Abstract
Ppt1 is the yeast member of a novel family of protein phosphatases, which is characterized by the presence of a tetratricopeptide repeat (TPR) domain. Ppt1 is known to bind to Hsp90, a molecular chaperone that performs essential functions in the folding and activation of a large number of client proteins. The function of Ppt1 in the Hsp90 chaperone cycle remained unknown. Here, we analyzed the function of Ppt1 in vivo and in vitro. We show that purified Ppt1 specifically dephosphorylates Hsp90. This activity requires Hsp90 to be directly attached to Ppt1 via its TPR domain. Deletion of the ppt1 gene leads to hyperphosphorylation of Hsp90 in vivo and an apparent decrease in the efficiency of the Hsp90 chaperone system. Interestingly, several Hsp90 client proteins were affected in a distinct manner. Our findings indicate that the Hsp90 multichaperone cycle is more complex than was previously thought. Besides its regulation via the Hsp90 ATPase activity and the sequential binding and release of cochaperones, with Ppt1, a specific phosphatase exists, which positively modulates the maturation of Hsp90 client proteins.
Keywords: enzyme regulation, heat-shock proteins, molecular chaperone, protein folding, Saccharomyces cerevisiae
Introduction
Ppt1 is a protein phosphatase from Saccharomyces cerevisiae that belongs to the PPP family of serine/threonine phosphatases (Becker et al, 1994; Chen et al, 1994; Chinkers, 1994). Although Ppt1 was discovered a decade ago, its function in yeast is still unclear. It does not appear to be essential for viability, as a ppt1Δ yeast strain shows no significant phenotype, even under a variety of stress conditions (Chen et al, 1994). The human homolog of Ppt1 is the phosphatase PP5, exhibiting a sequence identity of 63% (Chen et al, 1994). For both phosphatases, complex formation with the molecular chaperone Hsp90 was demonstrated in vitro and in vivo (Chen et al, 1996; Silverstein et al, 1997; Russell et al, 1999; Gavin et al, 2002; Ramsey and Chinkers, 2002; Yang et al, 2005). Interaction of PP5 and Hsp90 is mediated by the tetratricopeptide repeat (TPR) domain of PP5 (Chen et al, 1996; Silverstein et al, 1997; Yang et al, 2005). The presence of a TPR domain in PP5 and Ppt1 is unique among phosphatases (Cohen, 1997; Andreeva and Kutuzov, 1999; Chinkers, 2001). It consists of three motifs of degenerated 34-amino-acid repeats, each made up of two antiparallel α-helices connected by a short loop (Das et al, 1998). Consecutive motifs combine to a superhelical groove, which can bind TPR acceptor modules. The TPR domain interacts with a common TPR acceptor site, the C-terminal EEVD residues of Hsp90 (Silverstein et al, 1997; Scheufler et al, 2000; Ramsey et al, 2000; Yang et al, 2005). This is also the case in other Hsp90 partner proteins containing TPR domains like the large peptidyl prolyl cis/trans isomerase (PPIases) (Radanyi et al, 1994; Owens-Grillo et al, 1996) and the Hsp90/Hsp70 organizing protein Hop (Honore et al, 1992; Smith et al, 1993; Scheufler et al, 2000). In fact, PP5 and the large PPIases FKBP52 and Cyp40 compete for binding to mammalian heat-shock protein 90 (hHsp90) (Silverstein et al, 1997). The TPR domain resides in the N-terminal part of Ppt1/PP5, in front of the C-terminal catalytic domain (Chen et al, 1994; Yang et al, 2005).
The low basal phosphatase activity of PP5/Ppt1 is due to autoinhibition by the TPR domain and the C-terminal αJ subdomain (Chen and Cohen, 1997; Sinclair et al, 1999; Kang et al, 2001; Swingle et al, 2004). The recent determination of the three-dimensional structure of PP5 showed that the TPR domain of PP5 blocks the catalytic cavity (Yang et al, 2005). This autoinhibited conformation is stabilized by the C-terminal subdomain. Binding of a ligand to the TPR domain is thought to destabilize the TPR–phosphatase domain interaction and to ultimately lead to a complete activation of the phosphatase. For PP5, activation can be achieved by Hsp90, a C-terminal 12 kDa fragment of Hsp90 or an 8-amino-acid C-terminal peptide (Ramsey and Chinkers, 2002; Yang et al, 2005).
Hsp90 is a ubiquitous and abundant molecular chaperone (Young et al, 2001; Picard, 2002; Prodromou and Pearl, 2003; Pratt and Toft, 2003; Wegele et al, 2004; Whitesell and Lindquist, 2005). Under physiological conditions, a broad set of proteins depends on Hsp90 for reaching their native conformations. Many of these Hsp90 substrates are involved in signaling pathways, such as steroid hormone receptors and kinases (reviewed in Pratt and Toft (2003) and Wegele et al (2004)). Substrate activation by the Hsp90 multichaperone machinery is accompanied by the sequential binding and release of Hsp90 partner proteins or cochaperones, respectively (Smith, 1993).
The finding that a phosphatase is present in complexes of Hsp90 with its client proteins (Chen et al, 1996; Silverstein et al, 1997) raised the question of what the function of Ppt1 is. Competition between PP5 and the PPIases FKBP52 or Cyp40 for binding to Hsp90 suggests a role of PP5/Ppt1 in the late steps of the Hsp90 multichaperone cycle (Silverstein et al, 1997). Three different functions seemed plausible: (i) the dephosphorylation of components of the Hsp90 machinery: Many components of the Hsp90 multichaperone cycle are phosphoproteins: p23/Sba1 (Johnson et al, 1994), Cdc37 (Shao et al, 2003), Hop/Sti1 (Lassle et al, 1997; Longshaw et al, 2000) and FKBP52 (Miyata et al, 1997); (ii) the dephosphorylation of Hsp90: Hsp90 itself is phosphorylated at several serin/threonine residues (Garnier et al, 2001); and (iii) the unspecific dephosphorylation of proteins.
Here, we analyzed the function of Ppt1 in the yeast heat-shock protein 90 (yHsp90) chaperone system, both in vitro and in vivo. Our results show that Ppt1 specifically dephosphorylates yHsp90. Furthermore, ppt1Δ yeast cells exhibit an insufficient maturation of the several client proteins of Hsp90 investigated here. This is the first evidence that a phosphatase specifically modulates the chaperoning properties of Hsp90 by modifying its phosphorylation state.
Results
The TPR domain is required for complex formation between Ppt1 and yHsp90
In the first set of experiments, we determined whether Ppt1 interacts with Hsp90 in a similar way as mammalian PP5/Hsp90 (Chen et al, 1996; Russell et al, 1999; Ramsey et al, 2000; Gavin et al, 2002; Yang et al, 2005). Employing isothermal titration calorimetry (ITC) (Figure 1A), we obtained a dissociation constant KD for the complex of ∼670 nM with a binding stoichiometry of 0.82 molecules Ppt1 per yHsp90 monomer. With yHsp90 being a dimer and Ppt1, like PP5 (Das et al, 1998; Yang et al, 2005), being a monomeric enzyme (data not shown), the ITC data show that two Ppt1 molecules bind to a yHsp90 dimer.
Figure 1.
