Abstract
Yeast telomeres are anchored at the nuclear envelope (NE) through redundant pathways that require the telomere-binding factors yKu and Sir4. Significant variation is observed in the efficiency with which different telomeres are anchored, however, suggesting that other forces influence this interaction. Here, we show that subtelomeric elements and the insulator factors that bind them antagonize the association of telomeres with the NE. This is detectable when the redundancy in anchoring pathways is compromised. Remarkably, these same conditions lead to a reduction in steady-state telomere length in the absence of the ATM-kinase homologue Tel1. Both the delocalization of telomeres and reduction in telomere length can be induced by targeting of Tbf1 or Reb1, or the viral transactivator VP16, to a site 23 kb away from the TG repeat. This correlation suggests that telomere anchoring and a Tel1-independent pathway of telomere length regulation are linked, lending a functional significance to the association of yeast telomeres with the NE.
Keywords: nuclear organization, Sir4, telomerase, telomeres, yKu
Introduction
Chromosomes assume a nonrandom distribution in the eukaryotic nucleus, which helps regulate nuclear processes such as DNA replication, mRNA processing, transcription and DNA repair. In the interphase budding yeast nucleus, centromeres are clustered near the spindle pole body (SPB) opposite the nucleolus, whereas telomeres are distributed in perinuclear foci distal to the SPB (Bystricky et al, 2005). The 32 chromosome ends form only 4–8 foci and associate with the nuclear envelope (NE) in a reversible manner (Gotta et al, 1996; Hediger et al, 2002b). In addition, yeast telomeres nucleate a compact chromatin structure that silences nearby promoters, called the Telomeric Position Effect (TPE, Gottschling et al, 1990). TPE requires a complex of silent information regulators, Sir2, Sir3 and Sir4, that are recruited to telomeres by yKu70/80 and Repressor activator protein 1 (Rap1). The stable association of Sir proteins with nucleosomes requires the deacetylation of the histone H4 K16 by Sir2, which precedes the inward spreading of the Sir complex along nucleosomes (reviewed in Moazed et al, 2004). Importantly, enhanced silencing improves the efficiency of telomere anchorage, even though NE association can be mediated by a silencing-independent mechanism (Hediger et al, 2002b; Gartenberg et al, 2004; Taddei et al, 2004).
The analysis of yeast telomere position in mutant strains has shown that telomere anchoring depends on two parallel and partially redundant pathways. One requires the silencing factor Sir4 and a second the yKu heterodimer (Laroche et al, 1998; Hediger et al, 2002b; Taddei et al, 2004). Both yKu and Sir4 proteins bind telomeres in vivo, yet neither contains a transmembrane domain. Thus, integral components of the NE must also be involved. For Sir4, anchoring is achieved by binding Esc1, a large acidic protein that is found on the inner face of the NE in between nuclear pores (Andrulis et al, 2002; Gartenberg et al, 2004; Taddei et al, 2004). In S-phase cells, Esc1 is also needed for yKu80 association with the NE, although yKu binds another, Esc1-independent perinuclear site in G1 phase (Taddei et al, 2004). The second yKu anchorage site remains unknown, as elimination of its proposed ligands, Mlp1 and Mlp2, had no effect on telomere anchoring (Hediger et al, 2002a, 2002b).
To date, all yeast telomeres tested for perinuclear anchoring become randomly positioned in a double yku70Δ sir4Δ mutant (Hediger et al, 2002b). Nonetheless, the relative importance of the Sir- and yKu-mediated pathways is telomere-specific; some telomeres are very sensitive to loss of yKu, others are not (Tham et al, 2001; Hediger et al, 2002b). Consistently, natural telomeres show quite striking variations in the degree of silencing conferred on reporter genes inserted into subtelomeric regions (Pryde and Louis, 1999). Given such variation, we decided to examine whether the telomere anchoring efficiency might reflect subtelomeric sequence composition (Louis and Haber, 1992).
All yeast telomeres terminate with about 350 nt of a variable TG(1−3) repeat, which provides binding sites for ∼18 molecules of Rap1. Immediately centromere-proximal to this are three different types of subtelomeric elements (STEs; Figure 1A). These include the X core, the Subtelomeric repeat element (STR), and two forms of a larger, highly conserved repeat called Y′. All telomeres contain the X core element, most contain STR, and about 60–70% of chromosomal ends contain one or more copies of the Y′ repeat (Figure 1A; Louis, 1995). The core X element consists of a well-conserved sequence of ∼470 bp, which contains binding sites for ARS-binding factor 1 (Abf1; Diffley and Stillman, 1989), and the Origin Recognition Complex (ORC). Abf1 also binds multiple promoters throughout the genome, and both Abf1 and ORC contribute to silencer function at the HM loci. The STR repeat contains multiple copies of the vertebrate telomeric motif, T2AG3, within a series of short elements designated STR-A, -B, -C or -D. In yeast T2AG3 is recognized by the Myb-domain protein Tbf1, a protein closely related to the mammalian telomere repeat-binding factors, Trf1 and Trf2 (Brigati et al, 1993; Bilaud et al, 1996; Brun et al, 1997). Y′ elements also contain ORC and Tbf1 consensus, as well as an open reading frame for a stress-induced helicase (Chan and Tye, 1983; Louis and Haber, 1992; Brun et al, 1997; Yamada et al, 1998).
Figure 1.
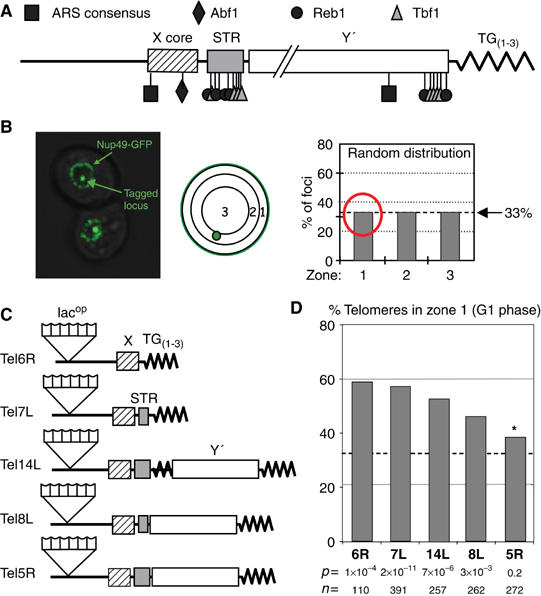
Telomere anchoring variability. (A) Yeast STEs X core, STR and Y′ with their factors and binding sites are shown. ORC binds the ARS consensus. (B) Focal section of two yeast cells bearing one lacops array and expressing fusions of GFP-lacI and GFP-Nup49 for visualizing the tagged locus and the NE. Position of the tagged locus is scored in 3D image stacks for three zones of equal surface, such that a stochastic distribution gives rise to 33% in each zone. (C) Telomeres 6R, 7L, 14L, 8L and 5R were tagged by inserting ∼150 lacops at 14, 18, 19, 11 and 16 kb from the chromosomal ends, respectively, in GA-1320 (Heun et al, 2001). STEs X core, STR and Y′ (indicated by indicated boxes) remain intact. Composition of STEs according to SGD database (Cherry et al, 1998) is represented for each telomere. (D) Position of telomeres was monitored in G1 cells on synthetic medium. The graph presents the percentage of indicated telomere detected in zone 1 for Tel6R, 7L, 14L, 8L and 5R (GA-1459, GA-2226, GA-1985, GA-1986 and YG-143). The P-values obtained by a proportional t-test (zone 1 comparison to 33%, dotted line), plus the number of G1 phase cells scored are indicated. S-phase data are in Supplementary Table 1. The asterisk indicates a value not statistically different from random.
