Abstract
We demonstrate that successive cleavage events involving regulated intramembrane proteolysis (Rip) occur as a function of time during the Caulobacter cell cycle. The proteolytic substrate PodJL is a polar factor that recruits proteins required for polar organelle biogenesis to the correct cell pole at a defined time in the cell cycle. We have identified a periplasmic protease (PerP) that initiates the proteolytic sequence by truncating PodJL to a form with altered activity (PodJS). Expression of perP is regulated by a signal transduction system that activates cell type-specific transcription programs and conversion of PodJL to PodJS in response to the completion of cytokinesis. PodJS, sequestered to the progeny swarmer cell, is subsequently released from the polar membrane by the membrane metalloprotease MmpA for degradation during the swarmer-to-stalked cell transition. This sequence of proteolytic events contributes to the asymmetric localization of PodJ isoforms to the appropriate cell pole. Thus, temporal activation of the PerP protease and spatial restriction of the polar PodJL substrate cooperatively control the cell cycle-dependent onset of Rip.
Keywords: cell division, differentiation, microarray analysis, pilus, proteolysis
Introduction
Proteolysis is integral to the regulatory networks of diverse biological systems (Gottesman, 2003; Jenal and Hengge-Aronis, 2003; Ehrmann and Clausen, 2004). In response to specific cues, proteases can stimulate, modulate, or eliminate the activities of their respective substrates. In the α-proteobacterium Caulobacter crescentus, proteolysis contributes to the defined fluctuations of many regulatory and structural proteins over the course of the cell cycle (Figure 1A and B) (Grunenfelder et al, 2001; Ryan et al, 2004; Skerker and Laub, 2004). The swarmer cell cycle begins with a chemotactic swarmer cell, which has pili and a single flagellum at one pole of the cell. After a period of motility, the swarmer cell differentiates into a sessile stalked cell, replacing these polar organelles with a stalk containing holdfast at its tip for surface adherence. The stalked cell prepares for cell division by replicating its chromosome and assembling a flagellum and the pili secretion apparatus at the pole opposite to the stalk. Asymmetric division then produces a stalked cell and a swarmer cell; the swarmer cell synthesizes pili and starts the swarmer cell cycle again, whereas the stalked cell initiates its cell cycle by immediately beginning another round of chromosome replication and division. Timely proteolysis is critical to the normal progression of these orchestrated processes. For example, degradation of the essential CtrA response regulator during the swarmer-to-stalked transition and in the stalked compartment of the dividing cell helps ensure that chromosome replication initiates at those points in the spatial–temporal matrix (Domian et al, 1997; Ryan et al, 2004). By varying the levels of target proteins, proteases can serve as regulatory switches for different physiological states.
Figure 1.
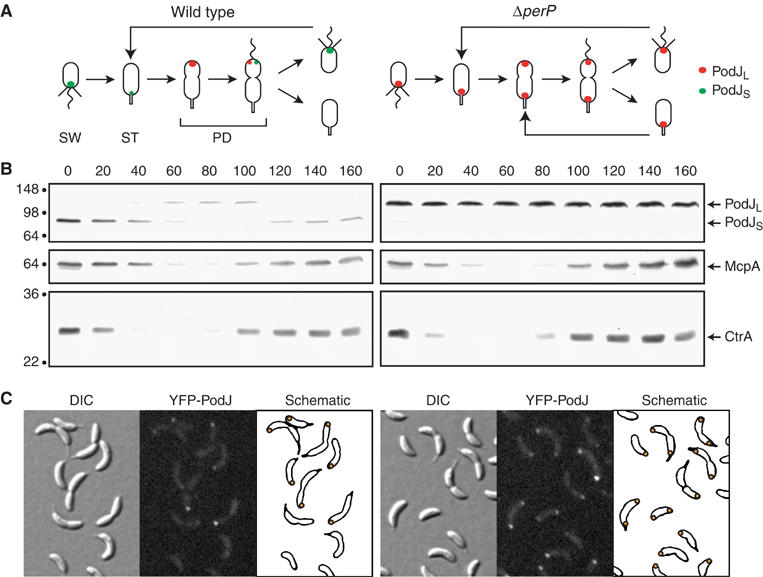
PodJ levels and localization vary over the cell cycle between wild-type (left) and ΔperP (right) strains. (A) Schematic diagrams depict PodJ localization during the Caulobacter cell cycle. Red circle indicates PodJL, whereas green circle indicates PodJS. SW, swarmer cell with polar pili (straight lines) and flagellum (wavy line); ST, stalked cell; PD, predivisional cell. (B) Cell extracts from a synchronous population of cells were analyzed for the presence of PodJL, PodJS, McpA, and CtrA by immunoblots. Samples were taken every 20 min, as indicated above the blots. Timing of the cell cycle corresponds to that depicted in panel A. Molecular mass standards, in kDa, are indicated to the left. (C) Cells with yfp-podJ replacing the endogenous podJ allele were examined by differential interference contrast (DIC) and fluorescence microscopy. Localization of YFP-PodJ in individual cells is represented in the schematic panels with orange circles.
The interplay of proteolysis and other regulatory mechanisms drives the developmental program integral to the Caulobacter cell cycle (Quardokus and Brun, 2003; Holtzendorff et al, 2004). The polarity determinant PodJ contributes a spatial component to this program (Figure 1A and B) (Wang et al, 1993; Viollier et al, 2002a; Hinz et al, 2003; Lawler et al, 2006). The podJ gene encodes a 974-aa protein with a cytoplasmic N-terminal domain, a single transmembrane segment, and a periplasmic C-terminal region. Transcription of podJ is controlled by the master regulators CtrA, GcrA, and DnaA, such that full-length PodJ (PodJL) is synthesized only in the early predivisional cell (Crymes et al, 1999; Holtzendorff et al, 2004; Hottes et al, 2005). PodJL localizes to the incipient swarmer pole (Figure 1A) and recruits factors required for pili biogenesis, including the PleC histidine kinase/phosphatase and components of the pili assembly machinery (Viollier et al, 2002a; Hinz et al, 2003; Lawler et al, 2006). As the cell divides, the periplasmic domain of PodJL is degraded, giving rise to a truncated form (PodJS) that stays attached to the membrane at the flagellated pole of the progeny swarmer cell (Viollier et al, 2002a; Hinz et al, 2003). PodJS is needed for chemotaxis and holdfast formation, presumably because, by analogy to PodJL, it recruits polar factors required for those processes (Viollier et al, 2002a; Lawler et al, 2006). PodJS is cleared from the cell pole during the swarmer-to-stalked transition, before PodJL is synthesized again and localized to the opposite pole (Viollier et al, 2002a; Hinz et al, 2003; Chen et al, 2005). This proteolytic clearance of PodJS depends on the functionally conserved membrane metalloprotease MmpA (Chen et al, 2005). MmpA and its homologs, in both prokaryotes and eukaryotes, participate in regulated intramembrane proteolysis (Rip), which releases membrane-bound substrates from the membrane and has been adapted as a control mechanism in diverse biological systems (Brown et al, 2000; Urban and Freeman, 2002; Weihofen and Martoglio, 2003). In Caulobacter, Rip by MmpA contributes to the asymmetric distribution of PodJ: loss of MmpA inhibits PodJS degradation and leads to bipolar localization of PodJ isoforms (Chen et al, 2005). Thus, the combination of transcriptional regulation, protein localization, and proteolysis restricts the presence of PodJ isoforms to a specific spatial and temporal pattern during the Caulobacter cell cycle.