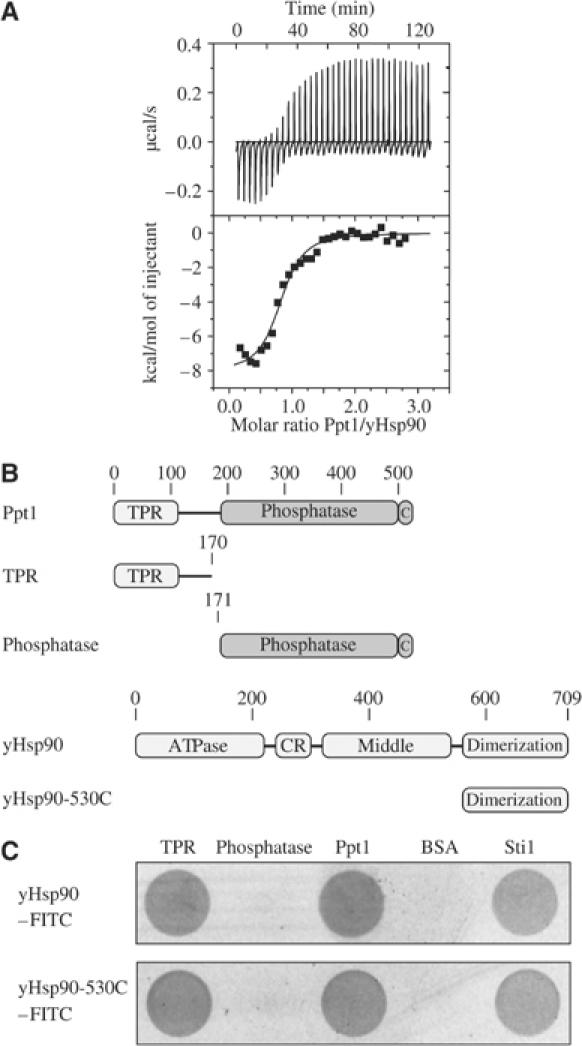
Interaction of Ppt1 and yHsp90. (A) Calorimetric analysis of the Ppt1–yHsp90 interaction. ITC was performed at 25°C. Data were analyzed using the Origin software package provided by the manufacturer. The upper panel shows the change in heat upon injection of Ppt1 (273 μM in the syringe) to yHsp90 (16 μM) in the cell. The lower panel shows the derived binding curve. The filled squares correspond to the integration of the peaks of the upper panel. (B) Schematic representation of the Ppt1 and yHsp90 fragments used. Ppt1 consists of two domains, the TPR (amino acids 12–113), and the catalytic domain (amino acids 188–513). The C-terminal subdomain is denoted as C. The TPR fragment ranges from amino acids 1 to 170, the phosphatase fragment from amino acids 171–513. (C) ELISA-based analysis of the interaction of the full-length Ppt1 and its fragments with the full-length yHsp90 (top panel) and the C-terminal yHsp90 fragment 530C (bottom panel). Proteins denoted in the top line were coated on the ELISA plate as described in Materials and methods. FITC-labeled yHsp90/530C was employed to detect interactions by recording the FITC fluorescence. BSA and Sti1 served as negative and positive control, respectively.
PP5 was found to bind the common TPR acceptor site of Hsp90, the C-terminal amino acids of Hsp90, and competes with other TPR-containing Hsp90 partner proteins (Chen et al, 1996; Silverstein et al, 1997; Ramsey et al, 2000; Yang et al, 2005). For a further analysis of the interaction of Ppt1 and yHsp90, fragments of Ppt1 were generated (Figure 1B) and used in a modified ELISA to determine the association of Ppt1 and Ppt1 fragments with yHsp90 (Figure 1C). Both Ppt1 and the TPR fragment retained FITC-labeled yHsp90. No interaction was observed for the phosphatase fragment, suggesting that the first 170 amino acids of Ppt1 are sufficient for the interaction of Ppt1 and yHsp90. For a truncation fragment of Hsp90, which contains only the C-terminal domain including the TPR acceptor site, the same result was observed as for the full-length yHsp90. This demonstrates that the C-terminal domain of yHsp90 is sufficient for the interaction with Ppt1. A longer fragment of Ppt1 (amino acids 118–513), exclusively lacking the TPR domain, showed no interaction with Hsp90 in a co-immunoprecipitation experiment (data not shown).
Therefore, the binding of Ppt1 and Hsp90 is, like that of PP5, based on the TPR domain binding to the C-terminal end of Hsp90. In a yeast two-hybrid experiment, the interaction of Hsp90 with the TPR fragment of Ppt1 was also confirmed in vivo (data not shown).
Enzymatic properties of autoinhibited Ppt1 and its catalytic domain
The enzymatic properties of Ppt1 were analyzed using the artificial phosphatase substrate p-nitrophenylphosphate (pNPP) (Ramsey and Chinkers, 2002). The pH optimum of Ppt1 was at pH 7.8 (data not shown), which is close to the published pH optimum of PP5 (Ramsey and Chinkers, 2002). The KM value of Ppt1 for pNPP was determined by recording the kinetics at increasing substrate concentrations. A representative example is shown in Figure 2A. The KM value derived from the substrate titration curve was calculated to be ∼27 mM and the turnover number kcat was 7 min−1 (Figure 2B). These values are similar to data obtained for PP5 (Skinner et al, 1997; Kang et al, 2001). The high KM value mirrors the problem that pNPP is an artificial substrate, which does not mimic all aspects of phosphorylated Ser/Thr residues in the context of a protein. Specific protein substrates of Ppt1 are likely to be dephosphorylated with higher affinity.
Figure 2.
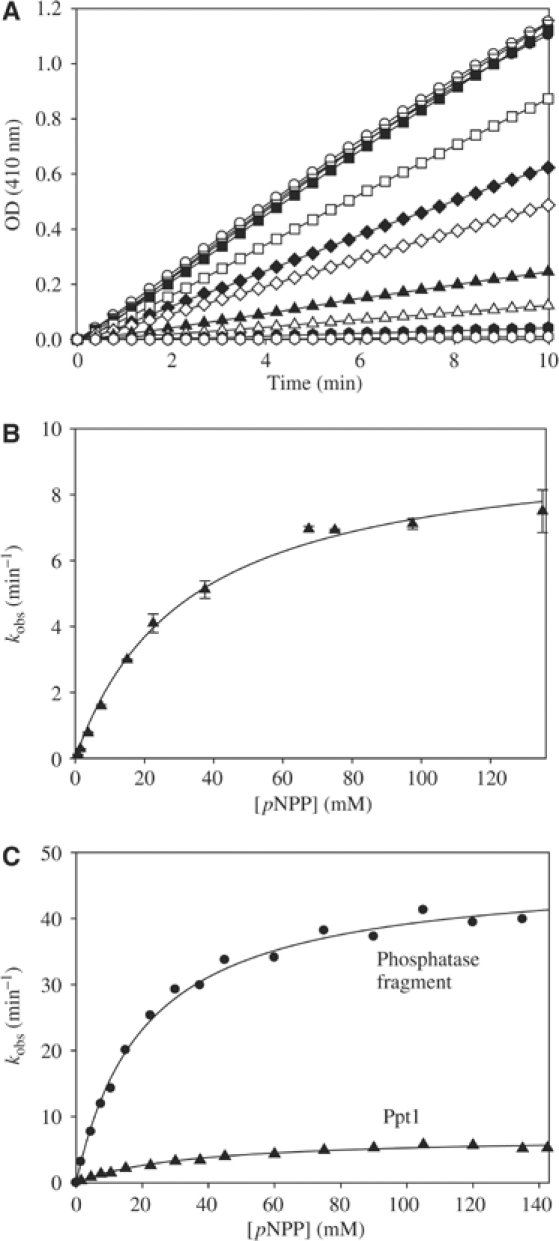
Analysis of the enzymatic properties of Ppt1 for the substrate pNPP. (A) Titration of Ppt1 with increasing concentrations of pNPP. The substrate titration was performed at the optimal pH value of 7.8 as described in Materials and methods. pNPP was added up to 150 mM. The symbols in (A) correspond to increasing concentrations of pNPP: open hexagon: 0.75 mM pNPP; filled hexagon: 1.5 mM pNPP; open triangle up: 3.75 mM pNPP; filled triangle up: 7.5 mM pNPP; open diamond: 15 mM pNPP; filled diamond: 22.5 mM pNPP; open square: 37.5 mM pNPP; filled square 67.5 mM pNPP; open triangle down: 75 mM pNPP; filled triangle down: 97.5 mM pNPP; open circle: 135 mM pNPP; and filled circle: 148 mM pNPP. This kind of substrate titration was used to obtain the reaction kinetics of Ppt1 as shown in (B). (B) Reaction kinetics of Ppt1 (1 μM) with pNPP. Shown is the average of a triple experiment (▴) together with the standard error (bars). The black curve corresponds to the regression of the Michaelis–Menten equation. The formation of pNP was recorded photometrically at 410 nm to obtain the reaction velocity as described in Materials and methods. (C) Comparison of the reaction kinetics of Ppt1 (▴) and the phosphatase fragment (•). Both titrations were performed as in (A). The kcat of the phosphatase fragment (∼47 min−1) is approximately seven-fold higher than that of the full-length Ppt1 (∼7 min−1).
