Abstract
Cell cycle progression in Caulobacter is driven by the master transcriptional regulators CtrA and GcrA. The cellular levels of CtrA and GcrA are temporally and spatially out-of-phase during the cell cycle, with CtrA repressing gcrA transcription and GcrA activating ctrA transcription. Here, we show that DnaA, a protein required for the initiation of DNA replication, also functions as a transcriptional activator of gcrA, which in turn activates multiple genes, notably those involved in chromosome replication and segregation. The cellular concentration of DnaA is cell cycle-controlled, peaking at the time of replication initiation and gcrA induction. Regulated proteolysis of GcrA contributes to the cell cycle variations in GcrA abundance. We propose that DnaA couples DNA replication initiation with the expression of the two oscillating regulators GcrA and CtrA and that the DnaA/GcrA/CtrA regulatory cascade drives the forward progression of the Caulobacter cell cycle.
Keywords: Caulobacter crescentus, CtrA, DnaA, GcrA, proteolysis
Introduction
Caulobacter crescentus divides asymmetrically in each cell cycle, giving rise to two different progeny cells with distinct morphological features and cell fates: a stalked cell that can immediately initiate DNA replication and a swarmer cell that cannot initiate DNA replication until it differentiates into a stalked cell (Figure 1A). Caulobacter, therefore, must not only coordinate DNA replication with cell division, but also with morphogenetic events. Multiple cell cycle events, such as the G1/S transition, flagellar and pili biogenesis, DNA methylation, chromosome segregation and cytokinesis are mediated by the GcrA and CtrA master regulators, which oscillate out of phase during the cell cycle (Holtzendorff et al, 2004). These events are coordinated, in part, by an interdependent feedback loop between GcrA and CtrA and by the differential methylation state of the chromosome during the cell cycle (Reisenauer and Shapiro, 2002; Holtzendorff et al, 2004). Here, we show that DnaA is a critical coordination factor, functioning to allow replication initiation while activating the expression of gcrA, with the consequent expression of ctrA and genes involved in chromosome replication and segregation.
Figure 1.
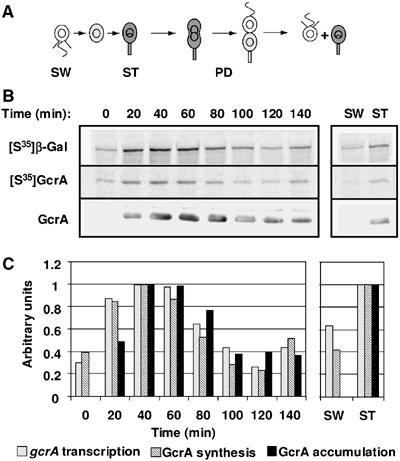
Cell cycle expression of gcrA. (A) Schematic of the Caulobacter cell cycle. Gray indicates accumulation of GcrA. Theta structures indicate replicating DNA. SW, swarmer cell; ST, stalked cell; PD, predivisional cell. (B) From the same synchronized LS4220 (gcrA+ gcrAP(−507…+92)-lacZ) strain culture, aliquots were taken at the times indicated and were either pulse-labelled with [35S]methionine to follow β-galactosidase synthesis or GcrA synthesis using either anti-β-galactosidase or anti-GcrA immunoprecipitation, or immunobloted with anti-GcrA. Autoradiograms from the immunoprecipitation of labelled β-galactosidase ([35S]β-Gal) and GcrA ([35S]GcrA), and immunoblots with anti-GcrA (GcrA) are shown. Also shown are autoradiograms and immunoblots from cell extracts from newly divided stalked and swarmer cells harvested at the end of the cell cycle. (C) Histograms of the relative rates of gcrA transcription (β-galactosidase synthesis), GcrA synthesis and GcrA accumulation as measured by phosphorimaging or densitometric scanning of immunoblots. Values were normalized to the maximum value of each experiment (at 40 min from 0 to 140 min and in stalked progenies).
CtrA is an essential transcription factor and a selective chromosome replication inhibitor in Caulobacter (Quon et al, 1996, 1998). CtrA accumulates in swarmer cells, where it directly binds to five sites within the Caulobacter origin of DNA replication (Cori) and thereby blocks replication initiation. CtrA is cleared from the cell during the swarmer-to-stalked cell differentiation, which is functionally analogous to the eukaryotic G1-to-S transition of the cell cycle. Concurrently, as CtrA is proteolyzed, the dnaA and the himA genes, which encode the DNA replication initiator protein DnaA and the α-subunit of the ‘histone-like' integration host factor (IHF), are transcribed (Zweiger and Shapiro, 1994; Laub et al, 2000). The DnaA protein, conserved in most eubacteria, directly binds to DNA at DnaA boxes (Fuller et al, 1984). Five DnaA boxes and an exceptional AT-rich region are found in the Caulobacter Cori (Marczynski and Shapiro, 1992). DnaA binding to Cori has been proposed to unwind the AT-rich region and provide an entry site for protein components of the replisome that then initiates chromosome replication (Bramhill and Kornberg, 1988; Gille and Messer, 1991). An IHF-binding site overlaps one of the CtrA-binding sites in Cori. IHF binding to the Cori may help displace CtrA from the Cori sequence and bend DNA, promoting the initiation of replication (Siam et al, 2003).
Many functions needed for normal progression of the Caulobacter cell cycle are regulated by discrete transcription patterns: genes are activated at the time when their function is needed. Whole-genome analysis of messenger RNA (mRNA) accumulation during the Caulobacter cell cycle has revealed that the expression of about 19% of the Caulobacter genes (∼550 out of 3760) are regulated in a cell-cycle-dependent manner (Laub et al, 2000). More than 15% of these cell-cycle-regulated genes are directly regulated by CtrA, which binds to a conserved motif in the promoters of 55 operons (Quon et al, 1996; Laub et al, 2002). CtrA's targets include genetic modules for cell division, DNA methylation, flagellum and pili biosynthesis, chemotaxis and metabolism (Laub et al, 2002). GcrA is another essential regulatory protein controlling 49 cell-cycle-regulated genes, of which eight are also regulated by CtrA (Holtzendorff et al, 2004). GcrA notably controls polar morphogenesis and multiple components of the chromosome replication and segregation machinery. The cellular levels of GcrA are cell-cycle-dependent, both temporally and spatially out-of-phase with CtrA ((Holtzendorff et al, 2004) and Figure 5). Functional complementarities of the conserved CtrA and GcrA proteins allow time- and cell type-specific transcriptional regulation of key cell cycle events, thus providing a motive force for cell cycle progression. The expression and the activity of the CtrA and GcrA master regulators must therefore be tightly regulated.
Figure 5.
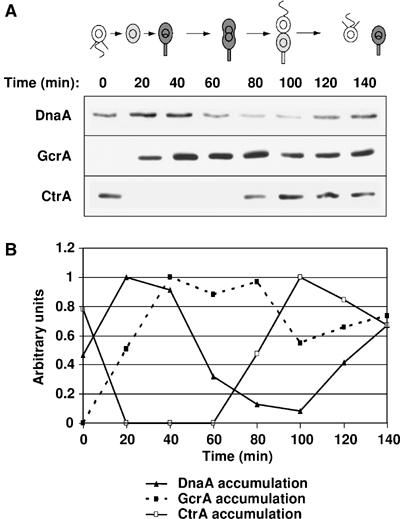
DnaA is cell cycle regulated, accumulating before GcrA. (A) Immunoblots of cell extracts from a synchronized NA1000 culture using DnaA, GcrA and CtrA antibodies at the indicated times in the cell cycle. Cultures were adjusted to the same A660 for all lanes of each gel. (B) DnaA (plain triangles, plain line), GcrA (plain squares, dashed line) and CtrA (open squares, plain line) protein levels at the indicated times of the cell cycle from densitometry graphs of the DnaA, GcrA and CtrA immunoblots. Values were normalized so that the maximum value of each immunoblot equals 1. In the schematic of the Caulobacter cell cycle, gray indicates accumulation of GcrA.