Changes in PodJ's protein levels and localization during the cell cycle correlate with its functions in polar organelle development. PodJ is required to localize the PleC histidine kinase/phosphatase (Viollier et al, 2002a; Hinz et al, 2003), which, together with the DivJ histidine kinase and the DivK response regulator, constitute the basic components of a signal transduction system that monitors cytokinesis (Figure 2A, left) (Sommer and Newton, 1991; Matroule et al, 2004; McGrath et al, 2004). In the predivisional cell, the DivJ kinase is located at the stalked pole, where it phosphorylates the DivK single-domain response regulator; diffusion of phosphorylated DivK to the swarmer pole results in its dephosphorylation by PleC at that pole (Hecht et al, 1995; Wu et al, 1998; Wheeler and Shapiro, 1999; Jacobs et al, 2001; Lam et al, 2003; Matroule et al, 2004). Hence, DivJ and PleC act in opposition, spatially and catalytically, on DivK's phosphorylation state between the two cell poles (Matroule et al, 2004; McGrath et al, 2004). Cytokinesis, resulting in the formation of a diffusion barrier (Judd et al, 2003), traps dephosphorylated DivK in the swarmer compartment, leading to induction of a swarmer-specific developmental program in that compartment (Matroule et al, 2004). If cell compartmentalization is blocked, dephosphorylated DivK fails to accumulate, and the swarmer-specific program does not initiate (Matroule et al, 2004). PodJ's recruitment of PleC to the correct pole is central to the functionality of this DivJ–PleC–DivK signaling system. Null alleles of podJ and pleC cause similar developmental defects: for example, pili synthesis, a component of swarmer progeny development that occurs after cell division, is abolished in both pleC and podJ null mutants (Wang et al, 1993; Viollier et al, 2002a, 2002b; Hinz et al, 2003).
Figure 2.
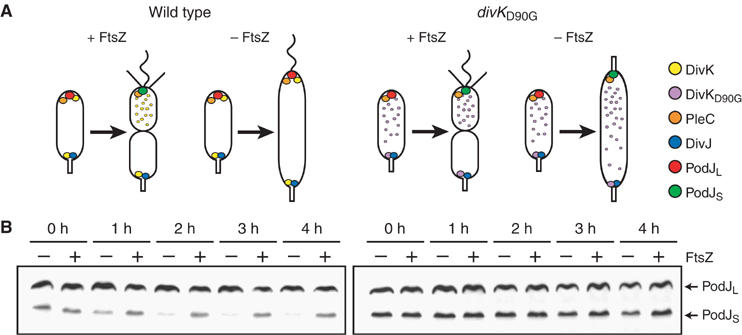
Conversion of PodJL to PodJS depends on compartmentalization, as signaled by DivJ, PleC, and DivK. (A) Schematic diagrams depict the subcellular locations of DivK, PleC, DivJ, and PodJ molecules in wild-type (left) and divKD90G (right) strains. Cytokinesis occurs in the presence of the essential cell division protein FtsZ, but is inhibited in its absence. Wild-type DivK requires compartmentalization for release from the swarmer pole, whereas DivKD90G bypasses that requirement, allowing the swarmer pole to develop a stalk even when division is blocked (Matroule et al, 2004). (B) Immunoblots show contrasting PodJ levels in wild-type (left) and divKD90G (right) cells in the presence (+) or absence (−) of FtsZ. Asynchronous cultures of cells with ftsZ under the control of a chromosomal xylose-regulated promoter were grown in the presence of xylose at 30°C. The cells were then washed and resuspended in media containing glucose or xylose to inhibit or induce FtsZ production, respectively. Samples were taken every hour, starting immediately after the wash.
We report here that proteolytic processing of PodJL to PodJS is an integral component of the swarmer development program, initiated by the DivK response regulator after completion of cytokinesis. We also show that the DivJ–PleC–DivK monitoring system controls the expression of a newly identified gene, perP, which encodes a periplasmic protease required for efficient truncation of PodJL. Cell cycle-dependent transcription of perP plays a critical role in regulating its function, as untimely expression of perP prevents PodJL accumulation and, consequently, inhibits pili formation. In addition, perP expression is activated by the CtrA response regulator, reinforcing the hypothesis that CtrA integrates signals from DivJ, PleC, and DivK (Wu et al, 1998; Sciochetti et al, 2002; Ausmees and Jacobs-Wagner, 2003). Because PodJ is required for PleC localization, polar PleC activity affects perP expression, and PerP promotes PodJL processing, we propose a novel regulatory mechanism in which a sequential proteolytic pathway involving Rip is under temporal and spatial control.
Results
Proteolytic conversion of PodJL to PodJS depends on DivK signaling the completion of cytokinesis
Given that proteolytic conversion of PodJL to PodJS occurs at the time of cell division, and that PodJS is the only isoform present in swarmer cells (Viollier et al, 2002a; Hinz et al, 2003), we asked if PodJL processing is a component of the swarmer cell-specific developmental program. First, as determined by Quardokus and Brun (2004), we confirmed that PodJL processing depends on cytoplasmic compartmentalization (Figure 2B, left). Steady-state levels of PodJ isoforms were monitored after inhibition of cytokinesis using a strain in which the essential cell division gene ftsZ is under the control of a xylose-inducible promoter (Wang et al, 2001). In the presence of xylose, cells divided normally, and PodJ levels stayed constant. When xylose was removed, depleting FtsZ and blocking cell division, PodJS levels dropped dramatically, suggesting that PodJL was no longer being converted to PodJS, while any remaining PodJS disappeared because it continued to be degraded.
Knowing that the D90G mutation (divK341 allele) in the DivK response regulator can relieve the cytokinesis dependence of swarmer cell development (Figure 2A, right) (Sommer and Newton, 1991; Hecht et al, 1995; Hung and Shapiro, 2002; Matroule et al, 2004), we next examined PodJ levels in the divKD90G mutant when FtsZ was depleted. In the mutant, levels of PodJL and PodJS in non-dividing cells remained comparable to the levels in dividing cells (Figure 2B, right), indicating that PodJL processing bypassed the requirement for compartmentalization. Thus, proteolytic processing of PodJL appears to be a developmental event that occurs in response to DivK signaling the completion of cytokinesis and the creation of separate swarmer and stalked progeny compartments.
