Abstract
The passage from proliferation to terminal differentiation is critical for normal development and is often perturbed in malignancies. To define the molecular mechanisms that govern this process during erythropoiesis, we have used tagging/proteomics approaches and characterized protein complexes nucleated by TAL-1/SCL, a basic helix–loop–helix transcription factor that specifies the erythrocytic lineage. In addition to known TAL-1 partners, GATA-1, E2A, HEB, LMO2 and Ldb1, we identify the ETO2 repressor as a novel component recruited to TAL-1 complexes through interaction with E2A/HEB. Ectopic expression and siRNA knockdown experiments in hematopoietic progenitor cells show that ETO2 actively represses erythroid TAL-1 target genes and governs the expansion of erythroid progenitors. At the onset of erythroid differentiation, a change in the stoichiometry of ETO2 within the TAL-1 complex activates the expression of known erythroid-specific TAL-1 target genes and of Gfi-1b and p21Cip, encoding two essential regulators of erythroid cell proliferation. These results suggest that the dynamics of ETO2 recruitment within nuclear complexes couple cell proliferation to cell differentiation and determine the onset of terminal erythroid maturation.
Keywords: differentiation, erythropoiesis, ETO2, hematopoiesis, proliferation, TAL-1
Introduction
The variety of mature hematopoietic cells that typically express a set of highly specialized proteins are generated from a hematopoietic stem cell (HSC) that undergoes lineage specification and differentiation. This process is coordinated at the molecular level by evolutionary conserved transcription factors, as typified by the importance of GATA factors and their ortholog srp in blood cell development in vertebrates and in Drosophila, respectively (Cantor and Orkin, 2002). These sequence-specific DNA-binding factors typically act as transcriptional activators or can modulate gene expression through long-range interaction. One such example is the regulation of erythroid-specific gene expression by SCL/TAL-1, a member of the helix–loop–helix family of transcription factors, that either directly activates target gene expression (Xu et al, 2003; Lahlil et al, 2004) or is involved in opening the α-globin locus via binding to far upstream regulatory sequences without driving transcription (Anguita et al, 2004). In this context, definitive transcriptional activation requires extended chromatin remodeling and could only be attained in mature hematopoietic cells upon additional binding of stage-specific transactivators and/or alleviation of an active repression that maintains these genes inactive prior to differentiation while keeping the locus transcriptionally competent (Smale, 2003).
The composition of transcription factor complexes bound to lineage-specific genes undergoes dynamic change as HSCs differentiate (Bottardi et al, 2003; Anguita et al, 2004). In multipotent progenitors, the mouse α-globin gene cluster is first activated by protein complexes containing GATA-2, NF-E2 and TAL-1 at two DNAse I-hypersensitive sites (Anguita et al, 2004). As erythroid differentiation proceeds, additional DNAse I-hypersensitive sites are observed in which GATA-1 takes over GATA-2 to nucleate protein complexes that again contain NF-E2 and TAL-1. Interactions between these regulatory sequences and the α-globin genes promoters result in an erythroid-specific domain of histone hyperacetylation where active transcription can occur. The stoichiometry of the transcription factor complexes is also of critical importance for lineage-specific gene expression. For example, members of the basic helix–loop–helix (bHLH) family form homo- and heterodimers and the dosage of these proteins (E2A, HEB and E2-2) as well of TAL-1 has been shown to shape biological outcome in lymphoid lineages (Lecuyer and Hoang, 2004).
The production of erythrocytes is the largest quantitative output of the hematopoietic system with estimated production rates of 2 × 1011 erythrocytes per day. Yet, the frequency of committed granulocyte–macrophage progenitors exceeds by three-fold that of committed erythroid progenitors in the bone marrow. This imbalance is compensated by a high proliferative index in the late transitional stages of erythroid development (Iscove, 1977; Gregory and Eaves, 1978). The cell cycle time of pro-erythroblasts is estimated to be 6–7 h, and this property under steady-state conditions is unique in the adult and more closely resembles that of embryonic cells. How cell differentiation and activation of specific gene expression programmes are coordinated with cell growth remains to be documented, despite a detailed knowledge of the transcription factor network that governs lineage gene expression (Cantor and Orkin, 2002).
TAL-1 is necessary for the establishment of the hematopoietic system (Shivdasani et al, 1995; Robb et al, 1996; Porcher et al, 1999; Ravet et al, 2004). Conditional deletion of the tal-1 gene in the mouse reveals its critical importance in the formation of the erythroid and megakaryocytic lineages (Hall et al, 2003; Mikkola et al, 2003). At the molecular level, TAL-1 can activate or repress transcription (Begley and Green, 1999), but the factors that are recruited in TAL-1-nucleated complexes to mediate these opposing functions remain largely unknown. TAL-1 has been shown to assemble a multifactorial complex containing E2A, LMO2, Ldb1 and GATA-1 (Wadman et al, 1997) in erythroid-committed cells to activate the expression of terminally expressed erythroid-specific genes (Xu et al, 2003; Lahlil et al, 2004), while in CD34+ hematopoietic progenitors, a GATA-2 variant of this TAL-1 complex activates c-kit transcription (Lecuyer et al, 2002). Furthermore, the recruitment of Ldb1 in TAL-1-dependent transcriptional regulation (Xu et al, 2003) links TAL-1 complexes to the establishment of chromatin domains through Ldb1 protein partners (Torigoi et al, 2000).
To determine how TAL-1 can regulate gene expression as hematopoietic progenitors progress through the erythroid lineage, we have undertaken proteomic approaches to isolate and characterize TAL-1 complexes. We show here that the ETO2 repressor is a bona fide participant of the TAL-1-containing complex that can repress its transcriptional activity. Furthermore, our functional studies indicate that ETO2 dynamic fluctuations within the TAL-1 complex contribute to the precise timing of erythroid-specific gene expression, and that the ETO2 expression level regulates the expansion of erythroid progenitors.
Results
ETO2 is part of a nuclear TAL-1-containing complex in erythroid cells
To identify the proteins that are associated with nuclear TAL-1 in erythroid cells, we purified TAL-1 complexes using in vivo biotinylation (de Boer et al, 2003). The TAL-1–biotin tag was expressed from a lentiviral vector in which the EGFP-BirA fusion protein was translated via an IRES (Supplementary data, Figure 1), thus allowing for the selection of cells expressing the TAL-1–biotin tag at a level similar to the endogenous TAL-1 protein using cell sorting based on EGFP. The TAL-1–biotin tag retained its nuclear localization, specific DNA-binding and trans-activation properties (data not shown). This TAL-1–biotin tag protein was expressed in mouse erythroleukemic (MEL) cells. Nuclear extracts were prepared from noninduced or induced MEL cells under mild conditions and proteins bound to the biotinylated TAL-1 protein were characterized in two different experiments.