In addition to Tbf1 and ORC, an essential rDNA enhancer binding factor called Reb1 (Chasman et al, 1990) binds within STR and Y′ elements. Reb1 has been shown to play an important role in RNA Pol I- and Pol II-mediated transcription, and helps specify termination of rRNA transcripts as well (see Wang and Warner, 1998). Moreover, like Abf1 and Rap1, Reb1 is thought to contribute to nucleosome organization (Chasman et al, 1990). Consistently, Reb1 was shown to serve as a boundary factor at telomeres (Fourel et al, 2001).
One of the physiological roles of STEs and their clusters of factor binding sites may be to regulate the propagation of TPE. The insertion of a URA3 reporter gene at variable distances from the TG(1−3) sequence in natural telomeres, showed that Sir-mediated repression was weakened when a reporter was positioned within the Y′ element (Pryde and Louis, 1999). In contrast, repression was reinforced for reporters placed at the X core. Silencing efficiency dropped again as the reporter was moved more centromere proximal, such that only background levels of silencing existed 2 kb from the X core. It was concluded that although the X element could promote repression, it also seemed to limit its propagation inwards. Such fluctuations were not seen at truncated telomeres that lack STEs, in which case repression decreases linearly with the reporter gene's distance from the TG repeat (Renauld et al, 1993).
Using other silencing assays, it could be shown that the insertion of subdomains of the STR and Y′ sequences immediately adjacent to a telomere repeat, insulate against the inward spread of TPE (Fourel et al, 1999). This insulator function could also be achieved by directly targeting the N-terminal domains of either Reb1 or Tbf1 to equivalent sites, suggesting that these proteins provide or recruit the barrier function that prevents inward spreading of silent chromatin (Fourel et al, 2001). The X core element, on the other hand, was able to enhance silencing, an effect partially dependent on Abf1 and ORC (Lebrun et al, 2001).
Although STEs are not essential for the telomere capping function of the TG repeats, it has been proposed that STEs may influence telomere length regulation (Craven and Petes, 1999; Brevet et al, 2003). The number of Rap1/Rif1-2 complexes bound to telomeric DNA negatively regulates telomerase by reducing the probability that it will elongate the TG(1−3) repeat (Marcand et al, 1997; Levy and Blackburn, 2004; Teixeira et al, 2004). This mechanism depends on the yeast ATM kinase homologue, Tel1 (Craven and Petes, 1999; Ray and Runge, 1999). A second, Rap1-independent pathway of telomere length regulation was detected in tel1Δ cells (Brevet et al, 2003), and this appears to be sensitive to the factors that bind subtelomeric repeats (Berthiau et al, 2006).
We report here an unexpected effect of STEs on the anchoring of telomeres to the NE. We find that deletion of STEs can enhance the efficiency with which weakly bound telomeres associate with the nuclear periphery. Moreover, the antagonistic action of STEs can be mimicked by targeting domains of Reb1 and Tbf1 to truncated telomeres. This argues that proteins bound to native subtelomeric regions either relocalize telomeres to nonperipheral sites, or, more likely, interfere with the interactions that anchor telomeres at the NE. The antagonism we monitor can also be achieved by targeting the transactivator domain of the viral protein VP16. Most importantly, we see an intriguing correlation between the delocalization of telomeres from the nuclear periphery and a reduction in their steady-state length in tel1Δ strains, upon targeting Tbf1, Reb1 or VP16 to sites distal from the TG(1−3) repeat.
Results
The efficiency of binding to the NE is telomere-specific
Live imaging of intact cells bearing GFP-tagged telomeres, shows that most yeast telomeres are spatially positioned throughout interphase within a narrow peripheral rim of the nucleus, roughly 200 nm wide. Nonetheless, even yeast telomeres are subject to constant random movement, which can vary through the cell cycle (Heun et al, 2001). Therefore, an accurate comparison of telomere position requires the statistical analysis of a population of cells, grouped by their cell cycle stage. We do this by taking a high-resolution 3D stack of fluorescence images, and scoring the position of a telomere-proximal lacop array (visualized by the binding of a GFP-lac repressor fusion) relative to a nuclear pore marker (Nup49-GFP). Both markers are monitored in living cells under normal growth conditions, and cell cycle stages are determined (Hediger et al, 2002b). The distance between a telomere and the nearest point on the NE is scored relative to the diameter of the nucleus in the focal plane containing the lacop array. A tagged telomere's position is scored for several hundred data points binned into three equal concentric circles, and its distribution can be compared with a stochastic or random distribution. Preferential positioning of a given telomere would result in a value significantly higher than 33% for any one zone. Because our interest is to quantify perinuclear association, we perform statistical analyses by comparing the frequency with which a telomere occupies the outermost zone (zone 1) to a theoretical random value, or to another empirical value, using the Student's t-test (Figure 1B; see Supplementary Table 1 and Materials and methods).
Tel6R has been extensively analysed in earlier studies. It is reproducibly detected in the outermost zone (zone 1) in ∼59% of G1-phase cells (Hediger et al, 2002b; Figure 1 and Supplementary Table 1). This number indicates that at a given time point Tel6R abuts the NE-defining Nup49-GFP fluorescence in almost 60% of the cells. Time-lapse imaging of the same telomere shows that despite this highly significant enrichment, Tel6R moves continuously, both along the inner NE and towards the nucleoplasm (Heun et al, 2001; Hediger et al, 2002b). Previous analyses of yeast telomeres showed that Tel14L was bound to the same degree as Tel6R, whereas Tel8L was more randomly distributed (Hediger et al, 2002b). Because both Tel8L and Tel14L have Y′ elements, we extended our study to two additional telomeres, Tel7L and Tel5R, one without and one with Y′ elements, to see whether NE association rates correlate with STE composition (Figure 1C). The subtelomeric sequence composition is based on the sequenced strain S288c (Saccharomyces cerevisiae Genome Database, http://www.yeastgenome.org/; Louis and Haber 1992), but restriction fragment size and PCR mapping indicates the same STE organization in the W303 background used here, for these telomeres (data not shown).
Consistent with previous results, we found variable degrees of perinuclear anchorage for these telomeres. In G1-phase as well as S-phase cells (Figure 1D and Supplementary Table 1), we detect values that range from 38% in zone 1 for Tel5R, to a highly significant enrichment (60–65% in zone 1). Tel6R, Tel14L and Tel7L showed the highest degree of association, Tel8L was intermediate (46%), whereas the value scored for Tel5R showed no significant enrichment in zone 1 by the proportional t-test (Figure 1D and Supplementary Table 1). Intriguingly, whereas Tel5R position was random in G1 phase, it binds efficiently in S-phase cells (65%, P=2 × 10−5), although Tel8L becomes less tightly bound in S phase (Supplementary Table 1). The variability could not be correlated in a simple manner with STE composition, nor was there a clear correlation with the length of the corresponding chromosomal arm (6R=122 kb, 8L=105 kb, 14L=628 kb, 7L=497 kb and 5R=423 kb). Nonetheless, we note that the two telomeres with the highest levels of NE association (Tel6R and Tel7L) lack Y′ elements and have abbreviated STR repeats. This prompted us to remove these native STEs to see whether they affect the anchoring of yeast telomeres in cis.