The distinct phosphorylation states of DivK in the two progeny compartments, generated by the antagonistic effects of the DivJ kinase at the stalked pole and the PleC phosphatase at the swarmer pole, form the basis of the cytokinesis signal (Matroule et al, 2004; McGrath et al, 2004). To confirm that the DivJ–PleC–DivK monitoring system regulates PodJL processing, we compared steady-state levels of PodJL in cells with mutations in divK, divJ, or pleC to that in wild-type cells (Figure 3A, left). In the divKD90G and ΔdivJ mutants, PodJL levels were reduced relative to the wild-type standard, as expected: both mutations tilt the equilibrium toward premature initiation of the swarmer cell-specific program, including PodJL processing. DivKD90G itself does not localize to the flagellar pole, while dephosphorylated DivK predominates in the absence of DivJ and also fails to localize (Wheeler and Shapiro, 1999; Jacobs et al, 2001; Lam et al, 2003; Matroule et al, 2004). These conditions mimic the situation when dephosphorylated DivK is released from the flagellar pole in the swarmer compartment following cell division (Jacobs et al, 2001; Matroule et al, 2004).
Figure 3.
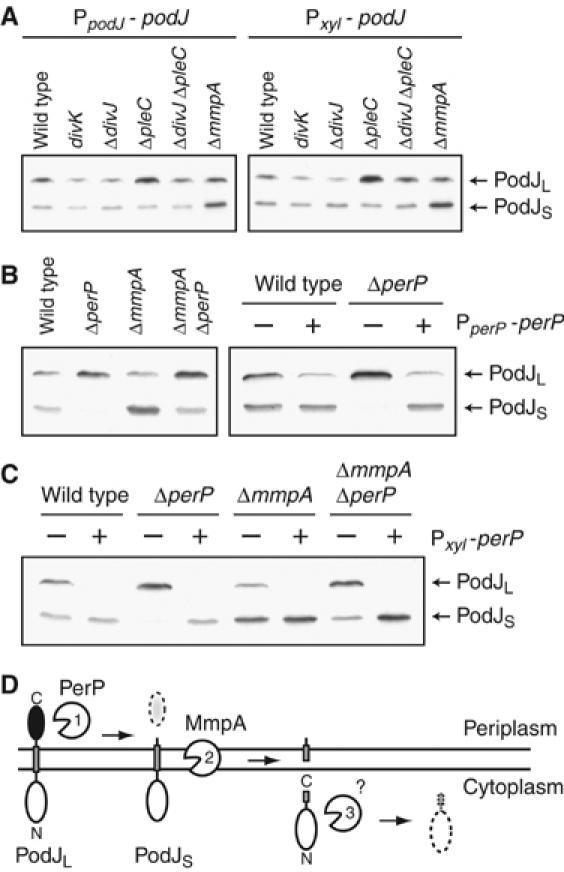
Proteolytic processing of PodJ is affected in multiple mutants. (A) Immunoblots show that PodJL levels in divKD90G, ΔdivJ, and ΔpleC mutants differ from that in wild-type cells, regardless of whether podJ is expressed from its native promoter (PpodJ-podJ) or from a xylose-inducible promoter (Pxyl-podJ) on the chromosome. Cells were grown at 30°C. (B) Efficient conversion of PodJL to PodJS requires perP. Immunoblot on the left compares steady-state levels of PodJL and PodJS in wild-type, ΔperP, ΔmmpA, and ΔmmpA ΔperP strains. Immunoblot on the right shows PodJ levels in wild-type and ΔperP strains when they carry a complementing plasmid (+) or the vector alone (−). The complementing plasmid contains perP under the control of its own promoter (PperP-perP). (C) Constitutive perP expression prevents PodJL accumulation. perP was placed under the control of a xylose-inducible promoter on the chromosome (Pxyl-perP). Wild-type, ΔperP, ΔmmpA, and ΔmmpA ΔperP strains carrying the construct were grown in the presence (+) or absence (−) of xylose and harvested for immunoblot analysis. (D) Schematic diagram depicts sequential proteolytic processing of PodJ, first by PerP, then by MmpA, and finally by an unknown cytoplasmic protease.
On the other hand, the ΔpleC mutation leads to accumulation of phosphorylated DivK, preventing its release from the flagellated pole and initiation of swarmer cell development (Wheeler and Shapiro, 1999; Jacobs et al, 2001; Matroule et al, 2004). As a consequence, PodJL processing should be inhibited. Indeed, we observed accumulation of PodJL in ΔpleC cells (Figure 3A). This elevated level of PodJL was reduced when the ΔdivJ mutation was combined with the ΔpleC mutation (Figure 3A), because DivJ and PleC act in opposition on DivK. We also compared PodJS levels in the various mutants to that in wild-type and ΔmmpA cells (Figure 3A). As previously reported (Chen et al, 2005), PodJS degradation was inhibited in the ΔmmpA mutant, leading to higher steady-state levels, but it did not differ significantly from wild type in strains with mutations in divK, divJ, or pleC.
To eliminate the possibility that variation in PodJL levels resulted from changes in podJ transcription, we placed podJ under the control of a xylose promoter on the chromosome and examined protein levels when the gene was constitutively induced (Figure 3A, right). Similar results were obtained as when podJ was under the control of its own promoter. Therefore, the DivJ–PleC–DivK signaling pathway does not modulate PodJL levels by affecting podJ expression; rather, it may regulate post-transcriptional events such as proteolytic processing of PodJL.
A periplasmic protease participates in PodJL processing
We postulated that the DivJ–PleC–DivK system controls the expression of factors that regulate PodJL processing. To find such factors, we performed microarray analysis on pleC, podJ, and divJ mutants, comparing their steady-state mRNA levels to that in wild-type cells (Figure 4). We identified a set of 26 genes whose transcript levels appeared reduced in pleC and podJ and elevated in divJ mutants. In addition, the expression patterns of these genes during the Caulobacter cell cycle were consistent with participation in the swarmer progeny program (Figure 4) (Laub et al, 2000): their mRNA levels were elevated in swarmer cells, dropped during the swarmer-to-stalked transition, and stayed low until the time of cell division. The expression patterns of these swarmer-specific genes were also similar over the course of a modified cell cycle in the divKD90G mutant (Figure 4). The divKD90G mutant was isolated as a cold-sensitive extragenic suppressor of a pleC mutation (Sommer and Newton, 1991) and, at the restrictive temperature, fails to degrade the master response regulator CtrA and to initiate chromosome replication (Hung and Shapiro, 2002). When divKD90G swarmer cells were allowed to start the cell cycle synchronously at the restrictive temperature, transcript levels of these swarmer-specific genes stayed elevated, as the cell cycle was stalled due to the continual presence of active CtrA. Only when the cells were shifted to the permissive temperature, allowing CtrA to be degraded and the cell cycle to progress, did the transcript levels drop. Thus, the DivJ–PleC–DivK pathway appears to trigger expression of this set of genes in the swarmer compartment.
