Abstract
In the budding yeast Saccharomyces cerevisiae, transcription of genes encoding for the high-affinity iron (FET3, FTR1) and copper (CTR1) transporters does not occur in the absence of heme. We show that the Aft1p binding region of the FET3 promoter or the Mac1p binding region of the CTR1 promoter is necessary and sufficient to mediate heme-deficient repression. Transcription is repressed in the absence of heme, and a genetic screen identified Tup1p and Hda1p as being required for transcriptional repression. In contrast to FET3 and CTR1, Aft1p target genes ARN1 and FIT1 are transcribed in the absence of heme. A 14 bp sequence in the ARN1 promoter is necessary and sufficient to permit transcription in the absence of heme. Transcription in the absence of heme required the presence of Cti6p to overcome the effect of Tup1p, and Cti6p was recruited to the ARN1 promoter in the absence of heme. We hypothesize that transcription of the siderophore transporter ARN1 permits yeast to accumulate iron in the absence of oxygen and to deny iron to competing organisms.
Keywords: Aft1p, Cti6p, heme, iron, Tup1p
Introduction
Many of the biochemical reactions that require the redox active transition metals iron and copper do not occur under anaerobic conditions. For example, the requirement of copper for the reduction of oxygen (cytochrome oxidase) and for the dismutase of superoxide anion (SOD1) would be dispensable during anaerobiosis. Similarly, the requirement of iron for heme synthesis as well as for reactions that use heme to either bind oxygen or transfer electrons to oxygen is unnecessary under anaerobic conditions, as oxygen–heme interactions would not occur. In the absence of oxygen, the requirement for these transition metals is reduced and accordingly there is decreased expression of both the high-affinity copper and iron transport systems.
Whereas humans sense oxygen and coordinate gene expression directly through the hypoxia-inducible factor (HIF-1α) (Bardos and Ashcroft, 2005), the budding yeast Saccharomyces cerevisiae uses an indirect method for detection of oxygen tension. Aerobiosis is detected in yeast not by sensing oxygen directly, but rather by the presence of heme, a molecule that requires the presence of oxygen for biosynthesis (for review, see Kwast et al, 1998; Kastaniotis and Zitomer, 2000; Rosenfeld and Beauvoit, 2003). The yeast protein Hap1p (heme activator protein) is a transcriptional activator in the presence of heme, inducing the expression of genes required for respiration and for controlling oxidative damage. Hap1p activation also induces expression of Rox1p, a protein required for repression of anaerobic genes. Loss of heme results in the formation of a high-molecular-weight cytosolic complex containing Hap1p, resulting in the subsequent loss of expression of target aerobic genes (Becerra et al, 2002).
Previously, we showed that decreased expression of the high-affinity iron and copper transport systems during heme deficiency results from loss of transcription. In the absence of heme, the iron-sensing transcription factor Aft1p shows a normal response to iron deprivation: Aft1p translocates from the cytosol to the nucleus, and occupies the promoter region of target genes such as FET3 (Crisp et al, 2003). Transcription of FET3, however, does not occur and the lack of transcription is independent of the Hap/Rox/Mot system. In this report, we analyze the promoter region of FET3 to define regions required for the transcriptional response to heme deficiency. Our data suggest that the lack of transcription results from the presence of a repressor. Through a genetic screen, we show that the general repressor, Tup1p (Smith and Johnson, 2000), and the histone deacetylase, Hda1p (Wu et al, 2001), are required for heme-deficient repression of FET3, FTR1, and CTR1. Further, we demonstrate that within the family of Aft1p-regulated genes, not all members are inhibited by the absence of heme. Using chimeric reporter constructs, we define the sequences that are responsible for the relief of repression, and demonstrate that relief of heme-deficient repression can be overcome by retention of Cti6p, a PHD finger-containing protein (Papamichos-Chronakis et al, 2002).
Results
Identification of promoter elements required for heme-deficient repression of FET3
To study the loss of FET3 transcription under heme-deficient conditions, we utilized a yeast strain in which the gene (HEM1) that encodes the first enzyme in the porphyrin biosynthesis pathway, aminolevulinic acid synthase, had been disrupted. Normal cellular heme was restored by supplementing the growth media with the product of the enzyme, δ-aminolevulinic acid (ALA). As heme-deficient yeast become auxotrophic for unsaturated fatty acids, ergosterol and methionine, media were supplemented with these products to achieve normal cellular growth during heme deficiency (Tween 80, ergosterol, methionine (TEM)).
We generated a series of reporter constructs with different regions of the FET3 promoter and transformed them into a Δhem1 strain to identify sequences required for the transcriptional response to heme deficiency (Figure 1A). For these and other experiments, low-iron conditions were achieved by addition of an iron chelator, bathophenanthroline disulfonate (BPS), to the growth media. The wild-type FET3 reporter construct (pFET3-wt) was only induced by low iron when heme was present. A reporter construct that contained the Aft1p binding region and preserved the spacing of this element relative to the TATA box was also induced by low iron only when heme was present (Figure 1A, pFET3-RC07). Truncation of the promoter region upstream of the Aft1p binding site in this construct led to a decrease in induction. The decrease may be due to loss of several non-consensus Aft1p binding sites that are within this region. Even though maximal induction decreased, both iron-sensitive transcription and the loss of transcription in the absence of heme were maintained.
Figure 1.
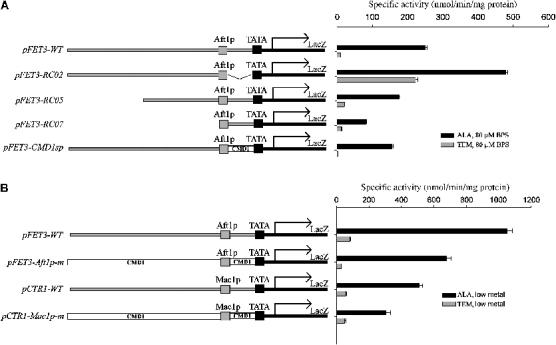
The Aft1p binding site in FET3 is necessary and sufficient for repression. (A) The Δhem1 strain was transformed with the indicated reporter constructs. Strains were induced in 80 μM BPS in the indicated media. Cells were grown for 6 h, harvested, and a β-galactosidase assay was performed. (B) The Δhem1 strain was transformed with the indicated reporter constructs. Strains with FET3 constructs were induced in 80 μM BPS, and strains with CTR1 constructs were induced in the presence of 50 μM CuSO4 or 40 μM bathocuproine sulfonate (BCS). Cells were grown for 6 h, harvested, and a β-galactosidase assay was performed.
