Abstract
Stabilization of telomere length in germline and highly proliferative human cells is required for long-term survival and for the immortal phenotype of cancer-derived cells. This is achieved through expression of telomerase reverse transcriptase (TERT), which synthesizes telomeric repeats through reverse transcription of its tightly associated RNA template (TR). The telomeric repeat binding factor TRF1 inhibits telomerase at telomeres in cis in a length-dependent manner to achieve telomere length homeostasis. Here we manipulate telomerase activity over a wide range in cancer and primary cells. Concomitant overexpression of TERT and TR was necessary and sufficient to substantially increase telomerase activity. Upon overexpression, more telomerase associated with telomeres and telomeres elongated at a constant rate (up to 0.8 kb/population doubling (PD)) in a length-independent manner. Thus, in less than 50 PDs, the length of telomeres increased 3–8-fold beyond physiological size, while telomere-bound TRF1 and TRF2 increased proportionally to telomere length. Thus, long telomeres do not permanently adopt a structural state that is non-extendible. A low cellular concentration of telomerase is critical to achieve preferential elongation of short telomeres and telomere length homeostasis.
Keywords: cancer, immortalization, ribonucleoprotein, telomerase, telomere length homeostasis
Introduction
Telomere shortening, which accompanies each round of chromosome replication in the absence of telomerase, limits the replicative potential of many primary human cells and is thought to provide powerful tumor-suppressive function. Conversely, telomerase expression in the germline, in somatic cells during early embryogenesis and in cancer-derived immortal cells allows telomere length homeostasis (Harley et al, 1990; Kim et al, 1994; Wright et al, 1996; Ulaner and Giudice, 1997; Bodnar et al, 1998; Vaziri and Benchimol, 1998). Many stem or stem-like cells in the adult human also exhibit some telomerase activity when stimulated to divide, which is thought to slow, but not prevent, telomere shortening. Telomere length regulation and telomerase expression in multicellular organisms may have evolved by opposing selective pressures to suppress tumor formation on one side, while not promoting premature cellular senescence in highly proliferative tissues on the other side.
Telomere length in telomerase-positive immortal cell lines and unicellular organisms is maintained within a narrow range that is specified by species or cell type (Smogorzewska and de Lange, 2004). This is mediated through protein complexes that bind along the telomeric tract, forming part of a negative feedback loop inhibiting telomere elongation in cis (van Steensel and de Lange, 1997; Ancelin et al, 2002; Smogorzewska and de Lange, 2004). In Saccharomyces cerevisiae, the number of Rap1 protein complexes binding along the telomeric tracts is proportional to telomere length, providing a so-called protein-counting mechanism (Marcand et al, 1997). The counting mechanism in yeast involves telomeres switching between telomerase-extendible and non-extendible states to mediate telomere length homeostasis (Teixeira et al, 2004). In humans, a similar counting mechanism is provided by the TRF-complexes bound along the telomere tract (van Steensel and de Lange, 1997; Smogorzewska et al, 2000; Ancelin et al, 2002). The single-strand telomeric DNA-binding protein POT1, which is recruited via the TRF-complex, can prohibit telomerase activity when bound to the chromosome 3′ end, and may transduce telomere length control from the TRFs to telomerase (Loayza and De Lange, 2003; Kelleher et al, 2005; Lei et al, 2005). However, it is not known whether, as in budding yeast, the frequency, rather than the extent of telomere elongation is the regulated feature in vivo.
Telomerase is a large (more than 1 MDa) ribonucleoprotein particle whose composition is not fully determined. The catalytic core of telomerase consists of the telomerase reverse transcriptase (TERT) and the RNA subunit (TR), which provides the template (Greider and Blackburn, 1989; Lingner et al, 1997; Weinrich et al, 1997; Beattie et al, 1998). Other telomerase-associated proteins may play a role in biogenesis and regulation. While hTERT is expressed only in telomerase-positive cells, hTR and other telomerase-associated proteins are ubiquitously expressed and are often considered as house-keeping genes (Feng et al, 1995). However, hTR steady-state levels are higher in human cancer cells (Avilion et al, 1996; Yi et al, 1999). Here we establish a system to increase telomerase activity in a wide range in primary and telomerase-positive cell lines, and analyze the consequences on telomere length homeostasis.
Results
Restriction of telomerase activity by both hTR and hTERT in telomerase-positive cell lines
To increase telomerase activity in telomerase-positive cells, we transiently transfected 293T cells with vectors carrying hTR under control of the U1 promoter and hTERT under control of the CMV promoter (Figure 1A and B). At 2 days after transfection, cell extracts were prepared and telomerase activity was quantitatively measured by RQ-TRAP, a modified version of the TRAP assay, in which telomere elongation products are amplified by quantitative RT–PCR (Wege et al, 2003). As shown in Figure 1C, telomerase activity was not considerably affected when either hTR or hTERT was individually overexpressed. In contrast, when both of these subunits were transiently co-overexpressed, a >200-fold increase in telomerase activity was observed. Similar results were obtained in HeLa cells (Supplementary Figure 1). To confirm this result, we performed direct telomerase assays in which a telomeric primer is extended by telomerase in the presence of [α-32P]dGTP and unlabeled dNTPs, and extension products are analyzed without prior amplification by PCR. As expected, telomerase activity in crude extracts derived from untransfected cells or from cells that individually overexpressed either hTR or hTERT was not sufficient to detect activity in this assay (Figure 1D, lanes 2–4). Telomerase activity from human telomerase-positive cells can only be detected upon PCR amplification of the products, as done in TRAP assays (Kim et al, 1994), or upon enrichment of telomerase (Morin, 1989). However, robust primer extension was observed upon concomitant overexpression of hTR and hTERT (Figure 1D, lane 5). Thus, both hTR and hTERT, but apparently no other telomerase subunits, limit in vitro telomerase activity in these cells. For simplicity, we refer below to the massive telomerase activity conferred by co-overexpression of hTR and hTERT as ‘super-telomerase'. This term does not imply that super-telomerase has different properties, but only that it is highly expressed and to distinguish it from cells in which hTERT alone is overexpressed.
Figure 1.
