Abstract
The p14 subunit of the essential splicing factor 3b (SF3b) can be cross-linked to the branch-point adenosine of pre-mRNA introns within the spliceosome. p14 stably interacts with the SF3b subunit SF3b155, which also binds the 65-kDa subunit of U2 auxiliary splicing factor (U2AF65). We combined biochemical and NMR techniques to study the conformation of p14 either alone or complexed with SF3b155 fragments, as well as an interaction network involving p14, SF3b155, U2AF65, and U2 snRNA/pre-mRNA. p14 comprises a canonical RNA recognition motif (RRM) with an additional C-terminal helix (αC) and a β hairpin insertion. SF3b155 binds to the β-sheet surface of p14, thereby occupying the canonical RNA-binding site of the p14 RRM. The minimal region of SF3b155 interacting with p14 (i.e., residues 381–424) consists of four α-helices, which are partially preformed in isolation. Helices α2 and α3 (residues 401–415) constitute the core p14-binding epitope. Regions of SF3b155 binding to p14 and U2AF65 are nonoverlapping. This allows for a simultaneous interaction of SF3b155 with both proteins, which may support the stable association of U2 snRNP with the pre-mRNA. p14–RNA interactions are modulated by SF3b155 and the RNA-binding site of the p14–SF3b155 complex involves the noncanonical β hairpin insertion of the p14 RRM, consistent with the β-sheet surface being occupied by the helical SF3b155 peptide and p14 helix αC. Our data suggest that p14 lacks inherent specificity for recognizing the branch point, but that some specificity may be achieved by scaffolding interactions involving other components of SF3b.
INTRODUCTION
The accurate removal of introns from eukaryotic pre-mRNAs is mediated by a large ribonucleoprotein (RNP) complex called the spliceosome (Burge et al. 1999). In the first catalytic step of splicing the 2′ hydroxyl of the branch point, adenosine carries out a nucleophilic attack on the 5′ splice-site phosphate, generating a free 5′ exon and the intron-3′ exon in the form of a lariat structure. In the subsequent step, the 3′ hydroxyl of the 5′ exon attacks the 3′ splice site leading to intron release and ligation of the 3′ and 5′ exons.
The major (U2-dependent) spliceosome is formed via the ordered interaction of U1, U2, U5, and U4/U6 snRNPs (small nuclear RNPs) and numerous splicing factors with RNA elements located at the 5′ and 3′ ends of a pre-mRNA intron (Burge et al 1999). The minor U12-dependent spliceosome catalyzes splicing of relatively rare U12-type introns that are found only in some metazoans and is formed by the ordered interaction of the U11/U12, U4atac/U6atac, and U5 snRNPs (Patel and Steitz 2003).
While splicing catalysis occurs in the so-called C complex, the splice-site elements are initially recognized at the onset of spliceosome assembly. During formation of the initial E complex, U1 snRNP binds to the 5′ splice site. The 3′ splice-site elements are recognized by cooperative binding of splicing factors to cognate RNA sequences. Specifically, the large and small subunits of U2AF (U2 snRNP auxiliary factor; U2AF65 and U2AF35) (Zamore et al. 1992) bind to the polypyrimidine (Py) tract (Zamore et al. 1992; Singh et al. 1995) and the downstream AG dinucleotide, respectively (Zamore et al. 1992; Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). The branch-point sequence (BPS) is recognized by splicing factor 1 (SF1, or mammalian branch point binding protein, mBBP) (Krämer 1996; Berglund et al. 1997). Binding of SF1 to the BPS is weak (KD ~1 μM, (Liu et al. 2001), but its affinity is significantly increased by a simultaneous interaction with the third RNA recognition motif (RRM3) of U2AF65 (Berglund et al. 1998; Selenko et al. 2003).
Subsequently, the U2 snRNP interacts with the branch-point sequence displacing SF1. In this so-called A complex, a duplex is formed between the BPS and the U2 snRNA. The invariant branch-point adenosine that acts as the nucleophile in the first catalytic step is bulged out in this branch-site helix (Query et al. 1994; Berglund et al. 2001). A phylogenetically conserved pseudouridine in the U2 snRNA, located opposite the branch site, may induce a unique conformation of the branch-point adenosine that primes it for attack at the 5′ splice site (Newby and Greenbaum 2002). In addition, U2AF and several subunits of the U2 snRNP-associated splicing factors (SF) 3a and 3b contact the pre-mRNA near the BPS (Staknis and Reed 1994; Gozani et al. 1996, 1998; Query et al. 1996; Krämer et al. 1999; McPheeters and Muhlenkamp 2003). For example, the 155-kDa subunit of SF3b (SF3b155) can be cross-linked to RNA sequences up- and downstream of the BPS. An N-terminal region in SF3b155 also binds the third RNA recognition motif (RRM) of U2AF65 (Gozani et al. 1998). The U2AF65 RRM3–SF3b155 interaction replaces the U2AF65–SF1 interaction and contributes to the recruitment of the U2 snRNP to the branch site (Gozani et al. 1998).
In the minor spliceosome, the U11 and U12 snRNPs form a stable U11/U12 di-snRNP that binds to pre-mRNA during assembly of the minor A complex. U12 snRNA base pairs with the BPS and the branch-point adenosine is also bulged out (Hall and Padgett 1996). Interactions between the U11 snRNP and the 5′ splice site are required to stabilize the U12 branch-site interaction (Frilander and Steitz 1999). As U12-type introns lack a polypyrimidine tract, U2AF does not appear to facilitate association of U12 with the branch site. In contrast, SF3b appears to take part in both splicing pathways, as it is an integral part of both the U2 snRNP and the U11/U12 di-snRNP (Will et al. 1999, 2004).
Together with all other SF3b subunits (i.e., SF3b155, SF3b145, SF3b130, SF3b49, SF3b14b, and SF3b10), SF3b14a/p14 (denoted p14 hereafter) is present in both the major and minor spliceosomes (Will et al. 1999, 2001, 2004). p14 can be cross-linked to the branch-point adenosine (Query et al. 1997; Will et al. 2001) first after the integration of the U2 snRNP into the major prespliceosome, and at least until the formation of complex C (MacMillan et al. 1994). Thus, p14 is located near the catalytic center responsible for the first catalytic step of the splicing reaction and the presence of p14 in both the minor and major spliceosome suggests a common mechanism of branch-point selection in mammals. Like all other SF3b subunits, p14 is highly conserved among higher eukaryotes (Will et al. 2001), suggesting that it plays an important role in the splicing process.
A cryo-electron microscopic (EM) investigation of isolated SF3b (Golas et al. 2003) indicated that the particle forms a spherical structure with a hollow interior. It was suggested that p14 is located inside the central cavity, where it would be largely inaccessible to the branch-point adenosine. However, the EM structure of the U11/12 di-snRNP, which contains SF3b (Golas et al. 2005), revealed a more open conformation of SF3b, suggesting that it may undergo a conformational rearrangement that makes p14 accessible for pre-mRNA interaction.
Previous studies revealed a tight interaction between p14 and SF3b155, and roughly mapped the region of human SF3b155 that contacts p14 to amino acids 255–424 (Will et al. 2001). Intriguingly, this region also includes residues mediating SF3b155′s interaction with U2AF65 (Gozani et al. 1998). Here, we have analyzed the conformation of p14 alone or complexed with a fragment of SF3b155 using NMR. In addition, we have defined the p14/SF3b155 interaction interface and the binding site of SF3b155 with U2AF65 using NMR and peptide scanning experiments. Finally, we have studied the binding of p14 alone, or as part of either a p14/SF3b155 heterodimer or the entire SF3b complex, with the pre-mRNA BPS, U2 snRNA, or duplexes thereof. These studies delineate the protein and RNA interaction network involving p14, and thus provide insights into molecular interactions near the catalytic core of the spliceosome.
RESULTS
Analysis of the interaction between p14 and SF3b155
Residues 381–424 of SF3b155 constitute a minimal binding epitope for p14
Previous GST pull-down experiments have demonstrated that a region comprising residues 255–424 in the N-terminal region of SF3b155 preceding its HEAT repeats binds to p14 (Will et al. 2001) (cf. Fig. 1 ▶). We found that SF3b1551–459 can be coexpressed and copurified with p14 through a series of affinity and ion-exchange chromatographic steps (data not shown). We used the SF3b155282–424 polypeptide, which, in contrast to other subfragments, could be expressed and purified also in the absence of p14, to determine the binding affinity of SF3b155 and p14. Using isothermal titration calorimetry, a dissociation constant (KD) of 9.6 nM was determined with a 1:1 p14–SF3b155282–424 stoichiometry, indicating a strong enthalpically driven interaction between SF3b155 and p14 (Table 1 ▶).
