Abstract
Trypanosome RNA editing is massive post-transcriptional U-insertion and U-deletion, which generates mature mRNA coding regions through cycles of endonuclease, terminal U transferase (TUTase) or 3′-U-exo, and ligase action. Both types of editing are thought to be catalyzed by distinct sets of proteins of a multiprotein complex, and no enzymatic activity of wild-type editing complex had been shown to function in both forms of editing. By examining the individual steps of the U-deletion cycle using purified editing complex, traditional mitochondrial extract, and rapidly prepared cell lysate, we here demonstrate that TbMP57 TUTase of U-insertion can act efficiently within a U-deletion cycle. When physiological UTP levels are provided, it adds U’s to the upstream cleavage fragment after U-deletional endonuclease and 3′-U-exo action, but before rejoining by the U-deletional ligase, generating partial U-deletion products. TUTase activity in U-deletion was not previously appreciated since its detection requires UTP, which is not normally added to in vitro U-deletion reactions. Fractionation and RNAi analyses show this U-addition in U-deletion requires TbMP57 TUTase be present and competent for U-insertion; such U-addition does not occur with another mitochondrial TUTase that is separate from the basic editing complex. Efficient TbMP57 action in both U-insertion and U-deletion suggests these two editing forms may be less separate than generally envisioned. Should such promiscuous TUTase action also occur in vivo, it could explain why editing utilizes substantially fewer U-deletional than U-insertional events and why partial editing appears preferential in U-deletion.
Keywords: editing, RNA, terminal U transferase, trypanosome, TUTase
INTRODUCTION
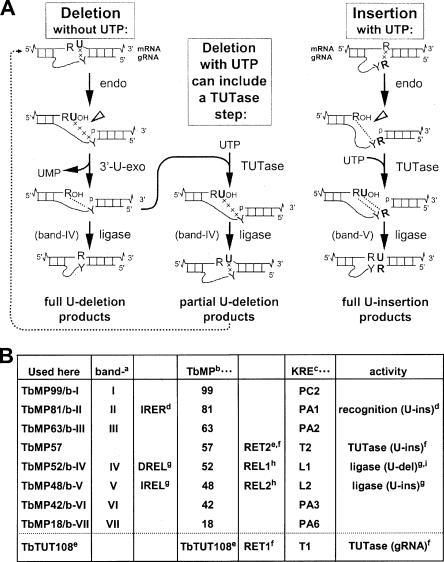
Trypanosome RNA editing is a bizarre form of RNA processing in which uridylates (Us) are inserted into and, less frequently, deleted from the coding regions of mitochondrial transcripts, creating up to 80% of the codons of mature mRNAs (for reviews, see Gott and Emeson 2000; Madison-Antenucci et al. 2002; Stuart and Panigrahi 2002; Simpson et al. 2003). The editing is directed by short, trans-acting mitochondrial transcripts, called guide RNAs (gRNAs), that are complementary to segments of the mature mRNA and thus mismatch the pre-mRNA at each insertion and deletion site. The actual editing reactions occur sequentially across the pre-mRNA, always abutting an “anchor duplex” that forms between the already mature sequence in the pre-mRNA and the gRNA (see left and right columns of Fig. 1A ▶; Blum et al. 1990). Each U-deletion or U-insertion cycle involves endonuclease cleavage of the mRNA at the upstream end of the anchor duplex, then U removal by a 3′ U exonuclease (3′-U-exo) or U-addition by a terminal U transferase (TUTase) on the upstream cleavage product, and finally religation of the mRNA (see left and right columns of Fig. 1A ▶; Blum et al. 1990; Cruz-Reyes and Sollner-Webb 1996; Kable et al. 1996; Seiwert et al. 1996). These steps together form the complete editing cycle and can be assayed individually using partial editing reactions (e.g., Cruz-Reyes et al. 1998b, 2002; Igo et al. 2000, 2002a, Igo et al. b; Huang et al. 2001).
FIGURE 1.
The reactions and proteins of the editing complex. (A) RNA editing mechanism. The left- and right-hand pathways depict the traditional view of editing cycles, U-deletion (without UTP) and U-insertion (with UTP), as discussed in the text. The central pathway illustrates the conclusions from this study, that in the presence of UTP, U-addition can occur within a U-deletion cycle, generating partial U-deletion. The dotted arrow indicates its re-editing (Cruz-Reyes and Sollner-Webb 1996). Since partial U-deletions do not precisely base pair with the gRNA (shown by the connecting line of x’s), their ligation is assuredly by the U-deletional TbMP52/b-IV (band IV), which has relaxed pairing requirements, while TbMP48/b-V (band V) ligates U-insertional product with a precisely base-paired bridge (the dotted connecting lines) (Cruz-Reyes et al. 2002; Igo et al. 2000, 2002a). (B) Multiple nomenclatures for the proteins of the basic editing complex. (a) Rusche et al. 1997; (b) Panigrahi et al. 2001b, 2003b; (c) Stuart et al. 2005; (d) IRER indicates U-Insertional RNA Editing Recognition; Law et al. 2005; (e) Ernst et al. 2003; (f) Aphasizhev et al. 2003b; (g) DREL indicates RNA Editing Ligase required for U-Deletion; IREL indicates RNA Editing Ligase specific for U-Insertion; Cruz-Reyes et al. 2002; (h) Schnaufer et al. 2001; (i) Huang et al. 2001. Below the dotted line is shown another mitochondrial TUTase that is separate from the basic editing complex.
Editing reactions are catalyzed in vitro using traditional mitochondrial extract or its 20S fractions (Pollard et al. 1992; Seiwert and Stuart 1994; Kable et al. 1996; Seiwert et al. 1996), purified editing complex (Rusche et al. 1997; Cruz-Reyes et al. 1998c, 2001, 2002; Panigrahi et al. 2003b), and rapidly prepared trypanosome lysate (see Materials and Methods; Huang et al. 2002; O’Hearn et al. 2003; Law et al. 2005). The first preparation of purified editing complex has seven major staining proteins (Rusche et al. 1997; Sollner-Webb et al. 2001) and catalyzes full cycle U-deletion and U-insertion with apparently the highest efficiency yet reported (Cruz-Reyes et al. 1998c, 2001, 2002). (The editing proteins have generated several alternate nomenclatures [Fig. 1B ▶] and will be identified here using a combined format.) A related purification yields 13–15 major proteins and catalyzes U-insertion (Madison-Antenucci et al. 1998; McManus et al. 2000). Two other preparations contain about 20 major staining proteins (Panigrahi et al. 2001a, 2003a,Panigrahi et al. b; Aphasizhev et al. 2003a,b; Schnaufer et al. 2003), including the earlier-reported seven (Fig. 1B ▶). These preparations catalyze the full U-deletion cycle (Panigrahi et al. 2001a, 2003b) and actively catalyze “precleaved editing”, which bypasses the cleavage step (Igo et al. 2000, 2002a; Panigrahi et al. 2001b, 2003a). A TbMP57 TUTase that adds the U’s in U-insertion (Aphasizhev et al. 2003b; see also Ernst et al. 2003) appears as one of the major proteins in those latter preparations (Panigrahi et al. 2003b) but appears substantially substochiometric in the preparation exhibiting seven major proteins (Rusche et al. 1997).
Theoretically, the comparable steps of U-deletion and U-insertion could have used the same enzymes (Hajduk 1997; Stuart et al. 1997). However, the initial cleavages in U-deletion and U-insertion exhibit different responses to ADP (and related nucleotides; Cruz-Reyes et al. 1998b) and to gRNA features (Cruz-Reyes et al. 2001), raising the possibility of different endonucleases. Similarly, in the second step, the 3′-U-exo is not the reverse reaction of the TUTase, since the latter consumes UTP and generates pyrophosphate (PPi), while the former generates UMP and does not consume PPi (Cruz-Reyes and Sollner-Webb 1996; Rusche et al. 1997). At the third reaction step, the two ligases of the editing complex are also utilized differently: TbMP52/b-IV is required for U-deletion, while TbMP48/b-V is specific for U-insertion (Huang et al. 2001; Cruz-Reyes et al. 2002; Gao and Simpson 2003; Schnaufer et al. 2003). Thus U-deletion and U-insertion were hypothesized to use distinct sets of enzymes, comprising different regions of the editing complex (Cruz-Reyes et al. 1998b,c, 2002; Huang et al. 2001).