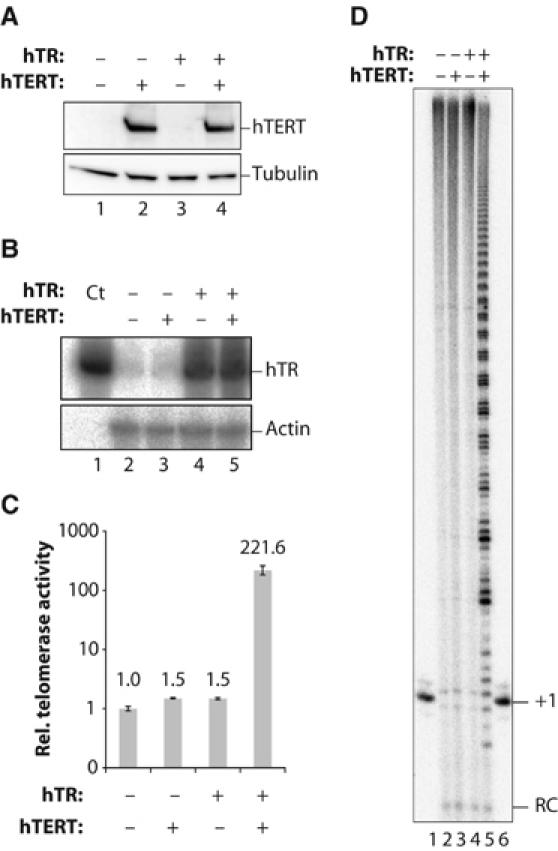
Massive increase of telomerase activity upon transient coexpression of hTR and hTERT in HEK293T cells. (A) Immunoblot with anti-hTERT R484 antibody (upper panel) or with anti-tubulin antibody (lower panel) as a loading control. (B) Northern blot with an anti-hTR probe (upper panel) or actin probe as a loading control (lower panel). Ct contains in vitro transcribed hTR. (C) Telomerase activity measured by RQ-TRAP. RNase treatment of extracts reduced the signal to below the detection limit (not shown). Values indicate mean telomerase activity and standard deviation obtained from three parallel transfections. Transfection efficiency was >90%, as estimated from a separate transfection performed with a GFP-encoding plasmid. (D) Telomerase activity was determined in a direct telomerase assay using (TTAGGG)3 as a telomerase substrate. Lanes 1 and 6, 32P-ddATP 3′-labeled primer as +1 marker. RC, recovery control (see Materials and methods). The periodicity is somewhat irregular, presumably due to the presence of nucleases in the crude extract that may attack extended and nonextended primers from both ends. Upon enrichment of this telomerase over one column, we obtained a very clear and regular six-nucleotide pattern (unpublished results).
High levels of telomerase activity result in continuous telomere elongation
In budding yeast, overexpression of the TR subunit TLC1 together with the TERT subunit Est2p also results in higher telomerase activity levels in cell extracts (Teixeira et al, 2002). However, this has no dramatic effects on telomere length, suggesting that other telomerase-associated components are limiting in vivo, or that telomere length homeostasis in this organism is largely controlled at the telomere, by the equilibrium between extendible and non-extendible states (Teixeira et al, 2004). To test the role of telomerase concentration for telomere length homeostasis in human immortal cell lines, we stably transduced HeLa cells with hTR and hTERT retroviral vectors and analyzed populations of cells for telomerase activity and telomere length. hTERT was expressed from the retroviral LTR promoter and hTR was expressed as above from the U1 promoter. As a control, cells were transduced with empty vectors and grown in parallel. As above, cotransduction with hTR and hTERT vectors resulted in substantially increased telomerase activity levels (23-fold; Figure 2A and B). However, in contrast to the transient expression experiments, stable overexpression of hTERT alone also resulted in an increase of activity (seven-fold). To determine whether the discrepancy between transient and stable expression of hTERT was due to increased levels of endogenous hTR upon stable expression of hTERT, we quantified hTR levels by qRT–PCR (data not shown; see also Figure 2A). However, hTR levels were not affected by ectopic expression of hTERT in this cell line. hTR overexpression on its own also led to a moderate increase of telomerase activity (five-fold).
Figure 2.
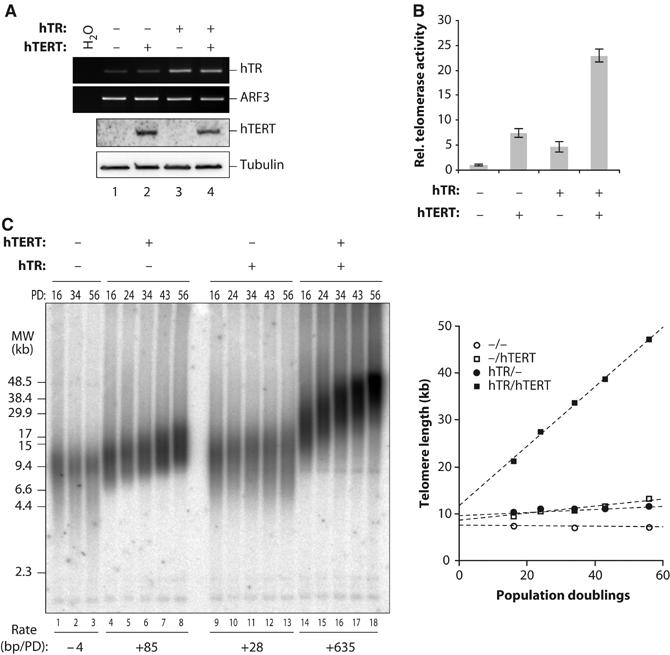
Massive telomere elongation upon long-term super-telomerase expression in HeLa cells. HeLa cells were transduced with retroviral vectors encoding hTR or hTERT or the respective empty vectors. Cell populations were analyzed after double selection. (A) Overexpression of telomerase core components. RT–PCR amplification (upper panels) of hTR or ARF3 as a loading control. RT–PCR conditions were ensured to be in the dynamic range and RT-dependent (not shown). Immunoblot (lower panels) with anti-hTERT antibody or with anti-tubulin antibody as a loading control. (B) Telomerase activity was determined by RQ-TRAP. RNase treatment of extracts reduced the signal to below the detection limit (not shown). Each bar represents the mean±s.d. of at least three measurements. (C) Genomic blot of transduced HeLa cell populations at the indicated PDs. TRFs were separated by PFGE. The rate of elongation is indicated below in bp/PD for this gel. The graph shows the median telomere lengths at the indicated cell PD. Telomere lengths were deduced by subtracting 2 kb for subtelomeric sequences from the median TRF length obtained from the gel.