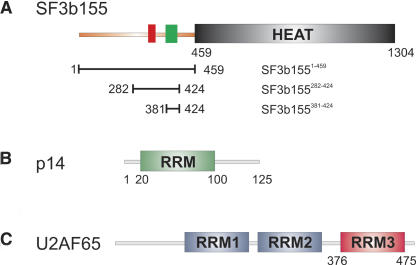
FIGURE 1.
SF3b155, p14, and U2AF65 domain composition. Domain composition and protein fragments used for NMR and biochemical experiments for (A) SF3b155, (B) p14, and (C) U2AF65.
TABLE 1.
Isothermal titration calorimetry
| Kd | ΔH [kcal mol−1] | ΔS [cal K−1 mol−1] | Stoichiometry (n) | |
| SF3b155282–424-p14 | 9.6 nM | –12.70 | –6.1 | 1.00 |
| SF3b155317–357-U2AF65-RRM3 | 6300 nM | –1.13 | –14.61 | 1.05 |
The EM structure of the native SF3b complex does not provide any information about the structure of the N-terminal region of SF3b155, while the C-terminal HEAT repeats could be clearly visualized (Golas et al. 2003). Secondary structure predictions (http://bip.weizmann.ac.il/fldbin/findex) suggest that the entire N-terminal region of SF3b155 (residues 1–459) could be largely unstructured. We therefore reasoned that p14 could recognize short, linear peptide epitopes within the SF3b155 N-terminal portion and used NMR titrations and peptide scan experiments to map the binding epitope.
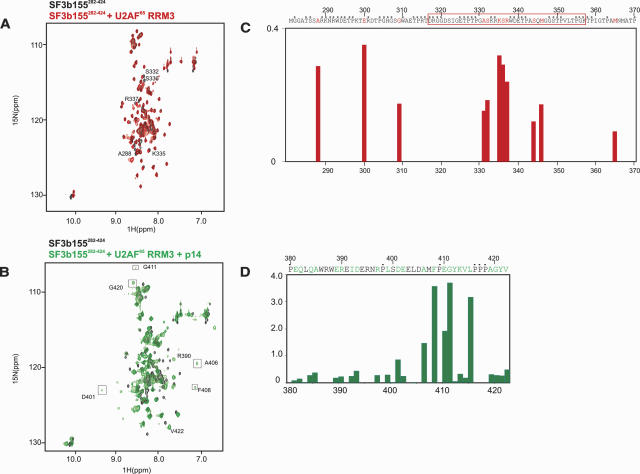
In order to define residues within SF3b that participate in the binding interface with p14, we analyzed NMR spectra of 15N-labeled SF3b155 upon addition of p14 (Fig. 2A ▶). Within the SF3b155282–424 region, only NMR signals of residues 380–424 are affected by the interaction with p14 (Fig. 2B ▶). Moreover, the NMR signals of a 15N-labeled 43-residue SF3b155381–424 peptide that is bound to p14 are very similar to the corresponding residues within the SF3b155282–424–p14 complex, indicating identical binding interfaces (Fig. 2A ▶). We next monitored changes in 1H,15N correlation NMR spectra of 15N-labeled p14 upon addition of unlabeled SF3b155282–424 (Fig. 3A ▶). As expected, large chemical-shift changes confirm the tight interaction of SF3b155 with p14. Likewise, the addition of the SF3b155282–424 or SF3b155381–424 peptides to 15N-labeled p14 affects the same set of residues in p14, and the resulting spectra are very similar (Fig. 3A ▶). These data demonstrate that the 43-residue SF3b155381–424 fragment contains all determinants required for p14 binding.
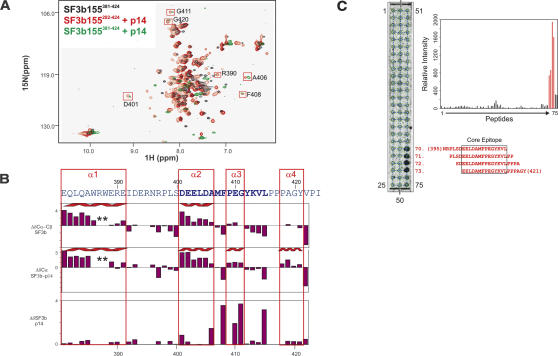
FIGURE 2.
Secondary structure and p14 binding site of SF3b155. (A) 1H,15N correlation spectrum of 15N-labeled SF3b282–424 (black) and in the presence of unlabeled p14 (red). 1H,15N correlation spectrum of SF3b381–424 after addition of p14 (green). Addition of p14 to SF3b282–424 or SF3b381–424 affects the same set of residues in both spectra. Residues with large chemical-shift perturbations belong to the minimal binding epitope for the p14 interaction and are boxed in red. (B) 13C secondary chemical shifts of free SF3b155282–424 and when bound to p14 (row 1 and 2), and chemical-shift perturbation of SF3b155282–424 upon addition of p14. Secondary structure elements are indicated on top. (C) Peptide scan analysis of the SF3b155–p14 interaction. (Left) Autoradiogram obtained by incubating the cellulose-bound peptide array with [35S]methionine-labeled p14. Peptide numbers for the terminal spots are indicated. Sequences for peptides 70–73, which constitute the central binding epitope of SF3b155 for p14, are indicated in red (starting and ending residue numbers in parentheses). (Right) Bar graph of the quantified relative affinities of the SF3b155 peptides 1–75 toward p14 (represented by the radioactivity at each spot). The response of the peptides 70–73, comprising the central binding epitope, is shown in red.
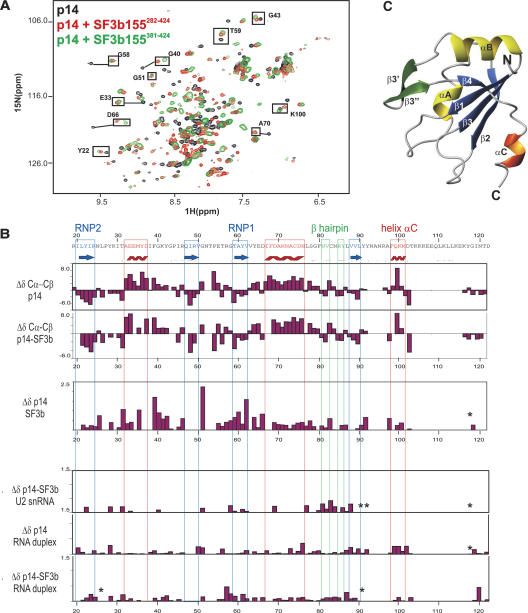
FIGURE 3.
Secondary structure and binding site of p14 with SF3b155 and RNA. (A) 1H,15N correlation NMR spectra of 15N-labeled p14 alone (black) and in the presence of unlabeled SF3b282–424 (red) or SF3b381–424 (green). Both SF3b peptides induce very similar effects on p14 as indicated for some residues (black boxes). (B) 13C secondary chemical shifts of p14 alone and when bound to SF3b282–424. Secondary structure elements are shown on top. Chemical-shift changes of p14 amide groups upon addition of SF3b282–424, and chemical-shift perturbation on p14 and p14–SF3b155282–424 upon addition of the U2 oligo and the U2/BPS RNA duplex. (*) Signals that are strongly affected but could not be assigned in the bound state. (C) Homology model of p14 based on the secondary NMR chemical shifts and a sequence alignment with the U1A RRM.
Additional evidence mapping the p14-SF3b155 interaction interface was obtained by performing peptide scanning experiments. An array of overlapping 20-mer peptides covering amino acids 258–424 of SF3b155 was chemically synthesized and tested for binding to p14. In vitro translated p14, labeled with [35S]methionine, bound strongly to four consecutive peptides of the array representing amino acids 395–421 of SF3b155. The smallest peptide fragment common to all four peptides is 402EELDAMFPEGYKVL415 (Fig. 2C ▶). Notably, these residues experience the largest chemical-shift perturbation upon addition of p14 to SF3b155381–424, indicating that this region comprises an important part of the interface.
