Abstract
Telomeres are regulated by a homeostatic mechanism that includes telomerase and telomeric repeat binding proteins, TRF1 and TRF2. Recently, it has been hypothesized that telomeres assume distinct configurations in a cell-cycle-dependent manner, although direct biochemical evidence is lacking. Here we demonstrated that Xenopus TRF1 (xTRF1) associates with telomere chromatin specifically in mitotic Xenopus egg extracts, and dissociates from it upon mitotic exit. Both the N-terminal TRF-homology (TRFH) domain and the linker region connecting the TRFH domain and the C-terminal Myb domain are required for this cell-cycle-dependent association of xTRF1 with chromatin. In contrast, Xenopus TRF2 (xTRF2) associates with chromatin throughout the cell cycle. We showed that Polo-like kinase (Plx1) phosphorylates xTRF1 in vitro. Moreover, the mitotic xTRF1–chromatin association was significantly impaired when Plx1 was immunodepleted from the extracts. Finally, high telomerase activities were detected in association with replicating interphase chromatin compared with mitotic chromatin. These results indicate that telomere chromatin is actively regulated by cell-cycle-dependent processes, and provide an insight for understanding how telomeres undergo DNA metabolisms during the cell cycle.
Keywords: Polo-like kinase, telomerase, telomere, TRF1, TRF2
Introduction
Eukaryotic chromosomes terminate in nucleoprotein complexes, termed telomeres, that are composed of arrays of telomeric repetitive sequences and telomere-binding proteins (reviewed in de Lange, 2005). One strand of telomeric DNA, the 3′-end of which faces the DNA end, is G-rich (TTAGGG repeats in vertebrates, G-strand), whereas the other strand is C-rich (C-strand). At the extreme end of telomeric DNA, the G-strand extends its 3′-terminus as a single-stranded DNA, a structure called the G-tail (Wellinger et al, 1993). Telomeres protect the ends of chromosomes from noxious reactions such as end-to-end fusion and degradation. Telomeric DNA, together with specific and nonspecific telomere-binding proteins, participates in forming higher ordered structures that protect chromosomal ends (reviewed in de Lange, 2005).
Telomeric DNA becomes shortened every time cells divide, due to incomplete DNA replication at the most terminal lagging-strand synthesis (the end replication problem (Harley, 1991; Ohki et al, 2001)). Telomerase, a specialized form of reverse transcriptase containing its own template RNA, counteracts telomere shortening (reviewed in Cech, 2004; Chan and Blackburn, 2004). Two components, namely, the catalytic subunit, TERT (telomerase reverse transcriptase), and the RNA template, TR (telomerase RNA), are essential for telomerase activity measured in vitro. It is only in some specific types of normal human cells, including germ cells, actively dividing embryonal cells and adult progenitor cells of proliferating tissues, that telomerase activity has been detected.
There is mounting evidence that telomerase activity is regulated in cis at individual telomeres by the number of associated telomeric-DNA-binding proteins (the protein-counting model) (Marcand et al, 1997). Such proteins include Rap1p in the budding yeast, Saccharomyces cerevisiae; Taz1 in the fission yeast, Schizosaccharomyces pombe; and TRF1 (TTAGGG repeat binding factor 1) in mammals. These proteins share the Myb-like domain, and Taz1 and TRF1 form homodimers that bind to telomeric DNA through the TRF-homology (TRFH) domain (Bianchi et al, 1997; Spink et al, 2000). TRF1 negatively regulates telomere length, as evidenced by observations that telomere lengths are decreased and increased by the overexpression of wild-type and dominant-negative mutants, respectively (van Steensel and de Lange, 1997). TRF2, a paralog of TRF1, also binds to telomeric DNA as a homodimer through the Myb-like domain (Bilaud et al, 1997; Broccoli et al, 1997). TRF2 plays an important role in protecting chromosomal ends, rather than controlling telomerase reaction, because the dominant-negative mutant expression induces chromosomal end-to-end fusion (van Steensel et al, 1998). It has been proposed that these telomere-binding proteins regulate the accessibility of telomerase as well as protect chromosomal ends by determining the configuration of the telomere complex. Specifically, it has been proposed that TRF1 and TRF2 collaborate to form a terminal loop structure called the t-loop, thereby concealing the 3′-end of the G-tail from the action of telomerase or other enzymatic activities (Griffith et al, 1999; de Lange, 2004). One recent study of budding yeast has demonstrated that short telomeres have a higher probability of being extended by telomerase than long telomeres, suggesting the presence of two telomere configurations, one accessible and the other inaccessible to telomerase (Teixeira et al, 2004). In addition, another study has provided evidence implicating the presence of an intermediate state of telomere configuration that contains TRF2 but lacks TRF1, thereby protecting the chromosomal ends, yet allowing access by telomerase (Chang et al, 2003). However, it is not known how and when in the cell cycle these distinct telomere configurations transit among each other. As the G-strand synthesis by telomerase is intimately coupled with the C-strand synthesis by the conventional lagging-strand DNA synthesis (Nakamura et al, 2005; reviewed in Price, 1997), the telomerase reaction should proceed coordinately with DNA replication in S phase. Indeed, budding yeast telomerase elongates telomeres in late S phase (Marcand et al, 2000). This argument suggests that telomere configuration is regulated in a cell-cycle-dependent manner, such that it becomes accessible to telomerase only transiently within the narrow window of S phase when telomere replication occurs.
Here we investigated whether telomere chromatin is dynamically regulated during the cell cycle through biochemical approaches using unfertilized Xenopus egg extracts, in which high levels of telomerase and cyclin B–Cdc2 kinase activities are retained and cyclin B destruction is triggered by the addition of Ca2+ (Murray, 1991; Mantell and Greider, 1994). We found that Xenopus TRF1 (xTRF1) dynamically associates with mitotic telomere chromatin and actively dissociates from interphase telomere chromatin in Xenopus egg extracts. Moreover, we demonstrated that Polo-like kinase (Plx1), a Xenopus member of the Plx1 family, plays a critical role in regulating the dynamic behavior of TRF1. We also provide evidence that Xenopus TRF2 (xTRF2) is bound to telomere chromatin throughout the cell cycle and phosphorylated in a cell-cycle-dependent manner. These results provide a new avenue for understanding the regulatory mechanism of telomere configurations.
Results
Identification of xTRF1 and xTRF2
We searched the EST database for the Xenopus homologs of TRF1 and TRF2 (xTRF1 and xTRF2) and obtained Xenopus EST sequences having high homology with human TRF1 (hTRF1) or TRF2 (hTRF2) (accession nos. BG552350 and BJ638816, respectively). Several independent clones containing the EST sequence were isolated, and the composite full-length xTRF1 and xTRF2 cDNA sequences encoding open reading frames of 420 and 468 amino acids, respectively, were deduced (Supplementary Figure 1). We noticed that whereas mouse and human TRF1 and TRF2 proteins possess amino-terminal regions that are rich in acidic and basic amino acids, respectively, xTRF1 and xTRF2 lack such regions. We demonstrated that recombinant xTRF1 and xTRF2 produced in reticulocyte lysates or Escherichia coli form homodimers (data not shown) and bind to double-stranded telomeric DNA in electrophoretic mobility shift assay (EMSA) as human TRF proteins do (Supplementary Figure 2).