STEs have an antianchoring effect on poorly anchored telomeres
The impact of subtelomeric composition on telomere positioning was analysed in telomeres Tel6R, Tel8L and Tel5R by the introduction of a TG repeat at a site centromere-proximal to the STEs (Figure 2A and Materials and methods). The strains maintained the same subtelomeric lacop array for quantifying telomere position GFP-lacI repressor association, and none of the genes removed by the truncations is essential or is even expressed under normal growth conditions (SGD database; Cherry et al, 1998). We detected no significant change in the location of Tel6R or Tel8L after truncation, although a strong repositioning towards the nuclear periphery was detected for Tel5R (Figure 2B). Upon removal of its STE, Tel5R shifted from a near random distribution to a distribution that shows significant enrichment in the outermost zone 1 (51%; P=4.3 × 10−3 comparing natural to truncated distributions). We note that this does not reflect a trans effect nor impaired transcription of a subtelomeric gene, as Tel5R truncation does not affect the position of any other telomere nor alter detectable transcript levels. Furthermore, under conditions in which the native Tel5R is efficiently bound, that is, in S-phase cells, the removal of its STEs has no effect (see Supplementary Table 1). The results described here argue that on native Tel5R, the STEs negatively influence anchoring in cis, and that their removal compromises this negative effect, improving anchoring efficiency.
Figure 2.
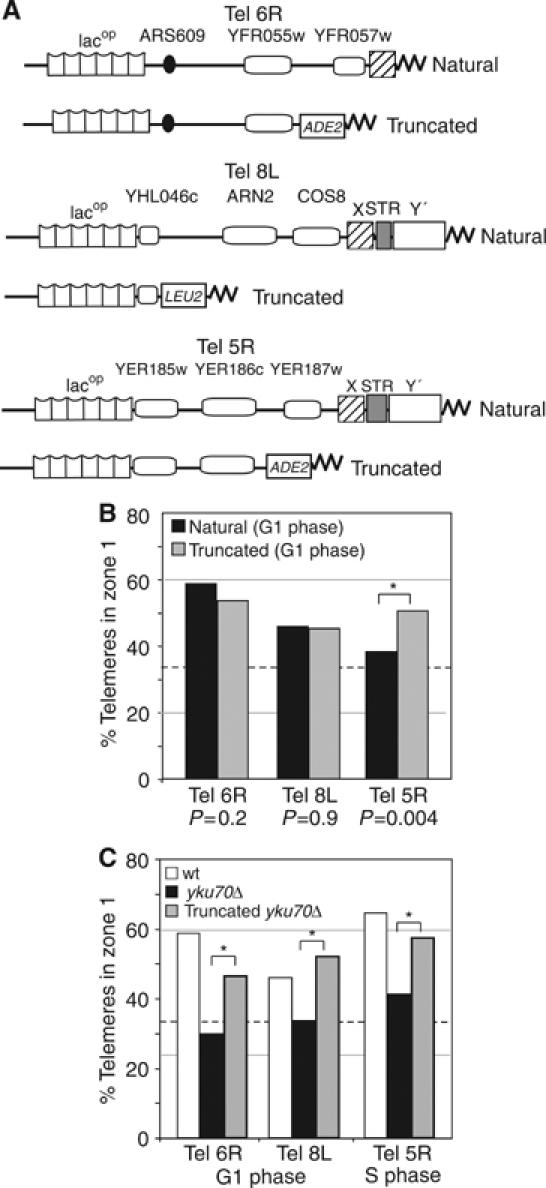
Localization of truncated telomeres. (A) Scheme of natural telomeres with lacop array and truncated telomeres after STEs are replaced by a gene marker (ADE2 for Tel6R and 5R, LEU2 for Tel8L). Bar graph as in Figure 1D, presents the percentage of foci in zone 1 for natural or truncated telomeres 6R, 8L and 5R (GA-1917, GA-2256 and YG-138) in G1 cells. Here asterisks indicate significantly different distributions (P-values reflect a proportional comparison between natural and truncated telomeres). (B) Telomere position was monitored for natural or truncated telomeres in the indicated strain background. (C) Bar graph as in (B) with zone 1 values for natural Tel6R, 8L and 5R in wt (open boxes) or ku70Δ trains (black boxes, GA-1489, GA-1916, GA-195) or for truncated forms in ku70Δ (grey boxes, GA-1948, GA-2804, GA-149). For P-values see Supplementary Table 1.
We suspected that the effects of STE deletion might be more pronounced on Tel5R because it shows weak association in G1 phase cells. To test whether the effect of STE removal can be detected on other randomly positioned telomeres, we deleted STEs in strains that bear mutations that compromise one of the two anchoring pathways; that is, pathways that depend on the telomere-bound factors yKu and Sir4, respectively. Elimination of just one of the two pathways compromises anchoring to different degrees for different telomeres (Hediger et al, 2002b, and Supplementary Table 1). To see if a weakened NE association could be suppressed by removal of STEs, we monitored the position of truncated, lacop-tagged Tel6R, Tel8L and Tel5R in a yku70Δ background. As previously shown, native Tel6R and Tel8L are randomly distributed in the absence of yKu (Figure 2C, black bars), and by deleting the STEs of these telomeres we restored perinuclear association to a significant degree (Figure 2C, grey bars; P=2.8 × 10−4, 1.3 × 10−4, respectively, comparing natural to truncated telomeres). The residual association is Sir4-dependent; as these telomeres assume a random distribution in yku70 sir4 double mutants (Hediger et al, 2002b). For the truncated Tel5R, we see that the peripheral association detected in S phase is significantly reduced in the absence of the yKu-mediated anchoring pathway, but again anchoring could be restored by eliminating the STEs (Figure 2C; P=1.1 × 10−2, comparing natural to truncated in S phase). The simplest interpretation of this phenomenon is that truncation eliminates an element that antagonizes anchoring for at least three telomeres and throughout interphase.
Reb1 and Tbf1 recapitulate the antianchoring effect of STEs
STE elements contain multiple protein binding sites for the factors Reb1 and Tbf1 (Figure 1A), which have been previously shown to mimic the insulator effects of STEs (Fourel et al, 1999, 2001). To see whether the effect of STEs on telomere position is related to this function, we next tried to restore the effects of STE sequences on anchoring by targeting the characterized insulator domains of Reb1 or Tbf1 to the truncated telomeres. This could be performed on the same strains for which the initial localization was analysed, because the inserted array of lacop sequence includes a cluster of four lexA-binding sites. By expressing lexA-Reb1N and lexA-Tbf1N fusions at low levels in strains bearing a tagged Tel6Rtr, we asked whether the presence of these protein domains in a subtelomeric context would be sufficient to counteract anchoring. When targeted to Tel6Rtr in a wild-type background, only the lexA-Tbf1N fusion significantly reduced telomere anchoring in G1-phase cells (Figure 3A; P=8.8 × 10−3, comparing lexA to lexA-Tbf1N).
Figure 3.