SF3b155381–424 adopts an α-helical conformation that is partially induced upon p14 binding
We assigned the 1H,15N and 13C backbone chemical shifts of the SF3b155282–424 fragment using standard methods (Sattler et al. 1999). The 13Cα and 13Cβ secondary chemical shifts in the free SF3b155282–424 peptide indicate two helical regions between residues 381–391 (helix α1) and 401–406 (α2) (Fig. 2B ▶). Upon binding to p14, a helical conformation is further induced for residues 410–414 (α3) and residues 419–423 (α4) as indicated by the 13Cα secondary chemical shifts (Fig. 2B ▶). Thus, the NMR data show that the p14-binding epitope of SF3b155 adopts a conformation comprised of four helices, of which helix α1 and α2 are preformed, while helices α3 and α4 are induced upon binding to p14. The core binding motif (residues 401–415) is mainly helical and comprises helices α2 and α3.
Secondary structure and a homology model of the p14 RRM
p14 is a highly conserved 125-residue protein. Primary sequence signatures indicate that the protein contains an RNA recognition motif (RRM) (Will et al. 2001). We have assigned the NMR resonances of almost all backbone atoms of the p14 RRM in isolation. The flanking N- (residues 1–20) and C-terminal regions (residues 100–125) are unstructured, and only partial chemical-shift assignments could be obtained due to signal overlap and line broadening. The analysis of the 13C secondary chemical shifts, Δδ (Cα-Cβ), confirms the expected β1-αA-β2-β3-αB-β4 topology of the canonical RRM fold (Fig. 3B ▶). Notably, this fold is extended by an additional β hairpin (β3′-β3”) between helix αB and β4, and the p14 RRM also comprises a short C-terminal helix αC (A97-K100). A helix C exists only in few RRM structures, where it usually packs against the β-sheet surface. In the U1A RRM1 domain, helix αC can move away in order to allow RNA binding by the RNP motif residues in the center of the β sheet (Maris et al. 2005). On the basis of the secondary NMR chemical shifts, which define the secondary structure elements, and a sequence alignment with the U1A RRM (39%/56% sequence identity/homology), we generated a homology model of the p14 RRM (Fig. 3C ▶).
Mapping the SF3b155-binding site on the structural model of p14
NMR titrations of p14 with SF3b155 were analyzed to map the chemical-shift perturbations observed upon SF3b155 binding onto the structural model of the p14 RRM (Fig 6A ▶, below). The most pronounced chemical-shift changes are observed for residues located at the β-sheet surface. Most notably, residues within the RNP1 and RNP2 motifs are strongly affected by the addition of the SF3b155282–424 peptide. In addition, residues in the β1-αA and the αA-β2 loops experience large chemical-shift changes upon SF3b155 binding (Fig. 3B ▶). Based on the secondary 13C chemical shifts of free and bound p14, the secondary structure of p14 is not altered by binding of SF3b155 (Fig. 3B ▶). Specifically, the C-terminal helix αC is present in both free and SF3b155-bound p14.
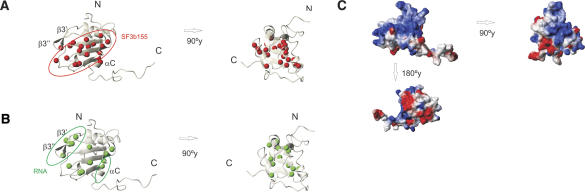
FIGURE 6.
Binding site of p14 with SF3b155 and RNA. (A) Ribbon representation of the homology model of p14. Red balls indicate amides of residues with large chemical-shift perturbations upon binding to SF3b155282–424. The SF3b155-binding site is circled. (B) Residues of p14 in the p14–SF3b155282–424 complex, which are affected by the addition of the U2/BPS RNA duplex are indicated by green balls. The main RNA-binding site is highlighted. (C) Surface representation of the p14 homology model, shown in the same orientation as in A and B, colored by electrostatic surface potential. The surface was calculated with MOLMOL (Koradi et al. 1996).
A surprising observation is that the β-sheet surface, including residues in the RNP1 and RNP2 motifs, which mediate RNA binding in canonical RRMs, forms the binding interface with the SF3b155 peptide. This indicates that—in addition to helix αC—the helical SF3b155 peptide (presumably helices α2 and α3 within the core binding motif) binds to the β-sheet surface, and thus renders RNP1 and RNP2 motif residues inaccessible for RNA binding. Therefore, RNA binding of the p14 RRM either would require a conformational rearrangement where the helical SF3b155 peptide and helix αC move away from the p14 β-sheet surface or involves a different site of the p14 RRM. To clarify this question, we next investigated the RNA-binding properties of p14 and the p14–SF3b155 complex.
RNA binding of p14 and the p14–SF3b155 complex
UV-induced cross-linking of RNA to p14 and the p14–SF3b155282–424 complex
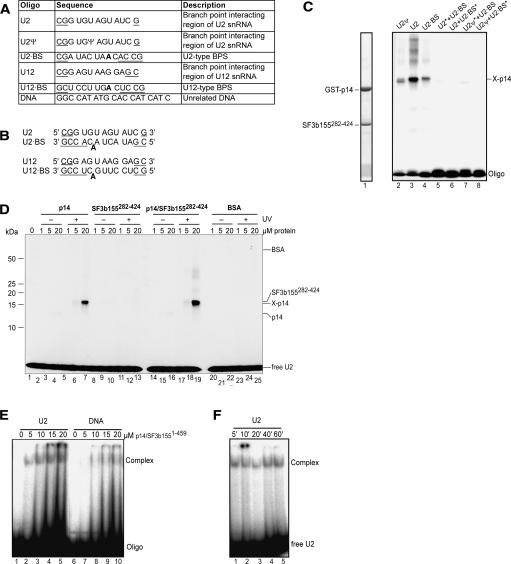
As p14 could previously be cross-linked to the branch-point adenosine of the pre-mRNA during splicing (MacMillan et al. 1994; Query et al. 1996), we investigated the RNA-binding properties of p14, fragments of SF3b155, and p14–SF3b155 complexes by UV-induced cross-linking or by performing electrophoretic gel mobility shift assays (EMSA). The branch-point adenosine of the pre-mRNA is bulged out from a double-helical segment formed by the pre-mRNA BPS and a short stretch of U2 snRNA that exhibits sequence complementarity with the U2-type BPS, respectively (Query et al. 1994; Berglund et al. 2001) (Fig. 4B ▶). We thus investigated the interaction of the p14 and SF3b155 with single-stranded RNA oligonucleotides comprising the region of the human U2 snRNA that binds the BPS, or oligos containing a consensus U2-type BPS, as well as U2-BPS duplexes (Fig. 4A,B ▶). The human U2 snRNA contains numerous post-transcriptionally modified residues, among which the pseudouridine at position 34 is of particular interest, as it may induce a unique structure in the U2–BPS duplex (Newby and Greenbaum 2002). We therefore tested both unmodified and pseudouridine-containing U2 snRNA mimics. Radiolabeled RNA oligomers or duplexes were incubated with proteins or protein complexes, exposed to 254 nm UV light, and cross-linked proteins were analyzed by SDS-PAGE followed by autoradiography (Fig. 4C,D ▶).
FIGURE 4.
RNA cross-linking and gel-shift assays of p14 and p14–SF3b155. (A) Sequence and description of RNA and DNA oligonucleotides. Additional nucleotides added to stabilize duplex formation are underlined. The branch point adenosine is shown in bold. (B) Duplexes formed by oligonucleotides shown in A. (C) UV cross-linking of p14–SF3b155282–424. RNP complexes were formed on the oligonucleotides or their duplexes (as indicated above the lanes). Following UV-irradiation, samples were separated on 12% SDS-PAGE and stained with Coomassie blue (lane 1) or visualized by autoradiography (lanes 2–8). The position of noncross-linked (input) proteins is indicated on the left and that of oligonucleotides and the RNA–p14 cross-link are indicated on the right. Duplexes contained only one 32P-labeled oligonucleotide, which is marked by an asterisk (*). Note that in this experiment, the p14 protein contained a GST tag. (D) UV-cross-linking of p14, SF3b155282–424, or p14–SF3b155282–424 to the U2 oligonucleotide. Increasing concentrations of protein (as indicated above each lane) were incubated with 32P-labeled U2 oligonucleotide and were either analyzed directly without UV irradiation (lanes 2–4, 8–10, 14–16, 20–22) or subjected to UV irradiation (lanes 5–7, 11–13, 17–19, 23–25) and analyzed as in C. BSA (lanes 20–25) was used as a control. (E) EMSA of the p14–SF3b1551–459 complex with oligonucleotides. Increasing concentrations of the p14–SF3b1551–459 protein complex (as indicated above each lane) were incubated with 32P-labeled U2 snRNA oligo (lanes 1–5) or unrelated DNA oligonucleotide (DNA) (lanes 6–10), separated on a native gel, and visualized by autoradiography. (F) Time course of U2 oligo-p14–SF3b1–459 complex formation. p14–SF3b1551–459 was incubated with 32P-labeled U2 oligo for the indicated time and RNP complex formation was analyzed as in E.
