Abstract
The parallel β-helix is an elongated β-sheet protein domain associated with microbial virulence factors, toxins, viral adhesins, and allergens. Long stacks of similar, buried residues are a prominent feature of this fold, as well as the polypeptide chain fold of an amyloid structure. The 13-rung, right-handed, parallel β-helix of the homotrimeric P22 tailspike adhesin exhibits predominantly hydrophobic stacks. The role of these stacked residues in the folding and stabilization of the protein is unclear. Through scanning alanine mutagenesis we have identified a folding spine of stacked residues in continuous contact along the length of P22 tailspike’s β-helix domain that is necessary for folding within cells. Nearly all chains carrying alanine substitutions of the 103 buried nonalanines were defective in folding in vivo at 37°C. However, the majority of these chains successfully reached a native state, stable to >80°C, when folded inside cells at low temperatures. Thus, nearly the entire buried core was critical for in vivo β-helix folding but negligible for stability. Folding at 18°C revealed the minimal folding spine of 29 nonglycine stack positions that were intolerant to alanine substitution. These results indicate that a processive folding mechanism, dependent on stacking contacts, controls β-helix formation. Such a stepwise folding pathway offers a new target for drug design against this class of microbial virulence factors.
Keywords: buried stacks, protein folding, folding mutants
The parallel β-helix, a structurally repetitive fold created by the coiling of the polypeptide backbone, is one of the topologically simplest β-sheet folds (1). Known β-helical proteins are predominantly associated with disease or infection. Examples include a number of viral adhesin proteins, virulence factors such as the pertactin and hemagglutinin of Bordetella pertussis, the major pollen allergen from the mountain cedar tree, and an antibiotic resistance protein that acts by means of DNA mimicry (1–5). P.69 pertactin from B. pertussis and hemoglobin protease are autotransporters whose folding and toxicity are coupled to membrane translocation (3, 6). The most common functional theme is the use of an elongated lateral surface to recognize an extended polysaccharide sequence (1). Multiple computational methods have predicted hundreds of additional β-helical proteins that are largely associated with infectious disease, pathogens, or allergens (7, 8). Given the rarity of β-helical host proteins, this fold represents a potential target for novel therapies.
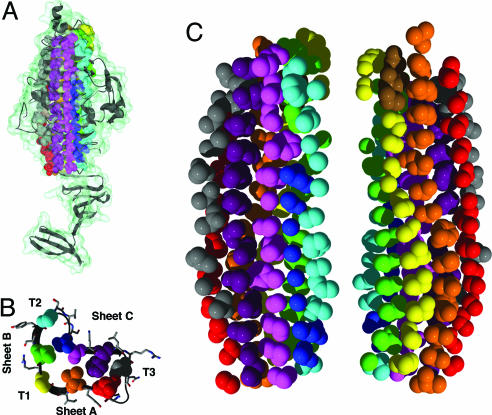
Each coil or rung of the canonical β-helix fold consists of three β-strands (A, B, and C) interrupted by variable turn or loop regions (T1, T2, and T3) (Fig. 1B) (9). Rungs are aligned, forming a cross-β structure of elongated β-sheets lying parallel to the helical axis. Structural repetition of coils creates a cylindrical hydrophobic core. A prominent feature common to all β-helical proteins is the existence of elongated buried core stacks composed of aligned, similar side chains with similar orientations to form “cupped stacks” of aliphatic residues, aromatic stacks, or polar zippers (1). Although stacks are composed of similar residues, structurally repeated rungs do not exhibit an overt sequence repeat, nor do they exhibit sequence similarity across β-helical proteins (7).
Fig. 1.
Side chain stacking in the core of the P22 tailspike β-helix. (A) The C-terminal portion (113–666) of a single chain of the P22 tailspike trimer is shown as a ribbon diagram and a transparent molecular surface. Backbone and side chain atoms of inward pointing, buried core residues are shown in space-fill and colored according to their stack. Solvent-exposed residues are not shown for clarity. (B) Rung 6 cross-section of the β-helix domain, demonstrating the locations of inward-pointing stacks (spheres) and outward-pointing residues (sticks). (C) Side chain atoms of the buried core. Single spheres represent glycines. (Left) The same orientation as A. (Right) Rotated by 180°.
The parallel β-helix has been proposed as a model structure for the chain fold in amyloid fibrils, a cross-β conformation associated with neurodegenerative diseases such as Alzheimer’s disease and prion diseases (10, 11). In the atomic structure of an amyloid fiber formed from a seven-residue peptide, the parallel β-sheets of the fiber prominently displayed stacked residues like those observed in the β-helices (12). Understanding the sequence-dependent role of side chain stacking in the β-helix fold may provide insight into the folding of elongated β-sheet proteins of considerable medical relevance.