Chromatin association of xTRF1 and xTRF2 during the cell cycle in Xenopus egg extracts
When Xenopus sperm (XSP) chromatin is incubated with extracts prepared from unfertilized Xenopus eggs arrested at meiotic metaphase II (CSF extracts for cytostatic-factor-arrested extracts), condensed mitotic chromatin is assembled (Murray, 1991). Therefore, CSF extracts recapitulate M phase events. The addition of Ca2+ to CSF extracts induces cyclin B destruction through the ubiquitin–APC–proteasome pathway, leading to the inactivation of cyclin B–Cdc2 kinase and a transition to interphase (interphase extracts) (Glotzer et al, 1991). Accordingly, upon the addition of Ca2+, the condensed chromatin in CSF extracts becomes decondensed, forms the nuclear envelope, and undergoes DNA replication. Using this system, we investigated how the chromosomal association of Xenopus TRF proteins is regulated during the cell cycle.
Despite repeated trials, we were unable to prepare anti-xTRF1 antibodies that are sufficiently sensitive to detect endogenous xTRF1. In contrast, endogenous xTRF2 was successfully detected by specific antibodies (see below). To facilitate the detection of xTRF1 in egg extracts, 35S-xTRF1 was in vitro translated in rabbit reticulocyte lysate (RRL) and added together with sperm chromatin to CSF or interphase extracts, and incubation was carried out for 1 h. Chromatin was isolated from the extracts by centrifugation, and chromatin-bound 35S-xTRF1 and endogenous xTRF2 were analyzed by autoradiography and immunoblotting, respectively. Similar amounts of histone H2B were recovered from both chromatins in CSF and interphase extracts, which served as an internal control of protein recovery (Figure 1A, bottom panel, lanes 3 and 4). We found that xTRF1 was specifically recovered with mitotic chromatin in CSF extracts, but not with interphase chromatin, as revealed by autoradiography (Figure 1A, top panel, lanes 3 and 4). Immunoblot analysis of endogenous xTRF2, in contrast, showed association with chromatin in both mitotic and interphase extracts, indicating that xTRF2 associates with chromatin throughout the cell cycle (Figure 1A, middle panel, lanes 3 and 4). We noticed that xTRF2 derived from mitotic chromatin migrated more slowly than that derived from interphase chromatin on SDS–PAGE (Figure 1A, middle panel, lanes 3 and 4). This is due, at least in part, to the phosphorylation of xTRF2, as will be described below.
Figure 1.
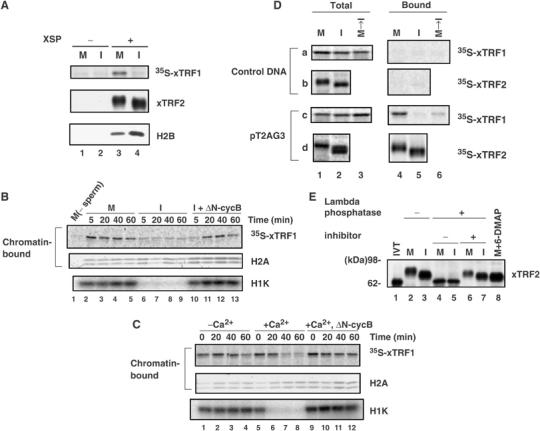
Chromatin binding of Xenopus TRF proteins during the cell cycle. (A) Chromatin binding of xTRF1 and xTRF2 in mitotic or interphase extracts. XSP chromatin was incubated with CSF (M, lanes 1 and 3) or interphase (I, lanes 2 and 4) extracts containing 35S-xTRF1 for 1 h. Chromatin was isolated by centrifugation on a sucrose cushion and the chromatin-bound proteins were analyzed for xTRF1 by phosphorimaging (top panel), and for xTRF2 and H2B by immunoblotting (middle and bottom panels). (B) Addition of nondegradable cyclin B to interphase extracts induces xTRF1 association with chromatin. Sperm chromatin was incubated with CSF (M, lanes 2–5) or interphase (I, lanes 6–13) extracts containing 35S-xTRF1. After 2 h incubation, the interphase extracts were supplemented with recombinant ΔN-cyclin B (ΔN-cycB) and incubated for an additional 2 h (I+ΔN-cycB, lanes 10–13). CSF extracts incubated with the labeled xTRF1 but not with sperm chromatin for 2 h were analyzed in lane 1. Aliquots were taken at the indicated time points during the incubation and chromatin-bound proteins were analyzed for xTRF1 by phosphorimaging (top panel), for histone H2A by immunoblotting (middle panel; the upper band is H2A.X, whereas the lower band is H2A) or for Cdc2 kinase activity (measured as histone H1 kinase activity, H1K, in the bottom panel). (C) xTRF1 dissociates from chromatin upon transition from M phase to interphase in egg extracts. Sperm chromatin was incubated with CSF extracts supplemented with 35S-xTRF1 for 10 min and the sample was divided into three parts. One part was further incubated for 60 min, chromatin was isolated at the indicated time points and bound proteins were analyzed as in A (lanes 1–4). The second part was supplemented with Ca2+ to make a final concentration of 0.4 mM (time 0), incubated for 60 min, and chromatin that was isolated at the indicated time points was analyzed. The third part was supplemented with ΔN-cyclin B (ΔN-cycB) 10 min prior to the addition of Ca2+ (time 0), and chromatin was analyzed (lanes 9–12). (D) xTRF1 and xTRF2 specifically recognize telomeric repeat sequence in Xenopus egg extracts. Linear plasmid DNAs harboring ∼800-bp telomeric repeats at one end (pT2AG3) or the control plasmid (control DNA) was conjugated to paramagnetic beads at the nontelomeric end, and incubated with CSF (M) or interphase (I) egg extracts supplemented with 35S-xTRF1 or -xTRF2 for 1 h. Chromatin-bound xTRF1 and xTRF2 were analyzed by SDS–PAGE, followed by phosphorimaging (panels a and c or panels b and d, lanes 4 and 5, respectively). Alternatively, DNA beads were incubated in CSF extracts with 35S-xTRF1 for 20 min, and then Ca2+ and CHX (at final concentrations of 0.6 mM and 100 μg/ml, respectively) were added to induce a transition from M phase to interphase. The chromatin was further incubated for 40 min, and bound proteins were analyzed as described above (lane 6). 35S-xTRF1 and -xTRF2 present in the total extracts are indicated in lanes 1–3 of each panel. (E) xTRF2 is a phosphoprotein. Paramagnetic beads conjugated with pT2AG3 were incubated with CSF (M, lanes 2, 4, 6 and 8) or interphase (I, lanes 3, 5 and 7) egg extracts. After 1 h incubation, the beads were magnetically isolated from the extracts and incubated at 30°C in the presence (lanes 4–7) or absence (lanes 2 and 3) of lambda phosphatase for 2 h. In lanes 6 and 7, phosphatase inhibitors (50 mM NaF, 10 mM orthosodium vanadate and 1 μM OA) were simultaneously added to the reaction. The DNA beads were isolated magnetically again and analyzed by immunoblotting with antibodies against xTRF2. In vitro translated xTRF2 was also analyzed (IVT, lane 1).