The cellular accumulation of CtrA is controlled by at least two mechanisms: transcriptional regulation and targeted proteolysis. The ctrA gene is transcribed from two promoters: P1 and P2. The ctrA P1 promoter can be methylated by the CcrM DNA methyltransferase at a GANTC site and a fully methylated ctrA P1 promoter cannot be transcribed (Reisenauer and Shapiro, 2002). At the initiation of replication, the ctrA P1 promoter is in the fully methylated state. The ctrA P1 promoter can only be transcribed after the passage of the replication fork that generates two hemi-methylated copies of the gene. Thus, ctrA transcription is coordinated with the progression of chromosomal replication. Once in the hemi-methylated state, transcription from the ctrA P1 promoter is activated by GcrA (Holtzendorff et al, 2004). As CtrA accumulates and is activated by phosphorylation, it represses the ctrA P1 promoter and activates the strong ctrA P2 promoter, leading to a burst of CtrA synthesis in late stalked cells and early predivisional cells (Domian et al, 1999), simultaneously repressing the gcrA promoter (Holtzendorff et al, 2004). Once the division plane establishes two cellular compartments prior to cell division, CtrA is rapidly proteolyzed by the ClpXP protease (Jenal and Fuchs, 1998) in the stalked compartment of the late predivisional cell and then at the swarmer-to-stalked cell transition, when the cell is preparing to initiate DNA replication (Domian et al, 1997; Judd et al, 2003).
Even though the cell cycle regulation of ctrA expression has been extensively studied, the mechanisms governing the oscillation of CtrA and GcrA concentrations during the cell cycle are not well understood, notably because little is known about the regulation of gcrA expression or GcrA stability. Previous microarray analyses showed that gcrA mRNA levels are maximal in stalked cells and decrease in predivisional cells (Laub et al, 2000), suggesting that gcrA transcription may be an important level of regulation of gcrA cell cycle expression. It was also shown that CtrA represses transcription of gcrA by directly binding to a CtrA-binding site within the gcrA promoter (Holtzendorff et al, 2004). In this study, we demonstrate that as DnaA accumulates during the swarmer-to-stalked cell transition, it directly activates gcrA transcription as the CtrA repressor is being cleared from the cell. Temporally regulated proteolysis further modulates GcrA cellular concentration during the cell cycle. The ‘dual-use' DnaA protein functions to both enable replisome formation at the origin and to control the transcription of gcrA. The accumulation of GcrA, in turn, activates the transcription of genes involved in DNA replication and chromosome segregation, and directly activates ctrA transcription. Thus, DnaA serves as a central coordinator of multiple cell cycle events.
Results
Cell cycle control of gcrA expression
To determine how GcrA protein levels are temporally regulated, each step of GcrA synthesis was analyzed during the Caulobacter cell cycle. To measure the transcription of gcrA, a gcrAP-lacZ transcriptional fusion (Holtzendorff et al, 2004) was integrated into the Caulobacter chromosome, at a site close to the natural gcrA locus (LS4220). Transcription and translation from gcrAP results in production of β-galactosidase. We measured newly synthesized β-galactosidase at different time points during the cell cycle. Synchronous cells were pulse-labelled with [35S]methionine and β-galactosidase was immunoprecipitated to assay the activity of the gcrA promoter (Figure 1). Transcription increased more than two-fold during the swarmer-to-stalked cell transition. The maximum level of transcription was reached in stalked cells and then decreased in the predivisional cells. The cell-cycle-dependent transcription of gcrA is comparable to the changes in the steady-state levels of gcrA mRNA during the cell cycle (Laub et al, 2000), suggesting that the increase in gcrA mRNA levels during the swarmer-to-stalked cell transition is the result of increased transcription, rather than stabilization of gcrA mRNA. The rate of gcrA transcription in separated populations of swarmer and stalked cells was also measured immediately after cell division (at ∼150 min) and was found to be significantly higher in stalked than swarmer progeny cells (Figure 1). Overall, these results demonstrate that the transcriptional regulation of gcrA contributes to the control of the cell-cycle-dependent accumulation of GcrA.
To measure the temporal synthesis of GcrA, we immunoprecipitated GcrA from the pulse-labelled cell samples from the same synchronous culture used to measure the temporal transcription of gcrA. We could thereby determine how much new GcrA protein was synthesized from the natural gcrA gene during the pulse (Figure 1). Synthesis of the GcrA protein increased during the swarmer-to-stalked cell transition, reached a maximum in stalked cells, decreased in predivisional cells and was much higher in the stalked than the swarmer progenies after cell division. Since the cell cycle variations of GcrA synthesis are comparable to the cell cycle variations of gcrA transcription, we conclude that gcrA mRNA translation and stability are not significantly cell cycle regulated, even though the 5′ untranslated region of the gcrA mRNA is surprisingly long (92 nucleotides long (Holtzendorff et al, 2004)).
The proteolysis of GcrA is cell cycle regulated
The accumulation of the GcrA protein was compared with the efficiency of its synthesis during the cell cycle by immunoblots using anti-GcrA antibodies (Figure 1). Cell-cycle-regulated synthesis of GcrA can approximately account for the increase of GcrA steady-state levels in the stalked cells. Nevertheless, the rapid decrease in GcrA protein levels in predivisional cells, approximately concurrent with the decrease in GcrA synthesis, indicates that GcrA is an unstable protein. To measure GcrA half-life in a mixed cell population, we performed pulse-chase experiments in the wild-type strain grown in exponential phase. We found that the half-life of GcrA is about 24 min (data not shown), which is substantially shorter that the 150 min cell cycle of wild-type Caulobacter cells.
The absence of detectable GcrA proteins in swarmer cells (at time=0 min and after isolation of the swarmer progeny at time=140 min), despite a basal level of GcrA synthesis (Figure 1), suggests that GcrA is particularly unstable in swarmer cells. If GcrA proteolysis changes during the cell cycle, accumulated levels of GcrA should vary even when GcrA is constitutively synthesized throughout the cell cycle. To test this hypothesis, we used a strain (LS3707) in which the sole copy of gcrA is under the control of the chromosomal xylose inducible promoter PxylX, yielding constitutive expression of gcrA in the presence of xylose (Holtzendorff et al, 2004). Swarmer cells from LS3707 grown in minimal media supplemented with xylose were isolated, and protein samples were collected at different time points throughout the cell cycle in the same medium for immunoblot analysis using anti-GcrA antibodies (Figure 2A). We observed that GcrA accumulation changed during the cell cycle, even under conditions when gcrA is transcribed constitutively throughout the cell cycle, with predominant accumulation in stalked and predivisional cells. This result suggests that GcrA is significantly more unstable in swarmer cells than other Caulobacter cell types. To ascertain that the weaker accumulation of GcrA in swarmer cells than in stalked cells was due to more rapid proteolysis of GcrA, we determined the half-life of GcrA in independent isolated populations of LS3707 swarmer and stalked cells (Figure 2B). We found that the half-life of GcrA is ∼10.5 min in swarmer cells, as compared to ∼43.7 min in stalked cells. Since the rate of synthesis of GcrA in wild-type swarmer cells is very low, we were not able to assess accurately the half-life of GcrA in swarmer cells from a wild-type strain. Nevertheless, we assessed the half-life of GcrA in stalked cells from a wild-type strain and found it was also about 42 min (data not shown), which confirms that proteolysis of GcrA in the LS3707 strain is comparable to proteolysis of GcrA in wild-type cells. Overall, these results show that GcrA proteolysis is cell cycle regulated, with GcrA being degraded approximately four-fold faster in swarmer than stalked cells.