Several additional lines of data indicate that U-deletion and U-insertion utilize different proteins within the editing complex. First, the TbMP52/b-IV ligase is held in the editing complex by binding TbMP63/b-III (Panigrahi et al. 2001b; Huang et al. 2002; Schnaufer et al. 2003), whereas the TbMP48/b-V ligase is held by binding TbMP81/b-II (Cruz-Reyes et al. 2002; O’Hearn et al. 2003; Schnaufer et al. 2003). Second, TbMP81/b-II is essential for specific protein and/or substrate recognition at every step of the U-insertion cycle but is not similarly important for any step of the U-deletion cycle (Law et al. 2005). Furthermore, overproduction of tagged ligases enables isolation of a subcomplex containing TbMP52/b-IV, TbMP63/b-III, TbMP99/b-I, and some TbMP18/b-VII that supports the 3′-U-exo and ligation steps of U-deletion or a subcomplex containing TbMP48/b-V, TbMP81/b-II, TbMP57, and possibly some TbMP18/b-VII that supports the TUTase and ligation steps of U-insertion (Schnaufer et al. 2003). This implies that the editing complex contains separate regions for U-deletion and U-insertion (Schnaufer et al. 2003), at least the last two steps. These aggregate data suggest the editing complex may be streamlined, with substrate RNAs acted upon by either the U-deletional or U-insertional enzymatic activities, but not by both, potentially preventing TUTase and 3′-U-exo from interfering in U-deletion and U-insertion cycles, respectively. Conversely, finding appreciable levels of such cross-cycle activity would imply the editing cycles are not so streamlined.
Curiously, U-deletion reactions performed in the presence of UTP appear to yield more partially edited products, and less completely edited product, than standard in vitro U-deletion reactions that lack UTP (Cruz-Reyes and Sollner-Webb 1996). Here we analyze the basis of this “UTP effect” using a variety of experimental approaches and find that the TbMP57 TUTase, an enzyme thought to act exclusively in U-insertion, can also function within the U-deletion cycle, after cleavage and 3′-U-exo but before U-deletional ligation (Fig. 1A ▶, middle column). These data indicate that the U-deletion and U-insertion editing reactions are less segregated than previously suggested.
RESULTS
UTP affects the U-deletion reaction
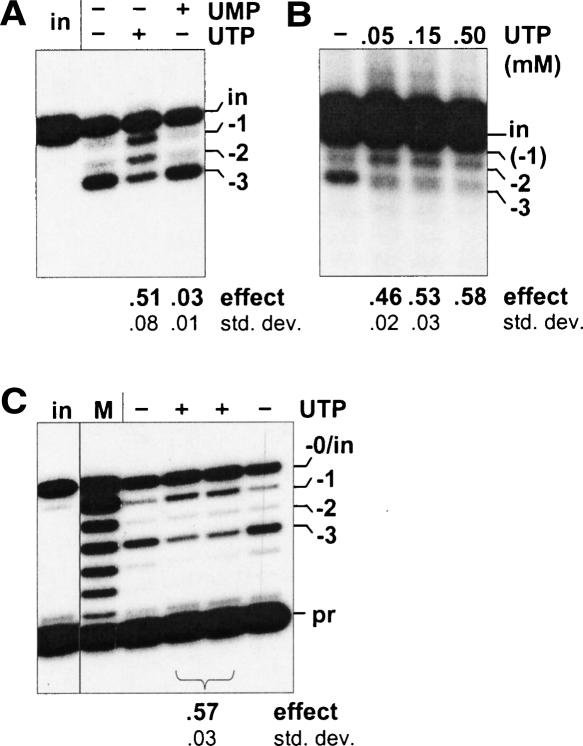
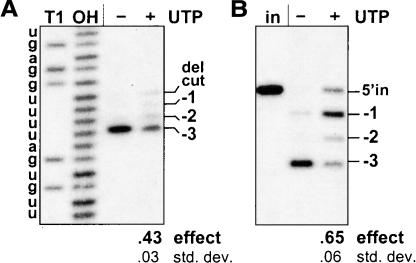
UTP is not normally added to in vitro U-deletion reactions, although it is essential in vivo and for in vitro U-insertion reactions. Intriguingly, in vitro U-deletion reactions catalyzed by UTP-supplemented mitochondrial extract produce more RNAs that migrate as if they are one or a few nucleotides longer than the complete U-deletion abundantly generated in the absence of UTP (Cruz-Reyes and Sollner-Webb 1996). We will refer to this as the “UTP effect” and have now examined whether it is due to the U-insertional TUTase acting within a U-deletion cycle or other possible scenarios. Figure 2A ▶ shows that UTP markedly alters the profile of U-deletion products catalyzed by our purified editing complex (the − and + UTP lanes), not only by cell extracts (Cruz-Reyes and Sollner-Webb 1996; also see ahead to Figs. 4C ▶, 6C,D ▶), suggesting the effect is caused by a component that purifies with that complex. The UTP effect occurs using natural (Fig. 2B ▶) or enhanced gRNAs (Fig. 2A ▶) and over a range of UTP concentrations (Fig. 2B ▶), including and below those generally used for in vitro U-insertion reactions (0.1–0.4 mM; Igo et al. 2000; Cruz-Reyes et al. 2002; Aphasizhev et al. 2003b). In all cases, UTP reduces the amount of the complete (− 3) U-deletion product and greatly increases products that are one or a few nucleotides longer. Additionally, the effect appears UTP specific, as it is not similarly directed by ATP, GTP, or UMP (Fig. 2A ▶; Cruz-Reyes and Sollner-Webb 1996). The latter result also suggests that the UTP effect does not arise from the extract generating UMP that causes end-product inhibition of the 3′-U-exo reaction (Rusche et al. 1997). Furthermore, assessment of full round U-deletion reactions using reverse transcriptase in poison primer extension analysis demonstrates that UTP causes products with an increased number of residues at the actual U-deletion site (Fig. 2C ▶). This result shows that UTP causes partial U-deletions and rules out the alternative possibility that the effect seen in Figure 2A,B ▶ could have reflected a “nonspecific” U-addition by TUTase (Bakalara et al. 1989) at the 3′ end of edited RNA molecules (see legend to Fig. 2C ▶), which in the standard assay using labeled substrate RNA would be indistinguishable from a partial U-deletion. Since the UTP effect occurs at the actual U-deletion site and is specific for UTP, it likely reflects either fewer U’s being removed or extra U’s being added at the U-deletion site, in the presence of UTP.
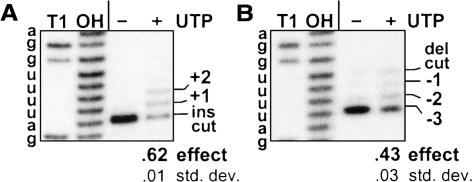
FIGURE 2.
Effect of UTP on full round U-deletion. Full-round reactions using A6 pre-mRNA (72 nt) and gRNA D32a, catalyzed by purified editing complex (see Materials and Methods; Rusche et al. 1997) and supplemented with no or 0.15 mM of the indicated nucleotide. The complete (−3) and partial (−2, −1) U-deletion products are indicated. The numbers below these and subsequent gels represent the UTP effect on U-deletion (effect; calculated as described in Materials and Methods and averaged over multiple experiments) and the standard deviation of this value (std. dev.). (A) U-deletion cycles with UTP. Input (in) pre-mRNA was 3′-end-labeled. (UMP, the end product of the 3′-U-exo reaction [Rusche et al. 1997], was also examined because it could be generated from UTP by phosphotransfer to the 3 mM AMP-CP that is added to facilitate U-deletional cleavage [Cruz-Reyes et al. 1998b).] (B) Effect of UTP concentration. Full round U-deletion reactions were as in A except with varying amounts of UTP and using the natural A6 gRNA g[2,1], which is ~30-fold less efficient than D32a gRNA (Cruz-Reyes et al. 2001). A duplicate experiment using D32a showed these same results, plus some UTP effect down to 1.5 μM UTP. (C) Analysis at the editing site. Reactions were as in A, except using nonradioactive input RNA and were assayed by poison primer extension (Seiwert and Stuart 1994), with synthesis extending just across the editing site. Assays of duplicate editing reactions are shown. The radiolabeled primer (pr; 25 nt) and extension products from input pre-mRNA (−0/in) and RNA with complete (−3) or partial (−2, −1) U-deletion at the editing site are indicated. The marker (M) is extension products from a partial hydroxide digest of the input RNA. (Preferential terminal U-addition to the 3′ end of mRNAs undergoing U-deletion could indeed be favored: Those 3′ ends are 28 nt from a U-deletion site bound to an editing complex, so its TUTase is tethered within ~12 nm of that 3′ end, making an effective concentration of ~200 μM relative to the TUTase, while the 3′ ends of unbound mRNAs are at 1.5 nM.)