We next examined the possibility that xTRF1 is recruited to and dissociates from chromatin upon the transitions from interphase to M phase, and from M phase to interphase, respectively. The N-terminally truncated form of cyclin B (ΔN-cyclin B) lacks the destruction box and is constitutively active. The addition of ΔN-cyclin B to interphase extracts leads to the activation of Cdc2 kinase and a transition to mitotic extracts (Glotzer et al, 1991). When recombinant ΔN-cyclin B was added to interphase extracts that had been incubated for 1 h with sperm chromatin and 35S-xTRF1, Cdc2 kinase activity (measured as histone H1 kinase activity) was increased (Figure 1B, bottom panel, lanes 10–13), as expected. Interestingly, xTRF1 became associated with the mitotic chromatin at 20 min and thereafter, indicating that the association of xTRF1 with chromatin is coupled with the transition from interphase to M phase. Conversely, when Ca2+ was added to CSF extracts in which xTRF1 had been incubated with chromatin for 20 min, the bound xTRF1 became dissociated from chromatin 40 min after the Ca2+ addition, following the inactivation of Cdc2 kinase 20 min after the Ca2+ addition (Figure 1C, lanes 5–8; note the 20-min lag between the start of Cdc2 kinase inactivation and that of xTRF1 dissociation). However, the dissociation did not occur when Cdc2 kinase activity was maintained by adding ΔN-cyclin B simultaneously with Ca2+ (lanes 9–12). These results indicate that the dissociation of xTRF1 from chromatin is coupled with the transition from M phase to interphase, and intimately related to the reduced Cdc2 kinase activity. Taken together, we concluded that xTRF1 actively associates with mitotic chromatin and dissociates from interphase chromatin with an apparently strong relationship with cyclin B–Cdc2 kinase activity.
xTRF1 and xTRF2 specifically bind to telomeric repeat sequence in Xenopus egg extracts
The experiments described above demonstrated that xTRF1 binds to chromatin in a cell-cycle-dependent manner, whereas xTRF2 constitutively associates with chromatin throughout the cell cycle. However, they did not reveal whether the association of xTRF1 and xTRF2 with chromatin is through specific binding to telomeric repeats or nonspecific binding to general chromatin. To this end, linear plasmid DNAs with or without an ∼800-bp telomeric repeat array were conjugated to paramagnetic beads, and incubated with Xenopus egg extracts and 35S-xTRF1 or 35S-xTRF2 for 1 h to allow for chromatin formation on the beads (Sandaltzopoulos and Becker, 1999). Reconstituted chromatin was isolated in a magnetic field and bound proteins were analyzed (Figure 1D). We found that 35S-xTRF1 was specifically recovered from the telomeric DNA beads incubated in CSF extracts, but not from those incubated in interphase extracts, or control DNA beads incubated in CSF or interphase extracts (panels a and c, lanes 4 and 5). Furthermore, when Ca2+ was added to telomeric DNA beads that had been incubated in CSF extracts for 20 min, the bound xTRF1 dissociated from the beads 40 min after the Ca2+ addition (lane 6). 35S-xTRF2 was also detected only in the telomeric DNA beads fraction and its binding activity was not affected by the cell-cycle stage (panels b and d), as observed in the chromatin-binding assay. We found that both 35S-xTRF1 and -xTRF2 proteins were very stable in Xenopus egg extracts even in the absence of sperm chromatin or DNA beads (data not shown), making it unlikely that the different half-lives of xTRF1 and xTRF2 were responsible for the distinct recoveries of the proteins. Moreover, when endogenous xTRF2 was analyzed using anti-xTRF2 antibodies, it behaved in a manner similar to 35S-xTRF2 (Figure 1E, lanes 2 and 3). These results clearly indicate that xTRF1 and xTRF2 specifically recognize and bind to telomeric repeat DNA in Xenopus egg extracts.
We noted that endogenous xTRF2 bound to telomeric DNA beads showed different mobility on SDS–PAGE (Figure 1E, lanes 2 and 3), as found in the chromatin-bound fractions (Figure 1A). When these fractions were treated with lambda phosphatase, not only mitotic xTRF2 but also interphase xTRF2 showed an increase in mobility in a manner sensitive to phosphatase inhibitors (lanes 4–7). This faster migrating form of xTRF2 showed the same mobility as the recombinant xTRF2 prepared in RRL (lane 1), and was also detected when CSF extracts were treated with 6-dimethylaminopurine (6-DMAP), an inhibitor of various protein kinases, including cyclin-dependent kinases (Blow, 1993) (Figure 1E, lane 8). Similar results were obtained for endogenous xTRF2 that was immunoprecipitated from CSF and interphase extracts without sperm chromatin (i.e., total xTRF2 present in the extracts, data not shown). We concluded that total xTRF2 in CSF and interphase extracts are hyperphosphorylated and phosphorylated, respectively. In addition, the fact that xTRF2 remains bound to telomeric DNA beads after lambda phosphatase or 6-DMAP treatment suggests that xTRF2 is recruited to telomere independent of phosphorylation.
Telomeric DNA binding of chimeric TRF proteins
As xTRF1 and xTRF2 lack the amino-terminal acidic or basic region that is present in mammalian TRF1 and 2 proteins, respectively, they have similar domain architecture, consisting of the amino-terminal TRFH domain, the carboxyl-terminal Myb domain and the linker region connecting those two domains. The Myb domain, the TRFH domain and the linker region show high, moderate and least sequence similarities, respectively, between xTRF1 and xTRF2 (Supplementary Figure 1B). The Myb and TRFH domains are involved in DNA binding and protein–protein interactions, including dimer formation, respectively.
