression of a selectable integrant. We showed in a parallel study (M.T.H., M.R.P., I. Roberts, W. O. F. Alazawi, A. E. Tescherndorff, X.-Y. Zhang, M.A.S., and N.C., unpublished data) that activation of antiviral genes does not inhibit transcription from integrated HPV16. Assuming that the substantially higher levels of E2 expressed in keratinocytes containing a “normal” episome content (100 copies per cell) exert more complete inhibition, our data suggest that episome loss is not only closely associated with selection of integrated HPV16 but is also a prerequisite event. Consistent with this, our parallel study (M.T.H., M.R.P., I. Roberts, W. O. F. Alazawi, A. E. Tescherndorff, X.-Y. Zhang, M.A.S., and N.C., unpublished data) demonstrated that “selectable” integrated HPV16 can exist in a minority of cells in a polyclonal population for long periods without exerting a selective growth advantage until episome loss is initiated. Indeed, we also found that multiple different integrants may exist in a polyclonal population before the emergence of a particular clone (M.T.H., M.R.P., I. Roberts, W. O. F. Alazawi, A. E. Tescherndorff, X.-Y. Zhang, M.A.S., and N.C., unpublished data). Our finding in the present study of a rare integrant (in clone W12.MRP28) that was not ultimately selected in W12.Series1 supports these observations.
Rapid loss of HPV16 episomes correlates closely with increased expression of antiviral genes (inducible by type I IFN; ref. 16) and genes in the TGF-β receptor signaling pathway, with expression levels returning to baseline after complete episome clearance and selection of integrated HPV16. In other studies, exogenous type I IFN produced a gradual reduction in episome load in cells containing bovine papillomavirus type 1 and HPV31 episomes (17, 18) and, in our parallel study (M.T.H., M.R.P., I. Roberts, W. O. F. Alazawi, A. E. Tescherndorff, X.-Y. Zhang, M.A.S., and N.C., unpublished data) of early passage W12 (containing HPV16 episomes), rapidly initiated episomal clearance and hastened the transition from episomal to integrated HPV16.
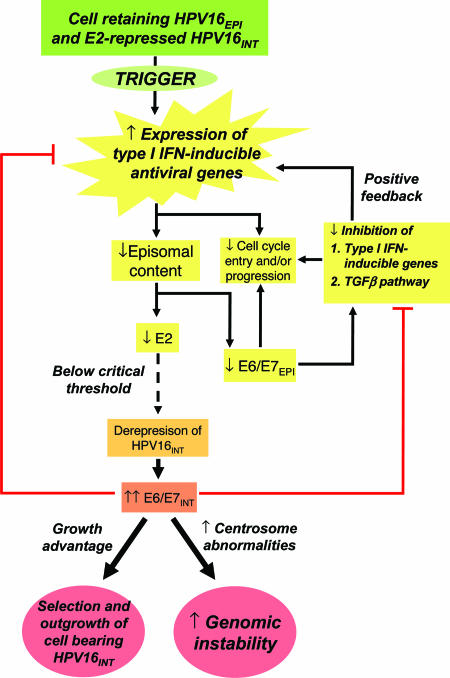
Taken together, our results are consistent with the scheme of events proposed in Fig. 4. Endogenous activation of antiviral genes in a cell containing a “selectable” HPV16 integrant induces a decrease in episome content, with proportionate reduction in episomal expression of E2, E6, and E7. Reduced E6 and E7 will lead to reduced inhibition of type I IFN-inducible genes (19–22) and the TGF-β pathway (23–25). These events would account for decreased expression of cell cycle genes (17, 26), and would also lead to rapid further reduction in episome levels in a positive feedback loop. Episome loss will eventually cause E2 expression to reach a critical low level, at which expression from the coexisting integrated HRHPV can no longer be inhibited. Deregulated expression of E6 and E7 from the integrant will confer a selective advantage and increase genomic instability, and at the same time inhibit type I IFN-inducible genes and activation of the TGF-β pathway. Thus, the expression levels of these host genes in emergent cells containing integrated HPV16 only will revert to baseline levels.
Fig. 4.
Proposed scheme of events leading to the selection of keratinocytes containing only integrated HPV16 in W12. E6/E7EPI and E6/E7INT refer to episome- and integrant-derived viral oncogene expression, respectively. HPV16EPI and HPV16INT refer respectively to episomal and integrated HPV16.