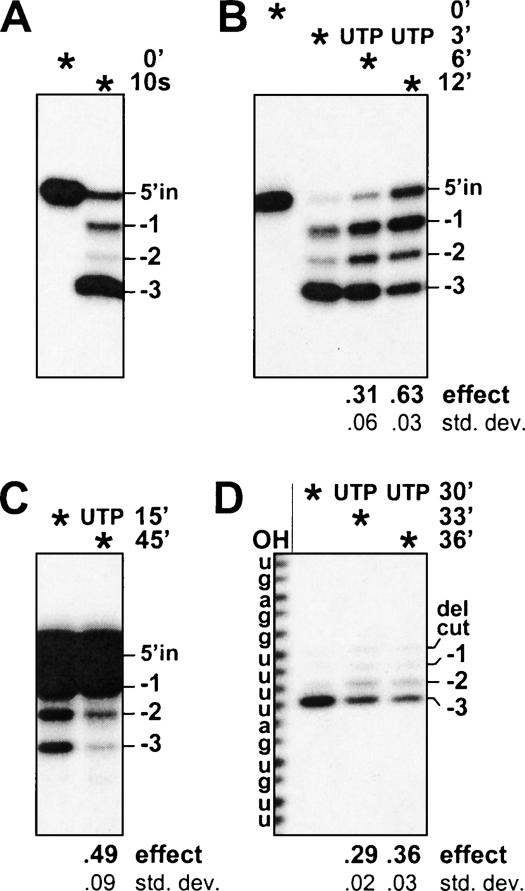
FIGURE 4.
UTP-mediated nucleotide addition following U removal by the 3′-U-exo. PPi-containing reactions used purified editing complex and were much as in Figure 3 ▶ except the UTP was added after a delay, as indicated by the time points on the right, next to the “UTP”. Asterisks indicate the time that the reactions were terminated. (A) 3′-U-exo reaction. Precleaved U-deletion reaction containing no UTP was incubated for only 10 sec. (B–D) UTP addition after a delay. (B) Precleaved U-deletion reactions. (C) Precleaved U-deletion reactions instead using rapidly prepared cell lysate (0.15 μg; see Materials and Methods) and 0.5 mM PPi. (D) Reactions using intact substrate, with the initial incubation without UTP for 30 min to allow endonuclease as well as 3′-U-exo action; after such an incubation, the TUTase activity is somewhat decreased.
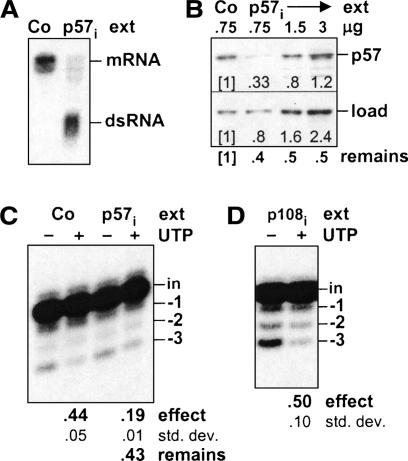
FIGURE 6.
RNAi analysis suggesting the UTP effect involves TbMP57. (A–C) RNAi to TbMP57. (A) Northern blot showing TbMP57 mRNA (mRNA) and TbMP57 double-stranded RNA (dsRNA) from TbMP57 RNAi (p57i) and control (Co; 29.13) cells at 24 h post-induction. (B) Western blot showing TbMP57 protein (p57) and a loading control protein (load) in various amounts of rapid cell lysate prepared from TbMP57 RNAi (p57i) and control (Co; 29.13) cells at 48 h post-induction. The numbers superimposed on the gel, under each lane, indicate the band intensity, relative to that of the control cells in the first lane. Below the gel is shown the fraction of TbMP57 protein remaining (remains) relative to the loading control protein for each lane. (C) UTP effect on the full round U-deletion reaction. Reactions were as in Figure 2A ▶, except using the rapid cell lysates of B. Below the final lane is shown the fraction of the UTP effect remaining in the TbMP57 RNAi (p57i) extract (remains) relative to that of the control (Co) extract. (D) UTP effect on full round U-deletion using TbTUT108 RNAi cells. Reactions were as in C, except using mitochondrial extract from cells in which TbTUT108 was depleted by RNAi (p108i).
Analyses in partial editing reactions
To start addressing at which step UTP affects U-deletion, we used 5′ and 3′ end-labeled substrates to visualize the upstream and downstream cleavage fragments, respectively, and PPi to inhibit ligation (Huang et al. 2001, 2002; Cruz-Reyes et al. 2002). In reactions with UTP, the size of the downstream mRNA fragment was not affected, but nearly half the upstream mRNA fragment became one or a few residues longer than in reactions without UTP, where the three terminal U’s are efficiently removed by the 3′-U-exo (Fig. 3A ▶; data not shown). Thus, UTP appears to act not by altering the position of the initial endonuclease cleavage but by altering the size acquired by the upstream cleavage fragment. Furthermore, UTP exerting its effect in the absence of ligation (Fig. 3A ▶) shows that this nucleotide does not cause partial U-deletions by favoring ligation before the 3′-U-exo has completed the U removal. It could have been that ATP (which is here added at 3 μM but decreases during the in vitro reaction) normally reaches levels that slow ligation and provide extra time for complete U removal, while an added 150 μM UTP could support phosphotransferase action to maintain the ~1 μM ATP concentration needed for maximal ligation (Cruz-Reyes et al. 2002). However, the result of Figure 3A ▶ eliminates the possibility that the UTP effect arises at the ligation step.
FIGURE 3.
The UTP effect scored without ligation and on cleaved RNA. (A) Assessing U-deletion after cleavage and before ligation. U-deletion reactions with purified editing complex were as in Figure 2A ▶ except using 5′-end-labeled A6 pre-mRNA (to examine the upstream RNA fragment) and were supplemented with 1.5 mM PPi. The upstream fragment following cleavage (del cut; 28 nt) and those upon subsequent removal of one, two, or all three of the 3′ U residues (−1, −2, −3) are indicated. Markers are the input mRNA treated with RNase T1 (T1; cleaves after G) or hydroxide (OH; cleaves after any nucleotide); they end in a 2′-3′ cyclic-P and therefore migrate slightly ahead of the experimental fragments, which end in 3′-OH (Cruz-Reyes et al. 1998a). (B) Analysis with precleaved U-deletion substrate. Reactions were as in A except using precleaved substrate and were incubated for 3 min. The input upstream oligoribonucleotide (5′ in; 16 nt) was 5′-end labeled; molecules that have lost one, two, or all three of the 3′-terminal U’s are indicated. The lane marked (in) contains input oligoribonucleotides without editing complex.
The effect of UTP on the upstream fragment can also be observed without the cleavage step, using an efficient U-deletional precleaved substrate (Law et al. 2005) in PPi-containing reactions. (Precleaved substrates mimic an editing site following endonuclease cleavage and consist of three annealed oligoribonucleotides corresponding to the upstream and downstream cleavage products plus the gRNA; Igo et al. 2000, 2002a, Igo et al. b.) The 5′-labeled upstream oligoribonucleotide ends in three unpaired U residues that are efficiently removed by the 3′-U-exo in the absence of UTP (Fig. 3B ▶, second lane), but with added UTP, its product is one or a few nucleotides longer (right lane). This experiment affirms that UTP affects the upstream fragment preceding ligation also with a precleaved U-deletion substrate. Thus the UTP effect results from either inhibition of the 3′-U-exo or stimulation of TUTase action at a U-deletion site.
The UTP effect results from TUTase action
Since UTP affects U-deletion without the ligation step, we can now add this nucleotide after a delay, to assess whether it causes 3′-U-exo inhibition or TUTase stimulation. The 3′-U-exo in the editing complex acts very quickly (Fig. 4A ▶), so precleaved U-deletion substrate (as used in Fig. 3B ▶) was first incubated for a short time with purified editing complex and PPi but no UTP (to support the 3′-U-exo but not activate TUTase or ligase), during which time >90% of the upstream oligoribonucleotide lost all three terminal U’s (Fig. 4B ▶, second lane). In parallel reactions conducted identically up to this time point, UTP was then added for a few minutes of incubation (Fig. 4B ▶, final two lanes). Notably, most of the shortened molecules then increased in size, first by an average of ~1 nt and later by ~2 nt. Thus, UTP can exert its effect after the 3′-U-exo of the editing complex has already acted. This demonstrates that its effect is not from inhibition of U removal but rather from stimulation of U-addition. Since the only activity of the basic editing complex known to add U’s is the TbMP57 TUTase (Aphasizhev at al. 2003b; O’Hearn et al. 2003; Schnaufer et al. 2003), this U-insertional TUTase appears to act also in U-deletion following the 3′-U-exo.
One could consider whether the lengthy protocol to purify the seven major protein complex might damage it in some manner that induces the TUTase to act within the U-deletion cycle. However, when we used very rapidly prepared trypanosomes lysate (Law et al. 2005), UTP causes lengthening of the upstream fragment following 3′-U-exo action and generates less of the complete U-deletion product (Fig. 4C ▶) in reactions like those of Figure 4B ▶. Thus, TUTase appears to act similarly in U-deletion whether catalyzed using purified complex or rapidly prepared extract.