The spacing between the Aft1p binding site and the TATA box was critical for the transcriptional repression in response to heme deficiency. Placement of the Aft1p binding site directly upstream of the TATA box resulted in iron-sensitive expression even in the absence of heme (Figure 1A, pFET3-RC02). Restoration of the spacing between the Aft1p binding site and the TATA box with a heterologous sequence from the CMD1 promoter region resulted in iron-sensitive expression under normal heme conditions and repression upon loss of heme (Figure 1A, pFET3-CMD1sp). As alteration of the spacing between the Aft1p binding region and the TATA box resulted in iron-sensitive transcription even in the absence of heme, we conclude that the lack of transcription resulted from a repressor that was relieved by heme, rather than the absence of a heme-dependent coactivator. Similar to the FET3 minimal reporter construct, a reporter construct consisting of the Mac1p minimal binding site from CTR1 showed copper-sensitive induction only when heme was present (Figure 1B).
Identification of Tup1p as the heme-sensitive repressor
To identify factors required for the repression of FET3 transcription in the absence of heme, we developed a genetic screen using the Δhem1 strain transformed with a low-copy plasmid harboring the constitutively active allele AFT1-1up, a high-copy plasmid containing FET3-lacZ, and a chromosomally integrated FET3-GFP chimera. When ALA was added to the medium, this strain expressed β-galactosidase and Fet3p-GFP independent of medium iron as a result of the constitutively active AFT1-1up allele (Yamaguchi-Iwai et al, 1995). In the absence of ALA, there was no expression of either β-galactosidase or Fet3p-GFP (Figure 2A). The strain was mutated by exposure to UV, and colonies that expressed β-galactosidase and Fet3p-GFP were identified. Positive clones were transformed with a centromeric library and colonies that no longer expressed β-galactosidase or Fet3p-GFP were selected. The transforming plasmid was rescued and sequenced.
Figure 2.
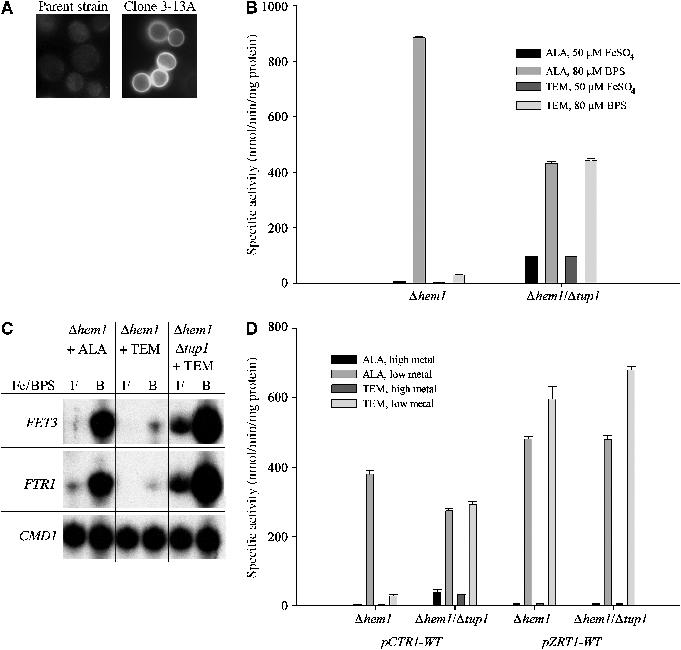
Loss of Tup1p relieves heme-deficient repression of FET3. (A) A strain was generated that contained an AFT1-1up plasmid, FET3-lacZ reporter plasmid, and a genomic FET3-GFP. The strain was UV mutagenized, and a β-galactosidase plate assay was performed. Blue colonies (positives) were collected, as they indicated loss of FET3-lacZ repression. To rule out mutations in the FET3-lacZ reporter construct, positives were then viewed by epifluorescence microscopy. The parent strain (repressed) and one representative positive clone, 3-13A, are shown. (B) A Δhem1 strain or a Δhem1/Δtup1 strain was transformed with a FET3-lacZ reporter construct. Strains were induced in the presence of 50 μM FeSO4 or 80 μM BPS in the indicated media. Cells were grown for 6 h, harvested, and a β-galactosidase assay was performed. (C) The Δhem1 strain or the Δhem1/Δtup1 strain was grown for 6 h in the media as indicated. Cells were harvested and total RNA was isolated. S1 nuclease protection assay was performed on FTR1 and FET3, with CMD1 as a positive control. (D) The Δhem1 or the Δhem1/Δtup1 strain was transformed with the indicated reporter constructs. Cells were grown in metal-replete (50 μM) or metal-depleted media for the respective construct in the indicated media. Cells were grown for 6 h, harvested, and a β-galactosidase assay was performed.
After screening approximately 50 000 colonies, we collected 12 strains that showed expression of FET3 in the absence of heme. Four of the strains were complemented by TUP1 on a centromeric plasmid, and three of the strains were complemented by a library clone that contained HDA1. We were unable to identify the remaining five strains as a result of their weak phenotype. We generated a TUP1 deletion in a Δhem1 strain and analyzed iron-sensitive expression of FET3-lacZ in the absence of heme. β-Galactosidase activity was iron sensitive regardless of heme status, indicating that the loss of Tup1p permitted heme-deficient expression of FET3 (Figure 2B). In addition to the relief of heme-deficient repression, the Δhem1/Δtup1 strain showed an increase in the transcription of FET3 under iron-replete conditions. These results were confirmed by S1 analysis in which expression of FTR1 (and FET3, data not shown) was now permitted in the absence of heme when cells were incubated in low-iron medium (Figure 2C). Further, when heme was depleted under iron-replete conditions, FTR1 message levels were increased to approximately 40% of that in iron-limited cells; the reporter construct data showed this to be approximately 25%. In heme-replete cells, deletion of TUP1 did not affect the copper-sensitive transcription of a CTR1-lacZ construct nor did it affect the zinc-sensitive transcription of a ZRT1-lacZ construct. Deletion of TUP1 permitted the expression of CTR1-lacZ in the absence of heme under copper-limiting conditions, whereas no effect was seen on transcription of ZRT1-lacZ (Figure 2D). Tup1p is a generic repressor, which is most often found in a complex with Ssn6p. Together, these proteins are responsible for repression of a wide variety of genes (Smith and Johnson, 2000). Deletion of SSN6, which acts as a corepressor with Tup1p, showed a modest (25%) but reproducible relief of repression (Figure 3A). This result was consistent with studies that suggested that in many cases Ssn6p was dispensable for repression of target Tup1p/Ssn6p promoters.
