Abstract
Although reducing the temperature slows most chemical reactions, freezing can stimulate some reactions by mechanisms that are only partially understood. Here we show that freezing stimulates the self-ligation (circularization) of linear forms of the hairpin ribozyme (HPR) containing 2′,3′-cyclic phosphate and 5′-OH termini. Divalent metal ions (M2+) are not required, but monovalent cations and anions at millimolar concentrations can have various effects on this reaction depending on the specific ion. Under optimal conditions, the observed rate of M2+-independent self-ligation reaches a peak (0.04 min−1) at −10°C with a yield of −60% after 1 h. In contrast, no ligation occurs either at above 0°C or in solutions that remain unfrozen when supercooled to subzero temperatures. Under freezing conditions, the cleavage–ligation equilibrium strongly favors ligation. Besides freezing, evaporation of the aqueous solvent as well as the presence of ethanol at levels of 40% or above can also induce M2+-independent HPR ligation at 25°C. We argue that partial RNA dehydration, which is a common feature of freezing, evaporation, and the presence of ethanol, is a key factor supporting HPR ligation activity at both above- and below-freezing temperatures. In the context of the RNA world hypothesis, freezing-induced ligation is an attractive mechanism by which complex RNAs could have evolved under conditions in which RNA was relatively protected against degradation.
Keywords: hairpin ribozyme, ligation, freezing, dehydration, ethanol, RNA world
INTRODUCTION
If the RNA world, where both information-carrying and catalytic capabilities were fulfilled by self-replicating RNA molecules (Gilbert 1986; Joyce and Orgel 1993), ever existed, the commonly assumed “warm and wet” prebiotic conditions are problematic. Because RNA is chemically unstable and can be quickly cleaved through transesterification (Pace 1991; Vlassov et al. 2004), it is hard to imagine how complex RNAs could have evolved and survived. Moreover, RNA cleavage is accelerated by the presence of divalent metal ions (M2+) (Dallas et al. 2004), which would have been abundant on the early earth (Monnard et al. 2002) and are usually considered as cofactors for RNA catalysis (Fedor 2002; Pyle 2002). Thus, RNA must have evolved under conditions in which either RNA synthetic reactions were efficient enough to outrun random degradation or random degradation was greatly reduced.
Lower temperatures reduce RNA cleavage, both non-enzymatic (Li and Breaker 1999; Dallas et al. 2004) and ribozyme-catalyzed (Tokumoto and Saigo 1992; Feig et al. 1998; Nesbitt et al. 1999), and some investigators have argued that early frozen oceans or lakes would be possible, even favorable, environments for the origin of life (Lazcano and Miller 1996; Levy et al. 1999; Bada and Lazcano 2002). While lowered temperature reduces diffusion and generally slows reactions, the onset of freezing can accelerate some chemical and enzymatic processes that may have relevance to the RNA world. Examples include dehydration of 5-hydro-6-hydroxydeoxyuridine (Prusoff 1963; Butler and Bruice 1964), mutarotation of glucose (Kiovsky and Pincock 1966), tetramerization of hydrogen cyanide to form synthetic precursors of purines (Sanchez et al. 1966; Levy et al. 1999), pyrimidine and purine synthesis from ammonium cyanide (Miyakawa et al. 2002), formation of dinucleotides from adenosine 2′,3′-cyclic phosphate (Renz et al. 1971), polymerization of activated nucleoside 5′-monophospate imidazolides (Stribling and Miller 1991; Monnard et al. 2002; Trinks et al. 2005), and ligation of phosphorimidazolides of short uridine oligomers (Sawai and Wada 2000). All of these reactions occur at temperatures moderately below the freezing point where solutes are not incorporated into the ice but become encapsulated and concentrated in liquid microinclusions between the ice crystals.
A polynucleotide length of at least 60–100 nt would probably be required to provide the necessary coding and catalytic capability for an RNA world (Carothers et al. 2004). The ability to ligate short RNA fragments generated by random polymerization of activated nucleotides would have provided a more efficient means of increasing RNA length and sequence complexity than polymerization alone (Vlassov et al. 2004). Naturally existing ribozymes such as the hairpin ribozyme (HPR) may be relics of the RNA world and may retain some catalytic features of their ancestors (Cech 1993; Hirao and Ellington 1995). The HPR can cleave RNA, generating fragments with 5′-hydroxyl (5′-OH) and 2′,3′-cyclic phosphate termini, and religate those same ends (see Fedor 2000). Previously, several investigators noticed slow spontaneous cleavage of different HPR species at freezing temperatures upon ethanol precipitation (Prody et al. 1986), repeated freezing and thawing (Donahue and Fedor 1997), or in partially hydrated dried films (Seyhan and Burke 2000). No ligation was reported in those studies. However, we have demonstrated that freezing induces M2+-independent ligation in trans by the HPR while disfavoring the reverse, cleavage reaction (Vlassov et al. 2004). To separate the concentration effect of freezing from any other effects, we have examined M2+-independent HPR ligation in cis under subfreezing temperatures, as well as at 25°C upon evaporation of solvent or in the presence of ethanol. This study extends our earlier observation that HPR catalyzes ligation in cis at −21°C, albeit very inefficiently (Kazakov et al. 1998), and provides evidence for the involvement of dehydration in the mechanism of HPR catalysis. HPR ligation in cis was found to be most efficient at −10°C and to be faster than an analogous reaction in trans under single turnover conditions. We argue that partial RNA dehydration, which is common to freezing, evaporation, and the presence of ethanol, is a key factor supporting HPR ligation activity at both above- and below-freezing temperatures.
RESULTS
Freezing induces HPR circularization
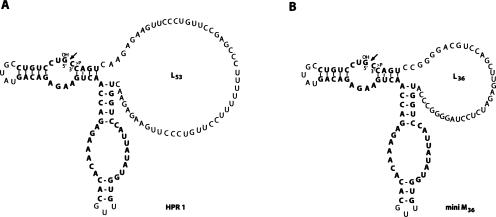
The hairpin ribozyme HPR1 (Fig. 1A ▶) is derived from a previously described minimal hairpin ribozyme, miniM36 (Feldstein and Bruening 1993; Fig. 1B ▶). During in vitro transcription from an appropriate DNA template, the primary transcript, pre-HPR1, undergoes self-processing at the 5′ and 3′ ends, forming semi- and fully processed linear (L) HPR1 species and a ligated, fully processed circular (C) form (Fig. 1A ▶). The circular form migrates in denaturing PAGE more slowly than the L form, as is usual for single-stranded RNAs longer than ~42 nucleotides (Harrison and Zimmerman 1984). The individual HPR species were gel-purified and characterized as previously described (Kazakov et al. 1998; see also Materials and Methods).
FIGURE 1.
Catalytically active structures of fully processed linear forms of hairpin ribozymes HPR1 (A) and miniM36 (B). The catalytic core is shown in bold letters. Self-ligation of the 5′-hydroxyl (5′-OH) and 2′,3′-cyclic phosphate (>p) ends at the arrow produces the circular form.
