Abstract
Metazoan replication-dependent histone mRNAs are not polyadenylated, and instead terminate in a conserved stem–loop structure generated by an endonucleolytic cleavage involving the U7 snRNP, which interacts with histone pre-mRNAs through base-pairing between U7 snRNA and a purine-rich sequence in the pre-mRNA located downstream of the cleavage site. Here we generate null mutations of the single Drosophila U7 gene and demonstrate that U7 snRNA is required in vivo for processing all replication-associated histone pre-mRNAs. Mutation of U7 results in the production of poly A+ histone mRNA in both proliferating and endocycling cells because of read-through to cryptic polyadenylation sites found downstream of each Drosophila histone gene. A similar molecular phenotype also results from mutation of Slbp, which encodes the protein that binds the histone mRNA 3′ stem–loop. U7 null mutants develop into sterile males and females, and these females display defects during oogenesis similar to germ line clones of Slbp null cells. In contrast to U7 mutants, Slbp null mutations cause lethality. This may reflect a later onset of the histone pre-mRNA processing defect in U7 mutants compared to Slbp mutants, due to maternal stores of U7 snRNA. A double mutant combination of a viable, hypomorphic Slbp allele and a viable U7 null allele is lethal, and these double mutants express polyadenylated histone mRNAs earlier in development than either single mutant. These data suggest that SLBP and U7 snRNP cooperate in the production of histone mRNA in vivo, and that disruption of histone pre-mRNA processing is detrimental to development.
Keywords: U7, SLBP, histone, Drosophila, mRNA processing, oogenesis
INTRODUCTION
Chromosome duplication during the cell cycle requires the production of histones during S phase to package newly replicated DNA into chromatin. Bulk histone production during S phase is achieved through the biosynthesis of replication-dependent histone mRNAs, which are cell-cycle regulated and accumulate only in S phase. In animal cells these histone mRNAs are unique: The 3′ end terminates in a conserved 26-nt sequence that forms a stem–loop rather than in a poly A+ tail (Marzluff 2005). As histone genes lack introns, the only processing step required for mature histone mRNA production is endonucleolytic cleavage of the pre-mRNA to form the 3′ end of the mRNA (Dominski and Marzluff 1999). Much of the cell-cycle regulation of histone mRNAs is post-transcriptional and is mediated by the 3′ end of the mRNA (Luscher et al. 1985; Harris et al. 1991; Zheng et al. 2003). Thus, a complete understanding of cell-cycle-regulated histone mRNA production requires a full understanding of the factors required for histone pre-mRNA processing.
The processing of histone pre-mRNAs requires two cis elements and a number of trans-acting factors. The cis elements are the stem–loop at the 3′ end of histone mRNA and a purine-rich region downstream of the cleavage site, termed the histone downstream element (HDE). A protein called stem–loop binding protein (SLBP) (Wang et al. 1996) or hairpin binding protein (HBP) (Martin et al. 1997) specifically binds the 3′ end of histone mRNA. SLBP is required for histone pre-mRNA processing in vivo (Sullivan et al. 2001; Kodama et al. 2002; Pettitt et al. 2002) and accompanies the mRNA to the cytoplasm (Erkmann et al. 2005), where it promotes the translation of the histone mRNA (Sanchez and Marzluff 2002; Whitfield et al. 2004). The HDE binds U7 snRNP by base-pairing with the 5′ end of U7 snRNA (Schaufele et al. 1986; Mowry and Steitz 1987; Cotten et al. 1988; Soldati and Schumperli 1988). In mammals, SLBP, the U7 snRNP, and a U7 snRNP-associated zinc finger protein called ZFP100 (Dominski et al. 2002) cooperate to recruit an endonuclease complex that cleaves the pre-mRNA. Recent evidence indicates that CPSF73, a component of the complex that mediates AAUAAA-directed cleavage prior to polyadenylation, is the likely endonuclease (Dominski et al. 2005a; Kolev and Steitz 2005). This revealed some unexpected overlap in the machinery carrying out histone pre-mRNA processing and canonical polyadenylation.
The U7 snRNA is a small RNA (55–70 nt) that, like the spliceosomal snRNAs, contains both a trimethyl guanosine cap and an Sm binding site, which is essential for its function (Grimm et al. 1993; Schumperli and Pillai 2004). The Sm site in these snRNAs stably binds a complex of seven related proteins of the LSm/Sm family to form the core snRNP particle. Proteins of the LSm/Sm family share a common tertiary structure called the Sm fold that assembles into hexameric or heptameric rings capable of binding single-stranded RNA. The U snRNPs contain a heptameric Sm ring, with each of the seven individual subunits making a specific contact with a residue in the Sm binding site of the snRNA (Khusial et al. 2005). The heptameric Sm ring of spliceosomal snRNPs contains the proteins SmB/B′, SmD1, SmD2, SmD3, SmE, SmF, and SmG. In contrast, the U7 snRNP contains five of these Sm proteins (B/B1, D3, E, F, G) and two novel Sm proteins called LSm10 and LSm11 that replace SmD1 and SmD2 of the spliceosomal snRNPs (Pillai et al. 2001, 2003). The Sm site found in U7 snRNAs is distinct from the Sm site in spliceosomal snRNAs and is responsible for incorporation of LSm10 and LSm11 into the U7 snRNP (Schumperli and Pillai 2004; Azzouz et al. 2005). In addition to the Sm fold that participates in ring formation, LSm11 contains an NH2 terminal extension that makes contacts with ZFP100 and possibly other components of the histone pre-mRNA processing machinery (Azzouz et al. 2005; E. Wagner and W.F. Marzluff, unpubl.).
The role of U7 snRNP in histone pre-mRNA processing has been examined primarily in nuclear extract systems that support the processing of synthetic histone pre-mRNAs, and by monitoring the processing of histone pre-mRNAs injected into Xenopus ooctyes (for review, see Dominski and Marzluff 1999). Complementary mutations in U7 snRNA and the HDE provided early evidence that base-pairing between the 5′ end of U7 and the HDE was an important part of U7 snRNP function (Schaufele et al. 1986; Bond et al. 1991). Furthermore, blocking the 5′ end of the U7 snRNA with a complementary oligonucleotide specifically inhibits processing of synthetic histone pre-mRNAs in nuclear extracts (Cotten et al. 1991; Dominski et al. 2003). However, the contribution of U7 snRNA to endogenous histone mRNA biosynthesis and whether this contribution is important for animal development have not been examined. To explore these issues, we have generated and characterized U7 snRNA mutations in Drosophila.
Drosophila provides an excellent model for examining the complexities of histone mRNA biosynthesis in a multicellular organism (Sullivan et al. 2001; Lanzotti et al. 2002, 2004a, Lanzotti et al. b; Marzluff and Duronio 2002). Drosophila has a single set of replication-dependent histone genes, which are present in ~100 tandem repeats of a 4.5-kB unit containing one copy of each of the five histone mRNAs (Lifton et al. 1978). This gene cluster is subject to multiple mechanisms of regulation that provide the histone proteins for the different types of cell cycles that occur during Drosophila development, including the early syncytial cell cycles that lack gap phases, the endocycles of polyploid tissues, and the canonical cycles of proliferating diploid cells in the CNS and imaginal discs.
Drosophila SLBP (Sullivan et al. 2001), U7 snRNA (Dominski et al. 2003), and U7 snRNP specific proteins Lsm10 and Lsm11 (Azzouz and Schumperli 2003), have all been identified, and we have begun to characterize them genetically. Mutations in the Drosophila Slbp gene block normal histone pre-mRNA processing during embryonic development and result in production of polyadenylated histone mRNAs as a consequence of read-through past the normal processing site (Sullivan et al. 2001; Lanzotti et al. 2002). This occurs because each of the five Drosophila histone genes contains cryptic polyadenylation sites downstream of the HDE that are utilized in the absence of SLBP (Lanzotti et al. 2002). Null mutations of Slbp cause lethality during larval and pupal stages, presumably because of the histone processing defects, although the precise cause of lethality is not known. Slbp mutant cells are capable of replicating chromatin, likely because the inappropriate polyadenylated mRNAs are translated. A hypomorphic Slbp mutant allele that produces reduced amounts of SLBP protein results in the production of both normal and poly A+ histone mRNAs during embryogenesis, but does not cause lethality. However, these viable mutant females lay eggs that contain reduced amounts of histone mRNA and protein and do not develop (Sullivan et al. 2001; Lanzotti et al. 2002). Thus, SLBP is required during both zygotic development and oogenesis.
Here we report the study of mutations in the U7 snRNA gene, and compare the resulting phenotypes with those caused by mutation of Slbp. Our results indicate that U7 snRNA is required for normal histone mRNA biosynthesis during Drosophila development and that, like Slbp mutations, loss of U7 snRNA results in the production of polyadenylated histone mRNAs. However, unlike Slbp null mutants, U7 null mutants are viable, but both males and females are sterile. This difference in terminal phenotype is most likely because the maternal supply of U7 snRNA delays the onset of the histone processing defect in U7 mutants relative to Slbp mutants, which do not have a significant maternal supply of SLBP protein. Both U7 and SLBP are required for normal histone mRNA biosynthesis in the female germ line, and mutation of either gene disrupts oogenesis. These data indicate that loss of SLBP and U7 cause similar molecular phenotypes in Drosophila and suggest that early expression of this molecular phenotype prevents normal development.
