Abstract
Small RNA-mediated gene silencing (RNA silencing) has emerged as a major regulatory pathway in eukaryotes. Identification of the key factors involved in this pathway has been a subject of rigorous investigation in recent years. In humans, small RNAs are generated by Dicer and assembled into the effector complex known as RNA-induced silencing complex (RISC) by multiple factors including hAgo2, the mRNA-targeting endonuclease, and TRBP (HIV-1 TAR RNA-binding protein), a dsRNA-binding protein that interacts with both Dicer and hAgo2. Here we describe an additional dsRNA-binding protein known as PACT, which is significant in RNA silencing. PACT is associated with an ∼500 kDa complex that contains Dicer, hAgo2, and TRBP. The interaction with Dicer involves the third dsRNA-binding domain (dsRBD) of PACT and the N-terminal region of Dicer containing the helicase motif. Like TRBP, PACT is not required for the pre-microRNA (miRNA) cleavage reaction step. However, the depletion of PACT strongly affects the accumulation of mature miRNA in vivo and moderately reduces the efficiency of small interfering RNA-induced RNA interference. Our study indicates that, unlike other RNase III type proteins, human Dicer may employ two different dsRBD-containing proteins that facilitate RISC assembly.
Keywords: Dicer, microRNA, PACT, RNAi, siRNA
Introduction
Small RNAs function as guide molecules in a wide range of regulatory pathways that are now collectively known as RNA silencing (Bartel, 2004; Baulcombe, 2005). Through specific base-pairing with mRNAs, these tiny ∼22 nt RNAs induce mRNA degradation and translational repression. Based on their origin, small RNAs can be grouped into two classes: microRNA (miRNA) or small interfering RNA (siRNA) (Kim, 2005b). miRNAs originate from endogenous transcripts harboring local hairpin structures, while siRNAs are generated from long dsRNAs. siRNAs can be further classified into transacting siRNA (tasiRNA), repeat-associated siRNA (rasiRNA), and small scan RNA (scnRNA).
miRNAs are transcribed from endogenous genes by RNA polymerase II (Cai et al, 2004; Lee et al, 2004a; Kim, 2005a). The hairpin embedded in the primary transcripts of miRNA (pri-miRNAs) are cropped into hairpin-structured precursors (pre-miRNAs) by the nuclear RNase III Drosha (Lee et al, 2003). Pre-miRNAs are then exported to the cytoplasm with the help of exportin-5 (Exp5) (Yi et al, 2003; Bohnsack et al, 2004; Lund et al, 2004) and subsequently processed into mature miRNA of ∼22 nt by another RNase III Dicer (Grishok et al, 2001; Hutvagner et al, 2001). Unlike miRNAs, siRNAs are generated from long double-stranded RNAs (dsRNAs) by Dicer (Bernstein et al, 2001; Ketting et al, 2001). Thus, Dicer serves as a common enzyme in the biogenesis of both miRNAs and siRNAs in mammalian cells.
The small RNAs are loaded onto the effector complex, known as the RNA-induced silencing complex (RISC) (Filipowicz, 2005; Sontheimer, 2005; Tomari and Zamore, 2005; Zamore and Haley, 2005). Argonaute proteins are the core components of the RISC (Hammond et al, 2001; Carmell et al, 2002; Liu et al, 2004; Meister et al, 2004; Sontheimer and Carthew, 2004). Argonaute proteins have three highly conserved regions: the PAZ domain at the N-terminus, the middle conserved domain, and the PIWI domain at the C-terminus. Structural and biochemical studies indicate that Argonaute proteins are capable of interacting directly with small RNAs through their PAZ domains (to the 3′ end) and through the PIWI and the middle domains (to the 5′ end) (Song et al, 2003, 2004; Yan et al, 2003; Lingel et al, 2004; Ma et al, 2004, 2005; Parker et al, 2005; Rivas et al, 2005; Yuan et al, 2005). A small RNA guides the RISC to its target RNA, which leads to mRNA degradation and/or to translational repression.
The mechanism of RISC formation has been intensively studied in several types of organisms, such as flies and humans (Filipowicz, 2005; Sontheimer, 2005; Tomari and Zamore, 2005; Zamore and Haley, 2005). In D. melanogaster, there are two types of Dicers, Dicer-1 and Dicer-2 (Lee et al, 2004b). Dicer-1 is responsible for miRNA biogenesis and possibly for the assembly of miRNA-incorporated RISC (miRISC), while Dicer-2 is able to process only siRNAs from long dsRNAs. Dicer-2 also appears to be required for loading a guide strand of siRNA duplex onto the RISC complex (siRISC). There is only one type of Dicer in humans, which generates both miRNA and siRNA. Besides the processing of miRNA, human Dicer has also been proposed to have a role in RISC assembly, because a depletion of Dicer results in a defect of RNA interference (RNAi) process mediated by siRNA duplex in human cell lines (Doi et al, 2003).
Several dsRNA-binding proteins are known to associate with RNase III proteins to promote miRNA processing or RISC programming. In the nucleus, Drosha associates with DGCR8 (in humans) or Pasha (in Drosophila and Caenorhabditis elegans), which contains tandem dsRNA-binding domains (dsRBDs), to form the microprocessor complex that processes pri-miRNAs into pre-miRNAs (Denli et al, 2004; Gregory et al, 2004; Han et al, 2004; Landthaler et al, 2004). Loquacious (Loqs) (also known as R3D1), which has three dsRBDs, is a partner of Drosophila Dicer-1 in flies (Forstemann et al, 2005; Jiang et al, 2005; Saito et al, 2005). The Dicer-1–Loqs heterodimer efficiently carries out pre-miRNA processing. When Loqs was depleted by RNAi in the Drosophila S2 cell line, the mature miRNA level was reduced, while pre-miRNAs accumulated, indicating that Loqs is required for the pre-miRNA cleavage event (Forstemann et al, 2005; Jiang et al, 2005; Saito et al, 2005). R2D2, another dsRNA-binding protein, is known to associate with Drosophila Dicer-2 to promote siRNA from entering into Ago2 (Liu et al, 2003). Dicer-2 and R2D2 were found in holo-RISC and in the RISC-loading complex (RLC) of the Drosophila cell extract (Pham et al, 2004; Tomari et al, 2004a). The heterodimer of Dicer-2 and R2D2 is capable of selecting a guide strand from siRNA duplex by sensing the thermodynamic stability of both ends of the siRNA duplex (Tomari et al, 2004b). Recently, TRBP (HIV-1 TAR RNA-binding protein), a human homolog of Loqs, was reported to interact with Dicer and human Ago2 (hAgo2). The depletion of TRBP by RNAi causes defects of siRNA- or miRNA-mediated RNA silencing processes in human cell lines (Chendrimada et al, 2005; Haase et al, 2005).