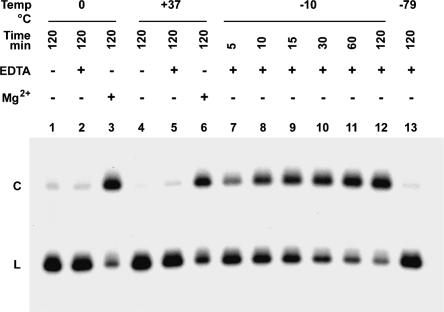
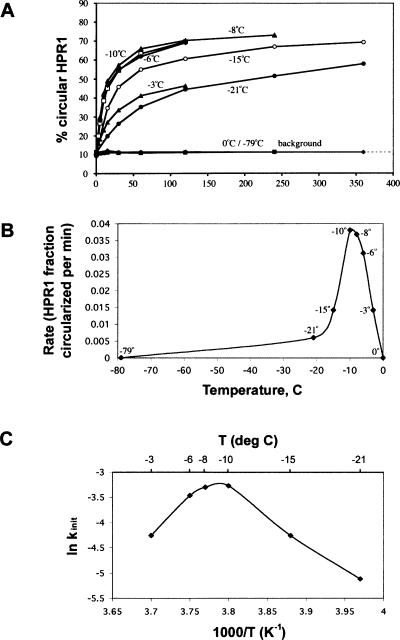
Under solution conditions typically used in studies of the hairpin ribozyme (i.e., 10 mM MgCl2, 40 mM Tris-HCl at pH 8.0), the gel-purified L form of HPR1 was partially converted to the circular form by self-ligation in unfrozen solutions at both 37°C and at 0°C (Fig. 2 ▶, lanes 3,6), but no circularization occurred in either deionized water or in the presence of the M2+-chelator Na3EDTA at pH 8.0 (henceforth referred to as EDTA) (Fig. 2 ▶, lanes 1–2,4–5). To further investigate our previous finding that the L form of HPR1 can undergo slow circularization upon freezing in the absence of M2+ at ~21°C (Kazakov et al. 1998), we measured the kinetics of HPR1 ligation at different subzero temperatures, after quick-freezing to −80°C followed by incubation at the desired temperature (see Materials and Methods). The only ligation product observed was the circular form of HPR1; no multimers were detected. The time course for the self-ligation reaction of radiolabeled HPR1 in the presence of 20 mM EDTA at −10°C is shown in Figure 2 ▶, lanes 7–12. The rate of circularization depends strongly on temperature (Fig. 3A ▶), with the maximum initial rate (kobs = 0.04 min−1) at ~ −10°C (Fig. 3B ▶). The maximum yield (at 24 h) was 65% over background, with the reaction nearly complete (60% yield) after 6 h (Fig. 3A ▶). No circularization was detected at either −79°C or 0°C (Fig. 2 ▶, lanes 2,13; Fig. 3 ▶) even after 24 h.
FIGURE 2.
Effect of Mg2+, EDTA, and freezing on circularization of HPR1. Solutions of the linear form of internally 32P-labeled HPR1 in either deionized H2O (lanes 1,4), 10 mM MgCl2/40 mM Tris-HCl at pH 7.9 (lanes 3,6), or 20 mM Na3EDTA at pH 8.0 (lanes 2,5,7–13) were incubated at various temperatures for the time periods (in minutes) indicated. Samples corresponding to lanes 7–13 were rapidly frozen and then incubated at −10°C (lanes 7–12) or −79°C (lane 13). Samples in lanes 1–6 were not frozen. The reaction products and control samples were analyzed by electrophoresis on a denaturing 6% polyacrylamide gel and quantified by phosphorimaging after subtracting the background level of circular HPR1 formed without freezing (lanes 1,2). L and C are linear and circular forms of HPR1, respectively.
FIGURE 3.
Time and temperature dependencies of HPR1 circularization in frozen solution. Solutions of the linear form of internally 32P-labeled HPR1 in 20 mM Na3EDTA at pH 8.0 were rapidly frozen, then incubated at the indicated temperatures, and analyzed as in Figure 2 ▶. The reaction products were analyzed as in Figure 2 ▶. Each experiment was done in three times, and average values are shown. Standard errors were <10%. (A) Ligation kinetics for various temperatures. (B) Observed initial rate as a function of temperature (based on the 10-min time point from A). (C) Arrhenius plot of the temperature dependence of the initial rate constants (derived from B).
HPR1 circularization did not occur in cases where the initial deep-freezing step (see Materials and Methods) was omitted and the HPR1 solution samples were supercooled to −8°C and −10°C for 1–24 h but did not freeze (data not shown), confirming our earlier conclusions (Kazakov et al. 1998; Vlassov et al. 2004) that freezing is absolutely required (supercooled solutions can remain unfrozen in poly-propylene tubes at temperatures down to −15°C; see Bruice and Butler 1964; Kanavarioti et al. 2001). When frozen solutions of HPR1 were incubated at −8°C and subsequently melted at various temperatures from 4°C to 95°C to provide a range of different melting rates, no variation in the ligation yield was observed (data not shown). Together with the lack of ligation observed for above-zero temperatures, these results indicate that ligation occurred in the frozen solution rather than during subsequent melting.
Temperature effects on the cleavage–ligation equilibrium
When solutions of gel-purified circular HPR1 containing 20 mM EDTA were frozen and incubated at either −8°C or −20°C for various time periods (from 1 to 24 h), very little of the linear form accumulated (<1% of the starting material; data not shown). A similar result was obtained for the cleavage reaction catalyzed by HPR in trans under the same conditions (Vlassov et al. 2004). These results are consistent with the finding that, under standard solution conditions, ligation (both in cis and in trans) is favored over cleavage for this ribozyme under single turnover conditions, and the preference for ligation increases with decreasing temperature over a broad temperature range (Fedor 1999; Nesbitt et al. 1999). This preference can be attributed to increased overall stabilization of the complex between the HPR and its ligation substrates with lower temperature (Esteban et al. 1997; Fedor 1999; Nesbitt et al. 1999). However, whereas in normal solution both ligation and cleavage rates drop with decreasing temperatures (Nesbitt et al. 1999), the absolute ligation rate in frozen solution increased dramatically as the temperature of incubation was lowered from −1°C to −10°C. After peaking at ~−10°C, the rate of the reaction in cis again fell with decreasing temperature (Fig. 3B ▶), as was seen for the reaction in trans, which rate peaks at −8°C (Vlassov et al. 2004). Thus, only for temperatures < −10°C the temperature dependence for HPR1 ligation is consistent with a classical activation barrier. An Arrhenius plot for the freezing ligation reaction (Fig. 3C ▶) in that temperature range gives an approximate value for the activation energy of 11 kcal/mol, which is similar to the range of 13–19 kcal/mol reported for the HPR cleavage and ligation reactions in normal solution (Hampel and Tritz 1989; Fedor 1999; Nesbitt et al. 1999).
Because the yield of circular HPR1 did not increase beyond 65% after 24 h incubation at −8°C, this yield and time point were taken as approximating equilibrium. The sum of the limiting yields for the cleavage and ligation reactions (65%–66%) does not equal 100%, suggesting that the balance of the HPR1 population, about one-third, is trapped in nonreactive conformations in the frozen solutions. Such conformational trapping of HPR in inactive structures has also been observed even at 37°C under normal solution conditions (Feldstein and Bruening 1993; Esteban et al. 1997).