As xTRF1 and xTRF2 bind to telomere chromatin during the cell cycle in distinct manners, we wished to pinpoint the region responsible for the difference in behavior. A series of chimeric proteins that are composed of the TRFH and Myb domains and the linker region derived from either xTRF1 or xTRF2 in all combinations were constructed (Figure 2A; in the construct names, a set of three numbers following ‘TRF' indicates the origin of the three domains; e.g., xTRF211 is a chimera of xTRF2-TRFH, xTRF1-linker and xTRF1-Myb). These proteins were produced as 35S-labeled recombinant proteins in RRL, and examined for their ability to bind to mitotic and interphase telomeric DNA beads (Figure 2B). All the chimeric proteins bound to mitotic telomeric DNA beads as efficiently as the wild-type protein, suggesting that they are functional. The finding that all the chimeric constructs bound to mitotic telomeric DNA beads suggests that binding to mitotic telomeres is a default property of xTRF1 and xTRF2, and that the dissociation of xTRF1 from interphase telomeres is a characteristic specifically conferred to xTRF1. The differential binding to mitotic and interphase telomeric DNA beads revealed in Figure 2B was scored and is summarized in Figure 2A. When the TRFH domain or the linker region of xTRF1 was swapped with the corresponding region of xTRF2 (xTRF211 and xTRF121), the interphase-specific dissociation was abolished, indicating that these two regions are required for the dissociation. In contrast, the xTRF1 Myb domain was dispensable for the dissociation (xTRF112). Substitution of the xTRF1 linker region in xTRF2 (xTRF212) conferred only a weak dissociation of the protein from interphase telomeric DNA. Similarly, xTRF2 with xTRF1-derived TRFH (xTRF122) showed robust binding to interphase telomeric DNA. It is difficult to interpret the results of the TRFH domain swapping because such mutant proteins may dimerize with endogenous xTRF protein having the cognate TRFH domain. Together, the combination of the TRFH domain and the linker region of xTRF1 is required and sufficient for the interphase-specific dissociation of xTRF1 from telomeres.
Figure 2.
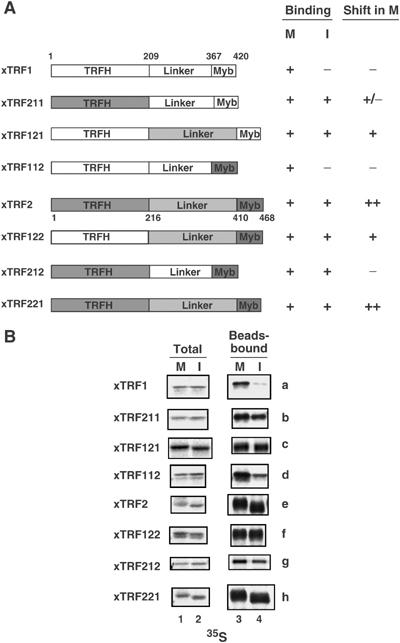
Telomeric DNA binding of chimeric xTRF proteins. (A) Schematic diagram depicting the chimeric xTRF proteins. Domains of xTRF1 and xTRF2 are represented by white and shaded boxes, respectively. The location of each domain is shown on the xTRF1 and xTRF2 structures. Telomere binding measured in mitotic or interphase extracts for each protein was scored from the results in (B) and is shown to the right of the diagrams (binding). The extent of mobility shift of chimeric proteins in mitotic extracts is also summarized (shift in M). (B) Telomeric DNA binding of chimeric xTRF proteins in Xenopus egg extracts. All the chimeric proteins were produced as 35S-labeled proteins in RRLs, and analyzed for telomeric DNA binding as described in Figure 1D.
Cdc2 kinase activity is not essential for association of xTRF1 with chromatin
One potential explanation for the mechanism of xTRF1 dissociation from chromatin in interphase extracts is that xTRF1 is passively excluded from chromatin during the initiation or progression of DNA replication. However, we found no effect of various DNA replication inhibitors, such as Geminin, p21 and aphidicolin, which inhibit the pre-replicative complex (pre-RC) assembly, DNA polymerase loading and fork progression, respectively, on the chromatin binding of xTRF1 (Supplementary Figure 3). Therefore, xTRF1 is not passively excluded from chromatin by events related to DNA replication.
We found that the amount of xTRF1 bound to mitotic chromatin was decreased when CSF extracts were treated with 6-DMAP (Supplementary Figure 3B), suggesting that the chromatin targeting of xTRF1 is regulated by a kinase/phosphatase balance in the extracts. One candidate molecule for regulating the M-phase-specific association of xTRF1 with chromatin is Cdc2 kinase. To investigate this possibility, we prepared cycloheximide (CHX)-treated interphase extracts. Sperm chromatin was first incubated with 35S-xTRF1 in CSF extracts and then Ca2+ and CHX were simultaneously added. In these extracts, endogenous B- and A-type cyclins are degraded during exit from M phase, whereas de novo cyclin synthesis is inhibited by CHX, resulting in complete inactivation of Cdc2 kinase. H1 kinase assay confirmed that no Cdc2 kinase activity was detectable in the CHX-treated interphase extracts (Figure 3A, lane 2). xTRF1 associated with mitotic chromatin (lane 1), but not with interphase chromatin in the presence of CHX (lane 2), as expected. It is known that the addition of okadaic acid (OA), an inhibitor of type1 and type 2A phosphatases, to interphase extracts in the presence of CHX activates M-phase-specific protein kinases other than Cdc2 kinase, thereby inducing a state mimicking M phase, as represented by Cdc25 activation and histone H3 phosphorylation (Sumara et al, 2000). Owing to the inability to synthesize cyclins due to the presence of CHX, Cdc2 kinase is inactive in these extracts (lane 3). Interestingly, we found that xTRF1 associated with chromatin in the OA-induced pseudo-M phase extracts (lane 3). These results indicate that although Cdc2 activation appears to be intimately related to xTRF1 association with chromatin in M phase (Figure 1), it is not essential for the association, as revealed in the OA-induced pseudo-M phase extracts (Figure 3A).
Figure 3.
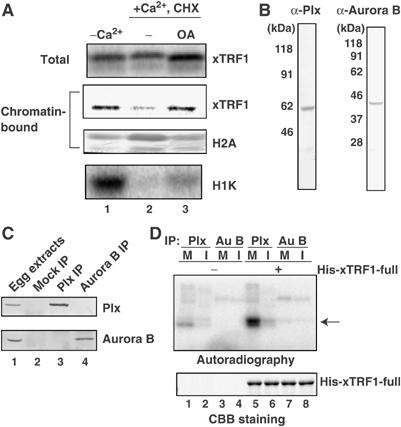
Plx1 phosphorylates xTRF1 in vitro. (A) XSP nuclei (4000 nuclei/μl) were incubated for 20 min at 22°C in CSF extracts with 35S-xTRF1, and then Ca2+ and CHX (100 μg/ml) were simultaneously added to release meiotic metaphase II arrest in the presence (lane 3) or absence (lane 2) of 1 μM OA. As control, the extracts were incubated without the addition of Ca2+ or CHX (lane 1). After 1 h incubation, total extracts (top panel) and chromatin-bound proteins (second panel) were analyzed as described in Figure 1B. (B) Immunoblot analysis of CSF extracts with antibodies against Plx1 or Xenopus Aurora B. The antibodies were raised against the carboxyl-terminal peptide sequences and affinity-purified. (C) Immunoprecipitation of Plx1 or Xenopus Aurora B. CSF extracts (lane 1) were incubated with normal rabbit IgG (lane 2), anti-Plx1 antibodies (lane 3) or anti-Xenopus Aurora B antibodies (lane 4). Immunoprecipitates were fractionated by SDS–PAGE, followed by immunoblotting with antibodies against Plx1 (top panel) or Xenopus Aurora B (bottom panel). Similar results were obtained when interphase extracts were used (data not shown). (D) Plx1 phosphorylates xTRF1 in vitro. Plx1 (Plx) and Aurora B (Au B) were immunoprecipitated with anti-Plx1 or anti-Xenopus Aurora B antibodies from CSF (M) or interphase (I) extracts. Immunoprecipitates were immobilized on protein A-Sepharose 4B beads and mixed with purified full-length His6-xTRF1 (His-xTRF1-full, lanes 5–8) or buffer only (lanes 1–4) in the presence of [γ-32P]ATP. Following incubation for 30 min at room temperature (RT), the reaction mixtures were analyzed by SDS–PAGE, followed by autoradiography (top panel). The CBB-stained gel is also shown (bottom panel). The arrow indicates the position of His6-xTRF1 as labeled by anti-Plx1 immunoprecipitates.