PACT is similar to TRBP in the domain structure, implicating that PACT may be another homolog of Loqs (Saito et al, 2005). PACT was initially known as a protein activator of PKR (Patel and Sen, 1998). Through its dsRBD1 and dsRBD2, PACT interacts with PKR (Peters et al, 2001). The dsRBD3 at the C-terminus of PACT functions as a PKR activation domain (Peters et al, 2001). Contrary to PACT, TRBP was proposed as an inhibitor of PKR. TRBP interacts with PKR in a very similar manner with PACT, except that its dsRBD3 functions as the PKR inhibition domain, instead of as the activation domain (Gupta et al, 2003). Thus, PACT and TRBP have opposite roles in the regulation of PKR, although their domain structures are very similar to each other.
In this study, we explore the role of dsRNA-binding proteins in RNA silencing and identify PACT as a novel component of human RISC.
Results
PACT interacts with Dicer
To identify the cofactor for Dicer in humans, various dsRNA-binding proteins were fused to V5 tag and coexpressed with FLAG-Dicer protein in HEK293T cells for coimmunoprecipitation and Western blot analysis. Among these proteins, we found that PACT and TRBP interacted efficiently with Dicer (Figure 1A), whereas DGCR8, SPNR, Staufen, and two hypothetical dsRNA-binding proteins (FLJ20036 and FLJ20399) did not bind significantly to Dicer (data not shown). Immunocytochemistry for PACT, TRBP, and Dicer fused to V5 tag indicated that they are localized primarily in the cytoplasm (Supplementary Figure 1). The localization patterns of Dicer, PACT, and TRBP are indistinguishable from each other.
Figure 1.
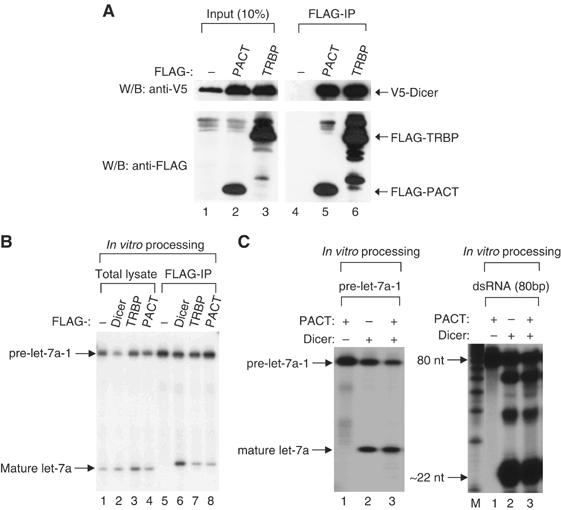
PACT interacts with Dicer, but does not facilitate the cleavage activity. (A) Immunoprecipitation followed by Western blotting. V5-Dicer protein was coexpressed in HEK293T cells with FLAG-PACT, or FLAG-TRBP protein. Immunoprecipitation was carried out using anti-FLAG antibody and the protein was visualized using either anti-V5 or anti-FLAG antibody. (B) In vitro pre-miRNA processing assay. Total HEK293 T extract (total lysate) or immunoprecipitates (FLAG-IP) of FLAG-Dicer, FLAG-TRBP, or FLAG-PACT were incubated with 5′ end radiolabeled pre-let-7a-1. The RNA was extracted by phenol extraction method, followed by separating on 15% urea-PAGE. (C) In vitro processing assay using recombinant human Dicer and PACT. In all, 200 ng of recombinant PACT prepared from E. coli and 0.25 U of recombinant human Dicer (Stratagene) were used in each reaction. Pre-let-7a-1 (left panel) and long dsRNA of 80 bp (right panel) were used as substrates.
When FLAG-PACT or FLAG-TRBP proteins were immunoprecipitated and incubated with pre-miRNA, the immunoprecipitates were capable of cleaving pre-let-7a-1 into ∼22 nt fragments (Figure 1B). This result demonstrates that PACT is associated with Dicer-processing activity.
To examine the role of PACT in processing reaction, recombinant human Dicer protein was incubated with pre-miRNA or long dsRNA in the presence or absence of recombinant PACT prepared from Escherichia coli (Figure 1C). The addition of recombinant PACT did not increase the processing activity of Dicer. Although it is possible that the recombinant protein used in this experiment was simply inactive, these data advocate that PACT may not be required for the Dicer-processing reaction itself and is instead involved in other step(s) along the RNA silencing pathway, as is the case for TRBP in humans (Chendrimada et al, 2005) and R2D2 in Drosophila (Liu et al, 2003).
Domains important for the interaction between Dicer and PACT
To examine which part of PACT is required for Dicer binding, we generated three point mutants (mDR1, mDR2, and mDR3) (Figure 2A). Point mutagenesis was carried out to change the highly conserved amino acids in dsRBDs. The dsRBD1 was mutated at the two conserved alanines (A91 and A92) to lysine residues in the mutant mDR1. Similarly, the conserved alanines at 185 and 186 of the dsRBD2 and the alanine residues of the dsRBD3 at 289 and 290 were converted to lysine residues in the mutants mDR2 and mDR3, respectively. These proteins were fused to the FLAG epitope at the N-termini and coexpressed with V5-Dicer for coimmunoprecipitation experiments. The PACT mutants, mDR1 and mDR2, efficiently bound to Dicer, while the C-terminal mutant (mDR3) was unable to interact with Dicer (Figure 2B). Therefore, the third dsRBD of PACT may be responsible for the interaction with Dicer. TRBP also behaves similarly in that only the third dsRBD mutant was affected in Dicer binding (data not shown), which agrees with the recent study on TRBP using yeast two hybrid assay by Haase et al (2005).
Figure 2.