To test whether Ppt1 is regulated by autoinhibition similar to PP5 (Chen and Cohen, 1997; Sinclair et al, 1999; Kang et al, 2001; Swingle et al, 2004), a fragment of Ppt1 lacking the TPR domain (Figure 1B) was analyzed. The kcat of this phosphatase fragment was ∼47 min−1 (Figure 2C), and thus, ∼7-fold higher than that of the full-length Ppt1. This corresponds to the stimulation observed for PP5. Binding of the full-length yHsp90 also resulted in a stimulation (data not shown) similar to that observed for PP5 (Yang et al, 2005).
Ppt1 specifically dephosphorylates Hsp90
The authentic in vivo substrate proteins of Ppt1 were unknown so far. The observed interaction of Ppt1 with yHsp90, however, suggested that components of the Hsp90 chaperone system could be potential Ppt1 substrates, especially as several are phosphoproteins. To assay whether Ppt1 was able to dephosphorylate these protein substrates, Hsp90 and its partner proteins were phosphorylated by casein kinase II (CKII) in the presence of [γ-32P]ATP. The phosphorylated proteins were then used as substrates for Ppt1. Remarkably, for Hsp90, we observed a specific removal of radioactive phosphate in the presence of Ppt1 (Figure 3A). In sharp contrast, addition of Ppt1 to all other radiolabeled proteins of the yHsp90 chaperone cycle did not result in a significant decrease of phosphorylation (Figure 3B).
Figure 3.
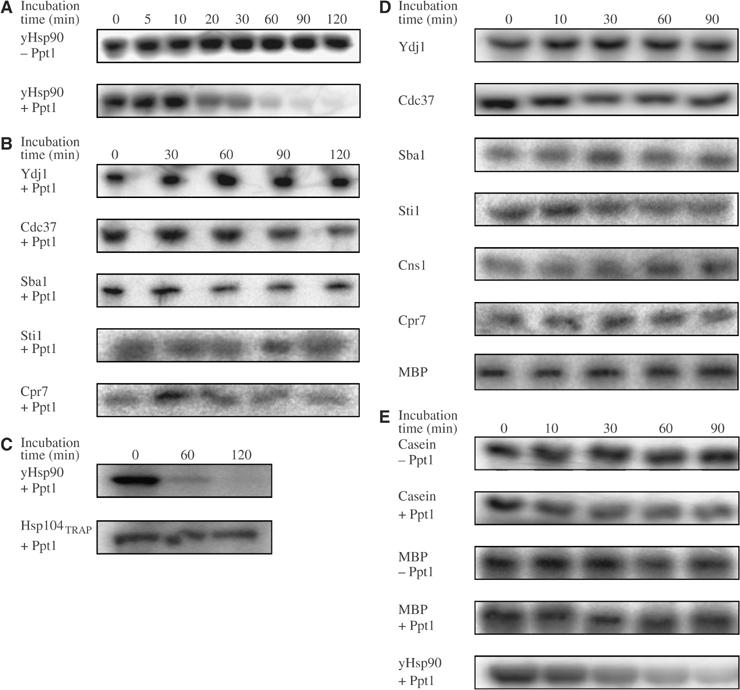
Analysis of the activity of Ppt1 towards phosphorylated proteins of the yHsp90 system. (A) Activity of Ppt1 (1 μM) towards yHsp90 (1 μM). Hsp90 was prepared using CKII and [γ-32P]ATP as described in Materials and methods. After labeling, apyrase was added to hydrolyze the remaining ATP before Ppt1 was added. Ppt1 addition was omitted in the experiment shown in the top panel of (A) to demonstrate the stability of the phosphorylation. (B) Activity of Ppt1 towards partner proteins of yHsp90. Hsp90 partner proteins were prepared as in (A). (C) Activity of Ppt1 towards Hsp104TRAP. The assay was performed as in (A). (D) Activity of Ppt1 towards partner proteins of yHsp90 in the presence of yHsp90. The assay was performed as in (B), except that prior to addition of Ppt1, yHsp90 was added. (E) Activity of Ppt1 towards casein and MBP. Proteins were labeled as described in (A). Ppt1 addition was omitted in the experiments shown in the top panels (denoted as −Ppt1). As a comparison, the yHsp90 dephosphorylation is shown in the lowest panel.
As Hsp90 is known to stimulate the phosphatase activity of PP5/Ppt1 (Ramsey and Chinkers, 2002; Yang et al, 2005), dephosphorylation of the Hsp90 cochaperones was further analyzed in the simultaneous presence of both Ppt1 and yHsp90. However, also under these conditions, Ppt1 was not able to dephosphorylate Hsp90 partner proteins (Figure 3D).
In addition, we tested the effect of Ppt1 on the phosphorylation state of Hsp104 and Ssa1, two chaperone proteins with C-terminal ends homologous to that of Hsp90. For Hsp104, we employed a mutant (Hsp104TRAP) lacking ATPase activity (Bosl et al, 2005). The Hsp70 protein Ssa1 could not be phosphorylated in vitro. Therefore, we used Ssa1 isolated from S. cerevisiae and phosphorylation specific stains. For both proteins, the addition of Ppt1 did not result in changes of the phosphorylation signal (Figure 3C; data not shown).
Finally, two standard substrate proteins for phosphatases, casein and myelin basic protein (MBP), that had previously been demonstrated to be dephosphorylated by Ppt1 were analyzed as targets for Ppt1 (Figure 3E). Consistent with published data (Jeong et al, 2003), we see a slight influence of Ppt1 on the phosphorylation state of casein and MBP. In contrast, dephosphorylation of Hsp90 performed in parallel was complete within 90 min. Again, dephosphorylation was also not observed in the presence of both Ppt1 and Hsp90 (Figure 3D and data not shown).
Taken together, these experiments demonstrate that, surprisingly, the phosphatase activity of Ppt1 is specific for Hsp90. The presence of Hsp90 does not render Ppt1 capable of dephosphorylating other proteins.
The TPR interaction enables Ppt1 to dephosphorylate Hsp90
In a next set of experiments, we wished to analyze whether the TPR interaction between Ppt1 and Hsp90 is required for the observed dephosphorylation of the chaperone. Therefore, we investigated a deletion mutant of yHsp90 lacking the C-terminal TPR-binding motif (ΔMEEVD-Hsp90), which, as in the mammalian system (Ramsey et al, 2000), is incapable of binding Ppt1 or its TPR fragment (data not shown). Upon addition of Ppt1, no change in phosphorylation could be observed for ΔMEEVD-Hsp90 (Figure 4A). Under the same conditions, wild-type (wt) yHsp90 was almost completely dephosphorylated. Furthermore, Ppt1 was found to compete for binding with other TPR proteins for the MEEVD site of yHsp90. The presence of excess Sti1, a yHsp90 partner protein known to bind to the MEEVD residues of yHsp90 (Carrello et al, 1999), prevented dephosphorylation by Ppt1 (Figure 4B). An equimolar mixture of Sti1 and Ppt1 led to a reduction of the dephosphorylation by Ppt1 (data not shown). Similar results were obtained when an isolated TPR fragment of Ppt1 was added to the dephosphorylation reaction (Figure 4C). With increasing concentrations of the isolated TPR domain, the dephosphorylation of yHsp90 decreases.
Figure 4.