The tailspike protein of Salmonella phage P22 has been one of the principal systems for studying protein folding in β-helices (1). Each 666-residue polypeptide chain in the homotrimeric tailspike contains a 13-rung, right-handed, parallel β-helix domain (residues 143–540), which includes the globular dorsal fin domain at turn T3 of rung 3 (Fig. 1A) (13). Tailspike’s β-helix is capable of recognizing the O-antigen of Salmonella’s cell surface lipopolysaccharide and cleaving it by means of an endorhamnosidase activity (14). Although a prokaryotic infectious agent, expression of the isolated β-helix of the P22 tailspike protein readily forms amyloid fibers (15).
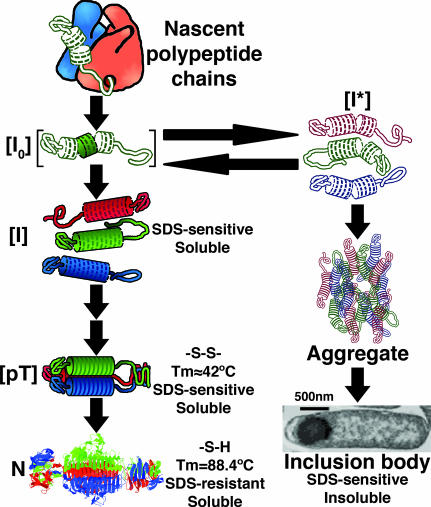
N-terminal to the β-helix domain is the head-binding domain, which is not required for β-helix folding and trimer formation but is necessary for attachment to the phage head (16). C-terminal to the β-helix domain, monomers wrap around each other to form an intertwined β-sheet domain where each chain’s residues contribute to a single buried hydrophobic core consisting of one rung of a triple β-helix (residues 541–557) and a triple β-prism structure (13, 14). The ultimate step in tailspike’s productive folding pathway (Fig. 2) coincides with the folding and assembly of this interdigitated region to form a molecular clamp that makes tailspike resistant to trypsin degradation and SDS denaturation and raises the melting temperature by ≈40°C to 88.4°C (17, 18).
Fig. 2.
In vivo folding pathway of the P22 tailspike homotrimer, as described in the text. The left branch of the folding pathway shows productive intracellular intermediates leading to the native state. Intermediates leading to the inclusion body state are on the right. [pT], protrimer.
Extensive studies of the in vivo folding pathway have revealed a number of long-lived, partially folded intermediates (Fig. 2) (19). The first of these intermediate structures, a β-helical conformer, [I0], rapidly develops while nascent chains are still attached to the ribosome (20). Upon release from the ribosome tailspike, thermolabile monomeric intermediates, [I], may oligomerize and further fold into the SDS-resistant trimeric tailspike, N (21). Mutations that block oligomerization or trimer maturation result in the accumulation of soluble, SDS-sensitive species (18, 22). Alternatively, the presence of any of >60 genetically isolated temperature-sensitive folding (tsf) mutations at temperatures in the higher range of cellular growth will generate a misfolded monomeric conformation, [I*], which specifically self-aggregates and accumulates in inclusion bodies (23). The tsf mutations, predominately located at surface loops and turns, appear to act by destabilizing the β-helical structure found in the thermolabile, monomeric folding intermediate (23, 24). Global suppressors (su) of the tsf mutations or lower temperatures stabilize the β-helical fold or otherwise inhibit aggregation (24, 25). The GroEL chaperone does not assist tailspike chains in efficiently folding or preventing aggregation in vivo (26). Partitioning between native and aggregation states is therefore a reflection of the folding state of the β-helix. As a result of the native tailspike’s SDS resistance, SDS/PAGE of whole lysates provides a sensitive and reliable measure of in vivo folding (17, 21).
The numerous surface site tsf mutations represent residues essential for the creation of the β-helical fold at higher temperatures but do not carry the essential information for β-helix folding, because they are unnecessary at lower temperatures. In contrast, five phenylalanines and one leucine in the hydrophobic core stacks are essential for the folding, but not stability, of tailspike at high and low temperatures (22). This finding suggests that stacked core residues contain the necessary sequence information to direct β-helix formation.
To investigate the contributions of all stacked, buried core side chains to the folding of the parallel β-helix, we carried out a high-throughput, scanning alanine mutagenesis study on the folding of the P22 tailspike.
Results
Stacking and Mutagenesis Target Selection.