Figure 3.
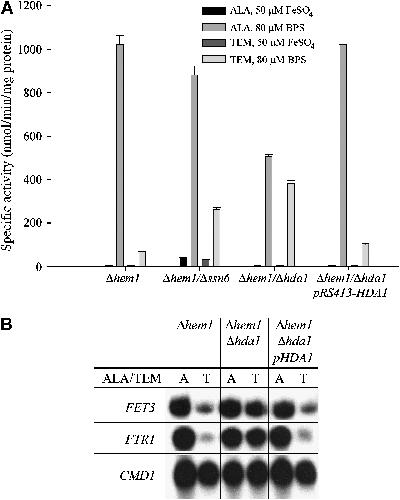
Loss of Hda1p relieves heme-deficient repression of FET3. (A) The Δhem1, Δhem1/Δhda1, and Δhem1/Δhda1/pRS413-HDA1 strains were transformed with a FET3-lacZ reporter construct. Strains were induced in the presence of 50 μM FeSO4 or 80 μM BPS in the indicated media. Cells were grown for 6 h, harvested, and a β-galactosidase assay was performed. (B) The Δhem1, Δhem1/Δhda1, and Δhem1/Δhda1/pRS413-HDA1 strains were grown for 6 h under low-iron conditions in the indicated media. Cells were harvested and total RNA was isolated. S1 nuclease protection assay was performed on FTR1 and FET3, with CMD1 as a positive control.
Some of the mutants identified in the genetic screen were complemented by HDA1-containing plasmids. To confirm that HDA1 was involved in FET3 repression in the absence of heme, we generated a deletion in HDA1 in our Δhem1 strain. A FET3-lacZ reporter construct was introduced into this strain and iron-sensitive expression was assayed in the presence and absence of heme (Figure 3A). The β-galactosidase assay was verified by S1 analysis, which showed expression of FTR1 under low-iron conditions in the Δhem1/Δhda1 strain regardless of heme status (Figure 3B). This was consistent with current literature suggesting that Tup1p can recruit Hda1p to deacetylate chromatin to make it less accessible to transcriptional machinery (Wu et al, 2001; Green and Johnson, 2004). Interestingly, although the deletion of HDA1 was able to mediate relief of repression under heme-deficient conditions, it did not result in increased basal expression of FET3 under iron-replete conditions, as was seen in the Δhem1/Δtup1 strain (Figure 2B and C). To examine whether other transcription factors known to affect repression were involved in heme-deficient repression of FET3, we generated deletions of HOS1, HOS2, HOS3, RPD3, SIN3, NHP6a/b, CTI6, and SRB10 in the Δhem1 strain, as these genes encode proteins involved in transcriptional repression or chromosome remodeling. All of these strains showed low-iron induction of FET3-lacZ under normal heme conditions as well as heme-deficient repression (data not shown).
Tup1p is a transcriptional activator/repressor that is not sequence specific; rather, it is recruited to a target promoter by another transcription factor (Smith and Johnson, 2000; Green and Johnson, 2005). We tested the ability of Tup1p to be recruited to Aft1p target promoters by chromatin immunoprecipitation. The Δhem1/Δtup1 strain was transformed with a low-copy plasmid encoding an HA epitope-tagged allele of TUP which complements the loss of Tup1p. In cells grown in high iron, there was little binding of Tup1p to the promoter regions of FET3 and FTR1. However, cells grown in iron-limited medium in the presence of heme showed significant recruitment of Tup1p (Figure 4). These results suggest that the binding of Tup1p requires the initial binding of Aft1p.
Figure 4.

Tup1p is recruited to iron-responsive promoters in response to changes in media iron. The Δhem1 strain was transformed with an epitope-tagged allele of TUP1 (pTUP1-HA). Cells were grown in the indicated media for 6 h and Tup1p-HA was crosslinked to DNA using formaldehyde treatment. Cells were harvested, disrupted, and chromatin was isolated. Chromatin was sheared using sonication to achieve an average fragment size of approximately 500 bp, with a range between 100 and 1500 bp. Chromatin was immunoprecipitated using the HA epitope of Tup1p. After crosslinking was reversed, PCR was performed using amplicons, which flank the Aft1p binding site of the indicated promoters, with CMD1 used as a positive control.
Differential response of the iron regulon to heme-deficient repression
Aft1p is responsible for the transcriptional activation of approximately 20 genes, collectively referred to as the iron regulon (Rutherford et al, 2003). We performed microarray analysis of iron-limited cells in the presence and absence of heme to determine whether transcription of all Aft1p-regulated genes was similarly affected by the absence of heme. As expected, transcription of both genes that encode the high-affinity iron transport system, FET3 and FTR1, was repressed by the absence of heme (Table I). Similarly, one of the heme-dependent reductases, FRE2, also was not expressed in the absence of heme. Surprisingly, other genes regulated by Aft1p could still be induced by low iron in the absence of heme, with some showing a higher level of expression than in the presence of heme. In particular, the cell wall mannoprotein, FIT1, and two siderophore transporters, ARN1 and ARN2, were expressed at high levels. The lack of heme-deficient repression of ARN1 was confirmed by S1 nuclease protection analysis (Figure 5A). Expression of ARN1 was sensitive to iron in both heme-replete and heme-depleted cells, with a slightly higher expression in the absence of heme (e.g. compare FTR1 in Figure 2C). An ARN1 reporter construct containing the Aft1p binding region with endogenous spacing to the TATA box permitted iron-dependent regulation in the presence and absence of heme (Figure 5B), indicating that the RNA profile seen in the S1 analysis was due to transcriptional activation rather than message stability. To determine whether heme- and iron-sensitive induction was Aft1p dependent, we examined the expression of ARN1 reporter constructs transformed into strains with deletions in AFT1, its paralogue AFT2, and strains with deletions in both AFT1 and AFT2. In the absence of Aft1p, there was no expression of either FET3 or ARN1 reporter constructs under low-iron conditions, regardless of cellular heme status (data not shown). These results indicate that Aft1p but not Aft2p was responsible for the heme-deficient as well as the heme-replete transcriptional activation of FET3 and ARN1. There were slight quantitative differences between the S1 and lacZ data. One possible explanation for these differences is that mRNA may be regulated post-transcriptionally. Two recent reports have shown post-transcriptional regulation for mRNA involved in iron metabolism (Lee et al, 2005; Puig et al, 2005).
