Abstract
1-Methyladenosine modification at position 58 of tRNA is catalyzed by a two-subunit methyltransferase composed of Trm6p and Trm61p in Saccharomyces cerevisiae. Initiator tRNA (tRNAiMet) lacking m1A58 (hypomethylated) is rendered unstable through the cooperative function of the poly(A) polymerases, Trf4p/Trf5p, and the nuclear exosome. We provide evidence that a catalytically active Trf4p poly(A) polymerase is required for polyadenylation of hypomethylated tRNAiMet in vivo. DNA sequence analysis of tRNAiMet cDNAs and Northern hybridizations of poly(A)+ RNA provide evidence that nascent pre-tRNAiMet transcripts are targeted for polyadenylation and degradation. We determined that a mutant U6 snRNA and an aberrant form of 5S rRNA are stabilized in the absence of Trf4p, supporting that Trf4p facilitated RNA surveillance is a global process that stretches beyond hypomethylated tRNAiMet. We conclude that an array of RNA polymerase III transcripts are targeted for Trf4p/ Trf5p-dependent polyadenylation and turnover to eliminate mutant and variant forms of normally stable RNAs.
Keywords: RNA surveillance, exosome, tRNA turnover, polyadenylation
INTRODUCTION
Gene transcription by eukaryotic RNA polymerases I, II, or III result in the formation of nascent pre-RNA transcripts that undergo a series of maturation steps involving exo-/endonucleolytic trimming, splicing, post-transcriptional modifications, and addition of 3′ polyadenylate tail. The addition of 3′ poly(A) tracts to mRNA is important in many aspects of mRNA metabolism. In eukaryotes, the conventional role of poly(A) tails has been in the stabilization, localization, and translation initiation of mRNA transcripts (Wahle and Ruegsegger 1999; Proudfoot 2004). In Xenopus laevis, polyadenylation of maternally derived cytoplasmic mRNAs stimulates their translation during oocyte maturation (Ballantyne et al. 1997; Dickson et al. 2001). In Escherichia coli, poly(A) tails are not normally present in mRNA, except when mRNA is under the process of turnover (Edmonds 2002).
This traditional view of polyadenylation in eukaryotic RNA metabolism was changed by our previous finding that it plays a role in the degradation of tRNAiMet lacking 1-methyladenosine (m1A58) modification in yeast (Kadaba et al. 2004) similar to a mechanism in E. coli where a mutant tRNATrp is degraded through polyadenylation (Li et al. 2002). In other instances, polyadenylated forms of other noncoding RNAs like SnRNA, SnoRNAs, and rRNAs have been observed in yeast mutants (Allmang et al. 1999; van Hoof et al. 2000b; Kuai et al. 2004).
The synthesis of m1A58 in the TΨC loop of tRNAs is catalyzed by the methyltransferase encoded by TRM6 and TRM61 (Anderson et al. 1998). The absence of the m1A58 modification from initiator tRNA (tRNAiMet) results in its degradation (Anderson et al. 1998; Kadaba et al. 2004). The selective degradation of tRNAiMet lacking m1A58 in trm6 and trm61 mutants is presumably due to a weakening of a tertiary interaction between D and T loops that is unique to tRNAiMet (Basavappa and Sigler 1991). Trf4p, a member of nucleotidyltransferase family of proteins (Aravind and Koonin 1999), and the exosome, a multiprotein complex composed of 3′–5′ riboexonucleases involved in various RNA degradation and processing (Mitchell et al. 1997; Mitchell 2001; Butler 2002), function cooperatively in a novel surveillance pathway to degrade tRNAiMet lacking m1A58 (hypomethylated tRNAiMet) (Kadaba et al. 2004).
Previous studies from our laboratory support that Trf4p is required for the polyadenylation of hypomethylated tRNAiMet (Kadaba et al. 2004). In accordance with this observation, recent in vitro studies have shown that Trf4p functions in an oligomeric complex composed of Mtr4p, a putative RNA helicase, and Air2p, a putative RNA binding protein, to polyadenylate in vitro synthesized RNA substrates and stimulate degradation by the exosome (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005). A limitation of these studies was that the function of Trf4p as a poly(A) polymerase of tRNA substrates was studied in vitro, which may or may not correlate with its actual function in cells. In the present study we show that polyadenylation of hypomethylated tRNAiMet in vivo is dependent on a functionally active Trf4p.
Another interesting aspect of RNA degradation by Trf4p and the exosome that remains to be elucidated is the preferential recognition of mutant forms of RNA by TRAMP and the exosome. In the present study, we show that nascent hypomethylated tRNAiMet with unprocessed 5′ leader and 3′ extension is the substrate for polyadenylation, thus indicating that polyadenylation by Trf4p and degradation by the exosome occur very early during the biogenesis of hypomethylated tRNAiMet.
Suggesting a more broadly utilized mechanism of RNA turnover, we have identified a mutant U6 SnRNA and a truncated 5S rRNAs that are stabilized by mutations in TRF4 and the exosome, supporting the idea that adenylation and degradation of aberrant RNAs is an important step in preventing their incorporation into the cellular macro-molecular machinery.
RESULTS
Trf4p is required for the polyadenylation of hypomethylated pre-tRNAiMet
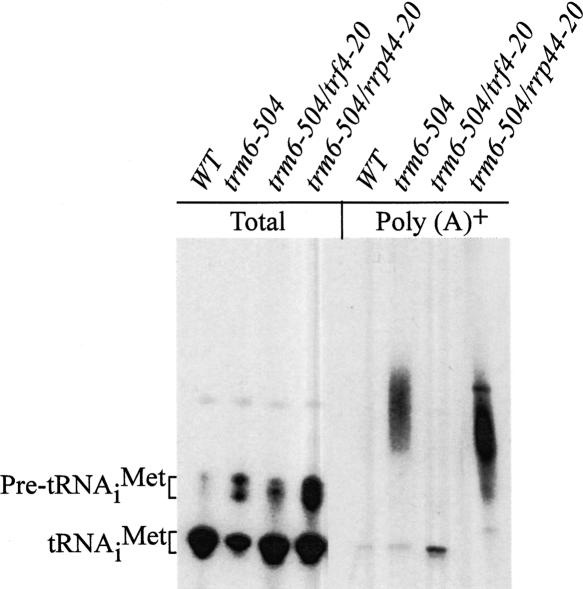
Previously, we proposed that polyadenylation of pre-tRNAiMet lacking m1A58 (hypomethylated) stimulates its degradation by the nuclear exosome based on the observation that tRNAiMet is polyadenylated in a trm6-504 rrp6Δ double mutant, where degradation of tRNAiMet is suppressed. To establish that polyadenylation of hypomethylated tRNAiMet is a consequence of m1A58 hypomethylation and not a pleiotropic effect of deleting RRP6, we conducted Northern blot analysis of poly(A)+ RNA from wild-type, trm6-504 and trm6-504 rrp44-20 strains. Using equivalent proportions of poly(A)+ RNA from the above strains we detected poly(A)+ tRNAiMet in a trm6-504 strain and to a greater extent in trm6-504 rrp44-20 strain, while at the same time there was no poly(A)+ tRNAiMet detectable in the wild-type strain (Fig. 1 ▶), indicating that tRNAiMet is polyadenylated in response to a loss of m1A58. Polyadenylation was unique to hypomethylated tRNAiMet and not triggered by a loss of m1A58, since we failed to detect poly(A)+ tRNAUAUILe or tRNAeMet (data not shown), both tRNAs that normally contain the m1A58 modification. Since the rrp44-20 mutant is defective for degradation of hypomethylated tRNAiMet (Kadaba et al. 2004), it is not surprising that poly(A)+ tRNAiMet accumulates to higher levels in the trm6- 504 rrp44-20 than in the trm6-504 mutant alone. From this we conclude that polyadenylation of hypomethylated tRNAiMet stimulates degradation by the exosome.