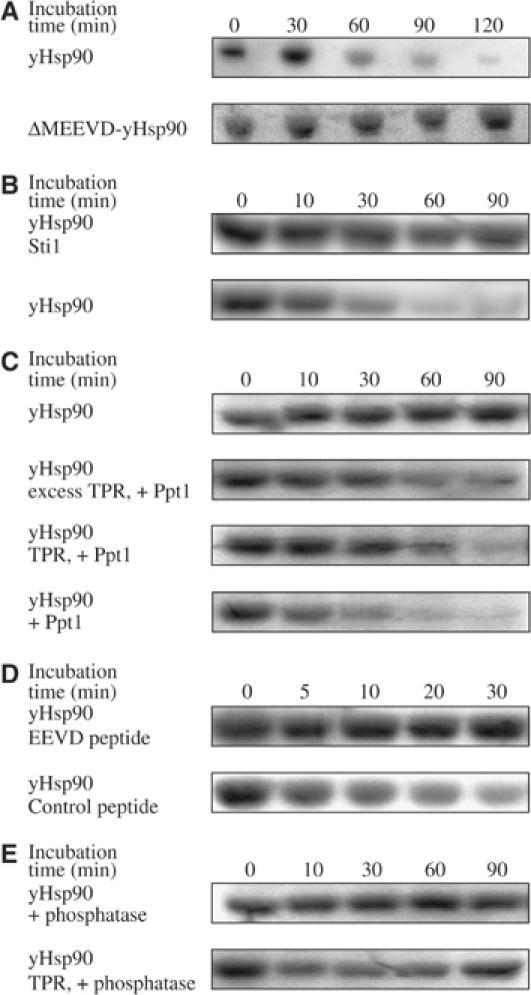
Investigation of the dependence of the TPR interaction in the yHsp90 dephosphorylation. (A) Activity of Ppt1 (1 μM) towards phosphorylated ΔMEEVD-yHsp90 (1 μM), a deletion mutant of the chaperone, lacking the C-terminal MEEVD residues necessary for binding of TPR domains. Phosphorylated proteins are always listed at the first position in the legend. (B) Competition of Ppt1 and Sti1 in binding to the common TPR acceptor site of yHsp90. The assay was performed as in (A). Prior to the addition of Ppt1 to phosphorylated yHsp90, Sti1 (10-fold excess) was added as indicated. (C) Inhibition of the phosphatase activity of Ppt1 in the presence of the TPR fragment of Ppt1 in different stoichiometries. In the experiment of the second panel, the TPR fragment was used in five-fold excess over Ppt1, while it was employed equimolar in the third panel. (D) Inhibition of the yHsp90 dephosphorylation by Ppt1 in the presence of an EEVD-containing peptide. Labeling of yHsp90 was performed as described in (A). Prior to addition of Ppt1, peptides were added in excess over yHsp90. Peptide sequences were PPAPEAEGPTVEEVD (EEVD peptide) and EVGLKRVVTKAMSSR (control peptide). (E) Analysis of the yHsp90 dephosphorylation by the phosphatase fragment of Ppt1 in the absence and presence of the TPR fragment.
Assays performed in the presence of a TPR-binding peptide showed that the peptide did not stimulate the dephosphorylation of yHsp90, but rather inhibited it (Figure 4D). This is best explained by saturation of the TPR domain of Ppt1 with the peptide and blocking of its interaction with yHsp90.
Finally, the activity of the isolated phosphatase domain towards yHsp90 was analyzed (Figure 4E). Although the phosphatase domain itself exhibits a ∼7-fold higher activity in the pNPP assay, it did not dephosphorylate yHsp90. Furthermore, the activity of the phosphatase domain was not restored when the isolated TPR fragment was added to the dephosphorylation reaction. These results are consistent with a model in which Ppt1 is only activated when bound to its substrate protein, Hsp90.
Hsp90 from ppt1Δ yeast cells shows difference in phosphorylation
In the next set of experiments, we investigated the relevance of Ppt1 for the Hsp90 system in vivo. A yeast strain with a genomic deletion of the ppt1 gene was used and compared to the otherwise genetically identical wt strain. The absence of Ppt1 was verified by immunoblotting (Figure 5A). Deletion of the ppt1 gene exhibits no obvious phenotype (Chen et al, 1994). In agreement with this observation, the ppt1Δ yeast cells show no morphological differences when compared to wt cells by scanning electron microscopy (Figure 5A). For investigation of the overall phosphorylation state of Hsp90, the chaperone was purified from both yeast strains, and subsequently phosphorylated with radioactive phosphate to uncover free phosphorylation sites. If Ppt1 is capable of dephosphorylating Hsp90 in vivo, Ppt1-sensitive phosphorylation sites of the chaperone should be occupied in yHsp90 derived from the ppt1Δ strain. Indeed, Hsp90 from the ppt1Δ strain contained only 60% of the radioactive phosphate detected for the protein from the wt strain (Figure 5B), strongly suggesting that Ppt1 dephosphorylates Hsp90 in vivo.
Figure 5.
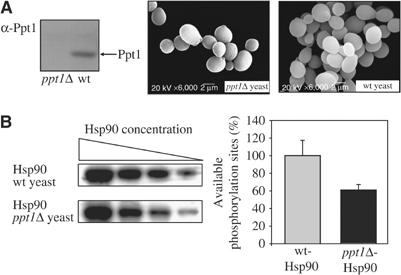
Analysis of the in vivo effect of ppt1Δ. (A) Immunoblot analysis for Ppt1 expression. Lysate of the ppt1Δ yeast strain (left) and the wt yeast (right) was investigated using a polyclonal rabbit serum directed against purified Ppt1 (α-Ppt1). The arrow indicates the position of Ppt1. The scanning electron microscopic images compare ppt1Δ yeast cells and wt yeast cells. (B) Radioactive phosphorylation of identical amounts of yHsp90 derived from a ppt1Δ yeast strain and the corresponding wt strain. Identical amounts of both proteins were treated with CKII and [γ-32P]ATP to reveal the availability of phosphorylation sites. After the phosphorylation reaction was allowed to take place for 2 h, yHsp90 was separated from free ATP by SDS–PAGE. Aliquots of different protein quantities were analyzed and the relative amount of radioactivity was calculated. The graph shows the mean value corrected for the respective protein concentration. Three independent experiments gave similar results.
Ppt1 positively modulates the Hsp90 chaperone machinery in vivo
Hsp90 hyperphosphorylation in a ppt1 knockout strain raised the question whether the phosphorylation state will affect the activity of yHsp90 in vivo. To investigate this, we employed several well-established in vivo assays using substrates of the Hsp90 system, for example, the glucocorticoid hormone receptor (GR) (Smith, 1993) and viral Src kinase (v-Src kinase) (Brugge et al, 1981). In yeast, as in mammals, GR and v-Src depend on the Hsp90 chaperone system to reach their active state (Xu and Lindquist, 1993; Nathan and Lindquist, 1995; Nathan et al, 1997). These proteins can therefore be used as reporters for the activity of the yeast Hsp90 system. To assay this quantitatively, we transformed ppt1Δ and wt yeast with a plasmid harboring a constitutively expressed GR gene and β-galactosidase under the control of a GR-regulated promotor (Nathan and Lindquist, 1995). Thus, the amount of β-galactosidase activity obtained after induction with the hormone derivative deoxycorticosterone (DOC) corresponds quantitatively to the functionality of the yHsp90 system. In the ppt1Δ yeast, GR activity was reduced to ∼55% of the wt activity, implying that the yHsp90 chaperone cycle requires Ppt1 for efficient maturation of GR in yeast (Figure 6A). To directly investigate whether the decrease of GR activity in ppt1Δ yeast is caused by the absence of the phosphatase activity of Ppt1, we reconstituted the expression of Ppt1 with an active site point mutant of Ppt1 (H311A). The active site of members of the PPP family of phosphatases is highly conserved. Among the residues involved in catalysis, histidine 311 is thought to function as a general acid (Swingle et al, 2004). Mutation of the corresponding histidine in various Ser/Thr phosphatases resulted in a complete loss of phosphatase activity (Zhuo et al, 1994; Shibasaki and McKeon, 1995; Zhang et al, 1996; Mondragon et al, 1997; Swingle et al, 2004). The levels of Ppt1 (H311A) expressed in ppt1Δ yeast cells were similar to Ppt1 in wt yeast (see also Figure 6C). However, the H311A mutant did not restore the GR activity to the level observed in wt yeast (Figure 6A).
Figure 6.