We have not yet identified the initial trigger for episome loss. Previous studies have suggested that the HRHPV E2 gene is required for a full antiviral response to be elicited by exogenous IFN (17). On the other hand, inhibition of type I IFN-inducible antiviral genes by the viral oncogenes that are also expressed by HRHPV episomes (19–22) suggests that there is a delicate balance between elicitation and inhibition of an antiviral response in cells containing HPV16 episomes. Perturbation of this balance may be the key to induction of the events shown in Fig. 4, and this in turn may be related to deregulation of virus or host gene expression due to genetic and/or epigenetic mechanisms.
Evidence from our parallel study (M.T.H., M.R.P., I. Roberts, W. O. F. Alazawi, A. E. Tescherndorff, X.-Y. Zhang, M.A.S., and N.C., unpublished data) that selectable integrated HPV16 can exist in a minority of cells in a polyclonal population for long periods without exerting a selective growth advantage argues against integration itself serving as the stimulus for episome loss. In an earlier study using W12.Series1 (8), we demonstrated increased expression (relative to an episome only population) of IFN-inducible genes in late passage integrant-only cells that had acquired high-level genomic instability after viral integration. These cells had undergone ≈40 population doublings after the selected integrant first became detectable, and had acquired many more chromosomal abnormalities than seen in any of the samples in the present study (Fig. 9, which is published as supporting information on the PNAS web site). This finding argues that high-level genomic instability in a HRHPV-infected cell may also activate genes inducible by the type I IFN pathway, despite the deregulated expression of the viral oncogenes.
It will now be important to assess whether the events that we have observed in W12 are also seen during spontaneous selection of other integrated HRHPV types. It may be possible to use other naturally infected keratinocytes, such as the HPV31b-containing cell line CIN612, for which episome-only and integrant-only forms are described (13). Given our evidence that episome loss and selection of integrated HPV16 occurs very rapidly in vitro, it will be difficult to detect similar events occurring in vivo. Nevertheless, our data are supported by the observation that most cervical carcinomas containing integrated HRHPV have little or no detectable episomal DNA (3, 27–31). Although some carcinomas do contain both episomal and integrated virus, in situ analysis has shown that regions containing only integrated HRHPV exist adjacent to regions containing apparently only episomes (32). Thus, in these cases, episome loss in the context of integrant selection is also applicable. On the other hand, ≈12.5% of cervical carcinomas appear to contain transcripts derived only from episomal HRHPV (3), suggesting an alternative pathway of episome-driven carcinogenesis that warrants further investigation.
We conclude from the W12 model system that induction of episome loss, associated with activation of antiviral response genes, is a key event in spontaneous selection of cells containing integrated HPV16. We propose that a revision of the current model of HPV16-induced cervical neoplasia is required. Progression of lesions in which HPV16 integration plays a role requires not only integration per se, but also loss of regulatory episomes.
Materials and Methods
Cell Culture.
Cell culture was as described (12, 33). Single-cell clones were generated by limiting dilution (34) from W12.Series1 at passage 11, when rapid episome loss was occurring (Fig. 1B). The single colonies generated were expanded in six-well plates for analysis of HPV16 physical state and levels of expression of host and viral genes. At ≈80% confluence, fibroblast feeder cells were removed, followed by extractions of genomic DNA as described (8) and of RNA using TRIzol (Invitrogen).
Southern Blot Analysis of the Physical State of HPV16.
Five micrograms of genomic DNA was restriction enzyme digested, electrophoresed through a 0.8% agarose gel, with appropriate copy number controls, and transferred to Hybond-N+ nylon membrane (Amersham Pharmacia) (8, 10). Probe was prepared by excision of full-length HPV16 DNA from the pspHPV16 plasmid (12), followed by labeling with [α-32P]dCTP by random priming. HPV16 was detected and quantified by using a PhosphorImager (Fuji).
Preparation and Hybridization of Probes for Microarray Analysis.
Total RNA from W12 samples was used to generate biotin-labeled cRNA for microarray analysis. Two technical replicates were performed for each sample to control for variation in labeling and hybridization efficiency. Double-stranded cDNA was synthesized by using SuperScript (Invitrogen), employing the (dT)24-T7 promoter primer. Biotin-labeled cRNA was then generated by Bioarray in vitro transcription (Enzo), and fragmented by metal-induced hydrolysis. Probe from each replicate was hybridized, washed, stained, and scanned by the Medical Research Council Geneservice (Cambridge, U.K.) using standard Affymetrix procedures. We used GeneChip HG-U133 Plus 2.0 Arrays (Affymetrix).