The effects of super-telomerase on telomere length were determined by pulse-field gel electrophoresis (PFGE) of TRFs, and in-gel hybridization. Our HeLa cell stock has a median TRF length of 9 kb, which, when the length of the subtelomeric region also resistant to HinfI and RsaI digestion is subtracted, corresponds to a median telomere length of approximately 7 kb (Figure 2C, lanes 1–3). As expected, telomere length was stable during cell propagation. In sharp contrast, telomeres in super-telomerase cells increased steadily to 50 kb after 56 population doubling (PD) (Figure 2C, lanes 14–18). Strikingly, the rate of elongation (415 and 635 base pairs (bp)/PD in two independent experiments) was constant in the analyzed size range and independent of telomere length. The initial elongation rate immediately after transduction (PD=0–16) may have been higher, since extrapolation of telomere length in super-telomerase cells to PD=0 gives longer telomeres than control cells (Figure 2E). The intermediate levels of telomerase activity, reached upon overexpression of only one of the two telomerase core components, gave slower telomere elongation rates, indicating that telomerase activity levels were limiting telomere length (Figure 2C). The effects of hTERT expression alone in HeLa cells are in agreement with previous observations (Baur et al, 2001).
Additive effects of hTR and hTERT overexpression on telomerase activity and telomere elongation were also observed in the fibrosarcoma-derived HT1080 cells (Supplementary Figure 2). In super-telomerase HT1080 cells, telomerase activity was increased 40 times and telomere length increased from 2.5 to 20 kb in 50 PD (eight-fold). In contrast to HeLa cells, overexpression of hTR alone in HT1080 cells resulted in a larger (nine-fold) increase of telomerase activity and telomere length than hTERT overexpression alone (three-fold). The low effects of hTERT expression in HT1080 are in agreement with previous observations (McChesney et al, 2000). Thus, hTR is more limiting in this cell line. In both cell lines, the rate of telomere elongation was directly related to the levels of telomerase activity (compare panels B and C in Figure 2 and Supplementary Figure 2).
To test if loss of telomere length control upon telomerase overexpression was an acquired feature of cancer cells, we also determined the effects of super-telomerase expression in primary telomerase-negative human lung fibroblasts (HLF) (Figure 3). Also in HLF, co-overexpression of hTERT and hTR most efficiently increased telomerase activity (Figure 3A and B). Telomere length increased at a constant rate (780 bp) to 31 kb after 28 PD, which corresponds to at least twice the physiological size of telomeres in embryos and six times the initial size at the time of infection. Thus, also in primary cells telomere overelongation was not prevented upon super-telomerase expression.
Figure 3.
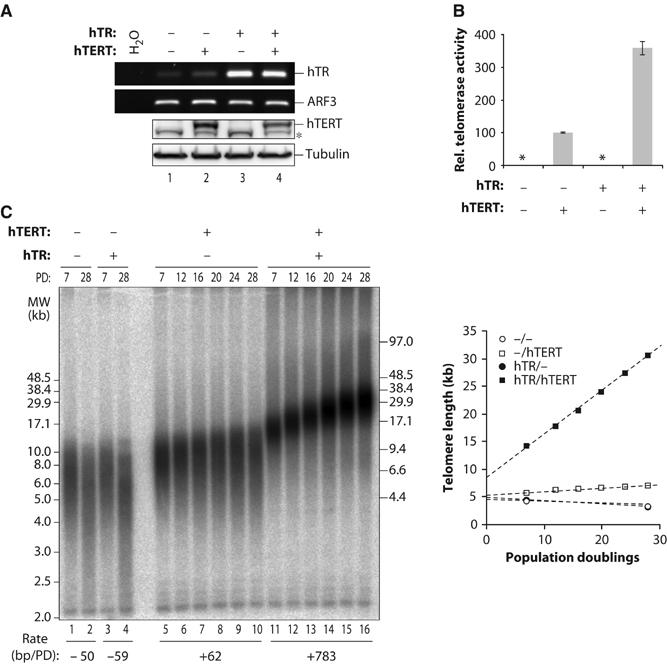
Cooperative effects on the telomere length of long-term hTR and hTERT overexpression in HLF. Cells were transduced with retroviral vectors encoding hTR or hTERT or empty vectors. Cell populations were analyzed after double selection. (A) Overexpression of telomerase core components. RT–PCR amplification (upper panels) of hTR or ARF3 as a loading control. RT–PCR conditions were ensured to be in the dynamic range and RT-dependent (not shown). Immunoblot (lower panels) with anti-hTERT R484 antibody or with anti-tubulin antibody as a loading control. The asterisk indicates a band that crossreacts with the R484 antibody. (B) Telomerase activity as determined by RQ-TRAP. The activity for hTERT-transduced cells was arbitrarily set to 100. RNase treatment of extracts reduced the signal to below the detection limit (not shown). Stars indicate signal below the detection limit. Each bar represents the mean±s.d. of at least three measurements. (C) Genomic blot of TRFs in transduced HLF cell populations at the indicated PDs separated by PFGE. The rate of elongation is indicated below in bp/PD for this gel. The graph shows median telomere length at the indicated cell PD. Telomere length was deduced by subtracting 2 kb for subtelomeric sequences from the median TRF length obtained from the gel.
Since telomerase only extends the G-rich 3′ end of the chromosome and incomplete C-strand synthesis has been observed in some telomere length mutants in budding yeast (Adams Martin et al, 2000), we tested if synthesis of the complementary C-rich telomeric strand was incomplete in super-telomerase cells. The 3′ G-rich overhang in telomeric DNA was detected by hybridization with a telomeric probe, following electrophoresis in gels without prior denaturation (Figure 4). After quantification of the signal, the DNA was denatured in the gel and rehybridized with the same telomeric probe. This analysis indicated that the length of the telomeric 3′ overhang was not affected by telomerase overexpression, and that there was no uncoupling between G- and C-rich strand synthesis. Collectively, these data suggested that steady-state telomere length requires restriction of telomerase levels and that telomere length is determined to a large extent by the absolute levels of telomerase activity and not solely by the balance between positive and negative regulatory forces.
Figure 4.