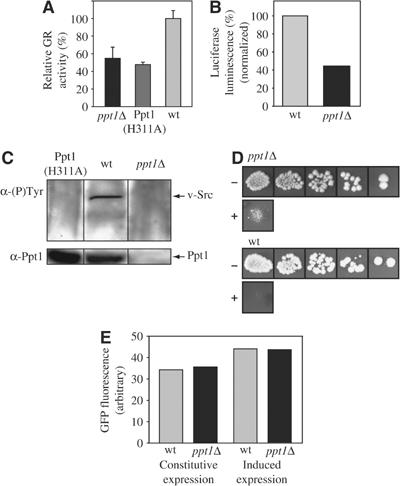
In vivo activation of yHsp90 client proteins. (A) In vivo activation of GR in ppt1Δ and wt yeast. The indicated yeast strains harboring a plasmid with the GR reporter β-galactosidase construct were grown in their logarithmic growth phase and identical cell numbers were induced with the GR ligand DOC (20 μM) for 90 min. Subsequently, cells were harvested and lysed. The β-galactosidase activity in the lysate, which corresponds to the GR activity, was determined with a chemoluminescence assay system as described in Materials and methods. The data shown represent the mean of five independent samples. The GR activity in the ppt1Δ yeast is reduced to ∼55% of the wt level and to ∼50% in ppt1Δ yeast cells expressing Ppt1 (H311A). (B) Analysis of luciferase activity in ppt1Δ and wt yeast cells. Identical numbers of yeast cells expressing firefly luciferase for 3 h were lysed and the lysate was supplemented with luciferin in a 96-well plate. Chemoluminescence was recorded using a Tecan Genios plate reader and normalized for the total protein concentration as determined by a Bradford assay (Bradford, 1976). Data shown are representative of multiple independent experiments. (C) Immunoblot analysis of v-Src activation in ppt1Δ, Ppt1 (H311A) and wt yeast. (Upper box) Immunoblot for autophosphorylated v-Src with a monoclonal antibody specific for phosphotyrosine residues (α-(P)-Tyr). The gel was loaded with identical quantities of cell lysate as indicated. The arrow points to the position of activated v-Src. (Lower box) Immunoblot of the lysates as shown above using a polyclonal serum directed against Ppt1. Data shown are representative of multiple independent experiments. (D) Growth behavior of ppt1Δ and wt yeast cells in response to the α-mating factor. Shown is a dilution series of identical numbers of yeast cells in the absence (−) and presence (+) of 5 μM α-factor in the media. (E) Analysis of eGFP production in ppt1Δ and wt yeast cells. Lysate of identical amounts of logarithmically grown yeast cells expressing eGFP constitutively or inducibly were prepared as described in Materials and methods. Induction of eGFP production was performed for 3 h. After eGFP oxidation, the fluorescence of the indicated cell lysates was recorded and normalized for the total protein concentration as determined by a Bradford assay (Bradford, 1976). Data shown are representative of multiple independent experiments.
To test whether the effect of Ppt1 is specific for GR maturation, firefly luciferase was employed as a further reporter of the yHsp90 system. Luciferase maturation in vivo depends on Hsp90 (Schneider et al, 1996; Nathan et al, 1997), and is easily detectable by the luciferase-catalyzed chemoluminescence of luciferin. Again, deletion of Ppt1 results in a marked decrease of luciferase activity to ∼43% of the activity in wt yeast cells (Figure 6B). As a third reporter, we utilized v-Src kinase. When the yHsp90 system is functional, v-Src passes through the chaperone cycle and reaches its native conformation, which can be monitored by the detection of the autophosphorylation of v-Src with an antibody specific for phosphotyrosine residues in an immunoblot experiment (Nathan and Lindquist, 1995). Maturation of this kinase depends highly on Hsp90 (Brugge et al, 1981; Xu and Lindquist, 1993; Nathan et al, 1997). Most interestingly, in this study, autophosphorylation of v-Src was only observed in wt yeast and not in the ppt1Δ yeast strain (Figure 6C). Again, expression of the active site mutant of Ppt1 did not restore the maturation of v-Src to levels found in wt yeast. Thus, in the absence of Ppt1, when Hsp90 is hyperphosphorylated, the yHsp90 system is not functional for this substrate and the protein does not reach its native conformation.
Finally, we analyzed an endogenous yeast-specific Hsp90 client protein involved in pheromone signaling. Pheromone signaling occurs in yeast as part of the mating behavior. Upon binding of the α-mating factor, a signal-transduction cascade is initiated by the kinase Ste11, leading to a cell cycle arrest (Rhodes et al, 1990). Yeast cells with mutations in Hsp90 fail to respond to α factor, which renders them capable of growth in the presence of the pheromone because Ste11 is unable to reach its signal-transducing conformation (Louvion et al, 1998). Wt yeast cells respond to the addition of pheromone with a tight cell cycle arrest (Figure 6D). In contrast, in ppt1Δ yeast cells, this growth arrest was alleviated.
To test whether protein synthesis and folding was generally impaired in the ppt1Δ yeast cells, maturation of enhanced green fluorescent protein (eGFP), a small and rapidly folding protein that does not seem to depend on the Hsp90 system (Coxon and Bestor, 1995), was studied. In contrast to the proteins described above, GFP activity was identical in ppt1Δ yeast cells and in wt cells (Figure 6E).
Discussion
The physiological role of many Hsp90 cochaperones is not yet understood well. Some of the cofactors, like Sti1 (Prodromou et al, 1999; Richter et al, 2003) and Hch1/Aha1 (Panaretou et al, 2002), are regulators of the ATPase activity of Hsp90 (Obermann et al, 1998; Panaretou et al, 1998), others (PPIases, p23/Sba1, Cdc37) share the ability to interact with unfolded proteins (Bose et al, 1996; Freeman et al, 1996; Kimura et al, 1997). The identification of the phosphatase PP5 as a component of the Hsp90 chaperone system raised the question of a functional contribution of PP5 to the Hsp90 system. The TPR interaction of PP5 resembles that of some large PPIases, enzymes that catalyze isomerization of peptidyl prolyl cis/trans bonds (Fischer et al, 1998). However, Ppt1 is devoid of PPIase activity in standard assays, and it also does not exhibit chaperone activity in in vitro assays (data not shown), suggesting that the function of Ppt1 must be restricted to its phosphatase activity.
The unique feature of the PP5/Ppt1 family of phosphatases is the regulation of the phosphatase activity by the TPR domain (Sinclair et al, 1999; Ramsey and Chinkers, 2002; Yang et al, 2005). Compared to most other cofactors associating with Hsp90 via a TPR domain (Mayr et al, 2000), the affinity between Ppt1 and Hsp90 is at least 10-fold lower. This result can be explained by the autoinhibitory mechanism of this family of phosphatases, in which the TPR domain was found to be partially occupied by the C-terminal end of PP5 (Yang et al, 2005). In consequence, the isolated TPR domains of PP5 and Ppt1 bind to Hsp90 with higher affinity (Prodromou et al, 1999; Yang et al, 2005; and data not shown).
Here, we demonstrate for the first time that Ppt1 specifically dephosphorylates Hsp90, while it does not exhibit significant phosphatase activity towards Hsp90 partner proteins or any other protein tested. Further characterization of the Ppt1/Hsp90 interactions showed that disruption of the TPR interaction (between the Ppt1 TPR domain and the C-terminal residues of Hsp90) abolishes dephosphorylation of Hsp90. Although ligand binding to the TPR domain of PP5/Ppt1 stimulates its phosphatase activity (Skinner et al, 1997; Sinclair et al, 1999; Ramsey and Chinkers, 2002; Yang et al, 2005), this activation does not enable Ppt1 to dephosphorylate proteins in trans. A direct link of Ppt1 to its substrate protein, Hsp90, seems to be required.
In line with our in vitro results on the substrate specificity of Ppt1, we found that Ppt1 mediates dephosphorylation of Hsp90 in vivo. Hsp90 was observed to be hyperphosphorylated in ppt1Δ yeast cells. Previous in vivo studies using phosphatase inhibitors gave contradictory results concerning effects of phosphatases on Hsp90 client protein maturation. For the heme-regulated eIF2α kinase, inhibition of PP5 led to a more active substrate (Shao et al, 2002). In contrast, for v-Src kinase, phosphatase inhibition reduced the activity of this particular Hsp90 client (Mimnaugh et al, 1995). Our experiments using four different Hsp90 client proteins gave a consistent picture, demonstrating unequivocally that the maturation and activation of each client protein tested was handicapped in a Ppt1 deletion strain. Each of the three heterologous client proteins analyzed was affected in a distinct manner, whereas GFP used as an Hsp90-independent control protein was not impaired. While the amount of active GR and luciferase folding was reduced to about one-half compared to the Ppt1 wt strain, maturation of v-Src kinase was completely blocked in ppt1Δ yeast cells. We also investigated the effect of the Ppt1 deletion on an authentic yeast client protein of yHsp90, the kinase Ste11, which is involved in the α-mating factor pheromone signaling cascade. Deletion of Ppt1 prevented the Ste11-dependent complete cell cycle arrest observable in ppt1 wt strains. It is also important to note that expression of an active site mutant of Ppt1 failed to restore client protein maturation, thus clearly demonstrating that the phosphatase activity of Ppt1 is specifically causative for the altered activation of Hsp90 client proteins.