Upon inspection of the structure of the tailspike protein [Protein Data Bank ID code 1TYU (27)] it became apparent that the majority of the β-helix buried core is composed of stacked residues whose side chains have similar conformations (Fig. 1). In total, we identified 10 buried core stacks containing 113 aa that composed the entirety of the β-helix domain’s core: six stacks participating in the structurally conserved β-sheets (red, orange, yellow, green, magenta, and purple stacks in Fig. 1), two pseudostacks located in T2 and T3 (cyan and gray), one short stack preceding sheet C running from rung 5 to rung 13 (blue), and a second shortened stack of three residues at the N terminus of the β-helix in the conserved turn T2 (brown). Each of the 103 buried, nonalanine residues was individually replaced with alanine and the mutant chain expressed in Escherichia coli.
Richardson and Richardson (28) suggested that capping regions exist in β-sheet structures to fulfill its hydrogen bonding potential and prevent aggregation. To investigate this in the case of the β-helix, all residues in the N-terminal capping region (122–146; Fig. 8A, which is published as supporting information on the PNAS web site) and the C-terminal region of the β-helix domain (521–545) were chosen as targets of mutagenesis irrespective of their solvent accessibility. Six further solvent-exposed residues located throughout the β-helix domain were added to the target list because of their proximity and potential chemical reactivity with cysteine residues. One disordered glycine, G511, was chosen for mutagenesis because of its potential location at a stacking position.
In total, 145 nonalanine sites were individually mutated to alanine, as confirmed by DNA sequencing, and assayed for their effect on folding relative to WT.
Protein Expression and Folding Analysis.
All single alanine mutants were individually overexpressed from a pET plasmid in E. coli at 37°C, 30°C, and 18°C to assess the contribution of the altered residue to the in vivo folding of the β-helix domain. Cells containing pET11a plasmids lacking any tailspike genes were induced as a negative control of tailspike expression. Positive controls included WT and ΔN tailspike, which lacks the N-terminal domain (1–108) (29). Mutant expression at each temperature was performed in triplicate. Induced samples were lysed and mixed with SDS sample buffer, and unheated samples were analyzed by SDS/PAGE. Sample gels are shown in Fig. 3.
Fig. 3.
Sample SDS/PAGE data of complete cellular lysates. (A) Positive (WT and ΔN) and negative (pET11a) control samples expressed at 37°C. WT and ΔN expression samples produce some native, SDS-resistant trimers (NT) as well as nonnative, SDS-sensitive chains that unfold (U) upon mixture with SDS. WT and mutant samples are shown for expression at 37°C (B), 30°C (C), and 18°C (D).
As can be seen in Fig. 3, background translational levels proved consistent across all samples. Mutant and WT lysates accumulated tailspike polypeptide chains as clearly visible, tailspike-specific bands. These observations proved true for all expression samples at all three temperatures. Thus, alanine substitutions at the targeted sites had no noticeable effect on translation or degradation of the tailspike polypeptide chain.
When complete E. coli lysate samples were electrophoresed through an SDS polyacrylamide gel, overexpressed tailspike chains partitioned between two bands in the gel (Fig. 3). Chains that reached the SDS-resistant, native tailspike structure do not dissociate in the presence of SDS (21). These chains appeared on the gel as a slowly migrating native trimer band (NT) of ≈210 kDa. In contrast, partially folded intermediates and inclusion body aggregates are dissociated and unfolded in detergent, causing these species to emerge as a fast moving, 72-kDa band (U). Quantification of the percent of tailspike polypeptide chains reaching the native state provided an accurate and precise measure of the in vivo partitioning between productive and nonproductive folding.
Negative control samples were used to subtract background cellular protein levels so that tailspike bands could be accurately quantified. Normalization of the percent folded measurement to the average WT value generated a useful measure, the percentage of WT folding efficiency. Comparison of these folding efficiencies allowed for an unbiased assessment of the effects of individual alanine substitutions with respect to a known standard, the WT sequence.
Quantitative analysis of the folding of all mutants at all three temperatures is shown in Fig. 4 and in Table 1, which is published as supporting information on the PNAS web site. Although a few mutant sequences were able to reach the native state at the presumed physiological temperature of the Salmonella phage P22, 37°C, the great majority exhibited severe folding defects. Even at the low temperature of 18°C, 31 mutants were unable to reach the native state.
Fig. 4.
Quantitative results of folding experiments. Average percentages of WT folding efficiencies are shown for all alanine mutants when expressed at 37°C (red circles), 30°C (brown triangles), and 18°C (blue squares). Colored lines indicate the standard deviations of triplicate experiments. Note that WT and ΔN average percentage of WT folding efficiencies and standard deviations are shown as Insets. The 75% threshold delineating WT-like folders and folding-deficient samples is shown as a dashed line.
At each temperature the distribution of folding efficiencies proved bimodal, allowing us to classify each mutant as a WT-like folder or as folding-deficient. Mutants whose folding efficiencies were <75% of WT’s folding efficiency were classified as folding-deficient. This 75% threshold was conservatively chosen based on the distribution of averages, examination of the statistical overlap of sample values with WT, and the expectation that a mutant classified as defective at a low temperature should be identically classified at higher temperatures.