When cross-linking was performed with a recombinant GST–p14–SF3b155282–424 complex, cross-links were observed between p14 and single-stranded U2 snRNA (both with and without pseudouridine) or single-stranded U2-type BPS RNA (Fig. 4C ▶, lanes 2–4). p14 alone, but not SF3b155282–424, could be cross-linked to the U2 snRNA oligomer lacking pseudouridine (Fig 4D ▶, cf. lanes 5–7 and 11–13). The cross-linking efficiency was lower with isolated p14 compared with p14 complexed with SF3b155282–424 (cf. lanes 5–7 and 17–19). In contrast, no cross-link was observed between the U2–BPS duplex RNA, either with p14, SF3b155282–424, or the p14–SF3b155282–424 complex (Fig. 4C ▶; data not shown).
In order to further test the specificity of the p14–RNA interactions, we examined cross-linking of a p14–SF3b1551–459 complex, in which the entire N terminus of SF3b155 is present. Both the U2 snRNA oligomer and an unrelated single-stranded DNA oligomer could be cross-linked with similar efficiency to this complex, with cross-links primarily formed with p14 (data not shown). A less-abundant cross-link of either nucleic acid to the longer SF3b1551–459 fragment was also observed (data not shown). Taken together, the above results suggest that p14–SF3b155 complexes exhibit general single-stranded nucleic acid-binding activity and that nucleic acid contacts are mediated primarily through p14. The cross-linking efficiency of nucleic acids to p14 is enhanced when p14 is complexed with SF3b155, and the extended N terminus of SF3b155 could contribute to nucleic acid binding.
RNA interaction of p14 and p14–SF3b155 by electrophoretic gel mobility shift assays
UV-induced cross-linking efficiency depends not only on binding affinity, but also on a productive orientation of the cross-linking moieties. Thus, the different conformation of the RNAs when singled-stranded versus in the form of a duplex, could hinder cross-linking to the latter, despite the formation of a protein–RNA complex. Indeed, UV-induced RNA–protein cross-links are typically not formed when nucleotides are engaged in base-pairing interactions. Therefore, we also assayed RNA binding by p14, SF3b155 deletion mutants, or p14–SF3b1551–459 by performing electrophoretic gel mobility shift assays. Increasing amounts of the aforementioned proteins were incubated with radiolabeled nucleic acids, and complex formation was analyzed on native polyacrylamide gels. SF3b155282–424 alone did not bind RNA or DNA at the protein concentrations tested (up to 100 μM) (data not shown). In contrast, nucleic acid interactions were detected with p14 and the p14–SF3b155282–424 complex when high concentrations of protein (100 μM) were used. However, only very diffuse bands were observed, indicating that the interactions are weak or governed by fast association and dissociation kinetics (data not shown). The binding appeared to be nonspecific, as all of the tested RNA and DNA oligos were bound in both cases (data not shown).
Distinct RNP complexes are, however, observed when EMSA was performed with p14 complexed with SF3b1551–459 (Fig. 4E ▶). A slightly higher affinity of p14–SF3b1551–459 is observed for single-stranded U2 RNA over DNA (Fig. 4E ▶, cf lanes 1–5 and 6–10). An even weaker interaction is observed with the U2/BPS RNA duplex (data not shown). The amount of complex formed between p14–SF3b1551–459 and the U2 snRNA oligo does not increase beyond 5 min of incubation (Fig. 4F ▶, lanes 1–5), indicating fast binding and dissociation kinetics of the protein–RNA complexes. Taken together, these results suggest that the interaction of p14 with SF3b1551–459 enhances the nucleic acid-binding capacity of p14, and is consistent with the notion that additional nucleic acid contacts are mediated by the extended N terminus of SF3b155.
Mapping the RNA-binding surface of p14 and p14/SF3b155
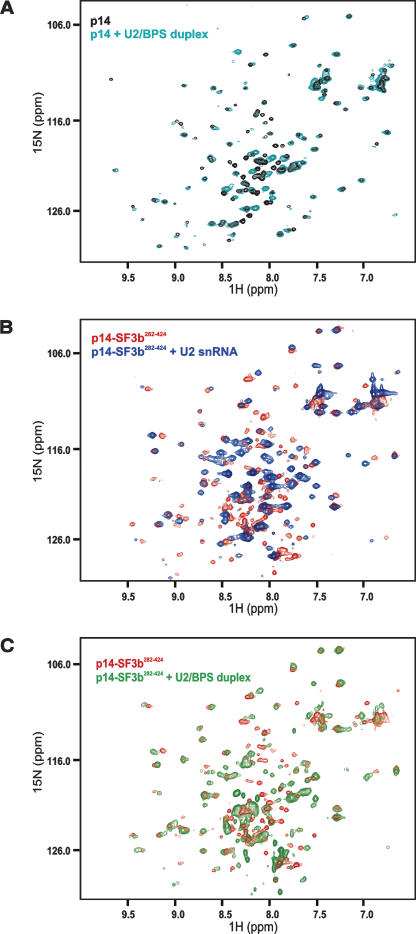
In order to map the RNA-binding surface of p14 and the p14–SF3b155 complex, we performed NMR titration experiments with the same RNA ligands that were used for cross-linking and gel-shift assays. Numerous chemical-shift changes were observed in the 1H,15N correlation spectra of p14 after addition of the single-stranded U2 RNA (data not shown). These effects are consistent with non-specific nucleic acid binding as also observed in the cross-linking and band-shift experiments, and may correlate with the extended positively charged surface of the p14 protein (Fig. 6C ▶, below). Smaller chemical-shift perturbations affecting only a subset of residues were observed upon addition of the U2 snRNA–BPS duplex, indicating a weaker interaction of p14 with the duplex RNA (Fig 5A ▶).
FIGURE 5.
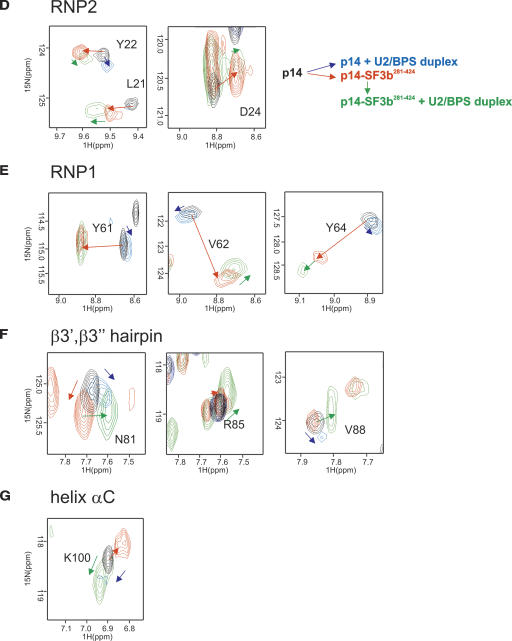
NMR RNA titration experiments with p14 and p14–SF3b155. (A) 1H,15N correlation spectra of 15N-labeled p14 alone (black) and upon addition of the U2/BPS RNA duplex (cyan). (B) 1H,15N correlation spectra of the p14 (15N labeled)–SF3b155282–424 (unlabeled) complex (red) and upon addition of the single-stranded U2 RNA oligonucleotide (blue) (C) 1H,15N correlation spectrum of the p14 (15N labeled)–SF3b155282–424 (unlabeled) complex (red) alone and upon addition of the U2/BPS RNA duplex (green) (D–G) Enlarged views of the spectra shown in A–D for residues located in RNP2, RNP1, the β3′, β3” hairpin, and helix αC. Colored arrows show the extent of chemical-shift perturbations between free p14 and the binary and ternary complexes with SF3b155 and the U2/BPS RNA duplex as indicated.
NMR titrations show that also the p14–SF3b155282–424 complex can bind to both the single-stranded U2 snRNA oligo and to the duplex RNA (Fig. 5B,C ▶). Residues that are affected by the addition of these RNAs precede the RNP1 motif (strand β3), or are located in the β3′, β3” hairpin, and helix αC (Figs. 3B ▶, 6B ▶). Notably, the residues in the RNP1 and RNP2 motifs do not experience very strong chemical-shift perturbations. This is in contrast to canonical RRMs, where large chemical-shift perturbations are observed for RNP residues that directly contact the RNA (see, for example, Howe et al. 1998).