Together, our results strongly imply that, in vivo, the efficacy of the Hsp90 chaperone machinery is significantly decreased in the absence of Ppt1 activity. Therefore, an additional layer of regulation has to be added to our current model of the Hsp90 chaperone system. It will be important to investigate the molecular details of the Hsp90 phosphoregulation in order to explain how Hsp90 phosphorylation determines the fate of different Hsp90 client proteins and how the phosphorylation state of Hsp90 is controlled. Since Ppt1 competes with other TPR domain proteins for binding to Hsp90, one could imagine that Hsp90 phosphorylation is governed by the ratio between TPR proteins, such as Sti1, and Ppt1 in the cells. Uncovering the molecular mechanisms underlying the Hsp90 phosphoregulation will be no easy task. The analysis is complicated by the sheer multitude of potential phosphorylation sites of Hsp90 (yHsp90 contains 45 serine and 40 threonine residues), and the possibility that several phosphorylation isoforms of Hsp90 might coexist in yeast cells.
Materials and methods
Materials
Kanamycin, pNPP, p-nitrophenolate (pNP), apyrase, casein and MBP were from Sigma (St Louis, MO), CKII from Roche Diagnostics (Mannheim, Germany), chloramphenicol from Roth (Karlsruhe, Germany), IPTG from ICN Biochemicals (USA), [γ-32P]ATP from Hartmann Analytics (Darmstadt, Germany), α-mating factor from Bachem (Weil am Rhein, Germany) and all other chemicals from Merck (Darmstadt, Germany). Peptides were synthesized by Professor S Modrow (University of Regensburg, Germany). Rabbit α-Ppt1 antiserum was produced with purified protein (Pineda, Berlin, Germany) and the antibody 4G10 (phosphotyrosine specific) was from Upstate (Hamburg, Germany).
Yeast and bacterial strains, culture media
ppt1Δ− and wt yeast strains were from Euroscarf (Frankfurt, Germany; ppt1Δ: YGR123c::kanMX4: Accession number Y14753; wt: Accession number Y10000). Corresponding Mat-a strains were used for the α-mating factor assay. Yeast cells were transformed and grown as described elsewhere (Ito et al, 1983; Nathan and Lindquist, 1995). Escherichia coli strain BL21 (DE3) Codon+ (Stratagene, La Jolla, CA) was used for recombinant protein expression as described elsewhere (Hainzl et al, 2004).
Proteins and constructs
Ppt1. The gene ppt1 was introduced for expression in pET28b (Novagen, Madison, WI). Fragments of Ppt1: pET28b-Ppt1 was used as template to amplify the TPR fragment (bp 1–510) and the phosphatase fragment (bp 511–1542) by PCR. Point mutant Ppt1 (H311A): Ppt1 was subcloned in pRS316CUP. Mutagenesis of Ppt1 was performed using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Each coding region was sequenced to confirm the absence of mutations.
Purification of Ppt1. Ppt1 was purified by Ni-NTA, ResourceQ anion exchange and Superdex 75 pg size-exclusion chromatography (Amersham Biosciences, Freiburg, Germany). Ppt1 was dialyzed against 40 mM Tris–Cl, pH 7.0, 150 mM KCl, 5 mM glycine, 2 mM EDTA, 3.5 mM DTT and stored at −80°C.
TPR fragment. The TPR fragment was purified similar to the full-length Ppt1, except for the use of a ResourceS cation exchange column (Amersham Biosciences, Freiburg, Germany).
Phosphatase fragment. The phosphatase fragment was expressed in inclusion bodies. These were solubilized as described elsewhere (Rudolph et al, 1997). Denatured protein (6 M GdmHCl) was applied on an Ni-IDA column (Amersham Biosciences, Freiburg, Germany). For renaturation, the phosphatase fragment was diluted in 700 mM Tris (pH 7.5), 250 mM L-arginine, 5 mM DTT. Afterwards, it was dialyzed against 50 mM HEPES, pH 7.5, 150 mM KCl and 5 mM DTT.
yHsp90, Ssa1 and Sti1 were purified as described elsewhere (Buchner et al, 1998; Prodromou et al, 1999; Panaretou et al, 2002; Wegele et al, 2003).
Hsp90 from ppt1D and the respective wt yeast strain. The cleared lysate was applied on a 5 ml Ni2+-loaded IDA-IMAC column (Amersham Biosciences, Freiburg, Germany). After extensive washing, the protein eluted without visible contaminants.
Spectroscopic phosphatase assay
The phosphatase activity of Ppt1 was assayed by monitoring hydrolysis of pNPP similar to that described by Ramsey and Chinkers (2002) at 410 nm in 300 mM Tris–Cl, pH 7.8, 150 mM KCl, 25 mM MgCl2, 5 mM glycine and 1 mM DTT. Reaction velocities at various c(pNPP) were used for the determination of the Michaelis–Menten parameters (Sigmaplot, SPSS Software, Chicago, IL).
Radioactive phosphatase assay
Substrate proteins were labeled with [γ-32P]ATP using 0.1 mU CKII at 37°C for 2 h. The remaining [γ-32P]ATP was hydrolyzed by treatment with apyrase for 45 min at 30°C. Dephosphorylation with Ppt1 was carried out at 30°C for up to 120 min. For competition experiments, competitors of Ppt1 were incubated with yHsp90 prior to the addition of Ppt1. Dephosphorylation was stopped by the immediate addition of Laemmli buffer (Laemmli, 1970) and by boiling at 95°C for 5 min. Proteins were separated from free radioactive phosphate on SDS–PAGE. The degree of protein phosphorylation was determined by phosphoimaging on a Typhoon 9200 Phosphoimager (Amersham Biosciences, Freiburg, Germany).
Determination of differences in free phosphorylation sites between Hsp90 derived from ppt1Δ and wt yeast cells: The Hsp90 batches were phosphorylated with identical aliquots of a CKII and [γ-32P]ATP-containing solution for 2 h. Equal quantities of Hsp90 were used. Protein concentrations were monitored by Coomassie-stained SDS–PAGE. This experiment was repeated multiple times to assure reproducibility. Nonradioactive analysis of protein phosphorylation was performed by phosphoprotein staining using the ProQ Diamond phosphostain (Invitrogen, Karlsruhe, Germany).
Isothermal titration calorimetry
The binding constant and stoichiometry of the Ppt1–yHsp90 complex were determined by ITC as described elsewhere (Richter et al, 2003). The buffer was 40 mM Tris–Cl (pH 7.0), 150 mM KCl, 5 mM glycine, 2 mM EDTA and 3.5 mM DTT.
Interaction of Ppt1 and fragments with yHsp90
To analyze the binding of Ppt1 and its fragments, we adopted an ELISA-based interaction assay (Muller et al, 2004). In brief, ELISA plates (Greiner-BioOne, Kremesmuenster, Austria) were coated with Ppt1, the TPR- or the phosphatase fragment (5 μM). After blocking and washing, 5 μM FITC-labeled yHsp90 or the C-terminal yHsp90 fragment 530C were added (provided by Lin Mueller) for 90 min (4°C). After a further washing step, FITC-labeled protein was visualized with a Typhoon 9200 Imager.
Assay for GR, v-Src and luciferase activity in ppt1Δ yeast cells
GR and the v-Src assays were performed as described elsewhere (Nathan and Lindquist, 1995). Luciferase activity was determined as described elsewhere (Nathan et al, 1997).
Assay for α-mating factor induced growth arrest
Growth arrest by α-mating factor was induced as described elsewhere (Abbas-Terki et al, 2000). For analysis of the effect of the α-mating factor upon Ste11-mediated cell cycle arrest, 106 cells/ml of the ppt1Δ or wt yeast (both Mat-a) were diluted stepwise and spotted on YPD plates with and without supplementation of 5 μM α-factor and grown at 30°C.