Because precleaved substrate might behave somewhat differently than substrate that goes through the cleavage step of the U-deletion cycle, we also examined the delayed UTP addition using uncleaved mRNA. U-deletion reactions using intact substrate RNA were first incubated for 30 min with PPi and no UTP (to allow mRNA cleavage and 3′-U-exo action but not TUTase or ligase activity, as was done in Fig. 3A ▶), but in this experiment, parallel reactions were supplemented with UTP only after the 30-min incubation without this nucleotide (Fig. 4D ▶). After the initial incubation, where the 5′-end-labeled mRNA becomes cleaved (“del cut”) and the upstream fragments lose virtually all terminal U’s (first full lane), the short additional incubations with UTP caused an increasing fraction of these upstream fragments to became longer by one or a few residues (final two lanes). Thus, UTP stimulates TUTase to act at a U-deletion site after its cleavage and 3′ U removal.
The TUTase of U-insertion is responsible for the UTP effect on U-deletion
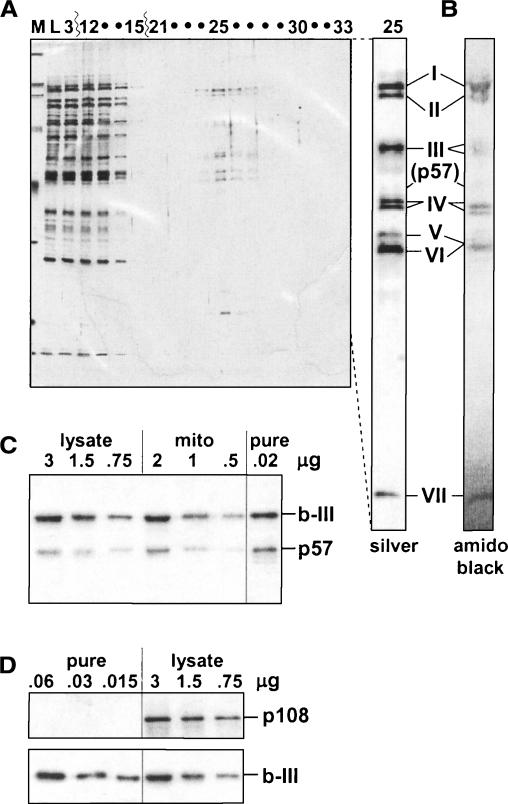
Figures 2 ▶–4 ▶ ▶ demonstrated that a TUTase adds U’s in U-deletion. Since the TbMP57 TUTase (Ernst et al. 2003), which adds the U’s in U-insertion (Aphasizhev at al. 2003b), is the only TUTase present in the purified editing complex, where the UTP effect is observed, this TUTase appears responsible for the U-addition in U-deletion. However, there is one potential caveat. In purified editing complex with seven major polypeptides, TbMP57 does not appear as a similarly major band, either by silver staining (Fig. 5A ▶; see also Rusche et al. 1997) or by amido black staining (Fig. 5B ▶) of SDS-PAGE gels. This raises the possibility that the U-insertional TUTase might be preferentially lost during our purification protocol (even though the resultant preparation catalyzes the U-insertion cycle relatively efficiently; Cruz-Reyes et al. 2002). To address this possibility, we examined rapidly prepared trypanosome lysate and purified editing complex as well as mitochondrial extract by Western blotting using antibodies against TbMP57 and TbMP63/b-III. TbMP63/b-III is used as a standard since it has been shown to remain at constant abundance relative to other members of the seven major protein editing complex throughout its purification (Huang et al. 2002; see also Brecht et al. 2005). Notably, the ratio of TbMP57 to TbMP63/b-III is approximately the same in all examined preparations (Fig. 5C ▶). We thus infer that our purified editing complex contains TbMP57 at its natural abundance relative to the seven major proteins, so this TUTase represents a viable candidate for the activity that adds the U’s in U-deletion.
FIGURE 5.
TbMP57 TUTase in the editing complex. (A) Proteins from the final purification step of the editing complex. A silver-stained 10% SDS-PAGE gel from the final DNA cellulose column in the purification of Rusche et al. (1997), showing a marker (M), the column load (L), fractions 3 and 12–15 representing the flow-through, and fractions 21–33 from the 30–350 mM KCl gradient elution. The editing complex peaks in fraction 25 (~100 mM KCl), and no other regions in this elution exhibited detectable protein signal. The separate lane at the right was loaded with more of fraction 25 and similarly stained. (The TbMP48/b-V protein stains brown rather than black and is considerably darker than appears in the photograph.) The positions of TbMP99/b-I (I) to TbMP18/b-VII (VII) are indicated, as is the region of TbMP57 (p57), identified by Western blotting. (B) Amido black staining. The SDS-PAGE gel of editing complex sent for sequence analysis (by Edman degradation; TbMP48/b-V and TbMP42/b-VI comigrated, presumably due to a slightly greater gel porosity; Rusche et al. 2001). (C) Western blot analysis of TbMP57. This TUTase (p57) is scored relative to TbMP63/b-III (b-III) using the indicated amounts of rapidly prepared cell lysate (lysate), mitochondrial extract (mito), and purified editing complex (pure). (D) Western blot analysis of TbTUT108. This TUTase (p108) is scored relative to TbMP63/b-III (b-III) using the indicated amounts of purified editing complex (pure) and rapid cell lysate (lysate).
Several lines of evidence support that the U’s are added in the U-deletion cycle by the TbMP57 TUTase of the editing complex that functions in U-insertion (Aphasizhev at al. 2003b) and not by the other trypanosome mitochondrial TUTase, TbTUT108, that adds the 3′ oligo-U tail to gRNAs and is separate from the basic editing complex (Aphasizhev at al. 2002, 2003b; Ernst et al. 2003). Indeed, our purification enriches by several hundred-fold the examined editing proteins (Rusche et al. 1997; O’Hearn et al. 2003), including TbMP57 (Fig. 5C ▶), while removing >99% of unrelated proteins (see also Sollner-Webb et al. 2001), including TbTUT108, which becomes undetectable (Fig. 5D ▶). Yet, as shown above, the UTP effect occurs at least as efficiently with this purified editing complex (Figs. 2 ▶–4 ▶ ▶) as with trypanosome lysate (Fig. 4C ▶) or whole mitochondrial extract (Cruz-Reyes and Sollner-Webb 1996), implying that the enzyme responsible for the U-addition is purified with the editing complex. This suggests that TbMP57 and not TbTUT108 mediates the UTP effect in purified complex.
Additional evidence that the TbMP57 TUTase catalyzes the UTP effect in U-deletion derives from cells induced to silence TbMP57 by RNAi. As previously reported (Aphasizhev at al. 2003b), RNAi of this TUTase markedly depletes its mRNA (Fig. 6A ▶) and is lethal. In our cultures, growth stops ~48 h post-induction, and Western blotting shows TbMP57 protein is reduced (Fig. 6B ▶). Importantly, TbMP57 RNAi cell extract also exhibits a reduced UTP effect, as shown in the full-cycle U-deletion reaction of Figure 6C ▶. Quantitation of the Western blots, normalized for a loading control, demonstrates that 40%–50% of TbMP57 protein remains, relative to control cells (Fig. 6B ▶), and quantitation of the full round U-deletion reactions reveals a similar fraction of the UTP effect remains in TbMP57 RNAi cell extract, relative to parallel control cell extract (Fig. 6C ▶). This suggests that TbMP57 TUTase is critical in adding U’s during U-deletion.
Conversely, the UTP effect is actively observed in extract from cells expressing RNAi to the TbTUT108. Figure 6D ▶ shows this in the full round U-deletion reaction, using extract where the TbTUT108 TUTase was depleted to a similar extent (not shown) as was the TbMP57 TUTase in Figure 6A–C ▶. While Figure 6D ▶ reports what is effectively a negative result, and thus cannot be used to provide proof, it certainly is consistent with the above conclusion that TbTUT108 is unimportant for the UTP effect.