Table 1.
Expression of iron-depleted cells grown under heme-deficient conditions relative to normal heme conditions
| Target gene | Log(2) expression | Function |
|---|---|---|
| FET3 | −2.07 | Multicopper oxidoreductase |
| FTR1 | −2.86 | Iron permease |
| FRE1 | 0.06 | Metalloreductase |
| FRE2 | −1.31 | Metalloreductase |
| FRE3 | −0.13 | Ferric reductase |
| FRE5 | 0.21 | Ferric reductase (putative) |
| FRE6 | 0.80 | Ferric reductase (putative) |
| FIT1 | 3.73 | GPI-anchored cell wall mannoprotein |
| FIT2 | 0.93 | GPI-anchored cell wall mannoprotein |
| FIT3 | −1.39 | GPI-anchored cell wall mannoprotein |
| ARN1 | 1.69 | Siderophore-iron uptake |
| ARN2 (TAF1) | 1.17 | Siderophore-iron uptake |
| SIT1 (ARN3) | 0.68 | Siderophore-iron uptake |
|
ENB1 (ARN4) |
0.36 |
Siderophore-iron uptake |
| The value shown is Log(2) expression measured in two experiments. For each experiment, labeling was reversed and replicates were combined. The Log(2) expression represents the average of four different microarray points. Full microarray data have been deposited with Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE3470. | ||
Figure 5.

Aft1p-regulated genes respond differently to heme deficiency. (A) The Δhem1 strain was grown for 6 h in the media as indicated. Cells were harvested and total RNA was isolated. S1 nuclease protection assay was performed on ARN1 and FIT1 (not shown), with CMD1 as a positive control. (B) Δhem1 cells were transformed with the indicated reporter constructs. Strains were induced in the presence of 50 μM FeSO4 or 80 μM BPS in the indicated media. Cells were grown for 6 h, harvested, and a β-galactosidase assay was performed. (C) Chromatin samples were prepared as in Figure 4. PCR was performed using amplicons, which flank the Aft1p binding site of the indicated promoters, with CMD1 used as a positive control.
Lesuisse et al (2001) showed that deletion of TUP1 and/or SSN6 led to an increase in siderophore transport activity in iron-containing medium. Their observation suggested a loss of transcriptional repression, although mRNA levels were not measured. We observed that while Δhem1 cells grown in ALA-containing medium showed a slight increase in ARN1 mRNA in the presence of iron, iron-sensitive transcription was maintained in both heme-replete and heme-depleted wild-type and TUP1-deleted strains (data not shown). As deletion of Tup1p was required for repression of FET3/FTR1 in the absence of heme, we examined whether Tup1p was recruited to the ARN1 and FIT1 promoters, using chromatin immunoprecipitation. When Aft1p was activated by low iron in the presence of heme, Tup1p was found on the promoters of both ARN1 and FIT1 (Figure 5C). Under heme-deficient conditions when iron was limited, however, Tup1p was unable to immunoprecipitate the Aft1p binding region from the promoter of either ARN1 or FIT1. These results suggest that Tup1p was not required for iron-sensitive expression but was required for repression of a set of genes in the absence of heme. The absence of Tup1p at the promoters of iron-sensitive genes defines whether these genes are expressed in the absence of heme.
To identify features of the ARN1 sequence responsible for the differential response to heme loss, we converted sequences in the endogenous ARN1 promoter to those found in FET3 by site-specific mutagenesis (Figure 6A). Changing 14 bp immediately downstream of the consensus Aft1p binding site in the full-length ARN1 construct to that found in FET3 retained iron-sensitive expression but led to loss of expression in the absence of heme. To determine if this sequence was sufficient to permit expression in the absence of heme, we converted the corresponding sequence in the full-length FET3 promoter to that found in the ARN1 promoter. Replacement of the 14 bp immediately downstream of the Aft1p binding site in the wild-type FET3 promoter with those found in ARN1 permitted expression of FET3 in the absence of heme under low-iron conditions. Similarly, replacement of the sequence immediately downstream of the Mac1p binding site of CTR1 with the ARN1 sequence increased the expression of CTR1 under heme-deficient conditions (Figure 6B). Thus, the ARN1 sequence was both necessary and sufficient to permit transcription in the absence of heme.
Figure 6.
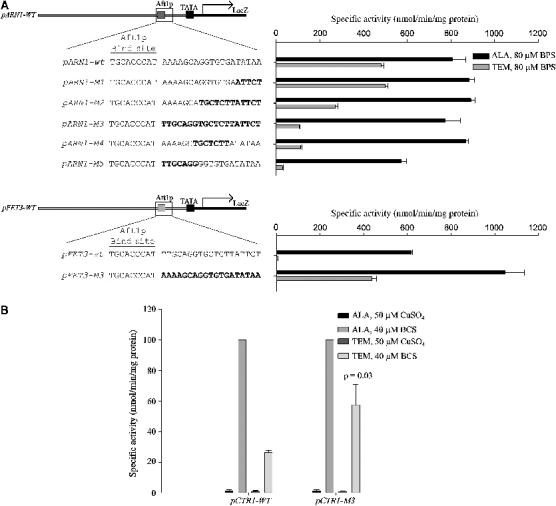
Mapping the differential response to heme deficiency. (A) Site-directed base-pair changes were made in the full-length wild-type ARN1 and FET3 constructs as shown. The Δhem1 strain was transformed with the indicated reporter constructs. Strains were induced in the presence of 50 μM FeSO4 or 80 μM BPS in the indicated media. Cells were grown for 6 h, harvested, and a β-galactosidase assay was performed. (B) Δhem1 cells were transformed with the indicated reporter constructs. Cells were induced in the presence of 50 μM CuSO4 or 40 μM BCS. Cells were grown for 6 h, harvested, and a β-galactosidase assay was performed.
