Abstract
Polypyrimidine tract binding protein (PTB or hnRNP I) has several known functions in eukaryotic cells, including exon exclusion during alternative splicing events, mRNA stabilization, and regulation of viral translation and replication. PTB contains four RNA Binding Domains (RBDs, or RRMs), all of which can potentially bind RNA, but their roles in the various biological functions of PTB are not clear. We investigate the properties of the complexes formed by human PTB1 on two target RNAs: the rat GABAA receptor γ2 subunit pre-mRNA and the Hepatitis C Virus 3′ NonTranslated RNA. The GABA RNA contains four polypyrimidine tracts in the intron and exon, while the HCV NTR contains a 75-nt U-rich tract and a highly structured 3′-terminus. Electrophoretic mobility shift assays show that PTB1 protein first binds to both RNAs with nanomolar affinities, but subsequent protein addition leads to formation of higher-order complexes. Stoichiometry experiments show that the ultimate complexes contain up to eight PTB1 proteins per RNA strand. Protein constructs containing two tandem RBDs also bind the two RNAs, but with different affinities and stoichiometries. Nuclease protection assays show that PTB1 protects the polypyrimidine tracts in the GABA RNA, as does a construct consisting of RBD3 and RBD4; however, a construct containing RBD1 and RBD2 enhances cleavage of bound RNA. The binding mechanisms of PTB1 are unique to the full-length protein; these modes appear to include direct association with the RNA as well as weaker intermolecular protein associations.
Keywords: PTB1, polypyrimidine tract binding protein, HCV RNA, GABA pre-mRNA
INTRODUCTION
Polypyrimidine tract binding protein (PTB, also known as hnRNP I) is ubiquitously expressed in eukaryotic cells, and found in both the nucleus and cytoplasm. It functions in pre-mRNA alternative splicing, where it acts as a regulator of 3′-splice site selection (Valcarcel and Gebauer 1997; Wagner and Garcia-Blanco 2001; Black 2003). PTB protein appears to act most often as a splicing repressor (resulting in exon exclusion). The mechanism of repression by PTB is not clear, and is complicated by the presence of three PTB isoforms (PTB1, PTB2, and PTB4) (Wollerton et al. 2001) and specific cellular proteins, all of which are differentially expressed in tissues and developmental stages of the cell.
PTB is a modular protein, with four noncanonical RNA binding domains (RBDs, or RNA Recognition Motifs, RRM) that are tethered together by conserved linker domains. There are NMR structures of all four RBDs (Conte et al. 2000; Simpson et al. 2004). RBD1 and RBD4 have the standard β αβ β αβ secondary structure and adopt an α/β sandwich tertiary fold with a four-stranded antiparallel β-sheet, characteristic of RBDs. RBD2 and RBD3 have an unusual structure, for they have a fifth β-strand connected by a loop that passes over the surface of the β-sheet. Since many RBDs use the β-sheet as the RNA-binding surface, the presence of this loop could impede access by the RNA, especially in RBD2, where the loop appears in close proximity to a portion of the β-sheet (Simpson et al. 2004). It is not known if the RBDs function independently or synergistically, nor is it understood how they are positioned with respect to each other in the free protein or on an RNA substrate. Back et al. (2002) report that during poliovirus infection, the viral protease 3CDpro cleaves PTB within the linker between RBD2 and RBD3; the PTB isoforms are cleaved at different times during infection, and with different efficiencies. This separation of PTB into two fragments suggests that each could be used as independent units by the virus, and perhaps also by the cell.
As models of PTB interactions with different alternatively spliced mRNAs, Wagner and Garcia-Blanco (2001) illustrate schemes for PTB association with α-tropomyosin, α-actinin, GABAA γ 2, and c-src. Where possible, their schemes incorporate other proteins that have been implicated in specific exon inclusion, but all models assume that PTB binds directly to RNA, then PTB:PTB interactions stabilize a complex, creating what these authors call a “zone of silencing”; that is, refractory to splicing. Models of exon exclusion propose either that PTB proteins bound to polypyrimidine tracts in flanking introns interact with each other to position the excluded exon in a loop; or, that PTB proteins bind in a long linear arrangement to cover both intron and exon. A recent study of c-src N1 exon regulation by PTB demonstrated that not only were flanking binding sites required for exon repression, but downstream weak binding sites were required for conversion to a multimeric complex (Amir-Ahmady et al. 2005). The length, composition, and location of polypyrimidine tracts in alternatively spliced RNAs greatly vary, and will certainly lead to different numbers of PTB proteins bound, as well as different conformations of the complexes. The idea that several PTB proteins bind to these pre-mRNAs to form a large complex is common to most models, but the specific interactions between RNA and RBDs and the sites of protein:protein interaction remain to be elucidated.
PTB does not function only in alternative splicing, and does not exclusively reside in the nucleus. Among many functions (Wagner and Garcia-Blanco 2001), it has been shown to stabilize insulin mRNA (Tillmar and Welsh 2002; Fred and Welsh 2005), interact with the EMCV (encephalomyocarditis virus) Internal Ribosome Entry Site (IRES) (Witherell et al. 1993), and, in HCV (hepatitis C virus) is variously reported as required for translation and replication (Ali and Siddiqui 1995; Chung and Kaplan 1999; Ito and Lai 1999; Anwar et al. 2000; Tischendorf et al. 2004). A single model of complex formation may not account for the properties of all PTB:RNA complexes.
In this work, two RNAs that are normally bound by PTB are used as model systems to investigate the properties of the complexes. One RNA is the GABAA receptor γ 2 pre-mRNA intron/exon; the other is the 3′-NTR (NonTranslated Region) of HCV. These RNAs differ in the sequence and length of their polypyrimidine tracts, and thus represent two potentially different binding modes of the protein.
Alternative splicing of the GABAA receptor γ 2 subunit (Ashiya and Grabowski 1997) is modulated in part by interactions between PTB and pre-mRNA including sequences within both intron and exon. The sequence of the rat GABAA pre-mRNA is shown in Figure 1A ▶. Like many alternatively spliced pre-mRNAs, it has a complex primary sequence around its 3′-splice site. In addition to the (U/C)11 polypyrimidine tract between the 3′-splice site and the branch-point A, there is another upstream: a (U/C)34 tract, separated from (U/C)11 by an An sequence (n = 15 in rat). Additional (U/C)7 and (U/C)9 tracts occur in the exon. Secondary structure predictions of GABA RNA show the intron to be predominantly single stranded (Fig. 1B ▶), but the exon could have significant secondary structure, including a UUCG tetraloop.
FIGURE 1.
(A) The GABAA receptor γ 2 pre-mRNA used in binding and stoichiometry experiments. A is the branchpoint adenosine; AG102/CU is the 3′-splice site at the intron/exon junction; AG127/GU is the 5′-splice site at the exon/intron junction. Putative UCUU PTB-binding sites are in bold, and the polypyrimidine tracts mentioned in the text are framed. (B) Predicted lowest free-energy secondary structure (bioweb.pasteur.fr/seqanal/tmp/mfold) of the GABA γ 2 intron/exon sequence used in these experiments. Note the presence of a UUCG tetraloop at nucleotide G110.
HCV is a positive-strand RNA virus, and contains a complex 3′-NTR that is required for replication (Kolykhalov et al. 1996, 2000; Friebe and Bartenschlager 2002). Kolykhalov et al. (1996) characterized the 3′-NTR sequence, and showed that it could be subdivided: a hypervariable region with no sequence conservation; a variable length polypyrimidine tract (typically 70–120 nt); and an invariant sequence (“X”) (Fig. 2B ▶). PTB is clearly important to HCV in the cell: in Huh7 cells (a human hepatocyte cell line) transfected with the HCV replicon, the endogenous PTB goes from the nucleus to the cytoplasm, where it colocalizes with the viral NS5B protein (Domitrovich et al. 2005). PTB has been reported to bind to both the U-rich polypyrimidine tract (Gontarek et al. 1999) and to X RNA (Tsuchihara et al. 1997), and is thought to regulate replication (Domitrovich et al. 2005). Because the HCV 3′-NTR contains predominantly uridines, it represents another class of PTB-binding sites, and therefore the binding, orientation, and global topology of PTB proteins could be quite different from other RNA sequences.
FIGURE 2.
(A) HCV 3′-NTR sequence. NS5B sequences (in brackets), X sequences (underlined), and the different X RNA constructs used in these experiments. Putative UCUU PTB-binding sites are in bold. (B) Predicted lowest free-energy secondary structure (bioweb.pasteur.fr/seqanal/tmp/mfold) of the HCV 3′-NTR sequence.
Here, we compare binding affinity, stoichiometry, and footprinting of full-length PTB1 protein and its three tandem RBD constructs (PTB1:12, PTB1:23, and PTB1:34) to the 3′-NTR of HCV and to a construct of the rat GABAA receptor γ 2 pre-mRNA intron/exon.
RESULTS
PTB1 and GABAA receptor γ2 pre-mRNA intron/exon
The GABAA receptor γ 2 (GABA) intron/exon (I/E) RNA contains ~75 nt of the upstream intron including the branch site, all of the 24-nt alternatively spliced exon, and 29 nt of the downstream intron (Ashiya and Grabowski 1997).