Expression and analysis of eGFP in yeast cells
Yeast cells expressing eGFP were lysed in the presence of a protease inhibitor. To allow complete oxidation of the eGFP chromophore, the cleared lysate was agitated at 4°C over night. After centrifugation, fluorescence spectra were recorded (excitation at 395 nm, emission 430–560 nm) and normalized for the total protein concentration as determined by a Bradford assay (Bradford, 1976).
Scanning electron microscopy
Scanning electron microscopy was performed as described elsewhere (Haslbeck et al, 2004).
Acknowledgments
This work was supported by grants of the Deutsche Forschungsgemeinschaft (SFB 594) and the Fonds der Chemischen Industrie (FCI) to JB. SKW was supported by a fellowship of the Fonds der chemischen Industrie. Cns1, Cpr7 and ΔMEEVD-yHsp90 were a kind gift of Otmar Hainzl. Sba1 was provided by Martin Haslbeck and Hsp104TRAP by Benjamin Bösl. Yeast plasmids pCH3Src (v-Src), pHCH/GRGZ (GR) and pRS316CUP were kindly provided by Dr Susan Lindquist and plasmids for eGFP by Martin Haslbeck. We thank Lin Muller for aid with computer graphics, Bettina Richter for electron microscopy, Klaus Richter for ITC and acknowledge the practical assistance of Eva-Katharina Stäblein and Martin Strehle. We also thank Heike Eberl, Otmar Hainzl and Marco Rezlaff for carefully reading the manuscript.
References
- Abbas-Terki T, Donze O, Picard D (2000) The molecular chaperone Cdc37 is required for Ste11 function and pheromone-induced cell cycle arrest. FEBS Lett 467: 111–116 [DOI] [PubMed] [Google Scholar]
- Andreeva AV, Kutuzov MA (1999) RdgC/PP5-related phosphatases: novel components in signal transduction. Cell Signal 11: 555–562 [DOI] [PubMed] [Google Scholar]
- Becker W, Kentrup H, Klumpp S, Schultz JE, Joost HG (1994) Molecular cloning of a protein serine/threonine phosphatase containing a putative regulatory tetratricopeptide repeat domain. J Biol Chem 269: 22586–22592 [PubMed] [Google Scholar]
- Bose S, Weikl T, Bugl H, Buchner J (1996) Chaperone function of Hsp90-associated proteins. Science 274: 1715–1717 [DOI] [PubMed] [Google Scholar]
- Bosl B, Grimminger V, Walter S (2005) Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J Biol Chem 280: 38170–38176 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brugge JS, Erikson E, Erikson RL (1981) The specific interaction of the Rous sarcoma virus transforming protein, pp60src, with two cellular proteins. Cell 25: 363–372 [DOI] [PubMed] [Google Scholar]
- Buchner J, Bose S, Mayr C, Jakob U (1998) Purification and characterization of prokaryotic and eukaryotic Hsp90. Methods Enzymol 290: 409–418 [DOI] [PubMed] [Google Scholar]
- Carrello A, Ingley E, Minchin RF, Tsai S, Ratajczak T (1999) The common tetratricopeptide repeat acceptor site for steroid receptor-associated immunophilins and hop is located in the dimerization domain of Hsp90. J Biol Chem 274: 2682–2689 [DOI] [PubMed] [Google Scholar]
- Chen MS, Silverstein AM, Pratt WB, Chinkers M (1996) The tetratricopeptide repeat domain of protein phosphatase 5 mediates binding to glucocorticoid receptor heterocomplexes and acts as a dominant negative mutant. J Biol Chem 271: 32315–32320 [DOI] [PubMed] [Google Scholar]
- Chen MX, Cohen PT (1997) Activation of protein phosphatase 5 by limited proteolysis or the binding of polyunsaturated fatty acids to the TPR domain. FEBS Lett 400: 136–140 [DOI] [PubMed] [Google Scholar]
- Chen MX, McPartlin AE, Brown L, Chen YH, Barker HM, Cohen PT (1994) A novel human protein serine/threonine phosphatase, which possesses four tetratricopeptide repeat motifs and localizes to the nucleus. EMBO J 13: 4278–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinkers M (1994) Targeting of a distinctive protein-serine phosphatase to the protein kinase-like domain of the atrial natriuretic peptide receptor. Proc Natl Acad Sci USA 91: 11075–11079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinkers M (2001) Protein phosphatase 5 in signal transduction. Trends Endocrinol Metab 12: 28–32 [DOI] [PubMed] [Google Scholar]
- Cohen PT (1997) Novel protein serine/threonine phosphatases: variety is the spice of life. Trends Biochem Sci 22: 245–251 [DOI] [PubMed] [Google Scholar]
- Coxon A, Bestor TH (1995) Proteins that glow in green and blue. Chem Biol 2: 119–121 [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D (1998) The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein–protein interactions. EMBO J 17: 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, Tradler T, Zarnt T (1998) The mode of action of peptidyl prolyl cis/trans isomerases in vivo: binding versus catalysis. FEBS Lett 426: 17–20 [DOI] [PubMed] [Google Scholar]
- Freeman BC, Toft DO, Morimoto RI (1996) Molecular chaperone machines: chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science 274: 1718–1720 [DOI] [PubMed] [Google Scholar]
- Garnier C, Lafitte D, Jorgensen TJ, Jensen ON, Briand C, Peyrot V (2001) Phosphorylation and oligomerization states of native pig brain HSP90 studied by mass spectrometry. Eur J Biochem 268: 2402–2407 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Hainzl O, Wegele H, Richter K, Buchner J (2004) Cns1 is an activator of the Ssa1 ATPase activity. J Biol Chem 279: 23267–23273 [DOI] [PubMed] [Google Scholar]
- Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, Buchner J (2004) Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J 23: 638–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honore B, Leffers H, Madsen P, Rasmussen HH, Vandekerckhove J, Celis JE (1992) Molecular cloning and expression of a transformation-sensitive human protein containing the TPR motif and sharing identity to the stress-inducible yeast protein STI1. J Biol Chem 267: 8485–8491 [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JY, Johns J, Sinclair C, Park JM, Rossie S (2003) Characterization of Saccharomyces cerevisiae protein Ser/Thr phosphatase T1 and comparison to its mammalian homolog PP5. BMC Cell Biol 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JL, Beito TG, Krco CJ, Toft DO (1994) Characterization of a novel 23-kilodalton protein of unactive progesterone receptor complexes. Mol Cell Biol 14: 1956–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Sayner SL, Gross KL, Russell LC, Chinkers M (2001) Identification of amino acids in the tetratricopeptide repeat and C-terminal domains of protein phosphatase 5 involved in autoinhibition and lipid activation. Biochemistry 40: 10485–10490 [DOI] [PubMed] [Google Scholar]
- Kimura Y, Rutherford SL, Miyata Y, Yahara I, Freeman BC, Yue L, Morimoto RI, Lindquist S (1997) Cdc37 is a molecular chaperone with specific functions in signal transduction. Genes Dev 11: 1775–1785 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lassle M, Blatch GL, Kundra V, Takatori T, Zetter BR (1997) Stress-inducible, murine protein mSTI1. Characterization of binding domains for heat shock proteins and in vitro phosphorylation by different kinases. J Biol Chem 272: 1876–1884 [DOI] [PubMed] [Google Scholar]
- Longshaw VM, Dirr HW, Blatch GL, Lassle M (2000) The in vitro phosphorylation of the co-chaperone mSTI1 by cell cycle kinases substantiates a predicted casein kinase II-p34cdc2-NLS (CcN) motif. Biol Chem 381: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Louvion JF, Abbas-Terki T, Picard D (1998) Hsp90 is required for pheromone signaling in yeast. Mol Biol Cell 9: 3071–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Richter K, Lilie H, Buchner J (2000) Cpr6 and Cpr7, two closely related Hsp90-associated immunophilins from Saccharomyces cerevisiae, differ in their functional properties. J Biol Chem 275: 34140–34146 [DOI] [PubMed] [Google Scholar]
- Mimnaugh EG, Worland PJ, Whitesell L, Neckers LM (1995) Possible role for serine/threonine phosphorylation in the regulation of the heteroprotein complex between the hsp90 stress protein and the pp60v-src tyrosine kinase. J Biol Chem 270: 28654–28659 [DOI] [PubMed] [Google Scholar]