In addition to all known TAL-1 partners, novel protein interactions were identified (Figure 1A). Three members of single-stranded DNA-binding proteins (SSDP) were pulled down by the TAL-1 bait, SSDP1, 2 and 3. Interestingly, ssdp1 in Drosophila physically and functionally interacts with Chip, the Ldb1 ortholog, in wing development, suggesting that SSDP1 may also associate with Ldb1 to regulate the function of the TAL-1 complex in hematopoiesis. Two members of the ETO (821) family were also identified, ETO2 and MTGR1. ETO2 is a transcriptional repressor that recruits several members of the HDAC family (Melnick et al, 2000) and, consistent with these interactions, we found HDAC1 and HDAC2 associated with the TAL-1–biotin tag protein in undifferentiated MEL cells only (Figure 1A). As ETO2 could modulate the transcriptional activities of numerous DNA-binding proteins, we further investigated the function(s) mediated by ETO2 in TAL-1 nuclear complex(es).
Figure 1.
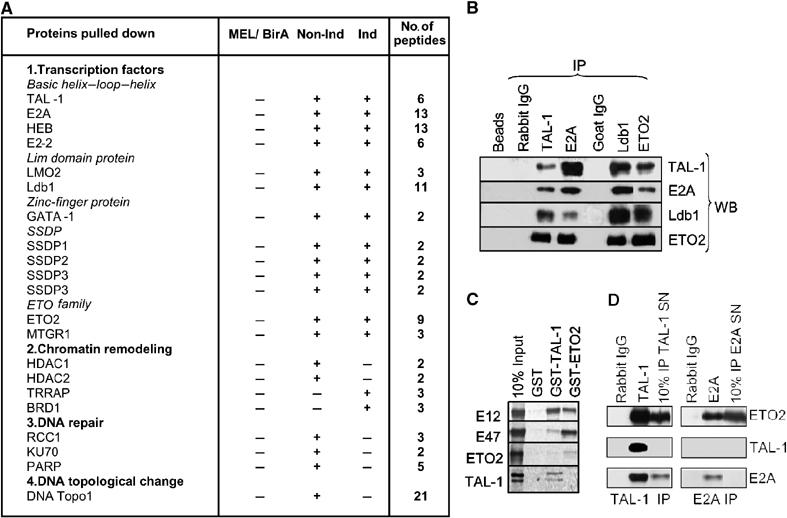
TAL-1/ETO2 interaction in erythroid cells. (A) Proteins identified by LC-MSMS in Bir-TAL-1 pull-down experiments in noninduced (Non-Ind) and induced (Ind) MEL cells. MEL/BirA indicated that none of the proteins identified was obtained when a pull down of MEL cells expressing only BirA was carried out. The number of peptides identified by mass spectrometry is indicated. (B) Co-immunoprecipitation of TAL-1, E2A, Ldb1 and ETO2. Nuclear extracts from MEL cells were immunoprecipitated with the indicated antibodies (IP) and the precipitated proteins were revealed by immunoblotting using antibodies shown (WB). (C) ETO2 interacts in vitro with E12 or E47, but not with TAL-1. GST-pull-down analysis was performed using the indicated GST and 35S-labeled TAL-1, E12, E47 or ETO2. The signal obtained with 10% of the input is shown. (D) ETO2 is associated with TAL-1/E2A and with E2A in distinct complexes. MEL cell nuclear extracts were subjected to sequential immunoprecipitation using first TAL-1 antibodies (TAL-1 IP), and second, E2A antibodies (E2A IP). Preimmune rabbit IgG serve as negative controls. Both the indicated immunoprecipitates and 10% of the respective supernatants were used for Western blot analysis.
ETO2 is associated with E2A in TAL-1-containing nuclear complexes
We studied the specificity of TAL-1 interactions using co-immunoprecipitation experiments. TAL-1-, E2A-, Ldb1- and ETO2-specific antibodies were used to immunoprecipitate MEL nuclear extracts and the immunoprecipitated proteins were analyzed by Western blotting. As shown in Figure 1B, Ldb1, ETO2, E2A and TAL-1 co-immunoprecipitate, suggesting that they could be part of the same nuclear complex. In GST pull-down assays, ETO2 interacts with E2A and HEB, but not TAL-1 (Figure 1C). These results indicate that ETO2 is part of a TAL-1 nuclear complex in erythroid cells through E2A or HEB interaction. Finally, to determine how ETO2 is partitioned in TAL-1- and non-TAL-1-containing complexes, we performed sequential immunoprecipitation of MEL cell nuclear extracts under conditions where both TAL-1 and E2A immunodepletion were complete with their respective antibodies. As illustrated in Figure 1D, part of ETO2 is associated with E2A in both TAL-1-containing complex(es) and TAL-1-independent complexes in erythroid cells.
The ratio of activator to repressor determines the transcriptional output of the TAL-1 complex during erythroid differentiation
To define the composition of the TAL-1 complex bound to DNA, we optimized a DNA-binding assay based on immobilized DNA templates, using sequences of the GPA promoter that were shown to be necessary and sufficient to recruit a TAL-1-containing complex (Lahlil et al, 2004). In this assay, TAL-1, E2A, HEB and ETO2 association with the DNA template was comparable, whereas E2-2 binding was lower and Ldb1 binding was higher (Figure 2A). We also found high GATA-1 binding, consistent with its role in tethering the TAL-1/GATA-1 complex to DNA and with the requirement in the integrity of the two GATA binding sites and the E box for recruitment of the TAL-1 complex to the DNA template (Figure 2A). In summary, our results indicate that the ETO2 is recruited to the GPA promoter by the TAL-1 complex.
Figure 2.
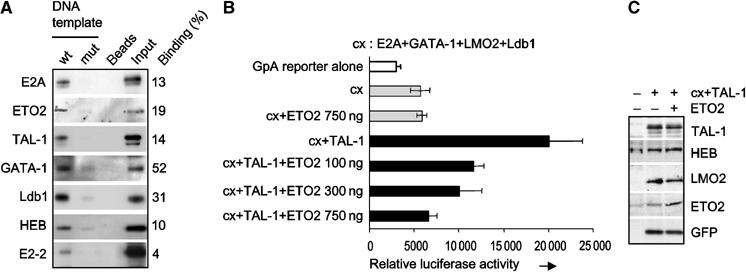
ETO2 and known TAL-1 partners bind the GPA promoter and ETO2 represses the transcriptional activity of TAL-1 on this promoter. (A) Nuclear extracts from TF-1 cells were incubated with the immobilized wild-type GPA promoter (wt), with its E box-GATA mutant version (mut), or with beads alone as a negative control. Bound proteins were eluted and revealed by Western blot. Input represents 30% of each binding assay. (B) A GPA reporter plasmid was transfected with expression plasmids for E2A, GATA-1, Ldb1 and LMO2 alone or with a TAL-1 expression vector in NIH 3T3 cells. ETO2 was cotransfected with the above plasmids at the indicated concentrations. Data are the average of three independent experiments and error bars denote s.d. (C) The expression level of transfected proteins was verified by Western blotting.
