Abstract
The RNA World model for prebiotic evolution posits the selection of catalytic/template RNAs from random populations. The mechanisms by which these random populations could be generated de novo are unclear. Non-enzymatic and RNA-catalyzed nucleic acid polymerizations are poorly processive, which means that the resulting short-chain RNA population could contain only limited diversity. Nonreciprocal recombination of smaller RNAs provides an alternative mechanism for the assembly of larger species with concomitantly greater structural diversity; however, the frequency of any specific recombination event in a random RNA population is limited by the low probability of an encounter between any two given molecules. This low probability could be overcome if the molecules capable of productive recombination were redundant, with many nonhomologous but functionally equivalent RNAs being present in a random population. Here we report fluctuation experiments to estimate the redundancy of the set of RNAs in a population of random sequences that are capable of non-Watson-Crick interaction with another RNA. Parallel SELEX experiments showed that at least one in 106 random 20-mers binds to the P5.1 stem–loop of Bacillus subtilis RNase P RNA with affinities equal to that of its naturally occurring partner. This high frequency predicts that a single RNA in an RNA World would encounter multiple interacting RNAs within its lifetime, supporting recombination as a plausible mechanism for prebiotic RNA evolution. The large number of equivalent species implies that the selection of any single interacting species in the RNA World would be a contingent event, i.e., one resulting from historical accident.
Keywords: fluctuation analysis, RNA evolution, RNA World, non-Watson-Crick binding, SELEX
INTRODUCTION
The existence of RNAs that are both catalysts and templates for replication forms the basis of a consistent model for the origin of biochemical information (Gesteland et al. 1999). In the simplest of these RNA World scenarios, RNA template-catalysts are presumed to have been selected from pools of random sequence and length. SELEX serves as an analogue for this selection (Ellington and Szostak 1990; Tuerk and Gold 1990). RNA populations of high complexity can be selected for ligand binding or enzymatic functions in vitro, suggesting that the RNA World could emerge through the successive steps of variation, selection, and replication seen in other evolving systems.
Limitations inherent in polymer synthetic systems, however, make it unlikely that sufficient sequence and structural diversity for this evolution could be achieved in abiotic RNA populations. In the absence of a processive replicator, short RNAs will dominate a population of synthetic RNAs. The statistical distribution of the chain length of polymers in a population synthesized by a distributive mechanism depends on the efficiency of the synthetic mechanism by which monomers are joined together. The mean chain length, X̄, of a random polymer is X̄ = 1 / (1 − p), where p is the probability of reaction for each monomer. Thus, for example, if 90% of precursors have reacted, the mean chain length would be 10 (Flory 1936). RNA polymerization in available models of the RNA World is considerably less efficient than needed to effect functional diversity. For example, an RNA-extending ribozyme has a variable processivity ranging from near zero to ~90% at each step, depending on the strength of template-primer binding (Lawrence and Bartel 2003), but the processivity comes at the cost of reduced overall polymerization since the extended product does not dissociate readily from the ribozyme. Activated nucleoside monophosphates can be polymerized in a surface reaction on montmorillonite clay, but the mean chain length of the resulting population is still near 20 nucleotides (nt) (Huang and Ferris 2003). It is likely that the molecular population available for evolution of an RNA World would be dominated by small RNAs.
The dominance of short species in prebiotic RNA synthesis has implications for the generation of prebiotic RNA ensembles. In particular, it disfavors direct evolution of large, structurally complex RNAs. RNAs are modular structures and the structural complexity of the species within an RNA population increases with chain length (Knight and Yarus 2003; Gevertz et al 2005). This has functional consequences: The affinity of aptamers for their cognate ligands depends on the chain length of the RNA population subjected to SELEX (Sabeti et al. 1997; Carothers et al. 2004), and the activities of ribozyme active sites are enhanced greatly by the presence of phylogenetically variable auxiliary domains that do not directly participate in catalysis (Darr et al. 1992; Ikawa et al. 1999; Penedo et al. 2004; Westhof and Massire 2004; Kazantsev et al. 2005; Torres-Larios et al. 2005). Since a prebiotic RNA population would be dominated by species as short as or shorter than a minimal active site (e.g., a three-way junction with 12 conserved residues in the hammerhead), there would be few if any nonessential residues in an active-site molecule that could be mutated to enhance efficiency.