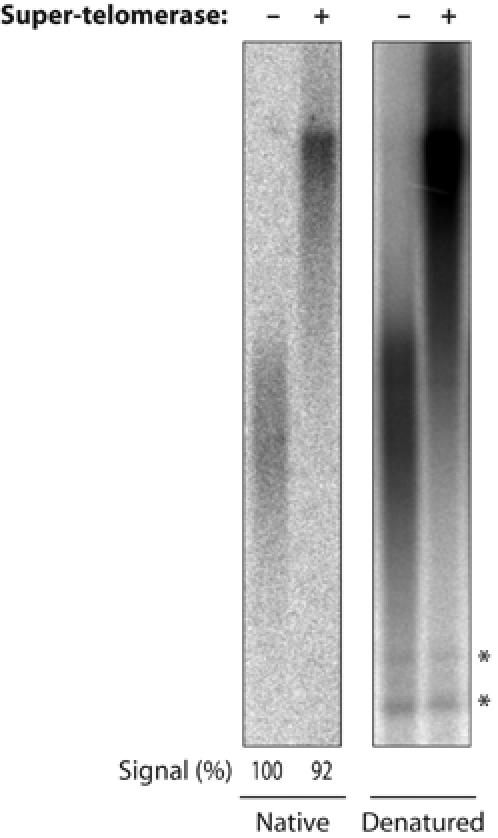
The G- and C-strands are both elongated in super-telomerase cells. Telomeric overhang was analyzed by in-gel hybridization of TRFs from control- or super-telomerase-expressing HeLa cells (same as in Figures 2 and 5). Hybridization to native DNA (left) detecting telomeric overhang or to denatured DNA (right) detecting the total telomeric DNA in the same gel re-hybridized with the same probe after denaturation. The signal of internal restriction fragments containing telomeric repeats (asterisks) shows equal loading of DNA.
Telomere-bound TRF1 and TRF2 are not limiting
The elongation of telomeres in super-telomerase cells far beyond physiological size indicated that telomere length regulation by TRF1 and TRF2 does not involve locking overelongated telomeres permanently in a non-extendible state. Alternatively, it was conceivable that TRF1 and TRF2 levels became limiting at very long telomeres, thus not allowing any more efficient repression of telomerase activity. To test this possibility, TRF1 and TRF2 binding was assessed by confocal immunofluorescence (IF) microscopy using TRF1- and TRF2-specific antibodies. It was previously shown in two HeLa cell clones with different telomere lengths that the amount of TRF1 and TRF2 at telomeres correlates with telomere length (Smogorzewska et al, 2000; Loayza and De Lange, 2003). Staining and image acquisition were performed under identical conditions in parallel, and TRF1 and TRF2 staining are shown without contrast adjustment (Figure 5A). TRF1 and TRF2 staining gives a punctate nuclear pattern corresponding to telomeres. In super-telomerase cells, nuclear dots were much brighter than in control cells, as expected for longer telomeres that bind more TRF1 and TRF2. We quantified TRF1 and TRF2 IF signals in nuclear foci (Supplementary Table 1), as described in Supplementary Methods. The TRF1 signal was increased 11-fold in super-telomerase cells and the TRF2 signal nine-fold at 87 PD. This increase is proportional to the increase in telomere length from 7 to 60 kb (nine-fold).
Figure 5.
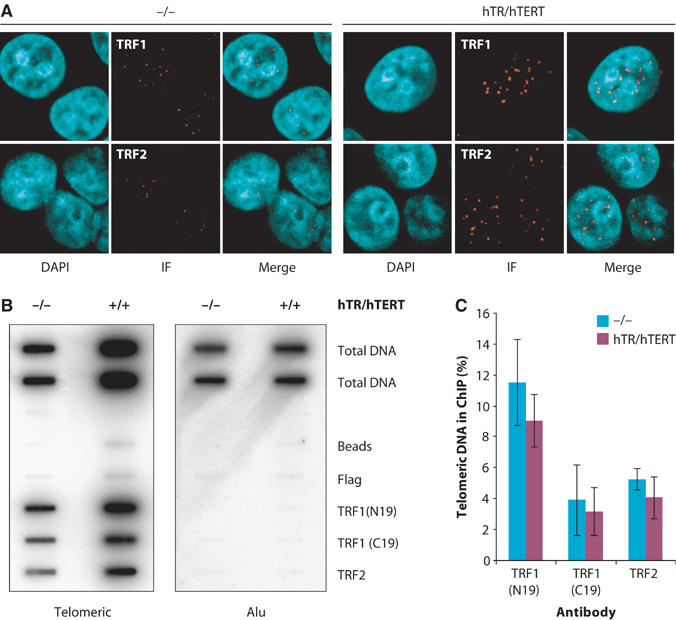
Telomere length sensing by TRF1 and TRF2 is operating in super-telomerase cells. (A) Confocal IF to detect TRF1 and TRF2 in control (−/−) and super-telomerase (hTR/hTERT) cells. Staining and image acquisition of the two cell populations were executed in parallel and IF pictures are presented without any adjustment. During acquisition, the laser beam was set such that the signal in super-telomerase cells was not saturating the CCD camera. (B) ChIP of telomeric DNA by TRF1 and TRF2 in control cells (−/−) or super-telomerase cells (+/+). The antibodies used for IP are indicated on the right. Duplicate dot blots were probed for telomeric or Alu repeats. (C) Quantification of the data in (B) representing per cent TTAGGG repeat DNA recovered in each ChIP. Averaged duplicate signals obtained with total DNA samples were used as 100% value for the quantification. As the histograms represent the percentage of input telomeric DNA, they are corrected for telomere length changes.
To determine more accurately the relative amounts of TRF1 and TRF2 per telomeric DNA, we performed chromatin immunoprecipitation (ChIP) experiments (Figure 5B and C). The percentage of telomeric DNA bound to TRF1 and TRF2 was determined in slot blots using a telomeric probe. Crosslinked extracts that were immunoprecipitated with M2 Flag antibody or no antibody were used as negative controls. Two different TRF1 antibodies immunoprecipitated 12 and 4% of total telomeric DNA, respectively, and a TRF2 antibody immunoprecipitated 5% of total telomeric DNA in control HeLa cells. Since these values are expressed as percentage of total telomeric DNA, they take into account telomere length. In super-telomerase cells, the quantity of total telomeric DNA immunoprecipitated by the two TRF1 antibodies and the TRF2 antibody was not significantly different. Thus, TRF1 and TRF2 are not limiting in super-telomerase cells. They bind to the very long telomeres at a density similar to that measured for the telomeres of control cells. Together with the IF experiments, our data show that increased telomere length in super-telomerase cells results in a directly proportional increase in telomere-bound TRF1 and TRF2 proteins.