ETO2 is known as a transcriptional repressor, while TAL-1 is a positive regulator of erythroid gene expression. Thus, the presence of ETO2 within the TAL-1 complex might indicate an active repression of TAL-1 target genes during the early stages of erythropoiesis. In transient assays using a reporter gene driven by the GPA promoter (Lahlil et al, 2004), there was an eight-fold activation of the GPA promoter by the TAL-1 complex and ETO2 imposed a dose-dependent repression of this activity (Figure 2B) without interfering with the expression levels of the proteins present in the TAL-1 complex (Figure 2C). These data indicate that ETO2 represses the transcriptional activity of the TAL-1 complex bound to the GPA promoter, and that this repression depends on the relative dosage of TAL-1 and its partners over that of ETO2.
To assess the dynamics of TAL-1-associated proteins on DNA during erythroid differentiation, we used TF-1 cells that remain undifferentiated in the presence of GM-CSF. When these cells are switched to an Epo-containing medium, GPA mRNA levels increased on day 1 and subsequently remained high (Figure 3A), coinciding with increased TAL-1 and HEB protein levels, while ETO2 protein levels were not induced on day 1 and showed modest variations thereafter (Figure 3A). We determined the kinetics of binding of TAL-1, ETO2 and HEB to GPA regulatory sequences using our in vitro binding assay on immobilized DNA templates (Figure 3B) and in vivo chromatin immunoprecipitation (ChIP) (Figure 3C). Coinciding with the timing of upregulation of GPA mRNA, the TAL-1/ETO2 ratio was reproducibly increased more than six-fold on day 1 of Epo exposure, and remained constant afterwards (Figure 3B). In contrast, the TAL-1/HEB ratio did not fluctuate significantly within the same time frame (Figure 3B). Together, these data indicate a strong correlation between the ratio of activator to repressor and the expression of the endogenous GPA target gene.
Figure 3.
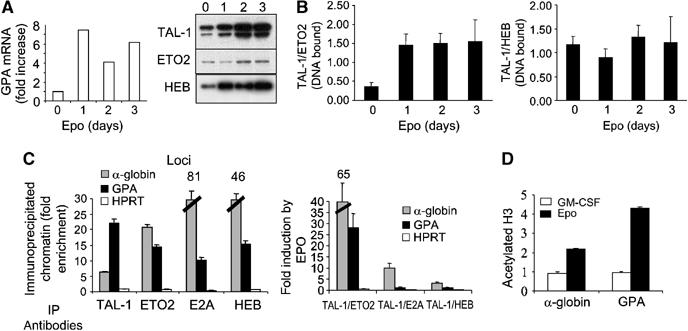
Stoichiometry of ETO2 and TAL-1 binding to the GPA promoter during erythroid differentiation. (A) Kinetics of GPA mRNA levels, assessed by real-time RT–PCR (left panel) and of TAL-1, HEB and ETO2 protein levels (right panel) following TF-1 cell exposure to Epo. (B) The ratio of TAL-1 (activator) over ETO2 (repressor) bound to the immobilized GPA promoter increases with erythroid differentiation in Epo-induced TF-1 cells, while the ratio of TAL-1 over HEB (activator) remains unchanged. (C) TF-1 cell chromatin extracts were subjected to immunoprecipitations with the indicated antibodies (IP) and species-matched control IgG. DNA from immunoprecipitated chromatin was subjected to PCR analysis to detect the presence of the GPA and HPRT promoter sequences, as well as the core sequence of the α-globin HS-40. Fold enrichments were calculated as described in Materials and methods. The left panel shows ChIP results in uninduced cells and the right panel illustrates ratio changes in favor of activator (TAL-1 and E2A, HEB) over-repressor (ETO2) bound to GPA and α-globin HS-40 during Epo stimulation of TF-1 cells. (D) Acetylated histone H3 is induced by Epo at the GPA promoter and the α-globin HS-40. All ChIP data are typical of two independent experiments.
We next quantitated the occupancy of the GPA promoter by TAL-1, ETO2 and the extent of acetylated histone H3 in TF-1 cells cultured with GM-CSF or Epo. We also assessed the HS-40 of the human α-globin gene, since members of the TAL-1 complex were previously shown to occupy the corresponding murine HS-26 sequence in undifferentiated cells, prior to α-globin gene expression (Anguita et al, 2004). In the presence of GM-CSF, GPA promoter and α-globin HS-40 sequences were efficiently brought down by antibodies against TAL-1, ETO2, HEB or E2A (Figure 3C, left panel), but not acetylated H3 (Figure 3D). We then calculated the ratio of DNA-bound proteins when TF-1 cells were switched to Epo-containing medium (Figure 3C, right panel). The ratio of TAL-1 to ETO2 at the GPA promoter was increased twenty-fold, while the TAL-1/HEB or TAL-1/E2A ratio did not vary significantly. We finally studied local histone H3 acetylation as it correlates with the induction of gene expression (Reinke and Horz, 2003), and found that, on erythroid differentiation, H3 acetylation increased at the α-globin HS-40 and, to a greater extent, at the GPA promoter (Figure 3D). These ChIP experiments indicate that TAL-1, HEB, E2A and ETO2 are specifically associated with the GPA promoter and the α-globin locus in cells when these genes are poorly transcribed. The induction of erythroid differentiation with Epo changes the ratio of TAL-1/HEB to ETO2 occupancy of these regulatory sequences in favor of TAL-1/HEB, together with increased histone H3 acetylation as well as increased transcription of the GPA and α-globin genes.
Overexpression of ETO2 represses the expression of endogenous TAL-1 target genes during erythroid differentiation
To address the functional impact of the stoichiometry of ETO2 and TAL-1/HEB or TAL-1/E2A, we first overexpressed ETO2 in three model cell lines that mimic different steps of erythropoiesis and in primary hematopoietic cells. TF-1 and UT-7 cell lines are dependent on Epo for differentiation (TF-1) or proliferation (UT-7) and MEL cells are considered as a cellular model of terminal erythroid differentiation (Figure 4). In these models, we studied the expression of the GPA and the band 4.2 gene as these genes are known to be direct TAL-1 target genes, whereas the functional effect of TAL-1 on α-globin gene expression is not documented.
Figure 4.
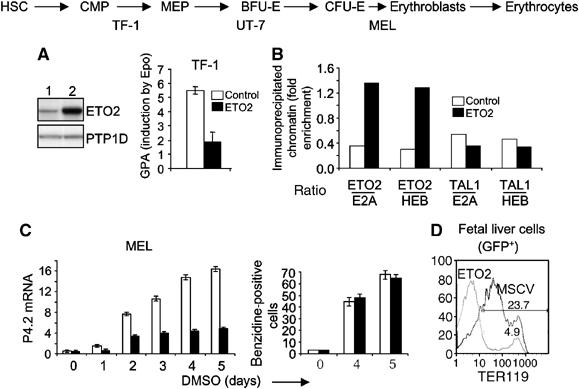
ETO2 overexpression inhibits the expression of TAL-1 target genes. (A) ETO2 overexpression in TF-1 cells. Western blot indicates a 5.4-fold higher ETO2 level compared to the endogenous level. GPA mRNA levels were quantified in TF-1 cells before or 5 days after Epo addition. Open columns, mock-infected TF-1, and black columns, ETO2-overexpressing TF-1. (B) ETO2 overexpression results in an imbalance of ETO2 over E2A or HEB without affecting the ratio of TAL-1 over E2A or HEB at the GPA locus. ChIP results are expressed as in Figure 3C. (C) MEL cells expressing ETO2 or the control GFP vector were induced by DMSO to undergo terminal erythroid differentiation. Band 4.2 mRNA levels in control cells (open columns) and ETO2-expressing cells (black columns) harvested at 24 h intervals were quantified (left panel). In parallel, hemoglobinization was assessed by benzidine staining (right panel). Data shown are the average of three independent experiments and error bars denote s.d. (D) Fetal liver cells were transduced with the empty MSCV-GFP vector (thin line) or an ETO2-expressing vector (thick line). Cells were harvested immediately after infection and analyzed for TER119 fluorescence in the GFP+ fraction (right panel), which was in the range of 20–50% (not shown). Data are typical of three independent experiments.