RESULTS
U7 snRNA null mutants are viable but sterile
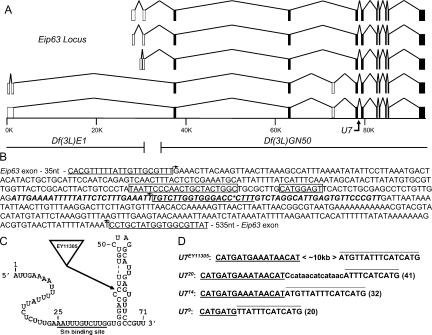
To examine the contribution of U7 snRNA to histone mRNA production during animal development, we generated null alleles in the Drosophila U7 gene. The U7 locus is located within an intron of the differentially spliced Eip63E gene (Dominski et al. 2003), which encodes a CDK-like Ser/Thr kinase that is induced in response to the fly hormone ecdysone (Stowers et al. 2000; Rascle et al. 2003; Fig. 1A ▶). We obtained from the Drosophila Gene Disruption Project (Bellen et al. 2004) a stock carrying a P element transposon (EY11305) that inserted 8 nt downstream of the Sm protein binding site between nucleotides 43 and 44 of the 71-nt U7 snRNA (Fig. 1B,C ▶). U7EY11305 homozygous animals develop into adult flies with no overt morphological defects. However, both males and females of this genotype are completely sterile. Precise excision of the EY11305 P element restored male and female fertility, indicating that the U7EY11305 homozygous adult sterility was caused by the P element insertion.
FIGURE 1.
Mutation of the Drosophila U7 gene. (A) Schematic of the Eip63 locus. The five identified Eip63 mRNAs species are shown, with open boxes indicating variably spliced exons and closed boxes indicating invariant exons. The scale bar indicates genomic DNA in kilobase pairs. The U7 gene is located at position ~79K. The right breakpoint of Df(3L)E1 and the left breakpoint of Df(3L)GN50, located at position ~31K and ~34K, respectively, are shown below the scale bar. (B) Sequence of the genomic fragment used to rescue the U7 mutant phenotype. The U7 snRNA transcript is indicated in bold italic. The asterisk indicates the location of the ~10-kb EY11305 P element. The large and the small boxes, respectively, indicate the PSEA and PSEB essential snRNA promoter elements (Zamrod et al. 1993). The bent arrows indicate primers used to analyze the excision mutations. Note that the middle and top primers were able to detect a product in U723 homozygous mutant animals. (C) Putative U7 snRNA secondary structure indicating the location of the EY11305 insertion. The underlined sequence indicates the Sm protein binding site. (D) Molecular characterization of U7 imprecise excision mutations. Each sequence shown (total length in parentheses) is located at the EY11305 insertion site indicated in panels B and C. The underlined and overlined sequence is derived from the 5′ and 3′ end, respectively, of the P element inverted terminal repeat (ITR). The lowercase sequence in U720 is not derived from the ITR, and its origin is unknown.
We generated five additional U7 alleles by genetically mobilizing the EY11305 P element (see Materials and Methods). Three of these (U79, U714, and U720) contain an insertion ranging from 20 to 41 nt, including sequence from both the 5′ and 3′ ends of the P element (Fig. 1D ▶). The insertions are located downstream of the Sm site and after the first 6 nt of the terminal stem. The other two alleles (U71 and U723) retain a larger portion of the P element that we did not precisely determine. These events result from imprecise repair of the double-strand break induced by transposase during P-element excision (Adams et al. 2003). Like the original U7EY11305 mutation, each of the five imprecise excision mutations is viable when homozygous but causes complete male and female sterility.
To test whether these mutations specifically disrupt U7 and not Eip63 function, we performed genetic complementation analyses with various deletions of the Eip63 locus. Df(3L)E1 deletes the 5′ third of Eip63 but not U7, and Df(3L)GN50 deletes the entire U7 gene and the 3′ two-thirds of Eip63 (Fig. 1A ▶). Df(3L)E1/Df(3L) GN50 heterozygotes are lethal because of the loss of Eip63 function (Stowers et al. 2000). As expected if our mutations do not affect Eip63, each of the six U7 mutant alleles complements the Df(3L)E1 lethality, and the resulting adult male and female flies are fertile. In contrast, all six mutations result in viable, sterile flies when present in trans to Df(3L)GN50 because it removes U7. In addition, a P element transgene containing only the U7 gene (Fig. 1B ▶) rescues the sterility of U714/U7EY11305 mutant females and males. We conclude from these data that all of our mutant alleles affect U7 function without affecting Eip63, consistent with the U7 snRNA being derived from an independent transcript distinct from that producing Eip63E mRNA.
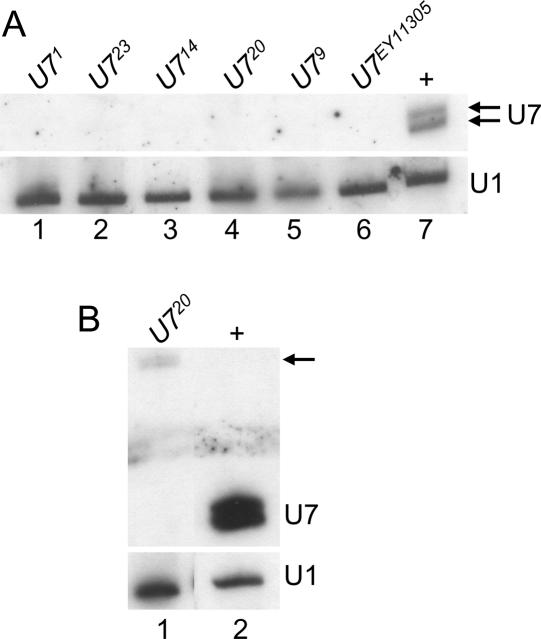
To determine whether the U7 mutants represent null alleles, total RNA was extracted from 1- to 2-d-old adult female flies and subjected to Northern blot analysis. In wild-type female flies, the U7 probe detects a characteristic doublet representing RNAs of 71 and 74 nt that result from differential processing of the 3′ end of U7 snRNA (Dominski et al. 2003; Fig. 2A ▶, lane 7). None of the mutant samples contained detectable levels of either wild-type U7 snRNA (Fig. 2A ▶, lanes 1–6). Interestingly, the U720 allele produced very low levels of an RNA that hybridizes with the U7 probe, but that is larger in size, consistent with the 41-nt insertion (Fig. 1D ▶; arrow in Fig. 2B ▶, lane 1). Based on the lack of production of wild-type U7 snRNA, the sterility phenotype, and the histone mRNA molecular phenotypes described below, we conclude that all of these alleles are null. The short insertions in U79, U714, and U720 are in a region of the U7 snRNA that does not have a defined function, and they likely affect the assembly and/or stability of the U7 snRNP.
FIGURE 2.
Northern analysis of U7 mutants. (A) (Top) Total RNA extracted from 1- to 2-d-old adult females was fractionated through an 8% polyacrylamide-7 M urea gel, transferred to nitrocellulose, and probed with 32P-labeled U7 RNA. (Lanes 1–6) RNA from the indicated homozygous mutant genotypes. (Lane 7) w1118 control RNA. Note the lack of detectable U7 snRNA in the mutant samples. (Bottom) The same blot hybridized with a 32P-labeled U1 probe as a loading control. (B) Long exposure of mutant samples. The arrow indicates the low abundance, aberrantly large mutant U7 snRNAs produced by the U720 excision mutant.
U7 snRNA is essential for histone pre-mRNA processing in Drosophila
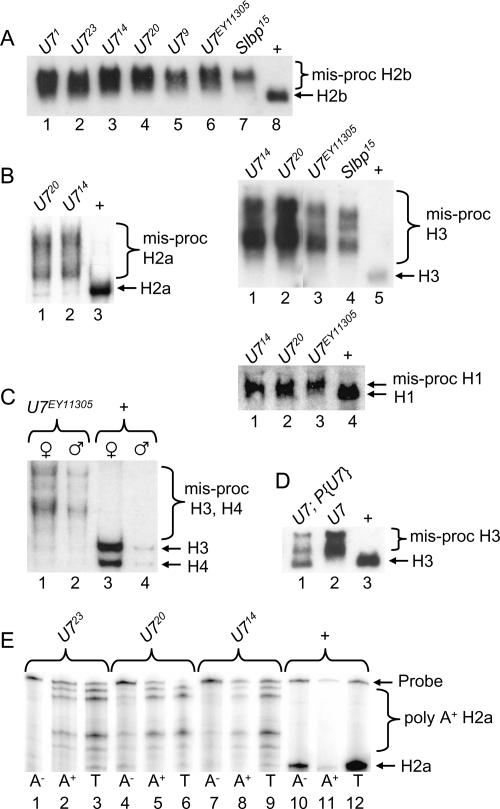
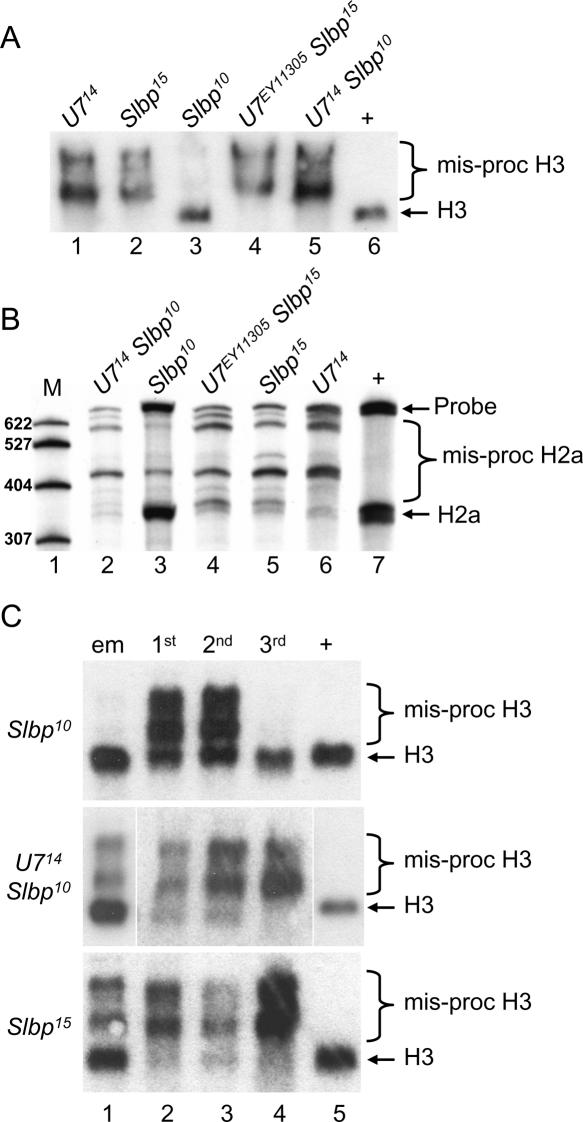
To determine how the loss of U7 snRNA affects the biosynthesis of histone mRNA, total RNA isolated from wild-type or mutant adult female flies was subjected to Northern blot analysis. A probe derived from the histone H2b gene hybridizes with a single mRNA in wild-type control RNA samples (Fig. 3A ▶, lane 8). In contrast, H2b mRNAs that migrate more slowly are detected in all of the U7 mutants (Fig. 3A ▶, lanes 1– 6). An identical phenotype is observed in Slbp15 null mutant larvae (Lanzotti et al. 2002; Fig. 3A ▶, lane 7), indicating that the longer mRNAs in the U7 mutants likely represent inappropriately polyadenylated histone mRNAs. Each of the U7 mutants produces a similar pattern of aberrant H2b mRNAs (Fig. 3A ▶), suggesting that they all disrupt histone pre-mRNA processing to the same extent. Longer, aberrantly processed mRNAs are also detected in U7 mutants with probes derived from each of the other four replication-associated histone genes (Fig. 3B,C ▶). Aberrantly processed histone mRNA is detected in U7 mutant males as well as females, consistent with the sterility of both genders (Fig. 3C ▶). Proper histone H3 processing is partially restored in U7 mutant flies containing a P element transgene harboring the wild-type U7 gene (Fig. 3D ▶, lane 1), and these flies are no longer sterile. The incomplete restoration of H3 processing is likely because the transgene does not restore wild-type levels of U7 snRNA (see Fig. 4C ▶, lanes 8–10). These data indicate that the U7 locus we have identified is required for processing all five of the replication-dependent histone mRNAs and thus likely represents the only U7 snRNA encoded in the Drosophila genome.