Figure 2.
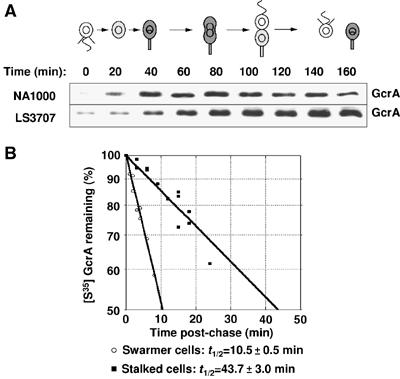
Proteolysis of GcrA is cell cycle regulated. (A) GcrA accumulation when gcrA is transcribed constitutively. Immunoblots of cell extracts from a synchronized NA1000 and LS3707 (ΔgcrA PxylX∷gcrA) culture using GcrA antibodies at the indicated times of the cell cycle. In the schematic of the Caulobacter cell cycle, gray indicates accumulation of GcrA in the NA1000 cells. (B) Stability of GcrA synthesized in swarmer and stalked LS3707 cells. A synchronized population of LS3707 swarmer cells was allowed to proceed through the cell cycle. At 5 (swarmer cells) and 30 min (swarmer-to-stalked cell transition), aliquots of the culture were pulse-labelled with [35S]methionine and chased for increasing amounts of time. Aliquots were taken at the times indicated and the remaining radiolabelled GcrA was determined by immunoprecipitation using GcrA antibodies, followed by SDS–PAGE and phosphorimaging. The t1/2 corresponds to the calculated half-life of GcrA in each cell type, along with the calculated standard deviation from three independent LS3707 populations.
Characterization of the gcrA promoter
A DNA fragment from −507 to +92 relative to the gcrA transcript initiation site (Holtzendorff et al, 2004) was used to construct the transcriptional fusion to lacZ that we used to study the temporal transcription of gcrA shown in Figure 1. We show (in Supplementary data) that all significant regulatory sequences should be included in the −78 to +1 region of the gcrA promoter. This sequence notably contains the previously characterized CtrA-binding site (Holtzendorff et al, 2004), in addition to a putative DnaA box and two putative DNA methylation sites (Figure 3A).
Figure 3.
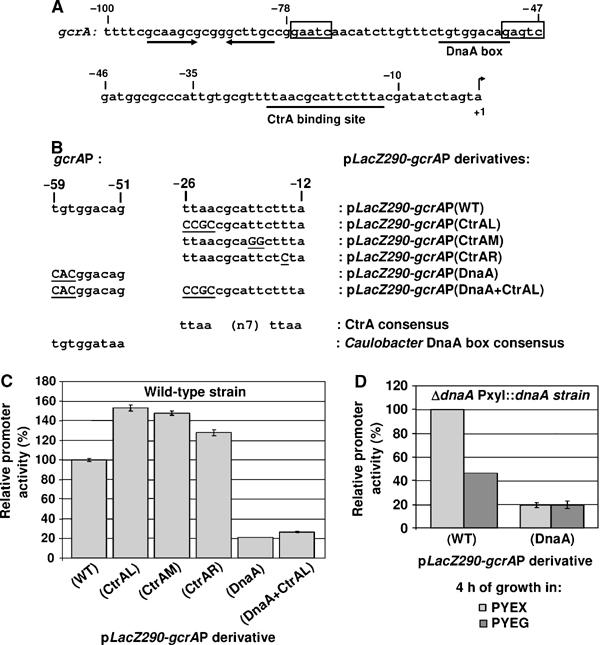
Opposite effects of CtrA and DnaA on gcrA transcription. (A) The nucleotide sequence of the gcrA promoter is shown. The transcriptional +1 start site (Holtzendorff et al, 2004), the CtrA-binding site (underlined) (Holtzendorff et al, 2004), the two putative DNA methylation sites (boxes), the putative DnaA box (underlined) and a putative hairpin structure (two arrows) are indicated. (B) The sequence of the CtrA-binding site and the DnaA box within the gcrA promoter (gcrAP) are shown and compared to the consensus CtrA-binding site and DnaA box in Caulobacter. Mutations introduced in the gcrAP are underlined and in capital letters. The names of the pLacZ290 derivatives carrying the unmodified and the mutated gcrAP fused to lacZ are also shown. (C) The graph shows the relative β-galactosidase activities from the six plasmids in (B) in an unsynchronized NA1000 strain. (D) The graph shows the relative β-galactosidase activities from pLacZ290-gcrAP(WT) and pLacZ290-gcrAP(DnaA) in an unsynchronized GM2471 (ΔdnaA∷Ω PxylX∷dnaA) strain, upon depletion of DnaA 4 h after a shift from PYE+xylose (PYEX) to PYE+glucose (PYEG). Activities in Miller units were normalized so that the activity of the gcrAP(WT) equals 100% in PYE (NA1000) or PYEX (GM2471), to facilitate comparison. Errors bars indicate the standard deviations (when they were more than 1%).
CtrA is a negative regulator of gcrA transcription
A previous study from our laboratory, assessing the activity of the gcrA promoter in a ctrA401ts mutant shifted to restrictive temperature, showed that CtrA represses gcrA transcription (Holtzendorff et al, 2004). Even though CtrA directly binds to the gcrA promoter, the activation of gcrA transcription at restrictive temperature in the ctrA401ts strain could partially be an indirect effect of the cell cycle arrest caused by the CtrA depletion. To quantitatively assess the effect of CtrA on gcrA transcription in a wild-type strain, we analyzed the activity of three mutant gcrA promoters, carrying mutations in the CtrA-binding site in the gcrA promoter fused to lacZ carried on the low copy number plasmid pLacZ290 (Figure 3B). The consensus DNA sequence for CtrA binding in Caulobacter is TTAA-N7-TTAA (Marczynski et al, 1995) and both the CtrA-binding half-sites and the N7 spacing are critical for CtrA-mediated transcriptional regulation (Ouimet and Marczynski, 2000). The first mutant gcrA promoter we constructed (gcrAP(CtrAL)) carries a 4 base-pair mutation in the left portion of the CtrA-binding site, the second (gcrAP(CtrAM)) carries a double base-pair mutation in the middle part of the CtrA-binding site, and the third (gcrAP(CtrAR)) carries a single base-pair mutation in the right portion of the CtrA-binding site. All mutated promoters were cloned in pLacZ290 and introduced into a wild-type Caulobacter strain to compare their activities with the wild-type promoter (gcrAP(WT)) by β-galactosidase assays. In all three cases, the activities of the mutated promoters were higher than that of the wild-type gcrA promoter, with a 25–50% difference in the activity (Figure 3C). To check whether the mutations we created in the gcrA promoter render the gcrA promoter independent of CtrA protein levels in the cell, we also introduced these constructs in a ctrA401ts strain. After a shift from permissive temperature to restrictive temperature, the activity of the gcrAP(CtrAL) did not change significantly, unlike the activity of the wild-type gcrA promoter, which increased significantly (data not shown). We conclude that the mutation we introduced in the left portion of the CtrA-binding site of the gcrA promoter (gcrAP(CtrAL)) renders it independent of CtrA protein levels in the cell. The activities of the gcrAP(CtrAR) and the gcrAP(CtrAM) in a ctrA401ts strain at restrictive temperature slightly increased, but more moderately than the wild-type promoter (data not shown), showing that these two mutated promoters are still slightly sensitive to CtrA protein levels in the cell, which probably explains why the effect of these two mutations on gcrA transcription are more moderate than the mutation in the left portion of the CtrA-binding site (Figure 3C). Overall, these results confirm that CtrA is a significant repressor of gcrA transcription. Yet, β-galactosidase assays using lacZ transcriptional fusions measure transcriptional levels in log phase cultures, but not temporal control.