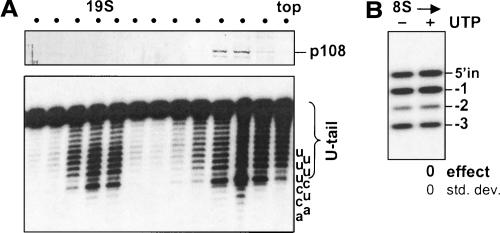
To examine whether a UTP effect on U-deletion could be generated by TbTUT108 when it is present in the reaction (even though this TUTase is not essential for the UTP effect with purified editing complex), we utilized a mitochondrial extract of control cells that had been resolved by velocity centrifugation. As expected, TbTUT108 sediments at ~5–10S (Aphasizhev at al. 2003b; O’Hearn et al. 2003; Fig. 7A ▶, upper panel), and the ~20S basic editing complex exhibits 3 ▶′-U-exo activity (Rusche et al. 1997; Fig. 7A ▶, lower panel). However, there is also substantial 3′-U-exo activity in upper fractions of the gradient, including in those that contain the TbTUT108 (Fig. 7A ▶, lower and upper panels). When assayed using precleaved U-deletion substrate, these ~5–10S fractions exhibit editing-specific 3′-U-exo activity (Fig. 7B ▶, first lane), but the product is not perceptibly affected by addition of UTP (Fig. 7B ▶, second lane). This result occurs despite these fractions being enriched in TbTUT108 (Fig. 7A ▶, upper panel), and contrasts with the substantial UTP effect exhibited by complex containing TbMP57 but not TbTUT108 (see Figs. 3B ▶, 4B ▶). The lack of a UTP effect from TbTUT108 suggests that this TUTase may not act on products generated by the 3′-U-exo, unlike the TbMP57 TUTase of the editing complex.
FIGURE 7.
The UTP effect does not result from TbTUT108. Glycerol gradient fractionation of mitochondrial extract from control cells. (A) (Upper panel) The fractions were assessed by Western blotting for TbTUT108 protein (p108) (see O’Hearn et al. 2003). (Lower panel) The same fractions were assayed for nonspecific 3′-U-exo activity. The substrate ends in a 3′ oligo-U tail, and the last nucleotide removed to generate each band is shown at the right. (B) UTP effect assayed as in Figure 3B ▶, except using the ~8S glycerol gradient fraction from A (the fraction 3 from the top) and 0.5 mM PPi. (As is traditional in the editing field, sedimentation coefficients are inferred relative to thyroglobulin [19S] and catalase [11S] in editing reaction buffer.) The specific assay of the 3′-U-exo appears less robust when this activity is separate from the ~19S editing complex.
TbMP57 TUTase acts in an editing-specific manner in U-deletion
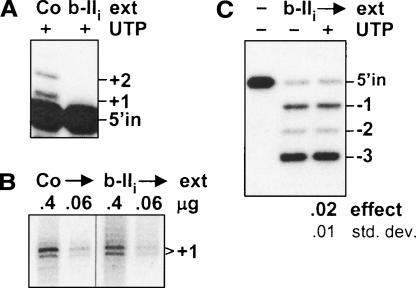
Does TbMP57 action in U-deletion involve its editing-specific recognition, as is needed in the U-insertion cycle, or only editing-independent end addition? To discern this, we have utilized cells in which TbMP81/b-II is silenced by RNAi (O’Hearn et al. 2003), since that protein is needed for specific recognition at each step of U-insertion (Law et al. 2005), including the TUTase step (Fig. 8A ▶), but is unimportant for editing-independent end addition by this TUTase (Fig. 8B ▶, as scored in its original assay; Bakalara et al. 1989). TbMP81/b-II is also unimportant for the normal U-deletion cycle (Law et al. 2005). Interestingly, in TbMP81/b-II RNAi extract, there is no discernable UTP effect on U-deletion (Fig. 8C ▶). Thus the action of the U-insertional TUTase in U-deletion appears dependent on an editing-specific recognition, rather than merely editing-independent addition by this TUTase.
FIGURE 8.
The UTP effect involves specific action of the TbMP57 TUTase. (A–C) RNAi to TbMP81/b-II. (A) Assay of the specific TUTase step in U-insertion, using precleaved U-insertion substrate (Igo et al. 2000) and 0.25 mM PPi, catalyzed by rapid cell lysates of TbMP81/b-II RNAi cells (b-IIi; Law et al. 2005) and control cells (Co; 29.13). The (5′-end-labeled) upstream input oligoribonucleotide (5′ in; 18 nt) and the guided +1 and +2 U-additions are indicated. (B) Assay of nonspecific U-addition activity of the TbMP57 TUTase in the lysates of A (see Law et al. 2005). In this experiment, the unlabeled input RNA is a mixture of two size classes, and the two indicated bands both represent molecules that acquired one U from the added radioactive [α-32P]UTP (Law et al. 2005), while nonspecific U-addition activity of the TbTUT108 should instead generate a ladder of longer fragments (Aphasizhev et al. 2002, 2003b; Ernst et al. 2003). (C) UTP effect assayed in precleaved U-deletion reactions as in Figure 3B ▶, except using the same lysates as in A and B, with 0.25 mM PPi.
Reinforcing the conclusion that the UTP effect derives from U-addition by TbMP57 and not TbTUT108, TbMP81/b-II RNAi extract retains the TbTUT108 (O’Hearn et al. 2003) and 3′-U-exo activity (Law et al. 2005) but did not show a UTP effect (Fig. 8C ▶). Therefore, the aggregate data indicate that UTP-induced partial U-deletions are due to U-addition by the functional TbMP57 TUTase of the editing complex, and not TbTUT108, acting within the U-deletion cycle.
TUTase exhibits similar efficiency in both forms of editing
We wanted to compare this cross-editing activity of the TbMP57 TUTase to its action in normal U-insertion reactions. Figure 9 ▶ shows that TUTase acts on U-insertional substrates following their cleavage (panel A) only slightly more efficiently than it acts on U-deletional substrates following their cleavage and 3′-U-exo action (panel B). Furthermore, we showed above that U’s appear to be added at least as quickly in U-deletion (Fig. 4B,D ▶) as in U insertion (Igo et al. 2000; data not shown). Thus, in vitro analysis demonstrates that the TUTase of the editing complex can act nearly as robustly at U-deletion sites as at U-insertion sites.
FIGURE 9.
Comparison of TUTase efficiency. (A) TUTase activity in U-insertion. PPi-containing reactions using intact U-insertional substrate were as in Figure 3A ▶, except using gRNA I47G and no AMP-CP (see Materials and Methods). The positions of the 5′-end-labeled mRNA after cleavage at the U-insertion site (ins cut; 32 nt) and when extended by one or two U nucleotides (+1, +2) are indicated. (B) TUTase activity in U-deletion. PPi-containing reactions using intact U-deletional substrate, reproduced from Figure 3A ▶.
DISCUSSION
We show that the majority of in vitro U-deletion cycles conducted on the standard ATPase 6 editing substrate in the presence of UTP includes a U-addition step that is catalyzed by the TbMP57 TUTase of the editing complex. Until now this enzyme has been thought to function only in U insertion (Fig. 1A ▶). However, during U-deletion, we show that the TUTase acts following cleavage and 3′-U-exo action, but before the ligation step (Fig. 4 ▶), to create partial U-deletion products. Observing this effect over the UTP concentration range reported to be physiological in other systems (~0.1–0.5 mM; Traut 1994; Moore et al. 1997; Casey et al. 1999) and commonly used for in vitro U-insertion reactions (0.1–0.4 mM; Igo et al. 2000; Cruz-Reyes et al. 2002; Aphasizhev et al. 2003b), as well as at lower UTP concentrations (Fig. 2B ▶), implies the phenomenon does not arise from abnormal levels of UTP. Furthermore, the UTP effect appears not to be from one peculiar cell line or unusual ionic conditions, as it is observed using extracts from multiple trypanosome lines (including TREU667 cells [Fig. 2A ▶] and derivatives of 427 cells [Fig. 6C,D ▶]) and using reaction buffers with salt concentrations (10 mM or 60 mM KCl, plus 10 mM MgCl2; Fig. 2A ▶; Cruz-Reyes and Sollner-Webb 1996) that span the range commonly used in editing reactions (Seiwert and Stuart 1994; Kable et al. 1996; Cruz-Reyes et al. 2002; Aphasizhev et al. 2003a,b; Schnaufer et al. 2003). The UTP effect also does not arise from aberrant guiding, as it occurs with natural (Fig. 2B ▶) and enhanced (Fig. 2A ▶) gRNAs, as well as with precleaved substrate (Figs. 3B ▶, 4B,C ▶). Importantly, UTP similarly affects U-deletion when catalyzed by purified complex (Fig. 2A ▶) or rapidly prepared lysate (Figs. 4C ▶, 6C ▶), suggesting the effect is due to activities of the editing complex, not to extraneous components and not to complexes that might have been damaged during lengthy purification. We therefore conclude that the UTP effect is an inherent property of the TUTase present in the basic editing complex. The reason that this TUTase action in U-deletion is observed here, but not in most studies, is that normal in vitro U-deletion reactions contain no UTP, which is needed for TUTase activity.