FIGURE 1.
Detection of polyadenylated tRNAiMet in trm6-504 and trm6-504 suppressor mutants. Total RNA and poly(A)+ RNA were isolated from wild-type (Y200), trm6-504(Y190), trm6-504 trf4-20 (sup1), and trm6-504 rrp44-20(sup2) strains grown in YPD at 30°C as detailed in Materials and Methods. Total RNA (5 μg) and poly(A)+ RNA (2–4 μg) were separated on a 6% polyacrylamide (19:1) 8 Murea gel. Membrane-bound tRNAiMet was detected using a radiolabeled oligonucleotide JA11 followed by autoradiography.
Previously we had observed that overexpression of Trf4p causes an increase in the length distribution of poly(A) tails on hypomethylated tRNAiMet (Kadaba et al. 2004), indicating that Trf4p positively regulates or functions as the poly(A) polymerase that adenylates hypomethylated tRNAiMet. Next we wanted to determine whether Trf4p is required for the polyadenylation of hypomethylated pre-tRNAiMet in vivo. To do this, we tested whether a mutant Trf4p eliminates polyadenylation of tRNAiMet in a trm6-504 strain. Northern hybridization of poly(A)+ selected RNA from trm6-504 trf4-20 detecting tRNAiMet revealed no poly(A)+ tRNAiMet (Fig. 1 ▶), supporting the hypothesis that Trf4p functions as a poly(A) polymerase to polyadenylate only hypomethylated tRNAiMet.
Since Trf4p contains a well-characterized nucleotidyltransferase domain, we wanted to determine whether the trf4-20 lesion represents a mutation that would predictably disrupt the poly(A) polymerase function of Trf4p. To this end, the entire TRF4 open reading frame (ORF) of 1752 nt was amplified from genomic DNA isolated from trm6-504 trf4-20 using PCR. Plasmid DNA from two independent bacterial clones was analyzed by DNA sequencing, and it was found that each clone contained a single adenosine nucleotide insertion that had occurred at nucleotide 795 of the TRF4 ORF, which is predicted to cause a +1 translational frame shift during translation of the TRF4 mRNA to produce a truncated Trf4p (~33 kDa) that is approximately half the size of the full-length Trf4p (~66 kDa). To determine whether the production of a truncated Trf4p was characteristic of other suppressors identified in our original screen and assigned to the TRF4 complementation group, we randomly chose seven independent isolates that carry mutant TRF4 alleles for isolation of genomic DNA, amplification and cloning of the TRF4 ORF, and DNA sequence analysis. From this analysis, we identified three insertion mutants at the same position as trf4-20, one single adenosine nucleotide deletion mutant at position 794 of the TRF4 ORF, which is predicted to cause a −1 frame shift in translational reading frame, and two nonsense mutations: one at position 1375, a T → A mutation that forms an opal codon and one at position 573 a C → A mutation that forms a ochre codon. Each of these changes should result in premature translation termination of TRF4 mRNA, thus predicted to produce truncated Trf4p. We conclude that lesions arising in TRF4 that result in trm6-504 suppression represent a loss of Trf4p function through a failure to make full-length Trf4p.
Mutation of the polymerase catalytic core in Trf4p eliminates polyadenylation of tRNAiMet
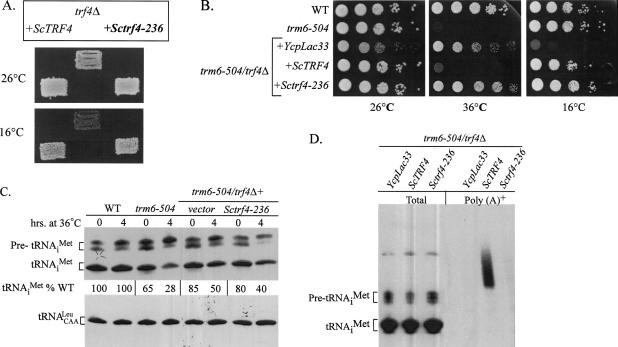
Truncation of Trf4p most likely results in a decreased level or a complete loss of Trf4p, but this does not distinguish between two possible explanations for the role of Trf4p in tRNAiMet degradation: (1) Trf4p functions as a poly(A) polymerase and adenylates tRNAiMet, or (2) Trf4p is required as a positive effector of an unrelated poly(A) polymerase. In order to distinguish between these two possibilities and explore the enzymology of Trf4p, site-directed mutations were made in TRF4 to change the conserved polymerase core DXD motif to AXA. Mutation of the DXD motif of Trf4p (trf4-236) has been shown to lead to diminished DNA and poly(A) polymerase activity in vitro (Wang et al. 2000; Vanacova et al. 2005; Wyers et al. 2005). Since the trf4-236 mutations do not alter the expression of Trf4p proteins (Wang et al. 2002), we tested if this mutant protein can complement the cold-sensitive phenotype of a trf4Δ strain. The trf4-236 allele in a single-copy number plasmid was used to transform a trf4Δ strain, and transformants were tested for growth at 16°C (Fig. 2A ▶). Surprisingly, the trf4-236 mutant supported growth of trf4Δ at 16°C, indicating that the polymerase function of Trf4p is not required to complement the Cs− phenotype of a trf4Δ mutant, but stable expression of Trf4p is required for complementation (Fig. 2A ▶) and also supports that the trf4-236 protein is expressed at normal levels.
FIGURE 2.