Divalent metal ions are irrelevant for the freezing-induced ligation
The presence of EDTA during gel purification of HPR species as well as in the freezing reaction buffer makes it unlikely that the HPR1 activity was catalyzed by initial trace amounts of divalent metal ions that become concentrated during freezing. However, there remained the possibility that the observed HPR circularization, rather than being truly M2+-independent, was instead catalyzed by divalent metal ions tightly bound to HPR1 RNA. To remove tightly bound divalent metal cations (and to protect the RNA samples against accidental contamination with M2+ from subsequently added components), we adapted a procedure originally described for tRNA (Stein and Crothers 1976). Samples of gel-purified HPR1 species were incubated at 85°C for 10 min in the presence of 10 mM EDTA, cooled to room temperature, and subjected to gel filtration through columns pre-equilibrated with 10 mM EDTA (see Materials and Methods). In subsequent use, EDTA was maintained at a concentration of 10 mM or greater. Assuming that these treatments permit equilibration of M2+ ions between the 10 mM EDTA and the heat-denatured HPR1 (whose estimated concentration in the 32P-labeled samples was <1 nM), and taking into account the known affinities of Mg2+ to EDTA and RNA molecules (for details, see Vary and Vournakis 1984), it is unlikely that M2+ ions would remain bound to HPR1 under these conditions. We found that depletion of Mg2+ by the above procedure had no effect on the HPR1 circularization reaction at either −10°C (this study) or −21°C (Kazakov et al. 1998).
In normal solution, divalent metal ions can either promote or inhibit HPR activity according to whether they stabilize active or inactive conformations (Chowrira et al. 1993; Feldstein and Bruening 1993; Fedor 2000). Lowering the temperature can enhance such conformational trapping. Previously, we showed that 10 μM Mn2+, Co2+, Cu2+, and Zn2+ ions neither inhibited nor promoted circularization of HPR1 at −21°C when added as chlorides to solutions buffered with sodium acetate (pH 6.0) (Kazakov et al. 1998). This concentration (10 μM) is well above what could be expected from any contaminating divalent cations present in salts of monovalent cations at the moderate concentrations (≤150 mM) used in our experiments (Geyer and Sen 1997). In the present study, we examined the influence of Mg2+ at −10°C in acetate-phosphate-borate buffer (APB), and found that the addition of from 1 μM to 1 mM MgCl2 to Mg2+-depleted solutions had no effect on either the rate or the overall yield of the ligation reaction (data not shown). However, at elevated concentrations of Mg2+ (5 and 10 mM), up to 15% more circular HPR1 was observed than without added Mg2+ (data not shown). The additional ligation product may have resulted from reaction in solution prior to freezing.
Salt and pH effects on the freezing ligation reaction
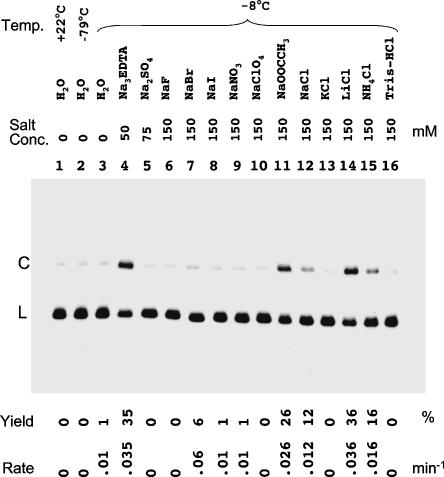
In contrast to divalent cations, salts of monovalent cations were found to have significant influence on freezing-induced circularization of HPR1, either stimulatory or inhibitory, depending on the type of counterion. A minimum level of certain salts is required for optimal HPR ligation activity under freezing conditions for both the cis (Kazakov et al. 1998) and trans reactions (Vlassov et al. 2004). In this study we examined these salt effects in greater detail and identified salt compositions that provide optimal HPR1 activity. The observed ligation rate for HPR1 (measured at −8°C at pH 8.0) increases with sodium ion concentration, reaching a plateau at ~ 20 mM Na+ when complemented by an APB anion mixture, or at 60 mM Na+ in the form of Na3H-EDTA (Fig. 4A ▶). Among the salts tested (at a cation ion concentration of 150 mM), the sodium salts of APB, EDTA, and acetate along with LiCl were most stimulatory of freezing ligation. NaCl was moderately stimulatory, and NaF, NaBr, NaI, NaNO3, and NaClO4 either had no effect on or inhibited the ligation reaction (Fig. 5 ▶). Among various chlorides of monovalent cations, the order of efficacy for freezing-induced circularization of HPR1 was LiCl > NH4Cl > NaCl >> KCl ~ Tris-Cl (Fig. 5 ▶). Tris-HCl tested across the concentration range 1–50 mM and the pH range 7.2–8.5 failed to support freezing ligation. Moreover, addition of Tris-HCl (pH 8.0) to HPR solutions containing 30 mM Na3EDTA or 100 mM LiCl (salts which otherwise strongly support freezing ligation) was inhibitory, with 50 mM Tris-HCl reducing the ligation yield by ~95% (Fig. 4B ▶). The fact that 40–50 mM Tris-HCl is commonly used as a buffer in studies of HPR catalysis under normal solution conditions might explain why ligation has not been routinely observed when HPR solutions were frozen.
FIGURE 4.
Effect of monovalent cation concentration on the initial rate of HPR1 circularization at −8°C. Solutions of linear, internally 32P-labeled HPR1 containing the indicated salts in the presence of 1 mM Tris-EDTA at pH 8.0, were rapidly frozen, then incubated for 10 min at −8°C and analyzed as in Figure 3 ▶. (A) Stimulatory effect of Na+ ions tested either in the form of Na3EDTA at pH 8.0 (▴) or Na-APB at pH 8.0 (▪), where APB is the universal buffer mixture of acetate, phosphate, and borate. (B) Inhibitory effect of Tris-HCl at pH 8.0 in the presence of either 30 mM Na3EDTA at pH 8.0 (▴) or 100 mM LiCl (▪).
FIGURE 5.
Effect of different cations and anions on the initial rate of HPR1 circularization at −8°C. Solutions of linear, internally 32P-labeled HPR1 containing 1 mM Tris-EDTA at pH 8.0 and the indicated salts were rapidly frozen (with the exception of the sample in lane 1), then incubated for 10 min at the indicated temperatures and analyzed as in Figure 2 ▶. Lanes 4–16 all contain in total 150 mM of the indicated cation.
To examine the effect of pH on the freezing ligation reaction in a relatively uniform ionic environment, we used the “universal” buffer APB, which functions over a wide range of pH, with a constant Na+ concentration of 60 mM for all pH values tested. The initial rate of HPR1 ligation upon freezing increases with pH, rising about fourfold over the pH range 4.5–9.0 (data not shown). A similar pH effect is seen for HPR ligation in the normal solution reaction at 25°–37°C (Hampel and Tritz 1989; Nesbitt et al. 1997; Fedor 1999), although direct comparison of pH in normal and in frozen solutions is not possible due to the different solubilities of various salt and buffer components at subzero temperatures as well as the dependence of pH on temperature, buffer concentration, and water activity (Butler and Bruice 1964; Fink and Geeves 1979).
Effect of initial RNA concentration on the freezing ligation reaction
Freezing-induced concentration of the RNA may either promote HPR ligation through formation of active intermolecular complexes or inhibit this reaction because of RNA aggregation and precipitation (see Discussion). We found that addition of nonradioactive HPR1 transcript to gel-purified 32P-labeled, linear HPR1 prior to freezing resulted in a dose-dependent inhibition of the formation of circular HPR1 upon freezing (data not shown). This inhibition was as much as 50% at 0.3 mg/mL HPR1 for incubation at −8°C for 10 min. A similar inhibitory effect was seen upon addition of an unrelated RNA (unfractionated tRNA) instead of HPR1. The only ligation product observed in these experiments was circular HPR1; no multimers were detected.