Cti6p is required for the differential response to heme deficiency
The absence of Tup1p on the promoters of ARN1 and FIT1 was consistent with a lack of repression of these genes in the absence of heme. The question that arises is what promotes the loss of (or prevents the binding of) Tup1p from these promoters. Studies have suggested that Cti6p plays a role in both iron metabolism (Puig et al, 2004) and in overcoming Tup1p-mediated repression at selected genes (Papamichos-Chronakis et al, 2002). Deletion of CTI6 had no effect on iron-sensitive transcription of FET3 or copper-sensitive transcription of CTR1 either in the absence or presence of heme. Deletion of CTI6, however, led to transcriptional repression of an ARN1-lacZ reporter construct in the absence of heme (Figure 7A, pARN1-lacZ). Further, deletion of CTI6 resulted in the loss of expression of the FET3-lacZ hybrid reporter construct with the 14 bp from ARN1 inserted after the Aft1p binding site (Figure 7A, pFET3-M3). This result indicates that Cti6p was required to overcome the inhibitory effect of Tup1p.
Figure 7.
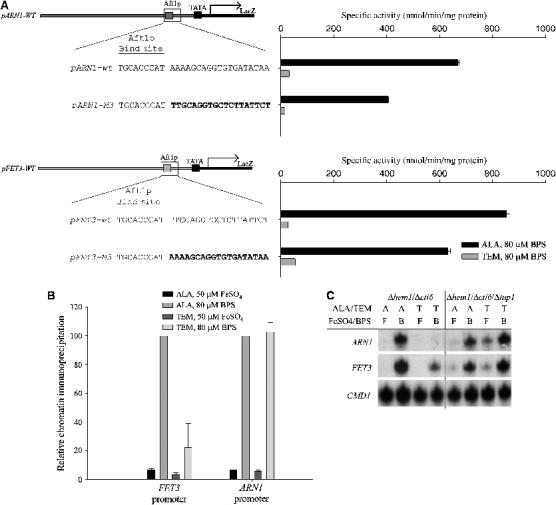
Cti6p mediates induction of ARN1 when heme biosynthesis is disrupted. (A) The Δhem1/Δcti6 strain was transformed with the indicated reporter constructs. Strains were induced in the presence of 50 μM FeSO4 or 80 μM BPS in the indicated media. Cells were grown for 6 h, harvested, and a β-galactosidase assay was performed. (B) The Δhem1/Δcti6 strain was transformed with a myc epitope-tagged Cti6p. Chromatin was prepared as in Figure 4. PCR was performed using amplicons, which flank the Aft1p binding site of FET3 and ARN1, with CMD1 used as a positive control. (C) Strains were induced in the indicated media for 6 h. Cells were harvested and total RNA was isolated. S1 nuclease protection assay was performed on FET1 and FTR1, with CMD1 as a positive control.
To characterize the recruitment of Cti6p to Aft1p-regulated promoters, we generated a c-myc epitope-tagged allele of CTI6. This construct complemented the repression of ARN1 expression in the Δcti6 strain (data not shown). The CTI6-myc construct was transformed into a Δhem1/Δcti6 strain, and chromatin immunoprecipitation was performed on cells under iron-replete or iron-limiting conditions in the presence and absence of heme (Figure 7B). Under iron-replete conditions, Cti6p showed minimal promoter occupancy of the Aft1p binding site of both FET3 and ARN1. Iron limitation under heme-replete conditions led to an increase in recruitment of Cti6p to the promoter of both genes, coincident with the recruitment of both Aft1p (data not shown) and Tup1p (Figures 4 and 5C). When iron was depleted under heme-deficient conditions, there was a differential recruitment of Cti6p: the FET3 promoter showed a decrease in recruitment of Cti6p, whereas the amount of Cti6p on the ARN1 promoter was unaffected by the absence of heme. The presence of Cti6p on the promoter of ARN1 under heme-depleted conditions was consistent with previous data suggesting that Cti6p was necessary to overcome repression.
For the expression of ARN1 under heme-depleted conditions, our data suggested that Tup1p was dispensable whereas Cti6p was required. Cti6p is thought to interact with Tup1p to overcome repression at certain Tup1p/Ssn6p target promoters (Papamichos-Chronakis et al, 2002). To test whether the loss of Tup1p can override the ARN1 repression that was mediated by the Cti6p mutation (Figure 7A), we generated a strain with deletions in HEM1, TUP1, and CTI6 and examined both ARN1 and FET3 expression by S1 analysis (Figure 7C). The Δhem1 and the Δhem1/Δtup1 strain confirmed our previous work and showed the predicted message profile for both FET3 and ARN1 (data not shown). Decreased expression of both FET3 and ARN1 under heme-depleted conditions occurred in Δhem1/Δcti6 cells, consistent with our β-galactosidase reporter construct data. The triple deletion Δhem1/Δtup1/Δcti6 strain showed inducible expression of both FET3 and ARN1 under heme-depleted conditions, identical to that seen in the Δhem1/Δtup1. As ARN1 expression under heme-depleted conditions in the triple deletion was epistatic to the Δhem1/Δcti6 strain, we conclude that the repression that was mediated by loss of Cti6p is dependent on the presence of Tup1p.
Discussion
Acquisition of iron and copper is reduced in yeast grown under anaerobic conditions owing to a decrease in the transcription of genes that encode both high-affinity iron and copper transporters (Crisp et al, 2003). In this communication, we define the mechanism underlying the lack of transcription and demonstrate that the lack of transcription was due to repression. Through a genetic screen, we identified Tup1p as the repressor that responds to heme deficiency: the absence of Tup1p lead to transcription of both FET3 and CTR1 in the absence of heme. Reintroduction of Tup1p into either the mutant identified by the screen or a Δhem1/Δtup1 deletion strain restored repression of FET3 transcription. Finally, chromatin immunoprecipitation analysis demonstrated that Tup1p occupies the FET3 and FTR1 promoters. The effect of Tup1p is mediated through chromosome remodeling, as suggested by the fact that deletion of HDA1, which encodes a histone deacetylase, also relieves the heme-deficient repression. Previous studies have shown that Tup1p-Ssn6p is required for the repression of anaerobic genes, as it is recruited to anaerobic genes by Rox1p and Mot3p (Kastaniotis and Zitomer, 2000). This is the first study that shows that this repressor complex is required for transcriptional inhibition of genes that are not utilized under anaerobic conditions.