In TF-1 and UT-7 cells, ETO2 overexpression nearly abrogated the induction of GPA (TF-1) or its expression (UT-7) on Epo exposure (Figure 4A and data not shown). Decreased expression was associated with an imbalance of ETO2 over E2A or HEB in favor of ETO2 at the GPA locus, while the ratio of TAL-1 over E2A or HEB remained unchanged (Figure 4B), as revealed by ChIP. In MEL cells, ETO2 overexpression greatly reduced the induction of the band 4.2 gene by DMSO (Figure 4C). These cells nonetheless underwent terminal maturation and became fully hemoglobinized (Figure 4C, right panel). Finally, we expressed ETO2 in normal fetal liver cells taken at E12.5 and found that the expression of the GPA complex recognized by the TER119 monoclonal antibody was nearly abrogated (Figure 4D). While we cannot exclude the possibility that ETO2 interferes with the differentiation of fetal liver cells, studies in MEL cells suggest that decreased band 4.2 gene expression was due to a direct inhibition of TAL-1 transcriptional activity by ETO2 and not to differentiation blockade.
Inhibition of ETO2 expression accelerates the onset of expression of TAL-1 target genes during terminal erythroid differentiation
Conversely, ETO2 knockdown using siRNA in UT-7 cells enhanced GPA mRNA levels (Figure 5A). In purified mouse primary erythroid cell populations, TAL-1 and ETO2 are coexpressed in pro-erythroblasts (E1). TAL-1 mRNA levels then increase two-fold with differentiation, while ETO2 mRNA levels sharply drop as the cells engage to the next stage (basophilic erythroblast, E2), indicating that ETO2 expression is switched off with terminal maturation in the erythroid lineage (Supplementary data, Figure 2). This decrease in ETO2 expression was also observed during terminal erythroid differentiation of human erythroid progenitors, while TAL-1 expression showed little variation and E2A expression was increased (Figure 5B).
Figure 5.
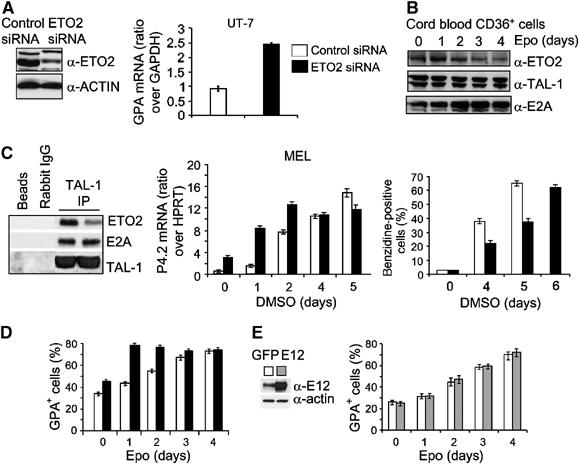
The relative amount of ETO2 in the TAL-1-containing complex determines the timing of expression of TAL-1 target genes. (A) An siRNA directed against human ETO2 decreased ETO2 protein levels in UT-7 cells (left panel, lane 2) as compared to cells expressing a control siRNA (left panel, lane 1), without affecting α-actin levels. GPA mRNA levels were quantified in control cells (right panel, open column) and in cells expressing the ETO2 siRNA (right panel, black column). (B) Kinetics of ETO2, TAL-1 and E2A protein levels during terminal erythroid differentiation of human CD36+CD34− cells as revealed by immunoblotting. (C) The amounts of ETO2 and E2A associated with TAL-1 in control or ETO2-directed siRNA expressing MEL cells were assessed by Western blot analysis after TAL-1 immunoprecipitation (left panel). Preimmune rabbit IgG and protein G beads serve as negative controls. MEL cells expressing a control (open columns) or an ETO2 siRNA (black columns) were induced to undergo terminal erythroid differentiation. Band 4.2 mRNA level was assessed every day (middle panel) and hemoglobinization determined as in Figure 4C (right panel). (D) Human CD34−CD36+ erythroid progenitors expressing an ETO2 (black columns) or a control (open column) siRNA were induced to terminal erythroid differentiation with Epo and analyzed for GPA+ cells. (E) Human CD34−CD36+ erythroid progenitors expressing E12 (gray columns) or GFP (open column) were induced to terminal erythroid differentiation with Epo and analyzed for GPA+ cells. Data shown in (C)–(E) are the average of three independent experiments and error bars denote s.d.
In MEL cells, the ETO2 siRNA decreased the amount of ETO2 within the TAL-1 complex without affecting the levels of E2A bound to TAL-1 (Figure 5C, left panel). Decreased ETO2 level was associated with enhanced band 4.2 expression on day 1, that is, when the transcriptional activity of band 4.2 gene is low in control cells (Figure 5C, middle panel). However, when band 4.2 gene is normally activated from days 2 to 4 of DMSO exposure, coinciding with decreased ETO2 expression (not shown), its expression was not affected by further lowering ETO2 levels. Similarly, in primary human erythroid progenitors, ETO2 siRNA delivery accelerated the kinetics of upregulation of GPA by Epo without further elevating its final levels (Figure 5D). Contrary to the effects of ETO2 siRNA, E12 overexpression in CD36+ cells did not accelerate GPA expression (Figure 5E), suggesting that E12 is not limiting and that E2A is not sequestered by ETO2. Together, these data indicate that the relative amount of ETO2 within the TAL-1 complex determines the outcome in the transcriptional activity of essential TAL-1 target genes and the timing of their expression.
ETO2 regulates the Epo-dependent proliferation of erythroid cells and the expansion of human erythroid progenitors
During erythropoiesis, a high proliferation rate is observed in erythroid precursors (CFU-Es) and early erythroblasts (E1/E2) (Gregory and Eaves, 1978), while proliferation decreases sharply in late erythroblasts (E3) and ceases with final maturation (E4) (Asari et al, 2005). Strikingly, the ETO2 siRNA induced growth arrest in UT-7 cells (Figure 6A, left panel) and decreased the proliferation of differentiating MEL cells (Figure 6, right panel) in the absence of apoptotic features (Supplementary data, Figure 3). Decreased growth in MEL cells resulted in delayed hemoglobinization, which was offset by one day compared to control cells, despite accelerated band 4.2 expression (Figure 5C, middle and right panels). These observations indicate that ETO2 may regulate the proliferation of erythroid cells. We therefore investigated the molecular pathways that link ETO2/TAL-1 to cell proliferation.