The DXD motif of Trf4p is crucial for its poly(A) polymerase activity and the degradation of tRNAiMet. (A) Sctrf4-236 complements the cold sensitivity of trf4Δ. A strain bearing chromosomal trf4Δ was transformed with YCplac33, ScTRF4 (B254), or Sctrf4-236 (B316), and the transformants were grown on SC-URA at 30°C, then replica-printed onto SC-URA plates and incubated at 16° and 26°C for 3 d and 1 d, respectively. (B) trf4-236 suppresses the Ts− phenotype of a trm6-504 mutant. A trm6-504 trf4Δ strain (Y301) was transformed with YCpLac33, ScTRF4 (B254), and Sctrf4-236 (B316); wild type (Y200) and trm6-504 (Y190) were transformed just with YCpLac33. Individual transformants were grown to saturation in SC-URA and 10-fold serial dilutions were spotted on SC-URA plates and incubated at 26°, 36°, and 16°C for 2, 3, and 4 d, respectively. (C) Northern hybridization of total RNA (5 μg) from wild type (Y200), trm6-504(Y190), trm6-504 trf4Δ (sup1) with YCpLac33, or Sctrf4-236 strains. Total RNA (5 μg) was separated on a 6% polyacrylamide (19:1) 8 Murea gel and transferred to a membrane and probed with JA11 to detect tRNAiMet and JA 151 to detect tRNALeu. The tRNAs were visualized by autoradiography and the quantities were determined by phosphorimage analysis using a STORM 640 and ImageQuant software (GE Healthcare). The quantity of tRNAiMet in each sample is shown as a percentage of tRNAiMet in the wild-type strain after normalization with tRNACAALeu. This experiment has been repeated and the quantity of tRNAiMet was found to be similar each time. (D) The DXD motif is important for its poly(A) polymerase activity in vivo. Total RNA (5 μg) and poly(A)+ RNA (2–4 μg) isolated from the transformants described in B were separated, and a Northern hybridization was conducted to detect tRNAiMet using a radiolabeled oligonucleotide (JA11) and autoradiography.
We previously showed that suppression of the temperature- sensitive (Ts−) phenotype of a trm6-504 strain by trf4- 20 is accompanied by an increase in the steady-state levels of hypomethylated tRNAiMet (Anderson et al. 1998; Kadaba et al. 2004). In order to determine if trf4-236 can function to complement trf4Δ and restore the (Ts−) phenotype of trm6-504, trf4-236 on a single-copy number plasmid (Sctrf4-236) was introduced into a trm6-504 trf4Δ strain and individual transformants carrying ScTRF4, Sctrf4-236, or empty vector (YcpLac33) were plated on selective media and tested for growth at 26°, 36°, and 16°C (Fig. 2B ▶). Comparable growth of Sctrf4-236 trm6-504 trf4Δ and YcpLac33 trm6-504 trf4Δ transformants at 36°C indicated that trf4-236 is unable to rescue the trf4Δ mutant to restore the trm6-504 Ts− phenotype; thus trf4-236 functions as a suppressor of trm6-504.
To correlate the inability of trf4-236 to complement trf4Δ in a trm6-504 background with the steady-state level of tRNAiMet, Northern hybridizations were done using equal amounts of total RNA from wild-type, trm6-504, trm6- 504 trf4Δ, and trm6-504 trf4Δ + Sctrf4-236 at 26° and 36°C. As seen previously, trm6-504 contains levels of mature tRNAiMet that are 65% and 28% of that seen in a wild-type strain at 26° and 36°C, respectively (Fig. 2C ▶). The level of mature tRNAiMet in trm6-504 trf4Δ and trm6-504 trf4Δ + Sctrf4-236 was increased to 80%–85% of wild type at 26°C and 40%–50% of the wild type at 36°C. Since trf4- 236 resulted in a ~1.5–2-fold stabilization of hypomethylated tRNAiMet at permissive and nonpermissive temperatures, we conclude that mutation of the DXD motif in Trf4p causes it to lose the ability to participate in the degradation of hypomethylated tRNAiMet, presumably due to a lack of its poly(A) polymerase activity.
To determine whether trf4-236 can function as a poly(A) polymerase to polyadenylate hypomethylated tRNAiMet in vivo, poly(A)+ RNA was isolated from trm6-504 trf4Δ, trm6-504 trf4Δ + ScTRF4, and trm6-504 trf4Δ + Sctrf4- 236 and subjected to Northern hybridization. While the strain containing a ScTRF4 plasmid contained poly(A)+ tRNAiMet, we were unable to detect poly(A)+ tRNAiMet in the strain bearing Sctrf4-236 (Fig. 2D ▶). Thus, we conclude that Trf4p is the poly(A) polymerase that polyadenylates hypomethylated tRNAiMet.
Trf4p and Trf5p have redundant functions in tRNAiMet turnover
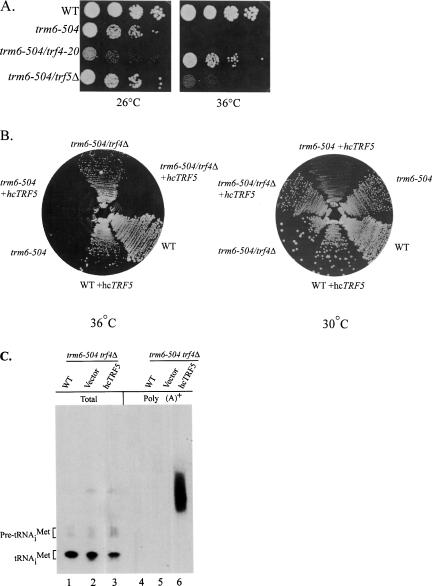
Trf4p has a structural homolog, Trf5p, that is 55% identical to Trf4p at the amino acid level, and it has been shown genetically that TRF4 and TRF5 are functionally redundant (Wang et al. 2000). We wanted to know whether Trf5p plays a role in the degradation of hypomethylated tRNAiMet along with Trf4p. First we tested whether deleting the TRF5 gene could suppress the Ts− phenotype of trm6-504 mutant. We found that deletion of TRF5 in a trm6-504 strain leads to a slight increase in growth rate at 36°C (Fig. 3A ▶), demonstrating that loss of Trf5p partially suppressed the trm6-504 mutant phenotype. To determine whether Trf5p and Trf4p have redundant functions in the degradation of hypomethylated tRNAiMet, we overexpressed Trf5p from a high-copy number plasmid in a trm6-504 trf4Δ mutant and tested growth at 30° and 36°C. If Trf5p is capable of replacing Trf4p function to polyadenylate hypomethylated tRNAiMet, then Trf5p overexpression should restore the Ts− phenotype of the trm6-504 mutant. We observed comparable growth levels between a trm6-504 trf4Δ strain overexpressing Trf5p and a trm6-504 mutant grown at 30°C and negligible growth of both strains at 36°C (Fig. 3B ▶), demonstrating that high-level Trf5p expression replaces Trf4p function in the degradation of hypomethylated tRNAiMet. In addition to complementing trf4Δ, hcTRF5 also leads to an exacerbation of the slow-growth phenotype of a trm6-504 mutant at 30°C (Fig. 3B ▶ trm6- 504 + hcTRF5) like what was seen previously when Trf4p was overexpressed in the trm6-504 mutant (Kadaba et al. 2004).