A more detailed analysis of the chemical-shift perturbation of residues in p14 upon addition of SF3b155282–424 and the U2–BPS duplex RNA provides additional insight (Fig. 5D–G ▶). Residues located in the RNP1 and RNP2 motifs within the central part of the β sheet experience the largest perturbations upon addition of the SF3b155282–424 peptide, while smaller shifts are observed for these residues upon addition of the RNA duplex to either p14 or the p14–SF3b155282–424 complex (Fig. 5D,E ▶; red arrows). These observations suggest that even upon addition of the RNA, the helical SF3b155 peptide remains bound at the β-sheet surface. In contrast, the NMR signals of residues located in the β3′ β3” hairpin or helix αC are only weakly affected by the addition of SF3b155282–424 (Fig. 5F,G ▶), and are mainly shifted by the addition of the RNA duplex to the p14–SF3b155282–424 (Fig. 5F,G ▶; green arrows). Notably, the extent and directionality of the chemical-shift changes observed for these residues upon addition of the RNA duplex to p14–SF3b155282–424 are very different (Fig. 5F,G ▶; green arrows) compared with the addition of SF3b155282–424 to p14 (red arrows), and rather small changes are seen upon addition of the RNA duplex to p14 alone (blue arrows).
These data suggest that the binding site of the p14–SF3b155282–424 complex with the RNA involves a region flanking the central β sheet, involving mainly the β3′, β3” hairpin. Most notably, the RNP residues appear not to be involved in direct RNA interactions. The overall small chemical-shift perturbations indicate a weak interaction and argue against any large conformational changes upon RNA binding. Thus, most likely, helix αC remains bound at the surface of the β sheet, even in the presence of RNA. Thus, the β-sheet surface, which represents the canonical RNA-binding surface in RRM domains, would be occupied by helix αC as well as the helical SF3b155 peptide in the p14–SF3b155 complex.
We also tested whether residues of SF3b155282–424 within the p14–SF3b155282–424 complex may mediate direct RNA contacts by monitoring spectral changes for residues of 15N-labeled SF3b155282–424 bound to unlabeled p14 upon addition of the U2 snRNA/BPS RNA duplex. The NMR spectrum of SF3b155282–424 bound to p14 does not show any additional spectral changes upon addition of the RNA duplex, indicating that SF3b155282–424 does not directly contact the RNA (data not shown).
Taken together, the NMR studies indicate that both p14 and the p14–SF3b155282–424 complex bind weakly and with poor specificity to RNA. Our data show that within the p14–SF3b155282–424 complex, the p14 RRM interacts with the RNA in a noncanonical fashion. The β-sheet surface of the RRM is occupied by the helical SF3b155 peptide and presumably helix αC, while contacts to the RNA mainly involve the β3′ β3” hairpin. The distinct binding interfaces of the p14 RRM with SF3b155 and RNA are highlighted in Figure 6, A and B ▶.
Binding of the native SF3b particle to single-stranded and double-stranded RNA
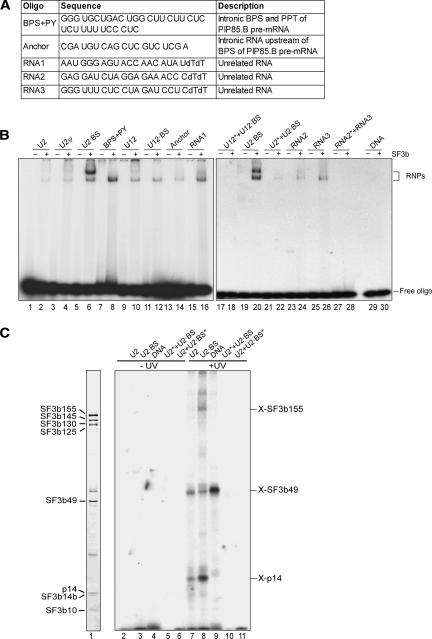
Although p14 has been UV cross-linked to the branch site in spliceosomal complexes (MacMillan et al. 1994; Query et al. 1996), we did not detect any cross-links between recombinant p14–SF3b155 complexes and a U2 snRNA/BPS duplex. Thus, it is possible that the nucleic acid-binding properties of p14 are modulated upon its incorporation into the heteromeric SF3b protein complex. To test this possibility, we immunoaffinity purified native SF3b complexes from HeLa nuclear extract and assayed for their interaction with several nucleic acids using EMSA and UV cross-linking. In addition to the U2 snRNA and U2-type BPS oligomers and their duplex and a DNA oligomer (DNA) (see Fig. 4A ▶), the following RNAs were tested for SF3b binding: the region of the human U12 snRNA that binds the U12-type BPS (U12), the consensus U12-type BPS (U12 BPS), a U12 snRNA-U12-type BPS duplex, a 34-mer intronic sequence containing a U2-type BPS and adjacent polypyrimidine tract (BPS+PY), an RNA encompassing 19 nucleotides upstream of a U2-type BPS (Anchor), unrelated 21-mer RNAs (RNA1, RNA2, and RNA3), and an unrelated RNA duplex formed between RNA2 and 3 (Fig. 7A ▶).
FIGURE 7.
Binding of nucleic acids to the native SF3b particle. (A) Sequence and description of additional RNA oligonucleotides used in binding studies. (B) EMSA of native SF3b protein complexes with oligonucleotides or their duplexes. The various single- or double-stranded oligo-nucleotides (indicated above each lane) were incubated without (odd-numbered lanes) or with SF3b (even-numbered lanes) and analyzed as in Figure 4E ▶. Duplexes contained only one 32P-labeled oligonucleotide that is marked by an asterisk. (C) UV-cross-linking of the native SF3b complex. SF3b was incubated with single- or double-stranded oligonucleotides (indicated above each lane) and nonirradiated samples (lanes 1–6) or UV-irradiated samples (lanes 7–11) were analyzed by SDS-PAGE and visualized by autoradiography (lanes 2–10) or by Coomassie blue staining (lane 1). The position and apparent identity of RNA–protein cross-links are indicated on the right.
SF3b did not bind DNA in these experiments, suggesting that, in contrast to the recombinant p14–SF3b155 complexes, the native SF3b complex preferentially binds RNA over DNA (Fig. 7B ▶, lane 30). In the presence of RNA, a slower and faster migrating complex was observed, which could reflect different SF3b conformations. The faster migrating complex formed on all single-stranded RNAs tested (Fig. 7B ▶), including the unrelated RNAs 1, 2, and 3, suggesting that SF3b exhibits general RNA-binding activity. Formation of this complex was considerably weaker with the U2 snRNA/BPS RNA duplex or did not form at all in the case of the U12 snRNA/BPS (Fig. 7B ▶, lane 18) and control duplexes (lane 28). A distinct, slower migrating complex was efficiently formed with the U2-type BPS RNA (Fig. 7B ▶, lanes 6,20), but only weakly with most of the other RNAs tested. A double band was observed with the 34-mer intronic sequence containing a U2-type BPS and adjacent polypyrimidine tract (Fig. 7B ▶, lane 8). The upper band could be similar to the slower migrating complex observed with the U2-type BPS alone, the difference in migration behavior resulting from the larger size of the BPS+PY oligomer.
To determine which subunits of SF3b contact RNA or DNA, UV-induced cross-linking analyses were performed. Similar to results obtained with recombinant p14 or p14 complexed with an N-terminal fragment of SF3b155, p14 within the SF3b complex appeared to be cross-linked to the U2 snRNA oligomer and U2-type BPS RNA (Fig. 7C ▶, lanes 7–8). The U2-type BPS RNA was also additionally cross-linked to SF3b155 (lane 8). This observation is consistent with the notion that the extended N terminus of SF3b155 might mediate additional RNA contacts (see above). Furthermore, the fact that SF3b155 was efficiently cross-linked solely to the U2-type BPS RNA is in agreement with the more efficient formation of a distinct, slower migrating SF3b–RNA complex in the gel-shift experiments upon addition of this RNA (see above). Of the remaining SF3b subunits, only SF3b49, which contains two RNA recognition motifs, was clearly cross-linked to the single-stranded but not duplex RNAs tested (Fig. 4C ▶, cf. lanes 7–11). In addition, a strong cross-link between SF3b49 and DNA (but not other SF3b components) was observed (Fig 4C ▶, lane 9). However, as no DNA–SF3b complexes were observed in our gel-shift assays, this likely represents a relatively weak interaction. Taken together, these results suggest that purified SF3b exhibits general RNA binding, with some preference for U2-type branch-point sequences.