Plx1 promotes mitotic xTRF1 association with chromatin in vitro
The effect of OA on the promotion of xTRF1 association with chromatin suggested that protein phosphorylation either directly or indirectly facilitates xTRF1 recruitment to chromatin. Moreover, we observed free xTRF1 in extracts, which appeared as doublets on SDS–PAGE, and only the band with reduced mobility was detected in the chromatin-bound fraction (data not shown), suggesting that the chromatin-bound xTRF1 specifically undergoes protein modification, such as phosphorylation. As Cdc2 kinase is dispensable for the xTRF1 recruitment to chromatin, we focused on two mitotic kinases, Plx1 and Aurora B. These kinases associate with chromatin throughout the cell cycle in Xenopus egg extracts and are activated in M-phase or OA-induced pseudo-M phase extracts (Losada et al, 2002), making them good candidates for modifying xTRF1.
We first examined whether Plx1 or Aurora B phosphorylates xTRF1 in vitro. We prepared antibodies against Xenopus Plx1 and Aurora B that specifically recognized proteins in immunoblot and immunoprecipitation experiments (Figure 3B and C). We also prepared purified recombinant His6-tagged-xTRF1, and incubated the protein with immunoprecipitated Plx1 or Aurora B kinase immobilized on protein A-Sepharose 4B beads in the presence of [γ-32P]ATP (Figure 3D). Plx1 and Aurora B were detected in immunoprecipitates derived from mitotic extracts (Figure 3C), as well as from interphase extracts (data not shown). Significant labeling of xTRF1 was observed only when the protein was incubated with anti-Plx1 immunoprecipitates derived from mitotic extracts (Figure 3D, lane 5). In contrast, anti-Aurora B immunoprecipitates derived from mitotic or interphase extracts failed to label xTRF1 (lanes 7 and 8). We also confirmed that Plx1, but not Cdc2 kinase, was activated in the OA-induced pseudo-M-phase extracts (data not shown), consistent with the idea that Plx1 plays a role in the xTRF1 recruitment to chromatin in the OA-treated extracts.
To confirm that Plx1 is responsible for the mitotic xTRF1 recruitment to chromatin, Plx1 was immunodepleted from CSF extracts using anti-Plx1 antibodies (Figure 4A). The efficiency of immunodepletion was >95% in all cases, as judged by quantitative immunoblotting (data not shown). The amount of Aurora B and the level of histone H1 kinase activity in the Plx1-depleted extracts were indistinguishable from those in the mock-depleted extracts. Sperm chromatin was incubated with CSF or interphase extracts that had been mock-depleted or depleted of Plx1. Aliquots were taken from each sample and chromatin fractions were analyzed. We found that the depletion of Plx1 reproducibly resulted in significantly, but not completely, reduced loading of xTRF1 to mitotic chromatin (32% of control), whereas the mock depletion had no effect at all (Figure 4B; similar results were obtained in three independent experiments). We also observed that xTRF2 remained bound to chromatin in the Plx1-depleted extracts as well as the mock-depleted extracts (Figure 4B). Taken together, we concluded that Plx1 is required for xTRF1 loading to mitotic chromatin, and that Aurora B has no noticeable role in this process.
Figure 4.
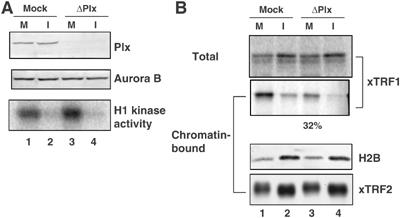
xTRF1 binds inefficiently to telomeric chromatin in mitotic extracts depleted of Plx1. (A) CSF extracts were immunodepleted with control IgG (mock, lanes 1 and 2) or with antibodies against Plx1 (ΔPlx1, lanes 3 and 4) and analyzed by immunoblotting using anti-Plx1 (top panel) or anti-Xenopus Aurora B antibodies (middle panel). The level of H1 kinase activity was also examined (bottom panel). (B) Telomeric chromatin binding of xTRF1 in the absence of Plx1. Sperm chromatin was incubated in either mock-depleted (lane 1) or Plx1-depleted (lane 3) CSF extracts at 22°C for 1 h. Half of these extracts were induced to transit into interphase by the addition of Ca2+ prior to the incubation (lanes 2 and 4). After incubation, chromatin fractions were isolated from the extracts and bound proteins, as well as total proteins, were analyzed by autoradiography (top and second panels) or immunoblotting using antibodies against histone H2B and xTRF2 (third and bottom panels).
Cell-cycle-dependent chromatin-bound telomerase activity in Xenopus egg extracts
Given that TRF1, which negatively regulates telomerase from acting on telomeres (van Steensel and de Lange, 1997), is recruited to telomeric DNA in a cell-cycle-dependent manner, it is interesting to investigate whether telomerase is also recruited to chromatin depending on the cell-cycle stage. We first measured the total telomerase activity present in Xenopus CSF and interphase extracts. As reported previously (Mantell and Greider, 1994), similar levels of telomerase activity were detected in both mitotic and interphase extracts (Figure 5A, lanes 1 and 2). We next analyzed the telomerase activity associated with sperm chromatin reconstructed in CSF and interphase extracts. Isolated sperm chromatin derived from either extract was directly analyzed by the stretch PCR assay (Tatematsu et al, 1996). This analysis revealed that a significantly higher level of telomerase activity was detected in association with interphase chromatin than with mitotic chromatin (Figure 5A, lanes 5 and 6). When the CSF extract was induced to transit into interphase by Ca2+ addition, the chromatin-associated telomerase activity started to increase after the Ca2+ addition (Figure 5B).
Figure 5.