Ethanol induces Mg2+-independent HPR ligation in unfrozen solution
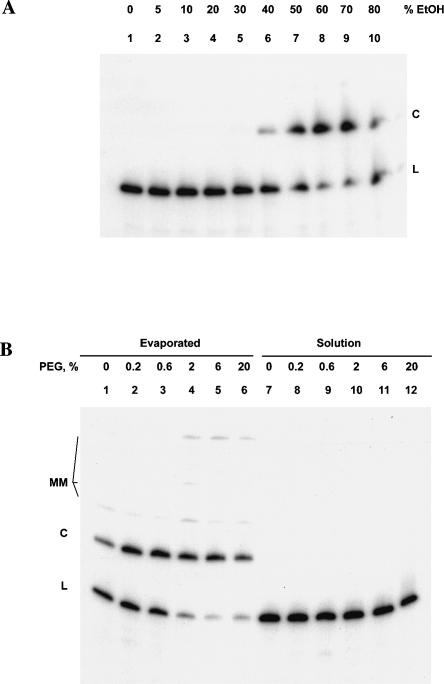
To see if conditions of reduced water activity unrelated to freezing would promote similar Mg2+-independent circularization of HPR1, we first studied water–ethanol mixtures. In the absence of Mg2+, we found that the presence of ethanol at concentrations above ~40% induces very rapid circularization (t1/2 < 1 min) of 32P-labeled HPR1 at 25°C in the presence of 25 mM sodium acetate (pH 6.0) (Fig. 6A ▶). The reaction occurs equally well in the presence or the absence of 10 mM EDTA (data not shown), further demonstrating that the ethanol-induced ligation reaction does not require Mg2+ ions. The yield of circular HPR1 is maximal (~70%) in 60% ethanol, falling off at higher alcohol concentrations perhaps due to rapid aggregation and precipitation of the RNA. In contrast to the freezing reactions, the dissolved salts do not undergo concentration or precipitation in aqueous solutions containing ≤70% ethanol at 25°C (Fink and Geeves 1979).
FIGURE 6.
Dehydration-induced HPR1 circularization without freezing. Solutions of linear, internally 32P-labeled HPR1 in 25 mM Na-acetate and 10 mM Na3EDTA at pH 8.0 were incubated for 1 h at 25°C under the conditions indicated and analyzed as in Figure 2 ▶. (A) Effect of ethanol. (B) (Lanes 1–6) Effect of evaporation under vacuum of solution samples containing the indicated concentrations of poly(ethylene glycol). (Lanes 7–12) Same as 1–6 but without evaporation. MM, putative multimers.
Evaporation also induces Mg2+-independent HPR ligation
To further examine whether low water activity (or dehydration) was the key to Mg2+-independent ligation, we evaporated the water from dilute aqueous solutions of linear HPR1 containing 25 mM sodium acetate and 10 mM EDTA (pH 8.0). Evaporation was carried out under vacuum for 1 h at 25°C while making sure that there was no freezing of the samples. When the resulting film-like residue was dissolved in water and analyzed by PAGE, circular HPR1 was seen along with slower migrating species that were presumed to be multimeric products of intermolecular ligation (Fig. 6B ▶, lanes 1–6). This is in contrast to HPR1 ligation reactions induced both by freezing (Figs. 2 ▶, 5 ▶) and ethanol (Fig. 6A ▶), where no such multimeric species were observed. The total yield of ligation products was greatly increased by the presence of 2%–6% poly(ethylene glycol) (PEG) (Fig. 6B ▶, lanes 4–6) in the HPR1 solutions before evaporation. However, PEG did not promote HPR ligation in solutions that had not been evaporated, frozen, or mixed with ethanol (Fig. 6B ▶, lanes 7–12). The ability of PEG to exclude water and induce molecular crowding may promote circularization and multimerization of HPR1 upon evaporation in the same way that it promotes ligation of oligoribonucleotides by T4 RNA ligase under normal solution conditions (Harrison and Zimmerman 1984). PEG also may slow the rate of evaporation (Prestrelski et al. 1993), thus prolonging the period during which water activity is at a level conducive to catalysis. The idea that HPR ligation is favored by a low but nonzero water activity is consistent with the observation of Seyhan and Burke (2000) that HPR samples that were completely dried and maintained under vacuum showed little or no catalytic activity. However, in the hydrated films they observed Mg2+-independent HPR cleavage and not ligation, and the reaction was very slow (Kobs ~ 2 d−1). In contrast, we found that the ligation reaction under vacuum was substantially complete within 1 h after all the solvent appeared to have evaporated.
DISCUSSION
Our key conclusion from the results presented is that lowered water activity leading to partial RNA dehydration is a likely explanation for the divalent metal-independent stimulation of HPR ligation by freezing, solvent evaporation, and addition of ethanol. Because concentration of solutes has been the favored mechanism of freezing-enhanced catalysis in most studies in the past, we examine below how concentration could also play a role. We then discuss whether this reaction might have relevance to other ribozymes and to prebiotic evolution.
What role does concentration of RNA or salts play in freezing-induced ligation?
As the temperature is lowered below the freezing point, water molecules crystallize into ice while the concentrations of solutes in the remaining liquid phase increase. The final concentration of RNA and salts in the unfrozen liquid inclusions depends on the freezing temperature, the specific nature of those solutes, and their initial (prefreezing) concentration. The concentration effect can account for the increase in rate of intermolecular reactions as well as the decrease in rate caused by the addition of salts, buffers, and other solutes that lower the freezing point and thereby limit the extent to which RNA is concentrated (Pincock 1969; Vajda 1999).
The circularization of the HPR (ligation in cis) is a unimolecular reaction, and its rate is therefore not expected to be concentration-dependent. However, there is the formal possibility that the concentrating effects of freezing promote the formation of dimeric complexes of HPR1 that can circularize by a bimolecular mechanism in which the catalytic domain of one molecule catalyzes intramolecular ligation in a separate molecule. There is some evidence for such a process. Freezing can indeed promote the formation of nucleic acids complexes (Zimmerman and Coleman 1972; Elghanian et al. 1997). Also, complementation experiments with mutant HPRs that fail to self-ligate (due to their inability to fold properly) but were catalytically active in intermolecular complexes suggest the possibility of catalysis in trans in the normal solution reaction (Feldstein and Bruening 1993; Komatsu et al. 1993). However, we found instead that either increasing the concentration of HPR1 or addition of unrelated RNA before freezing had inhibitory effects on ligation. These results, although not conclusive, argue that HPR1 circularization in frozen solution is indeed a unimolecular reaction, and suggest that increased HPR1 concentration is not the factor stimulating its catalytic activity in the frozen solutions. One possible explanation for the inhibiting effect of added RNA is the increased formation of inactive RNA aggregates or precipitates upon freezing. Alternatively, if the ligation reaction is promoted by HPR1 contact with the surface of ice or precipitated salt crystals, the added RNA may compete with HPR1 for surface binding sites.
If concentration of RNA does not account for freezing-induced ligation of HRP1, what about concentration of salts? Our results demonstrate that salts of monovalent cations have significant influence on freezing-induced circularization of HPR1. Millimolar levels of certain cations (e.g., Na+) are required for optimal HPR1 ligation upon freezing (see Fig. 4A ▶). The concentration of these cations should dramatically increase upon freezing, with lower temperatures causing greater concentration in liquid micro-inclusions. For example, freezing of NaCl solutions results in NaCl concentrations in unfrozen liquid inclusions of ~1 M at −3°C, ~2 M at −6°C, ~2.7 M at −9°C, ~4 M for −15°C, reaching ~5 M at the eutectic point (−21°C) (Dean 1985; Stribling and Miller 1991). Depending on the initial concentration, salts can be concentrated >1000-fold by freezing at appropriate temperatures (Kiovsky and Pincock 1966).