In order to understand the role of CtrA in the regulation of gcrA transcription during the cell cycle, we integrated the apparently CtrA-independent gcrAP(CtrAL)-lacZ transcriptional fusion into the Caulobacter chromosome. As was done for the wild-type promoter (Figure 1), transcription and translation from the gcrAP(CtrAL) at different time points of the cell cycle were compared by pulse-labelling cell samples from a synchronous culture with [35S]methionine and immunoprecipitating the β-galactosidase (Figure 4A). Surprisingly, we found that transcription from the mutated gcrAP(CtrAL) promoter is still cell cycle regulated. We also immunoprecipitated GcrA from the same pulse-labelled samples, which allowed us to accurately compare GcrA synthesis from the wild-type gcrA promoter to β-galactosidase synthesis from the mutated promoter, during the same synchronization experiment. This experiment shows that the level of transcription from the mutated gcrAP(CtrAL) is still cell cycle controlled. We conclude that the degradation of the CtrA repressor is not solely responsible for the increase in gcrA transcription during the swarmer-to-stalked cell transition, suggesting that other regulatory elements are also involved in the temporal control of gcrA transcription.
Figure 4.
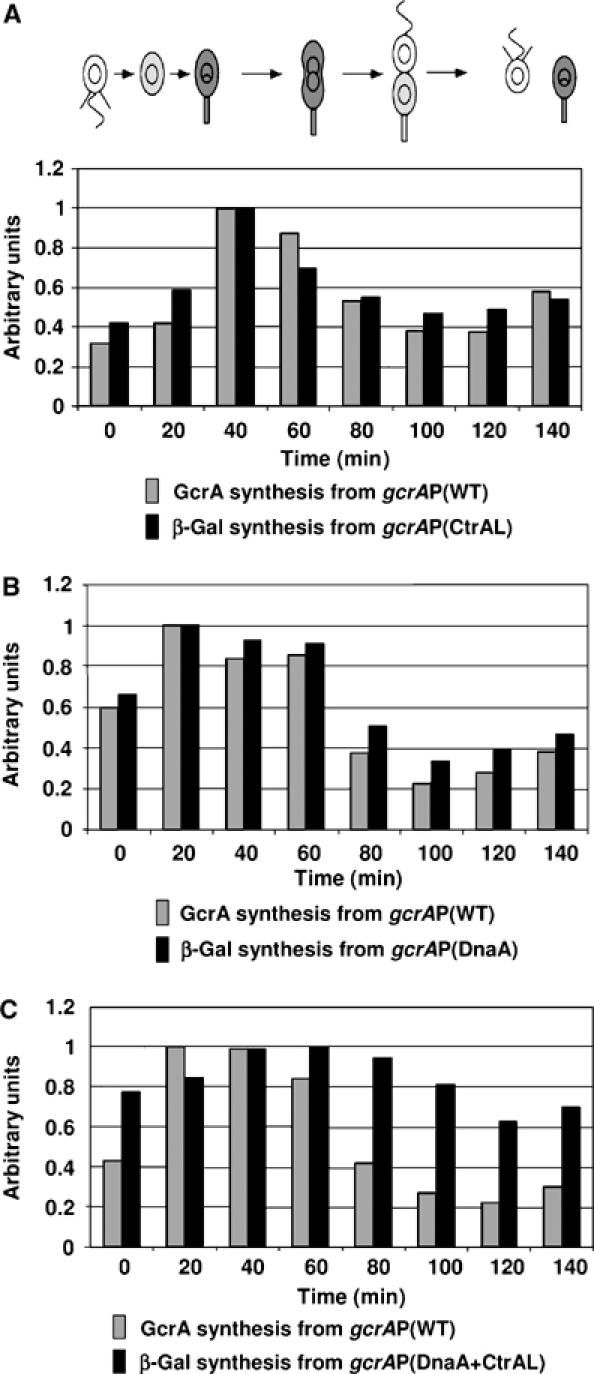
Effects of CtrA and DnaA on gcrA cell cycle transcription. Cell cycle activities of the mutated gcrA promoters were measured by immunoprecipitation of β-galactosidase from pulse-labelled samples of synchronized Caulobacter cultures. To compare with the activity of the gcrAP(WT), radiolabelled GcrA was also immunoprecipitated from the same synchrony. Relative rates of gcrA transcription from the mutated gcrA promoter and GcrA synthesis from the wild-type gcrA promoter at the indicated times of the cell cycle were measured by phosphorimaging and were normalized so that the maximum value of each experiment equals 1. The averages of the results of multiple independent experiments are shown. In the schematic of the Caulobacter cell cycle, gray indicates accumulation of GcrA in the NA1000 cells. (A) The LS4224 (gcrA+ gcrAP(CtrAL)-lacZ) strain was used to compare the cell cycle activity of the gcrAP(CtrAL) with the cell cycle activity of the gcrAP(WT). (B) The NA1000 (gcrA+) strain carrying the pLacZ290-gcrAP(DnaA) plasmid was used to compare the cell cycle activity of the gcrAP(DnaA) with the cell cycle activity of the gcrAP(WT). (C) The NA1000 (gcrA+) strain carrying the pLacZ290-gcrAP(DnaA+CtrAL) plasmid was used to compare the cell cycle activity of the gcrAP(DnaA+CtrAL) with the cell cycle activity of the gcrAP(WT).
DnaA is critical for the induction of gcrA transcription
The gcrA promoter has a putative DnaA box about 50 base pairs upstream of its +1 transcriptional start site (Figure 3A), which matches seven of the nine base-pair Caulobacter consensus DnaA box sequence (Marczynski and Shapiro, 1992). DnaA is a dual function protein: it is essential for the initiation of DNA replication and it acts as a transcription factor in Escherichia coli (for a review, see Messer and Weigel, 1997), Bacillus subtilis (Goranov et al, 2005) and Caulobacter (Hottes et al, 2005).
To determine whether DnaA is a regulator of gcrA transcription, we used a strain (GM2471) in which the sole copy of dnaA is under the control of the chromosomal xylose-inducible promoter PxylX. The gcrAP-lacZ fusion on plasmid pLacZ290-gcrAP(WT) was assayed in strain GM2471 (Figure 3D). At 4 h after this strain was shifted from PYEX to PYEG to deplete DnaA, the activity of the gcrA promoter decreased by more than 50%. Thus, DnaA is directly or indirectly involved in the activation of the gcrA promoter.
To test whether the activation of gcrA transcription by DnaA requires the presence of the DnaA box in the gcrA promoter, we analyzed the transcription of a mutant gcrA promoter (gcrAP(DnaA)), in which the DnaA box has been disrupted (Figure 3B). The gcrAP(DnaA) was cloned in the pLacZ290 and introduced in a wild-type Caulobacter strain to compare its activity with the wild-type promoter (gcrAP(WT)) by β-galactosidase assays. We observed a striking 80% loss of activity of the gcrAP(DnaA) as compared to the wild-type promoter (Figure 3C). Similarly, when the construct was introduced in the GM2471 strain grown in PYEX, the activity of the gcrAP(DnaA) was also five-fold lower than the activity of the wild-type promoter (Figure 3D). Since the mutated gcrAP(DnaA) has the same activity in GM2471 grown for 4 h in either xylose or glucose containing media, we conclude that the mutations we introduced in the DnaA box renders the mutated gcrAP(DnaA) independent of the DnaA protein levels in the cell. The dependence of gcrA transcription on DnaA and on the presence of an intact DnaA box in the gcrA promoter provides evidence that the induction of gcrA transcription by DnaA is direct and requires binding of DnaA to the gcrA promoter.