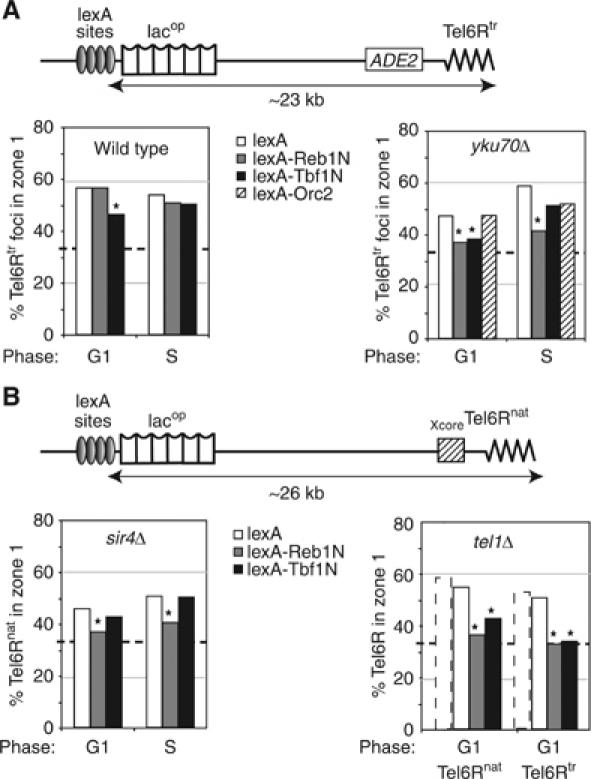
Role of Reb1 and Tbf1 in telomere anchoring. (A) The indicated LexA fusions were targeted to four lexA-binding sites located ∼23 kb from the end of truncated Tel6R. Reb1 and Tbf1 domains encompass the transactivation domain (aa 1–405). Orc2 is a full-length fusion. Position of Tel6Rtr was monitored in cells expressing lexA alone (white boxes), lexA-Reb1N (grey boxes), lexA-Tbf1N (black boxes) or a lexA-Orc2 fusion (hatched boxes) in G1 cells of isogenic wild-type (GA-1917) or yku70Δ (GA-1918) strains. (B) As in (A) except that where indicated strains bear the native Tel6R (Tel6Rnat; GA-1459) with either sir4Δ (GA-1867) or tel1Δ (GA-3453). Tel6Rtr is also monitored in a tel1Δ train (GA-3052). Stipled bars indicate values for the same telomeres with lexA in a wild-type background. Symbols are as in Figure 2, and S phase and P-values are in Supplementary Table 1.
We reasoned that this partial effect might be due to anchorage pathway redundancy, as this was also observed for the presence of STE. We therefore targeted lexA-Tbf1N or lexA-Reb1N to truncated Tel6R in the yku70Δ background, which weakens its NE association. In this case, the targeting of Reb1N efficiently delocalized Tel6Rtr in both G1- and S-phase cells (Figure 3A; P=5 × 10−2 and 2.8 × 10−2, respectively, comparing lexA to lexA-Reb1N). The targeted Tbf1N again efficiently reduced Tel6Rtr-NE association in G1-phase cells, although it had a weaker effect in S phase (Figure 3A; P=8.7 × 10−2 and 0.3 respectively, comparing lexA to lexA-Tbf1N). We conclude that the targeting of the insulator domains of Reb1 and Tbf1 mimics the antianchoring effect of the STE.
As a control for the specificity of the Reb1 and Tbf1 effect, we targeted a third STE-binding factor, Orc2, which is a subunit of the Origin Replication Complex. LexA-Orc2 complements the orc2 deletion strain for DNA replication function (K Shimada, personal communication) and can help nucleate silencing by cooperating with protosilencer elements. However, the targeting of lexA-Orc2 had no significant delocalizing effect on Tel6Rtr (Figure 3A; P=0.9 and 0.4 in G1 and S phase, comparing lexA to lexA-Orc2).
Reb1N and Tbf1N can also counteract yKu-mediated anchoring of telomeres
In order to rule out the possibility that the antagonism of telomere anchoring documented in Figure 3A might reflect a feature unique to the yku background, we tested the effects of targeting lexA-Tbf1N and lexA-Reb1N in other backgrounds that partially compromise anchoring efficiency. Notably, these fusions were targeted to the lexA-binding sites near a native Tel6R in sir4Δ and tel1Δ strain backgrounds. As previously reported, native Tel6R remains 46% in zone 1 in sir4Δ cells, which is reduced from 59% in wild-type cells. However, the targeting lexA-Reb1N in this background in either G1- or S-phase cells, led to a significant shift of the telomere from the outermost zone (Figure 3B). The effect of Tbf1N is less pronounced (Figure 3B). It is noteworthy that this antianchoring effect is manifest not only in wild-type cells, but also in strains that lack TPE altogether (i.e. both sir4 or yku mutants).
Finally, we tested the effects of targeting Reb1N or Tbf1N in a strain lacking the ATM kinase homologue, Tel1. The loss of Tel1 does not seriously compromise TPE (10 × down) nor perinuclear anchoring (Laroche et al, 1998; and see dotted versus open bars, Figure 3B), although the length-maintenance pathway that counts Rap1-binding sites in cis is compromised (Craven and Petes, 1999; Ray and Runge, 1999). However, the short TG tracts of tel1Δ strains are stable (at ∼150 bp), arguing that a second pathway of length maintenance exists. In the tel1Δ strain, we again target lexA-Tbf1N or lexA-Reb1N to the subtelomeric lexA sites at either native or truncated Tel6R (Figure 3B). Quantitation of telomere position showed that these domains strongly antagonized the NE-association of either end (Figure 3B), with Reb1N effects being more pronounced than those of Tbf1N (P=1.4 × 10−3 (Reb1N, natural), 2.6 × 10−2 (Tbf1N, natural), 3.4 × 10−3 (Reb1N truncated), 1.3 × 10−2 (Tbf1N, truncated) when compared to their lexA counterparts).
Reb1 has been reported to bind the spacer region of ribosomal DNA and to influence both activation and termination of RNA Pol I-mediated transcription (Morrow et al, 1989; Butlin and Quincey, 1991; Kulkens et al, 1992). In order to test whether Reb1N antagonizes perinuclear anchoring by relocating chromatin to the nucleolus, lexA-Reb1N was expressed in a strain bearing a fluorescent nucleolar marker (Nop1-CFP) and an internal lacop-tagged locus in the middle of the Chr 6 right arm (ARS607). Neither expression of lexA nor the lexA-Reb1N fusion altered the localization of ARS607, which remained randomly distributed and coincided rarely with the nucleolus (<7%; Figure 4). This is consistent with the reported granular distribution of Reb1 throughout the nucleoplasm and makes it unlikely that the antagonism because of Reb1N targeting is due a strong affinity for sites in the rDNA (Kumar et al, 2002).
Figure 4.
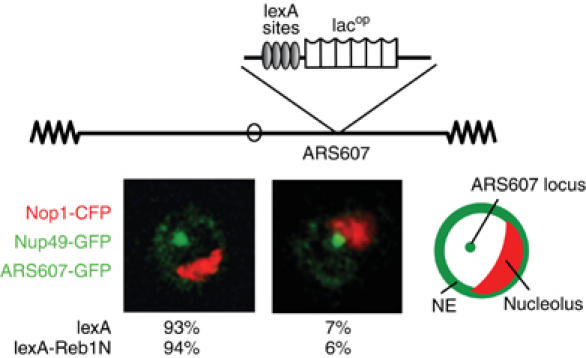
Tethering activity of Reb1. GA-1461 (Taddei et al, 2004) contains lacops and four lexA-binding sites at ARS607 located in the middle of the right arm of Chr 6. GA-1461 bears GFP-lacI, GFP-Nup49 and Nop1-CFP (pGVH45) along with either plexA or plexA-Reb1N. Colocalization of the ARS607 locus with the nucleolar Nop1 signal was scored in confocal sections as distinct (left image) or colocalizing (right image).
VP16 targeting releases telomeres from the NE
One interpretation of the preceding results is that the ultimate localization of a telomere in interphase yeast cells is determined by opposing forces that arise either from factors that promote association with the NE (mediated by yKu and Sir4) or factors that counteract this (mediated by STE-binding factors), leading to telomere release into the nuclear interior. Both Reb1N and Tbf1N function as transcriptional activators, and it has been argued that their insulator function correlates with histone and nucleosome modifications that precede transcriptional initiation. To test whether the antianchoring effect of Reb1 and Tbf1 domains can be replaced by another transactivator, we targeted lexA fused to the acidic domain of the viral protein, VP16. VP16-AD induces transcription in yeast in part by recruiting the SAGA histone acetylation complex and the SWI2/SNF2 nucleosome remodeller (Sadowski et al, 1988). To assay its effects on telomere position, lexA-VP16 was expressed in the tagged Tel6Rtr strain, containing lexA sites upstream of a subtelomeric gene HXK1, 23 kb from the TG repeat (Figure 5A).