It is known that high concentrations (>1.5 M) of monovalent cations can substitute for M2+ in supporting the catalytic activity of the HPR under normal solution conditions (Nesbitt 1997; Murray et al. 1998; Scott 1999). This apparent stabilization of the active conformation by concentrated monovalent salts raises the question of whether the same mechanism is at work under freezing conditions, with freezing-induced concentration providing the high monovalent cation levels. However, most salts (including those used in our experiments) are not concentrated by the presence of 40%–70% ethanol (Fink and Geeves 1979), whereas such levels induce rapid circularization of HPR1 (Fig. 6A ▶, lanes 7–9). Our results therefore suggest that it is the dehydrating effect, produced by highly concentrated salts (Anderson and Record 1995; Barciszewski et al. 1999) or ethanol, that is the key factor promoting HPR activity, rather than electrostatic shielding.
Dehydration results in displacement of water from RNA molecules (Kuntz et al. 1969). The first hydration shell of an RNA molecule, whose water molecules interact primarily with hydrogen bond donor and acceptor groups of nucleotide residues, plays an important role in RNA structure (Westhof et al. 1988; Rozners and Moulder 2004). Both intramolecular folding and intermolecular interactions require that competing water molecules be removed before nucleotide residues can form hydrogen-bonding interactions with other residues (Israelachvili and Wennerström 1996). How much dehydration occurs depends on the degree to which salts are concentrated, which in turn depends on the solubility of the salt in question under freezing conditions. It is not clear whether solubility differences can account for the fact that both cations and anions can influence the degree of HPR1 circularization in frozen solution. For example, different sodium salts provide dramatically different stimulating effects on the ligation reaction (Fig. 5 ▶, lanes 4–12), as do different chlorides (Fig. 5 ▶, lanes 12–16). Other possible roles of salts in this reaction include preventing the solution phase (which seems to be required for the reaction) from completely freezing through freezing point lowering, and preventing concentration of the RNA molecules to the point of aggregation or precipitation.
Dehydration of RNA by concentrated salts could also play the dominant role in the enhancement of HPR ligation upon evaporation of the aqueous solvent (Fig. 6B ▶). In the case of ethanol, dehydration is believed to be the mechanism by which it affects the conformation and stability of nucleic acids (Vollenweider et al. 1978; Beneventi and Onori 1986). For example, ethanol can stabilize folded RNA molecules by promoting the formation of A-form helices (and thereby loop structures defined by adjacent helical segments), which are less hydrated than unstructured single strands (Beneventi and Onori 1986; Hanna and Szostak 1994). It was previously demonstrated that ethanol at concentrations <30% could significantly increase the Mg2+-dependent activity of different ribozymes in normal (unfrozen) solution, whereas ≥40% ethanol in the presence of Mg2+ diminishes this activity, apparently owing to rapid aggregation of the RNA (Gardiner et al. 1985; Hanna and Szostak 1994; Feig et al. 1998). In the absence of Mg2+, a condition which should reduce the tendency for aggregation, the optimum enhancement of catalysis occurs at >40% ethanol. Because higher starting concentrations of RNA, which could accelerate aggregation, are inhibitory rather than stimulatory in the freezing reaction, aggregation is probably not favorable to catalysis. Thus, it appears that lowered water activity leading to dehydration is the key factor in the divalent cation-independent catalysis induced by freezing, evaporation, and alcohol addition.
Is freezing-induced ligation unique to the hairpin ribozyme?
In general, the size and the sequence of the loops flanking the HPR core such as L53 in HPR1 (see Fig. 1A ▶) can affect the propensity of the ribozyme to fold into catalytically active conformations as well as the equilibrium between its ligated and its cleaved forms (Feldstein and Bruening 1993; Hegg and Fedor 1995; Komatsu et al. 1995). However, we have observed efficient freezing-induced circularization of other HPR derivatives that were catalytically active under normal solution conditions. These include ribozymes having inserts of various sequences and sizes in the loops (e.g., miniM36) (Fig. 1B ▶) or sequence alterations in the catalytic core (data not shown). These results, as well as the fact that HPR derivatives lacking such loops retain the ability to ligate in trans under freezing conditions (Vlassov et al. 2004), suggest that Mg2+-independent ligation induced by freezing is a general feature of catalytically active HPR derivatives. It is important to note that HPR ligation under freezing conditions yields normal 3′-5′ internucleotide linkages (Vlassov et al. 2004).
It remains to be seen whether ribozymes unrelated to the HPR also possess M2+-independent freezing-induced ligation capabilities. The proportion of time the termini spend in an orientation allowing ligation to proceed will affect the position of equilibrium between ligation and cleavage. For example, with small ribozymes that are relatively floppy, cleavage is often entropically favored in normal solution conditions. Under conditions that stabilize an appropriate orientation of the ends, ligation may prevail over cleavage (Tokumoto and Saigo 1992; Stage-Zimmermann and Uhlenbeck 2001). The freezing environment could provide such stabilization. Indeed, heavily fragmented and mutated versions of the HPR that are inactive under normal solution conditions can efficiently ligate RNA fragments (in trans) in frozen solution (Vlassov et al. 2004) but cleave only slowly. Given that there is likely to be more than one way in which RNA–RNA interactions can align and orient these reacting termini to catalyze their ligation (Usher and McHale 1976; Sharmeen et al. 1989), there may be other ribozymes besides the HPR capable of ligating 2′,3′-cyclophosphate and 5′-hydroxyl RNA ends under freezing conditions. The hammer-head ribozyme seems not to be one of them, since the rate of ligation in trans (which is much slower than cleavage in normal solution conditions) is not stimulated in frozen, M2+-free solutions (Tokumoto and Saigo 1992; data not shown). In vitro selection methods, which were successfully used for the isolation of ribozymes ligating 3′-OH and 5′-ppp termini under normal solution conditions (Ekland et al. 1995; Joyce 2002), may be the best approach to finding new ribozyme ligases that are active in frozen solutions.
Possible significance for prebiotic evolution
The finding that ribozyme catalysis, particularly ligation yielding natural 3′-5′ phosphodiester internucleotide linkages (Vlassov et al. 2004), can occur in frozen solutions provides a possible “missing link” for the RNA world hypothesis. Previously we suggested that freezing-induced HPR ligation might be an example of a primitive synthetic reaction that could increase RNA length and complexity by combining short fragments generated by other processes (Vlassov et al. 2004). One such process might have been the nonenzymatic oligomerization of nucleoside 2′,3′-cyclic phosphates, which may have been among the most abundant chemically activated nucleotide derivatives in the prebiotic world (Verlander and Orgel 1974; Usher and Yee 1979).
Besides building up size and sequence complexity, the ligation reaction might have also contributed to the integrity of RNA-encoded genetic information during prebiotic evolution through its ability to create circular RNAs. Circular RNAs have several features that could have been useful at an early stage of evolution: (1) They remain in one piece after a single random cleavage event, allowing recovery of their structure through religation of their ends; (2) they can serve as a template for rolling circle replication and amplification; and (3) they have fewer conformational degrees of freedom compared to their linear counterparts, and therefore can bind to other molecules (or fold) with less entropic cost.