To understand the effect of DnaA on the temporal control of gcrA transcription, we analyzed the transcription and translation from the gcrAP(DnaA) at different time points of the cell cycle by pulse labelling cell samples from a synchronous culture and immunoprecipitating the β-galactosidase (Figure 4B). We then compared GcrA synthesis from the wild-type gcrA promoter to β-galactosidase synthesis from the DnaA-independent gcrAP(DnaA) promoter during the same synchronization experiments. This experiment shows that the low-level activity from the DnaA-independent promoter is still cell cycle controlled, parallel to our results with the CtrA-independent promoter and with the wild-type gcrA promoter.
Caulobacter integrates the negative control by CtrA and the positive control by DnaA to regulate gcrA cell cycle transcription
We observed that the temporal regulation of the mutated gcrAP(CtrAL) and gcrAP(DnaA) promoters are equivalent to the wild-type gcrA promoter. This result suggests that CtrA and DnaA both contribute to the control of the temporal transcription of gcrA. Another possibility is that a third regulator is responsible for gcrA temporal regulation. To discriminate between these two possibilities, we constructed a gcrA promoter double mutant (gcrAP(DnaA+CtrAL)) that is mutated in both the left portion of the CtrA-binding site and in the DnaA box (Figure 3B). This mutated promoter was cloned in pLacZ290 and introduced into a wild-type Caulobacter strain and its activity was compared to strains with single promoter mutants: β-galactosidase assays were carried out in all three strains (Figure 3C). Like the gcrAP(DnaA), we observed that the activity of the gcrAP(DnaA+CtrAL) is much lower than the activity of the wild-type promoter when introduced in a wild-type Caulobacter strain (Figure 3C), showing that DnaA can activate the CtrA-independent gcrAP(CtrAL). Yet, the activity of the gcrAP(DnaA+CtrAL) is about 26% higher than the activity of the gcrAP(DnaA) (Figure 3C), showing that CtrA can repress the basal transcription from the DnaA-independent gcrAP(DnaA). These observations suggest that CtrA and DnaA independently contribute to the level of expression of the gcrA promoter but that their activities may function together to provide correct temporal regulation.
To test this hypothesis, we compared the temporal activity of the gcrAP(DnaA+CtrAL) double mutant fused to lacZ with that of the wild-type gcrA promoter by immunoprecipitation experiments from samples pulse labelled at different time points in a synchronized cell culture (Figure 4C). Unlike that observed with the single mutants gcrAP(CtrAL) (Figure 4A) or gcrAP(DnaA) (Figure 4B), the cell cycle regulation of the double mutant gcrAP(DnaA+CtrAL) is significantly attenuated as compared to the wild-type promoter (Figure 4C). Indeed, we observed that the maximum change in the activity of the gcrAP(DnaA+CtrAL) over the course of a cell cycle is about 1.5-fold, versus about five-fold for the maximum change in the activity of the gcrAP(WT) over the course of a cell cycle. This result argues that Caulobacter integrates the activation of gcrA transcription by DnaA and the repression of gcrA transcription by CtrA to maintain strong cell cycle regulation of gcrA transcription. Although these experiments show that either DnaA or CtrA is required for the strong temporal control of gcrA transcription, other regulatory elements may also contribute to this control, since transcription from the gcrAP(DnaA+CtrAL) double-mutant promoter is not totally constant throughout the cell cycle.
If DnaA, in combination with CtrA, contributes to the temporal control of gcrA transcription, then active DnaA should accumulate during the swarmer-to-stalked cell transition and before GcrA accumulates. Indeed, we observed that DnaA protein levels are cell cycle regulated and that DnaA starts to accumulate at the swarmer-to-stalked cell transition about 20 min before GcrA (Figure 5). Since DnaA functions to allow DNA replication initiation at this same time period (Zweiger and Shapiro, 1994), DnaA likely carries out a dual function at that time in the cell cycle.
The two methylation sites in the gcrA promoter are minor contributors to gcrA regulation
In addition to a DnaA box and a CtrA consensus sequence, the gcrA promoter contains two GANTC sequences (Figure 3A) that are the recognition motifs for the CcrM DNA methyltransferase in Caulobacter (Zweiger et al, 1994). We explored the possibility that the methylation state of these two sites affects the activity of the gcrA promoter, as it is the case for the ctrA P1 promoter (Reisenauer and Shapiro, 2002). Because the native gcrA gene is located next to the terminus of the Caulobacter chromosome, it remains in the fully methylated state till near the end of chromosome replication (Figure 6B). To determine whether the position of gcrA on the chromosome, and thus its methylation state, affects the level of gcrA transcription, we integrated the gcrAP-lacZ reporter at a site next to the origin of replication on the chromosome (at the hrcA locus, named site 2, in strain LS4221) and compared its activity to the same reporter integrated at a site next to the terminus of replication on the chromosome (at the trpE locus, named site 1, in strain LS4220). Previous studies have demonstrated that DNA methylation at the two sites used in this study varies during the cell cycle (Stephens et al, 1996; Marczynski, 1999). GANTC sites near Cori (site 2) become hemimethylated soon after the initiation of DNA replication and remain hemimethylated until the end of S phase when the DNA methyltransferase CcrM is present and active. In contrast, GANTC sites near the terminus (site 1) are hemimethylated only for a short period at the end of S phase. β-Galactosidase assays showed that the activity of the wild-type gcrA promoter fused to lacZ integrated near the terminus (site 1) is ∼35% lower than its activity when integrated near Cori (site 2) (Figure 6C). To determine what proportion of this 35% difference in activity reflects simple changes in the copy number of the reporter during DNA replication, we also constructed a mutant gcrA promoter that cannot be methylated [gcrAP(UM)], by generating a single base pair mutation in each of the two GANTC sequences in the gcrA promoter (Figure 6A). The activity of the mutated gcrAP(UM)-lacZ reporter integrated near the terminus (site 1) was ∼20% lower than its activity when integrated near Cori (Figure 6C), revealing the difference due to change in the copy number between the two integration sites. These results suggest that the difference in the timing of methylation of the two GANTC sequences in the gcrA promoter near the Cori or the terminus, results in a 15% difference (35% total difference minus 20% difference due to change in the copy number) in the activity of the gcrA promoter. Overall, we propose that full methylation of the gcrA promoter partially represses gcrA transcription.
Figure 6.
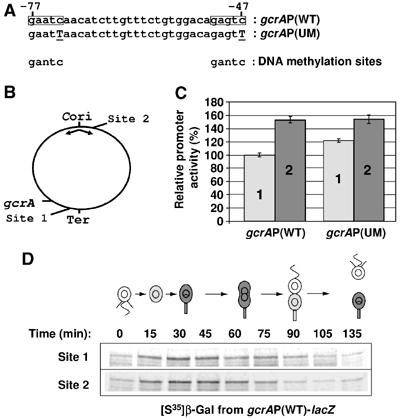
Effect of gcrAP methylation on gcrA transcription. (A) The sequences of the two putative methylation sites (boxes) within the gcrA promoter (gcrAP) are shown and compared to the consensus DNA methylation sites in Caulobacter. Mutations introduced in the gcrAP(−507…+92) methylation sites are underlined and in capital letters. (B) Diagram of the Caulobacter chromosome showing the locations of the origin of replication (Cori), the terminus region (Ter), the gcrA gene and the trpE (site 1) and hrcA (site 2) integration sites. (C) Activities of the wild-type gcrAP(WT) fused to lacZ and integrated at site 1 or site 2 (strains LS4220 and LS4221, respectively) and the unmethylatable gcrAP(UM) fused to lacZ integrated at site 1 or site 2 (strains LS4222 and LS4223, respectively) were determined by β-galactosidase assays. The activities of the control LS3321 and LS3323 strains were subtracted from the activities of the gcrAP-lacZ constructs. Activities were normalized so that the activity of the gcrAP(WT) at site 1 equals 100%. Errors bars indicate the standard deviations from three independent experiments. (D) Cell cycle gcrAP(WT) activities at site 1 and site 2 were measured by immunoprecipitation of β-galactosidase from pulse-labelled samples of synchronized LS4220 and LS4221 strains cultures, respectively. Autoradiograms from the immunoprecipitations of labelled β-galactosidase are shown. In the schematic of the Caulobacter cell cycle, gray indicates accumulation of GcrA in the NA1000 cells.