Wild-Type and ΔN in Vivo Folding.
For WT chains expressed at 18°C, 94% (average of 22 expression samples) of the polypeptide chains reached the SDS-resistant, trimeric native conformation. When renormalized to this average, the standard deviation of these samples was 2%. When expressed at 30°C, 87 ± 6% (21 samples) of WT chains reached the native conformation. Expression at 37°C yielded 62 ± 10% (22 samples) of WT chains reaching the native state. The deviations at each temperature were well within the 75% threshold for a WT-like folder (Fig. 4 Insets and Table 1). The small standard deviations observed at each expression temperature illustrate the precision of this SDS/PAGE-based assay.
ΔN chains folded with WT-like efficiency at 37°C: 95 ± 13% for 28 samples. At lower temperatures, ΔN folded significantly better than WT, with a folding efficiency of 109 ± 3% for 26 samples at 30°C and 104 ± 1% for 28 samples at 18°C. It has been reported that tailspike chains lacking the N-terminal head-binding domain fold faster than the full WT sequence (29, 30). The ability to detect the effects of this accelerated folding rate validates the precision and accuracy of this assay.
Mutant in Vivo Folding at 37°C.
In contrast to the insignificant effect that removal of >100 aa had on the folding of the ΔN tailspike protein, alanine substitution of any one of 95 of the 103 buried core sites significantly hindered the tailspike chains’ ability to reach the native state when expressed at 37°C. One of these mutants, F308A, was previously shown to fail to fold at high and low temperatures (22). Our high-throughput system was able to accurately reproduce the published data for this residue as well as all other β-helix residues investigated by Betts et al. (22). Of the remaining 42 predominantly surface-exposed sites, 20 had defective folding phenotypes. When mapped to the native structure of the P22 tailspike (Fig. 5), the vast majority of defective folding sites were in the cylindrical buried core. These sites were intolerant to alanine substitution when folded at 37°C in vivo.
Fig. 5.
Folding efficiencies mapped onto the native structure of the tailspike β-helix. Mutated positions are colored according to the folding efficiency of the residue’s alanine mutant. Positions that are glycine in the WT sequence or fold like WT are shown by stick representation. Folding-deficient positions are shown as a surface volume.
Conversely, the 30 WT-like folding alanine mutations map almost entirely to surface-exposed residues, to the termini of the β-helix, or to the shortened stack that precedes sheet C. V386A and V455A, located in a valine stack in sheet B, were the only exceptions to this. G511A, located in a disordered loop in the WT crystal structure, had a folding efficiency of 86 ± 9% at 37°C and remained a WT-like folder at lower temperatures.
Mutant in Vivo Folding at 30°C.
At 30°C, alanine substitutions at 47 sites prevented folding, as compared to 95 sites at 37°C. Thus, a drop of 7°C in expression temperature rescued 48 defective folding mutants back to WT-like levels. Among these rescued sites are nearly all of the remaining surface-exposed residues. Thirty-three of the rescued positions were in the buried core stacks, so only 62 of the 103 stacking residues were intolerant to alanine substitution at 30°C. Intolerant sites localized to the central region of the β-helix opposite the dorsal fin (Fig. 5), whereas more tolerant positions clustered near the termini of the β-helix. The fourth and fifth rungs of the β-helix were completely intolerant to alanine substitutions of core residues at 30°C.
Mutant in Vivo Folding at 18°C.
In contrast to the necessity of 92% of the buried core residues at 37°C, only 31 of 103 alanine mutations were absolutely incapable of achieving WT-like folding efficiencies at 18°C. The sites of these completely intolerant residues were located in every rung of the β-helix except rung 7. Rungs 6 and 8 have the most number of intolerant positions, four and five, respectively. Three glycines could not reach the native state when mutated to alanine, most likely because of inaccessible backbone conformations.
When nonglycine positions completely intolerant to alanine substitution are mapped onto the native structure of tailspike’s β-helix, a continuous network of 29 interacting side chains is revealed that spans the length of the β-helix (Fig. 5). This chain of mostly hydrophobic residues, which we call the “folding spine,” is composed of short stacks that cluster around variable turn regions, such as the dorsal fin. In the case of the dorsal fin domain, a cluster of 6 aa is essential for folding where the chain detours from the β-helix and into the dorsal fin. At the center of this cluster, two residues (M195 and L262) pack against each other, as if stapling together the meandering backbone (Fig. 9, which is published as supporting information on the PNAS web site).
Two asparagines, N310 and N319, and the only histidine in the buried core, H426, were present in the folding spine. In the native structure, these side chains interact with resolved, buried water molecules that coordinate the backbone of loop regions. These necessary residues highlight the complexity of the folding process in vivo.