Figure 5.
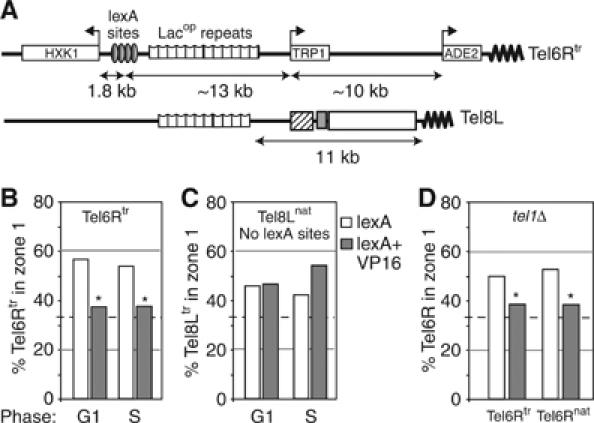
VP16 counteracts telomere anchoring in cis. (A) Telomeres used in the VP16 targeting assay. (B) Tel6Rtr position was monitored for G1 and S phase cells (GA-1917) expressing a lexA or a lexA-VP16 fusion as indicated. (C) As (B), but for Tel8Lnat (GA-1986), which lacks lexA sites. (D) As (B), but positions of Tel6Rnat (GA-3453) and Tel6Rtr (GA-3052) were monitored in tel1Δ G1-phase cells. Zone 1 percentages are compared and asterisks represent values that are not significantly different from random.
The targeting of VP16 induced a complete delocalization of Tel6Rtr in both G1- and S-phase wild-type cells, in a cis-acting manner (Figure 5B). There was no delocalization of Tel8L, which does not contain the lexA-binding sites (Figure 5C), ruling out an effect in trans. The effect of VP16 binding is the strongest of the domains tested, for it is able to delocalize both native and truncated Tel6R in both wild-type and tel1Δ backgrounds (Figure 5D). In addition to compromising perinuclear attachment, the binding of lexA-VP16 enhances the mobility of a tagged chromosomal locus: quantitative mean square displacement analysis of time-lapse movies showed that the radius of constraint increases from 0.5 to 0.7 μm (FR Neumann and SM Gasser, personal communication). Not surprisingly, VP16 targeting to Tel6Rtr could be correlated with a five-fold increase in expression of the gene immediately adjacent to the lexA-binding sites, HXK1, although the more telomere-proximal TRP1 gene is not induced and the subtelomeric ADE2 marker still shows a partially variegating phenotype under these conditions (Supplementary Figure 2). On the other hand, Tel6R delocalization can also be achieved by inducing a subtelomeric TRP1 gene by its normal mechanism, which demonstrates that the antianchoring effect is not due to nonphysiological aspects of the VP16 domain (H Schober and SM Gasser, data not shown).
Remote tethering of Reb1, Tbf1 and VP16 leads to telomere shortening
In the accompanying manuscript, Berthiau et al (2006) present data that implicate Tbf1 and Reb1 in the downregulation of a Tel1-independent pathway of telomere length maintenance. It is argued that this secondary pathway of length control nonetheless acts through telomerase, as the coupling of Tbf1 targeting with an est2 deletion, does not accelerate telomere shortening. In view of this result, we examined whether the effect we document above of the targeted lexA-fusions on telomere localization, correlates with changes in telomere length at a native chromosomal ends. Using the same tagged Tel6Rnat telomere described above, we targeted lexA-Reb1N, -Tbf1N and -VP16 to lexA sites placed 26 kb from the chromosomal end. The length of the Tel6R-specific terminal TG repeats was determined by a Southern blot. To control for general nonspecific effects, we monitored the TG repeat length on all Y′ telomeres in the same strain. The binding of the lexA fusions did not change the length of the terminal repeats in wild-type or Tel1+ cells (data not shown), yet they induced a significant shortening of Tel6Rnat in the tel1Δ strain by roughly 50 bp (Figure 6B and C). Importantly, this effect requires the targeted protein in cis; for in the same tel1Δ cells, we did not observe length changes at Y′ telomeres, which lack the lexA target sites (Figure 6B). The shortening induced by Reb1, Tbf1 and VP16 targeting is not observed in sir4Δ cells, although telomere position is affected by these factors. This argues that the loss of NE association per se does not compromise telomere length regulation as long as the Tel1-dependent counting mechanism is intact. In tel1Δ cells, the remote targeting of Tbf1, Reb1 and VP16 led to telomere displacement and at the same time seemed to render the telomeres less likely to be extended by telomerase, such that they become constitutively shorter. The shortening of Tel6Rnat (Figure 6B and C) is completely consistent with the shortening observed at Tel7Ltr when Tbf1 and Reb1 are tethered at more telomere proximal sites (Berthiau et al, 2006). Overall, we conclude that there is a significant correlation between a loss of peripheral position and telomere shortening, in the absence of the Tel1-mediated pathway for telomerase regulation.
Figure 6.

Tethering of Reb1, Tbf1 and VP16 at a remote subtelomeric location reduces telomere length in tel1Δ cells. (A) Schematic representation of Tel6Rnat and lexA sites, indicating the XhoI site (X) used to measure telomere length. (B) Examples of Southern blots used for the telomere length quantifications shown in (C). Genomic DNA was cut with XhoI and probed with a 6R or a Y′-specific probe as indicated. Three independent plasmid-bearing transformants are tested for each condition. (C) The plot represents the mean length of the terminal XhoI fragment of the Tel6Rnat (see B and Materials and methods). Values and error rates represent an average of two measurements of three independent clones of isogenic strains: wild-type (GA-1459), tel1Δ (GA-3453) and sir4Δ (GA-1867).
Discussion
Telomere anchoring variability
Live time-lapse monitoring of GFP-tagged yeast telomeres indicates that telomeres associate with the nuclear periphery in a rapidly reversible manner, typical of interactions that depend on multiple short-lived protein–protein contacts (Hediger et al, 2002a, 2002b). We confirm here that the probability with which a telomere is located at the NE varies both among chromosomal ends and during the cell cycle. Nonetheless, out of a total of 10 GFP-tagged, nontruncated telomeres analysed, only Tel5R shows no significant enrichment in the narrow 200 nm rim of G1-phase nuclei (Supplementary Table 1; Bystricky et al, 2005). Because the yKu and Sir4 proteins known to mediate interaction between telomeres and the NE are associated with all yeast TG repeats, including Tel5R, we examined whether subtelomeric sequences contribute to the variability in telomere anchoring, and Tel5R's unusual mobility. Indeed, elimination of the STEs, X, STR and Y′, increased the affinity of Tel5R for the NE. This observation was extended to Tel6R and Tel 8L, under conditions that partially compromise the normally redundant anchorage sites (yku70; Figure 2B). This suggests that the factors that bind STEs can antagonize NE interactions.
We note that only a subset of telomeres becomes delocalized upon elimination of yKu, whereas others are sensitive to loss of Sir4 (Hediger et al, 2002b). Notably, the truncated forms of Tel6R and Tel14L both remain tightly anchored in the absence of yKu through the Sir4–Esc1 interaction, although the native, STE-containing forms of these same telomeres do not (Hediger et al, 2002b). This observation argued that STEs are able to antagonize the Sir pathway of telomere-NE interaction. Importantly, here we show that we can reconstitute the antianchoring effect of STEs by targeting the insulator domains of the STE-binding factors Reb1 or Tbf1, to truncated telomeres (Figure 3). Furthermore, Reb1N delocalized telomeres in a sir4Δ strain, indicating that STE factors antagonize either anchoring pathway.