EMSA assays
Binding to GABA RNAs by PTB1 and three PTB1 constructs (PTB1:12, PTB1:23, and PTB1:34) was observed using electrophoretic mobility shift assays (EMSA) in native 8% polyacrylamide gels, run at 4° C. An increasing concentration of protein was added to a constant picomolar RNA concentration in a binding buffer containing 100 mM NaCl, 1 mM MgCl2, 10 mM sodium cacodylate (pH 7.0), 20 μg/mL BSA, and 10 μg/mL tRNA. Experiments were also done in 50 mM NaCl and 250 mM NaCl; binding was observed in both conditions, but to prevent nonspecific association at the low salt, and gel artifacts at the higher salt, standard conditions of 100 mM NaCl were used throughout the experiments.
All PTB1 constructs bound to the GABA Intron/Exon (I/E) RNA, albeit with different affinities, as shown in Figure 3 ▶. Note that while bands are fairly well-resolved in the PTB1:RNA gel, the bands from PTB1:34 binding are more smeared, suggesting that the complexes are either dissociating during electrophoresis, or that these complexes are heterogeneous, with PTB1:34 binding at different positions on the RNA. Such smearing made quantification difficult, since bands could not be cleanly separated; the same behavior was observed in EMSA experiments with c-src RNA and PTB1:34 by Simpson et al. (2004). EMSA experiments of Liu et al. (2002) also showed binding of PTB1 and PTB1:12 to GABA I/E RNA (there called rGγ 57Bam), but they did not observe binding by PTB1:34, using a virtually identical protein construct. Perhaps the low crosslinking ratio of their gel (80:1) did not allow good resolution of these diffuse bands.
FIGURE 3.
PhosphorImager EMSA data for binding to GABAA receptor γ 2 intron/exon (I/E) RNA; 100 mM NaCl, 1 mM MgCl2, 10 mM sodium cacodylate (pH 7), 20 μg/mL BSA, and 10 μg/mL tRNA.
Because there are potentially four independent polypyrimidine tracts in GABA RNA, the sequence was subdivided for binding experiments, in order to understand how each sequence contributed to the total complex (Table 1 ▶). Binding of PTB1 and its constructs to these RNAs is summarized in Table 2 ▶. Because most titrations produced several (super)shifted bands, for comparison, the protein concentration is given at which the first shifted band was observed.
TABLE 1.
GABA RNA constructs
| I/E ---(A)15—A---□-- | 5′-AUUUGUCUUAUUUUGUUUCUCUUUCUCUCCUUUCCUUUUCCUUCUUCUUAUUAAAAAAAAA AAAAAACUACGCAAUUCUCUUUUCUGUCUACAAAUCCAAAG/CUUCUUCGGAUGUUUUCCUUCA AG/GUAUACUGUUUUUGGAAUGGGCAUUUCACG-3′ |
| I ----(A)15 —A--- | 5′-AUUUGUCUUAUUUUGUUUCUCUUUCUCUCCUUUCCUUUUCCUUCUUCUUAUUAAAAAAAAAA AAAAACUACGCAAUUCUCUUUUCUGU-3′ |
| I34 ----(A)15 | 5′-AUUUGUCUUAUUUUGUUUCUCUUUCUCUCCUUUCCUUUUCCCUCUUCUUAUUAAAAAAA AAAAAAAACUA-3′ |
| I11—[ | 5′-GGCGCAAUUCUCUUUUCUGUCUACAAAUCCAAAG/GUAAGU-3′ |
| E44---□ | 5′-GGACAAAUCCAAAG/CUUCUUCGGAUGUUUUCCUUCAAG/GUAAGU-3′ |
| E □ | 5′-AG/CUUCUUCGGAUGUUUUCCUUCAAG/GUAAGU-3′ |
| I11/E --A—□ | 5′-GGACUACGCAAUUCUCUUUUCUGUCUACAAAUCCAAAG/CUUCUUCGGAUGUUUUCCUUCAAG/GUAAGU-3′ |
I11/Em--A—
|
5′-GGACUACGCAAUUCUCUUUUCUGUCUACAAAUCCAAAG/CUUCUUCAGAUGUUUUCCUUCAAG/GUAAGU-3′ |
GABA RNA constructs used in EMSA and stoichiometry experiments. I/E is the Intron/Exon; I34 contains the intron (U/C)34 tract and A15; I11/E contains the intron (U/C)11 and the exon; I11 contains the intron (U/C)11 polypyrimidine tract; E is the exon; E44 contains extra nucleotides from the intron; I11/Em contains a G→A mutation in the UUCG tetraloop.
TABLE 2.
EMSA binding of PTB1 constructs to GABA RNAs
| I/E | I | I34 | I11 | I11/E | Exon | I11/Em | |
| PTB1 | 2 nM (4) | 1 nM (3) | 1 nM (3) | 3 nM (1) | 15 nM (3) | 0 | 3 nM (3) |
| PTB1:12 | 7 nM (3) | 70 nM (3) | 16 nM (2–3) | 0 | ~500 nM (1) | 0 | 190 nM (2) |
| PTB1:23 | 90 nM (3) | 450 nM (1–2) | nd | 0 | 90 nM (2) | 0 | nd |
| PTB1:34 | 1 nM (4) | 5 nM (2)* | 8 nM (3) | 84 nM (1) | 4 nM (3–4) | 0 | 4 nM (>3)* |
Protein concentrations are given that correspond to the first appearance of a shifted band on the gel; they are not dissociation constants. (nd) Not determined; (0) no binding observed at protein concentrations from nanomolar to 3μM. Numbers in parentheses are the number of discrete shifted bands observed. (*) Bands very diffuse, with no clear boundaries.
An unexpected result was that there was no binding observed to the exon (E) alone, by any PTB1 construct, despite the presence of two polypyrimidine tracts. The UCUU sequence, which is thought to be a high-affinity binding site (Perez et al. 1997b), is part of the (U/C)7 tract, which in the secondary structure prediction is part of a UUCG tetraloop structure; apparently the remaining UUUUCCUUC sequence [the (U/C)9 tract] in the exon is insufficient for stable PTB1 binding, since an RNA oligo- nucleotide consisting of the (U/C)9 sequence (plus a 5′-gggac T7 polymerase start sequence) was not bound by PTB1 in EMSA experiments (data not shown). This exceptionally stable UUCG element could sequester the (U/C)7 site from PTB, and therefore was removed by mutation of its UUCG to UUCA in the background of the I11/E RNA construct. This mutant I11/Em RNA was bound by PTB1 and PTB1:12 proteins at three times lower protein concentrations than was the wild-type I11/E, while PTB1:34 binding properties did not change, implying that PTB binding is sensitive to RNA secondary structure.
Appearance of a first shifted band in EMSA experiments provides a qualitative indicator of the affinity of a PTB protein to RNA. As summarized in Table 2 ▶, PTB1:23 binding is weakest, followed by PTB1:12; PTB1:34 binding often shows a shift at a concentration similar to that of PTB1. In an attempt to rescue the weaker binding of PTB1:34 to the I11 RNA, the two PTB constructs, PTB1:34 and PTB1:12, were combined in an equimolar mixture, then added to the RNA; complexes were observed by EMSA. The band shifts observed were nearly identical to those of PTB1:34 alone, indicating that the addition in trans of the two halves of the protein could not increase PTB1:34 affinity (data not shown).
Estimation of apparent dissociation constants
To obtain a semiquantitative value of the binding affinity of PTB1 and its constructs to the various RNAs, the EMSA data were analyzed assuming that each band represented a distinct complex, but without any assumption of the stoichiometry. Bands were separated and quantified using PhosphorImaging software, and the RNA fraction bound in each complex as a function of PTB concentration was used as input to fit to a partition function for association (Origin 7.5; OriginLab, Northampton, MA) (Table 3 ▶). Semiquantitative analysis of EMSA data could not be performed with all complexes, and only those experiments with sufficient data points to measure initial complex formation were analyzed. In the fitting protocol, equilibrium association constants (K) and Hill coefficients (n) were allowed to float simultaneously, recognizing that these parameters are correlated. The values of n are interpreted to indicate that a binding event is cooperative when ni > 1, independent (noncooperative) when ni = 1, and anticooperative when ni < 1. (Goodness of fit of the function was determined by eye to agreement with the data points, using Occam’s razor to limit the number of parameters.) Because the number of bound proteins corresponding to each band shift is unknown, these values are apparent KAs and provide a relative comparison of the different PTB constructs and RNAs.
TABLE 3.
Dissociation constants for PTB1 constructs: GABA RNAs complexes
| Protein | I | I34 | I11 | I11/E |
| PTB1 |
KD1 = 5 ± 1 nM, n1 = 2 KD2 = 130 ± 5 nM, n2 = 1 |
KD1 = 2 ± 1 nM, n1 = 1 KD2 = 30 ± 5 nM, n2 = 1 |
KD = 15 ± 2 nM, n = 1.5 |
KD1 = 30 ± 5 nM, n1 = 0.5 KD2 = 100 ± 15 nM, n2 = 1.5 |
| PTB1:12 |
KD1 = 50 ± 5nM, n1 = 1 KD2 = 300 ± 20 nM, n2 = 1 |
KD1 = 50 ± 7 nM, n1 = 1 KD2 = 160 ± 10 nM, n2 = 1.5 |
||
| PTB1:34* |
KD1 = 40 ± 5 nM, n1 = 1.5 KD2 = 150 ± 10 nM, n2 = 3 |
KD = 50 ± 3 nM, n = 3 |
KD1 = 10 ± 3 nM, n1 = 0.5 KD2 = 50 ± 5 nM, n2 = 1 KD3 = 1 mM, n3 = 2.5 |
Data were obtained by allowing both n and K to vary simultaneously. (*) The values for I11/E are approximate, since resolution of individual bands in the gel was difficult. Only one band was observed for I11; 100 mM NaCl, 1 mM MgCl2, 10 mM sodium cacodylate (pH 7), 20 μg/mL BSA, 10 μg/mL tRNA, 4° C.