FIGURE 3.
Aberrant histone mRNA production in U7 mutants. Total RNA extracted from 1- to 2-d-old adult females was fractionated through agarose gels, transferred to nitrocellulose, and probed with 32P-labeled histone probes. (A) H2b probe. (Lanes 1–7) equal amounts of RNA from the indicated homozygous mutant genotypes. (Lane 8) WT control RNA. Since Slbp15 homozygotes are lethal at the pupal stage, Slbp15 mutant RNA samples were isolated from homozygous wandering third instar larvae identified as Tb+ progeny from Slbp15/TM6b Tb parents. (B) (Left) H2a probe. (Lanes 1,2) RNA from the indicated homozygous mutant genotypes. (Lane 3) WT control RNA. (Top right) H3 probe. (Lanes 1–4) RNA from the indicated homozygous mutant genotypes. (Lane 5) WT control RNA. (Bottom right) H1 probe. (Lanes 1–3) RNA from the indicated homozygous mutant genotypes. (Lane 4) WT control RNA. Note that because H1 mRNA is longer than the other histone mRNAs, and because the cryptic polyadenylation site is near the normal processing site (Lanzotti et al. 2002), there is a less pronounced difference in migration between the wild-type and polyadenylated mRNA. (C) A mixture of histone H3 and H4 probes hybridized to RNA isolated from adult males or females of the indicated genotypes. Note that the females contain more histone mRNA than males because of the large amounts of production during oogenesis. (D) Transgenic rescue of H3 processing. (Lane 1) RNA from U714/U7EY11305 adult females carrying a U7 P element transgene. (Lane 2) RNA from U714/U7EY11305 female siblings of those in lane 1. (Lane 3) RNA from w1118 control adult females. (E) H2a S1 nuclease protection of mRNA isolated from 1- to 2-d-old adult females. The RNA was separated into poly A+ and poly A− fractions prior to analysis. Note the multiple protected fragments in the mutant poly A+ RNA samples, representing the use of multiple cryptic polyadenylation signals located downstream of the normal pre-mRNA processing site (Lanzotti et al. 2002).
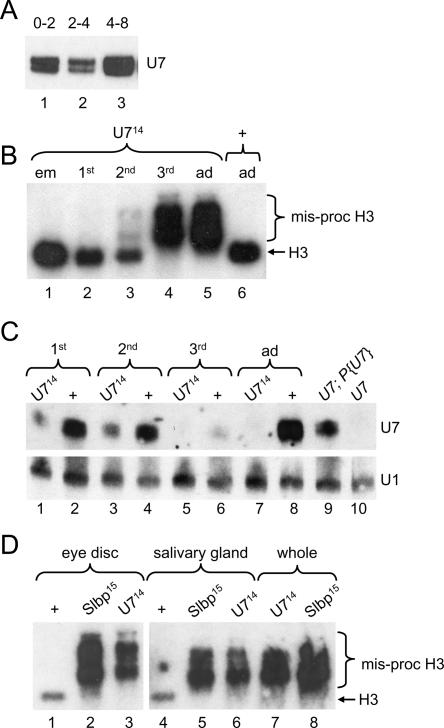
FIGURE 4.
Developmental onset of the U7 mutant phenotype. (A) RNA isolated from w1118 control embryos and subjected to Northern analysis with a 32P-labeled U7 RNA probe. (Lanes 1–3) equal numbers of 0- to 2-, 2- to 4-, and 4- to 8-h-old embryos, respectively. (B) Equal amounts of total RNA extracted from different stages of development and subjected to Northern analysis with a 32P-labeled H3 probe. (Lane 1) Embryos collected overnight from U714/TM3 heterozygous parents and allowed to age for 3 h. (Lanes 2–4) Homozygous first, second, and third instar U714 mutant larvae, respectively. (Lanes 5,6) 1- to 2-d-old homozygous U714 and w1118 control adult females, respectively. Note that the poly A+ H3 mRNA is first detectable in small amounts at the second larval instar stage, and that wild-type H3 mRNA is absent by the third larval instar stage. (C) U7 Northern analysis of RNA isolated from homozygous U714 or w1118 control larvae and adults. (Odd lanes) U714 homozygous mutants collected from U714/TM3 P[act-GFP] heterozygous parents. Note that the detectable U7 snRNA is maternally derived. (Even lanes) w1118 control animals. Hybridization of the blot with a U1 probe was used as a loading control. (D) H3 Northern analysis of RNA isolated from eye imaginal discs (lanes 1–3), salivary glands (lanes 4–6), or whole larvae (lanes 7,8) of the indicated genotypes.
To more precisely assess the contribution of U7 snRNA to histone pre-mRNA processing and to confirm that the aberrantly processed histone mRNAs in U7 mutants were polyadenylated, S1 nuclease protection analysis of H2a mRNA was performed with total RNA isolated from wild-type or U7 mutant females and separated into poly A+ and poly A− fractions. In wild-type flies a single protected fragment was observed that corresponds to the expected 3′ end of histone H2a mRNA (Fig. 3E ▶, lane 12), and that partitions primarily into the poly A− fraction (Fig. 3E ▶, lanes 10,11). In the U7 mutant RNA samples, multiple H2a mRNA protected fragments were detected, almost all of which are longer than the single species detected in wild-type samples (Fig. 3E ▶, lanes 3,6,9). All of the aberrant mRNAs were enriched in the poly A+ fraction (Fig. 3E ▶, cf. lanes 1,4,7 and 2,5,8). These data are consistent with the use of multiple, inefficient polyadenylation signals downstream of the normal site of H2a pre-mRNA processing, as we observed previously in Slbp mutants (Lanzotti et al. 2002). Importantly, very little, if any, normally processed H2a was detected in U7 mutant adult female RNA samples. We conclude from all of the data presented in Figure 3 ▶ that U7 snRNP is an essential histone pre-mRNA processing factor in Drosophila.
Loss of U7 and SLBP affects Drosophila development differently
Because zygotic mutation of Slbp and U7 results in a similar molecular phenotype with respect to histone mRNA biosynthesis, we expected that development of the animal would be similarly affected as well. However, Slbp null mutations cause lethality while U7 null mutations cause adult sterility. This difference in terminal phenotype may reflect differences in the maternal contribution of SLBP and U7. SLBP protein is not contributed maternally (as assessed by Western blotting) (Lanzotti et al. 2002), and zygotic expression of SLBP is required for processing of histone mRNAs transcribed in the early embryo. In contrast, there is a substantial amount of U7 snRNA stored in the egg, and the total amount of U7 snRNA changes relatively little during the first 8 h of development as assayed by Northern blotting (Fig. 4A ▶). This maternal U7 snRNA contribution may be sufficient to support normal histone mRNA synthesis during early development of U7 zygotic mutants, resulting in a delay in onset of the histone pre-mRNA processing defect compared to Slbp mutants. This later onset of the mutant phenotype likely explains the difference in developmental outcome.
To test this possibility we analyzed histone H3 mRNA production at different stages of development in U7 mutant animals (Fig. 4B ▶). Misprocessed (i.e., poly A+) histone H3 mRNA was not detected in U7 mutant embryos by in situ hybridization using a probe derived from sequences downstream of the normal H3 processing site (called H3-ds, for “H3-downstream”) that specifically detects poly A+ histone H3 mRNA in Slbp mutant embryos (data not shown; Lanzotti et al. 2002). Poly A+ H3 mRNA was also not detected in U7 mutant embryos or first instar larvae by Northern blotting (Fig. 4B ▶, lanes 1,2). We first detected aberrantly processed, poly A+ H3 mRNA in second instar U7 mutant larvae, although these larvae contain primarily normal and only small amounts of poly A+ histone mRNAs (Fig. 4B ▶, lane 3). By the wandering third larval instar stage only aberrant, poly A+ H3 mRNA was detected, and thus the molecular phenotype was identical to that observed in U7 mutant adult female flies (Fig. 4B ▶, lanes 4,5).