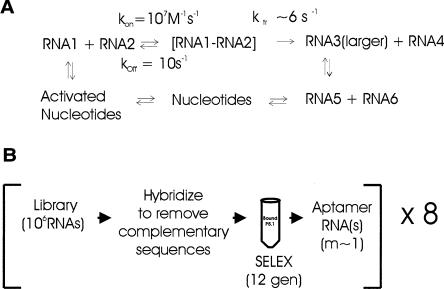
Larger, more proficient, RNAs potentially could be assembled from shorter oligoribonucleotides through recombination (Burke and Willis 1998; Schmidt 1999; Riley and Lehman 2003). In recombination-based scenarios (e.g., Fig. 1A ▶), the activity of a small RNA (either an aptamer or a ribozyme) would be enhanced by recombination with an auxiliary domain, both being present in the initial population of small random RNAs. The two RNAs could be recombined by ligation or more likely transesterification (Chetverin et al. 1997).
FIGURE 1.
(A) Kinetic scheme for recombination during RNA evolution (Schmidt 1999). The rate of interaction between Species 1 and 2, kon, is concentration-dependent and, therefore, depends on the total number of equivalent interacting species. The rate of recombination to produce a larger RNA 3 and smaller RNA 4 depends on the rate of transesterification (Herschlag and Cech 1990; Lawrence and Bartel 2003) within the RNA–RNA complex. If ligation between the ends occurs, the size of RNA 3 would be the sum of those of RNAs 1 and 2. The rate of degradation (Herschlag and Cech 1990) of the active RNA 3 is chain-length dependent. Eventually the RNAs will degrade to nucleotides. The scheme does not take RNA replication into account. (B) Parallel SELEX experiment. Each of eight experiments used an initial library of 106 species, derived by dilution and PCR from an initial pool containing all 420 (~1012) possible 20-mer sequences. All eight libraries had the same 5′ promoter sequence for T7 RNA polymerase; however, the terminal 5 nt of the 3′ were different for each library (Table 1 ▶). This latter attribute prevented cross-contamination of the parallel selections by “parasite” RNAs. The libraries were preselected to remove complementary sequences and selected for the ability to bind column-bound P5.1 RNA. RNAs were reverse-transcribed and cloned after 12 rounds of SELEX.
Evolution through recombination is in turn limited by the complexity of the RNA population that can productively recombine to give a more efficient aptamer or catalyst. Kinetically, if two RNAs have typical RNA–RNA affinity with KD = 10−6 M, the rate of dissociation, 10 sec−1, is given by the product of KD and the rate of bimolecular collision, ~107 M−1sec−1 (Hammes and Schimmel 1970). The rate constant for the chemical step of ribozyme-catalyzed transesterification or ligation is ~6 sec−1 (Herschlag and Cech 1990), approximately equal to the rate of dissociation. In this admittedly best-case scenario, recombination between two molecules of micromolar affinity could occur at approximately the same frequency as their dissociation. Thus, generalized recombination is a plausible mechanism for ribozyme evolution.
The difficulty with recombination-based scenarios such as in Figure 1A ▶ lies with the improbability of any two defined RNA molecules colliding in solution. The effective concentration of productive RNAs is reduced by a factor of r/4l, where r is the redundancy, the fraction of the 4l RNAs in the library that would be functional (Sabeti et al. 1997). The value of r could in principle vary between one and 4l. If r = 1, this single effective 20-mer would be present only at ~10−17 M in an equilibrium mixture of RNAs synthesized from 1 mM precursors by a reaction of 90% efficiency (see Supplemental Material at http://www.biochem.missouri.edu/schmidtf/Striggles2006supp.pdf). If the rate constant for bimolecular collision is ~107 M−1sec−1 (Hammes and Schimmel 1970), then another RNA would encounter this single activator only once every 1010 sec. Productive encounter under these conditions is unlikely because of degradation. The half-life of an RNA phosphodiester bond in solution is 2 × 108 sec (Herschlag and Cech 1990), meaning that the 20-mer would have a half-life of 107 sec. This implies that degradation of an RNA, e.g., a ribozyme active site, is 103 times more likely than is collision with a single defined auxiliary molecule.