To test whether the modest effect of DNA methylation on gcrA transcription affected the temporal regulation of gcrA transcription, we assayed transcription levels at multiple time points in a synchronized culture. We compared expression from the gcrA promoter integrated at site 1 to the promoter integrated at site 2, by pulse-labelling cell samples from the two synchronous cultures with [35S]methionine and immunoprecipitating the β-galactosidase (Figure 6D). We found that temporal transcription from the gcrA promoter fused to lacZ was not affected by the chromosomal position of the gcrA promoter reporter. However, the effect of methylation on the temporal regulation of gcrA transcription may be hidden by the strong temporal regulation of the wild-type gcrA promoter by DnaA and CtrA.
Discussion
By studying the regulation of gcrA expression, we have uncovered an unexpected and critical link between the oscillating GcrA and CtrA global cell cycle regulators and DNA replication control. We show that the DnaA replication initiation factor activates the transcription of gcrA, which encodes a central component of the regulatory circuit that drives the cell cycle (Figure 7). DnaA thereby acts to coordinate the initiation of DNA replication with the expression of genes required for DNA replication and segregation, cell division and cell differentiation.
Figure 7.
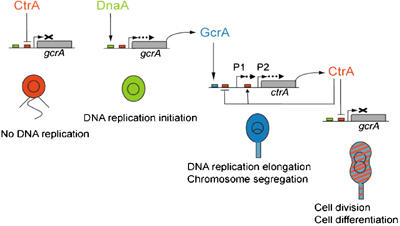
Model for the control of the Caulobacter cell cycle by sequential accumulation of the DnaA, GcrA and CtrA global cell cycle regulators. In swarmer cells, CtrA represses gcrA transcription and GcrA protein is very unstable, so GcrA does not accumulate. During the swarmer to stalked cell differentiation, CtrA is degraded and DnaA accumulates, which allows gcrA transcription to be turned on. Since GcrA is more stable in stalked cells, GcrA can accumulate efficiently. When cell division is initiated, ctrA transcription is turned back on (notably by accumulated GcrA), and DnaA is degraded and probably inactivated. Accumulation of CtrA and disappearance of active DnaA turns off gcrA transcription in predivisional cells. In the schematic of the Caulobacter cell cycle, red, green and blue indicate accumulation of CtrA, DnaA and GcrA, respectively.
Cell-cycle-dependent regulation of GcrA
Transcription of gcrA is very low in swarmer cells and increases during the swarmer-to-stalked cell transition. It reaches a maximum in the stalked cells, before it decreases to a low level again in predivisional cells. At the end of the cell cycle, transcription of gcrA rapidly resumes in the stalked progeny but remains very low in the swarmer progeny (Figure 1). The following evidence demonstrates that the CtrA cell cycle response regulator directly represses gcrA transcription: first, depletion of CtrA causes an increase in gcrA transcription (Holtzendorff et al, 2004). Second, CtrA directly binds to the gcrA promoter (Holtzendorff et al, 2004). Third, disruption of the CtrA-binding site in the gcrA promoter by targeted mutagenesis renders gcrA transcription non-responsive to CtrA depletion and causes an increase in gcrA transcription (Figure 3). Since CtrA is present and active in swarmer cells and in predivisional cells, but not in stalked cells (Domian et al, 1997, 1999), we hypothesized that the burst in gcrA transcription that we observed in stalked cells was due to the degradation of its repressor CtrA. However, here we show that CtrA is not the only regulator of gcrA cell cycle transcription since the mutant gcrA promoter that is not responsive to CtrA, is still cell cycle controlled (Figure 4A). Indeed, we demonstrate that transcriptional activation of gcrA by the DnaA protein, in concert with the negative effect of CtrA, regulates the temporal control of gcrA transcription. Transcription of gcrA is dramatically reduced when the DnaA protein is depleted (Figure 3D). Additionally, a mutant gcrA promoter, which is not responsive to DnaA cellular levels, has a transcriptional activity that is five-fold lower than the wild-type gcrA promoter (Figure 3). We observed that the DnaA cellular concentration is cell cycle regulated and is highest during the swarmer-to-stalked cell transition (Figure 5), when DNA replication also initiates. The model we propose for the temporal regulation of GcrA cellular levels during the Caulobacter cell cycle is presented in Figure 7. The low levels of the inducer DnaA and the presence of the repressor CtrA in the swarmer cells yield a basal level of gcrA transcription. Furthermore, the few GcrA proteins synthesized are rapidly proteolyzed to prevent any accumulation of GcrA in swarmer cells (Figure 2). During the swarmer-to-stalked cell transition, the repressor CtrA is removed and the inducer DnaA accumulates (Figure 5), leading to a significant increase in gcrA transcription. In addition, the turnover of the newly synthesized GcrA proteins in the stalked cells is four-fold slower than in swarmer cells (Figure 2B), which allows the rapid accumulation of GcrA proteins in stalked cells. As the cells elongate approaching cell division, GcrA accumulation turns on ctrA transcription. CtrA accumulation and concomitant reduction in DnaA levels together turn off gcrA transcription in the predivisional cell (Figure 7).
Multilayer systems tightly control the cell cycle
One striking feature of this model is that multiple pathways regulate GcrA accumulation and that they function in concert to affect the temporally controlled accumulation of GcrA.
First, the temporally regulated transcription of gcrA is controlled by the combined effects of CtrA and DnaA, integrating the negative control by CtrA and the positive control by DnaA (Figure 3). To affect the strong temporal regulation of gcrA transcription, both CtrA-mediated and DnaA-mediated regulation pathways have to be disrupted at the same time. Moreover, CtrA and DnaA may not be the only regulators of gcrA cell cycle transcription, since the cell cycle control of a DnaA and CtrA-independent gcrA promoter is greatly attenuated, but not abolished (Figure 4C). We present evidence that the methylation state of the gcrA promoter mildly influences the expression of gcrA (Figure 6C) and this may provide the additional regulation for complete cell cycle control.
Second, the cell cycle accumulation of GcrA is controlled both by transcriptional regulation and by regulated proteolysis. We showed that GcrA accumulation still exhibits cell cycle control when gcrA is transcribed constitutively (Figure 2A). This result suggests that the control by regulated gcrA transcription is partly redundant with the control by regulated GcrA proteolysis, to fine-tune the cellular amounts of GcrA during the cell cycle. Many other essential Caulobacter proteins are subject to multiple mechanisms of control. The master regulator CtrA (Domian et al, 1997, 1999; Reisenauer and Shapiro, 2002; Judd et al, 2003; Holtzendorff et al, 2004), the cell division protein FtsZ (Kelly et al, 1998), the replication initiator DnaA (Zweiger and Shapiro, 1994; Gorbatyuk and Marczynski, 2005) and the DNA methyltransferase CcrM (Stephens et al, 1995, 1996; Wright et al, 1996) are other examples of essential proteins regulated by differential transcription and differential proteolysis during the Caulobacter cell cycle. Since the proteolysis of DnaA and GcrA is coincident, occurring specifically in swarmer cells, a common mechanism may regulate their degradation.