Analysis of Microarray Data.
Data were analyzed by using genespring (Agilent Technologies). Initially, a “per chip” normalization to the 50th percentile was performed, followed by a “per gene” normalization using the mean expression values for each gene across the replicates of the reference W12p10EPI. Statistical analysis was performed in the “log of ratio” mode to give equal weighting to increases and decreases in expression level. Given that geometric mean values were used, this centered the expression values of all genes in the reference sample at or around 1, and gave expression values for the clones that equated to the expression ratio relative to the W12p10EPI reference. Stringent criteria were then applied to highlight significant gene expression changes. First, we filtered out genes with expression levels not altered by at least 1.5 fold in either of the clones relative to the reference. Second, to include OFF → ON and ON → OFF genes, we further considered genes flagged as present or marginal in both replicates of at least one of the three populations analyzed. Because low-range expression values may be unreliable, only genes with at least one raw value of >100 when flagged as present or marginal were analyzed. After filtering, a one-way ANOVA was performed to detect genes with significantly altered expression (P < 0.05) in at least one of the three populations analyzed. K means cluster analysis was then applied to the set of significantly altered genes. GO biological process analysis was performed by using the gominer tool (35). Significant enrichment of specific GO biological processes in each k means cluster was determined by using a one-sided Fisher’s exact test (35). P values were adjusted by using a false discovery rate correction (36).
Real-Time RT-PCR.
Quantitative PCR of cDNA was performed to quantify viral gene expression and validate changes in expression of selected host genes. We adapted SYBR green protocols and used four housekeeping genes to normalize expression levels (see Supporting Text).
Supplementary Material
Acknowledgments
We thank Ian Roberts and Emma Gooding (Medical Research Council Cancer Cell Unit) and Barry Zeeberg (National Cancer Institute, Bethesda) for help and advice. This work was supported by the Medical Research Council and Cancer Research UK.
Abbreviations
- HRHPV
high-risk human papillomavirus
- pn
passage n
- GO
Gene Ontology.
Footnotes
Conflict of interest statement: No conflicts declared.
Data deposition: Data from this study were deposited in the National Institutes of Health Gene Expression Omnibus database (accession no. GSE4289).
References
- 1.Longworth M. S., Laimins L. A. Microbiol. Mol. Biol. Rev. 2004;68:362–372. doi: 10.1128/MMBR.68.2.362-372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munger K., Baldwin A., Edwards K. M., Hayakawa H., Nguyen C. L., Owens M., Grace M., Huh K. J. Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klaes R., Woerner S. M., Ridder R., Wentzensen N., Duerst M., Schneider A., Lotz B., Melsheimer P., von Knebel Doeberitz M. Cancer Res. 1999;59:6132–6136. [PubMed] [Google Scholar]
- 4.Schwarz E., Freese U. K., Gissmann L., Mayer W., Roggenbuck B., Stremlau A., zur Hausen H. Nature. 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 5.Choo K. B., Pan C. C., Han S. H. Virology. 1987;161:259–261. doi: 10.1016/0042-6822(87)90195-4. [DOI] [PubMed] [Google Scholar]
- 6.Dowhanick J. J., McBride A. A., Howley P. M. J. Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeon S., Allen-Hoffmann B. L., Lambert P. F. J. Virol. 1995;69:2989–2997. doi: 10.1128/jvi.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alazawi W., Pett M., Arch B., Scott L., Freeman T., Stanley M. A., Coleman N. Cancer Res. 2002;62:6959–6965. [PubMed] [Google Scholar]
- 9.Duensing S., Munger K. Cancer Res. 2002;62:7075–7082. [PubMed] [Google Scholar]
- 10.Pett M. R., Alazawi W. O., Roberts I., Dowen S., Smith D. I., Stanley M. A., Coleman N. Cancer Res. 2004;64:1359–1368. doi: 10.1158/0008-5472.can-03-3214. [DOI] [PubMed] [Google Scholar]