Figure 4.

Microarray analysis of total RNA in pleC, podJ, divJ, and divKD90G mutants shows variations in expression profiles. For the pleC, podJ, and divJ strains, RNA was extracted from asynchronous populations of mutant cells and compared to that of wild-type CB15. Blue indicates a decrease and yellow indicates an increase relative to the CB15 reference. For the divKD90G mutant, swarmer cells were isolated and grown at the restrictive temperature (18°C) for 300 min, then shifted to the permissive temperature (33°C) for another 80 min. Samples were taken at the indicated times and compared to reference RNA from a mixed population of wild-type cells grown at 30°C. Expression profiles from the wild-type cell cycle are included for comparison (Laub et al, 2000; Hottes et al, 2005). For the wild-type and divKD90G cell cycles, yellow and blue indicate increase and decrease, respectively, relative to the average expression value for that gene during the cycle. Schematics depict stages of the cell cycle; circle or theta structure in the cell represents quiescent or replicating chromosome, respectively. HK, histidine kinase; RR, response regulator.
One member in particular among the set of DivK-regulated genes was a candidate for controlling PodJL processing. The CC1307 gene encodes a protein with a putative signal sequence or membrane anchor at its N-terminus (Bendtsen et al, 2004) and a conserved aspartic protease motif in its periplasmic domain (Marchler-Bauer and Bryant, 2004). As PodJL processing involves truncation of its periplasmic domain (see Figure 3D), we suspected that the CC1307 gene product is the perpetrating protease and named it perP, for periplasmic protease of PodJ. To confirm this, we constructed a strain with an in-frame deletion of perP and examined its PodJ levels by immunoblot analysis (Figure 3B). Compared to that in wild-type cells, PodJL levels were elevated in the ΔperP mutant, whereas PodJS levels were reduced to being almost undetectable, suggesting that conversion of PodJL to PodJS was inhibited in the ΔperP mutant. We were able to complement this PodJL processing defect in the ΔperP mutant by introducing a plasmid carrying perP under the control of its own promoter (Figure 3B, right). However, the complementing plasmid reduced steady-state levels of PodJL in both wild-type and ΔperP strains, raising the possibility that improperly regulated expression of perP from the plasmid affects PodJL accumulation (see below).
We also examined PodJ levels in a ΔmmpA ΔperP double mutant and found increased levels of PodJL, similar to that in the ΔperP mutant (Figure 3B). Notably, PodJS levels in the double mutant were similar to that in wild type and intermediate between those in the ΔmmpA and ΔperP single mutants. We interpret these results as follows: PodJL is normally truncated by PerP to produce PodJS, which is then cleaved by MmpA and released from the cell membrane for subsequent degradation in the cytoplasm (Figure 3D) (Chen et al, 2005). Sequential degradation of PodJ by the PerP and MmpA proteases forms the first two steps of the Rip pathway in Caulobacter. In the absence of PerP, conversion of PodJL to PodJS is inhibited, preventing PodJS accumulation. Nevertheless, PodJL may still be processed by proteases other than PerP, albeit significantly less efficiently, and this leaky processing may lead to some PodJS accumulation when PodJS degradation fails to occur in the absence of the MmpA protease.
To demonstrate that PerP participates in the proteolytic processing of PodJL, we performed pulse–chase analysis and compared PodJL stability in wild-type and ΔperP strains (Figure 5). In wild-type cells, newly synthesized PodJL stayed relatively stable for about 45 min and then rapidly disappeared, with a half-life of approximately 15 min. As previously reported (Hinz et al, 2003; Chen et al, 2005), the disappearance of PodJL corresponded to the concomitant appearance of PodJS, implicating proteolytic conversion. This stability profile is consistent with PodJL being synthesized in the early predivisional cell and remaining intact until cytoplasmic compartmentalization (Judd et al, 2003), when it is truncated. In the ΔperP mutant, PodJL was relatively stable for at least 120 min after the chase, with negligible appearance of PodJS. These results indicate that PerP is required for the efficient proteolytic conversion of PodJL to PodJS.
Figure 5.

Pulse–chase analysis shows inhibition of PodJL processing in the ΔperP mutant. Mixed populations of wild-type or ΔperP cells were pulse-labeled with [35S]methionine for 7.5 min and then chased with excess unlabeled methionine and casamino acids. Samples were taken at 15-min intervals after the chase and immunoprecipitated with antibodies to the N-terminal domain of PodJ. Immunoprecipitates were resolved by SDS–PAGE, as shown, to determine the relative levels of PodJL. Fraction of PodJL remaining over time was plotted on a logarithmic scale (base 2) and fitted according to exponential decay. A representative analysis is shown.
We determined that PerP directly truncates PodJL by performing in vitro cleavage assays with purified domains of PerP and PodJL (Figure 6). The periplasmic domain of PodJL (PodJPERI) was tagged with His6 at the N-terminus and purified as described previously (Viollier et al, 2002a). PerP, without its first 28 amino acids, including a hydrophobic leader sequence, was tagged with His6 at both termini to generate PerPΔLS. PerPΔLS was purified and cleavage reactions were carried out as described in Materials and methods. In the presence of PerPΔLS, the 33-kDa PodJPERI was cut into two fragments, PodJPERI-C (∼27 kDa) and PodJPERI-N (∼6 kDa); PodJPERI-N was not detectable by Coomassie blue staining but was visible by silver staining (data not shown). Cleavage of PodJPERI appeared to progress linearly with time, and while the assays shown in Figure 6 required long incubation times, there is precedent: in vitro cleavage of anti-sigma factor RseA by the DegS periplasmic protease required 4 h (Walsh et al, 2003). Furthermore, PerPΔLS was purified from inclusion bodies and may have a low fraction of active proteases. We assigned the 27-kDa fragment as the one containing the C-terminus of PodJPERI for two reasons. First, previous studies suggested that removal of 20–30 kDa from PodJL's C-terminus yields PodJS (Viollier et al, 2002a; Chen et al, 2005), and the size of the removed fragment is consistent with that of PodJPERI-C. Second, we examined Escherichia coli cells expressing a PodJL construct containing GFP at its N-terminus by immunoblotting (Supplementary Figure S6). The construct was cut into two fragments when the cells coexpressed full-length PerP: one fragment was only detectable with antibodies to GFP, and the other, exactly the same size as PodJPERI-C, was only detectable with antibodies to the C-terminal domain of PodJL. The 6-kDa fragment (PodJPERI-N) released in the in vitro assay, we believe, remains attached to the transmembrane domain in vivo. Our experiments thus suggest that PerP cleaves PodJL near the N-terminus of its periplasmic domain to produce membrane-bound PodJS and PodJPERI-C (Figure 3D). PodJPERI-C is then presumably cleared from the periplasm by other proteases in vivo.