FIGURE 3.
Trf4p and Trf5p have overlapping functions in the degradation of hypomethylated tRNAiMet. (A) Yeast strains wild type (Y200), trm6-504 (Y190), trm6-504 trf4-20 (sup1), and trm6-504 trf5Δ were grown to saturation in YPD and serial 10-fold dilutions were plated onto YPD plates and incubated at 26°C or 36°C for 3 d and 2 d, respectively. (B) The same strains from A transformed with empty vector (WT, trm6-504, and trm6-504 trf5Δ) or a high-copy number plasmid containing TRF5 (WT, trm6-504, and trm6-504 trf5Δ + hcTRF5) were streaked to synthetic complete media lacking uracil and incubated at 30°C and 36°C for 3 d. (C) Northern hybridization of total RNA (3 μg) and poly(A)+ RNA (4–5 μg) isolated from wild-type (Y200), trm6-504 trf4Δ and trm6-504 trf4Δ + hcTRF5. The blot was hybridized with a radiolabeled oligonucleotide (JA11) to detect tRNAiMet.
To determine if there is concomitant polyadenylation of tRNAiMet upon restoration of the Ts− phenotype of trm6-504 by overexpression of Trf5p, we isolated poly(A)+ RNA from wild-type, trm6-504 trf4Δ, and trm6-504 trf4Δ + hcTRF5 strains and probed the RNA by Northern hybridization to detect the presence of tRNAiMet. We did observe polyadenylated tRNAiMet in RNA from the trm6- 504 trf4Δ hcTRF5 strain, while it was not detectable in the trm6-504 trf4Δ strain bearing an empty plasmid (Fig. 3C ▶). Thus, we can conclude that Trf4p and Trf5p possess a common activity that is central to the degradation of tRNAiMet, but possibly differential expression levels or substrate preferences of Trf4p and Trf5p define how they are utilized in tRNA surveillance.
Polyadenylation of hypomethylated tRNAiMet occurs before 5′ or 3′ end processing
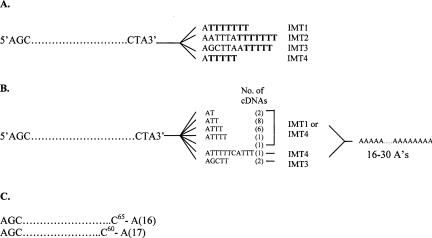
The first indication that hypomethylated pre-tRNAiMet is the target for degradation came during a high-copy suppressor analysis of a trm6-504 mutant (Anderson et al. 1998), and subsequently, support for this hypothesis was derived from an experiment in which it was shown that the treatment of poly(A)+ RNA from a trm6-504 rrp6Δ mutant with oligo-d(T) and RNaseH produced pre-tRNAiMet-sized species (Kadaba et al. 2004), leading to the conclusion that pre-tRNAiMet is polyadenylated in response to a loss of m1A58. Transcription termination by RNA polymerase III occurs when a stretch of 4–6 Ts is encountered (Cozzarelli et al. 1983); in tRNA this stretch of Ts usually begins 2–7 nt downstream of the tRNA coding sequence. To determine whether polyadenylated tRNAiMet were processed or unprocessed at their 3′ ends, we created and cloned cDNAs of tRNAiMet using oligo-d(T) primed reverse transcription (RT) of total RNA followed by specific amplification of tRNAiMet using PCR (see Materials and Methods). The DNA sequences of 23 cDNAs representing tRNAiMet are summarized in Figure 4 ▶. The predicted 3′ ends of pre-tRNAiMet derived from IMT1–IMT4 are shown with a partial sequence of tRNAiMet structural RNA (Fig. 4A ▶). The poly(A) tail length of the tRNAiMet cDNAs ranged from 16 to 30 adenylates, with the majority falling between 16 and 20 adenylates (Fig. 4B ▶). Nineteen of the 23 tRNAiMet cDNAs were derived from either IMT1 or IMT4 genes, and two were from IMT3. The majority of the IMT1 or 4 derived cDNAs contained 3′ ends ending in 2 or 3 Ts (Us in the RNA) just prior to the poly(A) tail. Three IMT1 or 4 derived cDNAs ended in a single T or no T, and one IMT4 cDNA extended beyond the expected termination site to a second shorter stretch of Ts (Fig. 4B ▶). Interestingly, none of the cDNAs contained the 3′ CCA sequence that is added post-transcriptionally and found only in mature tRNA, suggesting that polyadenylation is limited to 3′ end unprocessed hypomethylated tRNAiMet.
FIGURE 4.
Precursor form of hypomodified tRNAiMet is being polyadenylated. (A) An abbreviated and stylized representation of the tRNAiMet coding sequence with the predicted 3′ ends of each pre-tRNAiMet synthesized from IMT1–4 shown. The predicted transcription termination sites of pre-tRNAiMet are shown as 5–7 tandem Ts (Us in the transcript) are shown in bold. (B) The abbreviated coding sequence is followed by the exact sequence found at the 3′ end of each cDNA, and the poly(A) tail lengths are shown as a range (16–30), but the majority of the poly(A) tails were 16–19 As in length. The number of clones with the identical 3′ end sequence are shown in parentheses. (C) The abbreviated coding sequence for the truncated cDNAs found end with the last nucleotide of the tRNA (C65 or C60) followed by the poly(A) tail length for each.
The processing of pre-tRNAs includes, in a generally accepted temporal order, 5′ end leader removal, intron removal or 3′ end trailing sequence processing, and CCA addition (Hopper and Phizicky 2003). Base and nucleotide modifications can occur at any time during the processing steps of tRNA (Grosjean et al. 1997; Jiang et al. 1997), some conducted on nascent transcripts, others after addition of CCA. It is not known at which step in the processing of tRNA that m1A58 is added, but we have detected m1A58 in pre-tRNAs possessing an intron (J. Anderson, unpubl.). While the presence of unprocessed 3′ ends in the cDNAs supports the idea that pre-tRNAiMet is targeted for degradation, it does not address how early in a tRNA life the surveillance mechanism is employed.