N-Terminal Capping Residues.
Alanine substitutions resulting in folding-deficient phenotypes at 18°C and 30°C in the N-terminal capping region (122–146) of the β-helix highlight this region’s role in preventing aggregation (Fig. 8). This region is composed of a short β-strand, a turn, and a conserved α-helix. V141A, located at the C terminus of the conserved α-helix, is the only mutant in this region deficient in folding at all temperatures. This residue may act as a stop signal, ensuring that the α-helix is the proper length to cap the β-helix.
Only two capping mutants are rescued at 18°C, L133A and Y130A. L133 is located at the N terminus of the α-helix and points directly downward into the β-helix core, possibly anchoring the α-helix into its capping position. Y130 sits centrally over the core of the β-helix, potentially bridging the hydrophobic interior of the β-helix and the polar solvent. It is important to note that Y123A, as opposed to Y130A, displays completely WT-like folding phenotypes at all temperatures. Hence, the folding phenotypes observed in the N-terminal capping regions illuminate key roles of WT side chains that are not simply a reflection of the type of mutation.
Mutant Solubilities and Mobility Shifts.
To test the hypothesis that the observed folding phenotypes were due to an effect on the folding of the β-helical folding intermediate, we analyzed the aggregation state of SDS-sensitive tailspike species produced at 37°C by low-speed centrifugation and SDS/PAGE. As described, soluble SDS-sensitive species represent a defect in oligomerization or native-state maturation or stability. Substitutions affecting β-helix folding result in accumulation of chains as insoluble, aggregated inclusion bodies. All nonnative tailspike conformers in all 37°C samples accumulated in a consistent manner in the pelleted, insoluble fraction of cell lysates. Thus, these folding defects occurred at or before the monomeric, β-helical folding intermediate.
A number of native trimeric mutant chains displayed mobility shifts with respect to WT chains. These mutants primarily corresponded to the mutation of charged residues, although not all charge changes produced alterations in mobility. Two neutral changes (N499A and G462A) produced reproducible mobility shifts. The physical basis behind these shifts is not understood but may reflect an altered native conformation.
Tailspike Stability.
To test whether mutations acted by destabilizing the native state, the native mutant polypeptide chains in the soluble fractions of 18°C expression samples were assessed for thermostability. One hundred fourteen of the alanine mutants accumulated sufficient native trimers at low temperatures to be assayed. Lysates were heated to either 80°C or 90°C for 3 min in the presence of 2% SDS and analyzed for unfolding of the β-helix region. As described by Chen and King (31), WT proteins partially unfolded to a SDS-resistant, high-mobility species (UI) that maintains β-helical conformation but lacks structure in the N-terminal head-binding domain when heated to 80°C (Fig. 10, which is published as supporting information on the PNAS web site). Only 3.1 ± 1.7% of WT native chains fully denatured to an SDS-sensitive species at 80°C. Heating to 90°C resulted in nearly complete dissociation (89.9 ± 7.9%) to a denatured, monomeric SDS/polypeptide chain complex (Fig. 6 and Table 1). Heating of ΔN resulted in similar thermostability values, with only 4.5 ± 3.4% of chains unfolding at 80°C and 98.5 ± 3.7% of chains unfolding at 90°C.
Fig. 6.
Thermostability assay on 18°C WT-like folders. The percentage of native-tailspike chains completely unfolded in the presence of 2% SDS when heated to 80°C (blue circle) or 90°C (red square) is shown as the average of duplicate experiments. Lines represent standard deviations. WT and ΔN controls are shown in the Inset.
All trimeric mutant tailspikes consistently dissociated to monomeric SDS/polypeptide chain complexes when heated to 90°C (Fig. 6). All of these chains partially denatured similarly to WT when subjected to 80°C, except for F349A. F394A proved substantially destabilized when subjected to 80°C. These results show that the single alanine mutants have melting temperatures within the range of 80–90°C in the presence of 2% SDS. These environmental conditions are far harsher than the conditions experienced by mutant chains folding within cells at 37°C. The thermostability of native mutant proteins and the insolubility of nonnative species strongly support the notion that the effects of the alanine mutants explored here occur during the intracellular folding of the β-helix.
Discussion
The β-Helix Folds Processively.
Previous experiments on small, globular proteins have demonstrated that the amino acid folding code is highly degenerate and that, in at least one case, half of the residues can be replaced by alanines simultaneously (32–34). In the P22 tailspike β-helix the entire core is surprisingly sensitive to amino acid changes at a physiological temperature. If folding followed a global collapse mechanism, then the removal of two to eight atoms out of a core composed of 365 side chain atoms would not be expected to completely block the folding process. In contrast, a processive folding pathway, where a series of local folding events must occur in a specific order to reach a native structure, predicts these results. We conclude that the in vivo folding pathway of the tailspike’s β-helix occurs by means of a processive folding mechanism, such as the sequential winding of rungs (Fig. 7).