Figure 6.
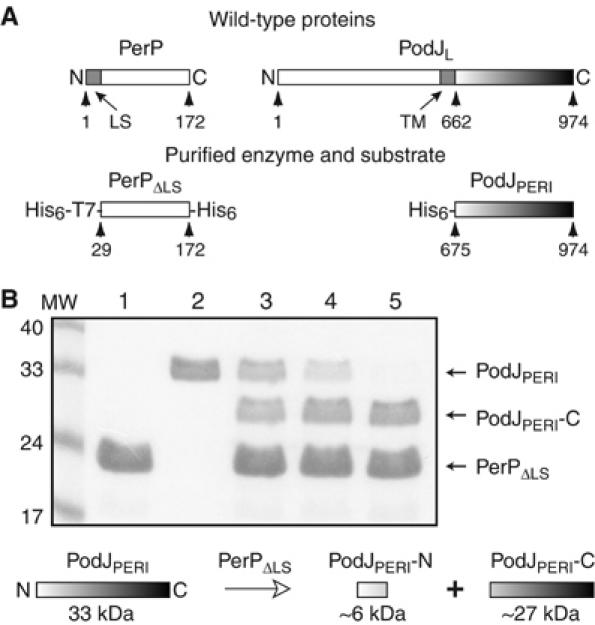
PerP cleaves the periplasmic domain of PodJ (PodJPERI) in vitro. (A) Schematics depict full-length PerP and PodJL and the purified fragments PerPΔLS and PodJPERI. PerPΔLS was tagged with His6 and T7 epitope at the N-terminus and with His6 at the C-terminus. PodJPERI was tagged with His6 at the N-terminus. Numbered arrows point to relevant amino-acid residues. LS, hydrophobic leader sequence; TM, transmembrane segment. (B) Reaction mixtures containing purified PerPΔLS (lane 1), PodJPERI (lane 2), or both (lanes 3–5) were incubated at 37°C for 4 (lane 3), 8 (lane 4), or 20 h (lanes 1, 2, 5). Samples were resolved by SDS–PAGE and visualized by Coomassie blue staining. The MW lane contains molecular weight markers, indicated on the left in kDa. Diagram below the gel shows cleavage of PodJPERI by PerPΔLS into PodJPERI-N and PodJPERI-C. The PodJPERI-N band is not visible here but is detectable by silver staining (data not shown).
PerP expression affects PodJL distribution and function
To ascertain that PerP is responsible for PodJL processing at a specific time during the Caulobacter cell cycle, we monitored PodJ levels over the course of the cycle in the ΔperP mutant (Figure 1B, right). As expected, PodJL was the only isoform detected in the mutant, and its level remained relatively constant throughout the cell cycle. This is in contrast to the situation in wild-type cells, where PodJL and PodJS levels varied in a defined pattern over the cycle (Figure 1B, left). We also examined other proteins whose levels fluctuated in a specific manner over the course of the cell cycle, including the CtrA response regulator (Domian et al, 1997) and the McpA chemoreceptor (Tsai and Alley, 2001) (Figure 1B), as well as the PleC kinase/phosphatase and pili components PilA and CpaE (Viollier et al, 2002b) (data not shown). Their patterns of cell cycle-dependent variation were the same in both wild-type and ΔperP cells. Thus, PerP appears to participate specifically in the proteolytic processing of PodJL.
The podJ gene has been shown to be required for pili biogenesis, holdfast formation, and efficient chemotaxis (Wang et al, 1993; Viollier et al, 2002a; Hinz et al, 2003), with PodJL specific for pili formation and PodJS for holdfast and chemotaxis functions (Viollier et al, 2002a). Because perP expression and PerP activity appeared to be stringently restricted to a specific time in the cell cycle, coincident with the completion of cytokinesis, we asked if altering the timing of perP expression would alter polar organelle phenotypes. Accordingly, we placed perP under the control of a xylose-inducible promoter on the chromosome, allowing perP to be expressed at all times in the cell cycle. In the presence of xylose, PodJL failed to accumulate (Figure 3C) and, as a consequence, pili synthesis failed to occur (data not shown). Thus, the timing of perP expression is critical for pili formation. Notably, constitutive conversion of PodJL to PodJS by PerP did not lead to immediate degradation by MmpA, and PodJS degradation was still blocked when MmpA was absent (Figure 3C). These results suggested that PodJS degradation is regulated by a mechanism independent of PerP, whereas PodJL processing is dependent on regulation of perP expression.
Previous analysis of the ΔmmpA protease mutant showed that PodJ degradation contributes to its asymmetric localization during the Caulobacter cell cycle (Chen et al, 2005). Thus, we predicted that inhibition of PodJL processing in the ΔperP mutant would lead to bipolar localization of PodJL because PodJL at the stalked pole would not be degraded before new PodJL is synthesized and localized to the incipient swarmer pole (Figure 1A). Assessment of YFP-tagged PodJ localization in ΔperP cells affirmed this prediction (Figure 1C). Cells expressing yfp-podJ as the only form of podJ from its native chromosomal locus were examined by fluorescence microscopy. YFP-PodJ localized to both poles in a subset of the ΔperP cells, whereas it was found at only one pole in wild-type cells. However, we did not observe a discernable alteration in polar organelle formation in ΔperP strains (data not shown), suggesting that, as is the case for the CtrA global regulator (Domian et al, 1997), multiple levels of control are operative on PodJ function.
Regulation of perP transcription
Because timely expression of perP is critical to PodJL accumulation and its associated functions, we examined the regulation of perP expression more closely. We created a reporter construct using a lacZ gene without its own start codon on a low-copy-number plasmid. The lacZ gene was transcriptionally fused to an approximately 350-bp sequence upstream of the perP gene and translationally fused to the first eight codons of perP (Figure 7A). The reporter plasmid was introduced into different strains to assess expression levels by β-galactosidase assays. We first measured β-galactosidase activities in strains with mutations in divJ, pleC, or divK (Figure 7B). In agreement with the microarray analysis, expression of the perP reporter construct was significantly lower in ΔpleC cells compared to wild type; lower perP expression correlated with reduced PodJL processing in the ΔpleC strain (Figure 3A). In contrast, expression was higher in the divKD90G strain at the permissive temperature, consistent with increased PerP activity and reduced PodJL accumulation in that strain. Although PodJL processing appeared enhanced in the ΔdivJ strain, perP expression did not differ significantly from the wild-type level. We do not have a definitive explanation for this, but one possibility is that the ΔdivJ mutation has different post-translational effects on PerP and LacZ. Regardless, PleC and DivJ did appear to act in opposition: expression of the lacZ reporter in the ΔpleC ΔdivJ double mutant was intermediate between those in the ΔpleC and ΔdivJ single mutants, consistent with the steady-state levels of PodJL in those mutants.