- Miyata Y, Chambraud B, Radanyi C, Leclerc J, Lebeau MC, Renoir JM, Shirai R, Catelli MG, Yahara I, Baulieu EE (1997) Phosphorylation of the immunosuppressant FK506-binding protein FKBP52 by casein kinase II: regulation of HSP90-binding activity of FKBP52. Proc Natl Acad Sci USA 94: 14500–14505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon A, Griffith EC, Sun L, Xiong F, Armstrong C, Liu JO (1997) Overexpression and purification of human calcineurin alpha from Escherichia coli and assessment of catalytic functions of residues surrounding the binuclear metal center. Biochemistry 36: 4934–4942 [DOI] [PubMed] [Google Scholar]
- Muller L, Schaupp A, Walerych D, Wegele H, Buchner J (2004) Hsp90 regulates the activity of wild type p53 under physiological and elevated temperatures. J Biol Chem 279: 48846–48854 [DOI] [PubMed] [Google Scholar]
- Nathan DF, Lindquist S (1995) Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol Cell Biol 15: 3917–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan DF, Vos MH, Lindquist S (1997) In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci USA 94: 12949–12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU (1998) In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol 143: 901–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens-Grillo JK, Czar MJ, Hutchison KA, Hoffmann K, Perdew GH, Pratt WB (1996) A model of protein targeting mediated by immunophilins and other proteins that bind to hsp90 via tetratricopeptide repeat domains. J Biol Chem 271: 13468–13475 [DOI] [PubMed] [Google Scholar]
- Panaretou B, Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH (1998) ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J 17: 4829–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, Millson SH, Clarke PA, Naaby-Hansen S, Stein R, Cramer R, Mollapour M, Workman P, Piper PW, Pearl LH, Prodromou C (2002) Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell 10: 1307–1318 [DOI] [PubMed] [Google Scholar]
- Picard D (2002) Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci 59: 1640–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Toft DO (2003) Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 228: 111–133 [DOI] [PubMed] [Google Scholar]
- Prodromou C, Pearl LH (2003) Structure and functional relationships of Hsp90. Curr Cancer Drug Targets 3: 301–323 [DOI] [PubMed] [Google Scholar]
- Prodromou C, Siligardi G, O'Brien R, Woolfson DN, Regan L, Panaretou B, Ladbury JE, Piper PW, Pearl LH (1999) Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J 18: 754–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radanyi C, Chambraud B, Baulieu EE (1994) The ability of the immunophilin FKBP59-HBI to interact with the 90-kDa heat shock protein is encoded by its tetratricopeptide repeat domain. Proc Natl Acad Sci USA 91: 11197–11201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey AJ, Chinkers M (2002) Identification of potential physiological activators of protein phosphatase 5. Biochemistry 41: 5625–5632 [DOI] [PubMed] [Google Scholar]
- Ramsey AJ, Russell LC, Whitt SR, Chinkers M (2000) Overlapping sites of tetratricopeptide repeat protein binding and chaperone activity in heat shock protein 90. J Biol Chem 275: 17857–17862 [DOI] [PubMed] [Google Scholar]
- Rhodes N, Connell L, Errede B (1990) STE11 is a protein kinase required for cell-type-specific transcription and signal transduction in yeast. Genes Dev 4: 1862–1874 [DOI] [PubMed] [Google Scholar]
- Richter K, Muschler P, Hainzl O, Reinstein J, Buchner J (2003) Sti1 is a non-competitive Inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the ATPase cycle. J Biol Chem 278: 10328–10333 [DOI] [PubMed] [Google Scholar]
- Rudolph R, Boehm G, Lilie H, Jaenicke R (1997) Folding proteins. In Protein Function, A Practical Approach, Creighton TE (ed) pp 57–99. Oxford, New York, Tokyo: IRL Press [Google Scholar]
- Russell LC, Whitt SR, Chen MS, Chinkers M (1999) Identification of conserved residues required for the binding of a tetratricopeptide repeat domain to heat shock protein 90. J Biol Chem 274: 20060–20063 [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I (2000) Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell 101: 199–210 [DOI] [PubMed] [Google Scholar]
- Schneider C, Sepp-Lorenzino L, Nimmesgern E, Ouerfelli O, Danishefsky S, Rosen N, Hartl FU (1996) Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc Natl Acad Sci USA 93: 14536–14541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Hartson SD, Matts RL (2002) Evidence that protein phosphatase 5 functions to negatively modulate the maturation of the Hsp90-dependent heme-regulated eIF2alpha kinase. Biochemistry 41: 6770–6779 [DOI] [PubMed] [Google Scholar]
- Shao J, Prince T, Hartson SD, Matts RL (2003) Phosphorylation of serine 13 is required for the proper function of the Hsp90 co-chaperone, Cdc37. J Biol Chem 278: 38117–38120 [DOI] [PubMed] [Google Scholar]
- Shibasaki F, McKeon F (1995) Calcineurin functions in Ca(2+)-activated cell death in mammalian cells. J Cell Biol 131: 735–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein AM, Galigniana MD, Chen MS, Owens-Grillo JK, Chinkers M, Pratt WB (1997) Protein phosphatase 5 is a major component of glucocorticoid receptor.hsp90 complexes with properties of an FK506-binding immunophilin. J Biol Chem 272: 16224–16230 [DOI] [PubMed] [Google Scholar]
- Sinclair C, Borchers C, Parker C, Tomer K, Charbonneau H, Rossie S (1999) The tetratricopeptide repeat domain and a C-terminal region control the activity of Ser/Thr protein phosphatase 5. J Biol Chem 274: 23666–23672 [DOI] [PubMed] [Google Scholar]
- Skinner J, Sinclair C, Romeo C, Armstrong D, Charbonneau H, Rossie S (1997) Purification of a fatty acid-stimulated protein-serine/threonine phosphatase from bovine brain and its identification as a homolog of protein phosphatase 5. J Biol Chem 272: 22464–22471 [DOI] [PubMed] [Google Scholar]
- Smith DF (1993) Dynamics of heat shock protein 90-progesterone receptor binding and the disactivation loop model for steroid receptor complexes. Mol Endocrinol 7: 1418–1429 [DOI] [PubMed] [Google Scholar]
- Smith DF, Sullivan WP, Marion TN, Zaitsu K, Madden B, McCormick DJ, Toft DO (1993) Identification of a 60-kilodalton stress-related protein, p60, which interacts with hsp90 and hsp70. Mol Cell Biol 13: 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingle MR, Honkanen RE, Ciszak EM (2004) Structural basis for the catalytic activity of human serine/threonine protein phosphatase-5. J Biol Chem 279: 33992–33999 [DOI] [PubMed] [Google Scholar]
- Wegele H, Haslbeck M, Buchner J (2003) Recombinant expression and purification of Ssa1p (Hsp70) from Saccharomyces cerevisiae using Pichia pastoris. J Chromatogr B 786: 109–115 [DOI] [PubMed] [Google Scholar]
- Wegele H, Muller L, Buchner J (2004) Hsp70 and Hsp90—a relay team for protein folding. Rev Physiol Biochem Pharmacol 151: 1–44 [DOI] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL (2005) HSP90 and the chaperoning of cancer. Nat Rev Cancer 5: 761–772 [DOI] [PubMed] [Google Scholar]
- Xu Y, Lindquist S (1993) Heat-shock protein hsp90 governs the activity of pp60v-src kinase. Proc Natl Acad Sci USA 90: 7074–7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Roe SM, Cliff MJ, Williams MA, Ladbury JE, Cohen PT, Barford D (2005) Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J 24: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Moarefi I, Hartl FU (2001) Hsp90: a specialized but essential protein-folding tool. J Cell Biol 154: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhang Z, Brew K, Lee EY (1996) Mutational analysis of the catalytic subunit of muscle protein phosphatase-1. Biochemistry 35: 6276–6282 [DOI] [PubMed] [Google Scholar]
- Zhuo S, Clemens JC, Stone RL, Dixon JE (1994) Mutational analysis of a Ser/Thr phosphatase. Identification of residues important in phosphoesterase substrate binding and catalysis. J Biol Chem 269: 26234–26238 [PubMed] [Google Scholar]