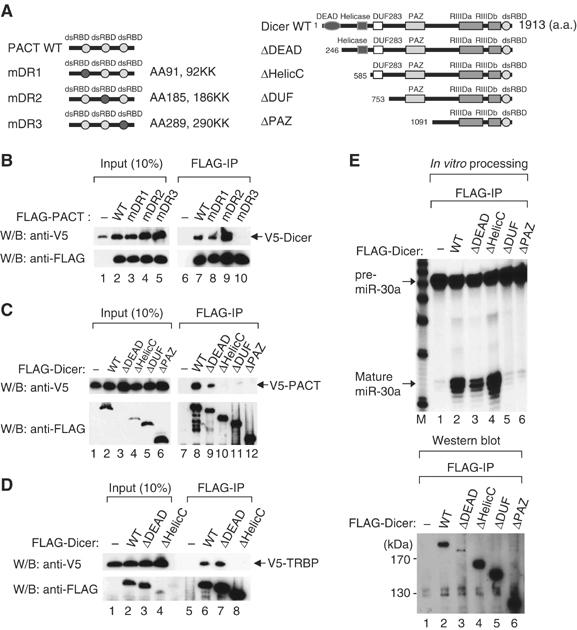
Domains responsible for the interaction of PACT and TRBP with Dicer. (A) Schematic representation of a series of mutants of Dicer and PACT. (B) Immunoprecipitation followed by Western blotting. FLAG-Dicer protein was coexpressed in HEK293T cells with PACT mutant proteins fused to a V5 tag. Immunoprecipitation was carried out using anti-FLAG antibody and the protein was visualized using either anti-V5 or anti-FLAG antibody. (C) The same experiment as described in (B), except for using FLAG-Dicer mutants and V5-PACT (wild type). (D) The same experiment as described in (A), except for using FLAG-Dicer mutants and V5-TRBP (wild type). (E) In vitro pre-miRNA processing assay using FLAG-Dicer mutants. Immunoprecipitates of FLAG-Dicer mutants were incubated with 5′ end radiolabeled pre-miR30a followed by RNA extraction and separation on 15% urea-PAGE (upper panel). The amount of FLAG-Dicer mutants used in in vitro pre-miRNA processing reactions was visualized using anti-FLAG antibody (lower panel).
Next, we looked for the region in Dicer that is required for binding to PACT or TRBP. Four deletion mutants were generated (ΔDEAD, ΔHelicC, ΔDUF, and ΔPAZ) (Figure 2A). The Dicer mutants, ΔHelicC, ΔDUF, and ΔPAZ, were unable to interact with either PACT or TRBP. Thus, the highly conserved helicase motif whose biochemical role in Dicer remains unknown may be critical for the interaction with PACT and TRBP (Figure 2C and D).
Interestingly, when the Dicer mutant proteins were immunoprecipitated and assayed for pre-miRNA processing activity, ΔHelicC mutant lacking the helicase motif was capable of processing (Figure 2E, lane 4). This result shows that the helicase motif is dispensable for processing and that PACT and TRBP may not be required for the Dicer cleavage reaction itself. It is noted that the DUF283 domain whose function remains unknown may be critical for processing, as the mutant lacking this domain (ΔDUF) lost the activity (Figure 2E, lane 5).
PACT binds to hAgo2
Both Dicer and TRBP have previously been shown to interact with hAgo2 (Chendrimada et al, 2005). To test whether PACT also associates with hAgo2, FLAG-PACT protein was coexpressed with V5-hAgo2 (Figure 3). Coimmunoprecipitation followed by Western blotting revealed that PACT is indeed associated with hAgo2 (Figure 3A). The dsRBD3 mutant of PACT is capable of binding to hAgo2, indicating that PACT may bind to hAgo2 independently of Dicer (Figure 3B). Like PACT, the mDR3 mutant of TRBP bound to hAgo2 efficiently (data not shown), suggesting that the mode of interaction between TRBP and hAgo2 is similar to that between PACT and hAgo2.
Figure 3.
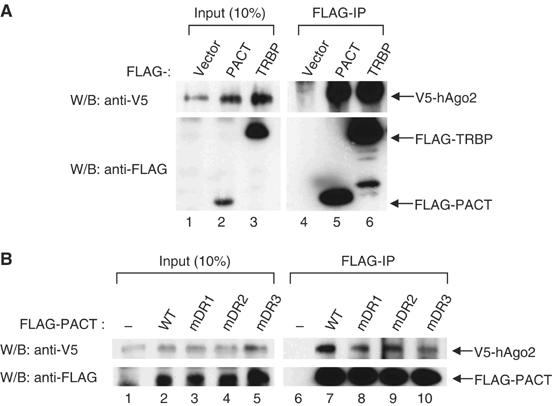
PACT binds to hAgo2. (A) Immunoprecipitation followed by Western blotting. FLAG-PACT or FLAG-TRBP proteins, V5-Dicer and V5-hAgo2 protein were coexpressed in HEK293T cells. Immunoprecipitation was carried out using anti-FLAG antibody and the protein was visualized using either anti-V5 or anti-FLAG antibody. (B) The same experiment as described in (A), except for using FLAG-PACT mutants and V5-hAgo2 (wild type). In this experiment, V5-Dicer was not coexpressed.
PACT, TRBP, and Dicer are associated with the slicer activity in vitro
We presented in Figure 1B that PACT and TRBP are associated with pre-miRNA processing activity, indicating that the complex(es) with Dicer may be preassembled prior to the pre-miRNA processing step. A similar in vitro processing experiment was performed by incubating immunoprecipitated FLAG-hAgo2 protein with 5′ endlabeled pre-miR-30a (Figure 4A, lane 2). Intriguingly, the FLAG-hAgo2 immunoprecipitate was also capable of executing pre-miRNA processing. This result supports the recent findings that hAgo2 may be preassembled in the pre-miRNA processing complex in human cells (Gregory et al, 2005; Maniataki and Mourelatos, 2005; Meister et al, 2005). Further supporting this notion, when the beads were washed after processing reaction, the miRNA product was found to be associated with hAgo2, Dicer, TRBP, and PACT (Figure 4A, lanes 6–10).
Figure 4.

Both PACT and TRBP are associated with the slicer activity in vitro. (A) In vitro pre-miRNA processing assay and the detection of mature miRNA bound to the RISC components. Dicer, hAgo2, TRBP, or PACT fused to FLAG was expressed in HEK293T cells and immunoprecipitated in RNAi reaction buffer. The immunoprecipitates were incubated with 5′-end radiolabeled pre-miR-30a. The left panel shows the RNA extracted from each reaction tube. After in vitro processing, the beads were washed with RNAi reaction buffer and treated with phenol to extract the RNA associated with the proteins (right panel). (B) In vitro RNAi assay. Dicer, hAgo2, TRBP, or PACT fused to FLAG was expressed in HEK293T cells and immunoprecipitated in RNAi reaction buffer. The immunoprecipitates were incubated with cold pre-miR-30a or 27mer siRNA. After washing, the immunoprecipitates were incubated with 5′-end radiolabeled target RNA that is complementary to miR-30a-5p.