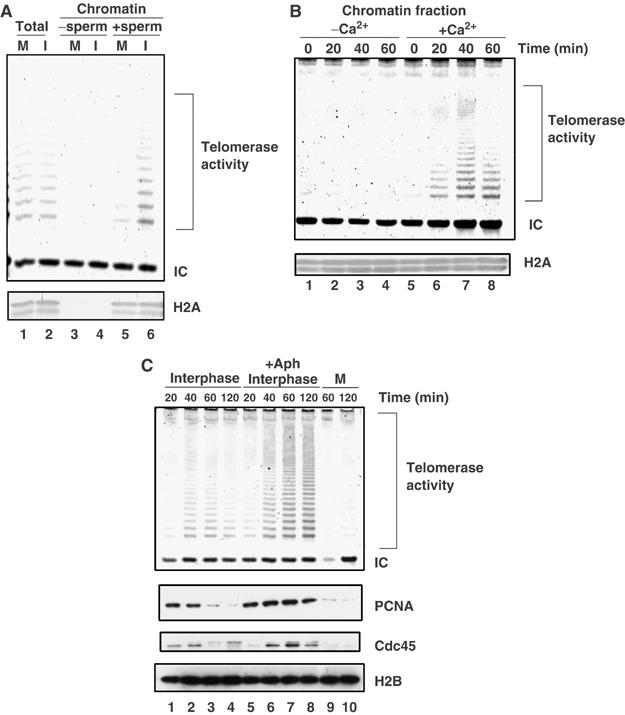
Chromatin-bound telomerase activities are increased in interphase. (A) High levels of telomerase activity are associated with interphase chromatin rather than with mitotic chromatin. Sperm chromatin was incubated in CSF extracts or interphase extracts at 22°C for 1 h. Chromatin fractions were isolated and associated telomerase activities were measured by the Telochaser system (lanes 5 and 6). CSF extracts or interphase extracts without sperm chromatin were treated in the same way as negative controls (lanes 3 and 4). Telomerase activity of total extracts was also examined (lanes 1 and 2). The internal control band (IC) monitored PCR efficiency. The same samples were also analyzed by immunoblotting with an antibody against histone H2A (bottom panel). (B) Time course of telomerase activity associated with chromatin during M phase to interphase transition. CSF extracts containing sperm chromatin were incubated at 22°C for 1 h in the absence (lanes 1–4) or presence (lanes 5–8) of 0.6 mM Ca2+ and 100 μg/ml CHX. Chromatin fractions harvested at the indicated time points were analyzed as described in (A). (C) Telomerase activity is associated with replicating chromatin. Sperm chromatin was incubated in interphase egg extracts in the absence (lanes 1–4) or presence (lanes 5–8) of 20 μg/ml aphidicolin (+Aph). Chromatin fractions were isolated at indicated intervals and analyzed for chromatin-bound telomerase activity. The same samples were analyzed by immunoblotting with antibodies against H2B, Cdc45 and PCNA.
We next examined the time course of telomerase activity associated with chromatin in interphase extracts. During the incubation of the sperm in interphase extracts, nuclei were formed in 15–20 min, and DNA replication was initiated at 20–40 min, and completed at 90 min (data not shown). Consistently, the levels of chromatin-bound Cdc45 and PCNA, two replication proteins present at the replication fork, peaked at 20–40 min (Figure 5C, lanes 1 and 2). We found that the chromatin-bound telomerase activity was detected at 40–60 min, and gradually decreased at 120 min (Figure 5C, lanes 1–4). These results suggest that telomerase associates with chromatin concomitantly with or slightly lagging behind the DNA replication reaction, and then dissociates gradually. The coupling of chromatin recruitment of telomerase with DNA replication was evident when we added aphidicolin to the reaction. Aphidicolin inhibits not the loading but the enzymatic activity of replicative DNA polymerases at the replication fork. As replication was not completed, PCNA and Cdc45 resided on chromatin for a prolonged period in the aphidicolin-treated reactions (Figure 5C, lanes 5–8). We found that the still higher telomerase activity than that found in control reactions was associated with and never dissociated from chromatin in the aphidicolin-treated reactions. Taken together, the results strongly suggest that telomerase is recruited to the replicating chromatin during S phase.
Discussion
Dynamic changes of xTRF1 chromatin binding during the cell cycle
The observations in this study apparently contradict the general belief in previous literature that TRF1 associates with telomere throughout the cell cycle including M phase. It is well documented that TRF1 is detected at both interphase and metaphase telomeres in human cells (Chong et al, 1995). The contradictory observations may stem from differences between the cell cycle operating in Xenopus early development and that in mammalian somatic cells. The Xenopus early embryos lack the G1 or G2 phase and alternate between S phase and M phase without cell growth. It is formally possible that the dynamic behavior of xTRF1 in the egg extracts is specific to the early developmental stages in Xenopus. Alternatively, the chromatin-binding assays for Xenopus cell-free extracts used in this study are more synchronous and sensitive than the conventional immunofluorescence (IF) assay used in cultured cells, and may have enabled the detection of cell-cycle-dependent changes of TRF1 binding that were not observed previously. Indeed, it has been reported that the entire population of telomere-associated human TRF1 and a major fraction (∼73%) of human TRF2 reside at telomeres with high turnover rates (∼25 s half-lives in both cases) (Mattern et al, 2004). These results suggest that, although it appears that TRF1 and TRF2 stably associate with telomeres in IF studies, they are, in fact, recruited to and dissociated from telomeres dynamically, presumably in a regulated manner. Interestingly, the same study demonstrated that a relatively minor but significant fraction (∼27%) of human TRF2 resides at telomeres stably (∼138 s half-life) (Mattern et al, 2004). The stable association of xTRF2 with chromatin observed in the egg extracts may reflect the presence of this stable population in human TRF2.
Analyses of chimeric protein mutants revealed that the TRFH domain and the linker region of xTRF1 are required and sufficient for the xTRF1-specific dissociation from interphase chromatin. TRFH domains are responsible for TRF1 and TRF2 homodimerizations and physical binding to TIN2 in hTRF1. Thus, it is possible that protein–protein interactions could be part of the regulatory mechanism modulating the affinity of xTRF1 for telomeric DNA in the interphase. Although it has been proposed that the flexible nature of the linker region of hTRF1 enables the hTRF1 homodimer to bind to two copies of telomeric repeats with extreme spatial variability (Bianchi et al, 1999), the precise role of the linker region is still unclear. Based on the change of the binding properties of chimeric xTRF1 protein possessing the xTRF2-derived linker region (xTRF212), the linker region may modulate the DNA-binding activity of xTRF1 in an unknown manner.