MATERIALS AND METHODS
Preparation of DNA templates and transcription
A 161-bp DNA encoding a T7 promoter and the sequence of pre-HPR1 was assembled from two synthetic oligodeoxynucleotides (overlapping where underlined): 5′-TGACAGTCCTGTCCGTATG ACAGAGAAGTCAACCAGAGAAACACACGTTGTGGTATATTACCTGGTCAAGAGAAG-3′ (75-mer) and 5′-AAACAGGACGGTC AGTTCTCTTCAAGGGACAAGGCTCGGGAAAAAAAGGAACAG GGAACTTCTCTTGACCAGG-3′ (73-mer), which were obtained from Midland Certified Reagent Co. The overlapped single strands were filled in by using the Klenow fragment of DNA polymerase (Invitrogen), and the resulting double-stranded DNA fragment was simultaneously extended and amplified by the polymerase chain reaction (PCR) using the oligodeoxynucleotide primers 5′-GGCTCGAATTCTAATACGACTCACTATAGGGTGACAGTCCTG TCCG-3′ (46-mer) and 5′-AAACAGGACGGTCAGTTC-3′ (18-mer) (Midland). This DNA template was then transcribed using T7 RNA polymerase (Promega) in the presence of either [α-32P]-labeled (NEN/Dupont) and/or unlabeled CTP (0.013 mM and 0.5 mM for radioactive and nonradioactive synthesis, respectively), as well as 0.5 mM GTP, ATP, and UTP (all NTPs from Sigma) in a buffer containing 40 mM Tris-HCl (pH 7.9), 6 mM MgCl2, 10 mM DTT, and 2 mM spermidine at 37°C.
Purification of linear and circular HPR1 forms
The transcription products were mixed with equal volumes of 2 × FLS (92% formamide, 10 mM EDTA, and 0.02% each of xylene cyanol and bromphenol blue), heated for 2 min at 95°C, and resolved by electrophoresis in denaturing 6% polyacrylamide gels containing 45 mM Tris-borate (pH 8.3), 1 mM EDTA, and 8 M urea. The individual RNA bands, corresponding to the unprocessed original transcript and products of self-processing, were localized by autoradiography or UV shadowing, excised, and extracted by 1 × TE buffer (10 mM Tris-HCl, 1 mM EDTA at pH 8.0). These RNA species were identified as previously described (Kazakov et al. 1998) using methods adapted from others (Buzayan et al. 1986; Saville and Collins 1991). The 117-nt linear form of HPR1 monomer (Fig. 1A ▶) spontaneously underwent partial conversion (≤10%) to its circular form during extraction from the gel. The use of 1 × TE provided less conversion than other elution buffers tested, such as 0.5 M ammonium acetate/1 mM EDTA/0.1% sodium dodecyl sulfate. The opposite process, partial (≤10%) circular-to-linear conversion during extraction of HPR from denaturing polyacrylamide gels, has been previously reported (Feldstein and Bruening 1993). However, no further interconversion was seen if the extracted HPR1 species were maintained in the elution buffer or in deionized water without freezing. After elution from the gel, the linear and the circular HPR1 species were desalted by gel filtration through MicroSpin G-25 columns (Amersham-Pharmacia Biotech) that had been prewashed with 1 mM EDTA and deionized water to remove residual M2+ ions. For the same purpose, nonradioactive HPR1 samples and total tRNA from Escherichia coli (Boehringer-Mannheim) were preincubated with 10 mM EDTA for 10 min at 85°C and subjected to gel-filtration as described above.
Reaction procedures and data analysis
To study the ligation of linear HPR1, 20-μL samples of the gel-purified, 32P-labeled linear form of HPR1 were incubated under various reaction conditions, including freezing, addition of ethanol, and evaporation of solvent under vacuum. To avoid variations in the rate of freezing of different samples due to incubation at different temperatures, we applied a quick freezing technique (Kazakov et al. 1998; Vlassov et al. 2004) that is an adaptation of methods previously described by others (Bruice and Butler 1964; Zimmerman and Coleman 1972). Briefly, the solution samples were frozen in 1.5-mL Eppendorf microcentrifuge tubes by immersing in a dry ice bath (~79°C) for 3 min. They were then transferred to a temperature-controlled ethanol–water bath (MGW Lauda RC3 Model B-2, Brinkman) for incubation at the desired (subfreezing) temperature. After incubation, the frozen samples were quickly melted in a 65°C bath for 1 min to stop the reactions. In a separate set of experiments, various proportions of ethanol were added, and the solutions were incubated at 25°C for either 1 min or 1 h (see figure legends for details). In yet another set of experiments, the solution samples were dried under vacuum on a Speed-Vac concentrator (Savant Instrument) for 1 h at 25°C with or without PEG 8000 (Sigma), and then dissolved in 20 μL of deionized water. At the end of the reactions, all samples were mixed with equal volumes of 2 × FLS, heated for 2 min at 95°C, and analyzed by denaturing electrophoresis through 6% polyacrylamide gels. Reaction products were quantified using a Molecular Imager FX (Bio-Rad) using its Molecular Analyst 2.1 software for Macintosh.
Reagents and buffers
Ultrapure absolute ethanol was from Quantum Chemical. All other chemicals were A.C.S. grade from J.T. Baker. Solutions were prepared using deionized, RNase-free water from Research Genetics. Stock solutions of a universal buffer mixture (APB) covering the pH range from 4.5 to 9.0 in 0.5 pH unit increments were prepared by titrating a mixture containing 40 mM acetic acid, 40 mM H3PO4, and 40 mM boric acid with 200 mM NaOH to the desired pH (Dean 1985). The concentration of Na+ in each universal buffer solution was then adjusted to 78 mM by the addition of an appropriate amount of 200 mM NaCl.
Acknowledgments
We thank Drs. Sidney Altman, Boris Belotserkovskii, Anthony Forster, Laura Landweber, Leslie Orgel, Norman Pace, Attila Seyhan, and Alexander Vlassov for useful comments on early versions of this manuscript. Special thanks to Alexander Vlassov for help in the manuscript editing. This research was partially supported by NASA grant no. NAG5-10904 and NSF grant no. MCB-0085627 to B.J.
NOTE ADDED IN PROOF Freezing-induced catalysis and the RNA world are discussed in a recent review (Vlassov et al. 2005a). Another article describing intermolecular ligation catalyzed by the HPR in solutions containing several alcohols has been accepted for publication (Vlassov et al. 2005b).
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2123506.