The gradual onset of misprocessed, poly A+ H3 mRNA during development is consistent with a gradual depletion of maternal U7 snRNA. To test this we analyzed U7 snRNA levels in wild-type and U7 mutant larvae by Northern analysis. U7 snRNA is clearly detectable but reduced relative to wild type in both first and second instar U7 mutant larvae, and is undetectable in third instar mutant larvae (Fig. 4C ▶). Unexpectedly, wild-type third instar larvae had greatly reduced amounts of U7 compared to younger larvae or adult females (Fig. 4C ▶, lane 6), suggesting that there may be transcriptional shutdown or active removal of U7 snRNA at this stage prior to the reaccumulation of U7 snRNA in the adult fly. These data indicate that both maternal stores of U7 as well as zygotic expression contribute to the total level of U7 snRNA in early larvae. The reduced amount of U7 snRNA in first and second instar U7 mutant larvae is sufficient to process all of the H3 pre-mRNA, indicating that U7 snRNA is normally present in excess. Taken together, our results suggest that the U7 mutant animals have a later onset (i.e., not until third instar) of the histone pre-mRNA processing defect than Slbp mutants because of maternal stores of U7 snRNA.
We used Northern analysis of RNA prepared from specific tissues as well as in situ hybridization to characterize the U7 larval mutant phenotypes in more detail, and to compare them to Slbp mutant phenotypes. Misprocessed, poly A+ H3 mRNA was present in RNA isolated from third instar eye imaginal discs and salivary glands dissected from both U7 and Slbp mutants (Fig. 4D ▶). This indicates that U7 snRNP and SLBP are required for histone pre-mRNA processing in both proliferating (imaginal discs) and endocycling (salivary glands) cells.
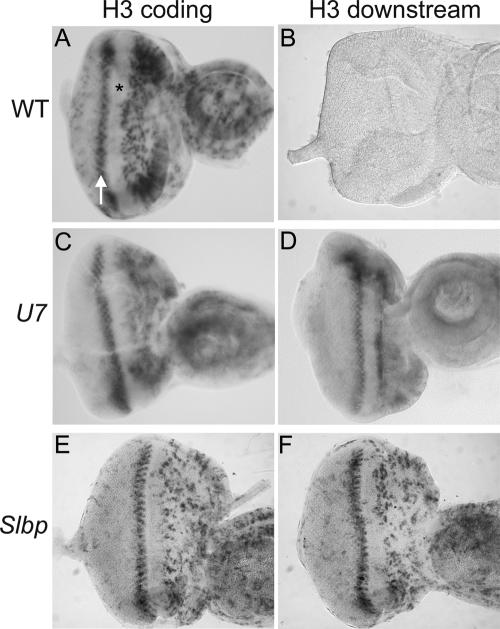
We next examined the accumulation of misprocessed H3 mRNA in the highly regulated cell cycles of the developing eye. During larval stages asynchronous cell proliferation supports eye imaginal discs growth. In late third instar larvae, a wave of differentiation called the morphogenetic furrow (MF) sweeps across the eye imaginal epithelium from posterior to anterior. All cells within the MF arrest in G1 phase (Fig. 5A ▶, asterisk). Certain of these cells remain arrested and begin to differentiate. The remaining cells synchronously enter a final, single cell division cycle called the second mitotic wave, which can be visualized with the H3 coding probe (Fig. 5A ▶, arrow), and then subsequently differentiate. Whereas control eye discs do not stain with the H3-ds probe (Fig. 5B ▶), in situ hybridization of U7 or Slbp mutant third instar eye imaginal discs with the H3-ds probe produced a staining pattern similar to that obtained using a histone H3 coding probe in wild-type discs (Fig. 5C–F ▶). This includes expression in the asynchronously proliferating cells anterior to the MF and the synchronously replicating cells of the second mitotic wave just posterior to the MF. Misprocessed, poly A+ H3 mRNAs were not detected in G1 arrested cells within the MF or in the G1 arrested terminally differentiating cells posterior to the MF. This essentially normal staining pattern suggests that poly A+ H3 mRNAs are regulated properly with respect to accumulation during the mitotic cell cycle, including efficient down-regulation in cells exiting the cell cycle in G1. However, we cannot exclude the possibility that in these cells the poly A+ mRNAs are not as rapidly down-regulated at the end of S phase as wild-type histone mRNAs (Lanzotti et al. 2002).
FIGURE 5.
U7 mutant eye imaginal discs display a wild-type pattern of polyadenylated H3 mRNA. Eye imaginal discs were dissected from w1118 control (A,B), U714 (C,D), or Slbp15 (E,F) mutant larvae and subjected to in situ hybridization with an H3 coding probe (A,C,E) or the H3-ds probe (B,D,F), which detects only misprocessed, poly A+ H3 mRNA. The asterisk indicates the morphogenetic furrow, which contains cells arrested in G1 phase. The arrow indicates S phase of the second mitotic wave. Asynchronously dividing, undifferentiated cells are located anterior of the MF (right of the asterisk), and differentiating cells that have exited the cell cycle and that will make up the adult eye are located posterior to the MF (left of the arrow).
SLBP and U7 snRNA cooperate in histone pre-mRNA processing
Synthetic histone pre-mRNA substrates are efficiently processed in mammalian nuclear extracts. When there is high complementarity between U7 snRNA and the HDE in the synthetic histone pre-mRNA, these extracts will support processing after SLBP depletion as well as the processing of mutant RNAs that cannot bind SLBP (Spycher et al. 1994; Dominski et al. 1999). This suggests that in vivo the U7 snRNP may be capable of supporting processing in the presences of very low amounts of SLBP. To examine this issue, we used the partially functional Slbp10 allele, which produces SLBP protein at ~10% the amount of wild type. As a result, Slbp10 mutant embryos produce a small amount of normally processed histone mRNA in addition to misprocessed mRNA (Lanzotti et al. 2002). In contrast, at the third larval instar stage Slbp10 mutants contain almost entirely normally processed histone H3 and H2a mRNA, as detected by Northern and S1 nuclease protection, respectively (Fig. 6A,B ▶, lane 3). To test whether production of normally processed histone mRNA in Slbp10 mutants was U7 dependent, we engineered U714 Slbp10 double mutants and analyzed histone H3 and H2a mRNA isolated from whole third instar larva, the earliest time during development at which the U7 mutant phenotype is fully expressed (see Fig. 4B ▶). As assayed by Northern blotting, U714 Slbp10 double mutant third instar larvae produced very little if any processed H3 mRNA, similar to U7 singlemutants (Fig. 6A ▶). This demonstrates that the H3 pre-mRNA processing occurring in Slbp10 mutants is U7-dependent. In contrast to the Slbp10 hypomorph, Slbp15 null mutant third instar larvae contain very little if any processed histone H3 mRNA (Fig. 6A ▶, lane 2), and mutating U7 in this background does not alter the phenotype as would be expected (Fig. 6A ▶, lane 4). Very similar results were obtained by analyzing H2a mRNAs in these different genotypes by S1 nuclease protection (Fig. 6B ▶). These data suggest that the U7 snRNP can stimulate histone processing with a reduced amount of SLBP but that in the absence of SLBP U7 snRNP cannot support processing.
FIGURE 6.
U7-Slbp double mutant analysis. RNA isolated from the indicated genotypes was subjected to Northern analysis with a [32P]-labeled H3 probe (A,C) or S1 nuclease protection analysis with a [32P]-labeled H2a probe (B). (A) H3 Northern of third instar larvae. (Lanes 1– 5) RNA from the indicated homozygous single or double mutant genotypes. (Lane 6) w1118 control RNA. (B) H2a S1 nuclease protection analysis of third instar larvae. (Lane 1) [32P]-labeled marker (M) RNAs of the indicated lengths. (Lanes 2–6) RNA from the indicated homozygous single or double mutant genotypes. (Lane 7) w1118 control RNA. (C) (Lane 1) RNA from embryos collected overnight from heterozygous parents. Note that one-quarter of the embryos will be homozygous mutant for the indicated genotypes. (Lanes 2–4) RNA from first, second, and third instar larvae, respectively, homozygous mutant for the indicated genotypes. (Lane 5) Control RNA from w1118 third instar larvae.
Interestingly, the U714 null and Slbp10 hypomorphic mutations display synthetic lethality: Whereas both U7 and Slbp10 single mutant animals develop into adults, the U7 Slbp10 double mutants do not eclose as adults. This result suggests that the severity of the processing defects may be enhanced as a result of limiting both SLBP and U7 snRNA, causing increased misprocessing of histone mRNA earlier in development. Indeed, U714 Slbp10 double mutants produce a greater proportion of misprocessed, poly A+ H3 mRNA at first and second larval instar stages than either of the Slbp10 or U714 single mutants (Figs. 6C ▶ and 4B ▶, respectively). In addition, the H3 mRNA expression profile of U714 Slbp10 double mutants is similar to that of Slbp15 null mutants, which are lethal (Fig. 6C ▶). This is most striking at embryonic stages, where there is a marked increase in poly A+ H3 mRNA in the U714 Slbp10 double mutants relative to either single mutant (lane 1 in Figs. 6C ▶, 4B ▶). Thus, the severity of the histone pre-mRNA processing defect is enhanced as a result of limiting both SLBP and U7 snRNA, and this increase in severity of the histone pre-mRNA processing defect may contribute to the lethality of Slbp10 U714 double mutants.