Interaction of SF3b155 and U2AF65
A region comprising residues 267–369 of SF3b155 has previously been shown to mediate its interaction with RRM3 of U2AF65 (Gozani et al. 1998). Based on the three-dimensional structures of the U2AF65 RRM3 (Selenko et al. 2003) and the U2AF35 RRM (Kielkopf et al. 2001) bound to short peptides, a consensus sequence for ligands of these atypical RRMs could be deduced. This consensus consists of an X-[R/K]-W tripeptide preceded by a stretch of positively charged amino acids. Within the N terminus of SF3b155, a number of such sequence motifs can be identified, which could potentially mediate binding to the U2AF65 RRM3. We therefore performed NMR-binding studies to identify which residues within SF3b155282–424 mediate the interaction with U2AF65 RRM3, and to test whether the SF3b155-binding epitopes for U2AF65 and p14 overlap.
Upon addition of unlabeled U2AF65 RRM3 to 15N-labeled SF3b155282–424, a limited number of residues were affected (Fig. 8A ▶). Notably, all of these residues precede the p14-binding epitope and large chemical-shift perturbations are observed for residues 331–346 (Fig. 8C ▶). The subsequent addition of p14 to the SF3b155–U2AF65 RRM3 complex does not alter the signals that are affected by the addition of U2AF65 RRM3 (Fig. 8B ▶). Moreover, the chemical perturbations observed upon addition of p14 to the SF3b155–U2AF65 RRM3 complex (Fig. 8B,D ▶) are virtually indistinguishable from the effects induced by addition of p14 to SF3b155282–424 alone (cf. Fig. 2A ▶). In ITC experiments, the binding affinity of a 40-residue fragment SF3b155317–357 to U2AF65 RRM3 is KD = 6.3 μM (Table 1 ▶). Thus, the affinity between this fragment of SF3b155 and U2AF65 RRM3 is smaller compared with the interaction with p14. This is also consistent with the significantly smaller chemical-shift perturbations observed in the NMR titration of the larger fragment SF3b155282–424 with U2AF65 RRM3 (Fig. 8 ▶, cf. C and D). Only one ligand consensus motif for binding to U2AF65 RRM3 is present between residues 317 and 357 in SF3b155, indicating that this KSRW338 motif strongly contributes to the binding affinity. Taken together, these data show that the consensus motif encompassing Trp338 of human SF3b155 mediates the U2AF65 interaction and that this region is non-overlapping with SF3b155′s p14 binding site. Our findings are thus compatible with the simultaneous binding to p14 and U2AF65 to SF3b155.
FIGURE 8.
Interaction of SF3b155, U2AF65, and p14. (A) 1H,15N correlation spectrum of 15N-labeled SF3b155282–424 (black) and after addition of U2AF65 RRM3 (red). (B) 1H,15N correlation spectrum of 15N-labeled SF3b155282–424 when bound to U2AF65 RRM3 (black) and after addition of p14 (green). (C) Chemical-shift perturbation for the amide signals of SF3b155282–424 upon addition of U2AF65 RRM3. Residues that could not be assigned in the N-terminal region of SF3b155 are marked by a dot. (D) Chemical-shift perturbation for the amide signals of SF3b155282–424 upon addition of p14. Residues that could not be assigned in the N-terminal region of SF3b155 are marked by a dot.
DISCUSSION
Structural features of the p14–SF3b155 interface
We studied the interaction of the spliceosomal p14 and SF3b155 proteins using biochemical and NMR techniques and analyzed their interaction with single-stranded RNAs comprising an intron branch-point sequence or the branch-site interacting region of U2 snRNA, as well as a branch-site RNA duplex formed by these oligonucleotides. NMR analyses demonstrated that p14 comprises an RRM domain, extended by a central β hairpin and a C-terminal helix αC, which is only found in a subset of RRM folds. We further showed that p14 interacts tightly with SF3b155 and that the binding interface involves the β-sheet surface of the p14 RRM fold. A 43-residue peptide (SF3b381–424) in the N-terminal part of SF3b155 defines a minimal binding epitope for p14, in which residues 401–415 constitute a core binding motif. NMR data revealed that the SF3b155 peptide binds to the p14 RRM in an α-helical conformation, which is partly preformed in the isolated SF3b155 protein.
Weak RNA binding to a noncanonical site of the p14 RRM
Based on available three-dimensional structures of other RRMs, the C-terminal helix αC of p14 presumably packs on top of the β-sheet surface, which is normally involved in RNA interaction in canonical RRM domains (Fig. 6 ▶). Additionally, a helical region in SF3b155 binds to the β sheet surface as indicated by the strongest chemical-shift perturbations for residues located in RNP1 (β3). Thus, the RRM β sheet in the p14–SF3b155 complex is most likely inaccessible for RNA binding. Consistent with this notion, the NMR chemical-shift perturbations map the binding site with RNA ligands to a region flanking the RRM β sheet. While SF3b155 binding induces the largest chemical-shift changes in residues within RNP1 and RNP2, RNA binding mainly affects residues in the β3′ β3” hairpin, especially in the p14–SF3b155 complex. Some caution is advised in the interpretation of the chemical-shift perturbation, since a priori it is not possible to distinguish between chemical shifts induced by direct RNA contacts and indirect effects, which may reflect small conformational changes upon RNA binding. It is possible that the small chemical-shift perturbations observed for residues in RNP1/2 motifs and helix αC upon addition of RNA to the p14–SF3b155 complex (see Fig. 6B ▶) reflect a combination of direct contacts and indirect effects. For example, direct contacts with the RNA at the β3′, β3” hairpin may slightly affect the conformation of the SF3b155 peptide and helix αC, resulting in small chemical-shift changes of residues in helix αC. Only few NMR signals could be analyzed for the unstructured regions flanking the RRM N- and C-terminally. Consistent with the NMR-binding data, UV cross-linking and gel-shift experiments indicated that both p14 and the complex of p14 with either SF3b155282–424 or SF3b1–459 bind only weakly and with little sequence specificity to single-stranded U2 snRNA or BPS RNA oligonucleotides and to duplexes formed between them.
RNA interaction of the p14–SF3b155 complex and SF3b
Taken together, the RNA-binding data show that p14 alone or complexed with SF3b155282–424 or SF3b1551–459 interacts weakly and nonspecifically with nucleic acids. However, NMR data suggest that the binding interface of the p14–SF3b155 complex with a U2 snRNA/BPS RNA duplex is distinct and more localized. Neither cross-linking experiments nor NMR-binding studies indicate that the SF3b155282–424 peptide engages in direct contacts with RNA. Nevertheless, RNA binding monitored by gel-shift experiments was improved when using a protein complex comprising the entire N terminus of SF3b155 (p14–SF3b1551–459). Moreover, RNA cross-links to SF3b155 were observed in the context of the native SF3b particle purified from HeLa cell nuclear extracts and also with the p14–SF3b1551–459 complex. These data suggest that some additional nucleic acid contacts ensue through the extended N terminus of SF3b155, which may explain the observed improvement in RNA-binding affinity. In contrast to the p14–SF3b1551–459 complex, the isolated SF3b particle did not bind DNA, suggesting that additional SF3b components or a different conformation of p14 and/or SF3b155 within this complex enhances RNA-binding specificity. Interestingly, p14 RNA cross-links were observed not only with p14 (or p14 complexed with SF3b155), but also with p14 within purified SF3b complexes. Previous EM studies suggested that p14 is located inside the central cavity of SF3b, where it would be largely inaccessible to the branch-point adenosine (Golas et al. 2003). Our data suggest that some regions of p14 may be accessible to RNA (at least to short oligoribonucleotides), even when p14 is complexed with all other subunits of SF3b.
p14 does not specifically recognize a U2 snRNA/BPS RNA duplex
Maybe the most surprising finding is that neither p14 nor the p14–SF3b155 complex recognize the branch-point RNA helix with high affinity and specificity. As p14 can be cross-linked to the branch-point adenosine during several steps of the pre-mRNA splicing process, from complex A to complex C (Query et al. 1997; Will et al. 2001), it might have been expected that p14 is able to specifically recognize the branch-point helix on its own or when complexed with SF3b155, its main interacting partner. We also found no evidence for improved binding affinity or specificity for a branch-point helix containing a pseudouridine-modified U2 snRNA, likewise indicating that there is no specific recognition of a bulged-out branch-point adenosine. Furthermore, isolated SF3b complexes also failed to recognize the branch-point RNA helix with high affinity and specificity. The apparently higher affinity for a single-stranded U2-type branch point sequence suggests that SF3b components, including p14, could potentially recognize the BPS prior to U2 snRNA base-pairing interaction and thus facilitate U2/BPS duplex formation. It is also possible that the association of SF3b with the U2 snRNP is accompanied by conformational changes in the p14–SF3b155 complex that impart enhanced branch-point helix-binding specificity on the proteins. Indeed, EM studies indicate that SF3b undergoes a significant conformational change upon association with the U11/U12 di-snRNP (Golas et al. 2005). On the other hand, p14 may simply be positioned at the BPS via the interaction of the U2 snRNP with this region of the pre-mRNA. In particular, additional pre-mRNA contacts involving other SF3b proteins as seen in our cross-linking experiments could restrain the BPS such that the branch-point adenosine is appropriately held in place for binding by p14.