The problem could be resolved if the frequency of collision between different RNAs were increased, essentially through an increase in the concentration of the productively recombining species. If the redundancy r > 1, the effective concentration of productive RNAs would be increased by the mole fraction of productive RNAs in a population of length l. A given RNA, e.g., a ribozyme active site, would therefore undergo more collisions with potential auxiliary RNAs during its lifetime. Some of these collisions could result in joining of two RNAs rather than their dissociation, depending on the rate of transesterification or ligation. Based on these considerations, we hypothesized previously that on the order of one in 106 random RNAs is capable of binding to other RNAs in vitro with affinities typical of contemporary RNA–RNA interaction (Schmidt 1999). We show here that this hypothesis is plausible through the use of fluctuation experiments (Luria and Delbrück 1943) to estimate the minimum frequency of RNAs within a random population that are capable of binding to a specific RNA target.
RESULTS AND DISCUSSION
In vitro selection
We used RNA–RNA SELEX (Cho et al. 1997; Soukup and Maher 1998) to select RNA aptamers that bound to a defined RNA. The target RNA for affinity selection was the column-bound P5.1 stem–loop element from Bacillus subtilis RNase P RNA (Table 1 ▶). P5.1 features a unique hexaloop structure that is required for the transactivation of a mutant (Δ5.1) form of P RNA (Kim et al. 2001; Leeper et al. 2002). It interacts with a second structural element in RNase P RNAs, P15.1 (Chen et al. 1998; Massire et al. 1998; Kazantsev et al. 2005). Because P5.1 and P15.1 elements do not interact through loop–loop complementarity, we chose to actively select against complementary RNAs (Cho et al. 1997). The criterion for success in each experiment was the identification of a winning RNA that bound to P5.1 RNA with an affinity close to that of the isolated P15.1 element for P5.1.
TABLE 1.
RNA characterization
| RNA source | RNA sequences | KD for P5.1 RNA, μM | |||
| P5.1 | ACGGUGCUGAGAUGCCCGUA | ||||
| P15.1 | CCUUCUAACGGAAUUCAACGAGGAGCC | 8 ± 2 | |||
| SELEX 1 | 1B | ggaauucuccua | UAUGACCUCGGAGUCCUGUG | uuccaggauccuuggccu | 1.4 ± 0.8 |
| SELEX 2 | 2A | ggaauucuccua | UCGACUACACGCUCCUCUGG | uuaguggauccuuggccu | 2.6 ± 0.5 |
| 2G | ggaauucuccua | AAAUAUACGGAUCUGGCGUG | uuaguggauccuuggccu | 14 ± 5 | |
| SELEX 3 | ggaauucuccua | None | ucuagggauccuuggccu | ||
| SELEX 4 | 4H | ggaauucuccua | CCCUCACCAUCAACGUGUCC | ugaugggauccuuggccu | 1.9 ± 0.7 |
| SELEX 5 | 5F | ggaauucuccua | AUUACCAAGUCGGACCGUG− | uggagggauccuuggccu | 1.7 ± 0.5 |
| 5H | ggaauucuccua | UCUUCCAUGACUUCACUGCC | uggagggauccuuggccu | 4 ± 1 | |
| SELEX 6 | ggaauucuccua | None | ugcugggatccuuggccu | ||
| SELEX 7 | 7B | ggaauucuccua | ACCCGGCGAUGAUGACCUCG | uccacggauccuuggccu | 6 ± 3 |
| 7C | ggaauucuccua | AGCCCAUGAAUCCUGCCGCC | uccacggauccuuggccu | 6 ± 1 | |
| SELEX 8 | ggaauucuccua | None | uccccggauccuuggccu | ||
Base-paired residues of P5.1 and P15.1 are underlined. Phylogenetically conserved residues in these RNAs are bolded. Bolded residues in P5.1 are essential for transactivation (Leeper et al. 2002). In the results from SELEX experiments, fixed (primer) residues are in lowercase; residues derived from the random sequences of the libraries are in uppercase. SELEX experiments 3, 6, and 8 did not yield aptamer clones.