REFERENCES
- Anderson, C.F. and Record Jr., M.T. 1995. Salt-nucleic acid interactions. Annu. Rev. Phys. Chem. 46: 657–700. [DOI] [PubMed] [Google Scholar]
- Bada, J.L. and Lazcano, A. 2002. Origin of life. Some like it hot, but not the first biomolecules. Science 296: 1982–1983. [DOI] [PubMed] [Google Scholar]
- Barciszewski, J., Jurczak, J., Porowski, S., Specht, T., and Erdmann, V.A. 1999. The decisive role of the water structure in changes of conformation of nucleic acids. Acta Biochim. Pol. 46: 133–144. [PubMed] [Google Scholar]
- Beneventi, S. and Onori, G. 1986. Effect of ethanol on the thermal stability of tRNA molecules. Biophys. Chem. 25: 181–190. [DOI] [PubMed] [Google Scholar]
- Bruice, T.C. and Butler, A.R. 1964. Catalysis in water and ice. II. The reaction of thiolactones with morpholine in frozen systems. J. Am. Chem. Soc. 86: 4104–4108. [Google Scholar]
- Butler, A.R. and Bruice, T.C. 1964. Catalysis in water and ice. A comparison of the kinetics of hydrolysis of acetic anhydride, β–propiolactone, and p-nitrophenyl acetate and the dehydration of 5-hydro-6-hydroxy-deoxyuridine in water and ice. J. Am. Chem. Soc. 86: 313–319. [Google Scholar]
- Buzayan, J.M., Gerlach, W.L., and Bruening, G. 1986. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature 323: 349–353. [Google Scholar]
- Carothers, J.M., Oestreich, S.C., Davis, J.H., and Szostak, J.W. 2004. Informational complexity and functional activity of RNA structures. J. Am. Chem. Soc. 126: 5130–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech, T.R. 1993. The efficiency and versatility of catalytic RNA: Implications for an RNA world. Gene 135: 33–36. [DOI] [PubMed] [Google Scholar]
- Chowrira, B.M., Berzal-Herranz, A., and Burke, J.M. 1993. Ionic requirements for RNA binding, cleavage, and ligation by the hairpin ribozyme. Biochemistry 32: 1088–1095. [DOI] [PubMed] [Google Scholar]
- Dallas, A., Vlassov, A.V., and Kazakov, S.A. 2004. Principles of cleavage of nucleic acids by metal ions. In Artificial nucleases (ed. M.A. Zenkova), Nucleic acids and molecular biology (ed. H.J. Gross), Vol. 13, pp. 61–88. Springer-Verlag, Heidelberg, Germany.
- Dean, J.A., ed. 1985. Lange’s handbook of chemistry, 13th ed. McGraw- Hill, New York.
- Donahue, C.P. and Fedor, M.J. 1997. Kinetics of hairpin ribozyme cleavage in yeast. RNA 3: 961–973. [PMC free article] [PubMed] [Google Scholar]
- Ekland, E.H., Szostak, J.W., and Bartel, D.P. 1995. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269: 364–370. [DOI] [PubMed] [Google Scholar]
- Elghanian, R., Storhoff, J.J., Mucic, R.C., Letsinger, R.L, and Mirkin, C.A. 1997. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science 277: 1078–1081. [DOI] [PubMed] [Google Scholar]
- Esteban, J.A., Banerjee, A.R., and Burke, J.M. 1997. Kinetic mechanism of the hairpin ribozyme. Identification and characterization of two non-exchangeable conformations. J. Biol. Chem. 272: 13629–13639. [DOI] [PubMed] [Google Scholar]
- Fedor, M. 1999. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry 38: 11040–11050. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Structure and function of the hairpin ribozyme. J. Mol. Biol. 297: 269–291. [DOI] [PubMed] [Google Scholar]
- ———. 2002. The role of metal ions in RNA catalysis. Curr. Opin. Struct. Biol. 12: 289–295. [DOI] [PubMed] [Google Scholar]
- Feig, A.L., Ammons, G.E., and Uhlenbeck, O.C. 1998. Cryoenzymology of the hammerhead ribozyme. RNA 4: 1251–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldstein, P.A. and Bruening, G. 1993. Catalytically active geometry in the reversible circularization of ‘mini-monomer’ RNAs derived from the complementary strand of tobacco ringspot virus satellite RNA. Nucleic Acids Res. 21: 1991–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink, A.L. and Greeves, M.A. 1979. Cryoenzymology: The study of enzyme catalysis at subzero temperatures. Methods Enzymol. 63: 336–370. [DOI] [PubMed] [Google Scholar]
- Gardiner, K.J., Marsh, T.L., and Pace, N.R. 1985. Ion dependence of the Bacillus subtilis RNase P reaction. J. Biol. Chem. 260: 5415– 5419. [PubMed] [Google Scholar]
- Geyer, C.R. and Sen, D. 1997. Evidence for the metal-cofactor independence of an RNA phosphodiester-cleaving DNA enzyme. Chem. Biol. 4: 579–593. [DOI] [PubMed] [Google Scholar]
- Gilbert, W. 1986. The RNA world. Nature 319: 618. [Google Scholar]
- Hampel, A. and Tritz, R. 1989. RNA catalytic properties of the minimum (–)sTRSV sequence. Biochemistry 28: 4929–4933. [DOI] [PubMed] [Google Scholar]
- Hanna, M. and Szostak, J.W. 1994. Suppression of mutations in the core of the Tetrahymena ribozyme by spermidine, ethanol and by substrate stabilization. Nucleic Acids Res. 22: 5326–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, B. and Zimmerman, S.B. 1984. Polymer-stimulated ligation: Enhanced ligation of oligo-and polynucleotides by T4 RNA ligase in polymer solutions. Nucleic Acids Res. 12: 8235–8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegg, L.A. and Fedor, M.J. 1995. Kinetics and thermodynamics of intermolecular catalysis by hairpin ribozyme. Biochemistry 34: 15813–15828. [DOI] [PubMed] [Google Scholar]
- Hirao, I. and Ellington, A.D. 1995. Re-creating the RNA world. Curr. Biol. 5: 1017–1022. [DOI] [PubMed] [Google Scholar]
- Israelachvili, J. and Wennerström, H. 1996. Role of hydration and water structure in biological and colloidal intertactions. Nature 379: 219–225. [DOI] [PubMed] [Google Scholar]
- Joyce, G.F. 2002. The antiquity of RNA-based evolution. Nature 418: 214–221. [DOI] [PubMed] [Google Scholar]
- Joyce, G.F. and Orgel, L.E. 1993. Prospects for understanding the origin of the RNA world. In The RNA world (eds. R.F. Gesteland and J.F. Atkins), pp. 1–25. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kazakov, S.A., Balatskaya, S.V., and Johnston, B.H. 1998. Freezing-induced self-ligation of the hairpin ribozyme: Cationic effects. In Structure, motion, interaction and expression of biological macromolecules (eds. R.H. Sarma and M.H. Sarma), Vol. 2, pp. 155–161. Adenine Press, Albany, NY.