Histone pre-mRNA processing is necessary for oogenesis
U7 mutant female flies are sterile and lay very few eggs, none of which hatch. These eggs are often smaller than wild type, some have a defective chorion (e.g., fused dorsal appendages), and most are desiccated (not shown). These data suggest that proper histone pre-mRNA processing is required during oogenesis, and we therefore analyzed histone mRNA expression in U7 and Slbp mutant ovaries.
The basic unit of Drosophila oogenesis is the egg chamber, which consists of a cyst of 16 interconnected germ cells surrounded by a single layer of somatic follicle cells (Spradling 1993). Fifteen of the germ cells differentiate into nurse cells, which become highly polyploid, and which synthesize and transport RNA and protein (including histones and histone mRNA) through cytoplasmic bridges into the single developing oocyte. The follicle cells contribute to oogenesis by synthesizing yolk and the eggshell, and by participating in dorsal/ventral and anterior/posterior axis formation. Both the follicle cells and the nurse cells become polyploid via endocycles, which consist of repeated rounds of S phase interrupted by a gap phase with no intervening mitosis (Lilly and Duronio 2005). Histone mRNA accumulation during endocycles correlates with S phase (Sullivan et al. 2001). Because the cells do not replicate in synchrony, this appears as a mosaic in situ hybridization staining pattern in both nurse cells and follicle cells using a histone H3 coding probe (Ambrosio and Schedl 1985; Ruddell and Jacobs-Lorena 1985; Walker and Bownes 1998; Fig. 7A ▶). Late in oogenesis, there is a burst of histone mRNA synthesis in the nurse cells that is not associated with DNA replication (Ambrosio and Schedl 1985; Ruddell and Jacobs-Lorena 1985). These mRNAs are transported into the oocyte and may be used to support early embryonic development (Sullivan et al. 2001; Lanzotti et al. 2002).
FIGURE 7.
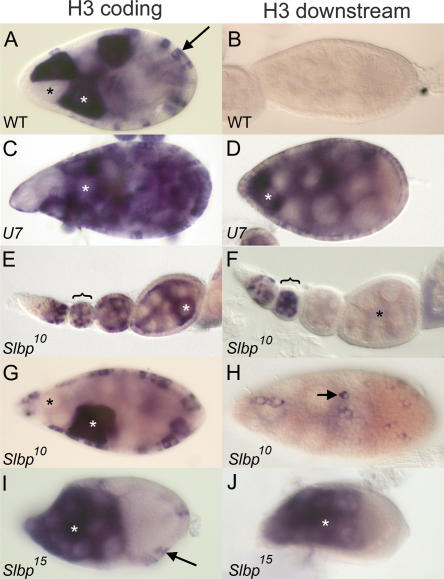
U7 and SLBP are required for histone pre-mRNA processing during oogenesis. (A) w1118 control egg chamber hybridized with a histone H3 coding probe. In this and subsequent panels, white and black asterisks indicate individual nurse cells with and without H3 mRNA, respectively, and arrows indicate follicle cells. (B) w1118 control egg chamber hybridized with an H3-ds probe. Note the lack of staining because H3-ds only detects misprocessed, poly A+ histone H3 mRNA. (C,D) U714 mutant egg chambers hybridized with H3 coding and H3-ds probes, respectively. (E,F) Early stage Slbp10 mutant egg chambers, each from an individual ovariole, hybridized with the H3 coding and H3-ds probes, respectively. The brackets indicate a stage 2 or 3 egg chamber with staining in the nurse cells undergoing endocycles. Note the absence of H3-ds probe in the older egg chambers (asterisk in F). (G) Slbp10 mutant egg chamber stained with the H3 probe indicating production of H3 mRNA in both nurse and follicle cells. (H) Slbp10 mutant egg chamber hybridized with an H3-ds probe, with focus on the follicle cells. The arrow indicates a follicle cell expressing misprocessed, poly A+ histone mRNA. (I,J) Mosaic egg chambers containing Slbp15 mutant germ cells hybridized with a histone H3 coding probe (I) or an H3-ds probe (J). Because this is a mosaic egg chamber, the follicle cells are phenotypically wild-type and do not stain with the H3-ds probe because they express only wild-type H3 mRNA (arrow in I). All egg chambers except E and F stage 9. Anterior at left and posterior at right in all panels.
To examine the cause of U7 mutant sterility, we dissected ovaries from U7 mutant females and examined egg chamber development. U7 mutant egg chambers appear morphologically similar to wild type until stage 10, as assessed by phalloidin and DAPI staining, which detect f-actin and DNA, respectively (not shown). However, few mature oocytes are produced and many egg chambers eventually degenerate, consistent with the lack of egg production. Hybridization of U7 mutant egg chambers with the histone H3 coding probe detects mRNA in a subset of follicle cells and nurse cells, both of which are endocycling at the stages shown (Fig. 7C ▶). Hybridization with the H3-ds probe, which detects only misprocessed, poly A+ H3 mRNA (Fig. 7B ▶), produces a similar staining pattern (Fig. 7D ▶). Thus, U7 snRNA is required for histone pre-mRNA processing in both germ line and somatic tissue. This is consistent with the Northern analysis described above, which showed that essentially all of the histone H3 mRNA produced in U7 mutant females is misprocessed (Fig. 3 ▶).
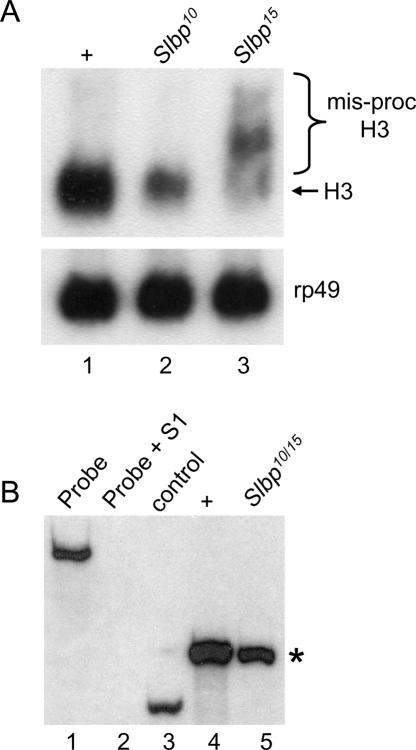
The U7 mutant sterility is reminiscent of that of Slbp10 hypomorphic mutants, which are also viable but female sterile (although Slbp10 males are fertile). In contrast to U7 mutants, Slbp10 mutant females lay normal numbers of eggs that are wild type in appearance. These eggs can be fertilized but do not hatch because of severe mitotic defects during the syncytial cycles (Sullivan et al. 2001). This is likely the result of reduced deposition of maternal histone mRNA (Sullivan et al. 2001; Lanzotti et al. 2002) and protein (P. Fort, W.F. Marzluff, and R.J. Duronio, unpubl.) into these eggs. While reduced ~10-fold in amount relative to wild type, the histone mRNA deposited into eggs by Slbp10 mutant females is processed normally, and we did not detect any poly A+ histone mRNAs (Sullivan et al. 2001; Lanzotti et al. 2002). To test whether this reflected a defect in histone mRNA synthesis during oogenesis, we analyzed RNA isolated from ovaries dissected from Slbp10 mutant females by Northern blotting and S1 nuclease protection. These assays revealed reduced amounts of normally processed H3 and H2a mRNA but no misprocessed H3 or H2a mRNA (Fig. 8A and B ▶, respectively).
FIGURE 8.
SLBP is required for histone pre-mRNA processing during oogenesis. (A) RNA was isolated from whole ovaries dissected from adult females and subjected to Northern analysis with 32Plabeled H3 (top) or rp49 (bottom) coding probes. (Lane 1) w1118 control. (Lane 2) Slbp10 homozygotes. (Lane 3) RNA from ovaries containing mitotic germ line clones of Slbp15 cells. Rp49 serves as a loading control. (B) Detection of H2a mRNA by S1 nuclease protection. (Lane 1) 3′ end-labeled 650-nt H2a probe. (Lane 2) H2a probe incubated with S1 nuclease and nonspecific yeast tRNA. (Lane 3) H2a probe + S1 and a synthetic, partial H2a mRNA that yields a protected 265-nt fragment that is shorter than the 340-nt protected fragment resulting from full-length H2a mRNA (asterisk). (Lane 4) H2a probe + S1 and total RNA isolated from ovaries dissected from wild-type females. Wild-type histone H2a mRNA protects a 340-nt fragment (asterisk). (Lane 5) H2a probe + S1 and total RNA isolated from ovaries dissected from Slbp10/Slbp15 mutant females.
To examine this in more detail, Slbp10 mutant egg chambers were dissected and hybridized with the H3 coding and H3-ds probes. Slbp10 mutant egg chambers are morphologically normal and produce H3 mRNA in both the nurse cells and the follicle cells in patterns similar to wild type (Fig. 7E,G ▶). In contrast, the H3-ds probe stained Slbp10 egg chambers more sporadically and much more weakly (Fig. 7F,H ▶). Strong staining was only usually detected in young (i.e., stages 2–3) egg chambers in nurse cells undergoing endoreduplication cycles (Fig. 7F ▶, bracket). In general, egg chambers older than stages 2–3 did not stain with the H3-ds probe (Fig. 7 ▶, cf. E and F, asterisks), although we could detect sporadic follicle cell staining and sometimes nurse cell staining in stage 9 egg chambers (shown for follicle cells in Fig. 7H ▶). These in situ data indicate that misprocessed H3 mRNA represents a small fraction of the total H3 mRNA that accumulates in Slbp10 mutant ovaries and are consistent with our inability to detect misprocessed H3 mRNA by Northern blot or S1 nuclease analysis. Taken together, our molecular and cytological analyses of Slbp10 mutant ovaries indicate that reduction of SLBP function during oogenesis causes a reduction of histone mRNA biosynthesis, likely because of defects in histone pre-mRNA processing.