Next, we asked if PACT and TRBP are present in the active RISC using the in vitro RNAi protocol that was recently developed by Maniataki and Mourelatos (2005) (Figure 4B). The FLAG-fused proteins were expressed in HEK293T cells and immobilized on anti-FLAG beads. The immunoprecipitates were then incubated with cold pre-miR-30a to allow pre-miRNA processing. As a control, 27-mer siRNA, which does not contain a complementary sequence to target RNA, was used. The beads were washed twice to remove excess pre-miRNA and then incubated with labeled target RNA, which is complementary to miR-30a-5p. Not only FLAG-hAgo2, but also FLAG-Dicer, FLAG-TRBP and FLAG-PACT mediated target cleavage reaction (Figure 4B). This result implies that PACT and TRBP are associated with hAgo2 in the functional RISC. Therefore, hAgo2, Dicer, PACT and TRBP may be preassembled prior to pre-miRNA processing and remain together to the final RISC, although the stoichiometry and the conformation of the complex are likely to change over various steps during processing and RISC assembly (Maniataki and Mourelatos, 2005).
We further characterized the PACT complex by fractionating the total cell extract from 293T cells that express FLAG-PACT. The FLAG-PACT protein was then immunoprecipitated from each fraction using anti-FLAG antibody (Figure 5). Next, the immunoprecipitates were analyzed by Western blotting with anti-TRBP antibody to examine the association of TRBP (Figure 5A). TRBP was coimmunoprecipitated in all fractions where PACT was present, indicating that PACT interacts with TRBP. We then assayed the activities of Dicer and hAgo2 by the coupled processing–RNAi assay described in Figure 4 (Maniataki and Mourelatos, 2005). The RISC activity was precipitated with PACT in fractions corresponding to ∼500 kDa (Figure 5B). Therefore, PACT associates with Dicer, hAgo2, and TRBP in a complex of ∼500 kDa that is capable of pre-miRNA processing and target cleavage. This complex may be comparable to the Dicer–TRBP–hAgo2 complex described recently by Shiekhattar and colleagues (Chendrimada et al, 2005; Gregory et al, 2005).
Figure 5.
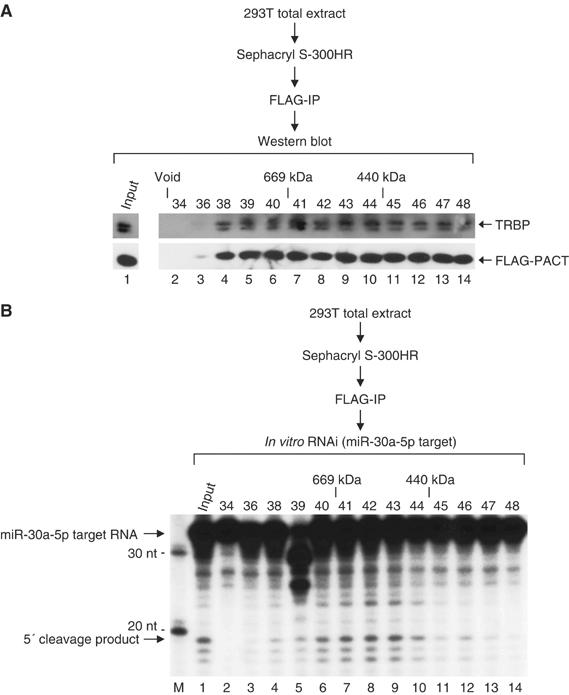
PACT and TRBP coexist in the ∼500 kDa RISC. Total extract was prepared from HEK293T cells transiently transfected with FLAG-PACT, V5-Dicer, and V5-hAgo2 expression vectors and fractionated through a Sephacryl-S300 HR column. Each fraction was subject to immunoprecipitation with anti-FLAG antibody. The immunoprecipitate was used for (A) Western blot analysis using anti-TRBP or anti-FLAG antibody, or (B) in vitro RNAi assay using miR-30a-5p target RNA. The fraction number and the protein molecular mass standard (Sigma) are indicated at the top of the figures.
PACT is required for siRNA-induced RNAi
To examine whether PACT is required for RNAi reaction induced by siRNA, we transfected siRNA against PACT, TRBP, and Dicer together with siRNA against luciferase and the luciferase reporter constructs (Figure 6A). siRNA against green fluorescence protein (siGFP) was used as a control. Depletion of PACT resulted in the reduction of the efficiency of RNAi against luciferase (Figure 6A). Although the effect of RNAi against TRBP was reproducibly stronger than that against PACT (Figure 6A), our data indicate that PACT may also be required for RNA silencing pathway.
Figure 6.
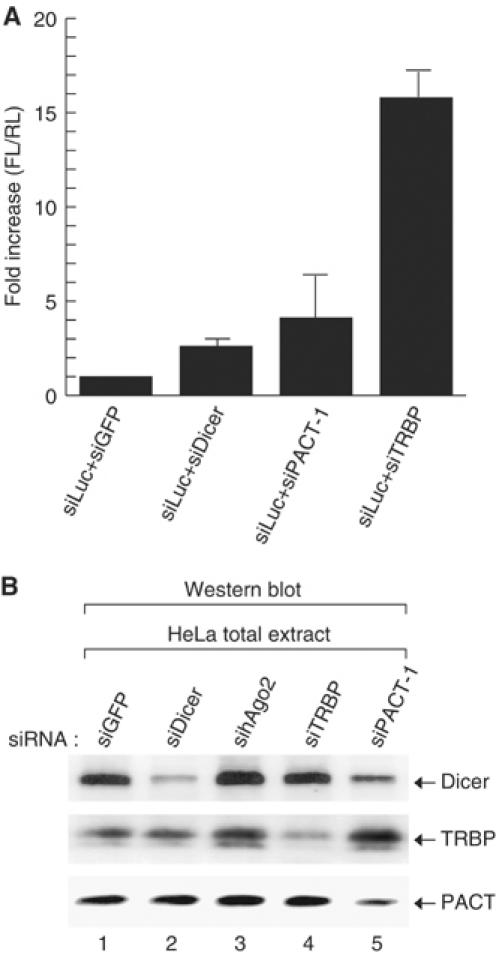
PACT is required for siRNA-induced RNAi. (A) HeLa cells were transiently transfected with siRNA targeting Dicer, PACT, or TRBP along with siRNA against firefly luciferase, firefly luciferase-expression plasmid, and renilla luciferase-expression plasmid. The firefly luciferase activity was normalized against renilla luciferase activity. This experiment was performed in triplicate. Error bars indicate standard deviations. (B) Western blot analysis. siRNA targeting GFP, Dicer, hAgo2, TRBP, or PACT was transfected into HeLa cells. Following transfection (48 h), total extract was prepared in lysis buffer (10 mM Tris, pH 7.4, 1 mM EDTA, 500 mM NaCl, 0.5% Triton-X100). In all, 120 μg of total extract was loaded on 10% SDS–PAGE. Endogenous Dicer, TRBP, or PACT was visualized using anti-Dicer, anti-TRBP, or anti-PACT antibody.