Figure 7.

perP transcription is regulated by signaling proteins that affect cell cycle progression. (A) A lacZ gene without its own start codon was translationally fused to the first eight codons of perP (as annotated in GenBank) and transcriptionally fused to a 350-bp upstream sequence. The fusion construct was placed on a low-copy-number vector to generate plasmid pJC327. Partial nucleotide sequence of the perP promoter is shown. Underlining indicates possible CtrA binding site (Ouimet and Marczynski, 2000), whereas box indicates a candidate methylation site (Chen and Shapiro, 2003). Bent arrow indicates the putative transcriptional start site, based on Affymetrix data (PT McGrath et al, unpublished). (B) β-Galactosidase assays indicate variations in perP expression in ΔpleC, divKD90G, ΔdivJ, and ΔpleC ΔdivJ backgrounds. Mixed populations of cells carrying pJC327 were grown at 30°C and harvested for analysis. β-Galactosidase activities were normalized according to that in wild-type cells. (C) Expression of the lacZ reporter construct was reduced in the ΔpodJ but not the ΔperP strain. (D) perP expression requires active CtrA. Wild-type, ctrA401 (Ts), and cckA1 (Ts) cells carrying pJC327 were grown at the permissive temperature (28°C) and then shifted to the restrictive temperature (37°C). Samples were harvested at 0, 2, and 4 h after the shift to measure β-galactosidase activities, which was normalized according to that in wild-type cells at 0 h. (E) CtrA is a positive regulator of perP transcription. Expression of the lacZ reporter construct was determined in strains carrying an empty vector or a plasmid with ctrA under the control of a xylose-inducible promoter (Pxyl-ctrA). Cells were grown in the presence of glucose and then shifted to media containing xylose. Samples were harvested 0, 2, and 4 h after the shift. Activities were normalized according to that at 0 h in cells carrying the vector alone. Error bars indicate standard deviations.
The opposing localization of DivJ and PleC is required for correct functioning of the DivJ–PleC–DivK signaling system (Matroule et al, 2004; McGrath et al, 2004). Because PodJ is necessary for PleC localization to the swarmer pole, deletion of the podJ gene disrupts many PleC-controlled functions (Viollier et al, 2002a; Hinz et al, 2003). Indeed, according to our microarray analysis, the ΔpodJ mutation affected Caulobacter transcriptional profile in much the same way the ΔpleC mutation did, albeit to a smaller magnitude (Figure 4). Examination of perP expression using the lacZ reporter construct confirmed this requirement for PodJ (Figure 7C). The ΔpodJ mutation reduced perP expression to about 43% of wild-type level, whereas the ΔpleC mutation reduced expression to about 10%. These effects are consistent with the idea that PleC can still dephosphorylate DivK in the ΔpodJ mutant, but the phosphatase activity is no longer concentrated at the swarmer pole. Although normal perP expression requires PleC localization by PodJ, it does not require PodJL processing by PerP: the lacZ reporter construct generated similar activity in the ΔperP mutant as in wild type (Figure 7C).
Previous studies suggested that DivJ, PleC, and DivK act through the CtrA master response regulator (Wu et al, 1998; Sciochetti et al, 2002; Ausmees and Jacobs-Wagner, 2003). In addition, data from previous microarray analyses indicated that the cell cycle-dependent expression of perP is regulated by CtrA (Laub et al, 2000, 2002). To confirm the transcriptional effect of CtrA on perP, we measured expression of the lacZ reporter in strains with temperature-sensitive mutation in ctrA or cckA, encoding a kinase that is required for the phosphorylation of CtrA (Jacobs et al, 1999, 2003) (Figure 7D). Even before shifting to the restrictive temperature (0 h), PperP-lacZ expression in either mutant was substantially lower, less than 10%, compared to wild type. Shifting to the restrictive temperature only further reduced the level of expression. On the other hand, when ctrA was constitutively expressed from a plasmid, expression of the lacZ reporter increased (Figure 7E). Therefore, CtrA appears to be a positive regulator of perP expression.
Discussion
Results presented here demonstrate that successive cleavage events involving Rip occur as a function of time during a bacterial cell cycle and are dependent on cytokinesis. The first proteolytic event in the series occurs following the establishment of two compartments (Judd et al, 2003) in the predivisional cell, when the newly identified PerP protease truncates PodJL to PodJS at the flagellated pole (Figure 3D). We have shown that this first cleavage step is temporally coordinated with cytokinesis via transcriptional regulation of PerP. Thus, integration of the temporal activation of the PerP protease and the spatial restriction of the polar PodJL substrate results in the controlled onset of the proteolytic sequence. The membrane metalloprotease MmpA then performs Rip on PodJS in the progeny swarmer cell as it differentiates into a stalked cell, releasing PodJS into the cytoplasm for further degradation (Figure 3D). The sequential actions of PerP and MmpA on PodJ illustrate how the Rip pathway has been adapted to regulate the asymmetric distribution of a polarity factor over the course of the Caulobacter cell cycle.
The proteolytic processing of the polarity determinant PodJL to PodJS occurs in the swarmer cell compartment of the predivisional cell when the DivJ–PleC–DivK phosphotransfer system signals the completion of cytokinesis. Correct accumulation and efficient truncation of PodJL both depend on the timely expression of perP, which encodes a periplasmic protease. We have shown both in vivo and in vitro that PerP clips the periplasmic domain of PodJL to produce PodJS. The DivJ–PleC–DivK system controls the timing of PodJL processing by ensuring that perP is transcribed at a specific time in the cell cycle. These results suggest a model of PodJ processing in which PodJ, PleC, and PerP form the components of a regulatory circuit (Figure 8). First, localized PodJL recruits the PleC histidine kinase/phosphatase to the incipient swarmer pole (Viollier et al, 2002a; Hinz et al, 2003; Lawler et al, 2006). Second, upon establishment of two compartments prior to cell separation, correct polar PleC localization controls the accumulation of the pili subunit PilA (Viollier et al, 2002a) and appears to be responsible for the monitoring system in which cytokinesis activates perP expression in the swarmer compartment. We propose that once a diffusion barrier forms between the two nascent daughter compartments, the phosphorylation level of DivK declines in the swarmer section, where PleC resides, activating perP transcription through the CtrA response regulator. Third, PerP converts PodJL to PodJS in the swarmer cell by proteolytic truncation. Thus, PodJL localizes PleC, PleC mediates the onset of PerP expression, and PerP cleaves PodJL (Figure 8).
Figure 8.