Figure 6.
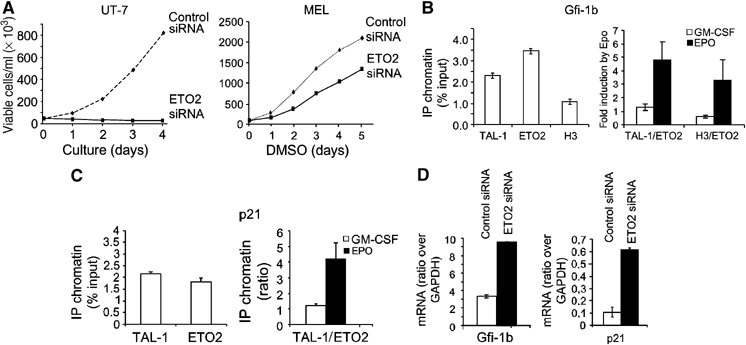
Gfi-1b and p21 are direct targets of the TAL-1/ETO2 complex during erythroid differentiation. (A) ETO2 knockdown with human or mouse ETO2 siRNA (thick line) abrogated the growth of UT-7 cells and decreased the growth of MEL cells compared to cells transduced with a control siRNA (dotted line). (B, C) TF-1 cells were grown in the presence of GM-CSF and chromatin extracts were subjected to immunoprecipitations with the indicated antibodies (IP) and species-matched control IgG. DNA from immunoprecipitated chromatin was subjected to PCR analysis to detect the presence of Gfi-1b (B, left panel) or p21 (C, left panel) promoter sequences. Ratio changes in favor of activator (TAL-1 and acetylated histone H3) over-repressor (ETO2) bound to the Gfi-1b (B, right panel) or p21 (C, right panel) promoters during Epo stimulation of TF-1 cells. All ChIP data are typical of two independent experiments. (D) Increased Gfi-1b and p21 mRNA in UT-7 cells one day after delivery of an ETO2-directed siRNA (black column) or a control siRNA (white column). Data shown are the average of three independent experiments and error bars denote s.d.
Gfi-1b is a zinc-finger repressor that governs the proliferation of erythroid progenitors in vitro (Garcon et al, 2005) and is essential for erythroid lineage cells in vivo (Saleque et al, 2002; Garcon et al, 2005). As Gfi-1b (Saleque et al, 2002) and TAL-1 deficiencies (Hall et al, 2003; Mikkola et al, 2003) similarly disrupt the formation of erythroid cells in vivo and as the Gfi-1b promoter harbors a TAL-1 DNA-binding sequence located at −245 (Huang et al, 2004), we hypothesized that Gfi-1b might be directly regulated by the TAL-1/ETO2-containing complex. ChIP reveals that the Gfi-1b promoter is occupied by TAL-1 and ETO2 and the stoichiometry of the complex changes in favor of TAL-1 after Epo induction (Figure 6B). Thus, the induction of erythroid differentiation with Epo changes the ratio of TAL-1 to ETO2 occupancy of the Gfi-1b promoter in favor of TAL-1, together with increased histone H3 acetylation (Figure 6B) and increased transcription (not shown).
P21, a cyclin-dependent kinase inhibitor and G1 checkpoint regulator, is upregulated in chromatophilic erythroblasts (E3) exactly when cell cycle decreases (data not shown). The p21 promoter contains several E boxes that could potentially recruit the ETO2/TAL-1 complex. As observed with Gfi1b, the p21 promoter is also occupied by TAL-1 and ETO2 (Figure 6C, left panel), and Epo induction changes the balance in favor of TAL-1. Similarly, acetylated histone H3 increases at this locus associated with increased transcription (not shown). In accordance with these results, an ETO2-directed siRNA increased Gfi-1b and p21 mRNA levels (Figure 6D). Together, our observations indicate that ETO2 favors the expansion of erythroid progenitors and suggest a molecular pathway involving Gfi-1b and p21.
Primary CD34+ cells isolated from umbilical cord blood undergo erythroid differentiation in a two-step culture system characterized by a 5-day period of expansion of erythroid progenitors in the absence of Epo, monitored by the expression of the CD34 and CD36 surface markers, followed by a 4-day period of erythroid differentiation in the presence of Epo, monitored by the expression of the CD36 and GPA (Figure 7A; Freyssinier et al, 1999). Similar to UT-7 cells, the delivery of ETO2 siRNA decreased by four-fold the expansion of the CD34−CD36+ population during the first phase and by two-fold during the second phase of culture (Figure 7B and C). This was associated with a 2.5- and eight-fold increased Gfi-1b and p21 mRNA levels during the first phase, respectively, and two-fold increased p21 mRNA levels during the second phase of culture (Figure 7B and C). Despite decreased growth, ETO2 knockdown in primary human erythroid cells did not affect terminal erythroid differentiation (Figure 7D). Together, these data indicate that ETO2 is required for the expansion of erythroid progenitors, but is dispensable for terminal maturation.
Figure 7.
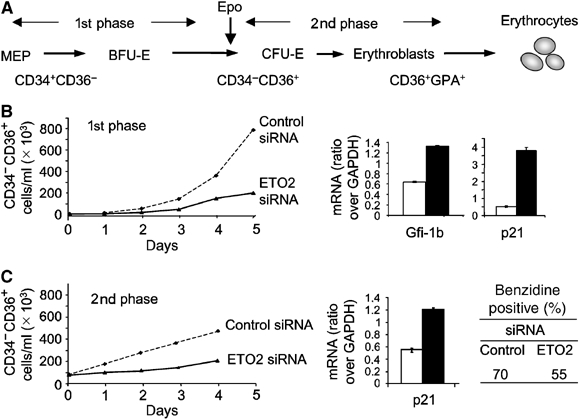
ETO2 regulates the proliferation of erythroid progenitors. (A) Schematical representation of the different stages of erythroid differentiation (MEP, BFU-E, CFU-E and erythroblasts) and of CD34 and CD36 expression within the two-step culture system used. (B) Left panel: Cumulative growth curves of CD34−CD36+ cells during the first phase of erythroid precursor amplification in the presence of an ETO2 (thick line) or a control siRNA (dotted line). Right panel: Gfi-1b and p21 mRNAs levels were quantified on day 3 of culture when the CD4−CD36+ erythroid progenitors represent the main population. (C) Left panel: Cumulative growth curves of CD34−CD36+ cells during the second phase of erythroid precursor amplification in the presence of an ETO2 (thick line) or a control siRNA (dotted line). Right panel: Quantification of p21 mRNA level. Hemoglobinization of CD36+ cells after 4 days of culture with Epo was monitored by benzidine staining. Data shown in (B) and (C) are the average of two independent experiments and error bars denote s.d.