RNAi against TRBP did not significantly affect the level of Dicer protein, which is consistent with the finding by Haase et al (2005) (Figure 6B). When PACT was depleted, the level of Dicer decreased slightly, suggesting that PACT, but not TRBP, may contribute to the stabilization of Dicer protein (Figure 6B).
PACT is required for the accumulation of mature miRNA
We then investigated whether PACT has any responsibility for miRNA biogenesis in vivo (Figure 7A). PACT, TRBP, Dicer, or exportin-5 was depleted by RNAi from a HeLa cell line that expresses pri-miR-30a from the tetracycline-inducible promoter. Reduction of the target mRNA (PACT, TRBP, Dicer, exportin-5) was verified by RT–PCR (Figure 7C). After incubation with siRNA, the cell line was exposed to doxycycline, the derivative of tetracycline, for induction of pri-miR-30a. Northern blotting on miR-30a-5p indicated that PACT may be required for the accumulation of newly synthesized miRNA. Interestingly, the depletion of PACT resulted in stronger effects on miRNA compared to that of TRBP. We also generated an additional HeLa cell line that expresses pri-miR-124a from the tetracycline-inducible promoter (Figure 7B). Accumulation of miR-124a was dependent on PACT in this cell line.
Figure 7.
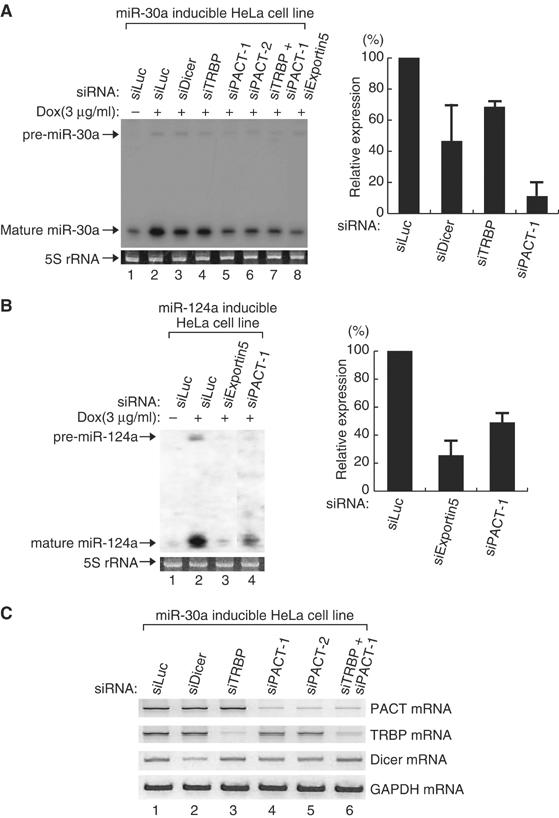
PACT is required for the accumulation of mature miRNA. (A) Northern blot analysis. siRNA targeting Dicer, TRBP, or PACT was transfected into a HeLa cell line that expresses primary miR-30a under the control of the tetracycline-inducible promoter. Following transfection (48 h), doxycycline (3 μg/ml) was added to the medium to induce the expression of miR-30a. After 24 h, the cells were harvested for Northern blotting (the left panel). After normalization for loading, the relative ratios for mature miR-30a were calculated and normalized to no doxycycline-treated experiment (lane 1). The average value obtained from two independent experiments was presented in the right panel. (B) The same experiment as described in (A), except for using pri-miR-124a inducible HeLa cell line. The left panel shows a representative Northern blot result. The right panel presents the mean value of relative expression level for mature miR-124a obtained from two independent experiments. (C) RT–PCR. Total RNA used in (A) was reverse-transcribed and amplified by PCR using gene-specific primers.
It is noted that the endogenous levels of miR-16 and let-7a in unmodified HeLa cells were changed only slightly after the depletion of PACT (data not shown). When TRBP was knocked down in the same condition, we did not observe significant changes in miRNA level (data not shown), which is consistent with the data of Filipowicz and colleagues (Haase et al, 2005). This also supports the previous observations that mature miRNA is highly stable in cells and therefore the changes in the steady-state level of mature miRNA may be difficult to detect (Lee et al, 2003; Yi et al, 2003; Lund et al, 2004).
Discussion
In this study, we present PACT as another dsRNA-binding protein that functions as a component of the human RISC. Human Dicer may associate with two different dsRNA-binding proteins, PACT and TRBP. Our in vitro pre-miRNA processing assay using a Dicer mutant lacking the helicase motif indicates that Dicer does not need a cofactor for pre-miRNA cleavage (Figure 2). However, the depletion of PACT by RNAi resulted in a reduction of mature miRNAs (Figure 7). These seemingly paradoxical data can be explained if PACT has a function in RISC formation. If a step of RNA loading into Argonaute protein is blocked, these small RNAs may become unstable. In fact, when PACT was depleted in HeLa cells, the decrease of mature miRNA, but not the accumulation of pre-miRNA, was monitored by Northern blot analysis (Figure 7). Our results suggest that PACT may play a role at a certain step of the RISC assembly, but not at the pre-miRNA cleavage step.
In flies, two different dsRBD proteins, Loqs/R3D1 and R2D2, interact with Dicer-1 and Dicer-2, respectively. Loqs/R3D1 facilitates pre-miRNA cleavage by Dicer-1, while R2D2 is not required for dsRNA cleavage, but instead participates in strand selection. The deletion of PRBP, a mouse homolog of TRBP, yields viable mice that often die at weaning, while knocking out Dicer results in embryonic lethality (Zhong et al, 1999; Bernstein et al, 2003), implicating that in mice, PRKRA, a mouse homolog of PACT, may compensate for PRBP during development (Chendrimada et al, 2005; Haase et al, 2005).