Increased telomerase association with chromosome ends in super-telomerase cells
As discussed above, telomere length control in telomerase-positive cancer cells by TRF proteins and POT1 may involve restricting telomerase access to chromosome ends in a length-dependent manner. Alternatively, these proteins may regulate the extent of telomere elongation by regulating, for example, telomerase processivity. To test if telomere association was enhanced upon super-telomerase expression, we performed ChIP experiments in stably transduced and transiently transfected HeLa cells (Figure 6). The experiment was performed as in Figure 5B, but antibodies against hTERT were used to assess telomerase–telomere interaction. No significant binding of hTERT with telomeres could be detected in empty-vector transduced or transfected cells. However, a clear association of hTERT with telomeric DNA could be detected in HeLa cells that either stably or transiently expressed super-telomerase. Since, upon transient transfection of HeLa cells, telomere length increased only slightly (data not shown, see also total and TRF1-bound telomere DNA signal in Figure 6, lower part) while telomerase–telomere binding was still enhanced (Figure 6, lower part), the increased association of hTERT with chromosome ends is not a secondary consequence of telomere elongation. Thus, increased telomerase binding to telomeres provides a possible molecular explanation for telomere overelongation in super-telomerase cells.
Figure 6.
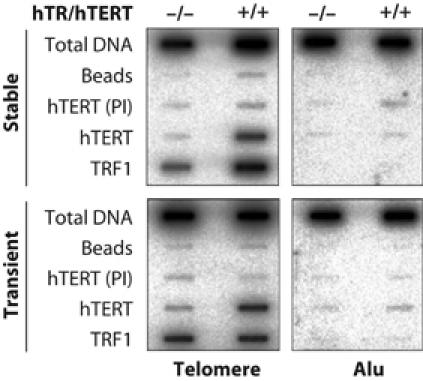
Enhanced, telomere length independent association of telomerase with telomeres in super-telomerase cells. ChIP with hTERT and hTR (+) or empty vector (−) stably transduced (upper) or transiently transfected (lower) HeLa cell populations. Transiently transfected cells were analyzed 72 h post-transfection (bottom panels). The used antibodies or preimmuneserum (PI) are indicated on the left. Duplicate dot blots were probed for telomeric (left) or Alu repeats (right).
Discussion
hTR and hTERT limit the amount of active telomerase RNP in telomerase-positive cells
Our work establishes a basis for obtaining unprecedented high levels of telomerase activity in cancer cell lines and primary cells. Our observations demonstrate that both hTR and hTERT restrict telomerase activity and telomere length in two cancer-derived and one primary cell line (Figures 1, 2, 3, and Supplementary Figures 1 and 2), while other telomerase subunits are not limiting. Upregulation of hTERT in tumors is required for telomerase activation and telomere length stabilization. hTR, which is ubiquitously expressed, is shown here to restrict telomerase activity in HT1080 and to a lesser extent in HeLa cells. Indeed, the hTR locus is amplified in some tumors, suggesting that hTR upregulation can be more important for telomerase activation than generally appreciated (Soder et al, 1997). hTR levels also limit telomerase activity in human haematopoietic cells, as seen in dyskeratosis congenita patients, in which decreased expression of hTR causes bone marrow failure (Vulliamy et al, 2001, 2004). Haploinsufficiency has also been reported in heterozygous mTR+/− mice (Hathcock et al, 2002; for a review, see Harrington, 2005). Our data reveal that telomere length homeostasis in the analyzed cancer cells is achieved with limiting amounts of telomerase, which results in stable but short telomeres. This observation could provide a simple molecular explanation for the apparent paradox that, despite the expression of telomerase in cancer cells, telomere length in these cells is often very short.
In Figure 2, we analyzed the fate of telomeres in super-telomerase HeLa cells up to PD=56, which corresponds to more than the estimated number of cell divisions needed to generate a human body (1016 mitoses; Strachan and Read, 1999). Telomere repeat addition was not inhibited in this time window. More recently, super-telomerase HeLa cells were followed up 87 PD, elongating their telomeres beyond 60 kb in a linear fashion (data not shown). Telomere length is seen here to largely depend on telomerase concentration. Since telomerase is downregulated during human embryogenesis in most tissues after 12–21 weeks (Ulaner and Giudice, 1997), our findings imply that both hTERT and hTR levels must be extremely tightly regulated during early human embryogenesis in order to ensure optimal telomere maintenance for the lifetime. However, we cannot exclude the possibility that overexpression of hTR and hTERT without overexpression of other telomerase components may result in formation of telomerase complexes that fail to respond to negative feedback regulation mechanisms.
Model for telomere length homeostasis: dynamic transitions between non-extendible, extendible and extending states
Super-telomerase-expressing cells elongated telomeres with a constant rate (0.25–0.8 kb/PD) far beyond physiological length, despite the fact that TRF1 and TRF2 increased at telomeres in proportion to the number of telomere repeats (Figure 5). To reconcile these findings with a model in which telomeres switch in a length-dependent manner between extendible and non-extendible states (Teixeira et al (2004) and references therein), we predict that long telomeres are not permanently locked in a non-extendible state, but that the switch between these two telomeric states is rather dynamic. In addition, we predict the existence of a distinct third telomeric state with extendible telomeres switching to an extending state, upon productive association with telomerase. In this model, the transition between the non-extendible and extendible states is length dependent, whereas the transition between extendible and extending states depends on the concentration of telomerase (Figure 7). In cells with low levels of telomerase activity (top panel), short telomeres oscillate between extendible and extending states and long telomeres would be mainly in the non-extendible state. In contrast, in cells with high levels of telomerase (such as super-telomerase cells, bottom panel), long telomeres would still be mostly in a non-extendible state, but the transition between the states would be frequent enough to permit continuous telomere elongation. We see increased telomerase binding to telomeres in super-telomerase cells (Figure 6). The mechanisms of telomerase recruitment in human are elusive. However, from studies in budding yeast, it is known that telomerase recruitment occurs through two distinct pathways (Evans and Lundblad, 1999; Pennock et al, 2001; Fisher et al, 2004). One is mediated by telomere-bound Cdc13p, which binds telomerase-associated Est1p. The other involves the interaction between telomere-binding Ku, which binds the TR subunit TLC1. Whatever the mechanism in human super-telomerase cells, we suspect that the increased local concentration of telomerase in these cells near its substrate may favor transition from the extendible to the extending state before the end is sequestered back into a non-extendible state (Figure 7).