Discussion
In the present study, we show a dual function for ETO2 in controlling erythroid lineage outcome, first by fine tuning the expression of TAL-1 target genes, and second, by controlling the expansion of erythroid progenitors. ETO2 seems to be recruited to TAL-1-containing complexes in undifferentiated CD34+ cells via physical interaction with E2A and inhibits the transcriptional activity of the complexes. Furthermore, the balance of ETO2 versus TAL-1/HEB/E2A in the complex determines the onset and the magnitude of erythroid differentiation in uncommitted progenitors.
ETO2 acts as a repressor of TAL-1 target genes
The ETO protein is a translocation partner of the acute myeloid leukemia (AML)-1 transcription factor in human AML containing the t(8;21) translocation, resulting in the expression of the AML-1-ETO fusion protein. The eto gene family consists of three members, ETO or MTG8, ETO2 or MTG16 (myeloid transforming gene chromosome 16) and MTGR1 (myeloid transforming gene-related protein-1) (Davis et al, 2003). This family is phylogenetically conserved and homologs are found in chicken (cETO) and Drosophila (nervy). The four nervy homology regions (NHR) are protein interaction motifs through which ETO and ETO2 can interact with HDAC family members (Gelmetti et al, 1998; Wang et al, 1998) and NCoR (Amann et al, 2001). Recently, ETO2 was shown to associate with E2A and HEB via its NHR1. This interaction precludes recruitment of p300/CREB-binding protein coactivators by E2A/HEB, thus inactivating their transcriptional properties (Zhang et al, 2004). We show here that ETO2 represses the activity of the TAL-1 complex on erythroid genes and that the ratio of TAL-1 to ETO2 increases during erythroid differentiation, coinciding with the timing of transcription of TAL-1 target genes.
Consistent with the repressor function of ETO2, we found HDAC1 and HDAC2 associated with the TAL-1 complex in undifferentiated cells. On induction of MEL cell differentiation, HDACs association decreased below the threshold of detection, while TRRAP and BRD1 were recruited to the TAL-1 complex. TRAPP is an essential member of several chromatin-modifying complexes with HAT activity (Herceg and Wang, 2005) and BRD1 is a bromodomain protein, a structure that has been implicated as an acetyl–lysine recognition module (Hassan et al, 2002). Thus, the exchange of ETO2-HDAC1/2 for TRRAP-BRD1 within the TAL-1 complex during differentiation potentially results in covalent modifications of histone tails that mark TAL-1 target loci for transcription. In support of this possibility, we found increased histone H3 acetylation at the GPA locus in cells undergoing erythroid differentiation, coinciding with increased GPA gene expression.
The dynamics and stoichiometry of the TAL-1 complex during erythroid differentiation
The TAL-1 transcription factor can activate transcription if and only if all of the appropriate partners are recruited to target DNA sequences. This all-or-nothing behavior that we refer to as coincidence detection allows for a tight control of gene expression (Lecuyer and Hoang, 2004). For example, obligatory TAL-1 partners include E2A or HEB bHLH transcription factors, LMO2, Ldb1 and GATA-1 or GATA-2 (Wadman et al, 1997; Cohen-Kaminsky et al, 1998; Lecuyer et al, 2002; Lahlil et al, 2004). Through unbiased mass spectrometry analysis, we identify here these proteins as components of the core TAL-1 complex, in which TAL-1 acts as a nucleation platform (Lecuyer et al, 2002). In this complex, TAL-1 is limiting since elevating TAL-1 dosage in the cell increases GPA expression (Hoang et al, 1996), while E2A is not, as shown here for E12. Given that E2A is partitioned in TAL-1 and non-TAL-1 complexes, these observations are not surprising. By these criteria, our functional studies suggest that ETO2 is limiting and therefore is not able to sequester E2A from the TAL-1 complex. Consistent with this view, elevating ETO2 levels does not cause an imbalance of TAL-1 over E2A or HEB at the GPA locus. We therefore surmise that ETO2 represses the activity of the TAL-1 complex.
During differentiation, the composition of transcription factor complexes varies through exchange of components (Lecuyer and Hoang, 2004), resulting in differential transcriptional output (Lahlil et al, 2004). Indeed, GATA-1 is recruited in late erythroblasts (MEL), while in erythroid progenitors (TF-1), GATA-2 recruitment is favored (Lahlil et al, 2004). We identify here another dynamic variation of the TAL-1 complex that does not involve partner substitution. We provide both biochemical and functional evidence that the stoichiometry of the ETO2 repressor with regard to the content of the TAL-1, E2A or HEB activators varies during the process of erythroid differentiation and that ETO2 levels determines the precise timing of the expression of a set of erythroid-specific TAL-1 target genes. It will be important to address the mechanisms that govern this stoichiometry.
Our observations also suggest a biphasic imbalance in repressor to activator during differentiation. In undifferentiated cells, the TAL-1 complex associates with erythroid-specific regulatory regions such as the GPA promoter or the HS-26 of the murine α-globin locus. This association might keep the chromatin in an open state, but transcription is repressed by ETO2. Local fluctuations in the association of ETO2 with the TAL-1 complex within single cells could trigger intermittent and low levels of transcription at these loci and account for the low level of lineage gene expression in multipotential progenitors, a process known as lineage priming (Orkin, 2003). At the initiation of erythropoiesis, an elevation in TAL-1 and E2A or HEB levels changes the ratio of TAL-1 to ETO2 in favor of TAL-1, which offsets the inhibitory activity of ETO2 and triggers sustained expression of TAL-1 marked erythroid genes.
Cell differentiation versus cell proliferation: implications in leukemias
Late erythroid progenitors (CFU-E), pro-erythroblasts and basophilic erythroblasts are the most actively cycling cells in the bone marrow (40–50%) (Iscove, 1977; Gregory and Eaves, 1978). Progression from the pro-erythroblast stage to the polychromatophilic erythroblast is associated with a sharp drop in cycling cells and an induction of p21 (Panzenbock et al, 1998). Thus, cell cycle arrest is intimately linked with terminal erythroid cell maturation, suggesting that the two processes need to be coordinately regulated, but the underlying mechanism was not known. In the present study, we provide evidence that the dynamics of the TAL-1/ETO2 complex controls the expression of two essential regulators of erythroid proliferation, Gfi-1b and p21, during terminal erythroid differentiation, as well as the expression of a subset of erythroid genes. Furthermore, ETO2 is required for the expansion of erythroid progenitors, but dispensable for terminal differentiation. Together, our observations are consistent with the view that growth cessation occurs when ETO2 is decreased in erythroid progenitors, concomitant with the expression of TAL-1 target genes, thereby pushing the cells towards terminal differentiation.
We therefore propose that the stoichiometry of ETO2 within the TAL-1 complex controls the transitional process from expansion to terminal differentiation, which may be viewed as an important checkpoint in normal development. Indeed, cell proliferation and differentiation are uncoupled in leukemias. It is possible that variants of the ETO2–TAL-1 complex control this critical transitional process in other lineages. Consistent with this hypothesis, the genes encoding several members of the ETO2–TAL-1 complex are rearranged or mutated in acute leukemias: ETO2 in t(16;21) AML, TAL-1/SCL and LMO2 in T-ALL, E2A in pre-B-ALL and GATA-1 in acute megakaryoblastic leukemias of Down's syndrome.