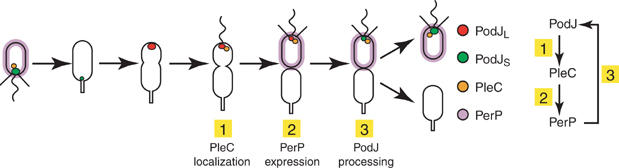
Model of PodJL processing includes a regulatory loop: (1) PodJL localizes PleC to the incipient swarmer pole; (2) compartmentalization, monitored in part by PleC, activates PerP expression; and (3) PerP cleaves PodJL.
After PodJL is converted to PodJS, a distinct mechanism appears to regulate the timing of subsequent PodJS proteolysis, as suggested by the continued presence of PodJS when perP was constitutively expressed (Figure 3C). This mechanism probably signals the MmpA membrane metalloprotease to initiate PodJS cleavage and release from the membrane during the swarmer-to-stalked cell transition (Chen et al, 2005). Notably, the CtrA master regulator is also cleared from the cell during this period of the Caulobacter cell cycle (Domian et al, 1997). Proteolytic removal of CtrA should stop further expression of perP, which is positively regulated by CtrA. Ending production of the PerP protease during the swarmer-to-stalked transition would enable PodJL to accumulate when it is synthesized in the early predivisional cell, beginning the cycle again. Therefore, our results demonstrate that regulated, sequential proteolysis contributes to the cell cycle-dependent variations in PodJL and PodJS levels.
Cell cycle-regulated expression of perP is critical for PodJL to perform its functions, because constitutive, untimely perP expression prevents PodJL accumulation and abolishes pili formation. However, the absence of an observable phenotype associated with the ΔperP mutation is puzzling. Clearly, PerP-mediated proteolysis contributes to the maintenance of polarity at the molecular level: PodJ loses its asymmetric distribution in the absence of PerP. There are at least two possible explanations for why this loss of molecular asymmetry does not translate into an overt physiological defect. First, although significantly less efficient, some degradation of PodJ does occur when PerP or MmpA (Chen et al, 2005) is absent. Residual degradation by other proteases may prevent sufficient PodJ accumulation for interference of cellular functions. The second and more likely explanation is that redundant mechanisms regulate PodJ activity, as is the case for CtrA. For example, inhibition of CtrA proteolysis alone does not prevent cell cycle progression; constitutive phosphorylation of the CtrA response regulator is also needed to have an adverse effect on cell viability (Domian et al, 1997). Similarly, post-translational modification of the PodJ protein, other than proteolysis, may modulate its function. PodJ activity may also be regulated via its interacting partners. PodJ requires other proteins, such as PleC and the CpaE pili assembly protein, to effect downstream functions (Viollier et al, 2002a), and those interacting proteins may be active or available only at specific times during the cell cycle. The full significance of sequential PodJ proteolysis may be masked until additional mechanisms that regulate PodJ and its downstream effectors are elucidated. However, the elaborate cell cycle-dependent mechanism that controls the sequential proteolysis of the PodJ polarity determinant provides a striking example of the integration of spatial and temporal elements to yield asymmetry.
Materials and methods
Bacterial strains, growth conditions, and molecular biology procedures
C. crescentus CB15, NA1000, and their derivatives were grown in PYE or M2 minimal media with appropriate supplements, as described previously (Chen et al, 2005). Unless noted otherwise, cultures were grown to mid-exponential phase at 30°C for subsequent analysis. Standard protocols were used for synchronization, electroporation, bacteriophage ΦCr30-mediated transduction, and plasmid mobilization from E. coli by conjugation (Ely, 1991). Assays to examine chemotaxis ability on semisolid media, pili biogenesis using bacteriophage ΦCbK, and holdfast synthesis by rosette formation have all been reported (Wang et al, 1993; Skerker and Shapiro, 2000; Viollier et al, 2002a; Chen et al, 2005). Standard techniques were used for the cloning, manipulation, and analysis of DNA (Ausubel et al, 1988). Detailed descriptions of strains and plasmids are provided in Supplementary Tables S1 and S2.
Immunoblots and pulse–chase analysis
Immunoblotting was performed using standard procedures (Ausubel et al, 1988): samples were collected at appropriate times, resolved by SDS–PAGE, and transferred to PVDF membrane for detection. Antibodies that detect CtrA, McpA, FliF, PleC, CpaE, PilA, GFP, or different domains of PodJ were used as described (Viollier et al, 2002a, 2002b; Chen et al, 2005). Pulse–chase analysis of PodJ stability was performed according to Chen et al (2005), except that the cells were pulse-labeled for 7.5 min and chased with 1 mM methionine and 2 mg/ml casamino acids.
Microarray analysis and β-galactosidase assay
RNA preparation and hybridization to microarrays were performed as described (Holtzendorff et al, 2004). Total RNA extracted from asynchronous cultures of CB15 ΔpleC, pleCH610A, ΔpodJ, ΔdivJ, or divJ::Tn5 strains were compared to that of wild-type CB15 grown in PYE. For the divKD90G mutant, swarmer cells were isolated, grown in M2G at the restrictive temperature (18°C), and then shifted to the permissive temperature (33°C). Samples from various time points were compared to reference RNA from an asynchronous culture of CB15N grown at 30°C in M2G. Data were filtered and normalized as described by Hottes et al (2004) and visualized using Cluster and Treeview (Eisen et al, 1998). Supplementary data contains the complete data sets and additional details about data processing. Assays for β-galactosidase were performed according to standard protocol (Ausubel et al, 1988), except that incubation with chromogenic substrate was at 25°C.
Microscopy
Live cells expressing YFP-PodJ were placed on 1% agarose pads and examined by differential interference contrast and fluorescence microscopy, as described previously (Viollier et al, 2002a; Chen et al, 2005).
Cleavage assays
The periplasmic domain of PodJ (residues 675–974) was purified from soluble E. coli cell extract as described previously (Viollier et al, 2002a). The periplasmic domain of PerP (residues 29–172) was tagged with His6 and T7 epitope at the N-terminus and with His6 at its C-terminus, overexpressed in E. coli, purified from inclusion bodies under standard denaturing conditions (Qiagen), and dialyzed into 50 mM Na2HPO4 (pH 8), 300 mM NaCl, and 5% glycerol. Cleavage reactions occurred at 37°C in 50 mM Na2HPO4 (pH 8), 180 mM NaCl, 60 mM KCl, 16.7 mM imidazole, 60 μM EDTA, and 7.5% glycerol. Each reaction contained 10 μg of PerPΔLS and/or 3 μg of PodJPERI. Samples were resolved using 12 or 15% SDS–PAGE and visualized by Coomassie brilliant blue or silver staining (Bio-Rad).
Supplementary Material
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Material
Supplementary Fig S6
Supplementary References
Acknowledgments
We thank Maliwan Meewan for help in performing the microarray experiments and the Brun lab for communicating results prior to publication. This work was supported by grants from the National Institutes of Health (GM032506 and F32 G067472) and the Department of Energy (DE-FG03-01ER63219).