Parallel systems regulate gcrA expression and the initiation of DNA replication
The regulation of gcrA expression is parallel to the regulation of the initiation of replication. First, chromosome replication is initiated coincident with the transcription of gcrA, during the swarmer-to-stalked cell transition. Second, Cori and the gcrA promoter contain CtrA-binding sites, DnaA boxes and DNA methylation sites (Figure 3A) (Marczynski and Shapiro, 2002; Holtzendorff et al, 2004). Third, the activities of Cori and the gcrA promoter are activated by DnaA and repressed by CtrA (Figure 3) (Marczynski and Shapiro, 2002; Holtzendorff et al, 2004). Thus, the cell is using similar mechanisms of regulation to coordinate the initiation of chromosome replication with the synthesis of GcrA. However, the regulation of gcrA is not identical to the regulation of the Cori. For example, the IHF protein activates the initiation of chromosome replication (Siam et al, 2003), but does not influence gcrA transcription (data not shown).
The sequential accumulation of three essential regulators control the Caulobacter cell cycle
GcrA concentration oscillates not only with CtrA concentration, but also with DnaA concentration during the Caulobacter cell cycle (Figure 5). CtrA notably acts as a silencer of the chromosome replication origin, an inhibitor of ftsZ transcription and a regulator of multiple genes required for pili and flagella biogenesis (Quon et al, 1996, 1998; Kelly et al, 1998; Laub et al, 2002). GcrA notably activates podJ and pleC that are required for polar morphogenesis and regulates genes needed for DNA replication elongation and chromosome partitioning, like dnaQ, dnaB, dnaC and gyrA (Holtzendorff et al, 2004). Further analysis now shows that GcrA does not significantly affect dnaA transcription, in contrast to previous results (Holtzendorff et al, 2004). Like CtrA, Caulobacter DnaA is an essential dual function protein (Gorbatyuk and Marczynski, 2001): it regulates the initiation of DNA replication and it is a transcription factor. The DnaA regulon in Caulobacter has recently been characterized showing that DnaA regulates the transcription of multiple genes, notably involved in nucleotide biosynthesis and DNA replication (Hottes et al, 2005). Overall, the DnaA/GcrA/CtrA cascade during the Caulobacter cell cycle defines the timing of expression of multiple genes encoding proteins with diverse functions needed for progress through the cell cycle (Figure 7). The activation of gcrA transcription by DnaA is a novel critical regulatory pathway, which coordinates DNA replication initiation with cell division and cell differentiation functions regulated by GcrA and CtrA.
Materials and methods
Bacterial strains, synchronization and growth conditions
Caulobacter strains were grown in peptone–yeast extract (PYE) complex media or M2 minimal salts+0.2% glucose (M2G) minimal media (Ely, 1991) at 30°C (or 28°C for experiments with LS3707). For GM1258 derivative strains, M2G was supplemented with 0.01% of tryptophan. When indicated, xylose (0.3% for GM2471 or 0.03% for LS3707) or glucose (0.2%) was added to the media to induce or repress the PxylX promoter, respectively. Strains and plasmids used are listed in Table I. Antibiotics used include oxytetracyclin (1 μg/ml), kanamycin (5 or 25 μg/ml), spectinomycin (25 μg/ml), streptomycin (5 μg/ml) and nalidixic acid (20 μg/ml). Plasmids were mobilized from E. coli S17-1 into Caulobacter by bacterial conjugation or introduced by transformation. Synchronized cultures of Caulobacter were obtained by centrifugation in a Ludox density gradient followed by isolation of swarmer cells (Evinger and Agabian, 1977). Swarmer cells were released into minimal medium for cell cycle studies.
Table 1.
Strains and plasmids used in this study
| Plasmids | Relevant characteristics or construction | Source or reference |
|---|---|---|
| pNPT228 | pLitmus28-derived integration vector | MRK Alley |
| pHP45Ω | Vector carrying a SpecR/StrepR cassette (Ω) | Prentki and Krisch (1984) |
| PBOR | 2-kb EcoRI fragment from pHP45Ω ligated into EcoRI-digested pBluescript | C Stevens |
| pLacZ290 | lacZ transcriptional fusion vector | Gober and Shapiro (1992) |
| pLacZ290-gcrAP(WT)=pLacZ290-gcrAP | gcrAP(−507…+92)-lacZ in pRKlac290 | Holtzendorff et al (2004) |
| pLacZ290-gcrAP′(WT) | gcrAP(−100…+92)-lacZ in pRKlac290 | This study |
| pLacZ290-gcrAP″(WT) | gcrAP(−78…+92)-lacZ in pRKlac290 | This study |
| pLacZ290-gcrAP(CtrAL) | gcrAP(CtrAL)-lacZ in pRKlac290 | This study |
| pLacZ290-gcrAP(CtrAM) | gcrAP(CtrAM)-lacZ in pRKlac290 | This study |
| pLacZ290-gcrAP(CtrAR) | gcrAP(CtrAR)-lacZ in pRKlac290 | This study |
| pLacZ290-gcrAP(UM) | gcrAP(UM)-lacZ in pRKlac290 | This study |
| pLacZ290-gcrAP(DnaA) | gcrAP(DnaA)-lacZ in pRKlac290 | This study |
| pLacZ290-gcrAP(DnaA+CtrAL) | gcrAP(DnaA+CtrAL)-lacZ in pRKlac290 | This study |
| pNPT228-gcrAP-lacZ | 4-kb BamHI-DraI fragment from pLacZ290-gcrAP ligated into BamHI–EcoRV-digested pNPT228 | This study |
| pNPT228-gcrAP-lacZ-Ω | 2-kb BamHI fragment from pBOR ligated into BamHI-digested pNPT228-gcrAP-lacZ | This study |
| pNPT228-gcrAP(UM)-lacZ | 4-kb BamHI-DraI fragment from pLacZ290-gcrAP(UM) ligated into BamHI–EcoRV-digested pNPT228 | This study |
| pNPT228-gcrAP(UM)-lacZ-Ω | 2-kb BamHI fragment from pBOR ligated into BamHI-digested pNPT228-gcrAP(UM)-lacZ | This study |
| pNPT228-gcrAP(CtrAL)-lacZ | 4-kb BamHI-DraI fragment from pLacZ290-gcrAP(CtrAL) ligated into BamHI–EcoRV-digested pNPT228 | This study |
| pNPT228-gcrAP(CtrAL)-lacZ-Ω | 2-kb BamHI fragment from pBOR ligated into BamHI-digested pNPT228-gcrAP(CtrAL)-lacZ | This study |
| Strains |
Relevant genotype |
Source or reference |
| E. coli | ||
| S17-1 | 294∷RP4-2(Tc∷Mu)(Km∷Tn7) | Simon et al (1983) |
| C. crescentus | ||
| NA1000 | Synchronizable derivative of CB15 | Evinger and Agabian (1977) |
| GM1258 | NA1000 trpE∷Tn5Ω-MP | Marczynski (1999) |
| LS3323 | NA1000 trpE∷Tn5Ω∷pNPT228-lacZ-Ω | Reisenauer and Shapiro (2002) |
| LS4220 | NA1000 trpE∷Tn5Ω∷pNPT228-gcrAP(−507…+92)-lacZ-Ω | This study |
| LS4222 | NA1000 trpE∷Tn5Ω∷pNPT228-gcrAP(UM)-lacZ-Ω | This study |
| LS4224 | NA1000 trpE∷Tn5Ω∷pNPT228-gcrAP(CtrAL)-lacZ-Ω | This study |
| LS2293 | NA1000 hrcAΩ | Roberts et al (1996) |
| LS3321 | NA1000 hrcAΩ∷pNPT228-lacZ-Ω | Reisenauer and Shapiro (2002) |
| LS4221 | NA1000 hrcAΩ∷pNPT228-gcrAP(−507…+92)-lacZ-Ω | This study |
| LS4223 | NA1000 hrcAΩ∷pNPT228-gcrAP(UM)-lacZ-Ω | This study |
| GM2471 | NA1000 ΔdnaA∷Ω PxylX∷dnaA | Gorbatyuk and Marczynski (2001) |
| LS3707 | NA1000 ΔgcrA∷Ω PxylX∷gcrA | Holtzendorff et al (2004) |
Site-directed mutagenesis and truncation of the gcrAP
See Supplementary data.