Materials and methods
Cell culture and erythropoietin induction
NIH 3T3, TF-1, UT-7 and MEL cells were cultured and stimulated to differentiate as described previously (Hoang et al, 1996; Lahlil et al, 2004). Human CD34+ cells were purified and differentiated as described (Freyssinier et al, 1999; Ravet et al, 2004).
Purification of TAL-containing complexes
The tagged full-length TAL-1 construct was obtained by the fusion of the TAL-1 cDNA to the BirA target peptide cDNA. BirA/EGFP fusion cDNA was obtained by the fusion of the two cDNAs. Both constructs were inserted in the TRIP-ΔU3-EF1α lentivirus vector and lentiviruses were produced and used to transduce MEL cells. Nuclear extract preparation, streptavidin binding and mass spectrometry were performed as described (de Boer et al, 2003). All acquired product ion mass spectra were searched against an NCBInr protein database using Mascot software (Matrix Science) and all potential matches were manually inspected.
Immunoprecipitations and immunoblot analysis
Nuclear extracts from the indicated cells were pre-cleared for 30 min at 4°C using Protein G–Sepharose beads in lysis buffer (150 mM KCl, 0.1 mM EDTA, 3 mM MgCl2, 10% glycerol and 0.3% NP-40) containing protease inhibitors. Immunoprecipitations were performed in lysis buffer overnight at 4°C using antibodies against E2A, ETO2 (both from Santa Cruz) or TAL-1. Beads were washed six times with the lysis buffer at 4°C and bound material was eluted by boiling at 95°C in 1 × Laemmli buffer. Immunoblot analysis was performed as described (Leroy-Viard et al, 1995).
Binding of transcription factors on immobilized promoters and transient transactivation assays
The −84 GPA promoter fragments, wild type and the mutant for GATA sites and E box (Lahlil et al, 2004), were biotinylated and subsequently immobilized on streptavidin-coated magnetic beads (Dynabeads M-280). Nuclear extracts from TF-1 cells, with or without Epo treatment, were incubated with immobilized −84 GpA promoters in binding buffer (20 mM Tris (pH 8.0), 10% glycerol, 6 mM MgCl2, 5 mM DTT, 0.1 mM EDTA, 0.01% NP-40, 10 μg/μl polydI–dC) in a final concentration of 100 mM. The binding reactions were incubated with rotation for 60 min at 30°C and washed twice with 0.3 ml of binding buffer. The proteins bound to the immobilized templates were recovered following a 5 min boiling step in SDS–PAGE sample buffer. Proteins were then resolved on SDS–PAGE and transferred to PVDF membranes for Western blot analysis. Band intensities were analyzed with the Multigauge program (Fuji).
Transactivation assays were performed in triplicates essentially as described previously (Lecuyer et al, 2002). Mouse ETO2 complete coding sequence was cloned into pCDNA3.1 vector.
Chromatin immunoprecipitation
ChIP assays were performed essentially as described previously (Lecuyer et al, 2002; Tremblay et al, 2003), using TF-1 cells.
For real-time quantitative PCR analysis of the immunoprecipitated chromatin, we used Sybergreen detection kits (Qiagen) according to the manufacturer's instructions and the Stratagene Mx4000 apparatus (Stratagene). Primer sequences used for specific promoter amplification are available upon request. A region of inactive chromatin was amplified as a negative control (c-kit +13 532) as described previously (Lecuyer et al, 2002). Threshold cycles (Ct) were determined as recommended by the manufacturer's software for a dilution of input chromatin extracts and for each immunoprecipitated chromatin extract. The occupancy of region X by protein p relative to input chromatin was obtained as described: p(X)=2[Ct(IP)−Ct(input)] (Geisberg and Struhl, 2004). Data are shown as fold enrichments over the values obtained for species-matched control Ig and the negative c-kit control region.
Viral vectors and cell transduction
For siRNA production, siDNAs were cloned downstream of the H1 promoter in a lentivirus vector that can also express the EGFP cDNA. For human ETO2 overexpression, the ETO2 coding sequence was cloned into the pTRIPΔU3-EF1α vector plasmid. Lentiviral vector supernatants were produced as described (Amsellem et al, 2002) and used for infections. Retroviruses encoding ETO2, or their MSCV control, were produced by transfection of the 293 VSV-G retroviral packaging cell line (Ory et al, 1996) and used for infections.
Flow cytometry
Fetal liver cells from embryonic day 12.5 were harvested and cultured overnight in the presence of retroviruses expressing either ETO2-GFP or the MSCV-GFP control vector. Cells were stained with PE-Cy7-TER119 antibodies and with 1 μg of propidium iodide to exclude dead cells and analyzed by flow cytometry with the LSR II (Becton-Dickinson). Erythroid precursors were purified as described previously (Socolovsky et al, 2001) using the FACSAria (Becton Dickinson).
RT–PCR
Preparation of cDNA and specific PCR amplifications were performed as previously described (Herblot et al, 2000). Oligonucleotide sequences are available upon request. PCR products were revealed with internal oligonucleotide probes and hybridation signals quantified through exposure to a PhosphorImager screen (Molecular Dynamics, Amersham Biosciences). For real-time quantitative PCR, we used Sybergreen detection kits (Qiagen) according to the manufacturer's instructions and the Stratagene Mx4000 apparatus (Stratagene).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Acknowledgments
We thank Drs Daniele Mathieu and Catherine Porcher for human or mouse TAL-1 antibodies, Dr Jeroen Krijgsveld for mass spectrometry analysis, B Izac for help in the preparation of the lentiviruses, R Lahlil for help with the VSV, MA Vinit, A Haman and A Perreault for technical help and Daniele Gagné for cell sorting. We thank all our colleagues for thoughtful discussions. NG was supported by studentships from the ARC and the French Hematology Society and JAL studentships from the National Science and Engineering Research Council, the Fonds de Recherche en Santé du Québec and the Canadian Institutes for Health Research (CIHR). This work was supported by grants from INSERM, CNRS, ARC, CIHR, the Cancer Research Society Inc. and the National Cancer Institute of Canada.