The number of potential winners in a SELEX experiment depends on the number of distinct species (complexity) in the random library and the stringency of the criteria that are judged to constitute success. If, as hypothesized earlier (Schmidt 1999), the frequency of binding RNAs in the population was ~10−6, then after adjustment of the number of RNA species in each library so that each experiment started with 106 species, each library would contain an average of one winning RNA. This assumption also implies that the probability of any single experiment being successful would be governed by a Poisson distribution with a mean of one event.
In order to evaluate this prediction, we carried out eight parallel SELEX experiments on separate libraries (Fig. 1B ▶). Each library contained a 20-mer random sequence bounded by a 5′ phage T7 promoter sequence and by a library-specific 3′ sequence (Table 1 ▶). The libraries therefore had a theoretical complexity of 420, ~1012, individual species, and the yield of the synthesis was large enough so that each possible sequence was expected to be present 102–103 times in the initial pool. We then diluted each library pool to a mass that could contain only 106 template DNAs prior to PCR amplification and transcription. The SELEX protocol used a secondary selection against RNAs that bound the target by extensive Watson-Crick interaction (Cho et al. 1997).
After 12 rounds of SELEX, five of eight experiments yielded a total of eight aptamer sequences (Table 1 ▶). This result was consistent with the prior estimation of the frequency of winning aptamers in a library of 20-mer species; i.e., the mean number of winning RNAs per experiment was close to one. The distribution of the winning RNA species from the parallel SELEX experiments was likewise consistent with a Poisson distribution (Luria and Delbrück 1943). Three of the libraries did not yield aptamer clones; only artifactual RNAs (e.g., primer-dimers) were identified in these experiments. Assuming that this was due to no binding RNAs being present in the original library, the Poisson mean of the winning aptamers per experiment was 0.98, the same as the observed average of 1.0.
RNA characterization
Table 1 ▶ shows the sequences of the winning RNAs that were selected from each library. There was limited or no sequence identity between the winning aptamers and the P15.1 domain that naturally interacts with P5.1 in B. subtilis RNase P RNA. This is consistent with the hypothesis that a large number of potential interacting RNAs are available in random populations.
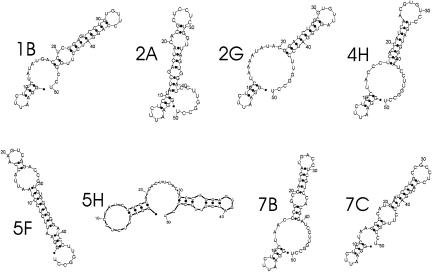
We mapped the winning RNAs structurally using limited nuclease digestion (Fig. 2 ▶; see Supplemental Material at http://www.biochem.missouri.edu/schmidtf/Striggles2006supp.pdf). There was no single predominant secondary structure, although all RNAs featured a stem–loop of varying length. Six of the winning RNAs (1B, 2A, 4H, 5F, 5H, and 7G) contained a 3′-terminal CU sequence in the loop. Two RNAs (2G, 7A) contained a UG dinucleotide in the loop. There appeared to be a bias against loops that contained extended sequences of A residues. The recent crystal structure of Bacillus stearothermophilus RNase P RNA shows that L15.1 distorts to insert the second of the four conserved loop A residues between the latter GA dinucleotide (G73-A74) of L5.1, with concomitant formation of A-minor contacts between the other three A residues, A290, A295, and A296 of L15.1 and the minor groove of L5.1 (Kazantsev et al. 2005). Similar residues are available in the predominant structures of RNAs 2G and 5F only; the other aptamers therefore are likely to bind to L5.1 by other, analogous, interactions.
FIGURE 2.
Structural characterization of winning aptamers. Nucleotides 13–32 (31 in 5F) in the RNAs are derived from the random sequence. RNAs (Table 1 ▶) were folded using Mfold (Zuker 2003). The structures were confirmed by limited nuclease digestion as described in the Supplemental Material (http://www.biochem.missouri.edu/schmidtf/Striggles2006supp.pdf).