The repression of FET3/FTR1 requires the recruitment of Tup1p to the Aft1p binding region (Figure 8), as shown by genetic analysis as well as the chromatin immunoprecipitation studies. These results imply that Tup1p is recruited by Aft1p and suggests a direct interaction between the two proteins. While Aft1p is a transcriptional activator, regulation of the homologous iron transport genes in the fission yeast Schizosaccharomyces pombe results from the repressive action of the iron-sensing transcription factor Fep1p, iron limitation resulting in the release of repression. Deletion of TUP11 (the S. pombe TUP1 homologue) resulted in constitutive expression of the iron regulon. A yeast two-hybrid analysis showed that the carboxy termini of Fep1p and Tup11p directly interacted (Znaidi et al, 2004). Thus, in both the budding and fission yeasts, Tup1 is required for repression.
Figure 8.

Model for heme regulation of FET3 and ARN1 expression. Under normal heme conditions, Aft1p binds to the promoters of both FET3 and ARN1, leading to the recruitment of Tup1p and Cti6p. Under low-iron conditions in the absence of heme, Tup1p is able to mediate repression of FET3 through the chromatin remodeling protein Hda1p. The sequence immediately downstream of the Aft1p binding site of ARN1 mediates retention of Cti6p under heme-depleted conditions, resulting in loss of Tup1p and expression of ARN1.
Tup1p and Ssn6p are required for the transcriptional activation of FRE2 and ARN2, both Aft1p-regulated genes, as no transcription was seen in the absence of either of the co-repressors (Fragiadakis et al, 2004). The study also reported that deletion of Tup1p did not prevent the transcriptional activation of other members of the iron regulon including FET3, FIT2, or FRE3. Using reporter constructs, we observed at best a two-fold decrease in transcriptional activation of FET3 in the absence of Tup1p, suggesting a modest role in transcriptional activation. We observed that Tup1p is bound to the FET3 promoter in both the presence and absence of heme. Studies have shown a specific role for the Hap/Rox/Mot proteins in the repression of anaerobic genes (Kwast et al, 1998). Hap1p is an activator of many genes that encode heme-containing proteins which can also act as a repressor. However, we found no effect of deletion of either HAP genes (HAP1, HAP2, HAP5) or MOT3, ROX1 on repression or activation of FET3. Currently, we do not know how Tup1p senses or responds to heme.
Fragiadakis et al (2004) first showed differential regulation of the iron regulon, as deletion of SSN6 affected the iron-sensitive regulation of some but not all Aft1p-regulated genes. We also observed differential regulation of Aft1p-regulated genes, as the absence of heme represses genes that encode for the oxygen-dependent iron transport system (FET3/FTR1), but not for the siderophore-iron transport system, which is oxygen independent. The transcriptional activation of ARN1 in the absence of heme is mediated by retention of Cti6p, overriding Tup1p-mediated repression (Figure 8). Cti6p was originally identified by its ability to overcome Tup1p/Ssn6p repression at the GAL1 and ANB1 promoters by recruiting the SAGA complex (Papamichos-Chronakis et al, 2002); however, subsequent work was unable to reproduce this result with the ANB1 promoter (Mennella et al, 2003). More recently, Cti6p was shown to be required for growth under low-iron conditions, through an interaction with the Sin3p/Rpd3p histone deacetylase complex (Puig et al, 2004).
Our data show that Cti6p is recruited to iron-responsive promoters in an Aft1p-dependent manner. A direct role for Cti6p was demonstrated by its ability to mediate expression of ARN1 under heme-deficient conditions. We also present data that show that a 14 bp sequence immediately adjacent to the conserved Aft1p binding site in ARN1 is able to mediate Cti6p retention, resulting in relief of Tup1p-mediated repression. Site-directed mutagenesis of this sequence in the ARN1 promoter results in repression in the absence of heme. Addition of this sequence to the promoter of FET3 or CTR1 permits their expression in the absence of heme. Thus, this sequence is both necessary and sufficient to overcome repression. The presence of Cti6p at the FET3 promoter under normal heme conditions shows that the ARN1 promoter sequence is not required for Cti6p recruitment. It does, however, show that the ARN1 sequence is required for retention of Cti6p, and subsequently expression, when heme is depleted. Our data suggest that in the absence of heme, Cti6p and the 14 bp sequence lead to the loss of Tup1p, permitting transcription in the absence of heme.
The high-affinity iron transport system comprised of Fet3p and Ftr1p is oxygen dependent, as Fet3p is a ferroxidase, which couples the oxidation of iron with the reduction of molecular oxygen (Askwith et al, 1994). Further, the substrate for Fet3p/Ftr1p is ferrous iron, which under aerobic conditions requires activity of heme-containing ferri-reductases. In the absence of oxygen/heme, elemental iron transport results from increased expression of Fet4p (Jensen and Culotta, 2002; Waters and Eide, 2002). Transcription of FET4 is regulated by not only iron but by Rox1p. Consequently, under heme-deficient conditions, FET4 expression is increased. The transport of siderophores, mediated by the ARN genes, however, is heme and oxygen independent (Lesuisse and Labbe, 1989). Saccharomyces cannot synthesize siderophores, but can accumulate siderophores secreted by other organisms (Yun et al, 2000). The presence of siderophores would lower available iron, resulting in the activation of Aft1p and expression of the siderophore transporter genes. The ability to accumulate siderophores under anaerobic conditions may permit Saccharomyces not only to accumulate iron for its own needs but also to deny iron to competing organisms.