To directly test whether the variable H3-ds staining in Slbp10 mutant ovaries reflected reduced but not absent SLBP function, we generated egg chambers containing Slbp15 null mutant germ cells using FLP/FRT-mediated mitotic recombination (see Materials and Methods). In contrast to the Slbp10 mutants, the H3 coding and H3-ds probes stained Slbp15 null mutant germ cells similarly, suggesting that a greater fraction of the H3 mRNA produced was misprocessed (Fig. 7I,J ▶). This result was confirmed by Northern analysis of RNA isolated from Slbp15 mosaic ovaries (Fig. 8A ▶, lane 3). These data indicate that SLBP function is required in the germ line to process histone mRNAs, and that the germ line is not intrinsically incapable of utilizing the cryptic polyadenylation signals downstream of the normal H3 pre-mRNA processing site. Females with Slbp15 mutant germ cells lay very few eggs that do not hatch and that have chorion defects, including fusion of the dorsal appendages, similar to the small number of eggs produced by U7 mutants (not shown). Taken together, our genetic analysis of U7 and SLBP function in ovaries suggests that histone pre-mRNA processing is necessary to complete oogenesis.
DISCUSSION
Probing the function of snRNAs using genetic approaches in metazoans has been difficult, since most of the snRNAs are encoded by multiple genes. Exceptions are the snRNAs in the minor spliceosome and U7 snRNA, each of which are present in a single copy in all organisms whose genome has been sequenced. The U12 snRNA has been disrupted in Drosophila and is essential for proper development (Otake et al. 2002). Here we report the first example of mutating a U7 snRNA gene as part of our studies aimed at understanding the in vivo function of factors involved in histone mRNA metabolism.
The role of U7 and SLBP in histone mRNA metabolism
We find that Drosophila U7 snRNA is essential for normal histone mRNA biosynthesis during development, and that all five replication-associated histone mRNAs are improperly processed in U7 mutant animals. The histone mRNAs produced in U7 mutants are longer than wild-type mRNAs and are polyadenylated, consistent with the use of cryptic downstream polyadenylation signals located within each histone gene. This phenotype is identical to the phenotype we previously described for mutations in Slbp (Sullivan et al. 2001; Lanzotti et al. 2002). The similar poly A+ histone mRNA phenotype in Slbp and U7 mutants suggests that neither the presence of SLBP in U7 mutants nor the presence of U7 snRNP in Slbp mutants impedes the ability of the polyadenylation machinery to utilize histone pre-mRNAs as a substrate. Polyadenylation occurs even though SLBP likely binds to the nascent transcripts in the U7 mutants. In addition, very recent evidence indicates that cleavage of histone pre-mRNA is carried out by some of the components that mediate pre-mRNA cleavage prior to polyadenylation of canonical mRNAs (Dominski et al. 2005a; Kolev and Steitz 2005). How then is polyadenylation of histone mRNAs normally prevented in Drosophila? In vitro studies of Drosophila histone gene transcription have suggested that RNA polymerase II pausing just 3′ of the normal processing site may contribute to efficient histone 3′ end formation (Adamson and Price 2003). The polymerase pauses before the cryptic polyadenylation signals are encountered and hence must resume transcription to allow the production of polyadenylated histone mRNAs. Since we do not see any polyadenylated histone mRNA in wild-type animals or cultured cells, it is possible that the presence of both SLBP and U7 snRNP promote both processing and transcription termination such that the downstream cryptic poly A signals are never encountered.
U7 snRNP likely acts in a catalytic fashion and is recycled after release from the downstream cleavage product, which is degraded in a U7-dependent reaction (Walther et al. 1998; Dominski et al. 2005a). U7 snRNA is present at very low levels in the cell and is largely confined to the Cajal bodies, nuclear organelles described in vertebrates that are responsible for snRNP maturation and function. Cajal bodies are often found in association with histone loci, although in vertebrates U7 snRNA is associated with all the Cajal bodies irrespective of whether they are close to histone loci (Wu and Gall 1993; Frey and Matera 1995; Gall 2003). U7 snRNP is thus normally in excess in the cell, and therefore cells may be able to tolerate a reduction in U7 snRNA levels. In Drosophila cells, the U7 snRNP also localizes to the histone locus (J. Gall, pers. comm.).
Consistent with the idea that U7 is present in excess, our data indicate that normal levels of H3 processing occur in first and second instar larvae that contain reduced amounts of wild-type U7 snRNA. U7 snRNA is then dramatically down-regulated in wild-type wandering third instar larvae, even though replication and histone gene expression are still occurring in many tissues, especially the imaginal discs (e.g., see Fig. 5 ▶). There is then a reaccumulation of U7 snRNA in wild-type adult females, most likely from production during oogenesis. Thus, Drosophila U7 snRNA is present in substantial excess over what is functionally required during much of zygotic development. SLBP may also be capable of acting efficiently in histone pre-mRNA processing in vivo even when it is present in reduced amounts. For instance, the majority of histone H3 and H2a mRNAs is processed in Slbp10 hypomorphic mutant third instar larvae even though the Slbp10 allele produces only 10% of the amount of wild-type SLBP protein (Sullivan et al. 2001). Perhaps a small nuclear pool of SLBP, possibly associated with Cajal bodies (Abbott et al. 1999), accumulates preferentially and is sufficient to carry out histone pre-mRNA processing.
Interestingly, the proportion of misprocessed:wild type H2a in Slbp10 embryos (Lanzotti et al. 2002) and first and second instar larvae is substantially higher than we observed in the third instar larvae (Fig. 6C ▶). This suggests that the demand for SLBP is greater in early development. Because the demand for histone mRNA is determined by the rate of DNA replication, the Slbp10 phenotype may reflect differences in replication rates and, hence, demand for histone proteins, between the early and the later stages of development. A similar situation may be occurring early during oogenesis when misprocessed H3 mRNAs preferentially accumulate in Slbp10 mutants. We envision that SLBP binds the nascent transcript and rapidly recruits the U7 snRNP to initiate processing very soon after transcription of the 3′ end (possibly while the polymerase is stalled at the processing site). In Slbp10 mutant cells with a high rate of histone gene transcription (e.g., early in zygotic development or oogenesis), there is likely not sufficient SLBP to recruit U7 snRNP to the transcripts, and read-through of many transcripts occurs. In the third instar larval cells of Slbp10 mutants, the rate of histone transcription may be less, and the small amounts of SLBP and U7 snRNP are sufficient to cooperate to stimulate a substantial amount of histone pre-mRNA processing. Recent in vitro studies indicate that both U7 and SLBP are required for processing in Drosophila nuclear extracts, as they are in the animal (Dominski et al. 2005b). Thus, in the absence of either SLBP or U7 snRNA, no accumulation of processed histone mRNA occurs in vivo or in vitro.
U7 and SLBP are required during oogenesis
During oogenesis there are two distinct modes of histone mRNA biosynthesis. The first is replication dependent and results in the accumulation of histone mRNA specifically during S phase. The second is a burst of replication-independent nurse cell expression late in oogenesis that generates the maternal mRNAs that are transported to the oocyte (Ambrosio and Schedl 1985; Ruddell and Jacobs-Lorena 1985; Walker and Bownes 1998). Both modes of expression are affected by mutation of Slbp. Slbp mutant nurse cells and follicle cells contain misprocessed, poly A+ H3 mRNA in replicating cells, and the total amount of histone mRNA produced during oogenesis is reduced relative to wild type. U7 mutants also contain misprocessed, poly A+ H3 mRNA in replicating nurse and follicle cells.
Mutation of either Slbp or U7 prevents the completion of oogenesis and results in the production of very few eggs, none of which develop. Although the egg chambers in each mutant ovary appear to form properly and contain the appropriate cell types, many do not progress to late stages. The U7 mutant egg chambers typically do not develop past the “dumping” stage where the nurse cell cytoplasmic components are rapidly transferred to the oocyte. The basis for this developmental defect is not known, and we have not pursued a detailed analysis of the phenotype.
Mutations in U7 and Slbp affect Drosophila development differently
Slbp null mutations result in lethality while U7 null mutations result in viable adults that are sterile. Because SLBP and U7 snRNP both play essential roles in the same molecular process, histone pre-mRNA processing, we expected that abolishing the function of these factors would cause very similar terminal phenotypes in the whole animal. However, there is a maternal pool of U7 snRNA that is sufficient to support histone pre-mRNA processing until the third larval instar. In contrast, there is very little if any maternal SLBP, and poly A+ histone mRNAs appear in Slbp mutants as soon as zygotic histone transcription begins (Lanzotti et al. 2002). Thus, the simplest explanation for the difference in terminal phenotypes between Slbp and U7 null mutants is the maternal pool of U7 snRNA, which results in a later onset of the mutant phenotype. The consequence of combining the viable Slbp10 hypomorphic allele with a viable U7 null mutation is consistent with this: Histone pre-mRNA processing is disrupted sooner in the double mutant than either single mutant, leading to lethality. The relative severity of the Slbp mutant phenotype compared to U7 may also result in part from the participation of SLBP but not U7 snRNP in aspects of histone mRNA metabolism other than pre-mRNA processing. SLBP is part of the mature histone mRNP (Sanchez and Marzluff 2002; Whitfield et al. 2004) and contributes to efficient histone mRNA translation (Sanchez and Marzluff 2002) and the rapid destruction of histone mRNAs after replication inhibition (Kaygun and Marzluff 2005). These processes do not involve U7 snRNP, and thus, loss of SLBP might cause more severe defects.