- Kiovsky, T.E. and Pincock, R.E. 1966. The mutarotation of glucose in frozen aqueous solutions. J. Am. Chem. Soc. 88: 4704–4710. [Google Scholar]
- Komatsu, Y., Koizumi, M., Sekiguchi, A., and Ohtsuka, E. 1993. Cross-ligation and exchange reactions catalyzed by hairpin ribozymes. Nucleic Acids Res. 21: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, Y., Kanzaki, M., Koizumi, A., and Ohtsuka, E. 1995. Modification of primary structures of hairpin ribozymes for probing active conformations. J. Mol. Biol. 252: 296–304. [DOI] [PubMed] [Google Scholar]
- Kuntz, I.D., Brassfield, T.S., Law, G.D., and Purcell, G.V. 1969. Hydration of macromolecules. Science 163: 1329–1331. [DOI] [PubMed] [Google Scholar]
- Lazcano, A. and Miller, S.L. 1996. The origin and early evolution of life: Prebiotic chemistry, the pre-RNA world, and time. Cell 85: 793–798. [DOI] [PubMed] [Google Scholar]
- Levy, M., Miller, S.L., and Oró , J. 1999. Production of guanine from NH(4)CN polymerizations. J. Mol. Evol. 49: 165–168. [DOI] [PubMed] [Google Scholar]
- Li, Y. and Breaker, R.R. 1999. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 121: 5364–5372. [Google Scholar]
- Miyakawa, S., Cleaves, H.J., and Miller, S.L. 2002. The cold origin of life: B. Implications based on pyrimidines and purines produced from frozen ammonium cyanide solutions. Orig. Life Evol. Biosph. 32: 209–218. [DOI] [PubMed] [Google Scholar]
- Monnard, P.A., Apel, C.L., Kanavarioti, A., and Deamer, D.W. 2002. Influence of ionic inorganic solutes on self-assembly and polymerization processes related to early forms of life: Implications for a prebiotic aqueous medium. Astrobiology 2: 139–152. [DOI] [PubMed] [Google Scholar]
- Murray, J.B., Seyhan, A.A., Walter, N.G., Burke, J.M., and Scott, W.G. 1998. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 5: 587–595. [DOI] [PubMed] [Google Scholar]
- Nesbitt, S., Hegg, L.A., and Fedor, M.J. 1997. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol. 4: 619–630. [DOI] [PubMed] [Google Scholar]
- Nesbitt, S.M., Erlacher, H.A., and Fedor, M.J. 1999. The internal equilibrium of the hairpin ribozyme: Temperature, ion and pH effects. J. Mol. Biol. 286: 1009–1024. [DOI] [PubMed] [Google Scholar]
- Pace, N.R. 1991. Origin of life—Facing up to the physical setting. Cell 65: 531–533. [DOI] [PubMed] [Google Scholar]
- Pincock, R.E. 1969. Reactions in frozen systems. Acc. Chem. Res. 4: 97– 103. [Google Scholar]
- Prestrelski, S.J., Arakawa, T., and Carpenter, J.F. 1993. Separation of freezing- and drying-induced denaturation of lyophilized proteins using stress-specific stabilization. II. Structural studies using infrared spectroscopy. Arch. Biochem. Biophys. 303: 465–473. [DOI] [PubMed] [Google Scholar]
- Prody, G.A., Bakos, J.T., Buzayan, J.M., Schneider, I.R., and Bruening, G. 1986. Autolytic processing of dimeric plant virus satellite RNA. Science 231: 1577–1580. [DOI] [PubMed] [Google Scholar]
- Prusoff, W.H. 1963. Low-temperature reversal of the ultraviolet photochemical reaction product of 2′-deoxyuridive. Biochim. Biophys. Acta 68: 302–310. [DOI] [PubMed] [Google Scholar]
- Pyle, A.M. 2002. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 7: 679–690. [DOI] [PubMed] [Google Scholar]
- Renz, M., Lohrmann, R., and Orgel, L.E. 1971. Catalysts for the polymerization of adenosine cyclic 2′, 3′-phosphate on a poly(U) template. Biochim. Biophys. Acta 240: 463–471. [DOI] [PubMed] [Google Scholar]
- Rozners, E. and Moulder, J. 2004. Hydration of short DNA, RNA and 2′-OMe oligonucleotides determined by osmotic stressing. Nucleic Acids Res. 32: 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, R., Ferris, J., and Orgel, L.E. 1966. Condition for purine synthesis: Did prebiotic synthesis occur at low temperatures? Science 153: 72–73. [DOI] [PubMed] [Google Scholar]
- Saville, B.J. and Collins, R.A. 1991. RNA-mediated ligation of selfcleavage products of a Neurospora mitochondrial plasmid transcript. Proc. Natl. Acad. Sci. 88: 8826–8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai, H. and Wada, M. 2000. Nonenzymatic template-directed condensation of short-chained oligouridylates on a poly(A) template. Orig. Life Evol. Biosph. 30: 503–511. [DOI] [PubMed] [Google Scholar]
- Scott, W.G. 1999. RNA structure, metal ions, and catalysis. Curr. Opin. Chem. Biol. 3: 705–709. [DOI] [PubMed] [Google Scholar]
- Seyhan, A.A. and Burke, J.M. 2000. Mg2+-independent hairpin ribozyme catalysis in hydrated RNA films. RNA 6: 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage-Zimmermann, T.K., and Uhlenbeck, O.C. 2001. A covalent cross-link converts the hammerhead ribozyme from a ribonuclease to an RNA ligase. Nat. Struct. Biol. 8: 863–867. [DOI] [PubMed] [Google Scholar]
- Stein, A. and Crothers, D.M. 1976. Conformational changes of transfer RNA. The role of magnesium(II). Biochemistry 15: 160–168. [DOI] [PubMed] [Google Scholar]
- Stribling, R. and Miller, S.L. 1991. Template-directed synthesis of oligonucleotides under eutectic conditions. J. Mol. Evol. 32: 289–295. [DOI] [PubMed] [Google Scholar]
- Tokumoto, Y. and Saigo, K. 1992. RNA–RNA and RNA–DNA ligation with the sTobRV(+) hammerhead ribozyme. Nucleic Acids Symp. Ser. 27: 21–22. [PubMed] [Google Scholar]
- Trinks, H., Schroder, W., and Biebricher, C.K. 2005. Ice and the origin of life. Orig. Life Evol. Biosph. 35: 429–445. [DOI] [PubMed] [Google Scholar]
- Usher, D.A. and McHale, A.H. 1976. Nonenzymatic joining of oligoadenylates on a polyuridylic acid template. Science 192: 53–54. [DOI] [PubMed] [Google Scholar]
- Usher, D.A. and Yee, D. 1979. Geometry of the dry-state oligomerization of 2′,3′-cyclic phosphates. J. Mol. Evol. 13: 287–293. [DOI] [PubMed] [Google Scholar]
- Vajda, T. 1999. Cryo-bioorganic chemistry: Molecular interactions at low temperature. Cell. Mol. Life Sci. 56: 398–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary, C.P.H. and Vournakis, J.N. 1984. RNA structure analysis using T2 ribonuclease: Detection of pH and metal ion induced conformational changes in yeast tRNAPhe. Nucleic Acids Res. 12: 6763–6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlander, M.S. and Orgel, L.E. 1974. Analysis of high molecular weight material from the polymerization of adenosine cyclic 2′, 3′-phosphate. J. Mol. Evol. 3: 115–120. [DOI] [PubMed] [Google Scholar]
- Vlassov, A.V., Johnston, B.H., Landweber, L.F., and Kazakov, S.A. 2004. Ligation activity of fragmented ribozymes in frozen solution: Implications for the RNA world. Nucleic Acids Res. 32: 2966–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov, A.V., Kazakov, S.A., Johnston, B.H., and Landweber, L.F. 2005a. The RNA world on ice: A new scenario for the emergence of RNA information. J. Mol. Evol. 61: 264–273. [DOI] [PubMed] [Google Scholar]
- Vlassov, A.V., Johnston, B.H., and Kazakov, S.A. 2005b. Hairpin ribozyme-catalyzed ligation in water-alcohol solutions. Oligonucleotides (in press). [DOI] [PubMed]
- Vollenweider, H.J., James, A., and Szybalski, W. 1978. Discrete length classes of DNA depend on mode of dehydration. Proc. Natl. Acad. Sci. 75: 710–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof, E., Dumas, P., and Moras, D. 1988. Hydration of transfer RNA molecules: A crystallographic study. Biochimie 70: 145–165. [DOI] [PubMed] [Google Scholar]
- Zimmerman, S.B. and Coleman, N.F. 1972. An ordered precipitate of polyadenylic acid formed by freezing at acidic pH. Biopolymers 11: 1943–1960. [DOI] [PubMed] [Google Scholar]