PTB1 binding to the GABA intron (I) RNA, with its two polypyrimidine tracts (U/C)11 and (U/C)34, gives three bands in EMSA experiments, only the first two of which were analyzed (the third band is seen at 2.5 μM [PTB1]) (see Fig. 4 ▶). The data were best fit by the expression
FIGURE 4.
Determination of the apparent binding constant for PTB1 and PTB1 constructs binding to GABAA receptor γ 2 intron RNA. (A,B,E,F) PhosphorImager EMSA data for PTB1, PTB1:12, PTB1:23, and PTB1:34, respectively, binding to GABA I RNA. Note that EMSA data for PTB1:23 show nonspecific association with large complexes in the wells of the gel. EMSA data for PTB1:34 do not show distinct bands. (C) Dissociation constants for PTB1 were calculated from the fit of the EMSA data to the partition function P = 1 + ( K1[P])n1 + (K1K2[P]2)n2, where n is the Hill coefficient and the Ks are the association constants, using Origin 7.5. (D) Dissociation constants for PTB1:12 were calculated from the fit of the EMSA data to the partition function P = 1 + ( K1[P])n1 + (K1K2[P]2)n2 (K1K22 [P]2)n3
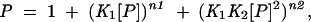 |
where K1 and K2 are association constants for the two bound species, and ni is the Hill coefficient. The first binding is characterized by a dissociation constant KD1 = 5 ± 1 nM, with n1 = 2, indicating that this is a cooperative transition. The second band corresponds to noncooperative binding with KD2 = 130 ± 5 nM. PTB1:12 binding to GABA I RNA produces three bands in EMSA experiments. Fitting these data was best done using the partition function
 |
where KD1 = 50 ± 5 nM, n1 = 1, and KD2 = 300 ± 20 nM, n2 = 1. In contrast to PTB1, binding by PTB1:12 does not exhibit cooperativity, indicating that its binding mechanism differs from that of the full-length protein.
GABA I RNA is bound by both PTB1:23 and PTB1:34. As shown in Figure 4E ▶, PTB1:23 binding is quite weak, and complicated by formation of complexes that do not enter the gel. These data were not analyzed further. In contrast, PTB1:34 binding to GABA I is characterized by broad and smeary bands (Fig. 4F ▶), making quantification unreliable. In toto, binding to the GABA I RNA serves to illustrate apparent variations in binding modes and affinities of each PTB1 construct.
PTB1 binding to GABA I11/E [containing the (U/C)11 tract in the intron and two shorter tracts in the exon] also showed three bands, but for analysis, only the two most rapidly migrating species were analyzed (data not shown; Table 3 ▶). Binding was best fit with the expression
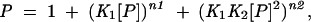 |
where KD1 = 30 ± 5 nM, n1 = 0.5, and KD2 = 100 ± 15 nM, n2 = 1.5. PTB1:34 binding is characterized by three or perhaps four diffuse bands in the gel. Binding data were analyzed assuming three complexes, using the function
 |
where KD1 = 10 ± 3 nM, n1 = 0.5, and KD2 = 50 ± 5 nM, n2 = 1, and KD3 = 1 μM, n3 = 2.5.
Formation of these complexes appears to occur via a first event that is anticooperative, followed by subsequent cooperative binding with lower affinity. Anticooperative binding is observed only on this RNA, and suggests that the exon sequences are somehow responsible.
The GABA I34 RNA [containing (U/C)34 and (A)15] is bound by PTB1, PTB1:12, and PTB1:34 proteins. Of the three bands observed upon PTB1 binding, only the two faster mobility complexes were analyzed (the third band at highest protein concentration is in the well) using the partition function
 |
The data were fit with KD1 = 2 ± 1 nM, n1 = 1, and KD2 = 30 ± 5 nM, n2 = 1. PTB1:34 binding to I34 RNA shows three resolved bands; data were fit with KD1 = 40 ± 5 nM, n1 = 1.5, and KD2 = 150 ± 10 nM, n2 = 3, indicating cooperative binding mechanisms for PTB1:34. PTB1:12 bound I34 RNA with affinities comparable to PTB1:34; weak binding is cooperative (n1 = 1.5), but tight binding is not. Despite the similar values of dissociation constants, PTB1:34 bound all the I34 RNA at 600 nM protein, whereas free RNA was still observed at 1.2 μM PTB1:12.
PTB1 and PTB1:34 binding to GABA I11 give only one band in EMSA experiments (Fig. 5 ▶). PTB1:12 does not detectably bind this RNA. PTB1 data were fit to P = 1 + (K[P])n, where KD = 15 ± 2 nM and n = 1.5, indicating that binding is tight and cooperative. Using the same expression for PTB1:34 affinity gives a highly cooperative binding event (n = 3) but a weaker dissociation constant (KD = 50 ± 3 nM). Again, these data indicate that PTB constructs exhibit distinct binding mechanisms.
FIGURE 5.
Determination of the apparent binding constant for PTB1 and PTB1:34 binding to GABA I11 RNA. (A,B) PhosphorImager EMSA data for PTB1 and PTB1:34, respectively. Dissociation constants for (C) PTB1 and (D) PTB1:34 were calculated from the fit of the EMSA data to the partition function P = 1 + (K[P])n.
Binding stoichiometry
The overall stoichiometry of the complex was determined using nitrocellulose filter binding assays, where the RNA concentration in an experiment was held constant while the protein was titrated. For these experiments, it is important that the RNA concentration be at or above the apparent dissociation constant (KD), as distinct from nitrocellulose filter binding assays conducted at low RNA concentrations that are designed to determine the dissociation constant (Lohman and Mascotti 1992). Estimates of the dissociation constants from EMSA data showed that weakest binding was often micro-molar, and therefore, the RNA concentration needed to exceed that value. The limitation of the stoichiometry experiments is that only the final state of the complex with the greatest number of bound proteins will be described, and not those intermediate states apparent from EMSA experiments. For each stoichiometry experiment, the RNA [R] concentration was varied, typically from 300 nM to 1 μM (always above the apparent KD) to ensure that the final ratio of proteins bound per RNA molecule did not depend on the total RNA concentration. The concentration of protein [P] was increased from 0 to 10–20 μM, to give a sufficient range of [P]/[R] to reach a plateau of binding; the value of [P]/[R] at the intersection of the two portions of the titration is the stoichiometry of protein binding to RNA. One example of the data is shown in Figure 6 ▶; all results from stoichiometry experiments are given in Table 4 ▶.
FIGURE 6.
Stoichiometry determination of PTB1:34 bound to GABA Intron/Exon RNA. [RNA] = 0.8 nM. Nitrocellulose filter binding (○) and fluorescence anisotropy (♦) data are superimposed.
TABLE 4.
Stoichiometry of PTB1 constructs bound to GABA RNAs
| RNA | |||||||
| Protein | I/E | I | I34 | I11 | I11/E | I11/Em | |
| PTB1 | (fb) | 8 ± 1 | 6 ± 1 | 6 ± 1 | 2 | 3 | 3 |
| (fa) | 7 ± 1 | ||||||
| PTB1:12 | (fb) | 9 ± 1 | 9 ± 1 | 8 ± 1 | 0 | 0 | nd |
| (fa) | 8 ± 1 | ||||||
| PTB1:23 | (fb) | 10 ± 1 | nd | nd | nd | nd | nd |
| PTB1:34 | (fb) | 10 ± 1 | 9 ± 1 | 8 ± 1 | 4 ± 1 | 5 ± 1 | 6 ± 1 |
| (fa) | 10 ± 1 | ||||||
Values are [P]/[R], determined by nitrocellulose filter binding titration of protein into a fixed concentration of RNA. (fb) Nitrocellulose filter binding; (fa) fluorescence anisotropy. [RNA] varied from 500 nM to 1μM. I11/Em has a mutation to disrupt the UUCG tetraloop. A value of 0 indicates no binding; (nd) not determined; 100 mM NaCl, 1 mM MgCl2 , 10 mM sodium cacodylate (pH 7), 20 μg/mL BSA, 10 μg/mL tRNA, 21° C.
The stoichiometry data give an indication of the potential size of the complex formed by PTB on this GABA I/E RNA, and how that complex is assembled. Clearly, the complex formed is not simply the sum of separate complexes formed on the isolated polypyrimidine tracts. An example of the nonadditivity is observed by a comparison of the 6 ± 1 PTB1 proteins bound to the intron (I) construct with the 6 ± 1 PTB1 proteins bound to I34 plus the two PTB1 proteins bound to I11 RNA. These data also show that there is not a 1:1 correspondence between number of UCUU sites and number of PTB1 proteins bound. UCUU was identified by Perez et al. (1997b) as a high-affinity binding site, and is colloquially used as an indicator of PTB association. In the GABA I/E RNA, there are four UCUU sequences within the intron/exon, but eight PTB1 proteins can bind.