The precise molecular and cellular basis for Slbp mutant lethality and U7 mutant sterility is not known. The correlation between the extent and time of onset of the histone pre-mRNA processing defects and the severity of the developmental phenotype strongly suggests that the developmental defects result from the disruption of histone mRNA metabolism. One possibility is that the presence of poly A+ histone mRNA is somehow detrimental or that the poly A+ mRNAs cannot produce the correct amount or correct stoichiometry of the canonical histones during S phase. However, we cannot exclude the possibility that the mutant phenotypes are due to the participation of SLBP and U7 snRNP in processes other than histone pre-mRNA processing. For instance, in the Xenopus oocyte germinal vesicle, some SLBP1 is found in Cajal bodies, as expected for a role in histone pre-mRNA processing, and some SLBP1 is also found in association with the genes encoding 5S rRNA and U1 and U2 snRNAs (Abbott et al. 1999). However, there is no direct evidence that SLBP1 is required for the function or biosynthesis of these RNA molecules. A continued examination of Slbp and U7 mutant phenotypes should contribute to a more complete understanding of the roles played by these factors during animal development.
MATERIALS AND METHODS
P-element excision
The EY11305 insertion site was determined by sequencing flanking genomic DNA obtained via PCR using a primer that hybridizes to the 5′-terminal repeat of the P element (SP1: 5′-ACA CAACCTTTCCTCTCAACA-3′) and a primer 5′ of the U7 coding region (U7F1; see Fig. 1B ▶). EY11305 third chromosome excisions were recovered over TM6b as white-eyed male progeny from yw; P[EY11305]/ry506 Sb P[ry+ Δ2–3] fathers. These single males were crossed to w; Df(3L)E1/TM3 Ser P[act-GFP] females. Df(3L)E1 removes Eip63E but not U7, and 17 balanced stocks were made from those excision events that complemented the lethality of Df(3L)E1. Twelve of the excision chromosomes were viable and five were lethal when homozygous. Seven of the 12 homozygous viable lines were fertile, and five were sterile. Sequencing of PCR products obtained using primers that flank the EY11305 insertion site (see Fig. 1B ▶) was used to precisely characterize the U714, U720, U79 excision alleles, and to confirm that the seven viable and fertile flies contained precise excision events. In the two cases where a PCR product using the flanking primers was not recovered (U71 and U723), primers SP1 and 5′-CAAGCATACGTTAAGTG GATGTC-3′, which hybridize to the 5′ and 3′ ends, respectively, of the P element, were paired with the flanking primers to assess if the P element ends remained intact. By this assay, the U71 allele retained both the 5′ and 3′ ends of the P element and likely sustained an internal deletion of EY11305 that inactivated the mini w+ gene. U723 similarly retained the 3′ end of EY11305 but sustained a deletion of the 5′ end of the P. However, very little if any U7 coding sequence was removed, since a primer just upstream of the EY11305 insertion site was able to amplify flanking DNA from U723 animals (Fig. 1B ▶). Each of the five homozygous lethal chromosomes contained a precise excision of EY11305, suggesting that in each case a lethal was induced elsewhere on the chromosome during transposition. The Slbp15 null and Slbp10 hypomorphic alleles are described in Sullivan et al. (2001). w1118 was used throughout as a U7 and Slbp wild-type control.
U7 transgenic rescue
DNA containing the U7 gene (Fig. 1B ▶) was amplified by PCR from adult female w1118 genomic DNA and subcloned into the pCaSpeR 4 transformation vector and confirmed by sequencing. A w1118; P[U7]/P[U7]; +/+ transgenic line was crossed to w; +/+; U714/TM3, and males of the genotype P[U7]/+; U714/+ were crossed to w; Df(3L)LY/TM6B females. Single P[U7]/+; (U714 or +)/TM6B male progeny were crossed to U7EY11305/TM3 females, and the nonbalanced progeny (i.e., those containing U7EY11305 and either + or U714) were analyzed for the presence or the absence of U714 by PCR. The w+ (i.e., containing P[U7]) and w− (i.e., not containing P[U7]) U7EY11305/U714 classes were then tested for fertility.
Northern analysis
For Northern blots, total cellular RNA was isolated using TRIzol Reagent (Gibco). For the analysis of adult U7 snRNA, 15 μg/lane of RNA denatured with 8M urea were subject to electrophoresis in 1× TBE through an 8% polyacrylamide gel containing 7 M urea. For the analysis of adult histone mRNA, 10 μg/lane of RNA were subject to electrophoresis in 1× TBE through a 1% agarose gel containing 0.01 M MOPS (pH 7.0) and 6.75% formaldehyde. Separated RNAs were transferred in 0.5× TBE to an N+ nitrocellulose membrane (Amersham) using a Genie Blotter (Idea Scientific). α[32P]-UTP was used for in vitro transcription reactions to label U7 or U1 anti-sense RNA probes. DNA containing histone or rp49 coding regions were labeled with α[32P]-dCTP using a random primer labeling kit (Stratagene). Hybridizations were performed at 58°C for snRNA probes and at 60°C for histone and rp49 probes.
Homozygous single or double mutant larvae were identified as the GFP negative class from heterozygous parents containing either TM3 Ser P[act-GFP] or TM3 Sb P[kr-GFP] balancer chromosomes. Agarose gel Northern analysis was performed as above, except that 2 μg of RNA per lane was used and the RNA was transferred via the wick method in 20× SSC. For the embryo analysis in Figure 4 ▶, a collection of eggs from U714/TM3 Ser P[act-GFP] parents was allowed to age 3 h and then subjected to RNA preparation. U7 mutant embryos were not selected because we previously demonstrated that we could detect polyadenylated H3 mRNA in a total embryo population where only one-quarter of the embryos are Slbp15 homozygous mutant (Lanzotti et al. 2002).
S1 protection analysis
A BspE I restriction fragment containing the histone H2a gene was end-labeled at 25°C for 20 min with Klenow (New England Bio-labs) in a reaction containing α-P32-dCTP. After removing unincorporated deoxynucleotides using a ProbeQuant G-50 Micro Column (Amersham), the labeled DNA was digested with HindIII. The resulting 650-nt, 3′-end-labeled fragment was recovered by gel extraction (Qiagen) after electrophoresis through 2% agarose. For the S1 nuclease protection assay, 5 μg of total RNA was hybridized at 52°C overnight with 1 μL of the labeled histone H2A probe in 40 mM PIPES (pH 6.4), 500 mM NaCl, 1 mM EDTA, 80% deionized formamide. Poly A RNA was purified from total RNA using oligo(dT) cellulose (Ambion). The hybridized samples were diluted 10-fold and treated with S1 nuclease (Promega) for 1.5 h at 25°C. The hybrids were recovered by ethanol precipitation, denatured in formamide, and subjected to electrophoresis through a 6% acrylamide gel in 1× TBE. The gel was dried using a SGD200 Slab Gel Drier (Savant) for 1.5 h at 80°C and subject to autoradiography.
In situ hybridization
Eye discs were dissected from wandering third instar larvae and fixed with either 4% formaldehyde for 15 min for Slbp mutants or 10% formaldehyde for 20 min for U7 mutants. Ovaries were dissected from 1- to 2-d-old females and fixed with 10% formaldehyde for 20 min. In situ hybridization was performed with digoxigenin-labeled riboprobes complimentary to the coding region of H3 or to the region downstream of the normal H3 pre-mRNA processing site (H3-ds) as previously described (Lanzotti et al. 2002).
Generation of germ line clones
Mosaic Slbp15 ovaries were generated using the dominant female sterile technique (Chou et al. 1993). Clones were induced by heat shocking P[hsFLP]/w; P[neoFRT]82B P[ovoD1–18]/P[neoFRT]82B Slbp15 larvae for 1 h at 37°C on the third and fourth days of development. Ovaries were dissected from the resulting adults and either fixed with 10% formaldehyde for 20 min for in situ hybridization analysis or subjected to Northern analysis as described above. Slbp15 mutant clones were identified because they developed past the ovoD1–18 developmental arrest point.
Acknowledgments
We thank the Bloomington Drosophila stock center for fly strains, Jeff Simms for technical help, and Joe Gall and Mark Peifer for comments on the manuscript. This work was supported by the NSF (MCB-0342847 to R.J.D.) and NIH (GM59214 to W.F.M.).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2270406.