RNA–RNA binding
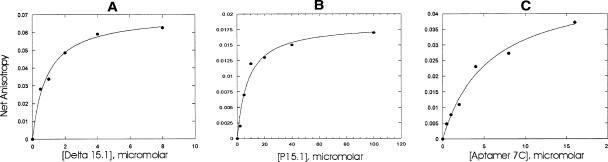
We tested the winning aptamers for their ability to bind P5.1 RNA in solution. Figure 3A ▶ shows the titration curve for the increased anisotropy of 5′ fluorescein-labeled P5.1 RNA as a function of the concentrations of Δ5.1 RNA, a deletion form of RNase P RNA that lacks this domain. The KD for the interaction of P5.1 with Δ5.1 RNase P RNA was 0.9 ± 0.1 μM (Fig. 3A ▶), consistent with the value for this interaction determined previously by transactivation kinetics and by electrophoretic mobility shift analysis (Kim et al. 2001). The KD for the interaction of P5.1 with its isolated cognate P15.1 element (derived from RNase P RNA) was 8 ± 2 μM (Fig. 3B ▶), approximately 10-fold weaker, presumably due to the unavailability of extra stabilization provided by the remainder of the RNase P RNA molecule. Aptamer 7C bound P5.1 with a KD of 6 ± 1 μM (Fig. 3C ▶), i.e., with an affinity equal to that of P15.1. Other winning aptamers had KD values of 2–14 μM (Table 1 ▶). Thus, the selected aptamers all bound to P5.1 with KD values within threefold of the KD between P5.1 and its naturally occurring partner, P15.1, and within 10-fold of the interaction between P5.1 and the remainder of the RNase P RNA molecule.
FIGURE 3.
Measurement of RNA–RNA binding. The increase in anisotropy of 5′ fluorescently labeled P5.1 RNA was measured as a function of the concentration of cognate RNAs. (A) Binding of P5.1 to Δ5.1 mutant RNase P RNA. (B) Binding of P5.1 RNA to its isolated cognate stem–loop, P15.1. (C) Binding of P5.1 RNA to aptamer 7C.
Information content and RNA frequency
The evolution of the RNA World is posited to proceed from a collection of random molecules that collectively contain little or no information. Information (reduction in uncertainty) results from the selection and propagation of a limited number of species from the ensemble of possible molecules (Schneider et al. 1986). The amount of information available in a single selected molecule is a function of its chain length. Thus, modeling the RNA World must account for both the generation of random RNA assemblages and for these RNAs being long enough to contain sufficient information for function. The latter property has been termed the functional information of a set of molecular solutions to a biological problem (Szostak 2003).
Fluctuation experiments allow an explicit estimate of the frequency at which redundant solutions exist in random populations (Luria and Delbrück 1943). We have applied this technique to a non-Watson-Crick RNA–RNA binding problem that is inherent in models of recombination in the RNA World and conclude that these RNAs occur at a frequency of 10−6 for a collection of 20-mers. This estimate does not account for the possible loss of winning species during PCR amplification or other steps of SELEX nor for those RNAs in the library that could bind P5.1 by Watson-Crick interaction, e.g., kissing loop formation, which were selected against in these experiments. Thus, we have determined a minimum estimate of interaction frequency in this model of an RNA World.
Previous combinatorial selection experiments have found novel functional RNA motifs that are not observed in contemporary RNAs, although there has been no explicit estimation of their frequency. Self-cleaving RNA species are commonly found in early stages of SELEX (Tang and Breaker 2000; Saleh-Ashtiani and Szostak 2001), and novel motifs capable of binding the GNRA tetraloop motif can be selected in vitro (Costa and Michel 1997). In contrast to comparative analysis of contemporary molecules, selection experiments admit solutions that are not found in extant biomolecules (Costa and Michel 1997); such is the case here.
Implications for RNA evolution
The presence of so many energetically equivalent, but relatively weak, RNA–RNA interactions in random populations makes chemical sense. Charge repulsion causes interaction between RNAs of limited complementarity to be weak even under optimal solution conditions (Benner 2004). For example, the KM for a pre-tRNA with RNase P RNA under conditions similar to those used presently is only 0.4 μM, indicating much weaker interaction than that of a protein enzyme for an RNA substrate. The KD for RNA–RNA interaction of P5.1 RNA with its cognate site on RNase P RNA has been measured by three independent means to be 0.6–0.9 μM (Kim et al. 2001; Fig. 3 ▶). Affinities of RNA-recognizing aptamers selected from an extensive library of 50-mers were similarly low (Cho et al. 1997). Apparently, there is a maximal strength allowed for RNA–RNA binding in the absence of facilitating proteins or extensive Watson-Crick base pairing. Under the solvent conditions used here, that limit is for KD values in the high nanomolar range.