References
- Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, Downing JR, Meyers S, Hiebert SW (2001) ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol 21: 6470–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsellem S, Ravet E, Fichelson S, Pflumio F, Dubart-Kupperschmitt A (2002) Maximal lentivirus-mediated gene transfer and sustained transgene expression in human hematopoietic primitive cells and their progeny. Mol Ther 6: 673–677 [PubMed] [Google Scholar]
- Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR (2004) Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J 23: 2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asari S, Sakamoto A, Okada S, Ohkubo Y, Arima M, Hatano M, Kuroda Y, Tokuhisa T (2005) Abnormal erythroid differentiation in neonatal bcl-6-deficient mice. Exp Hematol 33: 26–34 [DOI] [PubMed] [Google Scholar]
- Begley CG, Green AR (1999) The SCL gene: from case report to critical hematopoietic regulator. Blood 93: 2760–2770 [PubMed] [Google Scholar]
- Bottardi S, Aumont A, Grosveld F, Milot E (2003) Developmental stage-specific epigenetic control of human beta-globin gene expression is potentiated in hematopoietic progenitor cells prior to their transcriptional activation. Blood 102: 3989–3997 [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH (2002) Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21: 3368–3376 [DOI] [PubMed] [Google Scholar]
- Cohen-Kaminsky S, Maouche-Chretien L, Vitelli L, Vinit MA, Blanchard I, Yamamoto M, Peschle C, Romeo PH (1998) Chromatin immunoselection defines a TAL-1 target gene. EMBO J 17: 5151–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JN, McGhee L, Meyers S (2003) The ETO (MTG8) gene family. Gene 303: 1–10 [DOI] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J (2003) Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci USA 100: 7480–7485 (Epub 2003 June 7411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyssinier JM, Lecoq-Lafon C, Amsellem S, Picard F, Ducrocq R, Mayeux P, Lacombe C, Fichelson S (1999) Purification, amplification and characterization of a population of human erythroid progenitors. Br J Haematol 106: 912–922 [DOI] [PubMed] [Google Scholar]
- Garcon L, Lacout C, Svinartchouk F, Le Couedic JP, Villeval JL, Vainchenker W, Dumenil D (2005) Gfi-1B plays a critical role in terminal differentiation of normal and transformed erythroid progenitor cells. Blood 105: 1448–1455 [DOI] [PubMed] [Google Scholar]
- Geisberg JV, Struhl K (2004) Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res 32: e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA (1998) Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol 18: 7185–7191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory CJ, Eaves AC (1978) Three stages of erythropoietic progenitor cell differentiation distinguished by a number of physical and biologic properties. Blood 51: 527–537 [PubMed] [Google Scholar]
- Hall MA, Curtis DJ, Metcalf D, Elefanty AG, Sourris K, Robb L, Gothert JR, Jane SM, Begley CG (2003) The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis, erythropoiesis, and lineage choice in CFU-S12. Proc Natl Acad Sci USA 100: 992–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL (2002) Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379 [DOI] [PubMed] [Google Scholar]
- Herblot S, Steff AM, Hugo P, Aplan PD, Hoang T (2000) SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat Immunol 1: 138–144 [DOI] [PubMed] [Google Scholar]
- Herceg Z, Wang ZQ (2005) Rendezvous at mitosis: TRRAPed in the chromatin. Cell Cycle 4: 383–387 [DOI] [PubMed] [Google Scholar]
- Hoang T, Paradis E, Brady G, Billia F, Nakahara K, Iscove NN, Kirsch IR (1996) Opposing effects of the basic helix-loop-helix transcription factor SCL on erythroid and monocytic differentiation. Blood 87: 102–111 [PubMed] [Google Scholar]
- Huang DY, Kuo YY, Lai JS, Suzuki Y, Sugano S, Chang ZF (2004) GATA-1 and NF-Y cooperate to mediate erythroid-specific transcription of Gfi-1B gene. Nucleic Acids Res 32: 3935–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iscove NN (1977) The role of erythropoietin in regulation of population size and cell cycling of early and late erythroid precursors in mouse bone marrow. Cell Tissue Kinet 10: 323–334 [DOI] [PubMed] [Google Scholar]
- Lahlil R, Lecuyer E, Herblot S, Hoang T (2004) SCL assembles a multifactorial complex that determines glycophorin A expression. Mol Cell Biol 24: 1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Herblot S, Saint-Denis M, Martin R, Begley CG, Porcher C, Orkin SH, Hoang T (2002) The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood 100: 2430–2440 [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Hoang T (2004) SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp Hematol 32: 11–24 [DOI] [PubMed] [Google Scholar]
- Leroy-Viard K, Vinit MA, Lecointe N, Jouault H, Hibner U, Romeo PH, Mathieu-Mahul D (1995) Loss of TAL-1 protein activity induces premature apoptosis of Jurkat leukemic T cells upon medium depletion. EMBO J 14: 2341–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnick AM, Westendorf JJ, Polinger A, Carlile GW, Arai S, Ball HJ, Lutterbach B, Hiebert SW, Licht JD (2000) The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol Cell Biol 20: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola HK, Klintman J, Yang H, Hock H, Schlaeger TM, Fujiwara Y, Orkin SH (2003) Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421: 547–551 [DOI] [PubMed] [Google Scholar]
- Orkin SH (2003) Priming the hematopoietic pump. Immunity 19: 633–634 [DOI] [PubMed] [Google Scholar]
- Ory DS, Neugeboren BA, Mulligan RC (1996) A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA 93: 11400–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzenbock B, Bartunek P, Mapara MY, Zenke M (1998) Growth and differentiation of human stem cell factor/erythropoietin-dependent erythroid progenitor cells in vitro. Blood 92: 3658–3668 [PubMed] [Google Scholar]
- Porcher C, Liao EC, Fujiwara Y, Zon LI, Orkin SH (1999) Specification of hematopoietic and vascular development by the bHLH transcription factor SCL without direct DNA binding. Development 126: 4603–4615 [DOI] [PubMed] [Google Scholar]
- Ravet E, Reynaud D, Titeux M, Izac B, Fichelson S, Romeo PH, Dubart-Kupperschmitt A, Pflumio F (2004) Characterization of DNA-binding dependent and independent functions of SCL/TAL1 during human erythropoiesis. Blood 8: 8. [DOI] [PubMed] [Google Scholar]
- Reinke H, Horz W (2003) Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell 11: 1599–1607 [DOI] [PubMed] [Google Scholar]
- Robb L, Elwood NJ, Elefanty AG, Kontgen F, Li R, Barnett LD, Begley CG (1996) The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J 15: 4123–4129 [PMC free article] [PubMed] [Google Scholar]
- Saleque S, Cameron S, Orkin SH (2002) The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev 16: 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Mayer EL, Orkin SH (1995) Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373: 432–434 [DOI] [PubMed] [Google Scholar]
- Smale ST (2003) The establishment and maintenance of lymphocyte identity through gene silencing. Nat Immunol 4: 607–615 [DOI] [PubMed] [Google Scholar]
- Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF (2001) Ineffective erythropoiesis in Stat5a(−/−)5b(−/−) mice due to decreased survival of early erythroblasts. Blood 98: 3261–3273 [DOI] [PubMed] [Google Scholar]
- Torigoi E, Bennani-Baiti IM, Rosen C, Gonzalez K, Morcillo P, Ptashne M, Dorsett D (2000) Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc Natl Acad Sci USA 97: 2686–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M, Herblot S, Lecuyer E, Hoang T (2003) Regulation of pT alpha gene expression by a dosage of E2A, HEB, and SCL. J Biol Chem 278: 12680–12687 (Epub 2003 Feb 12683) [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH (1997) The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J 16: 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM (1998) ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA 95: 10860–10865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Huang S, Chang LS, Agulnick AD, Brandt SJ (2003) Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol Cell Biol 23: 7585–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG (2004) E protein silencing by the leukemogenic AML1-ETO fusion protein. Science 305: 1286–1289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3