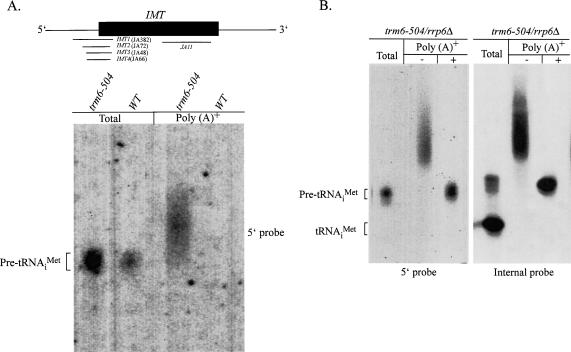
To better understand when during the processing of tRNA surveillance of hypomethylated tRNAiMet occurs, we tested whether poly(A)+ tRNAiMet contains the 5′ leader that is present in nascent pre-tRNAiMet transcripts by conducting Northern hybridizations on poly(A)+ RNA from trm6-504 and wild-type strains using probes that only recognize IMT1-4 pre-tRNAs containing the 5′ end leader sequence (Fig. 5A, 5 ▶′ IMT probes). We detected a significant amount of tRNAiMet using the 5′- specific IMT probes in poly(A)+ RNA from trm6-504 but not from wild type, supporting the idea that surveillance and polyadenylation of hypomethylated tRNAiMet occurs very soon after the nascent transcript is formed. Removal of the 3′ polyadenylate tail by oligo-d(T) hybridization and RNaseH treatment of poly(A)+ pre-tRNAiMet had no affect on detection of tRNAiMet by the 5′-end-specific probes (Fig. 5B ▶, lane 3), and the migration distance of 5′-end-containing tRNAiMet was identical to what we had seen previously (Kadaba et al. 2004), confirming that the polyadenylated tRNAiMet species contains unprocessed 5′ and 3′ ends. The specificity of the 5′ IMT probes was established by stripping and reprobing the membranes with a probe that hybridizes to an internal portion of tRNAiMet and detects both precursor and mature forms of tRNAiMet (Fig. 5B ▶, lanes 4–6). We conclude from these data that Trf4p and TRAMP preferentially polyadenylate the precursor form of hypomethylated tRNAiMet.
FIGURE 5.
Polyadenylated pre-tRNAiMet contains the 5′ leader sequence. (A) Northern hybridization analysis of total RNA (5 μg) and poly(A)+ RNA (2–4 μg) from trm6-504 (Y190) and wild type (Y200). (B) Northern hybridization of total RNA (5 μg) and poly(A)+ RNA (2–4 μg) from trm6-504 rrp6Δ (Y298) with (+) or without (−) pretreatment with oligo d(T) and RNaseH. The blots were hybridized with radiolabeled probes (JA382, JA72, JA66, and JA48) that recognize only pre-tRNAiMet containing the 5′ leader (A,B, lanes 1–3) or a radiolabeled probe (JA11) that is complementary to an internal portion of tRNAiMet (B, lanes 4–6). The point of migration of pre-tRNAiMet and mature tRNAiMet are shown for clarity.
Mutant U6 snRNA is degraded through the action of Trf4p and the exosome
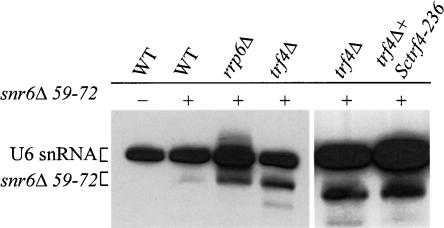
In order to determine if another non-tRNA mutant RNA might be degraded by polyadenylation and the nuclear exosome, we chose to analyze whether a mutant SNR6 gene that is rendered unstable by an internal deletion of 14 nt (snr6Δ59-72) (Eschenlauer et al. 1993) is targeted for degradation by Trf4p polyadenylation. SNR6 encodes the U6 SnRNA, which is a component of the spliceosome and required for pre-mRNA splicing. SNR6 has an internal A block element and a downstream B block element that are important for its transcription. It has been shown that deletion of residues 59–72 of SNR6 does not impair transcription in vitro but does result in an unstable U6 snRNA transcript in vivo (Eschenlauer et al. 1993). To test the idea that snr6Δ59–72 is destabilized by polyadenylation and degradation by the exosome, we isolated total RNA from wild-type, rrp6Δ, and trf4Δ yeast strains carrying the snr6Δ59–72 plasmid. Northern hybridization of total RNA from those strains showed that deletion of RRP6 or TRF4 resulted in a modest but significant accumulation of the snr6Δ59–72 compared to that detected in wild type (Fig. 6 ▶), demonstrating that Rrp6p and Trf4p are important for degradation of the snr6Δ59–72 transcript. To confirm that it is the poly(A) polymerase activity of Trf4p that was required to facilitate degradation of the mutant U6 snRNA, we conducted Northern hybridization of RNA from a trf4Δ containing the snr6Δ59–72 plasmid plus either Sctrf4-236. The accumulation of snr6Δ59–72 was similar in the trf4Δ and trf4Δ Sctrf4-236 strains (Fig. 6 ▶), supporting that the poly(A) polymerase function of Trf4p is required for degradation of mutant U6 snRNA. The fact that we did not observe levels of mutant U6 snRNA similar to that seen for wild-type U6 snRNA may be due to a lower level of transcription in vivo or another degradation pathway that is still actively degrading snr6Δ59–72 RNA in trf4Δ and rrp6Δ mutant strains.
FIGURE 6.
Internally truncated U6 snRNA is a substrate for Trf4p polyadenylation and degradation by the nuclear exosome. Wild type (F39), rrp6Δ (F23), trf4Δ (F22), and trf4Δ + Sctrf4-236 were transformed with a single-copy number plasmid containing a mutant SNR6 gene (snr6Δ59–72). Total RNA (5 μg) from single transformants and a wildtype strain bearing an empty plasmid were analyzed by Northern hybridization using a radiolabeled probe (JA242) that recognizes the endogenous and plasmid-borne U6 snRNAs. The migration positions of the endogenous and snr6Δ59–72 mutant U6 snRNAs are shown for clarity.