REFERENCES
- Abbott, J., Marzluff, W.F., and Gall, J.G. 1999. The stem–loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol. Biol. Cell 10: 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, M.D., McVey, M., and Sekelsky, J.J. 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267. [DOI] [PubMed] [Google Scholar]
- Adamson, T.E. and Price, D.H. 2003. Cotranscriptional processing of Drosophila histone mRNAs. Mol. Cell. Biol. 23: 4046–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio, L. and Schedl, P. 1985. Two discrete modes of histone gene expression during oogenesis in Drosophila melanogaster. Dev. Biol. 111: 220–231. [DOI] [PubMed] [Google Scholar]
- Azzouz, T.N. and Schumperli, D. 2003. Evolutionary conservation of the U7 small nuclear ribonucleoprotein in Drosophila melanogaster. RNA 9: 1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouz, T.N., Gruber, A., and Schumperli, D. 2005. U7 snRNP-specific Lsm11 protein: Dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100-independent function in histone RNA 3′ end processing. Nucleic Acids Res. 33: 2106–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen, H.J., Levis, R.W., Liao, G., He, Y., Carlson, J.W., Tsang, G., Evans-Holm, M., Hiesinger, P.R., Schulze, K.L., Rubin, G.M., et al. 2004. The BDGP gene disruption project: Single transposon insertions associated with 40% of Drosophila genes. Genetics 167: 761– 781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond, U.M., Yario, T.A., and Steitz, J.A. 1991. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes & Dev. 5: 1709–1722. [DOI] [PubMed] [Google Scholar]
- Chou, T.B., Noll, E., and Perrimon, N. 1993. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development 119: 1359–1369. [DOI] [PubMed] [Google Scholar]
- Cotton, M., Gick, O., Vasserot, A., Schaffner, G., and Birnstiel, M.L. 1988. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro 3′ RNA processing reaction. EMBO J. 7: 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton, M., Oberhauser, B., Brunar, H., Holzner, A., Issakides, G., Noe, C.R., Schaffner, G., Wagner, E., and Birnstiel, M.L. 1991. 2′-Omethyl, 2′-O-ethyl oligoribonucleotides and phosphorothioate oligodeoxyribonucleotides as inhibitors of the in vitro U7 snRNP-dependent mRNA processing event. Nucleic Acids Res. 19: 2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z. and Marzluff, W.F. 1999. Formation of the 3′ end of histone mRNA. Gene 239: 1–14. [DOI] [PubMed] [Google Scholar]
- Dominski, Z., Zheng, L.X., Sanchez, R., and Marzluff, W.F. 1999. Stem–loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19: 3561–3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Erkmann, J.A., Yang, X., Sanchez, R., and Marzluff, W.F. 2002. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem–loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes & Dev. 16: 58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Yang, X.C., Purdy, M., and Marzluff, W.F. 2003. Cloning and characterization of the Drosophila U7 small nuclear RNA. Proc. Natl. Acad. Sci. 100: 9422–9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Yang, X.C., and Marzluff, W.F. 2005a. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 123: 37–48. [DOI] [PubMed] [Google Scholar]
- Dominski, Z., Yang, X.C., Purdy, M., and Marzluff, W.F. 2005b. Differences and similarities between Drosophila and mammalian 3′ end processing of histone pre-mRNAs. RNA 11: 1835–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkmann, J.A., Sanchez, R., Treichel, N., Marzluff, W.F., and Kutay, U. 2005. Nuclear export of metazoan replication-dependent histone mRNAs is dependent on RNA length and is mediated by TAP. RNA 11: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, M.R. and Matera, A.G. 1995. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. 92: 5915–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall, J.G. 2003. The centennial of the Cajal body. Nat. Rev. Mol. Cell. Biol. 4: 975–980. [DOI] [PubMed] [Google Scholar]
- Grimm, C., Stefanovic, B., and Schumperli, D. 1993. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 12: 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, M.E., Bohni, R., Schneiderman, M.H., Ramamurthy, L., Schumperli, D., and Marzluff, W.F. 1991. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11: 2416–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun, H. and Marzluff, W.F. 2005. Regulated degradation of replication- dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 12: 794–800. [DOI] [PubMed] [Google Scholar]
- Khusial, P., Plaag, R., and Zieve, G.W. 2005. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem. Sci. 30: 522–528. [DOI] [PubMed] [Google Scholar]
- Kodama, Y., Rothman, J.H., Sugimoto, A., and Yamamoto, M. 2002. The stem–loop binding protein CDL-1 is required for chromosome condensation, progression of cell death and morphogenesis in Caenorhabditis elegans. Development 129: 187–196. [DOI] [PubMed] [Google Scholar]
- Kolev, N.G. and Steitz, J.A. 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes & Dev. 19: 2583–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti, D.J., Kaygun, H., Yang, X., Duronio, R.J., and Marzluff, W.F. 2002. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol. Cell. Biol. 22: 2267– 2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti, D.J., Kupsco, J.M., Marzluff, W.F., and Duronio, R.J. 2004a. string(cdc25) and cyclin E are required for patterned histone expression at different stages of Drosophila embryonic development. Dev. Biol. 274: 82–93. [DOI] [PubMed] [Google Scholar]
- Lanzotti, D.J., Kupsco, J.M., Yang, X.C., Dominski, Z., Marzluff, W.F., and Duronio, R.J. 2004b. Drosophila stem–loop binding protein intracellular localization is mediated by phosphorylation and is required for cell cycle-regulated histone mRNA expression. Mol. Biol. Cell. 15: 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifton, R.P., Goldberg, M.L., Karp, R.W., and Hogness, D.S. 1978. The organization of the histone genes in Drosophila melanogaster: Functional and evolutionary implications. Cold Spring Harbor Symp. Quant. Biol. 42: 1047–1051. [DOI] [PubMed] [Google Scholar]
- Lilly, M.A. and Duronio, R.J. 2005. New insights into cell cycle control from the Drosophila endocycle. Oncogene 24: 2765–2775. [DOI] [PubMed] [Google Scholar]
- Luscher, B., Stauber, C., Schindler, R., and Schumperli, D. 1985. Faithful cell-cycle regulation of a recombinant mouse histone H4 gene is controlled by sequences in the 3′-terminal part of the gene. Proc. Natl. Acad. Sci. 82: 4389–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, F., Schaller, A., Eglite, S., Schumperli, D., and Muller, B. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16: 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff, W.F. 2005. Metazoan replication-dependent histone mRNAs: A distinct set of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 17: 274–280. [DOI] [PubMed] [Google Scholar]
- Marzluff, W.F. and Duronio, R.J. 2002. Histone mRNA expression: Multiple levels of cell cycle regulation and important developmental consequences. Curr. Opin. Cell Biol. 14: 692–629. [DOI] [PubMed] [Google Scholar]
- Mowry, K.L. and Steitz, J.A. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA’s. Science 238: 1682–1687. [DOI] [PubMed] [Google Scholar]
- Otake, L.R., Scamborova, P., Hashimoto, C., and Steitz, J.A. 2002. The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila. Mol. Cell 9: 439–446. [DOI] [PubMed] [Google Scholar]
- Pettitt, J., Crombie, C., Schumperli, D., and Muller, B. 2002. The Caenorhabditis elegans histone hairpin-binding protein is required for core histone gene expression and is essential for embryonic and postembryonic cell division. J. Cell Sci. 115: 857–866. [DOI] [PubMed] [Google Scholar]
- Pillai, R.S., Will, C.L., Luhrmann, R., Schumperli, D., and Muller, B. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 20: 5470–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai, R.S., Grimmler, M., Meister, G., Will, C.L., Luhrmann, R., Fischer, U., and Schumperli, D. 2003. Unique Sm core structure of U7 snRNPs: Assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes & Dev. 17: 2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascle, A., Stowers, R.S., Garza, D., Lepesant, J.A., and Hogness, D.S. 2003. L63, the Drosophila PFTAIRE, interacts with two novel proteins unrelated to cyclins. Mech. Dev. 120: 617–628. [DOI] [PubMed] [Google Scholar]
- Ruddell, A. and Jacobs-Lorena, M. 1985. Biphasic pattern of histone gene expression during Drosophila oogenesis. Proc. Natl. Acad. Sci. 82: 3316–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, R. and Marzluff, W.F. 2002. The stem–loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 22: 7093–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele, F., Gilmartin, G.M., Bannwarth, W., and Birnstiel, M.L. 1986. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature 323: 777–781. [DOI] [PubMed] [Google Scholar]
- Schumperli, D. and Pillai, R.S. 2004. The special Sm core structure of the U7 snRNP: Far-reaching significance of a small nuclear ribonucleoprotein. Cell. Mol. Life Sci. 61: 2560–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati, D. and Schumperli, D. 1988. Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol. Cell. Biol. 8: 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A.C. 1993. Developmental genetics of oogenesis. In The development of Drosophila melanogaster (eds. M. Bate and A. Martinez- Arias), pp 1–70. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Spycher, C., Streit, A., Stefanovic, B., Albrecht, D., Koning, T.H., and Schumperli, D. 1994. 3′ End processing of mouse histone pre-mRNA: Evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 22: 4023–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers, R.S., Garza, D., Rascle, A., and Hogness, D.S. 2000. The L63 gene is necessary for the ecdysone-induced 63E late puff and encodes CDK proteins required for Drosophila development. Dev. Biol. 221: 23–40. [DOI] [PubMed] [Google Scholar]
- Sullivan, E., Santiago, C., Parker, E.D., Dominski, Z., Yang, X., Lanzotti, D.J., Ingledue, T.C., Marzluff, W.F., and Duronio, R.J. 2001. Drosophila stem–loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes & Dev. 15: 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J. and Bownes, M. 1998. The expression of histone genes during Drosophila melanogaster oogenesis. Dev. Genes Evol. 207: 535–541. [DOI] [PubMed] [Google Scholar]
- Walther, T.N., Wittop Koning, T.H., Schumperli, D., and Muller, B. 1998. A 5′-3′ exonuclease activity involved in forming the 3′ products of histone pre-mRNA processing in vitro. RNA 4: 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.F., Whitfield, M.L., Ingledue 3rd, T.C., Dominski, Z., and Marzluff, W.F. 1996. The protein that binds the 3′ end of histone mRNA: A novel RNA-binding protein required for histone pre-mRNA processing. Genes & Dev. 10: 3028–3040. [DOI] [PubMed] [Google Scholar]
- Whitfield, M.L., Kaygun, H., Erkmann, J.A., Townley-Tilson, W.H., Dominski, Z., and Marzluff, W.F. 2004. SLBP is associated with histone mRNA on polyribosomes as a component of the histone mRNP. Nucleic Acids Res. 32: 4833–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.H. and Gall, J.G. 1993. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc. Natl. Acad. Sci. 90: 6257–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamrod, Z., Tyree, C.M., Song, Y., and Stumph, W.E. 1993. In vitro transcription of a Drosophila U1 small nuclear RNA gene requires TATA box-binding protein and two proximal cis-acting elements with stringent spacing requirements. Mol. Cell. Biol. 13: 5918– 5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L., Dominski, Z., Yang, X.C., Elms, P., Raska, C.S., Borchers, C.H., and Marzluff, W.F. 2003. Phosphorylation of stem–loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol. Cell. Biol. 23: 1590–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]