On the other hand weak binding is advantageous for selection. Both calculation and experiment demonstrate that many more weakly interacting species than strong ones are available in a random mixture of potential ligands (Lancet et al. 1990). Thus, we suggest that models of pre-biotic evolution that allow many possible molecular solutions at early stages and the subsequent refinement of these solutions by mutation can account for many of the seeming anomalies in the abiogenic generation of complex structures. Weak binding increases the number of potential interacting species because so many energetically equivalent interactions exist. In other words, the fitness landscape for RNA–RNA interaction does not appear to be very rugged, having 106 equivalent peaks in a space of 1012 possible positions.
The large number of potential interacting RNA species seen in these and other selection experiments implies that contemporary RNA–RNA interactions represent only a limited subset of all possible interactions. This has implications for the evolution of an RNA World as a predecessor to biological systems. Because so many interactions are possible, the more limited repertoire of interacting domains seen in contemporary RNAs likely results from fixation of some molecular “solutions” from a large number of possibilities. This means that the interactions found in contemporary remnants of the RNA World may be evolutionarily contingent, i.e., the result of historical accident.
MATERIALS AND METHODS
Synthetic RNA libraries
Each of the eight synthetic DNA libraries (Fig. 1B ▶; IDT Technologies) contained 20 randomized nucleotides bounded by the T7 promoter sequences at the 3′ end and a unique primer at the 5′ end; primer sequences for each library are given in Table 1 ▶. The single-stranded DNAs were diluted so that each library contained 106 DNAs. The DNAs were amplified by PCR (95°C, 2 min; 25–40 cycles of 95°C, 30 sec; 51°C, 30 sec; 59°C, 1 min; 5 min, final extension at 59°C) using 3 U Taq DNA polymerase (Promega) in 50 μL containing 20 mM Tris-HCl, pH 9.0, 100 mM KCl, 3 mM MgCl2, 0.2% Triton X-100, 400 μM each dATP, dCTP, dGTP, and TTP, 0.8 μM of each primer. One microgram of each amplified low-complexity library was transcribed separately with T7 RNA polymerase (Epicentre). The transcripts were purified by denaturing polyacrylamide gel electrophoresis. The purified libraries in 0.1 mL 10 mM Tris-HCl, pH 8.0, 100 mM NaCl, were heated to 70°C for 10 min and slowly cooled to room temperature at which time the solution was made 25 mM in MgCl2.
Selex
Synthetic P5.1 stem–loop (1.45 microgram, Dharmacon) was dissolved in 100 μL of 0.1 M K-phosphate, pH 8.0. Then 50 μL of freshly prepared, ice-cold 20 mM NaIO4 were added and the solution kept on ice in the dark for 2 h. RNA was recovered by ethanol precipitation and redissolved in 0.1 mL of 0.1 M K-acetate, pH 5.0. The oxidized RNA was coupled to 0.5 mL of packed Sepharose-adipic acid hydrazide resin (Pharmacia) at 4°C overnight with gentle mixing.
Blocking deoxyoligonucleotides (5′-CTGAGATGCC-3′), (5′-ACGGTGCTGA-3′), and (5′-AGATGCCCGT-3′), each at a concentration 0.1× the library RNA concentration, were added to each library to occlude RNAs that could bind with P5.1 via Watson-Crick binding. The mixture was kept at room temperature for 1 h before selection. Each RNA library was pre-selected through a column (10 μL bed volume) of Sepharose-adipic acid hydrazide in P buffer (10 mM Tris-Cl, pH 8.0/100 mM NaCl/25 mM MgCl2) and then applied to a column (10 μL bed volume) of RNA-coupled resin. The affinity column was washed with 5 mL of P buffer and then the bound RNA population was eluted with two column volumes of 25 mM Na-EDTA, pH 8.0. The RNA was recovered by ethanol precipitation using 200 μg glycogen as carrier, reverse-transcribed (Superscript I, Bethesda Research Labs) and amplified by PCR. Amplified cDNAs after SELEX were digested with EcoR1 and BamH1 and ligated into pGEM–3Zf(+) vector (Promega) and transformed into MAX Efficiency DH5α competent cells (Invitrogen). DNA was prepared by QIAprep Miniprep procedures (Qiagen). Sequences of the inserted DNA were determined (MWG Biotech). Sequences were regarded as siblings and only counted once if 18 or more of the selected residues were identical.