Figure 7.
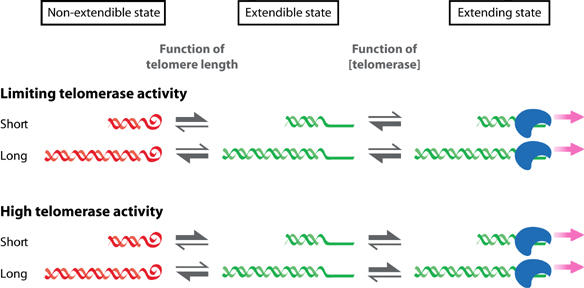
A three-state model for telomere length homeostasis. Telomeres are proposed to switch between non-extendible (red, curly end), extendible (green, straight end, middle) and extending states (green straight end associated with telomerase, right). The equilibrium between non-extendible and extendible states is a function of telomere length, while the equilibrium between extendible and extending state is a function of telomerase concentration. The top panel shows a situation where telomerase concentration is limiting, corresponding to the here-analyzed telomerase-positive cell lines. The bottom panel indicates the situation in super-telomerase cells.
The molecular nature of the putative non-extendible, extendible and extending states, as well as the molecular mechanisms that drive their transition are not well understood. The telomerase non-extendible state could correspond to the t-loop structure or the telomeric 3′ end bound to hPOT1 (Griffith et al, 1999; Loayza and De Lange, 2003). Our results imply that this state is not a permanent feature of long telomeres, as telomere elongation was still permitted with high levels of telomerase. In this respect, the chromatin structure of the non-extendible state appears very different from the telomere chromatin structure, which is responsible for telomere silencing. This latter chromatin structure is stably inherited over several cell cycles and rarely switches to the opposite state (Gottschling et al, 1990; Baur et al, 2001; Koering et al, 2002). Telomerase elongates telomeres in late S-phase at least in yeast (Marcand et al, 2000). This suggests that the transition from non-extendible to extendible state and/or extendible to extending state may occur only in S-phase of the cell cycle. Telomere transitions in S-phase may, for example, be enabled by passage of the replication fork, cell-cycle-dependent telomere binding of telomerase components, as documented in yeast (Fisher et al, 2004), or other cell cycle-dependent modifications. Finally, the finding that telomerase levels limit telomere length suggests that the main function of the negative feedback loop mediated by telomeric proteins is not to prevent telomere overelongation, but to favor extension of short telomeres when telomerase is limiting.
Materials and methods
Plasmid DNA
For transient expression, hTERT cDNA was cloned into the EcoRI site of pcDNA6/myc-His C (Invitrogen). The hTERT stop codon was upstream of the tags and therefore full-length untagged hTERT was expressed. hTR was cloned downstream of the U1 promoter in the EcoRI and HindIII sites of pBluescript II SK(+) (Stratagene). For stable infections, full-length hTERT cDNA was cloned in the EcoRI site of pBabe-Hygro downstream of the LTR-promoter and U1-hTR was cloned in the BglII and HindIII sites of pMND-Banshee retroviral vector containing a neomycin selection cassette (a kind gift of J Rossi).
Cell transfection and viral infection
Transient transfections were carried out using lipofectamine 2000 transfection reagent (Invitrogen). Retroviruses were prepared by transient transfection of vector-encoding plasmids into the Phoenix-amphotrophic encapsidation cell line (a kind gift of G Nolan). Cell supernatants were collected and filtered 48, 56 and 72 h after transfection. Target cells were infected three times in six-well plates with 2 ml of supernatant, 2 ml of fresh medium and 4 μl of 4 mg/ml polybrene. At 24 h after the last infection, cells were transferred to selection medium (DMEM containing 10% FCS, 1 mg/ml G418 and 250 μg/ml hygromycin B), at which time PD was arbitrarily set to 0. After 10–15 days, control uninfected cells were dead and selection was considered as complete. HLFs were first infected with the pMND-Banshee-U1-hTR or empty vector, selected with G418 and then with the pBabe-Hygro-hTERT or empty vector, and selected with hygromycin. PD was arbitrarily set to 0 at the start of hygromycin selection.
RQ-TRAP
Cells were collected, washed once in PBS and resuspended in 3-((3-cholamidopropyl)-dimethylammonio)-1-propane-sulfonate (Chaps) lysis buffer at 105 cells/ml. After 30 min incubation on a rotating wheel at 4°C, extracts were centrifuged at 14 000 g for 30 min at 4°C, the supernatants were quick-frozen and stored at −80°C. Protein concentration of extracts was determined with the Bradford protein assay (BioRad). The RQ-TRAP protocol was slightly modified and adapted to the LightCycler system (Wege et al, 2003). Reactions were carried out in 20-μl reactions containing the LC-FastStart Sybr Green II mix (Roche), 1 μl of undiluted or diluted cell extracts, 0.1 μg of telomerase primer TS, 0.05 μg of reverse primer ACX and 0.5 mM additional MgCl2. Using the LightCycler (Roche), samples were incubated for 30 min at 30°C, for 10 min at 95°C and amplified in 40 PCR cycles for 5 s at 95°C and 60 s at 60°C. The threshold cycle values (Ct) were determined automatically by the second-derivative maximum method using arithmetic baseline adjustment. Duplicate five-fold serial dilutions of the most active samples were used to draw a standard curve of the form log10(protein quantity)=aCt+b. Telomerase activity was expressed relative to this standard as the quantity of standard sample extract giving the same Ct value. Samples were serially diluted to verify the linearity of the RQ-TRAP reaction, and RNase-treated to verify that the amplification product was due to telomerase activity.