Fluorescence anisotropy and binding stoichiometry
Another method to determine the stoichiometry of binding to the GABA intron/exon (I/E) is to monitor the change in fluorescence anisotropy of a fluorophore attached to the RNA. Here, fluorescein is attached to d(T)16, which is hybridized to the A15 sequence in the middle of the GABA intron. No change in the total fluorescence intensity was observed during these experiments, indicating that the dye was not interacting with protein or RNA. The concentration of RNA used in these experiments was varied, always at concentrations above the KD. An example of the data is shown in Figure 6 ▶.
As shown in Table 4 ▶ and Figure 6 ▶, the stoichiometry of binding determined by fluorescence anisotropy and nitro-cellulose filter binding is in excellent agreement for all PTB1 constructs. Both methods show the same number of PTB1 proteins bound to the GABA intron/exon, within error: 8 ± 1 proteins bound by filter binding, and 7 ± 1 by fluorescence anisotropy. Similarly, there are 8 ± 1 PTB1:12 proteins and 10 PTB1:34 proteins bound per GABA I/E RNA. The agreement of these data shows that introduction of the 15-bp DNA:RNA duplex has not altered the binding mode of any protein.
Nuclease footprinting
Footprinting experiments with ribonuclease T1 and RNase A used PTB1, PTB1:12, and PTB1:34 bound to GABA I/E RNA, and PTB1 and PTB1:34 bound to I11/E RNA. Proteins at 10 nM, 100 nM, and 1 μM final concentrations were bound to RNAs in order to approximate the complexes observed in EMSA experiments; probing was done at 37° C. Dissociation constants were determined from reactions at room temperature and gels were run at 4° C, so that the KD values at 37° C will be weaker; however, at the PTB concentrations used, sequential formation of complexes should occur.
Binding of PTB1 to the GABA I/E showed progressive protection of polypyrimidine tracts (Fig. 7A ▶): at 10 nM PTB1, the intron (U/C)11 tract appeared to be protected, while partial cleavage of the flanking G87 and G72 was observed. At 100 nM protein, (U/C)11 and (U/C)34 in the intron were completely protected, as was (U/C)7 in the exon. No cleavage at G87 and G72 was seen at 100 nM PTB1. Strong cleavage of U48–U49 [just 5′ to (A)15 in the intron] was observed at 10 nM PTB1, but not at 100 nM protein. Nuclease T1 and RNase A cleavage within the exon was partially blocked at 100 nM PTB1. The cleavage patterns did not change in 1 μM PTB1. Liu et al. (2002) also footprinted PTB1 on this RNA (1 μM PTB1 with 20 nM RNA, using micrococcal nuclease as a probe), and noted the protection of the (U/C)11 sequence as well as protection of the tracts in the exon.
FIGURE 7.
Footprinting of PTB1 and PTB1 constructs on GABA RNAs. Lanes T1 and A correspond to digestion with RNase T1 and A, respectively. The control lanes are (—) no treatment; (OH) alkaline ladder; (T1 Ladder) RNase T1 under denaturating conditions. The G ladder is shown on the left side. On the right side a schematic representation of GABA Intron/Exon RNA is displayed, where strong and weaker protection sites by PTB1 are indicated by dark circles and open circles, respectively. (A) Footprinting of PTB1, PTB1:12, and PTB1:34 on GABA Intron/Exon RNA. Enzymatic hydrolysis of 5′-32P-GABA Intron/Exon RNA in the absence (free RNA) or in the presence of 10 nM, 100 nM, or 1 μM of PTB1, PTB1:12, or PTB1:34. (B) Footprinting of PTB1 and PTB1:34 on GABA I11/E RNA. Enzymatic hydrolysis of 5′-32P-labeled GABA I11/E RNA in the absence (free RNA) or in the presence of 10 nM, 100 nM, or 1 μM of PTB1 or PTB1:34. (C) Predicted lowest free-energy secondary structure of GABA Intron/Exon RNA. The (U/C)34, (U/C)11 tracts, and the (A)15 are shown. AG102/CU 3′-splice site and AG127/G128U 5′-splice site at the intron/exon junction are shown in bold characters. T1 cleavage sites (G) are indicated by the open arrows.
Footprinting of PTB1:12 on the GABA I/E did not show protection of the (U/C)34 tract at 10 nM or 100 nM protein, and only partial protection at 1 μM protein. Cleavage after G87 and G72 occurs at all protein concentrations, as did cleavage at U48–U49 (Fig. 7A ▶). Within the exon, the T1 cleavage patterns closely resembled those seen with denatured RNA, where cleavage occurs after all G residues.
PTB1:34 bound to GABA I/E RNA protects the intron (U/C)34 at 10 nM PTB1:34. Cleavage at the flanking U48–U49 occurs even at 1 μM PTB1:34. Nuclease T1 cleavage after G72 and G87 is diminished by addition of 1 μM protein, but is still present, as is cleavage of their flanking C68-U69 and U88-C89-U90 nucleotides.
Protection of exon sequences is more easily seen using the shorter I11/E RNA (Fig. 7B ▶). Here, PTB1 partially protects the two short tracts in the exon at 10 nM and 100 nM protein, and completely protects them at 1 μM protein. The G102 and G110 nucleotides flanking the (U/C)7 tract are partially protected by the full-length protein. PTB1:34 binding protects (U/C)11 but allows cleavage within the exon, even at 1 μM protein (Fig. 7B ▶).
These data reveal a difference in the binding modes of PTB1 and the two tandem constructs, as well as a difference between the two halves of the protein. Binding of PTB1 extends into regions of the RNA that are not polypyrimidine tracts, as shown by protection of the flanking purines and the exon sequences. PTB1:34 appears to protect polypyrimidine tracts in the intron, while PTB1:12 allows nuclease access even at 1 μM protein. This difference could be caused by the geometry of the RNA as it is bound to the constructs, or to a difference in off-rates of the proteins that allows more access to the RNA by the nucleases.
HCV 3′-NTR and PTB1
The full-length NTR construct used here contains 82 nt of the NS5B gene, 46 nt of the hypervariable region, (U/C)75, and ends with X. RNAs transcribed for these experiments include the 310-nt NTR, the 98-nt X sequence, XSLI (Stem–Loop I), XSLII/SLIII, and XSLIII.
Analysis of binding by EMSA
PTB1 and three PTB1 constructs (PTB1:12, PTB1:23, and PTB1:34) bind to the complete 3′-NTR as assessed by EMSA (Fig. 8 ▶). Reaction conditions were selected from experiments done over a range of NaCl concentrations from 50 to 500 mM and a pH range from 6 to 8. Binding was observed in all solution conditions although with weaker affinity at 300 and 500 mM NaCl. Standard conditions for all assays are 100 mM NaCl, 1 mM MgCl2, 10 mM sodium cacodylate (pH 7.0), 20 μg/mL BSA, and 10 μg/mL tRNA.
FIGURE 8.
PhosphorImager EMSA data for HCV 3′-NTR binding by PTB1 protein and PTB1:12, PTB1:23, and PTB1:34 protein constructs; 100 mM NaCl, 1 mM MgCl2, 10 mM sodium cacodylate (pH 7), 20 μg/mL BSA, 10 μg/mL tRNA, 4% polyacrylamide:bis 39:1 (50 mM tris/glycine at pH 8.3), 4°C.
The mode of binding of PTB1 and its constructs is not the same: not only does the apparent affinity of the full-length protein and its three constructs vary, but the migration patterns of the complexes are distinct (Fig. 8 ▶). The clearest difference is observed with PTB1:34 binding, where at low concentrations of protein, the mobility of the complex increases from that of the free RNA, consistent with a compaction of the hydrodynamic volume of the complex. PTB1:23 binding was characterized by formation of complexes that did not enter the gel. These complexes appeared at the lowest protein concentration used (1.4 nM), and became more pronounced as the protein concentration increased. In no experiment was there observation of discrete bands of 3′-NTR:PTB complexes.
PTB1, PTB1:12, and PTB1:23 binding to X RNA produced several discrete bands on the gel (Fig. 9 ▶). In contrast, a single shifted species was observed when only SLIII was bound. Binding to X and SLIII is quite weak (micromolar), but this is not nonspecific binding (PTB1 does not bind to SLII of the human U1 snRNA in these experiments) (data not shown). PTB1:34 did not bind to either X or SLIII, as measured by EMSA experiments. No construct bound to SLI, or to SLII/SLIII; the absence of binding to the latter can be best explained by the ability of this RNA to adopt secondary structures that block access by PTB1.
FIGURE 9.
Determination of the apparent binding constant for PTB1 and PTB1 constructs binding to the X RNA. (A,B,E,F) PhosphorImager EMSA data for PTB1, PTB1:12, PTB1:23, and PTB1:34, respectively, binding to X RNA. EMSA data for PTB1:23 show a nonspecific association; PTB1:34 does not bind. Dissociation constants for (C) PTB1 and (D) PTB1:12 were calculated from the fit of the EMSA data to the partition function P = 1 + (K1[P])n1 + (K1K2[P]2)n2.
Dissociation constants
To measure the affinity of binding to X RNA, the bands observed in the EMSA experiments were analyzed, and the data fit by the partition function
 |
Association constants (K) and the Hill coefficient (n) were allowed to float (see Table 5 ▶).
TABLE 5.