Plx1 immunodepletion experiments demonstrated that Plx1 is required either directly or indirectly for the efficient xTRF1 association with chromatin. The failure of complete inactivation of xTRF1 loading onto mitotic chromatin in Plx1-immunodepleted mitotic extracts may result from the presence of Plx1 paralogs Plk2 and Plk3, which may have escaped from immunodepletion in Xenopus egg extracts (Duncan et al, 2001). Alternatively, it is possible that features specific to mitotic chromatin, such as chromosome condensation, additionally contribute to xTRF1 recruitment. We have shown that Plx1 phosphorylates the linker region of xTRF1 in vitro (Supplementary Figure 4). However, mass spectrometry and mutation analyses showed that numerous serine or threonine residues are phosphorylated in this region, and it was not possible to construct mutants that lack all the potential phosphorylation sites. Therefore, unfortunately, we cannot conclude that the phosphorylation of xTRF1 by Plx1 is required for xTRF1 binding to mitotic chromatin at this moment. It is well established that Tankyrase 1 destabilizes TRF1 binding to telomeres (Smith and de Lange, 2000). It is therefore possible that Plx1 regulates the telomeric association of xTRF1 through an indirect effect on Tankyrase 1 or TIN2, a modulator of Tankyrase 1 (Ye and de Lange, 2004). Uncovering the target of Plx1 should be addressed in future studies.
Implications of cell-cycle-dependent association and dissociation of xTRF1
It has been previously proposed that the telomere chromatin structure where TRF1 is lost but TRF2 remains represents a semi-open configuration that simultaneously protects telomeres and allows access by telomerase (Chang et al, 2003). The interphase telomere chromatin in Xenopus egg extracts appears to correspond to this configuration (Figure 6).
Figure 6.
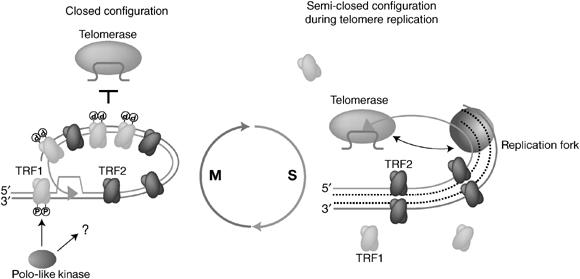
Cell-cycle-dependent regulation of telomere configurations. A hypothetical telomere structure is shown. xTRF2 is constitutively associated with telomere chromatin in Xenopus egg extracts throughout the cell cycle. Plx1 either directly or indirectly regulates the telomere association of xTRf1 upon the transition from interphase to M phase. The closed mitotic configuration harboring both xTRF1 and xTRF2 is inaccessible to telomerase. When the cell enters interphase, xTRF1 is dephosphorylated and released from telomere chromatin by unknown mechanisms. The thus-formed TRF2-associated and TRF1-lacking telomere assumes a semi-open configuration that allows access by telomerase, yet protects the chromosomal ends. Telomerase is recruited to the chromosomal ends, probably via intimate coordination with the DNA replication machinery. Note that the model does not suggest that the telomere shows the semi-open configuration throughout interphase. Rather, it is likely that the configuration is taken only transiently in the somatic cell cycle when the telomeres are to be replicated. The t-loop model is included in this figure although it is not directly demonstrated in this study.
One interesting implication of this study is that the semi-open configuration is induced in interphase telomeres to facilitate telomerase action. However, this study also indicated that the recruitment of telomerase activity to chromatin paralleled not only the lack of TRF1, but also the ongoing DNA replication reaction. When the progression of DNA polymerases was inhibited by aphidicolin, still higher telomerase activity remained associated with chromatin, suggesting that the dissociation of telomerase from chromatin in S phase requires the completion of DNA replication. We therefore propose that the recruitment of telomerase to telomeres is regulated by the dynamic modification of telomere chromatin occurring in S phase, which is characterized by the ongoing DNA replication and TRF1 dissociation. As we have shown that TRF1 retards the progression of the replication fork at telomeres (Ohki and Ishikawa, 2004), these two features of the semi-open configuration of replicating telomeres may be mechanistically related. It should be noted that we do not suggest the dissociation of TRF1 from telomeres throughout the interphase, particularly in the somatic cell cycle. Rather, it is likely that the semi-open configuration is assumed only transiently, for example, when telomeres are to be replicated. As individual telomeres appear to replicate independently of each other within a relatively narrow window in S phase, only a small fraction of telomeres in one cell may show this configuration at a given moment in S phase (Ohki and Ishikawa, 2004). This may explain why the dynamic behavior of telomeric chromatin in S phase has escaped observation in studies conducted to date. In this sense, the Xenopus egg extract is expected to serve as an invaluable tool to analyze the details of telomere chromatin, replication and telomerase reaction.
Materials and methods
Cloning of xTRF1 and xTRF2
Xenopus homologs of TRF1 and TRF2 cDNAs were isolated by RT–PCR and 5′-RACE. See Supplementary data for details.
Antibodies and recombinant proteins
Rabbit polyclonal antisera were raised against synthetic peptides corresponding to the C-terminal amino-acid sequences of Xenopus Plx1 (QSSKSAVAHVKASA), Aurora B (RRVLPPVYQSTQSK), INCENP (SNRHHLAVGYGLKY) and bacterial His6-fusion proteins containing peptide sequences (amino acids 1–410) from the Xenopus TRF2 protein. Immunization and affinity purification were performed as described (Hirano et al, 1997). Anti-human H2B antibody was purchased from Upstate. Anti-human Cdc45 and anti-human PCNA antibodies were purchased from Abcam. GST-tagged human p21 and Xenopus geminin were prepared as ΔN85cyclin B2 (ΔN-cyclin B) as described previously (Iwabuchi et al, 2000). The chimeric TRF proteins were created according to Warrens et al (1997).
In vitro transcription/translation
In vitro transcription/translation of TRF proteins were performed as described (Bianchi et al, 1997). See Supplementary data for details.
Preparation and immunodepletion of Xenopus egg extracts
CSF extracts were prepared according to Murray (1991) with minor modifications (Nishiyama et al, 2000). For the immunodepletion of Plx1, 100 μl of CSF extract was treated four times (15 min each, on ice) with 20 μl of protein A-Sepharose 4B beads (Amersham Pharmacia Biotech) conjugated with 7.5 μg of anti-Plx1 antibodies. Control depletions were performed using beads coated with nonimmune rabbit IgG.
Chromatin assembly
Preparation of the chromatin fractions was essentially performed as described (MacCallum et al, 2002). See Supplementary data for details.
Solid-phase chromatin assembly
Solid-phase DNA templates were prepared from the pT2AG3 plasmid containing ∼800-bp telomeric repeats (Ohki and Ishikawa, 2004) according to Sandaltzopoulos and Becker (1999). See Supplementary data for details.