Simultaneous binding of SF3b155 to p14 and U2AF65 in complex A
The interaction between U2AF65 and SF3b155 is essential for the recruitment of the U2 snRNP to the branch site (Gozani et al. 1998). We have shown that SF3b155 binds to the third RRM of U2AF65 in vitro, and that this interaction can occur simultaneously with the p14–SF3b155 interaction. Thus, U2AF65, when bound to the Py tract near the 3′-splice site of an intron, can recruit the U2 snRNP via binding to SF3b. The binding surfaces on U2AF65 for SF1 and SF3b155 proteins both involve RRM3 (Gozani et al. 1998) and are most likely structurally similar (Selenko et al. 2003). This suggests that the displacement of SF1 from RRM3 of U2AF65 upon transition of complex E to complex A may contribute to the stable integration of the U2 snRNP into the spliceosome. Interestingly, the region of SF3b155, which interacts with U2AF65 exhibits a number of potential phosphorylation sites (S/T-P-X). This raises the possibility that the U2AF/SF3b interaction might be regulated by phosphorylation.
MATERIALS AND METHODS
Recombinant protein production
A p14 expression construct has been described previously (Will et al. 2001). The expression level of p14 from this construct was variable, especially in labeling experiments. To get reliable yields, the gene was recloned into a modified pET24d vector including a GST carrier and a PreScission protease cleavage site. DNA fragments coding for SF3b155 fragments (SF5b-1551–459, SF3b155282–424, SF3b155282–381, SF3b155311–411, and SF3b155341–424) were PCR amplified from a human cDNA library and inserted via NcoI and Acc65I restriction sites into the expression vector pETM11 (EMBL). These proteins were produced as N-terminally His6-tagged variants with the tags removable via TEV protease cleavage. Transformed bacterial cultures were propagated in selective LB medium, induced by the addition of 1 mM IPTG, and maintained at 20°C overnight before harvesting.
Proteins were initially captured on glutathione–Sepharose beads (p14) or Ni-NTA resin (SF3b155 fragments). After affinity purification, tags were removed by PreScission or TEV protease cleavage. Proteins or complexes were further purified by anion exchange chromatography and gel filtration. Target fractions were identified by SDS-PAGE, pooled concentrated by ultrafiltration, and stored at −80 °C after flash-freezing in liquid nitrogen.
SF5b-1551–459 and SF3b155282–424 were either purified alone or in complex with p14. Other SF3b155 fragments were only produced in complex with p14. For complex production, Escherichia coli cells were transformed with both respective expression vectors. After coexpression, the complexes were copurified, making use of both affinity tags and sequential cleavage by the proteases.
Peptide array binding experiments
In order to delineate the minimal fragment of SF3b155 capable of interacting with p14, we synthesized overlapping 20-mer peptides (18-residue overlap) that covered the entire 258–424 region of human SF3b155. The N-terminally acetylated peptides were assembled as an array of spots tethered by their C-termini to a specially modified ACS-01 cellulose membrane (AIMS Scientific Products GmbH) as described (Frank and Overwin 1996). p14 was cloned into pETM10 (EMBL) via NcoI and Acc65I restriction sites and the N-terminally His6-tagged protein was produced by in vitro transcription/translation using rabbit reticulocyte lysate and [35S]methionine. The peptide array was incubated with the radio-labeled p14 translate, washed as described (Niebuhr et al. 1997), and bound p14 was visualized by autoradiography. The image was scanned with a white light scanner at 300 dpi and analyzed with Phoretix Array software. Integration of circular areas from the centers of the spots yielded the relative binding affinities shown in Figure 2C ▶.
Isothermal titration calorimetry
Thermodynamic parameters for complex formation between p14 and SF3b155282–424 were extracted from isothermal titration calorimetry (ITC) experiments on two MCS-ITC Instruments (MicroCal) at 25°C. Purified protein preparations were adjusted to concentrations of 0.23 mM (p14), 0.005 mM (SF3b155282–424), 0.032 mM (U2AF65), and 0.294 mM (SF3b155317–357). For the SF3b155–p14 interaction, heat releases following 11 injections of p14 of 8 μL each into 1.374 mL of SF3b155282–424 were recorded. For the SF3b155–U2AF65 RRM3 interaction, binding heats following 19 injections of SF3b155317–357 of 15 μL each into 1.414 mL of U2AF65 were measured. The data were analyzed with Microcal Origin 5.0 to extract the enthalpies and entropies of binding (ΔHa and ΔSa, respectively), the equilibrium dissociation constants (KD) and the interaction stoichiometries (n). In control experiments, no heat release was detectable during similar injections of protein or buffer into plain buffer.
UV-induced cross-linking of oligoribonucleotides to p14, p14/SF3b155282–424 and purified SF3b
RNA and DNA oligomers for interaction studies were chemically synthesized in-house or obtained from MWG, respectively. Ten-microliter reactions of preassembled purified protein complex p14–SF3b155282–424 (85 μM) or increasing concentrations (1, 5, and 20 μM) of p14, SF3b155282–424, p14–SF3b155282–424, or BSA were incubated in binding buffer (20 mM Tris-Hcl at pH 6.8, 40 mM NaCl, 2 mM DTT, 0.1 g/L tRNA) with radiolabeled oligonucleotides or preformed duplexes (0.1 μM) at 20°C for 30 min. Ten-microliter reactions containing purified SF3b complex (~0.1 μM) and radioactively labeled oligos or duplexes in buffer (20 mM HEPES-KOH at pH 7.9, 100 mM NaCl, 1.5 mM MgCl2, 5% glycerol, 0.1 g/L tRNA, 0.5 mM DTT, 0.5 mM PMSF) were incubated at 15°C for 20 min. Samples were subjected to UV irradiation at 254 nm for 2 min on a glass slide at 4°C. Cross-links were analyzed by SDS-PAGE and visualized by autoradiography.
Electrophoretic mobility shift assays of the p14–SF3b1551–459 complex
Increasing amounts of p14–SF3b1551–459 protein complex (5–20 μM) were incubated with 32P-labeled oligonucleotides in binding buffer containing 120 mM NaCl at 15°C for 20 min or the times indicated (see Results). Samples were separated on native 6% polyacrylamide gels (with acrylamide to bisacrylamide ratio of 80:1) and visualized by autoradiography.
Purification and nucleic acid binding of native SF3b complexes
Anti-SF3b155 monoclonal antibodies against peptide EQYDP-FAEHRPPKIA (positions 99–113 of SF3b155) were obtained from GBF. The affinity-purified antibodies reacted specifically with the SF3b155 protein on Western blots.
Native SF3b complex was purified from HeLa cell nuclear extract using the anti-SF3b155 monoclonal antibodies covalently coupled to protein A Sepharose essentially as described previously (Will et al. 2001). Native SF3b was incubated in buffer (20 mM HEPES-KOH at pH 7.9, 100 mM NaCl, 1.5 mM MgCl2, 5% glycerol, 0.1 g/L tRNA, 0.5 mM DTT, 0.5 mM PMSF) with radioactively labeled oligos or duplexes on ice for 1 h or at 15°C for 20 min. Complexes formed were analyzed on 4% native polyacrylamide gels and visualized by autoradiography.