Secondary structures were predicted by Mfold (Zuker 2003) and tested by partial nuclease digestion (Ambion). Data are given in the Supplemental Material (http://www.biochem.missouri.edu/schmidtf/Striggles2006supp.pdf).
RNA–RNA binding
Selected RNAs were transcribed from PCR-amplified DNA with T7 RNA polymerase. RNAs were diluted in binding buffer (50 mM Tris-acetate, pH 8.0, 0.1 mM EDTA, 50 mM Mg-acetate, 800 mM NH4Cl ), heated to 87°C and slowly cooled to room temperature. 5′-fluorescein-P5.1 stem–loop RNA (Dharmacon) was added to 200 μM. Fluorescence anisotropies at 320 nm were determined using L-geometry at 25°C (Lakowicz 1999). The increase in anisotropy of fluorescently labeled P5.1 as a function of aptamer RNA, P15.1 RNA, or Δ5.1 RNA concentration was fitted to a single-binding isotherm using Kaleidagraph (Synergy Software).
Acknowledgments
We thank the members of the MU Combinatorial Biotechnology group for helpful discussions. This work was supported in part by the NASA Exobiology program and the University of Missouri Research Board.
Data deposition: Aptamers except 5F (which is too short for acceptance) are deposited in GenBank, accession numbers DQ120097–DQ120103.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2204506.
REFERENCES
- Benner, S.A. 2004. Understanding nucleic acids through synthetic chemistry. Acc. Chem. Res. 37: 784–797. [DOI] [PubMed] [Google Scholar]
- Burke, D.H. and Willis, J.H. 1998. Recombination, RNA evolution, and bifunctional RNA molecules isolated through chimeric SELEX. RNA 4: 1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers, J.M., Oestreich, J.C., Davis, J.H., and Szostak, J.W. 2004. Informational complexity and functional activity of RNA structures. J. Am. Chem. Soc. 126: 5130–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J-L., Nolan, J.M., Harris, M.E., and Pace, N.R. 1998. Comparative photo-crosslinking analysis of the tertiary structures of Escherichia coli and Bacillus subtilis RNase P RNAs. EMBO J. 17: 1414–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverin, A.B., Chetverina, H.V., Demidenko, A.A., and Ugarov, V.I. 1997. Nonhomologous RNA recombination in a cell-free system: Evidence for a transesterification mechanism guided by secondary structure. Cell 88: 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, B., Taylor, D.C., Nicholas Jr., H.B., and Schmidt, F.J. 1997. Interacting RNA species identified by combinatorial selection. Bioorg. Med. Chem. 5: 1107–1113. [DOI] [PubMed] [Google Scholar]
- Costa, M. and Michel, F. 1997. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J. 16: 3289–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darr, S.C., Zito, K., Smith, D., and Pace, N.R. 1992. Contributions of phylogenetically variable structural elements to the function of the ribozyme ribonuclease P. Biochemistry 31: 328–333. [DOI] [PubMed] [Google Scholar]
- Ellington, A.D. and Szostak, J.W. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. [DOI] [PubMed] [Google Scholar]
- Flory, P.J. 1936. Molecular size distribution in linear condensation polymers. J. Am. Chem. Soc. 58: 1877–1885. [Google Scholar]
- Gesteland, R.F., Cech, T.R., and Atkins, J.F., eds. 1999. The RNA World. 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- Gevertz, J., Gan, H.K., and Schlick, T. 2005. In vitro RNA pools are not structurally diverse: A computational analysis. RNA 11: 853–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes, G.G. and Schimmel, P.R. 1970. Rapid reactions and transient states. In The Enzymes, 3rd ed., v. II. (ed. P.D. Boyer), pp. 67–114. Academic Press, New York.