Apparent dissociation constants calculated for PTB1 and PTB1:12: HCV X RNAs
| RNA | ||
| Protein | X RNA | X SLIII |
| PTB1 |
KD1 = 800 nM, n = 1 KD2 = 1 μM, n = 1 |
KD = 285 nM ± 120 nM |
| PTB1:12 |
KD1 = 800 nM, n = 1 KD2 = 1 μM, n = 1 |
KD = 1.6 μM ± 0.6 μM |
For protein binding to X RNA, dissociation constants were calculated from the relation P = 1 + (K1[P])n1 + (K1K2 [P]2)n2, where n is the Hill coefficient, using Origin 7.5 (OriginLab, Northampton, MA). Binding to X SLIII was measured using nitrocellulose filter binding, and the data were fit by Kaleidegraph to KD = ([R] – [PR])([P] – [PR])/[PR]; 100 mM NaCl, 1 mM MgCl2 , 10 mM cacodylate (pH 7), 20 μg/mL BSA, 10 μg/mL tRNA.
The dissociation constants of PTB1 and PTB1:12 to X RNA are weak and the same within error (800 nM and 1 μM;) (Fig. 9 ▶). The data were best fit with n = 1, indicating independent binding events. Binding to SLIII was determined by nitrocellulose filter binding, since the stoichiometry of association showed it to be 1:1. In these experiments, the affinities of PTB1 and PTB1:12 differed by a factor of ~6 [KD(PTB1) = 285 ± 120 nM; KD(PTB1:12) = 1.6 ± 0.6 μM].
Stoichiometry experiments
Upon demonstration of RNA:protein interaction via EMSA, and a qualitative determination of the apparent dissociation constant, the stoichiometry of the complex was determined using nitrocellulose filter binding. The results are given in Table 6 ▶.
TABLE 6.
Stoichiometry of PTB1 constructs bound to HCV 3′-NTR RNA constructs
| RNA | |||||
| Protein | 3′-NTR | X RNA | SLI | SLII/SLIII | SLIII |
| PTB1 | 6 ± 1 | 2 | 0 | 0 | 1 |
| PTB1:12 | 8 ± 1 | 2 | 0 | 0 | 1 |
| PTB1:23 | 2a | 0 | |||
| PTB1:34 | 15 ± 1 | 0 | 0 | 0 | 0 |
Binding of PTB1 protein constructs to the 3′-NTR of HCV and X constructs. Binding in 100 mM NaCl, 1 mM MgCl2, 10 mM sodium cacodylate (pH 7), 20 μg/mL BSA, 10 μg/mL tRNA.
aThis result is anomalous. EMSA experiments showed that most of the RNA was found in the well of the lanes at PTB1:23 concentrations >22 nM. This complex appears to have a tangled superstructure with an unknown stoichiometry.
Six PTB1 proteins, eight PTB1:12 proteins, and 15 PTB1:34 proteins can bind to the 3′-NTR. Only one PTB1 protein binds to SLIII, while two associate with X. The different stoichiometries illustrate that on this RNA, too, there is a difference in the binding modes of PTB1 and its two halves. Perhaps PTB1:34 proteins prefer Un sequences, whereas PTB1:12 RBDs prefer mixed U/C sequences. Alternatively, the physical binding modes of PTB1:34 and PTB1:12 could differ significantly, such that PTB1:34 is able to pack closely on the RNA.
Nuclease footprinting of X RNA
To map the sites on X that are protected by PTB1 binding, 1 μM PTB1 was used, based on the weakest KD of binding (Fig. 10 ▶). One protected region is within SLIII, C7-UC-C10-AUCU-U15; the other, U31-AGCUGU-G38, is in SLII. These protected sequences correspond to the loops of SLII and SLIII in the structure of X as mapped by Blight and Rice (1997).
FIGURE 10.
Footprinting of PTB1 and PTB1:12 on X RNA. Lanes T1 and A correspond to digestion with RNase T1 and A, respectively. The control lanes are (—) no treatment; (OH) alkaline hydrolysis ladder; (T1 Ladder) RNase T1 under denaturating conditions. The G ladder is shown on the left side. (B) Predicted lowest free-energy secondary structure of the X RNA. Positions cleaved by RNase T1 (open arrows) and by RNase A (dark arrows) are shown. Changes in cleavage intensity upon PTB1 and PTB1:12 proteins are indicated by a dark dot (protection) or an open dot (enhanced).
DISCUSSION
The results described here provide a new perspective on the complexes formed by PTB protein on its RNA substrates. First, formation of these complexes occurs sequentially: the first proteins bind with 1–10 nM affinity; then proteins with 100–400 nM affinity; and finally, there is a class of proteins that associates with micromolar affinity. Second, the physical mode of RNA binding by PTB1 differs from that of either PTB1:12 or PTB1:34, and the binding mode of PTB1:12 differs from that of PTB1:34. Third, stoichiometry of association shows that formation of the largest complexes involves multiple PTB proteins bound to an RNA.
Ordered assembly of PTB:RNA complexes
EMSA experiments with the various GABAA receptor γ2 RNA constructs show clear evidence of formation of large complexes through sequential addition of PTB proteins. The first PTB1 proteins to associate with the GABA RNAs do so with nanomolar affinity, certainly through direct association with the RNA. As nuclease protection assays show, the RNA is protected from cleavage at 10 nM PTB1 protein, a pattern that is not appreciably altered as more protein is added. This association presumably corresponds to the first shifted band in EMSA experiments, although there are currently no data that identify the stoichiometry of this tightly associated complex. Subsequent secondary addition of PTB1 is likely to occur through protein:protein interactions, although a weaker binding class of direct RNA:protein interactions cannot be ruled out. These complexes, observed as supershifted bands in EMSA experiments at 100–300 nM PTB1 concentrations, are stable in 100 to 250 mM NaCl in EMSA experiments at 4°C. There are no data to describe either the physical distribution of PTB1 proteins in these complexes, or to characterize the number of proteins associated. Heterogeneity of the physical morphology or binding stoichiometry could lead to broad bands in the gel, similar to those observed for many complexes. The final tertiary complexes occur at the highest (micromolar) concentrations of protein, and have considerably weaker dissociation constants; preliminary data show that these complexes are not stable at 250 mM NaCl, suggesting that the interactions are predominantly electrostatic. We suggest that this third class of binding occurs through protein:protein association, and is facilitated by a conformational change of PTB1 when it binds to RNA, thus exposing some portion of the protein for subsequent interactions. This hypothesis will be tested using fluorescence methods that monitor independent domains of PTB1.
The tandem RBD constructs of PTB1 also exhibit shifted and supershifted bands in EMSA experiments, indicative of sequential association of protein to the complexes. Bands of GABA RNAs bound to PTB1:12 are typically well resolved and reasonably sharp, suggesting a uniform distribution of complexes is present in a band. PTB1:23 complex formation is difficult to assess critically, since much of the RNA in EMSA experiments remains in the well of the gel. While such a result sometimes indicates formation of extremely large particles that cannot enter the gel matrix, these intractable PTB1:23–RNA complexes are observed at nanomolar concentrations of protein, suggesting some hydrodynamic property of these complexes interferes with their mobility. In contrast, PTB1:34 readily associates with RNAs, but typically results in shifted and supershifted bands that are broad and difficult to separate. While the first high-affinity complexes to appear on the gel are often well defined, subsequent shifted bands are diffuse. These data show that all PTB1 constructs are capable of forming hierarchies of complexes, but that the physical properties of those complexes are not identical.
There are currently no data to describe how many tandem RBD constructs are associated with any of the intermediate complexes observed in EMSA experiments. PTB1:34 binding to nearly all RNAs displays the greatest apparent heterogeneity in the composition and/or structure of its complexes, thus making them the most problematic to describe. Smeared bands can result from dissociation of the RNA during electrophoresis; structural heterogeneity of complexes with a constant stoichiometry, leading to variation in their electrophoretic mobility; or complexes that contain varying numbers of bound PTB1:34 proteins, leading to different charge to mass ratios, and hence to varying mobility. PTB1:34 has been shown to be generated through proteolytic cleavage by the polio protease 3CDpro (Back et al. 2002), the implication being that the virus has found a function for this construct. It is therefore especially important to identify the cause of the diffuse bands in PTB1:34–RNA gels, in order to correctly describe the properties of the complexes.
PTB1 and its subdomains do not share a binding mode
All RBDs are capable of association with short RNA oligomers, at least under conditions that favor electrostatic interactions (Oberstrass et al. 2005). Photochemical crosslinking is most commonly observed between RNA and RBD3 and RBD4 (Kaminski et al. 1995; Perez et al. 1997a; Oh et al. 1998; Charlet et al. 2002). In experiments with independent RBD constructs, RBD3 was shown to bind to polypyrimidine tracts (Oh et al. 1998; Perez et al. 1997a). Simpson et al. (2004) used a short RNA, CUUCUCUCU, to look for interactions with RBD1 and RBD2 by NMR chemical shift mapping (100 mM NaCl, 50 mM sodium phosphate at pH 6.5), and found chemical shift changes in both RBDs, the most dramatic being in RBD1. Amir-Ahmady et al. (2005) mapped UV crosslinks to RNP-2 of RBD4 and a portion of the linker between RBD3 and RBD4, and also detected 4-thiouridine crosslinks to RBD1. In addition, both RBD4 and RBD1 were crosslinked to the oligonucleotide CUU-CUCUCUCUG; those investigators suggest that both RBDs contact this RNA simultaneously. However, our experiments show that the physical context of the RBDs within PTB1 as well as the specific RNA sequence bound can and do lead to different binding modes of the different RBDs.