Fig. 7.
Model of the processive folding of the β-helix. Critical internal side chain contacts (cyan ovals) nucleate an initial set of well formed rungs. This nucleus acts as a template for other rungs to assemble by means of side chain stacking and/or tethering around loop regions. Capping of the N terminus likely prevents aggregation.
The contribution of WT side chains to productive folding was temperature-dependent. Folding analysis at low temperatures revealed an elongated folding spine of essential residues encoding critical contacts that allow for the in vivo folding of the tailspike protein. Although it cannot be ruled out that these residues represent positions that critically interact with the ribosome in a ribosome-assisted folding mechanism, it is unlikely that these residues are required for intermolecular interactions. Instead, we postulate the simpler model that these residues may control the folding of the β-helix by tethering key positions into a general cylindrical structure, thereby allowing β-sheets to form. At lower temperatures, certain packing interactions may tolerate an alanine replacement, but the interaction energy of an alanine mutant may not be sufficient to overcome added kinetic energies at higher temperatures.
Data at 30°C demonstrated that residues located centrally in the β-helix were more intolerant to substitution than at the termini. The proposed processive folding pathway predicts that inhibition of earlier folding events would have more drastic effects than later stages. This model suggests that β-helix formation in vivo initiates with the creation of a structural nucleus near the center of the β-helix and proceeds by means of a templating or tethering mechanism toward the termini. Folding data from mutants located in the N-terminal capping region suggest that folding of this domain concludes by means of formation of key capping contacts, which may stabilize the β-helix and prevent aggregation.
The Role of Stacking Residues.
Stacking of side chains plays a critical role in the folding of the β-helix. However, stacks that run the entire length of the β-helix are not necessary to direct folding at low temperatures. Instead, the folding spine spans numerous stacks. This finding supports the notion that side chain stacks are necessary to encode an elongated β-helix. This result is in contrast to the large number of previously isolated tsf mutations that represent solvent-exposed residues critical in folding at high temperatures but unnecessary at lower temperatures (22). The short core stacks may function to stabilize intermediate forms or to specifically prevent off-pathway aggregation.
The continuous nature of the folding spine indicates that a long-range network of interactions may act cooperatively to form the β-helix. Such networks have been previously proposed based on sequence conservation analysis of PDZ and POZ domains (35). Our data demonstrate the existence of such a network in the context of the folding of the β-helix domain. If the connectivity of short stacks did not act cooperatively, one might expect that these short stacks of necessary residues might be more randomly dispersed throughout the core. Instead, we believe that the presence of this continuous folding spine represents a cooperative network that directs the protein toward its native fold in vivo.
Understanding the full role key stacking positions play will require further investigations into the cooperativity of this network and identification of factors that link the amino acid sequence to its contribution toward folding. Seckler and coworkers have analyzed the site of the buried global suppressor, A334, in considerable detail (14). In contrast to the intolerance of side chain deletions presented here, β-helix folding tolerated bulkier substitutions at this site (24).
Targets of Drug Design.
A processive folding mechanism provides a new target for drug design against a number of diseases associated with β-helical proteins. The stepwise folding mechanism proposed here is the monomeric analog of the oligomerization-dependent formation of amyloid fibrils. Small molecules have already been discovered that prevent the nucleation and stepwise elongation of amyloid fibrils (36). It is reasonable that, instead of blocking the interactions between an infective β-helix and its recognition factor, drugs may be designed to hinder the folding process. For the autotransporter β-helices, folding occurs extracellularly and is linked to translocation (6), whereby inhibition would prevent maturation of toxicity. It is clear that a number of positions are highly sensitive to alterations, causing unrecoverable folding inhibition under physiological conditions. These positions represent an assortment of targets for inhibition of the stepwise folding of these proteins.
Because this class of proteins is rare and primarily specific to the microbial world, drugs of this type would benefit from specificity toward the infectious agent. Drugs capable of inhibiting β-helix formation may offer an effective mechanism for preventing selected microbial and viral infections.
Materials and Methods
Site-Directed Alanine Mutagenesis.
Gene 9, which encodes the Salmonella phage P22’s tailspike protein, was previously cloned into the vector pET11a (Novagen) as described (18). The resulting vector, pET(gene9), encoded the full-length WT tailspike protein.