Construction of the gcrAP-lacZ chromosomal integrants
Plasmids pNPT228-gcrAP-lacZ-Ω, pNPT228-gcrAP(UM)-lacZ-Ω and pNPT228-gcrAP(CtrAL)-lacZ-Ω were integrated into the chromosomal Ω cassette of strains LS2293 and/or GM1258 by a single integration event.
Promoter activity assays
The β-galactosidase activity (in Miller Units) of strains containing the promoter-lacZ plasmids or integrated promoter-lacZ transcriptional fusions was assayed in log-phase cultures (Miller, 1972). β-galactosidase activities represent the average of at least three independent assays.
Immunoblot analysis
GcrA and CtrA proteins were resolved on 15% SDS–PAGE and DnaA proteins were resolved on 8% SDS–PAGE (Sambrook et al, 1989), and electrotransferred to a polyvinylidene fluoride membrane (Millipore). Immunodetection was performed with polyclonal antibodies. Anti-DnaA, anti-CtrA and donkey anti-rabbit conjugated to horseradish peroxidase (Jackson ImmunoResearch) serums were diluted 1:10 000. Anti-GcrA serum was diluted 1:2000. A chemiluminescent reagent (PerkinElmer) and Kodak Bio-Max MR films were used. Images were processed with Photoshop (Adobe) and relative band intensities were determined using ImageQuant software (Molecular Dynamics).
Cell cycle transcription and synthesis analysis
Aliquots (1 ml) of cells grown in M2G were removed and labelled with 10 μCi of [35S]methionine (Amersham) for 2 or 5 min. Cells were collected and lysed. Equivalent counts of radiolabelled proteins were then used for immunoprecipitation using anti-GcrA or anti-β-galactosidase serums. Resulting samples were resolved by SDS–PAGE and labelled protein bands were quantified using the ImageQuant software. Their relative activity was normalized so that the maximum value equals 1. Details of protocols and composition of buffers used are in the Supplementary data.
GcrA half-life determination
The stability of GcrA was determined by pulse-chase experiments. Cells grown in M2G or M2GX were labelled with 10 μCi/ml of [35S]methionine for 3 or 5 min and then chased with 1 mM unlabelled methionine and 0.2 mg/ml casamino acids. Cells from 1 ml of culture were collected at the indicated times by centrifugation and freezing on dry ice. Labelled GcrA protein was immunoprecipitated as described above, resolved by 12.5 or 15% SDS–PAGE and quantified as described above. Data fitting to determine GcrA half-life is described in Supplementary data.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Alison K Hottes for critical reading of the manuscript. JC was the recipient of a Stanford Dean's Fellowship. This work was supported by a Department of Energy Grant DE-FG03-01ER63219 and by National Institutes of Health Grant GM32506.
References
- Bramhill D, Kornberg A (1988) Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52: 743–755 [DOI] [PubMed] [Google Scholar]
- Domian IJ, Reisenauer A, Shapiro L (1999) Feedback control of a master bacterial cell-cycle regulator. Proc Natl Acad Sci USA 96: 6648–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L (1997) Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90: 415–424 [DOI] [PubMed] [Google Scholar]
- Evinger M, Agabian N (1977) Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol 132: 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller RS, Funnell BE, Kornberg A (1984) The DnaA protein complex with the E. coli chromosomal replication origin (oriC) and other DNA sites. Cell 38: 889–900 [DOI] [PubMed] [Google Scholar]
- Gille H, Messer W (1991) Localized DNA melting and structural pertubations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J 10: 1579–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gober JW, Shapiro L (1992) A developmentally regulated Caulobacter flagellar promoter is activated by 3′ enhancer and IHF binding elements. Mol Biol Cell 3: 913–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goranov AI, Katz L, Breier AM, Burge CB, Grossman AD (2005) A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc Natl Acad Sci USA 102: 12932–12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT (2001) Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol Microbiol 40: 485–497 [DOI] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT (2005) Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol 55: 1233–1245 [DOI] [PubMed] [Google Scholar]
- Holtzendorff J, Hung D, Brende P, Reisenauer A, Viollier PH, McAdams HH, Shapiro L (2004) Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science 304: 983–987 [DOI] [PubMed] [Google Scholar]
- Hottes AK, Shapiro L, McAdams HH (2005) DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol 58: 1340–1353 [DOI] [PubMed] [Google Scholar]
- Jenal U, Fuchs T (1998) An essential protease involved in bacterial cell-cycle control. EMBO J 17: 5658–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd EM, Ryan KR, Moerner WE, Shapiro L, McAdams HH (2003) Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc Natl Acad Sci USA 100: 8235–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AJ, Sackett MJ, Din N, Quardokus E, Brun YV (1998) Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev 12: 880–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L (2000) Global analysis of the genetic network controlling a bacterial cell cycle. Science 290: 2144–2148 [DOI] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro L, McAdams HH (2002) Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci USA 99: 4632–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczynski GT (1999) Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J Bacteriol 181: 1984–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczynski GT, Shapiro L (1992) Cell-cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J Mol Biol 226: 959–977 [DOI] [PubMed] [Google Scholar]
- Marczynski GT, Shapiro L (2002) Control of chromosome replication in Caulobacter crescentus. Annu Rev Microbiol 56: 625–656 [DOI] [PubMed] [Google Scholar]
- Marczynski GT, Lentine K, Shapiro L (1995) A developmentally regulated chromosomal origin of replication uses essential transcription elements. Genes Dev 9: 1543–1557 [DOI] [PubMed] [Google Scholar]
- Messer W, Weigel C (1997) DnaA initiator – also a transcription factor. Mol Microbiol 24: 1–6 [DOI] [PubMed] [Google Scholar]
- Ouimet MC, Marczynski GT (2000) Analysis of a cell-cycle promoter bound by a response regulator. J Mol Biol 302: 761–775 [DOI] [PubMed] [Google Scholar]
- Prentki P, Krisch HM (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29: 303–313 [DOI] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT (1998) Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA 95: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT, Shapiro L (1996) Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84: 83–93 [DOI] [PubMed] [Google Scholar]
- Reisenauer A, Shapiro L (2002) DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J 21: 4969–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RC, Mohr CD, Shapiro L (1996) Developmental programs in bacteria. Curr Top Dev Biol 34: 207–257 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Siam R, Brassinga AK, Marczynski GT (2003) A dual binding site for integration host factor and the response regulator CtrA inside the Caulobacter crescentus replication origin. J Bacteriol 185: 5563–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1: 784–790 [Google Scholar]
- Stephens CM, Zweiger G, Shapiro L (1995) Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J Bacteriol 177: 1662–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C, Reisenauer A, Wright R, Shapiro L (1996) A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc Natl Acad Sci USA 93: 1210–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R, Stephens C, Zweiger G, Shapiro L, Alley MR (1996) Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev 10: 1532–1542 [DOI] [PubMed] [Google Scholar]
- Zweiger G, Shapiro L (1994) Expression of Caulobacter dnaA as a function of the cell cycle. J Bacteriol 176: 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweiger G, Marczynski G, Shapiro L (1994) A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J Mol Biol 235: 472–485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