A demonstration of the relation between RNA bound and RBD context is readily seen by our experiments with PTB binding to progressive truncations of the GABA intron RNA. All four protein constructs bound to the entire 100-nt intron. Although all four protein constructs also bound to the 70-nt intron [I34 containing the (U/C)34 tract but missing the (U/C)11 tract], binding by PTB1:12 and PTB1:34 was significantly weaker. Finally, the 38-nt intron sequence (I11) containing the (U/C)11 tract was bound by PTB1 and PTB1:34, but not by PTB1:12. These polypyrimidine tracts are predicted to be single stranded, and thus present a uniform structural context for protein binding, yet the different protein constructs selectively associate with RNAs.
Another demonstration of the interdependence of RBD context and RNA binding site comes from binding of the protein constructs to the highly structured 98-nt HCV X RNA. In previous reports of PTB1 binding to X, Tsuchihara et al. (1997) reported that PTB binds to the 5′-stem/loop (SLIII) of the conserved X structure. A similar conclusion came from work by Ito and Lai (1997) and Chung and Kaplan (1999), except that those investigators concluded that both SLII and SLIII were required. Our experiments partly support previous binding studies, but also quantify association: two PTB1 molecules bind weakly and independently to the complete X RNA; one PTB1 protein binds to SLIII RNA. Two PTB1:12 proteins also bind independently to X RNA, and only one PTB1:12 protein binds to SLIII. In contrast, PTB1:34 does not bind to either X or SLIII, indicating that these RNAs do not provide an appropriate target site for this construct.
Models of PTB1:34 binding to RNA
To reiterate our data briefly, we found that 15 (±1) PTB1:34 proteins, 8 (±1) PTB1:12 proteins, and only 6 (±1) PTB1 proteins are in the largest complex with HCV 3′-NTR. PTB1:34 binds to GABA I11 RNA with its (U/C)11 tract, but not to the HCV X hairpin or isolated SLIII. PTB1:34 at least partially protects bound polypyrimidine tracts from nuclease cleavage. We suggest that this construct preferentially binds single-stranded U-rich RNAs, using both RBDs to contact the RNA.
The structural form of the protein in physiological conditions will determine how it is situated on the RNA strand. Two examples of tandem RBDs that interact with RNA illustrate the variation possible. In a cocrystal of two RBDs of poly(A) binding protein bound to (rA)11, the RNA stretches along the β-sheet surface of the RBDs, and also makes contact with the nine-amino-acid linker between the two domains (Deo et al. 1999). In a structure of nucleolin determined by NMR, an RNA hairpin loop sits between two RBDs, making interactions with both β-sheet surfaces and the linker (Allain et al. 2000). The linkers between the PABP and nucleolin are short, facilitating RNA contact with both RBDs; the linker between PTB1 RBD3 and RBD4 is approximately three times longer. We predict that the disposition of RNA on the PTB1 RBDs will vary with the sequence of the RNA.
PTB1:34 is a monomer in solution, as shown by NMR (Conte et al. 2000) and equilibrium analytical ultracentrifugation (in 250 mM NaCl) (Simpson et al. 2004). In the 100 mM NaCl conditions of Conte et al.’s (1999) NMR experiments as well as in our 15N NMR experiments in 100 and 200 mM KCl (data not shown), the two RBDs of PTB1:34 tumble independently. However, in conditions of low salt (30 mM NaCl), Oberstrass et al. (2005) found interactions between the helices of RBD3 and RBD4, forcing the two β-sheet surfaces to face outward and away from each other. Based on our data, we propose that the dominant binding mode for this construct allows the two RBDs of PTB1:34 to close like a clam over the bound RNA, such that both β-sheet surfaces and the 23-amino-acid linker between the two domains make contact with the RNA. We will test this prediction using NMR and fluorescence methods, making use of specific mutations within the RBDs.
Models of PTB1:12 binding
Three properties of PTB1:12 binding to the HCV 3′-NTR differ fundamentally from PTB1:34 binding. First, only half the number of PTB1:12 proteins are bound; second, PTB1:12 binds to the X RNA; and third, bound RNA is exposed to nuclease digestion. Of the two RBDs in PTB1:12, previous experimental data clearly identify RBD1 as an RNA-binding site. Oh et al. (1998) demonstrated RBD1 binding to a portion of the EMCV 5′-NTR in EMSA experiments; NMR chemical shift mapping (Simpson et al. 2004) showed that RBD1 alone associates with a CUUCUCUCU oligomer; and Amir-Ahmady et al. (2005) crosslinked RBD1 to c-src RNA. RBD1, uniquely among the four RBDs in PTB, contains an aromatic amino acid in the conserved RNP1 sequence. Data supporting association of RBD2 with RNA are less compelling (e.g., Simpson et al. 2004; Oberstrass et al. 2005); its unusual structure includes a loop connecting β4 to β5 that lies on and partially occludes the β-sheet surface. Our current model for PTB1:12 binding is that RBD1 interacts directly with RNA, and can bind either a single strand or a loop structure. RBD2 or the linker between RBD1 and RBD2 may provide some electrostatic binding energy, but RBD2 is not the dominant site of RNA contact. Furthermore, we propose that there is a conformational change of the PTB1:12 construct upon RNA binding that facilitates intermolecular protein:protein interactions.
Model of PTB1 binding mode
On the basis of binding to the U-rich HCV RNA and the U/C GABA polypyrimidine tracts, we suggest that PTB1:12 requires both cytosine and uridine nucleobases (with perhaps some sequence preferences), while PTB1:34 binds to any pyrimidine tract of appropriate length. Because RBD3 and/or RBD4 have less stringent requirements of RNA length and RNA sequence, we propose that in the context of the intact PTB1 protein, PTB1:34 initiates RNA binding to single-stranded polypyrimidine tracts, bringing the RNA into proximity of PTB1:12, which then, in turn, binds RNA if the sequence is appropriate. It is possible that RBD3/RBD4 could bind to a site that has no contiguous binding site for RBD1/RBD2, thus forcing these RBDs to either remain free in solution, or bind to a distal RNA site. Assuming that the two halves of the protein have independent motion, facilitated by the ~90-amino-acid linker between RBD2 and RBD3, then this is a mechanism to tie two regions of an RNA together in a compact complex. Conversely, if a binding site is favored by RBD1/RBD2 (such as the stem–loops in X RNA), then RBD3/RBD4 is free to bind other RNA sites, facilitating intermolecular RNA association or intramolecular RNA looping.
PTB:RNA complexes can be large
The two RNAs used in these experiments have either one very long polypyrimidine tract or several juxtaposed tracts. PTB1 forms large, multimeric complexes on both RNAs, but the number of PTB1 proteins in the complex cannot be predicted from RNA length alone. If there were such a linear relationship, then the 75-nt U-rich tract in the HCV RNA should bind twice as many PTB1 proteins as the (U/C)34 tract in the GABA intron; in fact, six PTB1 proteins are found in both complexes. More generally, a minimum binding site size for PTB is not well defined. The sequence 5′-(U/G)CYGCCUG(Y/G)UGCYYYYCYYYYG(Y/G)CCC-3′ (Y is pyrimidine), was selected as a target in SELEX experiments with PTB1 (Singh et al. 1995). In contrast, a UCUU tetramer was identified as a PTB target by Perez et al. (1997b) but was not bound by PTB1:34 in NMR experiments (Yuan et al. 2002). Amir-Ahmady et al. (2005) found that a 38-nt RNA containing the 10-mer CUUCUCUCUC was a minimum length for binding by PTB1, which gave a single band in EMSA experiments. That RNA construct is similar in length and sequence to our GABA I11, which also gave a single band in EMSA experiments; however, our stoichiometry experiments showed it to contain two PTB1 proteins that bound cooperatively. Currently, we are unable to predict the number of PTB1 protein constructs that can ultimately associate with an RNA.
The global structure of the complexes could vary considerably, from long linear arrangements of PTB on contiguous polypyrimidine tracts to the looped out arrangements envisioned by Wagner and Garcia-Blanco (2001) and Amir-Ahmady et al. (2005). Models of PTB interaction with pre-mRNA have been proposed in which PTB complexes block access to bound RNA by steric exclusion (e.g., Wagner and Garcia-Blanco 2001), and on the basis of the stoichiometry measured here, the density of PTB proteins on the RNA could certainly occlude binding by other proteins. However, as the nuclease protection data indicate, RNAs with noncontiguous polypyrimidine tracts would have intervening uncovered RNA sequences, allowing access by other proteins.
Other cellular proteins are implicated in the modulation of PTB function. Some have been identified that appear to associate with PTB and act in synergy to form RNA:protein complexes: for example, hnRNP L (Hahm et al. 1998) and raver1 (Gromak et al. 2003a). In contrast, CELF proteins appear to compete with and/or displace PTB (Zhang et al. 1999; Gromak et al. 2003b). A neuronal homolog of PTB, nPTB, is reported to antagonize PTB binding (Markovtsov et al. 2000; Polydorides et al. 2000). Regulation by other proteins will depend on the relative affinities of protein:protein and RNA:protein interactions, and the relative concentrations of the proteins in the vicinity of the RNA. More detailed study of formation and dissociation of PTB:RNA complexes is necessary to determine how they can be stabilized by binding partners or destabilized by antagonists.