Direct telomerase assays
Conventional telomerase assays were carried out for 1 h at 30°C in 20-μl reactions containing 4 μg of 293T Chaps extracts, 50 mM Tris–HCl (pH 8.0), 50 mM KCl, 1 mM spermidine, 1 mM β-mercaptoethanol, 1 mM MgCl2, 0.5 mM dATP, 0.5 mM dTTP, 2 μM dGTP, 20 μCi of [α-32P]dGTP (3000 Ci/mmol) and 1 μM of telomeric primer (T2AG3)3. Reactions were stopped by the addition of 5 mM EDTA and 1% SDS. Trace amounts of a 32P-labeled 10-mer DNA oligonucleotide was added as recovery control (noted RC in Figure 1D). Products were ethanol-precipitated. DNA pellets were resuspended in 98% formamide containing 10 mM EDTA and 0.005% xylene cyanol, heated to 95°C for 1 min, and analyzed on 12% polyacrylamide-urea sequencing gels.
Chromatin immunoprecipitation
ChIP assays were performed as described (Loayza and De Lange, 2003) with the following modifications. For immunoprecipitations, 5 μg of M2 anti-Flag (Sigma), TRF1(N19), TRF1(C19)-X (Santa-Cruz), TRF2 (Upstate) antibodies or 25 μl of hTERT (R484, crude serum) or preimmune serum (PI) (Wenz et al, 2001) was used, followed by incubation for 6 h at 4°C with 15 μl of protein A/G-Sepharose bead mix (Amersham; 1:1 mix, preblocked overnight with 1 mg/ml bovine serum albumin). Crosslinks were reversed overnight at 65°C. In all, 20% of DNA was loaded for the detection of telomeric sequences using a 600-bp random-labeled TTAGGG probe, and 10% for detection of Alu sequences using a 5′-labeled oligonucleotide probe (5′-GTGATCCGCCCGCCTCGGCCTCCCAAAGTG- 3′).
Telomere assays
Genomic DNA was prepared with the Wizard genomic DNA kit (Promega). RsaI and HinfI digested DNA (2 μg) was fractionated on a CHEF-DRII PFGE apparatus (BioRad) in 1% agarose in 0.5 × TBE at 5.2 V/cm for 16 h at 14°C with switch times ramped from 0.5 to 6 s, followed by ethidium bromide staining. For the overhang assay, in-gel hybridization was performed directly without denaturation of DNA as described (Karlseder et al, 2002), using a 5′-end-labeled (C3TA2)4 oligonucleotide or a 600-bp random-labeled TTAGGG probe. For TRF analysis, gels were denatured first as described (Karlseder et al, 2002). Median telomere length was determined exactly as described (Li and de Lange, 2003). Note that median telomere length was obtained by subtracting 2 kb of subtelomeric sequences from the median TRF length.
Northern blot, RT–PCR and qRT–PCR
Total RNA was extracted with the RNeasy miniprep kit (Qiagen) using on-column DNase digestion (RNase-Free DNase Set, Qiagen). RNA concentration was determined by measuring the OD at 260 nm. For Northern blots, 2 μg of total RNA was separated by 6% PAGE under denaturing conditions (7 M urea, 1 × TBE) and transferred to a Hybond-N+ membrane. The hTR hybridization probe was prepared by random labeling of a NarI–StuI fragment of hTR cDNA, using the Invitrogen random primers DNA labeling kit. For β-actin mRNA detection, a 5′-end radiolabeled oligonucleotide (5′-AAGCTTATCAAAGTCCTCGGCCAC-3′) probe was used. For quantitative RT–PCR (qRT–PCR), cDNAs were prepared using SuperScript II reverse transcriptase (Invitrogen) followed by LightCycler qPCR, using the LC-FastStart Sybr Green II kit, primers hTR fw790 and R3c (Wenz et al, 2001), and 1 μl of cDNA, as indicated in the manufacturer's instructions. Serial dilutions of in vitro transcribed hTR were used as standard. hTR quantity was normalized to the amount of total RNA. As a control for DNA contaminations, RT–PCR without RT was performed in parallel for each sample, which did not result in detectable PCR products. SuperScript III one-step RT–PCR system with Platinum Taq DNA polymerase (Invitrogen) was used to perform RT–PCR. Initial control experiments were performed to ensure that amplification was in the dynamic range in the experimental conditions used (100 ng of total RNA, 15 min at 55°C, 2 min at 94°C (15 s at 94°C, 30 s at 57°C, 30 s at 68°C) repeated 24 times). For amplification of the house-keeping ARF3 mRNA, forward and reverse primers were 5′-TCACCACCATCCCTACCATT-3′ and 5′-AGGTGGCCTGAATGTACCAG-3′, respectively.
Immunoblots
In all, 5 μg of Chaps extracts (Figure 1) or 105 cells was directly boiled for 5 min in Laemmli sample buffer (Figures 2 and 3) and fractionated on 4–20% SDS–PAGE gradient gels. We performed immunoblots with standard protocols, using an affinity-purified rabbit polyclonal antibody against hTERT (1:10 000, R484 (Wenz et al, 2001)) or a mouse monoclonal antibody to β-tubulin (1:20 000, DM1A, Sigma). Secondary horseradish peroxidase-conjugated goat antibodies against rabbit or mouse were used (1:10 000, Promega). The AlphaInnotech chemoluminescence substrate and imaging system was used to detect bound antibodies.
Indirect IF
Cells were grown on glass coverslips, washed with cold PBS, and fixed with cold methanol for 15 min at −20°C. After two washes with cold PBS, cells were preblocked for 1 h in PBS containing 0.1% Tween-20 and 5% milk (preclarified by centrifugation for 30 min at 15 000 g) and incubated with primary antibody for 1 h at RT. For TRF1, a purified rabbit polyclonal antibody (370) was used (a kind gift from T de Lange) and diluted 1:500. For TRF2, a mouse monoclonal antibody (Upstate) was diluted 1:300. Cy3-conjugated secondary antibodies were used. Nuclei were stained with 0.1 μg/ml DAPI in PBS.