Our study supports this possibility that, in humans, the roles of dsRBD-containing Dicer cofactors may be partially redundant because RNAi experiments indicate that both PACT and TRBP may be involved in siRNA-induced RNAi and in miRNA accumulation. They also behave similarly in the modes of protein interaction: (1) two proteins share the common binding region in Dicer (246–584 a.a.), and (2) the third dsRBDs of PACT and TRBP are important for binding to Dicer.
However, our data also suggest that the roles of PACT and TRBP may be differentiated to some extent. The depletion of PACT resulted in a strong effect on miRNA accumulation (Figure 7) and a relatively moderate effect on RNAi reporter assay (Figure 6). In contrast, RNAi against TRBP had only limited effects on miRNA level, whereas the same siRNA induced clear effects on RNAi efficiency. One plausible possibility is that PACT is more suitable for the role in the transition step of miRNA duplex into single-strand miRNA, while TRBP is more active in the later stages of RISC formation and target cleavage. Thus, the roles of PACT and TRBP may be differentiated in normal conditions, but they may act redundantly to some extent in the absence of the other.
Our results from the coupled in vitro processing–RNAi assay support and extend the findings by Maniataki and Mourelatos (2005) that the protein components of the RISC complex seem to be preassembled prior to pre-miRNA processing in humans (Figure 4). Comparable results were reported recently by other groups (Gregory et al, 2005; Maniataki and Mourelatos, 2005; Meister et al, 2005). Gel filtration and coimmunoprecipitation experiments revealed that hAgo2, Dicer, TRBP, and PACT interact with one another, forming the RISC of ∼500 kDa (Figure 5). In Drosophila, the assembly of the RISC appears to be achieved in a stepwise manner. The Dicer-2–R2D2 heterodimer binds to siRNA duplex to form the R1 complex that is later converted into the intermediate complex R2/RLC (RISC loading complex). Subsequently, R2/RLC recruits Ago2 to form the mature RISC, which likely acts as the RNAi effector in Drosophila (Pham and Sontheimer, 2005). Our data indicate that the mechanism of human RISC formation may be different from that of flies. It is also possible that the siRISC assembly process may differ from that of miRISC.
PACT and TRBP are known as regulators of PKR. PKR is a dsRNA-dependent serine/threonine protein kinase, which phosphorylates eIF2α on Ser51 to cause a general reduction of protein synthesis (Taylor et al, 2005). Besides eIF2α, several proteins are known to be phosphorylated by PKR, such as NFAR-1, NFAR-2, and human protein phosphatase 2A (PP2A) regulatory subunit B56α (Xu and Williams, 2000; Saunders et al, 2001). It is possible that the component(s) of RISC may be regulated by PKR through phosphorylation and that PACT and TRBP may regulate PKR activity to control the RISC activity. Alternatively, a competition between the RNA silencing pathway and the PKR pathway may exist, because the third dsRBDs of both PACT and TRBP are responsible for their interaction with Dicer as well as for the regulation of PKR. It would be intriguing to investigate the possibility of the crosstalk between the RNA silencing pathway and the PKR pathway.
Materials and methods
Cloning of PACT and TRBP
PCR products of PACT and TRBP were subcloned into FLAG-pCK vector for expression in human cells, at the BamHI and XhoI sites. Primer sequences used for PCR are as follows. For PACT, 5′-GGATCCATGTCCCAGAGCAGGCACC-3′ (forward) and 5′-CTCGAGCTCCAGATTTACTTTCTTTCTGC-3′ (reverse) were used. For TRBP, 5′-GGATCCATGAGTGAAGAGGAGCAAGGCTC-3′ (forward) and 5′-CTCGAGTCAGGCAGTGAAGAGTCTGCTG-3′ (reverse) were used.
Mutagenesis of PACT and Dicer
Point mutagenesis of PACT was carried out by using a site-directed mutagenesis kit (Stratagene) according to the manufacturer's manual. Primer sequences used for mutagenesis are as follows. For PACT mDR1 mutant, 5′-CTGGCGAAACATAGAAAGAAGGAGGCTGCCAT AAAC-3′ (upper primer) and 5′-GTTTATGGCAGCCTCCTTCTTTCTATGTTTCG CCAG-3′ (lower primer) were used. For PACT mDR2 mutant, 5′-GCAAGCCAAAAGGAATAAGAAGGAGAAATTTC TTGCC-3′ (upper primer) and 5′-GGCAAGAAATTTCTCCTTCTTATTCCTTTTGG CTTGC-3′ (lower primer) were used. For PACT mDR3 mutant, 5′-GCAATGCACAAAGTGATAAGAAGCACAATGCT TTGCAG-3′ (upper primer) and 5′-CTGCAAAGCATTGTGCTTCTTATCACTTTGTG CATTGC-3′ (lower primer) were used.
To prepare Dicer deletion mutants, PCR products of Dicer deletion mutants were subcloned into FLAG-pcDNA3.1 vector (Invitrogen) for expression in human cells, at the HindIII and ApaI sites. Primer sequences used for PCR are as follows. For ΔDEAD, 5′-AAGCTTTATACTTCTCAGCCATGTGAG-3′ (forward) and 5′-CACAGTCGAGGCTGATCAG-3′ (reverse) were used. For ΔHelicC, 5′-AAGCTTATCTTGAGAAACAAGTGTTCC-3′ (forward) and 5′-CACAGTCGAGGCTGATCAG-3′ (reverse) were used. For ΔDUF, 5′-AAGCTTATTCCAGAGTGTTTGAGGGATAG-3′ (forward) and 5′-CACAGTCGAGGCTGATCAG-3′ (reverse) were used. For ΔPAZ, 5′-AAGCTTTACCCTAACTTAGACTTCG-3′ (forward) and 5′-CACAGTCGAGGCTGATCAG-3′ (reverse) were used.
Cell culture and transfection
HeLa cells and HEK293T cells were cultured in DMEM (WelGENE) supplemented with 10% FBS (WelGENE). Transfection was carried out by calcium-phosphate method.
Immunoprecipitation
HEK293T cells grown in a 10-cm dish were collected in 500 μl of ice-cold buffer D-K′100 (20 mM Tris, pH 8.0, 100 mM KCl, 0.2 mM EDTA, 0.2 mM PMSF) 48 h post-transfection. The cells were sonicated on ice and centrifuged at 13 200 r.p.m. for 15 min at 4°C. The supernatant was incubated with 10 μl of anti-FLAG antibody conjugated to agarose beads (anti-FLAG M2 affinity gel, Sigma) with constant rotation for 60 min at 4°C. The beads were washed five times in buffer D-K′100, drained, and used for Western blot analysis.