We consider three hypotheses that could account for the variability in native telomere positioning. First, because telomere anchoring is mediated by redundant pathways requiring either yKu or Sir4 (Taddei et al, 2004), anchoring efficiency could reflect a variable propagation of Sir-mediated silencing at different telomeres. Indeed, it was recently shown that silent chromatin alone is sufficient to mediate perinuclear anchorage of an excised HMR locus (Gartenberg et al, 2004). In support of this hypothesis, it has been shown that silencing at native telomeres is telomere-specific and shows discontinuity along the chromosomal arm (Pryde and Louis, 1999). Indeed, Sir proteins and Rap1 do not propagate inwards to the same extent on all telomeres (Lieb et al, 2001). However, we are unable to establish a significant correlation between the abundance of Sir protein binding and anchoring efficiency, based on published Chromatin IP data for Sir proteins (Lieb et al, 2001). Notably, the poorly anchored Tel5R appears to bind Sir factors more efficiently than the well-anchored Tel6R. Furthermore, as Tbf1 and Reb1 are able to delocalize telomeres when targeted to a site ∼23 kb from the TG repeats, that is, beyond the point to which Sir proteins spread, the antianchoring effect of these subtelomeric proteins is unlikely to reflect the blocking of inwardly spreading Sir factors. We can also rule out that weak telomere–NE association reflects a general chromosomal trait, as Tel5L, in opposition to Tel5R, is efficiently bound (Bystricky et al, 2005).
A second hypothesis suggests that the efficiency of telomere anchoring correlates with the level of transcription of subtelomeric genes. Highly transcribed domains have been shown to move out of their chromosomal territories in mammalian cells (e.g. Williams et al, 2002), and we show here that the induction of transcription in a subtelomeric domain influences the efficiency with which it is bound. The VP16-induced gene (HXK1) is 26 kb from the TG repeat and is transcribed towards the centromere, ruling out a disruptive effect of polymerase movement on the telosome (Sandell et al, 1994; Tham et al, 2001). Moreover, under conditions of HXK1 induction used here, the more telomere-proximal reporter, TRP1, shows no increase in transcript levels (Supplementary Figure 2). Although we confirm that the targeted VP16 fusion does antagonize the peripheral association of Tel6R in cis (Figure 5), subtelomeric transcription is unlikely to be responsible for the weak NE association of Tel5R in the absence of VP16 recruitment as none of the telomere-proximal genes on Tel5R (YER188w, YER187w, YER186w) are expressed to significant levels under normal growth conditions (SGD, Stanford database).
A third hypothesis to explain the variability in telomere anchorage is that the composition of factors bound to the STEs antagonizes the anchoring functions of yKu and Sir4 to different degrees. This is supported by our finding that not only STEs, but also the transactivation and insulator domains of Tbf1 and Reb1, reduce the efficiency with which telomeres are bound to the NE. Recognition sites for these factors are present at all yeast telomeres, but in widely varying numbers (Figure 1). The effect of Tbf1 or Reb1 binding could reflect changes in higher-order structures and possibly also trans interactions among chromosomal ends (Fourel et al, 1999; Pryde and Louis, 1999). We propose that variable STE factor binding alters higher-order folding of telomeres, which could account for both the insulator function of subtelomeric factors and their effect on anchoring efficiency. The folding of the telomere has also been implicated in the switch from an accessible to an inaccessible state for telomerase action (Teixeira et al, 2004).
Insulation and spatial location
We have shown that upon removal of STEs some telomeres become efficiently anchored at the NE (Figure 2), and that we can mimic the STE effect on anchorage by targeting domains of Reb1, Tbf1 and VP16 (Figure 3). The non-DNA-binding domains of Tbf1 and Reb1 also confer an insulator activity that protects subtelomeric genes Sir-mediated repression (Fourel et al, 2001). Although the activation domain of VP16 also antagonizes silencing, we cannot conclude that its mode of action with respect to telomere position, is the same as that provided by the N-termini of Reb1 and Tbf1, even though all three influence telomere length maintenance. It is well established that VP16 can promote the recruitment of histone acetylation complexes (NuA4, SAGA) and chromatin remodellers (SWI/SNF; Sadowski et al, 1988; Utley et al, 1998; Neely et al, 1999), which then lead to enhanced transcription (Figure 7). In other studies, we find that chromatin remodellers themselves can increase the mobility of chromatin within the nucleoplasm, independently of transcription (FR Neumann and SM Gasser, unpublished data), and we propose that the antianchoring effect of VP16 reflects the presence of these activities. Berthiau et al (2006) show that the Tbf1N domain is a relatively weak transcriptional activator, that it has little effect on subtelomeric nucleosomal organization. Thus, while nucleosome remodelling may provide one means to antagonize telomere anchorage, it is probably not the only one. The important point, however, is that binding of Tbf1N, Reb1N and VP16 to distal sites on both truncated and native telomere ends, is able to counteract telomere association with the NE, as depicted in the model (Figure 7). This displacement correlates with a reduction in telomere length, in the absence of the Tel1-regulated feedback mechanism for length regulation (Figure 6).
Figure 7.
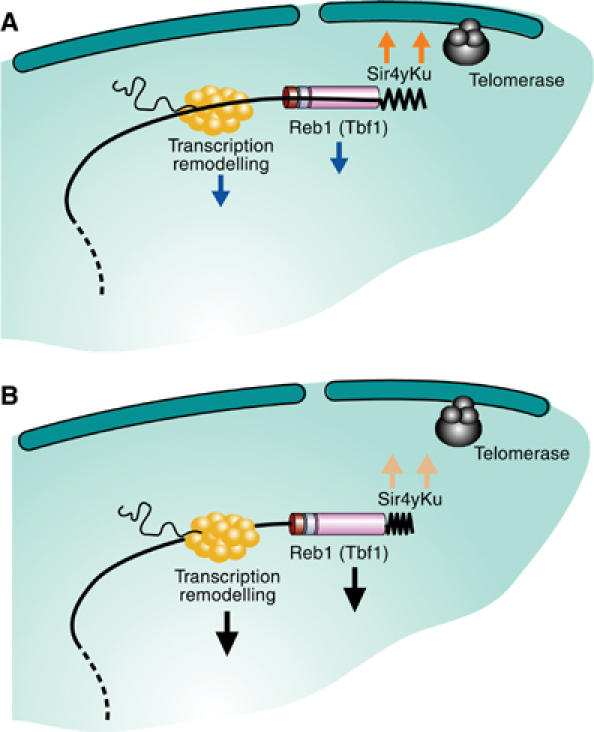
Model for telomere dynamics. (A) Two opposite types of forces are proposed to determine position and dynamics of yeast telomeres. Parallel anchoring pathways involving yKu70/80 and Sir4 can be challenged by STEs that provide binding sites for Reb1 and Tbf1. Other pulling forces may derive from active transcription, but if both anchoring pathways are intact, telomeres remain enriched at the NE. (B) When the antagonistic forces are strong enough to compromise anchoring, telomerase elongates short TG tracts less efficiently. This is detectable only if the Tel1-mediated counting mechanism is compromised, suggesting that this latter compensates for any drop in telomerase efficiency provoked by a change in telomere–NE interaction.
A role of telomere anchoring in telomerase regulation?
Our study clearly shows that the targeting of Tbf1, Reb1 and VP16 to sites remote from the TG repeat, compromises telomere length regulation in the absence of the ATM kinase homologue Tel1. This is fully consistent with the data reported by Berthiau et al (2006), which show that the binding of Tbf1 immediately proximal to the TG repeat of Tel7L, limits its TG tract length. This effect is most pronounced in the absence of Tel1 kinase. Genetic analysis further argues that the Tbf1 effect directly influences telomerase efficiency and not other mechanisms of maintenance, as Tbf1 targeting does not alter the rate with which telomeres shorten in an est2 telomerase mutant (Berthiau et al, 2006).