Methylated pET(gene9) DNA was purified to act as template DNA for mutagenesis from XL1-Blue cells (Stratagene) by using a Qiagen Plasmid Midi Kit. Scanning alanine mutagenesis was performed by using the PCR-based QuikChange mutagenesis procedure (Stratagene) in a parallelized, high-throughput manner. Sequences of mutagenic primers (Integrated DNA Technologies) are listed in Table 2, which is published as supporting information on the PNAS web site. Mutagenesis reactions were performed in parallel in 96-well plates to achieve high-throughput processing.
Clones encoding full-length, but potentially altered, tailspike were identified by means of overexpression in E. coli NovaBlue(DE3) cells (Novagen). Plasmid DNA was purified by using QIAprep Spin MiniPrep kits (Qiagen). DNA sequencing of a 400-bp region (20% of the tailspike gene) containing the targeted codon confirmed mutant clones. Of 241 samples sequenced, only one had a second-site mutation outside the primer region, resulting in an estimated 0.02 second-site mutations per gene.
Tailspike Expression and Cell Lysis.
Plasmid DNA encoding confirmed alanine mutants was transformed into E. coli BL21(DE3) by using a single-step procedure (37) performed in parallel in 2-ml 96-deep-well plates. Cultures were grown, induced for expression, and lysed according to standard procedures. Details on deviations to these procedures are described further in Supporting Text, which is published as supporting information on the PNAS web site.
SDS/PAGE and Optical Densitometry.
Lysate fractions were mixed with 2% SDS sample buffer as described (22). SDS/PAGE was run at 4°C on 7.5% 102-lane Triple Wide gels (C.B.S. Scientific) by using the discontinuous buffer system of King and Laemmli (38). Coomassie-stained gels were analyzed by optical densitometry, as described in Supporting Text.
Thermostability Assay.
After SDS/PAGE analysis, remaining supernatant samples in 2% SDS sample buffer from 18°C expression cultures were stored at −20°C. These samples were thawed on ice, and 10-μl aliquots were moved to new shallow-well plates. Plates were sealed with silicon cap lids clamped down by a glass plate and binder clips. The sealed plate was partially submerged in a heated water bath to either 80°C or 90°C for 3 min and then placed on ice until samples were analyzed by SDS/PAGE as described above.
Supplementary Material
Acknowledgments
We thank the laboratory of Prof. Robert Horvitz (Massachusetts Institute of Technology) for the use of their 96-well Thermal Cycler (MJ Research); Anne Robinson (University of Delaware, Newark) for the pET(ΔN) plasmid; and Peter Weigele, Cameron Haase-Pettingell, and Kristen Cook for their helpful discussions and technical assistance. This research was supported by National Institutes of Health Grant GM17980 (to J.K.).
Abbreviation
- tsf
temperature-sensitive folding.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Jenkins J., Pickersgill R. Prog. Biophys. Mol. Biol. 2001;77:111–175. doi: 10.1016/s0079-6107(01)00013-x. [DOI] [PubMed] [Google Scholar]
- 2.Clantin B., Hodak H., Willery E., Locht C., Jacob-Dubuisson F., Villeret V. Proc. Natl. Acad. Sci. USA. 2004;101:6194–6199. doi: 10.1073/pnas.0400291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emsley P., Charles I. G., Fairweather N. F., Isaacs N. W. Nature. 1996;381:90–92. doi: 10.1038/381090a0. [DOI] [PubMed] [Google Scholar]
- 4.Czerwinski E. W., Midoro-Horiuti T., White M. A., Brooks E. G., Goldblum R. M. J. Biol. Chem. 2005;280:3740–3746. doi: 10.1074/jbc.M409655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegde S. S., Vetting M. W., Roderick S. L., Mitchenall L. A., Maxwell A., Takiff H. E., Blanchard J. S. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 6.Otto B. R., Sijbrandi R., Luirink J., Oudega B., Heddle J. G., Mizutani K., Park S. Y., Tame J. R. J. Biol. Chem. 2005;280:17339–17345. doi: 10.1074/jbc.M412885200. [DOI] [PubMed] [Google Scholar]
- 7.Bradley P., Cowen L., Menke M., King J., Berger B. Proc. Natl. Acad. Sci. USA. 2001;98:14819–14824. doi: 10.1073/pnas.251267298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciccarelli F. D., Copley R. R., Doerks T., Russell R. B., Bork P. Trends Biochem. Sci. 2002;27:59–62. doi: 10.1016/s0968-0004(01)02046-1. [DOI] [PubMed] [Google Scholar]