The largest complexes are formed after consecutive addition of PTB proteins to the RNA. It is important to emphasize that the disposition of proteins in these complexes is not known, neither is it clear whether the large complexes ultimately formed are due to direct RNA:PTB binding only or include intermolecular protein interactions. The orientation of individual RBDs in the complexes is also unknown: two contrasting scenarios either allow contiguous RNA binding by all RBDs in a single PTB molecule or restrict RNA binding to a single RBD, leaving other RBDs free to associate with other proteins or other RNA sequences. The geometry and dynamics of the protein domains in these two limiting cases will be distinctive (and distinguishable by time-resolved fluorescence probing of labeled proteins), and would have consequences for the function of PTB as it associates with a specific RNA.
MATERIALS AND METHODS
Cloning
The human PTB1 gene cloned in pET 28A vector was a generous gift from Doug Black (UCLA). Standard PCR methods were used to generate each PTB1 RBD tandem construct (PTB1:12, PTB1:23, and PTB1:34, where PTB1 is the isoform and 12, 23, 34 designate the RBDs in the construct). The different PCR fragments were cloned in the pET28A vector using EcoRI and HindIII restriction sites. Each clone was verified by sequencing. Protein overexpression in Escherichia coli BL21(DE3) (Strata-gene) was checked by induction with 1 mM IPTG. PTB1:34 did not have a his-tag, and was cloned into a ptac2 vector with NcoI and HindIII.
PTB1 contains the N-terminal 70-amino-acid NLS, as does PTB1:12. PTB1:12 terminates at amino acid 300; PTB1:23 begins at amino acid 183 and terminates at amino acid 439; PTB1:34 begins at amino acid 334; numbering refers to the human PTB1 sequence.
Protein purification
Individual clones were grown overnight at 37°C in LB/Kanamycin (50 μg/mL) followed by 50-fold dilution into terrific broth (TB), and grown at 37°C to an OD600 of 0.7. IPTG was then added to a final concentration of 1 mM, and growth continued for 4 h. Cells were pelleted by centrifugation at 6500 rpm for 15 min and frozen at −80°C. The cell pellet was resuspended in 3 volumes of buffer A (50 mM sodium phosphate, 300 mM NaCl, 10% glycerol at pH 8). For lysis, cells were incubated at 4°C for 30 min with 5 mg/mL lysozyme, 1 mM PMSF, 5 μg/mL Pepstatin, and protease inhibitor cocktail (Sigma P2714) containing 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), trans-epoxysuccinyl-L-leucylamino(4-guanidino)butane(E-64), bestatin, leupeptin, aprotinin, and EDTA. Cells were then pulse-sonicated for 30 sec five times with cooling between. Cellular debris was removed by centrifugation at 10,000g for 30 min at 4°C. The supernatant was removed and mixed with the pre-equilibrated Ni-NTA resin (QIAGEN) in buffer A and rocked at 175 rpm for 1.5 h on ice. The resin was then washed four times for 10 min each with 25 mL of buffer A plus 20 mM imidazole followed by elution of the protein with a gradient of imidazole (0 mM–250 mM). PTB1:34 was purified using the protocol described in Showalter and Hall (2004). Peak fractions were collected, concentrated, and exchanged into storage buffer (20 mM sodium cacodylate, 100 mM KCl, 0.5 mM DTT, 0.5 mM EDTA, 10% glycerol) using a Vivaspin with the appropriate molecular weight cutoff.
Protein purifications were assessed by SDS-PAGE. Protein concentrations were determined by the absorption spectrum, using ɛ280 = 12,800 cm−1 M−1 (PTB1); 10,480 cm−1 M−1 (PTB1:12), 9080 cm−1 M−1 (PTB1:23), and 2560 cm−1 M−1 (PTB1:34). Protein secondary structure was measured by CD spectroscopy to confirm that the proteins were folded.
RNA transcription and purification
GABAA receptor γ2 pre-mRNA
Plasmid RG γ57 containing the GABAA receptor γ2 pre-mRNA construct was generously provided by Paula Grabowski (University of Pittsburgh, PA). The sequence was cloned downstream of a T7 RNA polymerase promoter in a pBS vector. Plasmids were linearized with either BamHI for the Intron/Exon or AccI for the Intron. Other RNAs were transcribed from oligodeoxynucleotides, using the method of Sampson and Uhlenbeck (1998).
HCV 3′-NTR RNA
The RNA sequence was cloned downstream of the T7 RNA polymerase promoter in a pBR322 derivative, pTET (Keril Blight, Washington University School of Medicine, St. Louis, MO). Plasmid DNA was linearized with either ScaI for the full-length HCV RNA and the X RNA, or SmaI for X RNA ΔSLI. X SLIII RNA was transcribed from oligodeoxynucleotides with T7 RNA polymerase.
In vitro transcription reactions were done using T7 RNA polymerase (Epicenter) in 40 mM Tris (pH 8.0), 1 mM spermidine, 6 mM MgCl2 with or without [α-32P]UTP and/or [α-32P]CTP. After phenol extraction and ethanol precipitation, RNAs were purified from denaturing polyacrylamide gels. The RNA was eluted from the gel by incubation in 0.3 M sodium acetate (pH 5.3) at 37°C overnight. Concentrations of unlabeled RNAs were determined spectrophotometrically using the approximation of 1 A260 = 40 μg/mL RNA. Each sample was heated at 95°C and snap-cooled on ice to generate the secondary structures.
Gel mobility shift assays (EMSA)
For gel mobility shift assays, folded [α-32P]RNA (~200 cpm) in 10 mM KCl and 10 μg/μL yeast tRNA (Boehringer) was mixed with the recombinant PTB proteins (1 nM–5 μM) in 10 mM sodium cacodylate, 50 mM–500 mM NaCl, 1 mM MgCl2, and 20 μg/mL BSA (pH 6–8). All reaction components were incubated for 30 min at room temperature. Glycerol was added to 10%, then reactions were loaded on 8% (GABA) or 6% (HCV) polyacrylamide gels (40:1 acrylamide:bis) in 50 mM Tris-HCl/Glycine buffer and run at 7 V/cm at 4°C for 3–4 h. Experiments were repeated at least three times, using different preparations of RNAs and different preparations of purified protein.
Filter binding stoichiometry
Nonradioactive RNA was mixed with a trace of [α-32P]RNA to a final concentration of 0.1 μM, 0.5 μM, or 1 μM RNA. The RNA was folded in solution by heating in water to 95°C, quenching on ice, then addition of KCl to a final concentration of 10 mM and yeast tRNA to a final 10 μg/μL tRNA. Increasing amounts of recombinant PTB1 proteins were incubated with the fixed concentration of RNA in 10 mM sodium cacodylate, 100 mM NaCl, 1 mM MgCl2, and 20 μg/mL BSA (pH 7). The reaction was incubated at room temperature for 30 min in a polypropylene microtiter plate (96 wells). Reactions were filtered through pre-soaked Schleicher and Schuell 0.45-μm nitrocellulose filters (Wong and Lohman 1997). Data were analyzed with Kaleidegraph (Synergy, Inc.).
Anisotropy stoichiometry measurements for GABAA receptor γ2 pre-mRNA (Intron/Exon)
GABAA receptor γ2 Intron/Exon pre-mRNA was mixed with 5′-fluorescein(dT)16 and annealed by heating at 95°C for 5 min in water. Samples were cooled to 65°C for 5 min, and buffer was added to give a concentration of 10 mM sodium cacodylate (pH 7), 50 mM NaCl, and 1 mM MgCl2 in 1 mL final volume. Samples were cooled slowly to room temperature. The annealed sample was added to 1× buffer (10 mM sodium cacodylate at pH 7, 50 mM NaCl, 1 mM MgCl2, 75 μg/mL BSA) in the fluorescence cuvette to give a final concentration of 520 nM for the GABA Intron/Exon RNA and 500 nM for the 5′-fluorescein(dT)16. Small aliquots of protein stock solution were then added to the fixed concentration of RNA.
Anisotropy measurements
Anisotropies were measured using the L-format method on an SLM 8100 spectrofluorometer. Anisotropy values were determined using the following equation:
 |
where I|| and I⊥ are the intensities of the fluorescence components polarized parallel and perpendicular, respectively, to the vertically polarized excitation beam. The G factor (here G = 1) corrects for the polarization-dependent properties of the detection system. For fluorescein RNAs, the anisotropy was measured with 490 nm excitation (bandwidth 2 nm) and polarization fluorescence was measured at 525 nm (bandwidth 4 nm). Before each acquisition, samples were allowed to equilibrate for 15 min at 25°C in the water-jacketed cuvette. Each anisotropy measurement was repeated until the standard deviation of the measurement was <0.001.
Nuclease footprinting
5′-32P-labeled RNAs were folded by heating to 95°C in water, quenching on ice, then adding KCl to a final concentration of 10 mM and tRNA to a final concentration of 20 μg/mL. To produce a base hydrolysis ladder, RNA was incubated in 0.1 M sodium carbonate (pH 9.3) at 100°C for 4 min, then quenched on ice. To produce a T1 nuclease ladder, RNA was incubated with T1 ribonuclease in 20 mM sodium acetate (pH 5), 1 mM EDTA, and 7 M urea at 50°C for 15 min, then quenched on ice. Samples were phenol-extracted and ethanol-precipitated, then resuspended in formamide for loading.