Materials and methods
Strains, plasmids, and growth media
The strains used in this study were derived from W303 and have been used in previous studies (Askwith et al, 1994). The Δhem1 strain was constructed by insertion of the LEU2 gene into the HEM1 locus of the DY1457 strain (MATα ura3-52, trp1-1, his3-11, ade6, can1-100(oc), hem1∷LEU2) (Crisp et al, 2003). The Δhem1 strain used in these studies has the following genotype: MATα ura3-52, trp1-1, his3-11, can1-100(oc), hem1∷LEU2. For all double deletion strains, the respective KanMX4 deletion cassette was PCR amplified from the homozygous diploid deletion collection (Research Genetics, Palo Alto, CA), and was transformed into the Δhem1 strain. Successful integrants were selected by resistance to G418, and verified by PCR amplification of the 5′ and 3′ KanMX4 integration sites. The Δhem1 strain was maintained in either YPD or CM media supplemented with 50 μg/ml ALA (Frontier Scientific, Logan, UT). To deplete heme, cells were washed three times in H2O, and re-inoculated into media containing a source of unsaturated fatty acids, sterols, and methionine (TEM media). 250 × Tween/ergosterol supplement was made by mixing 50:50 ethanol/Tween 80, followed by addition of ergosterol to a final concentration of 7.5 mg/ml. 500 × methionine stock was made at 27.8 mg/ml. Cells were grown for 6 h in TEM to deplete heme before being used in experiments. β-Galactosidase reporter constructs were generated by cloning out the indicated promoter region by PCR and insertion into the empty vector pJS128 (Crisp et al, 2003). A 5′ c-myc epitope-tagged allele of CTI6 was cloned into pRS314 (Trp) under the native CTI6 promoter. Constructs were sequenced before use in reporter assays. The YCp(23)TUP1-HA3 construct was a gift from Dr Richard Zitomer (State University of New York, Albany, NY). The pRS413-HDA1 construct was a gift from Dr David Stillman (University of Utah, Salt Lake City, UT).
Galactosidase reporter assay
The β-galactosidase reporter construct assay was performed as described previously (Crisp et al, 2003). A zinc-responsive galactosidase reporter construct containing the promoter sequence of the ZRT1 gene was a gift from Dr David Eide (University of Wisconsin, Madison, WI). A copper-sensitive β-galactosidase reporter construct containing the promoter sequence of CTR1 was a gift from Dr Dennis Winge (University of Utah, Salt Lake City, UT).
S1 nuclease protection assay
The S1 nuclease protection assay protocol was performed as described previously (Crisp et al, 2003). S1 probes used are as follows: CMD1−39+6: GGG CAA AGG CTT CTT TGA ATT CAG CAA TTT GTT CTT CGG GGG AGC; FET3−52+6: CAA GAA GGG CAC ACC GTC CAT AGA GGC GGT TCC GTT TTG GAA GAG ACC GTG GGG GCA T; ARN1−64+6: CTG AGG GAG ACC ATA TGA GTC GGA GTT GGA ATC GTT ATG AGT CTC AGG ATT CAA CTC GTG TTC CGC CTT T; FTR1−60+6: CCC CGA TTG CCT GTT TCA AAA ACG ATA GCA GCA CGG AAA TAA CAA TCA CTG CTT CCA AGC CAG AGA; FIT1−54+6: GGC GTA CCG GTC TTA CGC AGC CTT GAT TTA GTT GTG AGA AAG TTC TGC ATG GTG GTT CTT CCC GGA.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described (Crisp et al, 2003). Chromatin was immunoprecipitated using an antibody against an HA epitope-tagged Tup1p (Mennella et al, 2003) or a c-myc epitope-tagged Cti6p (this study). Rabbit polyclonal anti-HA antibody was a gift from Dr Tom Stevens (University of Oregon, Eugene, OR). The primers used for the Aft1p binding region of the FET3 promoter produced a 287 bp fragment: FET3-F GGA CCA ATA AAT GGT CTT CTG CGA, FET3-R CAT ACT CCC TCG AAG GAT CGA CT. The primers used for the Aft1p binding region of the ARN1 promoter produced a 326 bp fragment: ARN1-F CAT GTA AGC GAC CGC GAT GAA G, ARN1-R CTG TAC TCC GAT TTG CTT GGC. The primers used for the Aft1p binding region of the FTR1 promoter produced a 350 bp fragment: FTR1-F CTG AGA CAT CAG ACC AAC GTG TAG, FTR1-R CGA ACT ACA GCC CAT CGC TTC. The primers used for the Aft1p binding region of the FIT1 promoter produced a 298 bp fragment: FIT1-F GGA TCT CAC ATT AAT CGG TTT ATG CAT CGC, FIT1-R GCA CCC TTT CCT AGA ACT CGC. The primers used for CMD1 promoter produced a 252 bp fragment: CMD1-F CGC TTC CTC TCA ATT CCC AAA GT, CMD1-R GTG ATG TAG GAC ACT CTC CAA GG. Immunoprecipitated DNA was PCR amplified, run on a 2.5% agarose gel, and imaged using a Bio-Rad phosphorimager.
Microarray analysis
The Δhem1 strain was grown overnight in YPD media supplemented with ALA. Cells were washed three times with water to remove excess ALA. Cells were then inoculated into 500 ml cultures of low-iron YPD media (80 μM BPS) that was supplemented either with ALA or TEM. Cultures were induced for 6 h before cells were harvested and washed three times in DEPC water. The culture density at the time of harvest was approximately OD600=0.7. Total RNA was isolated using the protocol outlined in the S1 analysis (Crisp et al, 2003). mRNA was isolated using a Poly-A Tract mRNA isolation kit from Promega. For the microarray analysis, two different experiments were performed with reversed probe labeling. The Log(2) of the average of four independent hybridization measurements is presented in Table I. Microarray was performed by the University of Utah Microarray Facility, and the full data set has been deposited with Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE3470.