NMR spectroscopy and resonance assignments
For NMR studies, expression of p14, SF3b155282–424, and SF3b155381–424 was performed as described above, except that isotope labeling was obtained by expression in minimal medium containing [U-13C]-Glucose and/or 15 NH4Cl as sole carbon and nitrogen sources. The recombinant proteins were purified separately as described above and dissolved in a buffer comprising 20 mM phosphate buffer, 100 mM NaCl, and 2 mM DTT. NMR experiments were performed at 22°C with Bruker DRX600 or DRX500 spectrometers equipped with triple-resonance cryogenic probes and pulsed field gradients. Data were processed with NMRPIPE (Delaglio et al. 1995) and analyzed using NMRVIEW (Johnson and Blevins 1994). Chemical-shift assignments were obtained from standard three-dimensional triple resonance experiments recorded on 15N,13C-labeled samples of p14 and SF3b155282–242 (Sattler et al. 1999). For NMR-binding experiments, typically unlabeled protein or nucleic acid ligands were added to 15N-labeled p14 or SF3b155 peptides, and chemical-shift changes were monitored in two-dimensional 1H,15N correlation spectra. Chemical-shift perturbations are calculated as [Δδ1H)2-Δδ(15N)2]1/2.
Homology modeling
A homology model of the p14 RRM was calculated with the program MODELLER (Marti-Renom et al. 2000) based on a sequence alignment with the N-terminal RRM domain of U1A. The crystal structure of the U1A RRM1 (PDB accession 1FHT) was used as template since it has a high sequence similarity with the p14 RRM and shares the same secondary structure based on the secondary chemical shifts of the p14 RRM, including a β3′, β3” hairpin and a helix αC. Structures and surfaces are displayed using MOLMOL (Koradi et al. 1996).
Acknowledgments
We thank S. Daenicke (GBF, Braunschweig, Germany) for expert help in peptide array synthesis and J. Wehland and C. Erck (GBF, Braunschweig, Germany) for help in generating monoclonal antibodies against SF3b155. R.S. is grateful for a long-term EMBO fellowship. This work was supported by grant Lu 294/12-2 from the Deutsche Forschungsgemeinschaft (DFG) to R.L. and the Max-Planck-Society. M.S. acknowledges support from the DFG, the European Commission (FSG-V-RNA), and EMBL.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2271406.
REFERENCES
- Berglund, J.A., Chua, K., Abovich, N., Reed, R., and Rosbash, M. 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89: 781–787. [DOI] [PubMed] [Google Scholar]
- Berglund, J.A., Abovich, N., and Rosbash, M. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branch-point region recognition. Genes & Dev. 12: 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund, J.A., Rosbash, M., and Schultz, S.C. 2001. Crystal structure of a model branchpoint-U2 snRNA duplex containing bulged adenosines. RNA 7: 682–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosomes. In The RNA World, 2d ed. (R.F. Gesteland et al., eds), pp. 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Delaglio, F., Grzesiek, S., Vuister, G., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX Pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Frank, R. and Overwin, H. 1996. SPOT synthesis. Epitope analysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol. Biol. 66: 149–169. [DOI] [PubMed] [Google Scholar]
- Frilander, M.J. and Steitz, J.A. 1999. Initial recognition of U12-dependent introns requires both U11/5′ splice-site and U12/branchpoint interactions. Genes & Dev. 13: 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golas, M.M., Sander, B., Will, C.L., Lührmann, R., and Stark, H. 2003. Molecular architecture of the multiprotein splicing factor SF3b. Science 300: 980–984. [DOI] [PubMed] [Google Scholar]
- ———. 2005. Major conformational change in the complex SF3b upon integration into the spliceosomal U11/U12 di-snRNP as revealed by electron cryomicroscopy. Mol. Cell 17: 869–883. [DOI] [PubMed] [Google Scholar]
- Gozani, O., Feld, R., and Reed, R. 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes & Dev. 10: 233–243. [DOI] [PubMed] [Google Scholar]
- Gozani, O., Potashkin, J., and Reed, R. 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18: 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, S.L. and Padgett, R.A. 1996. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science 271: 1716–1718. [DOI] [PubMed] [Google Scholar]
- Howe, P.W., Allain, F.H., Varani, G., and Neuhaus, D. 1998. Determination of the NMR structure of the complex between U1A protein and its RNA polyadenylation inhibition element. J. Biomol. NMR 11: 59–84. [DOI] [PubMed] [Google Scholar]
- Johnson, B.A. and Blevins, R.A. 1994. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4: 603–614. [DOI] [PubMed] [Google Scholar]
- Kielkopf, C.L., Rodionova, N.A., Green, M.R., and Burley, S.K. 2001. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106: 595–605. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14: 51–55. [DOI] [PubMed] [Google Scholar]
- Krämer, A. 1996. The structure and function of proteins involved in nuclear pre-mRNA splicing. Annu. Rev. Biochem. 65: 367–409. [DOI] [PubMed] [Google Scholar]
- Krämer, A., Gruter, P., Groning, K., and Kastner, B. 1999. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell. Biol. 145: 1355–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Luyten, I., Bottomley, M.J., Messias, A.C., Houngninou-Molango, S., Sprangers, R., Zanier, K., Krämer, A., and Sattler, M. 2001. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294: 1098–1102. [DOI] [PubMed] [Google Scholar]
- MacMillan, A.M., Query, C.C., Allerson, C.R., Chen, S., Verdine, G.L., and Sharp, P.A. 1994. Dynamic association of proteins with the pre-mRNA branch region. Genes & Dev. 8: 3008–3020. [DOI] [PubMed] [Google Scholar]
- Maris, C., Dominguez, C., and Allain, F.H. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272: 2118–2131. [DOI] [PubMed] [Google Scholar]
- Marti-Renom, M.A., Stuart, A.C., Fiser, A., Sanchez, R., Melo, F., and Sali, A. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29: 291–325. [DOI] [PubMed] [Google Scholar]
- McPheeters, D.S. and Muhlenkamp, P. 2003. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol. Cell. Biol. 23: 4174–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino, L., Guth, S., Bilbao, D., Martinez, C., and Valcarcel, J. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841. [DOI] [PubMed] [Google Scholar]
- Newby, M.I. and Greenbaum, N.L. 2002. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat. Struct. Biol. 9: 958–965. [DOI] [PubMed] [Google Scholar]
- Niebuhr, K., Ebel, F., Frank, R., Reinhard, M., Domann, E., Carl, U.D., Walter, U., Gertler, F.B., Wehland, J., and Chakraborty, T. 1997. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 16: 5433–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, A.A. and Steitz, J.A. 2003. Splicing double: Insights from the second spliceosome. Nat. Rev. Mol. Cell. Biol. 4: 960–970. [DOI] [PubMed] [Google Scholar]
- Query, C.C., Moore, M.J., and Sharp, P.A. 1994. Branch nucleophile selection in pre-mRNA splicing: Evidence for the bulged duplex model. Genes & Dev. 8: 587–597. [DOI] [PubMed] [Google Scholar]
- Query, C.C., Strobel, S.A., and Sharp, P.A. 1996. Three recognition events at the branch-site adenine. EMBO J. 15: 1392–1402. [PMC free article] [PubMed] [Google Scholar]
- Query, C.C., McCaw, P.S., and Sharp, P.A. 1997. A minimal spliceosomal complex A recognizes the branch site and polypyrimidine tract. Mol. Cell. Biol. 17: 2944–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler, M., Schleucher, J., and Griesinger, C. 1999. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. NMR Spectrosc. 34: 93–158. [Google Scholar]
- Selenko, P., Gregorovic, G., Sprangers, R., Stier, G., Rhani, Z., Krämer, A., and Sattler, M. 2003. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell 11: 965–976. [DOI] [PubMed] [Google Scholar]
- Singh, R., Valcarcel, J., and Green, M.R. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173–1176. [DOI] [PubMed] [Google Scholar]
- Staknis, D. and Reed, R. 1994. Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol. Cell. Biol. 14: 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L., Schneider, C., Reed, R., and Lührmann, R. 1999. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 284: 2003–2005. [DOI] [PubMed] [Google Scholar]
- Will, C.L., Schneider, C., MacMillan, A.M., Katopodis, N.F., Neubauer, G., Wilm, M., Lührmann, R., and Query, C.C. 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 20: 4536–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L., Schneider, C., Hossbach, M., Urlaub, H., Rauhut, R., Elbashir, S., Tuschl, T., and Lührmann, R. 2004. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 10: 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., Romfo, C.M., Nilsen, T.W., and Green, M.R. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Patton, J.G., and Green, M.R. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355: 609–614. [DOI] [PubMed] [Google Scholar]
- Zorio, D.A. and Blumenthal, T. 1999. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 5: 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]