- 9.Jurnak F., Yoder M. D., Pickersgill R., Jenkins J. Curr. Opin. Struct. Biol. 1994;4:802–806. doi: 10.1016/0959-440x(94)90259-3. [DOI] [PubMed] [Google Scholar]
- 10.Govaerts C., Wille H., Prusiner S. B., Cohen F. E. Proc. Natl. Acad. Sci. USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wetzel R. Structure (London) 2002;10:1031–1036. doi: 10.1016/s0969-2126(02)00809-2. [DOI] [PubMed] [Google Scholar]
- 12.Nelson R., Sawaya M. R., Balbirnie M., Madsen A. O., Riekel C., Grothe R., Eisenberg D. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinbacher S., Seckler R., Miller S., Steipe B., Huber R., Reinemer P. Science. 1994;265:383–386. doi: 10.1126/science.8023158. [DOI] [PubMed] [Google Scholar]
- 14.Seckler R. J. Struct. Biol. 1998;122:216–222. doi: 10.1006/jsbi.1998.3974. [DOI] [PubMed] [Google Scholar]
- 15.Schuler B., Rachel R., Seckler R. J. Biol. Chem. 1999;274:18589–18596. doi: 10.1074/jbc.274.26.18589. [DOI] [PubMed] [Google Scholar]
- 16.Steinbacher S., Miller S., Baxa U., Budisa N., Weintraub A., Seckler R., Huber R. J. Mol. Biol. 1997;267:865–880. doi: 10.1006/jmbi.1997.0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldenberg D., King J. Proc. Natl. Acad. Sci. USA. 1982;79:3403–3407. doi: 10.1073/pnas.79.11.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreisberg J. F., Betts S. D., Haase-Pettingell C., King J. Protein Sci. 2002;11:820–830. doi: 10.1110/ps.3440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betts S., King J. Struct. Folding Des. 1999;7:R131–R139. doi: 10.1016/s0969-2126(99)80078-1. [DOI] [PubMed] [Google Scholar]
- 20.Clark P. L., King J. J. Biol. Chem. 2001;276:25411–25420. doi: 10.1074/jbc.M008490200. [DOI] [PubMed] [Google Scholar]
- 21.Goldenberg D. P., Berget P. B., King J. J. Biol. Chem. 1982;257:7864–7871. [PubMed] [Google Scholar]
- 22.Betts S., Haase-Pettingell C., Cook K., King J. Protein Sci. 2004;13:2291–2303. doi: 10.1110/ps.04676704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haase-Pettingell C., King J. J. Mol. Biol. 1997;267:88–102. doi: 10.1006/jmbi.1996.0841. [DOI] [PubMed] [Google Scholar]
- 24.Schuler B., Seckler R. J. Mol. Biol. 1998;281:227–234. doi: 10.1006/jmbi.1998.1944. [DOI] [PubMed] [Google Scholar]
- 25.Mitraki A., Fane B., Haase-Pettingell C., Sturtevant J., King J. Science. 1991;253:54–58. doi: 10.1126/science.1648264. [DOI] [PubMed] [Google Scholar]
- 26.Gordon C. L., Sather S. K., Casjens S., King J. J. Biol. Chem. 1994;269:27941–27951. [PubMed] [Google Scholar]
- 27.Steinbacher S., Baxa U., Miller S., Weintraub A., Seckler R., Huber R. Proc. Natl. Acad. Sci. USA. 1996;93:10584–10588. doi: 10.1073/pnas.93.20.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson J. S., Richardson D. C. Proc. Natl. Acad. Sci. USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller S., Schuler B., Seckler R. Protein Sci. 1998;7:2223–2232. doi: 10.1002/pro.5560071021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danner M., Fuchs A., Miller S., Seckler R. Eur. J. Biochem. 1993;215:653–661. doi: 10.1111/j.1432-1033.1993.tb18076.x. [DOI] [PubMed] [Google Scholar]
- 31.Chen B., King J. Biochemistry. 1991;30:6260–6269. doi: 10.1021/bi00239a026. [DOI] [PubMed] [Google Scholar]
- 32.Kuroda Y., Kim P. S. J. Mol. Biol. 2000;298:493–501. doi: 10.1006/jmbi.2000.3622. [DOI] [PubMed] [Google Scholar]
- 33.Sauer R. T. Folding Des. 1996;1:R27–R30. doi: 10.1016/S1359-0278(96)00015-6. [DOI] [PubMed] [Google Scholar]
- 34.He M. M., Wood Z. A., Baase W. A., Xiao H., Matthews B. W. Protein Sci. 2004;13:2716–2724. doi: 10.1110/ps.04875504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lockless S. W., Ranganathan R. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 36.Blanchard B. J., Chen A., Rozeboom L. M., Stafford K. A., Weigele P., Ingram V. M. Proc. Natl. Acad. Sci. USA. 2004;101:14326–14332. doi: 10.1073/pnas.0405941101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung C. T., Niemela S. L., Miller R. H. Proc. Natl. Acad. Sci. USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.King J., Laemmli U. K. J. Mol. Biol. 1971;62:465–477. doi: 10.1016/0022-2836(71)90148-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.