In vitro RNAi assay
HEK293T cells grown in two 10-cm dishes were collected in 500 μl of ice-cold buffer D-K′100 48 h post-transfection. The cells were sonicated on ice and centrifuged at 13 200 r.p.m. for 15 min at 4°C. The supernatant was incubated with 20 μl of anti-FLAG antibody conjugated to agarose beads (anti-FLAG M2 affinity gel, Sigma) with constant rotation for 2 h at 4°C. The beads were washed three times in buffer D-K′100, and were washed three times in RNAi reaction buffer (30 mM HEPES, pH 7.5, 40 mM potassium acetate, 5 mM magnesium acetate, 5 mM DTT). The processing reactions were performed in a total volume of 20 μl that consisted of 15 μl of the beads from immunoprecipitation, 1 μl of ribonuclease inhibitor (40 U/μl, TAKARA), 3 μl of RNAi reaction buffer, and 1 μl of synthetic pre-miR-30a (10 μM). As a control, 1 μl of 27mer siRNA (10 μM) was used. The sequences of 27mer siRNA are 5′-CCAUGUGAUCCGAAAGUUAGGCUdGdG-3′ (sense) and 5′-CCAGCCUAACUUUCGGAUCACAUGGUA-3′ (antisense). The reaction mixture was incubated at 37°C for 90 min. Before target RNA cleavage reaction, the beads were washed twice with RNAi reaction buffer. In all, 1 μl of ribonuclease inhibitor (TAKARA), 0.5 μl of the radiolabeled target RNA (miR-30a-5p target RNA, 1 × 104–1 × 105 c.p.m.), and 0.1 μl of yeast RNA (1 μg/μl, Ambion) were added to 20 μl of the washed beads. The reaction mixture was incubated at 37°C for 90 min. RNA was extracted from the reaction mixture by phenol extraction and analyzed on 15% denaturing urea polyacrylamide gel. The sequence of miR-30a-5p target RNA is 5′-AACCUUGCUUCCAGUCGAGGAUGUUUACACCA AG-3′. Pre-miR-30a, 27mer control siRNA and miR-30a-5p target RNA were purchased from Samchully Pham Co. Ltd. The miR-30a-5p target RNA was labeled at the 5′ end using T4 polynucleotide kinase (TAKARA) and [γ-32P]ATP.
Gel exclusion chromatography
Gel exclusion chromatography was carried out as described previously (Han et al, 2004), with the following modifications. HEK293T cells grown in 20 10-cm dishes were cotransfected with FLAG-PACT, V5-Dicer, and V5-hAgo2 expression vectors. Total cell extract was prepared in buffer D′-K100 by sonication and centrifugation, and then concentrated to the final volume of 1 ml using Centricon YM-30 (Millipore). This total extract was treated with 10 μl of 10 mg/ml RNase A and incubated at 4°C for 30 min. This extract was loaded on a Sephacryl S-300 HR (Sigma) column and fractionated. Each fraction was 1.65 ml in volume. In all, 1.6 ml of each fraction was taken for immunoprecipitation with anti-FLAG antibody, and the immunoprecipitate was used for Western blot analysis using anti-TRBP or anti FLAG antibody or for in vitro RNAi assay.
In vitro processing of pre-miRNAs
In vitro processing of pre-miRNAs using FLAG-immunoprecipitates or recombinant human Dicer (Stratagene) was carried out as described previously (Lee et al, 2002, 2003). Briefly, 30 μl of processing reaction contained 3 μl of 64 mM MgCl2, 1 U/μl of ribonuclease inhibitor (TAKARA), the radiolabeled pre-miRNA of 1 × 104–1 × 105 c.p.m., and 15 μl of the beads from immunoprecipitation. In Figure 1C, 0.25 U of recombinant Dicer and 200 ng of recombinant PACT were used instead of the beads from immunoprecipitation. The reaction mixture was incubated at 37°C for 90 min. RNA was extracted from the reaction mixture by phenol extraction and analyzed on 15% denaturing polyacrylamide gel. Pre-let7a-1 was prepared from in vitro processing of cold pri-let7a-1 using Drosha-FLAG followed by gel purification. Pre-miR-30a and pre-let7a-1 were labeled at the 5′ end using T4 polynucleotide kinase (TAKARA) and [γ-32P]ATP.
RNAi and luciferase assay
HeLa cells cultured in six-well plates were cotransfected with 720 ng of pGL3-CMV firefly luciferase, 80 ng of pRL-CMV renilla luciferase, and 20 nM of siRNA against firefly luciferase, and 100 nM of siRNA against GFP, Dicer, PACT or TRBP. Lipofectamine 2000 (Invitrogen) was used for transfections, using the manufacturer's protocols. Transfections were performed in triplicate. Cells were lysed 48 h after transfection and assayed for luciferase activity using the Dual-Luciferase Reporter system (Promega) as described by the manufacturer. All firefly luciferase activities were normalized to renilla luciferase activities to correct for transfection efficiency. The target sequence of siDicer has been described previously (Lee et al, 2003). The target sequences of siPACT-1, siPACT-2, siTRBP, siExportin5, sihAgo2, siGFP, and siLuciferase are 5′-GAACCAGCUUAAUCCUAUU-3′ (siPACT-1), 5′-AGGAAUGCUGCUGAGAAAU-3′ (siPACT-2), 5′-CACGUCAGGCUUACCUGGAUA-3′ (siTRBP), 5′-CUCGAUUGGAGAAGGUGUA-3′ (siExportin5), 5′-GCACGGAAGUCCAUCUGAAGU-3′ (sihAgo2), 5′-UGAAUUAGAUGGCGAUGUU-3′ (siGFP), and 5′-CUUACGCUGAGUACUUCGA-3′ (siLuciferase), respectively. All siRNAs were manufactured by Samchully Pham Co. Ltd.
Purification of recombinant proteins
To prepare recombinant PACT, cDNA was subcloned into pGEX-4T3 vector (Amersham), which has GST tag at the N-terminus, using BamHI and XhoI sites. The PACT expression clone was transformed to the E. coli BL21-RIL strain. The expression and purification of recombinant PACT was conducted according to the manufacturer's protocol. After purification of GST-PACT, GST was removed from GST-PACT by treatment with 1 U/ml of Thrombin at 16°C for 16 h. TRBP protein was prepared similarly. Both recombinant proteins were used for in vitro assays and for immunization to raise polyclonal antibodies in rabbits.