- Herschlag, D. and Cech, T.R. 1990. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry 29: 10159–10171. [DOI] [PubMed] [Google Scholar]
- Huang, W. and Ferris, J.P. 2003. Synthesis of 35–40 mers of RNA oligomers from unblocked monomers. A simple approach to the RNA World. Chem. Commun. 2003: 1458–1459. [DOI] [PubMed] [Google Scholar]
- Ikawa, Y., Shiraishi, H., and Inoue, T. 1999. Trans-activation of the Tetrahymena group I intron ribozyme via a non-native RNA–RNA interaction. Nucleic Acids Res. 27: 1650–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev, A.V., Krivenko, A.A., Harrington, D.J., Holbrook, S.R., Adams, P.D., and Pace, N.R. 2005. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl. Acad. Sci. 102: 13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H., Poelling, R.R., Leeper, T.C., Meyer, M.A., and Schmidt, F.J. 2001. In vitro transactivation of Bacillus subtilis RNase P RNA. FEBS Lett. 506: 235–238. [DOI] [PubMed] [Google Scholar]
- Knight, R. and Yarus, M. 2003. Finding specific RNA motifs: Function in a zeptomole world? RNA 9: 218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz, J.R. 1999. Principles of fluorescence spectroscopy, 2nd ed. Kluwer Academic/Plenum, New York.
- Lancet, D., Sadovsky, E., and Seidemann, E. 1990. Probability model for molecular recognition in biological receptor repertoires: Significance to the olfactory system. Proc. Natl. Acad. Sci. 90: 3715–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, M.S. and Bartel, D.P. 2003. Processivity of ribozyme-catalyzed RNA polymerization. Biochemistry 42: 8748–8755. [DOI] [PubMed] [Google Scholar]
- Leeper, T.C., Martin, M.B., Kim, H., Cox, S., Semenchenko, V., Schmidt, F.J., and Van Doren, S.R. 2002. Structure of the UGAGAU hexaloop that braces Bacillus RNase P for action. Nat. Struct. Biol. 9: 397–403. [DOI] [PubMed] [Google Scholar]
- Luria, S.E., and Delbrück, M. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massire, C., Jaeger, L., and Westhof, E. 1998. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 279: 773–793. [DOI] [PubMed] [Google Scholar]
- Penedo, J.C., Wilson, T.J., Jayasena, S.D., Khvorova, A., and Lilley, D.M.J. 2004. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA 10: 880–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, C.A. and Lehman, N. 2003. Generalized RNA-directed recombination of RNA. Chem. Biol. 10: 1233–1243. [DOI] [PubMed] [Google Scholar]
- Sabeti, P.C., Unrau, P.J., and Bartel, D.P. 1997. Accessing rare activities from random RNA sequences: The importance of the length of molecules in the starting pool. Chem. Biol. 4: 767–774. [DOI] [PubMed] [Google Scholar]
- Salehi-Ashtiani, K. and Szostak, J.W. 2001. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414: 82–84. [DOI] [PubMed] [Google Scholar]
- Schmidt, F.J. 1999. Ribozymes—why so many, why so few? Mol. Cells 9: 459–463. [PubMed] [Google Scholar]
- Schneider, T.D., Stormo, G.D., Gold, L., and Ehrenfeucht, A.I. 1986. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 188: 415–431. [DOI] [PubMed] [Google Scholar]
- Soukup, G.A. and Maher III, L.J. 1998. Selection and characterization of RNAs that relieve transcriptional interference in Escherichia coli. Nucleic Acids Res. 26: 2715–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak, J.W. 2003. Functional information: Molecular messages. Nature 423: 689. [DOI] [PubMed] [Google Scholar]
- Tang, J. and Breaker, R.R. 2000. Structural diversity of self-cleaving ribozymes. Proc. Natl Acad. Sci. 97: 5784–5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Larios, A., Swinger, K.K., Krasilnikov, A.S., Pan, T., and Mondragon, A. 2005. Crystal structure of the RNA component of bacterial ribonuclease P. Nature 437: 584–587. [DOI] [PubMed] [Google Scholar]
- Tuerk, C. and Gold, L. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510. [DOI] [PubMed] [Google Scholar]
- Westhof, E. and Massire, C. 2004. Evolution of RNA architecture. Science 305: 62–63. [DOI] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]