The exosome and Trf4p facilitate the degradation of 3′-end truncated 5S rRNA transcripts
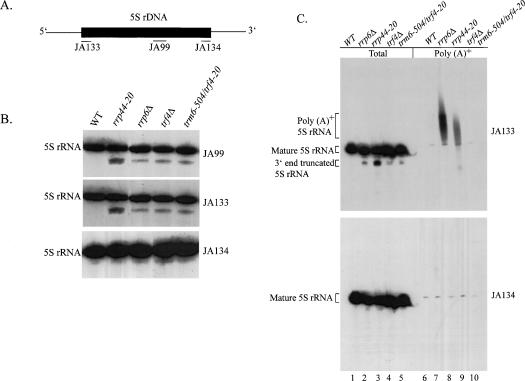
The nuclear exosome plays a central role in the precise 3′ end formation of many RNAs and is involved in the degradation of rRNA processing intermediates (Allmang et al. 1999; Raijmakers et al. 2004). Deletion of the gene encoding a nuclear-limited exosome component, Rrp6p, results in the accumulation of polyadenylated species of snRNAs, 5S rRNA, 27S rRNA, and snoRNAs (van Hoof et al. 2000b; Kuai et al. 2004). Whether these polyadenylated RNAs are aberrant transcripts that are being degraded is not known, although it has been proposed that 27S rRNA is polyadenylated and degraded in a 5′–3′ direction in an rrp6Δ mutant (Kuai et al. 2004). In conducting Northern hybridizations, we routinely use detection of 5S rRNA as an internal loading control, and we noted that trm6-504 suppressor mutants possessed a small fraction of 5S rRNA that was shorter by 8–20 nt and not detected in wild-type strains (Fig. 7B ▶, JA99). We hypothesized that this short form of 5S rRNA may be the result of aberrant processing and accumulated in trf4Δ, rrp6Δ and rrp44-20 mutants because the surveillance mechanism that normally eliminates it is defective. In order to establish the identity of the shorter form of 5S rRNA, we used oligodeoxynucleotide probes that selectively hybridized to 5′ or 3′ ends of mature 5S rRNA. A probe specific to the first 20 nt of 5S rRNA (Fig. 7B ▶, JA133) detected both the full-length mature and shorter 5S rRNA, whereas a probe that hybridizes to the last 21 nt of 5S rRNA (JA134) detected only the full-length 5S rRNA and not the faster migrating species (Fig. 7B ▶, JA134). This result demonstrates that the short 5S rRNA accumulating in exosome and trf4Δ mutants is truncated at the 3′ end.
FIGURE 7.
Trf4p dependent polyadenylation of 3′ end truncated 5S rRNA. (A) A schematic diagram of 5S rDNA showing the coding region (filled box) and the 5′ and 3′ intergenic regions (lines). The approximate positions (not to scale) complementary to the oligonucleotide probes used in this study are shown as lines beneath the coding region. (B) Total RNA (5 μg) isolated from wild type (F39), rrp6Δ (F23), rrp44-20 (Y303), trf4Δ (F22), and trm6-504/trf4Δ (Y301) strains grown at 30°C was used for Northern hybridization analysis. The membrane was probed with JA99 to detect mature 5S rRNA, JA133 to detect 5S rRNA containing the first 20 nt of 5S rRNA and JA134 to detect 5S rRNA containing the last 20 nt. The 5S rRNA was visualized by autoradiography. (C) Total RNA (5 μg, lanes 1–5) and poly(A)+ RNA (2–4 μg, lanes 6–10) isolated from the same strains described in B was used for Northern hybridization analysis using probes JA133 (top) and JA134 (bottom). Indicated are the migration positions of the mature 5S rRNA, 3′-end truncated 5S rRNA, and poly(A)+ 5S rRNA, as well as the probe used for each hybridization.
To determine whether the 3′ truncated 5S rRNA transcript is polyadenylated, and if Trf4p is responsible for its polyadenylation, total RNA and poly(A)+ RNA from wild-type, rrp44-20, rrp6Δ, trf4Δ, and trm6-504 trf4-20 strains were subjected to Northern hybridization analysis to detect 5S rRNA. The detection of poly(A)+ 5S rRNA in RNA from rrp44-20 and rrp6Δ using the 5′-specific probe (Fig. 7C ▶, JA133, lanes 7,8) but not the 3′-specific probe (Fig. 7C ▶, JA134, lanes 7,8) indicated that the polyadenylated 5S rRNA lacks the 3′ end as we had predicted. Moreover, the presence of 3′-end-shortened poly(A)+ 5S rRNA was found to be dependent on the expression of a functional Trf4p, since this species was undetectable in poly(A)+ RNAs isolated from trf4Δ or trm6-504 trf4-20 strains (Fig. 7C ▶, lanes 9,10). We conclude from these studies that mutant and truncated RNAs are recognized, polyadenylated, and degraded by the exosome.
DISCUSSION
We previously identified a novel tRNA surveillance pathway that utilizes polyadenylation to degrade hypomethylated tRNAiMet through the function of a new poly(A) polymerase, Trf4p, and the nuclear exosome (Kadaba et al. 2004). In the present study, we confirm this by demonstrating that a functional Trf4p is required for polyadenylation and degradation of hypomethylated tRNAiMet in vivo. We discovered that Trf5p is needed for optimal degradation of hypomethylated pre-tRNAiMet, but not to the same extent that Trf4p is required. Curiously, overexpression of Trf5p completely replaced Trf4p to cause efficient degradation of hypomethylated pre-tRNAiMet. This is in agreement with what was reported by LaCava et al. (2005), that overexpression of Trf5p in a trf4Δ mutant blocks the accumulation of U14 pre-snoRNA that is seen in a trf4Δ mutant, and Wyers et al. (2005), who showed that Trf5p accounts for a percentage of the degradation of intergenic mRNAs.
We have characterized the tRNAiMet substrate of Trf4p in vivo and found that polyadenylated tRNAiMet is unprocessed at its 5′ and 3′ ends, demonstrating that the integrity of tRNA is tested very early, possibly soon after transcription termination. We have expanded our understanding of what types of RNA are substrates for the tRNA surveillance mechanism. One such RNA, 5S rRNA, accumulates as a 3′- end-shortened polyadenylated species when the tRNA surveillance pathway is disrupted, suggesting that aberrations in 5S rRNA production or processing are rapidly eliminated in wild-type cells. This novel RNA surveillance pathway must serve a critical role in maintaining the structural or functional integrity of most or all circulating noncoding RNAs, in addition to its role in controlling the level of intergenic mRNA that was recently reported by Wyers et al. (2005).