In vitro phosphorylation of xTRF1 and H1 kinase assay
His6-tagged recombinant xTRF1 fragments and truncated GST-xTRF1 fragments were expressed using pTrcHisC (Invitrogen) and pGEX-5X-1 (Pharmacia) plasmid vectors, respectively. His6-tagged recombinant protein was purified using HisBind Resin (Novagen). GST-tagged recombinant proteins were purified as described above. Plx1 and Xenopus Aurora B were immunoprecipitated from 10 μl of CSF or interphase extracts using 1 μg of anti-Plx1 antibody or 0.8 μg of anti-Xenopus Aurora B antibody coupled to 10 μl of protein A-Sepharose 4B beads, followed by washing three times with β-GPEB containing 0.2% Triton X-100 and once with β-GPEB containing 1 mM DTT and 1 mM Mg2+-ATP. In all, 10 μl of beads bound to kinases was incubated with the purified fusion proteins in 30 μl of a reaction mixture containing 0.1 mM Mg2+-ATP, 1 mM DTT and 10 μCi [γ-32P]ATP (6000 Ci/mmol) in β-GPEB. After incubation at RT for 30 min, the reaction mixture was analyzed by SDS–PAGE, followed by CBB staining and autoradiography. H1 kinase assay was carried out according to Murray (1991), and the incorporation of [γ-32P]ATP was quantitated with an image analyzer (BAS 2000 Fuji Photofilm).
Assay for telomerase activity
For quantitative analyses of telomerase activity, the stretch PCR assay was performed using a Telochaser system according to the manufacturer's protocol (Toyobo) (Tatematsu et al, 1996). For analyses of chromatin-bound telomerase activity, XSP chromatin (2000 nuclei/μl) was incubated in 20 μl of egg extract at 22°C and isolated as described above. The isolated chromatin was assayed for telomerase activity by the Telochaser system.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Materials and methods
Acknowledgments
This work was supported by a COE Grant, Grants-in-Aid for Cancer Research and Grants-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science and Technology (FI). We thank Dr H Takisawa (Osaka University) for the gift of expression vector producing GST-xGeminin and critical reading of the manuscript, Dr M Nakanishi (Nagoya City University) for the gift of expression vector producing GST-p21, Dr N Sagata (Kyushu University) for the gift of cDNA of Plx1 and Dr T Hirano (Cold Spring Harbor Laboratory) for technical advice. We also thank M Tamura for technical assistance, and A Katayama, M Sakamoto, M Sasaki and K Fujimaki for excellent secretarial work. AN was supported by JSPS Research Fellowships for Young Scientists.
References
- Bianchi A, Smith S, Chong L, Elias P, de Lange T (1997) TRF1 is a dimer and bends telomeric DNA. EMBO J 16: 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Stansel RM, Fairall L, Griffith JD, Rhodes D, de Lange T (1999) TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J 18: 5735–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E (1997) Telomeric localization of TRF2, a novel human telobox protein. Nat Genet 17: 236–239 [DOI] [PubMed] [Google Scholar]
- Blow JJ (1993) Preventing re-replication of DNA in a single cell cycle: evidence for a replication licensing factor. J Cell Biol 122: 993–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet 17: 231–235 [DOI] [PubMed] [Google Scholar]
- Cech TR (2004) Beginning to understand the end of the chromosome. Cell 116: 273–279 [DOI] [PubMed] [Google Scholar]
- Chan SR, Blackburn EH (2004) Telomeres and telomerase. Philos Trans R Soc Lond B Biol Sci 359: 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Dynek JN, Smith S (2003) TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev 17: 1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T (1995) A human telomeric protein. Science 270: 1663–1667 [DOI] [PubMed] [Google Scholar]
- de Lange T (2004) T-loops and the origin of telomeres. Nat Rev Mol Cell Biol 5: 323–329 [DOI] [PubMed] [Google Scholar]
- de Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Duncan PI, Pollet N, Niehrs C, Nigg EA (2001) Cloning and characterization of Plx2 and Plx3, two additional Polo-like kinases from Xenopus laevis. Exp Cell Res 270: 78–87 [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW (1991) Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138 [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T (1999) Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Harley CB (1991) Telomere loss: mitotic clock or genetic time bomb? Mutat Res 256: 271–282 [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M (1997) Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89: 511–521 [DOI] [PubMed] [Google Scholar]
- Iwabuchi M, Ohsumi K, Yamamoto TM, Sawada W, Kishimoto T (2000) Residual Cdc2 activity remaining at meiosis I exit is essential for meiotic M–M transition in Xenopus oocyte extracts. EMBO J 19: 4513–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T (2002) Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev 16: 3004–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DE, Losada A, Kobayashi R, Hirano T (2002) ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol Biol Cell 13: 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantell LL, Greider CW (1994) Telomerase activity in germline and embryonic cells of Xenopus. EMBO J 13: 3211–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Brevet V, Mann C, Gilson E (2000) Cell cycle restriction of telomere elongation. Curr Biol 10: 487–490 [DOI] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- Mattern KA, Swiggers SJ, Nigg AL, Lowenberg B, Houtsmuller AB, Zijlmans JM (2004) Dynamics of protein binding to telomeres in living cells: implications for telomere structure and function. Mol Cell Biol 24: 5587–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW (1991) Cell cycle extracts. Methods Cell Biol 36: 573–597 [PubMed] [Google Scholar]
- Nakamura M, Nabetani A, Mizuno T, Hanaoka F, Ishikawa F (2005) Alterations of DNA and chromatin structures at telomeres, and genetic instability in mouse cells defective in DNA polymerase α. Mol Cell Biol 25: 11073–11088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Tachibana K, Igarashi Y, Yasuda H, Tanahashi N, Tanaka K, Ohsumi K, Kishimoto T (2000) A nonproteolytic function of the proteasome is required for the dissociation of Cdc2 and cyclin B at the end of M phase. Genes Dev 14: 2344–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki R, Ishikawa F (2004) Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res 32: 1627–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki R, Tsurimoto T, Ishikawa F (2001) In vitro reconstitution of the end replication problem. Mol Cell Biol 21: 5753–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM (1997) Synthesis of the telomeric C-strand. A review. Biochemistry (Moscow) 62: 1216–1223 [PubMed] [Google Scholar]
- Sandaltzopoulos R, Becker PB (1999) A solid-phase approach for the analysis of reconstituted chromatin. Methods Mol Biol 119: 195–206 [DOI] [PubMed] [Google Scholar]
- Smith S, de Lange T (2000) Tankyrase promotes telomere elongation in human cells. Curr Biol 10: 1299–1302 [DOI] [PubMed] [Google Scholar]
- Spink KG, Evans RJ, Chambers A (2000) Sequence-specific binding of Taz1p dimers to fission yeast telomeric DNA. Nucleic Acids Res 28: 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM (2000) Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol 151: 749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Nakayama J, Danbara M, Shionoya S, Sato H, Omine M, Ishikawa F (1996) A novel quantitative ‘stretch PCR assay', that detects a dramatic increase in telomerase activity during the progression of myeloid leukemias. Oncogene 13: 2265–2274 [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- van Steensel B, de Lange T (1997) Control of telomere length by the human telomeric protein TRF1. Nature 385: 740–743 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Warrens AN, Jones MD, Lechler RI (1997) Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186: 29–35 [DOI] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA (1993) Saccharomyces telomeres acquire single-strand TG1–3 tails late in S phase. Cell 72: 51–60 [DOI] [PubMed] [Google Scholar]
- Ye JZ, de Lange T (2004) TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat Genet 36: 618–623 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Materials and methods