Might TUTase be able to function within U-deletion cycles also in vivo? While trypanosomes do require accurately edited transcripts, >90% of Trypanosoma brucei steady-state pan-edited mRNAs are misedited (Decker and Sollner-Webb 1990), indicating that in vivo editing experiences complications. Assuredly U-deletion cycles are faster in vivo than in vitro, leaving less time between the 3′-U-exo and ligation steps for possible TUTase action, and in vivo editing conditions could impose additional restrictions, so one might expect TUTase to less commonly generate partial U-deletions in vivo than in vitro. In addition, experiments have shown that partial U-deletions induce rounds of re-editing until the complete U-deletion is achieved (Cruz-Reyes and Sollner-Webb 1996). Thus, if TUTase generates some partial U-deletions also in vivo, it should not prevent the trypanosome from ultimately achieving correctly edited RNA. Nonetheless, the requisite re-editing could well slow the U-deletion process (Cruz-Reyes and Sollner-Webb 1996). In agreement, based on an extensive study of Leishmania tarentolae mRNAs that had been partially edited in vivo, it has been deduced that correct editing of a U-deletion site takes longer than that of a U-insertion site in vivo (Sturm and Simpson 1990). Furthermore, blocks of aberrant editing that exist in vivo often begin at natural U-deletion sites where extra U’s have been inserted (Sturm and Simpson 1990), and these were almost assuredly added by a TUTase acting at what should have been U-deletion sites. These extra U’s could then favor binding of a gRNA that will direct a block of aberrant editing (Sturm et al. 1992; Kable et al. 1996). Finally, ~90% of editing sites being U insertional and only ~10% U-deletional (Feagin et al. 1988; Blum et al. 1990) likely reflects a selection, suggesting that U-deletions present some difficulty in vivo. We speculate that this in vivo selection against U-deletions is from a difficulty in achieving complete U-deletion due to such TUTase-mediated partial editing. Thus, TUTase may act within some U-deletion cycles also in the trypanosome.
The studies reported in this communication pursued the initial observation that UTP-supplemented U-deletion reactions generate longer products (Cruz-Reyes and Sollner-Webb 1996), asking whether it reflects inhibition of 3′-U-exo, activation of TUTase, or other viable possibilities. We find that the UTP effect occurs at the actual U-deletion site (Fig. 2C ▶), is not due to end-product inhibition of the 3′-U-exo (Fig. 2A ▶), affects editing after the cleavage step and before the ligation step (Fig. 3 ▶), and yields upstream cleavage fragments with only partial U removal (Fig. 3 ▶). This enabled supplementing U-deletion reactions with UTP after a delay, when the 3′-U-exo has already acted (Fig. 4 ▶), and thereby demonstrating that the UTP effect arises from addition by a TUTase, not from slowing the 3′-U-exo. These conclusions were reinforced by the absence of a UTP effect in various preparations that maintain the U-deletional activity of the 3′-U-exo (Figs. 7B ▶, 8C ▶). The UTP effect was then shown to involve the TbMP57 TUTase of the editing complex (Fig. 5C ▶), and not the other mitochondrial TUTase (TbTUT108), by separately depleting these two enzymes using biochemical fractionation (Figs. 2A ▶, 5C,D ▶, 7B ▶) and RNAi (Fig. 6C,D ▶) approaches. Indeed, the UTP effect appears diminished in proportion to TbMP57 (quantitation in Fig. 6B,C ▶). Interestingly, the lack of UTP effect in TbMP81/b-II RNAi extract (Fig. 8C ▶), which has previously been characterized (Drozdz et al. 2002; O’Hearn et al. 2003; Law et al. 2005), implies that TUTase action in U-deletion is a property of functional U-insertional machinery and is not merely due to nonspecific U-addition by TbMP57. (The TbMP81/b-II RNAi analysis is based on our recent demonstration that this extract is fully functional for U-deletion and retains the TbMP57 protein and its nonspecific end addition activity but not its specific recognition of the editing substrate; Law et al. 2005.) It is also notable that the U-insertional TUTase functions within a large fraction of in vitro U-deletion cycles (Fig. 2A ▶, 3A ▶, 4D ▶, 9B ▶), nearly as efficiently as in U-insertion cycles (Fig. 9A ▶), both assessed on their standard ATPase 6 editing substrates. Thus, the observed effect of UTP on U-deletion arises from the majority of active editing complexes, rather than from a small minority of possibly aberrant complexes. We conclude that TbMP57 TUTase adds U’s not only in U insertion but also within the U-deletion pathway, at least in vitro and potentially also in vivo.
Although TbMP57 in our purified editing complex stains much less intensely than the other seven major proteins, both with silver (Fig. 5A ▶; Rusche et al. 1997, 2001) and with amido black (Fig. 5B ▶), we here show that TbMP57 TUTase is as abundant, relative to the seven major proteins, in the purified editing complex as in a rapid T. brucei extract (Fig. 5C ▶). Although this suggestion that the TbMP57 TUTase is not selectively lost during our purification of the basic editing complex remains to be resolved with the reported similar intensity silver staining of TbMP57 and TbMP48/b-V in the editing complex preparation with ~20 proteins (Panigrahi et al. 2003b), there is general agreement that this TUTase is a key component of the editing complex.
Earlier studies on editing have indicated that U-deletional and U-insertional cycles use distinct activities, within distinct domains of the editing complex (as summarized in the Introduction), suggesting that progression through the two forms of editing may be streamlined to avoid potentially conflicting TUTase and 3′-U-exo activities. However, the data in the present study suggest a revised view of selectivity within the editing complex, where the U-insertional TUTase can also act efficiently within the U-deletional cycle (Figs. 1A ▶, 4 ▶–8 ▶ ▶ ▶ ▶). This implies that the U-deletional and U-insertional subcomplexes (Schnaufer et al. 2003) do not fully segregate the RNA editing substrate. Rather, U-deletional and U-insertional activities may compete for the RNA based on kinetics and/or binding affinity. While this is the first demonstration that an activity of the intact editing complex can function in both forms of editing, other activities might also function in that manner. For instance, TbMP52/b-IV, the ligase that seals U deletion, also seals U insertion when the U-insertion-specific ligase, TbMP48/b-V, is depleted by RNAi (Drozdz et al. 2002; Gao and Simpson 2003; O’Hearn et al. 2003); in fact, TbMP52/b-IV also fulfills this role when TbMP48/b-V is present but inactive (A. Zhelonkina and J. Cruz-Reyes, unpubl. observations). Further, it has been proposed that the 3′-U-exo might also act in U insertion, to remove extra U residues that may be added by the TUTase (Byrne et al. 1996; Alfonzo et al. 1997; McManus et al. 2000; Igo et al. 2002b). Thus, the editing pathway might turn out to be more complex than currently envisioned, with several enzymes functioning in both types of editing.
MATERIALS AND METHODS
Cells and extract preparation
We used T. brucei procyclic cells strain TREU-667 for experiments not involving RNAi and 29.13 cells (Wirtz et al. 1999) derived from strain 427 for RNAi experiments. 29.13 cells express T7 RNA polymerase and tetracycline (tet) repressor and were stably transfected with pZJM-based plasmids (Wang et al. 2000), which carry an insert flanked by T7 promoters and tet operators. The inserts were from the genes encoding TbMP81/b-II (O’Hearn et al. 2003; 712 bp), TbMP57 (509 bp), or TbTUT108 (886 bp), the latter two subcloned analogously to the former, using fragments PCR amplified with the following oligonucleotides:
TbMP57 (5′) XhoI: 5′-GATCTCGAGGTTGATGCATACCGCGCCATG-3′, TbMP57 (3′) HindIII: 5′-GAAAGCTTGCCTCCTCTTCGCTAAGTGGCG-3′, TbTUT108 (5′) HindIII: 5′-CCCAAGCTTTTTGTACAGCTCCTCCCGCTCTG-3′, and TbTUT108 (3′) XhoI: 5′-TCGCTCGAGCCGCTGGCATGATAAAGCACG-3′.
pZJM also directs phleomycin resistance and rDNA integration (Wang et al. 2000), enabling selection for ectopically integrated plasmid and then cloning lines from single cells by extreme dilution, as described (O’Hearn et al. 2003). Expression of the double-stranded RNA is induced by addition of tet (1 μg/mL) to the Cunningham’s culture media as described (Wang et al. 2000; Huang et al. 2002). Traditional mitochondrial extracts (Harris et al. 1990; Sabatini et al. 1998; ~2.5 × 1010 cell equivalents/mL), which contain bands I–VII as ~1/500 of total protein (Rusche et al. 1997; O’Hearn et al. 2003), and rapid cell lysates (Huang et al. 2002), which also catalyze the steps of editing (Law et al. 2005), were prepared. Editing complex with the seven major silver-staining proteins was purified from TREU-667 cells (Rusche et al. 1997; Sollner-Webb et al. 2001). Mitochondrial extract was also fractionated by glycerol gradient centrifugation (Rusche et al. 1997; Huang et al. 2001).