References
- Ausmees N, Jacobs-Wagner C (2003) Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu Rev Microbiol 57: 225–247 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1988) Current Protocols in Molecular Biology. New York: John Wiley & Sons [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Brown MS, Ye J, Rawson RB, Goldstein JL (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100: 391–398 [DOI] [PubMed] [Google Scholar]
- Chen JC, Viollier PH, Shapiro L (2005) A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol 55: 1085–1103 [DOI] [PubMed] [Google Scholar]
- Chen SL, Shapiro L (2003) Identification of long intergenic repeat sequences associated with DNA methylation sites in Caulobacter crescentus and other alpha-proteobacteria. J Bacteriol 185: 4997–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crymes WB Jr, Zhang D, Ely B (1999) Regulation of podJ expression during the Caulobacter crescentus cell cycle. J Bacteriol 181: 3967–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L (1997) Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90: 415–424 [DOI] [PubMed] [Google Scholar]
- Ehrmann M, Clausen T (2004) Proteolysis as a regulatory mechanism. Annu Rev Genet 38: 709–724 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B (1991) Genetics of Caulobacter crescentus. Methods Enzymol 204: 372–384 [DOI] [PubMed] [Google Scholar]
- Gottesman S (2003) Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol 19: 565–587 [DOI] [PubMed] [Google Scholar]
- Grunenfelder B, Rummel G, Vohradsky J, Roder D, Langen H, Jenal U (2001) Proteomic analysis of the bacterial cell cycle. Proc Natl Acad Sci USA 98: 4681–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht GB, Lane T, Ohta N, Sommer JM, Newton A (1995) An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J 14: 3915–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz AJ, Larson DE, Smith CS, Brun YV (2003) The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol 47: 929–941 [DOI] [PubMed] [Google Scholar]
- Holtzendorff J, Hung D, Brende P, Reisenauer A, Viollier PH, McAdams HH, Shapiro L (2004) Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science 304: 983–987 [DOI] [PubMed] [Google Scholar]
- Hottes AK, Meewan M, Yang D, Arana N, Romero P, McAdams HH, Stephens C (2004) Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J Bacteriol 186: 1448–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottes AK, Shapiro L, McAdams HH (2005) DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol 58: 1340–1353 [DOI] [PubMed] [Google Scholar]
- Hung DY, Shapiro L (2002) A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression. Proc Natl Acad Sci USA 99: 13160–13165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C, Ausmees N, Cordwell SJ, Shapiro L, Laub MT (2003) Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol Microbiol 47: 1279–1290 [DOI] [PubMed] [Google Scholar]
- Jacobs C, Domian IJ, Maddock JR, Shapiro L (1999) Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97: 111–120 [DOI] [PubMed] [Google Scholar]
- Jacobs C, Hung D, Shapiro L (2001) Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc Natl Acad Sci USA 98: 4095–4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Hengge-Aronis R (2003) Regulation by proteolysis in bacterial cells. Curr Opin Microbiol 6: 163–172 [DOI] [PubMed] [Google Scholar]
- Judd EM, Ryan KR, Moerner WE, Shapiro L, McAdams HH (2003) Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc Natl Acad Sci USA 100: 8235–8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Matroule JY, Jacobs-Wagner C (2003) The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell 5: 149–159 [DOI] [PubMed] [Google Scholar]
- Laub MT, Chen SL, Shapiro L, McAdams HH (2002) Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci USA 99: 4632–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L (2000) Global analysis of the genetic network controlling a bacterial cell cycle. Science 290: 2144–2148 [DOI] [PubMed] [Google Scholar]
- Lawler ML, Larson DE, Hinz AJ, Klein D, Brun YV (2006) Dissection of functional domains of the polar localization factor PodJ in Caulobacter crescentus. Mol Microbiol 59: 301–316 [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32: W327–W331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C (2004) Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell 118: 579–590 [DOI] [PubMed] [Google Scholar]
- McGrath PT, Viollier P, McAdams HH (2004) Setting the pace: mechanisms tying Caulobacter cell-cycle progression to macroscopic cellular events. Curr Opin Microbiol 7: 192–197 [DOI] [PubMed] [Google Scholar]
- Ouimet MC, Marczynski GT (2000) Analysis of a cell-cycle promoter bound by a response regulator. J Mol Biol 302: 761–775 [DOI] [PubMed] [Google Scholar]
- Quardokus EM, Brun YV (2003) Cell cycle timing and developmental checkpoints in Caulobacter crescentus. Curr Opin Microbiol 6: 541–549 [DOI] [PubMed] [Google Scholar]
- Quardokus EM, Brun YV (2004) A cell division checkpoint regulates the processing of the PodJ developmental regulator in Caulobacter crescentus. In American Society for Microbiology 104th General Meeting. New Orleans, LA: ASM Press (abstract I-104) [Google Scholar]
- Ryan KR, Huntwork S, Shapiro L (2004) Recruitment of a cytoplasmic response regulator to the cell pole is linked to its cell cycle-regulated proteolysis. Proc Natl Acad Sci USA 101: 7415–7420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciochetti SA, Lane T, Ohta N, Newton A (2002) Protein sequences and cellular factors required for polar localization of a histidine kinase in Caulobacter crescentus. J Bacteriol 184: 6037–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, Laub MT (2004) Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat Rev Microbiol 2: 325–337 [DOI] [PubMed] [Google Scholar]
- Skerker JM, Shapiro L (2000) Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J 19: 3223–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer JM, Newton A (1991) Pseudoreversion analysis indicates a direct role of cell division genes in polar morphogenesis and differentiation in Caulobacter crescentus. Genetics 129: 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Alley MR (2001) Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol 183: 5001–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban S, Freeman M (2002) Intramembrane proteolysis controls diverse signalling pathways throughout evolution. Curr Opin Genet Dev 12: 512–518 [DOI] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L (2002a) Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci USA 99: 13831–13836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Sternheim N, Shapiro L (2002b) A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J 21: 4420–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT (2003) OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113: 61–71 [DOI] [PubMed] [Google Scholar]
- Wang SP, Sharma PL, Schoenlein PV, Ely B (1993) A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc Natl Acad Sci USA 90: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jones BD, Brun YV (2001) A set of ftsZ mutants blocked at different stages of cell division in Caulobacter. Mol Microbiol 40: 347–360 [DOI] [PubMed] [Google Scholar]
- Weihofen A, Martoglio B (2003) Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell Biol 13: 71–78 [DOI] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L (1999) Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell 4: 683–694 [DOI] [PubMed] [Google Scholar]
- Wu J, Ohta N, Newton A (1998) An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc Natl Acad Sci USA 95: 1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Material
Supplementary Fig S6
Supplementary References