Supplementary Material
Supplementary Figure 1
Acknowledgments
We thank Julia Cooper, Moira Cockell and Claus Azzalin for comments on the manuscript, Nele Hug and Claus Azzalin for help with ChIP and immunostaining experiments, Claude Bonnard for writing the immunostaining quantification script, Marcel Allegrini for instruction on the Axioplan microscope, Colleen Kelleher, Claus Azzalin and Isabel Kurth for plasmids and reagents, John Rossi for the U1 promoter and the pMND-Banshee retroviral vector, Jack Griffith for the pRST5 plasmid and Titia de Lange for TRF1(370) antibody. Nicolas Fasel is thanked for lending the CHEF apparatus. This work was supported by an EMBO Long-Term Fellowship to GC and grants from the Swiss National Science Foundation, the Swiss Cancer League, the Human Frontier Science Program and the EU 6th Framework Programme to JL.
References
- Adams Martin A, Dionne I, Wellinger RJ, Holm C (2000) The function of DNA polymerase alpha at telomeric G tails is important for telomere homeostasis. Mol Cell Biol 20: 786–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, Ricoul M, Pommier JP, Sabatier L, Gilson E (2002) Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol Cell Biol 22: 3474–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Piatyszek MA, Gupta J, Shay JW, Bacchetti S, Greider CW (1996) Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res 56: 645–650 [PubMed] [Google Scholar]
- Baur JA, Zou Y, Shay JW, Wright WE (2001) Telomere position effect in human cells. Science 292: 2075–2077 [DOI] [PubMed] [Google Scholar]
- Beattie TL, Zhou W, Robinson MO, Harrington L (1998) Reconstitution of human telomerase activity in vitro. Curr Biol 8: 177–180 [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE (1998) Extension of life-span by introduction of telomerase into normal human cells. Science 279: 349–352 [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V (1999) Est1 and Cdc13 as comediators of telomerase access. Science 286: 117–120 [DOI] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, Le S, West MD, Harley CB, Andrews WH, Greider CW, Villeponteau B (1995) The RNA component of human telomerase. Science 269: 1236–1241 [DOI] [PubMed] [Google Scholar]
- Fisher TS, Taggart AK, Zakian VA (2004) Cell cycle-dependent regulation of yeast telomerase by Ku. Nat Struct Mol Biol 11: 1198–1205 [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337: 331–337 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW (1990) Telomeres shorten during ageing of human fibroblasts. Nature 345: 458–460 [DOI] [PubMed] [Google Scholar]
- Harrington L (2005) Making the most of a little: dosage effects in eukaryotic telomere length maintenance. Chromosome Res 13: 493–504 [DOI] [PubMed] [Google Scholar]
- Hathcock KS, Hemann MT, Opperman KK, Strong MA, Greider CW, Hodes RJ (2002) Haploinsufficiency of mTR results in defects in telomere elongation. Proc Natl Acad Sci USA 99: 3591–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J, Smogorzewska A, de Lange T (2002) Senescence induced by altered telomere state, not telomere loss. Science 295: 2446–2449 [DOI] [PubMed] [Google Scholar]
- Kelleher C, Kurth I, Lingner J (2005) Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol Cell Biol 25: 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW (1994) Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015 [DOI] [PubMed] [Google Scholar]
- Koering CE, Pollice A, Zibella MP, Bauwens S, Puisieux A, Brunori M, Brun C, Martins L, Sabatier L, Pulitzer JF, Gilson E (2002) Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep 3: 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Zaug AJ, Podell ER, Cech TR (2005) Switching human telomerase on and off with hPOT1 protein in vitro. J Biol Chem 280: 20449–20456 [DOI] [PubMed] [Google Scholar]
- Li B, de Lange T (2003) Rap1 affects the length and heterogeneity of human telomeres. Mol Biol Cell 14: 5060–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567 [DOI] [PubMed] [Google Scholar]
- Loayza D, De Lange T (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 423: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Mann C, Gilson E (2000) Cell cycle restriction of telomere elongation. Curr Biol 10: 487–490 [DOI] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- McChesney PA, Aisner DL, Frank BC, Wright WE, Shay JW (2000) Telomere dynamics in cells with introduced telomerase: a rapid assay for telomerase activity on telomeres. Mol Cell Biol Res Commun 3: 312–318 [DOI] [PubMed] [Google Scholar]
- Morin GB (1989) The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell 59: 521–529 [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V (2001) Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T (2004) Regulation of telomerase by telomeric proteins. Annu Rev Biochem 73: 177–208 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T (2000) Control of human telomere length by TRF1 and TRF2. Mol Cell Biol 20: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soder AI, Hoare SF, Muir S, Going JJ, Parkinson EK, Keith WN (1997) Amplification, increased dosage and in situ expression of the telomerase RNA gene in human cancer. Oncogene 14: 1013–1021 [DOI] [PubMed] [Google Scholar]
- Strachan T, Read AP (1999) Human Molecular Genetics 2. Oxford, NY: BIOS Scientific [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -non-extendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Forstemann K, Gasser SM, Lingner J (2002) Intracellular trafficking of yeast telomerase components. EMBO Rep 3: 652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulaner GA, Giudice LC (1997) Developmental regulation of telomerase activity in human fetal tissues during gestation. Mol Hum Reprod 3: 769–773 [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T (1997) Control of telomere length by the human telomeric protein TRF1. Nature 385: 740–743 [DOI] [PubMed] [Google Scholar]
- Vaziri H, Benchimol S (1998) Reconstitution of telomerase activity in normal human cells leads to elongation of telomeres and extended replicative life span. Curr Biol 8: 279–282 [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I (2001) The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413: 432–435 [DOI] [PubMed] [Google Scholar]
- Vulliamy T, Marrone A, Szydlo R, Walne A, Mason PJ, Dokal I (2004) Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat Genet 36: 447–449 [DOI] [PubMed] [Google Scholar]
- Wege H, Chui MS, Le HT, Tran JM, Zern MA (2003) SYBR Green real-time telomeric repeat amplification protocol for the rapid quantification of telomerase activity. Nucleic Acids Res 31: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet 17: 498–502 [DOI] [PubMed] [Google Scholar]
- Wenz C, Enenkel B, Amacker M, Kelleher C, Damm K, Lingner J (2001) Human telomerase contains two cooperating telomerase RNA molecules. EMBO J 20: 3526–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW (1996) Telomerase activity in human germline and embryonic tissues and cells. Dev Genet 18: 173–179 [DOI] [PubMed] [Google Scholar]
- Yi X, Tesmer VM, Savre-Train I, Shay JW, Wright WE (1999) Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol Cell Biol 19: 3989–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1