Characterization of editing complex
To evaluate protein constituents of the editing complex, samples were resolved on 10% SDS-PAGE gels and proteins detected by Western blotting (Huang et al. 2002) or staining, using silver (Rusche et al. 1997) or amido black (Bio-Rad) following the manufacturers’ instructions. TbMP63/b-III provides a loading control, as it has been shown to remain constant, relative to the unaffected editing proteins (and the mitochondrial protein lipoamide dehydrogenase) in parental cells (Huang et al. 2002), in TbMP81/b-II RNAi cells (O’Hearn et al. 2003), and in TbMP57 RNAi cells (Aphasizhev et al. 2003b). Polyclonal antibodies (Covance) were prepared using recombinant editing proteins expressed from cloned genes (as in Huang et al. 2002; O’Hearn et al. 2003), and antibody to lipoamide dehydrogenase was a gift from R.L. Krauth-Siegel (Heidelberg University Biochemistry Center, Germany). Northern blots were preformed as in O’Hearn et al. (2003), and nonspecific 3′-U-exo activity was assayed as in Huang et al. (2002) and Law et al. (2005).
Preparation of editing substrates
A6 pre-mRNA m[0,4] and gRNAs for U deletion (D32a and g[2,1]; Cruz-Reyes et al. 2001) and U insertion (I47G; Huang et al. 2001) were synthesized from PCR-generated templates, as described (Cruz-Reyes et al. 2001). The m[0,4] RNA was 3′-end labeled for direct analysis of full-round U deletion or 5′-end labeled for reactions with PPi (Cruz-Reyes and Sollner-Webb 1996; Cruz-Reyes et al. 1998b). The mRNA (~30 fmol) and gRNA (1.25 pmol) were pre-annealed for each reaction, as described (Cruz-Reyes et al. 1998b,c). With m[0,4], gRNAs D32a and g[2,1] direct deletion of three U’s at editing site 1 (Seiwert and Stuart 1994; Cruz-Reyes et al. 2001) while gRNA I47G directs insertion of two U’s at editing site 2 (Huang et al. 2001). The substrates for precleaved U deletion and precleaved U insertion were prepared and pre-annealed as described (Law et al. 2005 and Igo et al. 2000, respectively), using ~50 fmol of the (5′-end-labeled) oligoribonucleotide representing the upstream mRNA fragment, 1 pmol of oligoribonucleotide representing the downstream mRNA fragment, and 0.25 or 0.5 pmol of the complementary gRNA oligoribonucleotide. The oligoribonucleotides for precleaved U deletion comprise 16 nt upstream and 15 (or 18) nt downstream from the cleavage position in editing site 1 of m[0,4], plus gRNA D33′, and they direct deletion of three U’s (Law et al. 2005). The oligoribonucleotides for precleaved U insertion (Igo et al. 2000) utilize the A6 anchor sequence and an apparently novel duplexed tether sequence, and they direct insertion of two U’.
Full round U-deletion reactions and primer extension analyses
Full round U-deletion reactions (Cruz-Reyes et al. 1998b,c; 20 μL) were in 10 mM KCl-MRB buffer (Cruz-Reyes et al. 2002) supplemented with 3 mM AMP-CP plus 3 μM ATP, and, where indicated, also 0.15 mM UTP (or UMP). These reactions were catalyzed using ~15 ng of purified editing complex plus 0.05 μg/μL hexokinase (Rusche et al. 1997) or ~1.5 μg of mitochondrial extract protein plus 20 U RNase inhibitor (Promega) and 10 mM DTT, and were incubated at 28°C for ~1 h. The product RNAs were analyzed on a 1-m-long, 9% polyacrylamide/7.5 M urea gel in TBE. For poison primer extension analysis, U-deletion reactions used unlabeled m[0,4] pre-mRNA, and the product RNAs were annealed to 0.08 pmol of 5′-end-labeled A6-RT primer (Seiwert and Stuart 1994; 10 min at 70°C). After 1 h incubation at 48°C with 200 U of SuperScript II RNase H− reverse transcriptase (Invitrogen) in the manufacturer’s buffer supplemented with 10 mM DTT, 0.4 mM dATP and dCTP, plus 0.1 mM dideoxy-TTP (ddTTP, to stop synthesis just beyond the editing site), the product was resolved on a 20% polyacrylamide/7.5 M urea gel in TBE, as described (Seiwert and Stuart 1994).
Reactions without ligation
To inhibit ligation, U-deletion reactions were as above, except they lacked ATP and instead contained PPi, as specified in the figure legends and were for 30–45 mins. Reactions using precleaved U-deletion or U-insertion substrate or intact U-insertion substrate also lacked AMP-CP (Cruz-Reyes et al. 1998b,c, 2002; Huang et al. 2001; Law et al. 2005). Where indicated, catalysis instead used rapid cell lysates (1.2 μg protein) plus 20 U RNase inhibitor and 10 mM DTT. Recovered RNAs were resolved on 40-cm-long 15% (for precleaved assay) or 9% (for assays with cleavage) polyacrylamide/7.5 M urea gels in TBE. Markers were prepared from the labeled substrate, cleaved either with one U of RNase T1 (Ambion) following the manufacturer’s protocol or with NaOH as described (Cruz-Reyes et al. 1998a).
Data analysis
Autoradiograms of electrophoretically resolved RNA were scanned using a FluorChem 8000 Advanced Fluorescence, Chemiluminescence and Visible Light Imaging System with AlphaEaseFC software and analyzed within the linear range. The UTP effect, or increase of partial U-removal products when reactions contain UTP, is calculated as [(P/B)+ − (P/B)−]/[1 − (P/B) −] where the + and − subscripts indicate parallel reactions with and without UTP, and P and B represent the summed discrete band intensities from Partial U-removal products and from Both partial and complete U-removal products. This value can range from 0 to 1, meaning UTP caused none or all of the products to become partial U deletions. Shown below the gel lanes is the UTP effect, averaged over multiple replicated experiments and their standard deviation.
Acknowledgments
We thank Laura Rusche for purified editing complex and its gel analysis, R.L. Krauth-Siegel for antibody to lipoamide dehydrogenase, members of our laboratory for helpful discussions, and Paul Englund and Jeffrey Shaman for valuable suggestions on the manuscript. The work was funded by Public Health Service grant number GM34231 from the General Medical Science Institute.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2243206.