The surveillance of hypomethylated tRNAiMet requires two highly related poly(A) polymerases
We established that Trf4p-directed polyadenylation of hypomethylated tRNAiMet is required for its degradation by introducing mutations in Trf4p that disrupt its polymerase activity (Vanacova et al. 2005; Wyers et al. 2005) and demonstrating that hypomethylated tRNAiMet is no longer polyadenylated, nor is it degraded efficiently. This is strong support for the idea that polyadenylation initiates degradation or stimulates the nuclear exosome to degrade this RNA. Our conclusion is in agreement with that drawn from in vitro experiments showing that purified nuclear exosome alone has very little RNA degradation activity (Mitchell et al. 1997); however, the addition of an oligomeric complex composed of Trf4p, Mtr4p, and Air1p/Air2p (TRAMP) with RNA substrate and exosome led to polyadenylation of the RNA substrate and stimulated RNA degradation by the exosome (LaCava et al. 2005; Vanacova et al. 2005).
The closely related proteins Trf4p and Trf5p are required to achieve a high level of tRNAiMet degradation, but Trf5p is not sufficient to support degradation on its own (Fig. 3 ▶). The reason(s) for this is not known, but one explanation could be that Trf5p is expressed at a relatively low level compared to Trf4p, and only when both proteins are expressed optimally do adenylation and degradation occur efficiently. An alternative to this idea is that Trf4p and Trf5p retain the same function to adenylate tRNAiMet, but they work at different stages of degradation. It is possible that Trf4p is required for initial polyadenylation of tRNAiMet substrate and Trf5p plays a more important role during subsequent rounds of adenylation and degradation that has been suggested to occur (LaCava et al. 2005). Another possibility is that Trf4p and Trf5p reside in slightly different TRAMP complexes that show different binding affinities for hypomethylated tRNAiMet; Trf4p TRAMP would exhibit better recognition/binding to hypomethylated tRNAiMet than Trf5p TRAMP. Assuming that Trf4p and Trf5p are interchangeable in these TRAMP complexes, this would help explain why overexpression of Trf5p is required in a trf4Δ trm6-504 double mutant to restore efficient polyadenylation and degradation of the hypomethylated tRNAiMet (Fig. 3C ▶).
Degradation of aberrant pre-tRNAiMet and mutant U6 snRNA in vivo
We initially proposed that hypomethylated pre-tRNAiMet is a target for degradation after observing stabilization of mature tRNAiMet and rapid degradation of tRNAiMet precursor after treating trm6-504 mutant cells with an RNA polymerase III transcriptional inhibitor (Anderson et al. 1998). Additional support for this original model was obtained when we observed that polyadenylated tRNAiMet isolated from trm6-504 rrp6Δ exhibited a gel mobility identical to pre-tRNAiMet after removal of the poly(A) tail by treatment with oligo d(T) and RNaseH (Kadaba et al. 2004). Here we show that poly(A)+ tRNAiMet isolated from trm6-504 RNA retains the 5′ end leader and 3′ end trailer sequences, which strongly supports that the target for polyadenylation is nascent tRNAiMet transcript (Figs. 4 ▶, 5 ▶). It is generally accepted that tRNA processing begins with removal of the 5′-end leader, followed by either intron splicing or 3′-end processing and 3′-end CCA addition to follow (O’Connor and Peebles 1991; Hopper and Phizicky 2003). The temporal position of base and nucleotide modification during tRNA processing is not well characterized, but it is likely that these modifications occur throughout the production and processing of tRNA (Grosjean et al. 1997; Hopper and Phizicky 2003). Finding that polyadenylated tRNAiMet contains the 5′ leader indicates that mutant tRNAiMet has not undergone structural processing before it is polyadenylated and m1A58 modification probably occurs during or shortly after tRNA transcription termination.
The mechanism by which hypomethylated initiator tRNAMet is recognized for elimination is not yet known, but given that other tRNAs normally containing m1A58 are not adversely affected in its absence, it is unlikely that the surveillance mechanism detects the absence of m1A58. The dependence of tRNAiMet structure on m1A58 is strongly supported by the three-dimensional structure of yeast tRNAiMet (Basavappa and Sigler 1991) showing m1A58 forms N6:N7 hyrdrogen bonds with N1:N6 of adenosine 54; A54 is uniquely found in eukaryotic initiator tRNAs. This is different than the m1A58 N6:N7 with rT54 O2:N3, the so-called reverse Hoogstein base-pairing found in all elongator tRNAs where the three-dimensional structure is known (for review, see Arnez and Moras 1999). The reverse Hoosgstein base pair found in both the yeast tRNAPhe and tRNAAsp three-dimensional structures is intact regardless of the presence of 1-methylA58 (tRNAPhe) (Kim et al. 1974) or its absence (tRNAAsp) (Moras et al. 1980), but we believe that the tRNAiMet structure is sensitive to a loss of 1- methylA58. Interestingly, the rT/U replacement of A54 in tRNAiMet in S. cerevisiae failed to produce a functional tRNAiMet (von Pawel-Rammingen et al. 1992), supporting that the A54:m1A58 interaction in tRNAiMet is contextually different than the rT54:A58/m1A58 interaction found in elongator tRNAs. We favor a model in which tRNA that is to be degraded is recognized because of an aberrant structure, rather than one that identifies tRNAs lacking m1A58 or because the pre-tRNA is not efficiently processed.
A surveillance mechanism that recognizes and facilitates degradation of aberrant non-protein-encoding RNAs through polyadenylation might be considered protection against assembly of partial or nonfunctional RNPs. As such, the recognition of aberrant RNAs should occur prior to RNP incorporation, and this may explain why snr6Δ59–72 RNA is unstable (Fig. 6 ▶); mutant U6 snRNA fails to be incorporated into a U4/U6 U5 tri-snRNP. The incorporation of U6 into a U4/U6 snRNP is in part due to base-pairing between U4 and U6 (Hu et al. 1995), which would be severely impaired by deleting 13 of the 24 nt in U6 that form base pairs with U4 snRNA (Hu et al. 1995). The instability of snr6Δ59–72 RNA is partially suppressed after depletion of Trf4p or Rrp6p (Fig. 6 ▶). By partially eliminating mutant U6 degradation, some of the mutant might be incorporated into U4/U6 U5 tri-snRNP, which could cause a measurable defect in pre-mRNA splicing, although we see no evidence to support that stabilization of the mutant U6 causes a change in cell viability that would be expected if it were incorporated into a snRNP and perturbed splicing.
Global surveillance of nuclear RNAs
Since polyadenylation of noncoding RNAs (Piper et al. 1983; van Hoof et al. 2000b) and hypomethylated tRNAiMet (Kadaba et al. 2004) was reported, others have discovered that several noncoding RNAs (pre-snoRNA, pre-rRNA, and intergenic mRNA) are polyadenylated by the same surveillance machinery (LaCava et al. 2005; Wyers et al. 2005). The reason for polyadenylation of pre-snoRNA and 23S rRNA is unclear, since these precursor RNAs should be amenable to normal processing, ultimately resulting in the formation of functional, mature RNAs. It is formally possible that the polyadenylated forms of pre-snoRNAs and rRNAs observed were aberrantly structured or inaccurately processed, but this went undetected in these studies.