Genetic screen
The Δhem1 strain was transformed with a low-copy plasmid harboring the constitutive active allele AFT1-1up, a high-copy plasmid containing FET3-lacZ, and a chromosomally integrated FET3-GFP chimera. In the presence of ALA, this strain expresses β-galactosidase and Fet3p-GFP independent of media iron concentration as a result of the constitutively active AFT1-1up allele. In the absence of ALA, there is no expression of either β-galactosidase or Fet3p-GFP. Cells were grown overnight in media containing ALA followed by three washes in H2O. Cells were diluted and approximately 400 colony-forming units were plated onto TEM plates followed by UV treatment at a dose empirically determined to kill 50% of the colonies. Plates were grown in an anaerobic chamber for 72 h and colonies were examined by β-galactosidase lift assay (Breeden and Nasmyth, 1985). Positives were identified as blue colonies, and were verified by microscopy of FET3-GFP. Positives clones were crossed with a wild-type strain and tested for recessive/dominance. To identify recessive mutants, a low-copy library was transformed into the positive clones, and cells were plated onto TEM plates and grown anaerobically. Following β-galactosidase lift assay, positives with a complementing plasmid were identified as white colonies. Complementing plasmids were rescued and sequenced.
Acknowledgments
We thank Drs R Zitomer and D Stillman for their generous gift of plasmids and strains. This work was supported by National Institutes of Health Grants DK52380 (to JK) and by National Institutes of Health Training Grant T32 DK07115-29 (to RJC and EMA). Support for use of the Core Facilities was provided through Grant NCI-CCSG P30CA 42014 from the NCI, National Institutes of Health.
References
- Askwith C, Eide D, Van Ho A, Bernard PS, Li L, Davis-Kaplan S, Sipe DM, Kaplan J (1994) The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76: 403–410 [DOI] [PubMed] [Google Scholar]
- Bardos JI, Ashcroft M (2005) Negative and positive regulation of HIF-1: a complex network. Biochim Biophys Acta 1755: 107–120 [DOI] [PubMed] [Google Scholar]
- Becerra M, Lombardia-Ferreira LJ, Hauser NC, Hoheisel JD, Tizon B, Cerdan ME (2002) The yeast transcriptome in aerobic and hypoxic conditions: effects of hap1, rox1, rox3 and srb10 deletions. Mol Microbiol 43: 545–555 [DOI] [PubMed] [Google Scholar]
- Breeden L, Nasmyth K (1985) Regulation of the yeast HO gene. Cold Spring Harb Symp Quant Biol 50: 643–650 [DOI] [PubMed] [Google Scholar]
- Crisp RJ, Pollington A, Galea C, Jaron S, Yamaguchi-Iwai Y, Kaplan J (2003) Inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast. J Biol Chem 278: 45499–45506 [DOI] [PubMed] [Google Scholar]
- Fragiadakis GS, Tzamarias D, Alexandraki D (2004) Nhp6 facilitates Aft1 binding and Ssn6 recruitment, both essential for FRE2 transcriptional activation. EMBO J 23: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SR, Johnson AD (2004) Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol Biol Cell 15: 4191–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SR, Johnson AD (2005) Genome-wide analysis of the functions of a conserved surface on the corepressor Tup1. Mol Biol Cell 16: 2605–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LT, Culotta VC (2002) Regulation of Saccharomyces cerevisiae FET4 by oxygen and iron. J Mol Biol 318: 251–260 [DOI] [PubMed] [Google Scholar]
- Kastaniotis AJ, Zitomer RS (2000) Rox1 mediated repression. Oxygen dependent repression in yeast. Adv Exp Med Biol 475: 185–195 [PubMed] [Google Scholar]
- Kwast KE, Burke PV, Poyton RO (1998) Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J Exp Biol 201 (Part 8): 1177–1195 [DOI] [PubMed] [Google Scholar]
- Lee A, Henras AK, Chanfreau G (2005) Multiple RNA surveillance pathways limit aberrant expression of iron uptake mRNAs and prevent iron toxicity in S. cerevisiae. Mol Cell 19: 39–51 [DOI] [PubMed] [Google Scholar]
- Lesuisse E, Blaiseau PL, Dancis A, Camadro JM (2001) Siderophore uptake and use by the yeast Saccharomyces cerevisiae. Microbiology 147: 289–298 [DOI] [PubMed] [Google Scholar]
- Lesuisse E, Labbe P (1989) Reductive and non-reductive mechanisms of iron assimilation by the yeast Saccharomyces cerevisiae. J Gen Microbiol 135: 257–263 [DOI] [PubMed] [Google Scholar]
- Mennella TA, Klinkenberg LG, Zitomer RS (2003) Recruitment of Tup1-Ssn6 by yeast hypoxic genes and chromatin-independent exclusion of TATA binding protein. Eukaryot Cell 2: 1288–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Petrakis T, Ktistaki E, Topalidou I, Tzamarias D (2002) Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol Cell 9: 1297–1305 [DOI] [PubMed] [Google Scholar]
- Puig S, Askeland E, Thiele DJ (2005) Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120: 99–110 [DOI] [PubMed] [Google Scholar]
- Puig S, Lau M, Thiele DJ (2004) Cti6 is an Rpd3-Sin3 histone deacetylase-associated protein required for growth under iron-limiting conditions in Saccharomyces cerevisiae. J Biol Chem 279: 30298–30306 [DOI] [PubMed] [Google Scholar]
- Rosenfeld E, Beauvoit B (2003) Role of the non-respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae. Yeast 20: 1115–1144 [DOI] [PubMed] [Google Scholar]
- Rutherford JC, Jaron S, Winge DR (2003) Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J Biol Chem 278: 27636–27643 [DOI] [PubMed] [Google Scholar]
- Smith RL, Johnson AD (2000) Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci 25: 325–330 [DOI] [PubMed] [Google Scholar]
- Waters BM, Eide DJ (2002) Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J Biol Chem 277: 33749–33757 [DOI] [PubMed] [Google Scholar]
- Wu J, Suka N, Carlson M, Grunstein M (2001) TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol Cell 7: 117–126 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Dancis A, Klausner RD (1995) AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J 14: 1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CW, Ferea T, Rashford J, Ardon O, Brown PO, Botstein D, Kaplan J, Philpott CC (2000) Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae. Evidence for two pathways of iron uptake [In Process Citation]. J Biol Chem 275: 10709–10715 [DOI] [PubMed] [Google Scholar]
- Znaidi S, Pelletier B, Mukai Y, Labbe S (2004) The Schizosaccharomyces pombe corepressor Tup11 interacts with the iron-responsive transcription factor Fep1. J Biol Chem 279: 9462–9474 [DOI] [PubMed] [Google Scholar]