REFERENCES
- Alfonzo, J., Thiemann, O., and Simpson, L. 1997. The mechanism of U insertion/deletion RNA editing in kinetoplastid mitochondria. Nucleic Acids Res. 25: 3751–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev, R., Sbicego, S., Peris, M., Jang, S., Aphasizheva, I., Simpson, A., Rivlin, A., and Simpson L. 2002. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell 108: 637–648. [DOI] [PubMed] [Google Scholar]
- Aphasizhev, R., Aphasizheva, I., Nelson, R., Gao, G., Simpson, A., Kang, X., Falick, A., Sbicego, S., and Simpson, L. 2003a. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 22: 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev, R., Aphasizheva, I. and Simpson, L. 2003b. A tale of two TUTases. Proc. Natl. Acad. Sci. 100: 10617–10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalara, N., Simpson, A.M., and Simpson, L. 1989. The Leishmania kinetoplast-mitochondrion contains terminal uridylyltransferase and RNA ligase activities. J. Biol. Chem. 264: 18674–18686. [PubMed] [Google Scholar]
- Blum, B., Bakalara, N., and Simpson, L. 1990. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60: 189–198. [DOI] [PubMed] [Google Scholar]
- Brecht, M., Niemann, M., Schluter, E., Muller, U.F., Stuart, K., and Goringer, H.U. 2005. TbMP42, a protein component of the RNA editing complex in African trypanosomes, has endo-exoribonuclease activity. Mol. Cell 17: 621–30. [DOI] [PubMed] [Google Scholar]
- Byrne, E.M., Connell, G.J., and Simpson, L. 1996. Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J. 15: 6758–6765. [PMC free article] [PubMed] [Google Scholar]
- Casey, T., Dufall, K., and Arthur, P. 1999. An improved capillary electrophoresis method for measuring tissue metabolites associated with cellular energy state. Eur. J. Biochem. 261: 740–745. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes, J. and Sollner-Webb, B. 1996. Trypanosome U deletional RNA editing involves gRNA-directed endonuclease cleavage, terminal U-exonuclease, and RNA ligase activities. Proc. Natl. Acad. Sci. 93: 8901–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes, J., Piller, K., Rusche, L., Mukherjee, M., and Sollner-Webb, B. 1998a. Unexpected electrophoretic migration of RNA with different 3′ termini causes a RNA sizing ambiguity that can be resolved using nuclease P1. Biochemistry 37: 6059–6064. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes, J., Rusche, L., Piller, K., and Sollner-Webb, B. 1998b. T. brucei RNA editing: Adenosine nucleotides inversely affect U deletion and U insertion reactions at mRNA cleavage. Mol. Cell 1: 401– 409. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes, J., Rusche, L., and Sollner-Webb, B. 1998c. Trypanosoma brucei U insertion and U deletion activities co-purify with an enzymatic editing complex but are differentially optimized. Nucleic. Acids Res. 26: 3634–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes, J., Zhelonkina, A., Rusche, L., and Sollner-Webb, B. 2001. Trypanosome RNA editing: Simple guide RNA features enhance U deletion 100 fold. Mol. Cell. Biol. 21: 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes, J., Zhelonkina, A., Huang, C., and Sollner-Webb, B. 2002. Distinct functions of two RNA ligases in active T. brucei RNA editing complexes. Mol. Cell. Biol. 22: 4652–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C. and Sollner-Webb, B. 1990. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell 61: 1001–1011. [DOI] [PubMed] [Google Scholar]
- Drozdz, M., Palazzo, S., Salavati, R., O’Rear, J., Clayton, C., and Stuart, K. 2002. TbMP81 is required for RNA editing in T. brucei. EMBO J. 21: 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, N., Panicucci, B., Igo, R., Panigrahi, A., Salavati, R., and Stuart, K. 2003. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell 11: 1525–1536. [DOI] [PubMed] [Google Scholar]
- Feagin, J., Abraham, J., and Stuart, K. 1988. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell 53: 413–422. [DOI] [PubMed] [Google Scholar]
- Gao, G. and Simpson, L. 2003. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J. Biol. Chem. 278: 27570– 27574. [DOI] [PubMed] [Google Scholar]
- Gott, J. and Emeson, R. 2000. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34: 499–531. [DOI] [PubMed] [Google Scholar]
- Hajduk, S. 1997. Defining the editing “reaction”. Trends Microbiol. 5: 1–2. [DOI] [PubMed] [Google Scholar]
- Harris, M.E., Moore, D.R., and Hajduk, S.L. 1990. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 265: 11368–11376. [PubMed] [Google Scholar]
- Huang, C., Cruz-Reyes, J., Zhelonkina, A., O’Hearn, S., Wirtz, E., and Sollner-Webb, B. 2001. Roles for the ligases in the RNA editing complex of T. brucei: Band IV is needed for U-deletion and RNA repair. EMBO J. 20: 4694–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., O’Hearn, S., and Sollner-Webb, B. 2002. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 22: 3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo, R., Palazzo, S., Burgess, M., Panigrahi, A. and Stuart, K. 2000. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell. Biol. 20: 8447– 8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo, R., Lawson, S., and Stuart, K. 2002a. RNA sequence and base pairing effects on insertion editing in T. brucei. Mol. Cell. Biol. 22: 1567–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo, R., Weston, D., Ernst, N., Panigrahi, A., Salavati, R., and Stuart, K. 2002b. Role of uridylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Eukar. Cell 1: 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kable, M., Seiwert, S., Heidmann, S., and Stuart, K. 1996. RNA editing: A mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science 273: 1189–1195. [DOI] [PubMed] [Google Scholar]
- Law, J.A., Huang, C.E., O’Hearn, S., and Sollner-Webb, B. 2005. In Trypanosoma brucei RNA editing, band II enables recognition specifically at each step of the U insertion cycle. Mol. Cell. Biol. 25: 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison-Antenucci, S., Sabatini, R., Pollard, V., and Hajduk, S. 1998. Kinetoplast RNA editing associated protein 1 (REAP-1): A novel editing complex protein with repetitive domains. EMBO J. 17: 6368–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison-Antenucci, S., Grams, J., and Hajduk, S. 2002. Editing machines: The complexities of trypanosome RNA editing. Cell 108: 435–438. [DOI] [PubMed] [Google Scholar]
- McManus, M., Adler, B., Pollard, V., and Hajduk, S. 2000. T. brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Mol. Cell. Biol. 20: 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, A., Mercer, J., Dutina, G., Donahue, C., and Ryll, T. 1997. Effects of temperature shift on cell cycle, apoptosis and nucleotide pools in CHO cell batch cultures. Cytotechnology 23: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hearn, S., Huang, C., Hemann, M., Zhelonkina, A., and Sollner- Webb, B. 2003. Trypanosoma brucei RNA editing complex: Band II is structurally critical and maintains band V ligase, which is non-essential. Mol. Cell. Biol. 23: 7909–7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A., Gygi, S., Ernst, N., Igo, R., Palazzo, S., Schnaufer, A., Weston, D., Carmean, N., Salavati, R., Aebersold, R., et al. 2001a. Association of two novel proteins, TbMP52 and TbMP48, with the T. brucei RNA editing complex. Mol. Cell. Biol. 21: 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A., Schnaufer, A., Carmean, N., Igo, R., Gygi, S., Ernst, N., Palazzo, S., Weston, D., Aebersold, R., Salavati, R., et al. 2001b. Four related proteins of the T. brucei RNA editing complex. Mol. Cell. Biol. 21: 6833–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A., Schnaufer, A., Ernst, N., Wang, B., Carmean, N., Salavati, R., and Stuart, K. 2003a. Identification of novel components of Trypanosoma brucei editosomes. RNA 9: 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A.K., Allen, T.E., Stuart, K., Haynes, P.A., and Gygi, S.P. 2003b. Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J. Am. Soc. Mass. Spectrom. 14: 728–735. [DOI] [PubMed] [Google Scholar]
- Pollard, V., Harris, M., and Hajduk, S. 1992. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 11: 4429–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L., Cruz-Reyes, J., Piller, K., and Sollner-Webb, B. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16: 4069–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L., Huang C., Piller K., Hemann M., Wirtz E., and Sollner- Webb, B. 2001. The two RNA ligases of the Trypanosoma brucei RNA editing complex: Cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol. 21: 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini, R., Adler, B., Madison-Antenucci, S., McManus, M., and Hajduk, S. 1998. Biochemical methods for analysis of kinetoplastid RNA editing. Methods Companion Methods Enzymol. 15: 15–26. [DOI] [PubMed] [Google Scholar]
- Schnaufer, A., Panigrahi, A., Panicucci, B., Igo, R., Salavati, R., and Stuart, K. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291: 2159–2162. [DOI] [PubMed] [Google Scholar]
- Schnaufer, A., Ernst, N., Palazzo, S., O’Rear, J., Salavati, R., and Stuart, K. 2003. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell 12: 307–319. [DOI] [PubMed] [Google Scholar]
- Seiwert, S. and Stuart, K. 1994. RNA editing: Transfer of genetic information from gRNA to precursor mRNA in vitro. Science 266: 114–117. [DOI] [PubMed] [Google Scholar]
- Seiwert, S., Heidmann, S., and Stuart, K. 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84: 831–841. [DOI] [PubMed] [Google Scholar]
- Simpson, L., Sbicego, S., and Aphasizhev, R. 2003. Uridine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA 9: 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb, B., Rusche, L., and Cruz-Reyes, J. 2001. Ribonuclease activities of Trypanosome RNA editing complex directed to cleave specifically at a chosen site. Methods Enzymol. 341: 154–174. [DOI] [PubMed] [Google Scholar]
- Sturm, N. and Simpson, L. 1990. Partially edited mRNAs for cytochrome b and subunit III of cytochrome oxidase from Leishmania tarentolae mitochondria: RNA editing intermediates. Cell 61: 871–878. [DOI] [PubMed] [Google Scholar]
- Sturm, N., Maslov, D., Blum, B., and Simpson, L. 1992. Generation of unexpected editing patterns in Leishmania tarentolae mitochondrial mRNAs: Misediting produced by misguiding. Cell 70: 469– 476. [DOI] [PubMed] [Google Scholar]
- Stuart, K. and Panigrahi, A. 2002. RNA editing: Complexity and complications. Mol. Microbiol. 45: 591–596. [DOI] [PubMed] [Google Scholar]
- Stuart, K., Allen, T., Heidmann, S., and Seiwert, S. 1997. RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev. 1: 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, K.D., Schnaufer, A., Ernst, N.L., and Panigrahi, A.K. 2005. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 30: 97–105. [DOI] [PubMed] [Google Scholar]
- Traut, T. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140: 1–22. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Morris, J.C., Drew, M.E., and Englund, P.T. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275: 40174–40179. [DOI] [PubMed] [Google Scholar]
- Wirtz, E., Leal, S., Ochatt, C., and Cross, G.A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99: 89–101. [DOI] [PubMed] [Google Scholar]