5′-32P-RNA was allowed to incubate with and without added protein (final protein concentrations of 10 nM, 100 nM, and 1 μM) in 10 mM sodium cacodylate (pH 7.0), 100 mM KCl, for 30 min at room temperature, then enzyme was added. Digestion by nuclease T1 and RNase A was carried out under single-cleavage conditions, at 37°C for 5 min. Samples were diluted with water, phenol-extracted and ethanol-precipitated, resuspended in formamide, and run on 10% polyacrylamide/8 M urea gels. Gels were dried and imaged with a PhosphorImager. Probing was repeated with different RNA and protein preparations.
Acknowledgments
We thank Douglas L. Black for the His-PTB1 clone, Paula J. Grabowski for the GABAA pre-RNA clone, and Keril J. Blight for the HCV 3′-NTR RNA clones.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2178406.
REFERENCES
- Ali, N. and Siddiqui, A. 1995. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J. Virol. 69: 6367–6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain, F.H.T., Bouvet, P., Dieckmann, T., and Feigon, J. 2000. Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J. 19: 6870–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir-Ahmady, B., Boutz, P.L., Markovtsov, V., Phillips, M.L., and Black, D.L. 2005. Exon repression by polypyrimidine tract binding protein. RNA 11: 699–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar, A., Ali, N., Tanveer, R., and Siddiqui, A. 2000. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J. Biol. Chem. 275: 34231–34235. [DOI] [PubMed] [Google Scholar]
- Ashiya, M. and Grabowski, P.J. 1997. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA 3: 996–1015. [PMC free article] [PubMed] [Google Scholar]
- Back, S.H., Kim, Y.K., Kim, W.J., Cho, S., Oh, H.R., Kim, J.E., and Jang, S.K. 2002. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3C(pro). J. Virol. 76: 2529–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72: 291–336. [DOI] [PubMed] [Google Scholar]
- Blight, K.J. and Rice, C.M. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71: 7345–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlet, B.N., Logan, P., Singh, G., and Cooper, T.A. 2002. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 9: 649–658. [DOI] [PubMed] [Google Scholar]
- Chung, R.T. and Kaplan, L.M. 1999. Heterogeneous nuclear ribonucleoprotein I (hnRNP-I/PTB) selectively binds the conserved 3′ terminus of hepatitis C viral RNA. Biochem. Biophys. Res. Commun. 254: 351–362. [DOI] [PubMed] [Google Scholar]
- Conte, M.R., Grune, T., Ghuman, J., Kelly, G., Ladas, A., Matthews, S., and Curry, S. 2000. Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J. 19: 3132–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo, R.C., Bonanno, J.B., Sonenberg, N., and Burley, S.K. 1999. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98: 835–845. [DOI] [PubMed] [Google Scholar]
- Domitrovich, A.M., Diebel, K.W., Ali, N., Sarker, S., and Siddiqui, A. 2005. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology 335: 72–86. [DOI] [PubMed] [Google Scholar]
- Fred, R.G. and Welsh, N. 2005. Increased expression of polypyrimidine tract binding protein results in higher insulin mRNA levels. Biochem. Biophys. Res. Commun. 328: 38–42. [DOI] [PubMed] [Google Scholar]
- Friebe, P. and Bartenschlager, R. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76: 5326–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontarek, R.R., Gutshall, L.L., Herold, K.M., Tsai, J., Sathe, G.M., Mao, J., Prescott, C., and Del Vecchio, A.M. 1999. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3′NTR of the HCV RNA genome. Nucleic Acids Res. 27: 1457–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak, N., Rideau, A., Southby, J., Scadden, A.D., Gooding, C., Huttelmaier, S., Singer, R.H., and Smith, C.W. 2003a. The PTB interacting protein raver1 regulates α-tropomyosin alternative splicing. EMBO J. 22: 6356–6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak, N., Matlin, A.J., Cooper, T.A., and Smith, C.W. 2003b. Antagonistic regulation of α-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA 9: 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm, B., Cho, O.H., Kim, J.E., Kim, Y.K., Kim, J.H., Oh, Y.L., and Jang, S.K. 1998. Polypyrimidine tract-binding protein interacts with HnRNP L. FEBS Lett. 425: 401–406. [DOI] [PubMed] [Google Scholar]
- Ito, T. and Lai, M.M. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 71: 8698–8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1999. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology 254: 288–296. [DOI] [PubMed] [Google Scholar]
- Kaminski, A., Hunt, S.L., Patton, J.G., and Jackson, R.J. 1995. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA 1: 924–938. [PMC free article] [PubMed] [Google Scholar]
- Kolykhalov, A.A., Feinstone, S.M., and Rice, C.M. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70: 3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolykhalov, A.A., Mihalik, K., Feinstone, S.M., and Rice, C.M. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74: 2046–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Zhang, W., Reed, R.B., Liu, W., and Grabowski, P.J. 2002. Mutations in RRM4 uncouple the splicing repression and RNA-binding activities of polypyrimidine tract binding protein. RNA 8: 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman, T.M. and Mascotti, D.P. 1992. Nonspecific ligand–DNA equilibrium binding parameters determined by fluorescence methods. Methods Enz. 212: 424–458. [DOI] [PubMed] [Google Scholar]
- Markovtsov, V., Nikolic, J.M., Goldman, J.A., Turck, C.W., Chou, M.Y., and Black, D.L. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20: 7463–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass, F.C., Auweter, S.D., Erat, M., Hargous, Y., Henning, A., Wenter, P., Reymond, L., Amir-Ahmady, B., Pitsch, S., Black, D.L., et al. 2005. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science 309: 2054–2057. [DOI] [PubMed] [Google Scholar]
- Oh, Y.L., Hahm, B., Kim, Y.K., Lee, H.K., Lee, J.W., Song, O., Tsukiyama- Kohara, K., Kohara, M., Nomoto, A., and Jang, S.K. 1998. Determination of functional domains in polypyrimidine-tract-binding protein. Biochem. J. 331: 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez, I., Lin, C.H., McAfee, J.G., and Patton, J.G. 1997a. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA 3: 764–778. [PMC free article] [PubMed] [Google Scholar]
- Perez, I., McAfee, J.G., and Patton, J.G. 1997b. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry 36: 11881–11890. [DOI] [PubMed] [Google Scholar]
- Polydorides, A.D., Okano, H.J., Yang, Y.Y., Stefani, G., and Darnell, R.B. 2000. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. 97: 6350–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, J.R. and Uhlenbeck, O.C. 1998. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. 85: 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter, S.A. and Hall, K.B. 2004. Altering the RNA-binding mode of the U1A RBD1 protein. J. Mol. Biol. 335: 465–480. [DOI] [PubMed] [Google Scholar]
- Simpson, P.J., Monie, T.P., Szendroi, A., Davydova, N., Tyzack, J.K., Conte, M.R., Read, C.M., Cary, P.D., Svergun, D.I., Konarev, P.V., et al. 2004. Structure and RNA interactions of the N-terminal RRM domains of PTB. Structure 12: 1631–1643. [DOI] [PubMed] [Google Scholar]
- Singh, R., Valcarcel, J., and Green, M.R. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173–1176. [DOI] [PubMed] [Google Scholar]
- Tillmar, L. and Welsh, N. 2002. Hypoxia may increase rat insulin mRNA levels by promoting binding of the polypyrimidine tractbinding protein (PTB) to the pyrimidine-rich insulin mRNA 3′-untranslated region. Mol. Med. 8: 263–272. [PMC free article] [PubMed] [Google Scholar]
- Tischendorf, J.J., Beger, C., Korf, M., Manns, M.P., and Kruger, M. 2004. Polypyrimidine tract-binding protein (PTB) inhibits Hepatitis C virus internal ribosome entry site (HCV IRES)-mediated translation, but does not affect HCV replication. Arch. Virol. 149: 1955–1970. [DOI] [PubMed] [Google Scholar]
- Tsuchihara, K., Tanaka, T., Hijikata, M., Kuge, S., Toyoda, H., Nomoto, A., Yamamoto, N., and Shimotohno, K. 1997. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J. Virol. 71: 6720–6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcarcel, J. and Gebauer, F. 1997. Post-transcriptional regulation: The dawn of PTB. Curr. Biol. 7: 705–708. [DOI] [PubMed] [Google Scholar]
- Wagner, E.J. and Garcia-Blanco, M.A. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21: 3281–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherell, G.W., Gil, A., and Wimmer, E. 1993. Interaction of polypyrimidine tract binding protein with the encephalomyocarditis virus mRNA internal ribosomal entry site. Biochemistry 32: 8268–8275. [DOI] [PubMed] [Google Scholar]
- Wollerton, M.C., Gooding, C., Robinson, F., Brown, E.C., Jackson, R.J., and Smith, C.W. 2001. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB). RNA 7: 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, I. and Lohman, T.M. 1997. A two-site mechanism for ATP hydrolysis by the asymmetric Rep dimer P2S as revealed by site-specific inhibition with ADP-A1F4. Biochemistry 36: 3115–3125. [DOI] [PubMed] [Google Scholar]
- Yuan, X., Davydova, N., Conte, M.R., Curry, S., and Matthews, S. 2002. Chemical shift mapping of RNA interactions with the polypyrimidine tract binding protein. Nucleic Acids Res. 30: 456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Liu, W., and Grabowski, P.J. 1999. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA 5: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]