Northern blot analysis
To prepare the pri-miR-30a or pri-miR-124a inducible cell line, the insert containing pri-miR-30a genomic sequences, which was excised from pGEM-T-easy-pri-miR-30a (Lee et al, 2002) using NotI restriction enzyme, was subcloned into pTRE2-Hyg vector (Clontech) using NotI sites. Pri-miR-124a expression construct was cloned by PCR amplification of HeLa genomic DNA and inserted into the NotI site of the pTRE2-Hyg vector (Clontech). Tet-on HeLa cell line (Clontech) was transfected with the pri-miR-30a or pri-miR-124a inducible expression vector using Lipofectamine 2000 (Invitrogen). The selection of inducible cell line was conducted according to the manufacturer's protocol.
Pri-miR-30a or pri-miR-124a inducible cell line cultured in six-well plates was transfected with 100 nM of siRNAs using Lipofectamine 2000 (Invitrogen). In all, 3 μg/ml of doxycycline was added after 48 h. Then, total RNA was isolated using TRIzol reagent (Invitrogen) after 24 h. Total RNAs (25 μg) were resolved on 12.5% urea-polyacrylamide gels and transferred electronically to Zeta-probe membrane (BioRad). An oligonucleotide complementary to miR-30a-5p or miR-124a was endlabeled with [γ-32P]ATP and used as the probe for Northern.
RT–PCR
In all, 5 μg of HeLa total RNA was used for the first-strand cDNA synthesis with SUPERSCRIPT II and oligo-dT primers (Invitrogen). To detect the expression level of mRNAs, the following primers were used. For PACT, 5′-GGATCCATGTCCCAGAGCAGGCACC-3′ (forward) and 5′-CTCGAGCTCCAGATTTACTTTCTTTCTGC-3′ (reverse); for TRBP, 5′-GGATCCATGAGTGAAGAGGAGCAAGGCTC-3′ (forward) and 5′-CTCGAGTCAGGCAGTGAAGAGTCTGCTG-3′ (reverse); for Dicer, 5′-AAGCTTATTCCAGAGTGTTTGAGGGATAG-3′ (forward) and 5′-CACAGTCGAGGCTGATCAG-3′ (reverse); for GAPDH, 5′-CCCATCACCATCTTCCAGGAGTGAGTGGAAGA C-3′ (forward) and 5′-CGCCCCACTTGATTTTGGAGGGATCTCGCCTA CCG-3′ were used for PCR amplification.
Supplementary Material
Supplementary Figure 1
Acknowledgments
We thank Drs Zissimos Mourelatos, Roe Hyun Seong, and Joo Yong Lee, and the members of our laboratory for their input and participation in helpful discussions. This work was supported by the Basic Research Program (R02-2004-000-10173-0) from the Ministry of Education and Human Resources Development and by the National Research Laboratory Program (M1050000010905J000010910), the SRC program of KOSEF (R11-2005-009-01003-0), and the Molecular and Cellular BioDiscovery Research Program (2004-01749) from the Ministry of Science and Technology.
References
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Baulcombe D (2005) RNA silencing. Trends Biochem Sci 30: 290–293 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ (2003) Dicer is essential for mouse development. Nat Genet 35: 215–217 [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Doi N, Zenno S, Ueda R, Ohki-Hamazaki H, Ui-Tei K, Saigo K (2003) Short-interfering-RNA-mediated gene silencing in mammalian cells requires dicer and eIF2C translation initiation factors. Curr Biol 13: 41–46 [DOI] [PubMed] [Google Scholar]
- Filipowicz W (2005) RNAi: the nuts and bolts of the RISC machine. Cell 122: 17–20 [DOI] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD (2005) Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 3: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R (2005) Human RISC couples MicroRNA biogenesis and posttranscriptional gene silencing. Cell 123: 631–640 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Gupta V, Huang X, Patel RC (2003) The carboxy-terminal, M3 motifs of PACT and TRBP have opposite effects on PKR activity. Virology 315: 283–291 [DOI] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W (2005) TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 6: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN (2004) The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q (2005) Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev 19: 1674–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN (2005a) MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385 [DOI] [PubMed] [Google Scholar]
- Kim VN (2005b) Small RNAs: classification, biogenesis, and function. Mol Cells 19: 1–15 [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T (2004) The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167 [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21: 4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004a) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW (2004b) Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M (2004) Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol 11: 576–577 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X (2003) R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925 [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ (2004) Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429: 318–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ (2005) Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434: 666–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z (2005) A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev 19: 2979–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T (2005) Identification of novel Argonaute-associated proteins. Curr Biol 15: 2149–2155 [DOI] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D (2005) Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434: 663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RC, Sen GC (1998) PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J 17: 4379–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters GA, Hartmann R, Qin J, Sen GC (2001) Modular structure of PACT: distinct domains for binding and activating PKR. Mol Cell Biol 21: 1908–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ (2004) A Dicer-2-dependent 80 s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell 117: 83–94 [DOI] [PubMed] [Google Scholar]
- Pham JW, Sontheimer EJ (2005) Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J Biol Chem 280: 39278–39283 [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L (2005) Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC (2005) Processing of pre-microRNAs by the Dicer-1–Loquacious complex in Drosophila cells. PLoS Biol 3: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, Perkins DJ, Balachandran S, Michaels R, Ford R, Mayeda A, Barber GN (2001) Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J Biol Chem 276: 32300–32312 [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L (2003) The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol 10: 1026–1032 [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L (2004) Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437 [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ (2005) Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol 6: 127–138 [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Carthew RW (2004) Molecular biology. Argonaute journeys into the heart of RISC. Science 305: 1409–1410 [DOI] [PubMed] [Google Scholar]
- Taylor SS, Haste NM, Ghosh G (2005) PKR and eIF2alpha: integration of kinase dimerization, activation, and substrate docking. Cell 122: 823–825 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE, Zamore PD (2004a) RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116: 831–841 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD (2004b) A protein sensor for siRNA asymmetry. Science 306: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD (2005) Perspective: machines for RNAi. Genes Dev 19: 517–529 [DOI] [PubMed] [Google Scholar]
- Xu Z, Williams BR (2000) The B56alpha regulatory subunit of protein phosphatase 2A is a target for regulation by double-stranded RNA-dependent protein kinase PKR. Mol Cell Biol 20: 5285–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM (2003) Structure and conserved RNA binding of the PAZ domain. Nature 426: 468–474 [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ (2005) Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 19: 405–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B (2005) Ribo-gnome: the big world of small RNAs. Science 309: 1519–1524 [DOI] [PubMed] [Google Scholar]
- Zhong J, Peters AH, Lee K, Braun RE (1999) A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet 22: 171–174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1