Other lines of evidence also link the elongation of telomeres by the telomerase and their subtelomeric composition. It was shown that telomeres containing only the X element could be more efficiently elongated than Y′-containing telomeres in a tel1Δrif1Δ background (Craven and Petes, 1999). Indeed, Y′ elements contain several binding sites for the antianchoring factors Reb1 and Tbf1 and Tel5R anchoring is restored by a truncation that partially eliminates the Y′ element (Figure 2). Y′ elements, like Tbf1 and Reb1 binding, affect both anchoring efficiency and telomerase action. In our hands, the effects of targeted domains are only partial, leading to a shortening of 50–80 bp. This suggests that STEs simply reduce the efficiency with which telomerase acts on a given telomere. The common dependency upon a specific genetic context (tel1Δ) and the shared ability to induce a telomere shortening when tethered both at proximal and remote (23 kb) subtelomeric sites, argue that Tbf1, Reb1 and VP16 regulate telomere length by common means.
The effect of Tbf1, Reb1 and VP16 on Tel6R length in tel1Δ cells correlates with their ability to reduce telomere association with the nuclear periphery. We therefore propose that the Tel1-independent pathway of telomere length regulation is at least in part linked to NE association of the telomere. It is important to note, however, that the Tel1-dependent ‘Rap1 counting' mechanism is dominant over delocalization effects. In yku70Δ or sir4Δ cells, the counting pathway is functional and in this case the tethering of Reb1 or Tbf1 to a truncated Tel6R does not induce telomere shortening, despite their effects on telomere delocalization (Figure 6). Thus, the dissociation of telomeres from the NE can be compensated for in TEL1 cells by other mechanisms, which ensure that telomerase functions efficiently. The alternative mechanisms may differ between ku- and sir4-deficient strains, as yKu, unlike Sir4, can recruit telomerase.
Our paper provides novel evidence suggesting that telomerase action may be facilitated by the proximity of telomeres to the NE. We propose that a telomerase subcomponent, such as Est1 or Est3, which facilitate telomerase action, may be sequestered or activated by NE association. Alternatively, perinuclear enzymes, such as sumoylases, may help regulate telomere length, perhaps by acting on the replication machinery. Finally, given that telomerase is found at very low concentrations, telomere–NE association may simply increase the probability that enzyme and substrate interact. Further studies on induced telomerase action may help shed light on how peripheral positioning of short telomeres can favor TG tract elongation.
Materials and methods
Plasmids and yeast strains
Plasmids plexA-Reb1N and pTbf1N are derived from GBD-fusion plasmids described in Fourel et al (2001). The first 405 aa of Reb1N and Tbf1N were transferred in frame into the pAT4 vector (a 2 μm LEU2 vector expressing lexA from the ADH promoter; Taddei et al, 2004). The lexA-ORC2 fusion (kind gift of K Shimada, FMI, Basel) is expressed on a centromere containing LEU2 plasmid under control of a CYC1 promoter, and it complements an orc2-1 mutation at nonpermissive temperature. plexA-VP16 (pFN14) is a CEN LEU2 plasmid expressing 130 aa of VP16 fused to LexA from an ADH1 promoter. pGVH45 expresses a Nop1-CFP fusion from its natural promoter, on a CEN plasmid with an ADE2 marker.
All strains used are indicated in Table I. PCR-based complete gene deletions of yku70, tel1 and sir4 were checked by PCR and phenotypic assays. GA-2226 was obtained by transforming GA-1320 with pFH7 linearized with ClaI. This inserts four lexA sites and ∼10 kb of lacop repeats with a TRP1 marker 1 kb centromere-proximal from ADH4. YG-143 was obtained by transforming GA-1320 with pGVH22 linearized with PsiI. This inserts lacop repeats with the TRP1 marker into YER185w. GA-3032 was obtained by transforming GA2296 (S288C with an ADE2 complete deletion) with plasmid pFN15 linearized with BbsI. This inserts lexA sites and lacop repeats with the TRP1 marker 1.8 kb telomere-proximal of HXK1. pFN15 was also used to generate GA1459. Tel6R is truncated telomere-proximal of YFR055w with pFH1, containing 1 kb of Tel6R sequence, ADE2 and 81 bp of TG1−3 repeats. Tel8L is truncated telomere-proximal from YHL046c by inserting LEU2, a NES-GFP fusion under the URA3 promoter and TG repeats (GA-2556). Tel5R is truncated with plasmid pHR10-6 (Singer and Gottschling, 1994) which inserts ADE2 and TG repeats near the YER186c ORF (YG-138).
Table 1.
Yeast strains used in this study
| Name | Genotype | References |
|---|---|---|
| GA-1320 | MATa ade2-1 can1-100 his3-11,15∷GFP-LacI-HIS3 trp1-1 ura3-1 leu2-3,112 nup49∷NUP49-GFP- | Heun et al (2001) |
| GA-1459 | GA-1320 TEL6R∷lexA-lacO-TRP1 | Heun et al (2001) |
| GA-1985 | GA-1320 TEL14L∷lacO-TRP1 | Hediger et al (2002b) |
| GA-1986 | GA-1320 TEL8L∷lacO-TRP1 | Hediger et al (2002b) |
| GA-2226 | GA-1320 TEL7L∷lexA-lacO-TRP1 | This study |
| YG-143 | GA-1320 TEL5R∷lacO-TRP1 (into YER185W) | This study |
| GA-1917 | GA-1459 TEL6R∷lacO-lexA-TRP1-//-ADE2-TG1–3 | Hediger et al (2002b) |
| GA-2556 | GA-1986 TEL8L∷lacO-TRP1-//- LEU2-URA3p-NES-GFP-ADHt- TG1–3 | This study |
| YG-138 | YG143 TEL5R∷lacO-TRP1-//-ADE2- TG1–3 | This study |
| GA-1489 | GA-1459 hdf1∷URA3 | Hediger et al (2002b) |
| GA-1916 | GA-1986 hdf1∷URA3 | This study |
| GA-195 | YG-143 hdf1∷URA3 | This study |
| GA-1918 | GA-1917 hdf1∷URA3 | Hediger et al (2002b) |
| GA-2804 | GA-2556 hdf1∷URA3 | This study |
| GA-149 | YG-143 hdf1∷URA3 | This study |
| GA-1867 | GA-1459 sir4∷KanMx6 | Hediger et al (2002b) |
| GA-1461 | GA-1320 Chr6int∷lacO-lexA-TRP1 | Taddei et al (2004) |
| GA-3453 | GA1459 tel1∷kanMx6 | This study |
| GA-3052 | GA-1917 tel1∷kanMx6 | This study |
Live fluorescence microscopy
Cell growth, image acquisition and image analysis were performed as described in Taddei et al (2004). Images of Nop1-CFP/Nup49-GFP/ARS607-GFP were acquired with YFP/CFP settings (YFP/CFP filter, 432 and 504 nm excitation, 100–200 ms exposure) on the Zeiss LSM510 confocal microscope. Colocalization of ARS607 with the nucleolus was scored when the GFP focus was in or adjacent to the Nop1 signal.
Telomere length analysis
Telomere blotting and length measurement were performed as described in Brevet et al (2003). To prepare the Tel6R probe, a specific genomic PCR fragment has been amplified with primers SG355 (CACCGCCAAGCTTCCAATATCACG) and SG356 (GGAGGCATTATGGCTTTGTTACGC). Gels were quantified by two different experimenters, and the same samples were analysed on two different gels, allowing us to estimate that the standard error in telomere length measurements is less than 5%.