- 11.White A. E., Livanos E. M., Tlsty T. D. Genes Dev. 1994;8:666–677. doi: 10.1101/gad.8.6.666. [DOI] [PubMed] [Google Scholar]
- 12.Stanley M. A., Browne H. M., Appleby M., Minson A. C. Int. J. Cancer. 1989;43:672–676. doi: 10.1002/ijc.2910430422. [DOI] [PubMed] [Google Scholar]
- 13.Bedell M. A., Hudson J. B., Golub T. R., Turyk M. E., Hosken M., Wilbanks G. D., Laimins L. A. J. Virol. 1991;65:2254–2260. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bechtold V., Beard P., Raj K. J. Virol. 2003;77:2021–2028. doi: 10.1128/JVI.77.3.2021-2028.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosch F. X., Manos M. M., Munoz N., Sherman M., Jansen A. M., Peto J., Schiffman M. H., Moreno V., Kurman R., Shah K. V. J. Natl. Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 16.Der S. D., Zhou A., Williams B. R., Silverman R. H. Proc. Natl. Acad. Sci. USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang Y. E., Pena L., Sen G. C., Park J. K., Laimins L. A. J. Virol. 2002;76:8864–8874. doi: 10.1128/JVI.76.17.8864-8874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turek L. P., Byrne J. C., Lowy D. R., Dvoretzky I., Friedman R. M., Howley P. M. Proc. Natl. Acad. Sci. USA. 1982;79:7914–7918. doi: 10.1073/pnas.79.24.7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang Y. E., Laimins L. A. J. Virol. 2000;74:4174–4182. doi: 10.1128/jvi.74.9.4174-4182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nees M., Geoghegan J. M., Hyman T., Frank S., Miller L., Woodworth C. D. J. Virol. 2001;75:4283–4296. doi: 10.1128/JVI.75.9.4283-4296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronco L. V., Karpova A. Y., Vidal M., Howley P. M. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barnard P., McMillan N. A. Virology. 1999;259:305–313. doi: 10.1006/viro.1999.9771. [DOI] [PubMed] [Google Scholar]
- 23.Nees M., Geoghegan J. M., Munson P., Prabhu V., Liu Y., Androphy E., Woodworth C. D. Cancer Res. 2000;60:4289–4298. [PubMed] [Google Scholar]
- 24.Lee D. K., Kim B. C., Kim I. Y., Cho E. A., Satterwhite D. J., Kim S. J. J. Biol. Chem. 2002;277:38557–38564. doi: 10.1074/jbc.M206786200. [DOI] [PubMed] [Google Scholar]
- 25.Favre-Bonvin A., Reynaud C., Kretz-Remy C., Jalinot P. J. Virol. 2005;79:4229–4237. doi: 10.1128/JVI.79.7.4229-4237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herrera R. E. Prog. Mol. Subcell. Biol. 1998;20:11–24. [PubMed] [Google Scholar]
- 27.Cullen A. P., Reid R., Campion M., Lorincz A. T. J. Virol. 1991;65:606–612. doi: 10.1128/jvi.65.2.606-612.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das B. C., Sharma J. K., Gopalakrishna V., Luthra U. K. J. Gen. Virol. 1992;73:2327–2336. doi: 10.1099/0022-1317-73-9-2327. [DOI] [PubMed] [Google Scholar]
- 29.Durst M., Kleinheinz A., Hotz M., Gissman L. J. Gen. Virol. 1985;66:1515–1522. doi: 10.1099/0022-1317-66-7-1515. [DOI] [PubMed] [Google Scholar]
- 30.Kalantari M., Blennow E., Hagmar B., Johansson B. Diagn. Mol. Pathol. 2001;10:46–54. doi: 10.1097/00019606-200103000-00008. [DOI] [PubMed] [Google Scholar]
- 31.Tonon S. A., Picconi M. A., Bos P. D., Zinovich J. B., Galuppo J., Alonio L. V., Teyssie A. R. J. Clin. Virol. 2001;21:129–134. doi: 10.1016/s1386-6532(01)00155-x. [DOI] [PubMed] [Google Scholar]
- 32.Hopman A. H., Smedts F., Dignef W., Ummelen M., Sonke G., Mravunac M., Vooijs G. P., Speel E. J., Ramaekers F. C. J. Pathol. 2004;202:23–33. doi: 10.1002/path.1490. [DOI] [PubMed] [Google Scholar]
- 33.Coleman N., Greenfield I. M., Hare J., Kruger-Gray H., Chain B. M., Stanley M. A. Am. J. Pathol. 1993;143:355–367. [PMC free article] [PubMed] [Google Scholar]
- 34.Freshney R. I. Culture of Animal Cells: A Manual of Basic Technique. New York: Wiley-Liss; 1994. pp. 161–178. [Google Scholar]
- 35.Zeeberg B. R., Feng W., Wang G., Wang M. D., Fojo A. T., Sunshine M., Narasimhan S., Kane D. W., Reinhold W. C., Lababidi S., et al. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Behav. Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.