Statistics and error estimation
Distributions of tagged telomere position were compared either to the predicted random array or to another distribution with a Student's t-test comparing the proportion of scored telomeres in zone 1. Statistical significance was determined by using a 95% confidence interval. A χ2 test was used to compare three-zone distribution (instead of only zone 1), but this is only possible for comparison with a predicted random distribution and not when comparing two observed distributions.
Supplementary Material
Supplementary Data
Supplementary Figure 2
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation, the NCCR Frontiers in Genetics and by the Novartis Research Foundation (to SMG) and Ligue Nationale contre le Cancer, (to EG as ‘équipe labellisée'). We thank F Neumann and K Shimada for constructs and for communicating unpublished controls relevant for our study. M Vega-Palas (Sevilla, Spain) kindly created the strain bearing a tagged Tel8Ltr.
References
- Andrulis ED, Zappulla DC, Ansari A, Perrod S, Laiosa CV, Gartenberg MR, Sternglanz R (2002) Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol Cell Biol 22: 8292–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthiau AS, Yankulov K, Bah A, Revardel E, Luciano P, Wellinger RJ, Géli V, Gilson E (2006) Subtelomeric proteins negatively regulate telomere elongation in yeast. EMBO J (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilaud T, Koering CE, Binet-Brasselet E, Ancelin K, Pollice A, Gasser SM, Gilson E (1996) The telobox, a Myb-related telomeric DNA binding motif found in proteins from yeast, plants and human. Nucleic Acids Res 24: 1294–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevet V, Berthiau AS, Civitelli L, Donini P, Schramke V, Geli V, Ascenzioni F, Gilson E (2003) The number of vertebrate repeats can be regulated at yeast telomeres by Rap1-independent mechanisms. EMBO J 22: 1697–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigati C, Kurtz S, Balderes D, Vidali G, Shore D (1993) An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol Cell Biol 13: 1306–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun C, Marcand S, Gilson E (1997) Proteins that bind to double-stranded regions of telomeric DNA. Trends Cell Biol 7: 317–324 [DOI] [PubMed] [Google Scholar]
- Butlin M, Quincey R (1991) Activity of promoter mutants of the yeast ribosomal RNA gene with and without the enhancer. Yeast 7: 679–689 [DOI] [PubMed] [Google Scholar]
- Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM (2005) Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J Cell Biol 168: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Tye BK (1983) Organization of DNA sequences and replication origins at yeast telomeres. Cell 33: 563–573 [DOI] [PubMed] [Google Scholar]
- Chasman DI, Lue NF, Buchman AR, LaPointe JW, Lorch Y, Kornberg RD (1990) A yeast protein that influences the chromatin structure of UASG and functions as a powerful auxiliary gene activator. Genes Dev 4: 503–514 [DOI] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, Weng S, Botstein D (1998) SGD: Saccharomyces Genome Database. Nucleic Acids Res 26: 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RJ, Petes TD (1999) Dependence of the regulation of telomere length on the type of subtelomeric repeat in the yeast S. cerevisiae. Genetics 152: 1531–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Stillman B (1989) Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science 246: 1034–1038 [DOI] [PubMed] [Google Scholar]
- Fourel G, Boscheron C, Revardel E, Lebrun E, Hu YF, Simmen KC, Muller K, Li R, Mermod N, Gilson E (2001) An activation-independent role of transcription factors in insulator function. EMBO Rep 2: 124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J 18: 2522–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, Gasser SM (2004) Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119: 955–967 [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM (1996) The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type S. cerevisiae. J Cell Biol 134: 1349–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA (1990) Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762 [DOI] [PubMed] [Google Scholar]
- Hediger F, Dubrana K, Gasser SM (2002a) Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J Struct Biol 140: 79–91 [DOI] [PubMed] [Google Scholar]
- Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM (2002b) Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol 12: 2076–2089 [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM (2001) Chromosome dynamics in the yeast interphase nucleus. Science 294: 2181–2186 [DOI] [PubMed] [Google Scholar]
- Kulkens T, van der Sande CA, Dekker AF, van Heerikhuizen H, Planta RJ (1992) A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J 11: 4665–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, Cheung KH, Miller P, Gerstein M, Roeder GS, Snyder M (2002) Subcellular localization of the yeast proteome. Genes Dev 16: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM (1998) Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol 8: 653–656 [DOI] [PubMed] [Google Scholar]
- Lebrun E, Revardel E, Boscheron C, Li R, Gilson E, Fourel G (2001) Protosilencers in S. cerevisiae subtelomeric regions. Genetics 158: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Blackburn EH (2004) Counting of Rif1p and Rif2p on S. cerevisiae telomeres regulates telomere length. Mol Cell Biol 24: 10857–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO (2001) Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat Genet 28: 327–334 [DOI] [PubMed] [Google Scholar]
- Louis EJ (1995) The chromosome ends of S. cerevisiae. Yeast 11: 1553–1573 [DOI] [PubMed] [Google Scholar]
- Louis EJ, Haber JE (1992) The structure and evolution of subtelomeric Y′ repeats in S. cerevisiae. Genetics 131: 559–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Gilson E, Shore D (1997) A protein-counting mechanism for telomere length regulation in yeast. Science 275: 986–990 [DOI] [PubMed] [Google Scholar]
- Moazed D, Rudner AD, Huang J, Hopper GJ, Tanny JC (2004) Model for step-wise assembly of heterochromatin in yeast. Novartis Found Symp 259: 48–56 [PubMed] [Google Scholar]
- Morrow BE, Johnson SP, Warner JR (1989) Proteins that bind to the yeast rDNA enhancer. J Biol Chem 264: 9061–9068 [PubMed] [Google Scholar]
- Neely KE, Hassan AH, Wallberg AE, Steger DJ, Cairns BR, Wright AP, Workman JL (1999) Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell 4: 649–655 [DOI] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ (1999) Limitations of silencing at native yeast telomeres. EMBO J 18: 2538–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Runge KW (1999) Varying the number of telomere-bound proteins does not alter telomere length in tel1Δ cells. Proc Natl Acad Sci USA 96: 15044–15049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling D (1993) Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev 7: 1133–1145 [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M (1988) GAL4-VP16 is an unusually potent transcriptional activator. Nature 335: 563–564 [DOI] [PubMed] [Google Scholar]
- Sandell LL, Gottschling DE, Zakian VA (1994) Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc Natl Acad Sci USA 91: 12061–12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Gottschling D (1994) TLC1: template RNA component of S. cerevisiae telomerase. Science 266: 404–409 [DOI] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM (2004) Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J 23: 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner J (2004) Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117: 323–335 [DOI] [PubMed] [Google Scholar]
- Tham WH, Wyithe JS, Ko Ferrigno P, Silver PA, Zakian VA (2001) Localization of yeast telomeres to the nuclear periphery is separable from transcriptional repression and telomere stability functions. Mol Cell 8: 189–199 [DOI] [PubMed] [Google Scholar]
- Utley RT, Ikeda K, Grant PA, Cote J, Steger DJ, Eberharter A, John S, Workman JL (1998) Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394: 498–502 [DOI] [PubMed] [Google Scholar]
- Wang KL, Warner JR (1998) Positive and negative autoregulation of REB1 transcription in S. cerevisiae. Mol Cell Biol 18: 4368–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RR, Broad S, Sheer D, Ragoussis J (2002) Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp Cell Res 272: 163–175 [DOI] [PubMed] [Google Scholar]
- Yamada M, Hayatsu N, Matsuura A, Ishikawa F (1998) Y′-Help1, a DNA helicase encoded by the yeast subtelomeric Y′ element, is induced in survivors defective for telomerase. J Biol Chem 273: 33360–33366 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Figure 2