We identified a 3′-end-shortened 5S rRNA that is subjected to RNA surveillance. The shortened 5S rRNA was detected as a faster migrating species representing a small proportion of the total 5S rRNA produced in trf4Δ, rrp44-20, and rrp6Δ mutant strains. The 3′-end-shortened 5S rRNA is polyadenylated in rrp44-20 and rrp6Δ strains (Fig. 7C ▶) but not in a trf4Δ mutant, indicating that Trf4p is required for the polyadenylation. By approximating the size of this 3′-end-shortened 5S rRNA, we conclude that it is missing the final 8–20 nt. The processing of 5S pre-rRNA to remove nucleotides of the nascent transcript requires an endo- or a 3′–5′ exonuclease (van Hoof et al. 2000a) to trim the 3′ end trailing sequence, but because the mature 5′ end is formed during transcription initiation (Tekamp et al. 1980; Piper et al. 1983, 1987; Lee and Nazar 1997), no enzymatic alteration of the 5′ end is needed. The 5′ and 3′ termini form stem I, which is required for accurate processing of the 3′ end trailer (Tekamp et al. 1980; Piper et al. 1983; Lee and Nazar 1997). The absence of the last 8–20 nt from 5S rRNA would eliminate stem I and could destabilize the 5S rRNA. This 3′-end-shortened 5S rRNA may be a product of a variant 5S rDNA termed J2* that is normally turned over very rapidly. The J2* variant of 5S rDNA resides at the end of the tandem repeated ribosomal RNA genes on chromosome XII (McMahon et al. 1984), and it varies from the wild-type 5S rDNA at three positions near the 3′ end of the 5S coding region. These alterations would disrupt two Watson–Crick base pairs (G:C1,120 and U:A3,118) in stem I and replace them with G:U base pairs, which would predictably decrease the stability of the terminal end of stem I. The destabilization could render the 3′ end susceptible to a 3′–5′ exonuclease activity that could easily remove the 3′ end 9 nt that participate in stem I formation and result in a shortened 5S rRNA that is normally rapidly turned over. In support of this, several mutations have been described that, when introduced into stem I of a plasmid-borne copy of 5S rDNA, dramatically affect 5S rRNA stability in vivo (Lee and Nazar 1997) but not 5S rRNA transcription and processing in vitro (Brow 1987). Since the 3′-end-shortened 5S rRNA lacks the final 8–20 nt, we have no way of positively confirming it is a product of the J2* variant 5S rDNA, but it is either j2* 5S or incorrectly processed 5S rRNA that is usually rapidly degraded. This stands as a good example of how the surveillance mechanism that eliminates hypomethylated tRNAiMet is also utilized to eliminate aberrantly structured cellular RNAs.
Substrate recognition and an alternative degradation pathway
Understanding how Trf4p and TRAMP recognize these substrates is a critical question that remains unanswered. It is possible that an RNA chaperone like Lhp1p is involved in distinguishing between normal and abnormal RNA. Some support for this idea comes from a demonstration that the presence of Lhp1p positively influences the stability of a mutant tRNAArg by stabilizing the abnormal anti-codon stem (Chakshusmathi et al. 2003) and hypomethylated tRNAiMet that becomes extremely unstable in the absence of Lhp1p (Calvo et al. 1999; Chakshusmathi et al. 2003). Consistent with these observations, overexpression of Lhp1p partially suppressed degradation of hypomethylated tRNAiMet in trm6-504 and trm61-2 mutants (Anderson et al. 1998; Calvo et al. 1999). Whatever the mode of recognition, the elimination of aberrant RNAs is likely to have several faces and require many factors. For instance, Lhp1p is a good candidate for an RNA chaperone of tRNA, but the natural Lhp1p binding site oligo U (Stefano 1984) at the 3′ end of most RNA polymerase III transcripts (Campbell and Setzer 1992) is not found at the 3′ end of mRNA or rRNA. Thus, the action of Lhp1p may be limited to nascent RNA polymerase III transcripts. Expanding our understanding of what constitutes an aberrant RNA, the mode of recognizing those RNAs, and how the process is regulated will be an important focus of the research on nuclear RNA surveillance and turnover in the next decade.
MATERIALS AND METHODS
Yeast strains and media
Standard genetic techniques were employed as described (Sherman et al. 1974). Yeast strains used in this study are described in Table 1 ▶. Y303 was created by crossing trm6-504 rrp44-20 (S94) with F8 strain. The resulting diploid was sporulated in media containing 0.25% yeast extract; 1.5% potassium acetate; 0.05% glucose; 0.004% each of adenine, uracil, and tyrosine; 0.002% each of histidine, leucine, tryptophan, methionine, and arginine; 0.1% phenylalanine; and 0.35% threonine; and subjected to tetrad dissection. Haploids from the resulting tetrads were tested for Ts− and Cs− phenotypes at 37° and 16°C, respectively. rrp44-20 TRM6 was identified from a tetrad that showed 2:2 segregation of Ts−/Cs+ (trm6-504 RRP44) and Ts+/Cs− (TRM6 rrp44-20), and the presence of rrp44-20 in the Ts+/Cs− segregants was confirmed by complementation of the Cs− phenotype of rrp44-20 through introducing a single-copy number plasmid bearing wild-type RRP44. A trm6-504 trf5Δ (Y374) double mutant was created by gene replacement of TRF5 with the gene that encodes resistance to G418, KANMX, using PCR to create a TRF5:KANMX:TRF5 product as described (Longtine et al. 1998). The TRF5:KANMX:TRF5 PCR product was used to transform Y190 (trm6-504), and G418 resistant transformants were streaked to YPD plates supplemented with 0.2 mg/mL Gentecin sulphate. The replacement of TRF5 with KANMX was confirmed by PCR using the primer-specific external region of TRF5 ORF and to the KANMX gene.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference |
| Y190 | MATa trm6-504,gcn2-101,his1-29,ura3-52, ino1(HIS4:LacZ, ura3-52) | Kadaba et al. 2004 |
| Y200 | MATa TRM6, gcn2-101,his1-29,ura3-52, ino1(HIS4:LacZ, ura3-52) | Kadaba et al. 2004 |
| SUP1 | MATa trm6-504, trf4-20,gcn2-101,his1-29,ura3-52, ino1(HIS4:LacZ, ura3-52) | Kadaba et al. 2004 |
| SUP2 | MATα trm6-504, rrp44-20, gcn2-101,his1-29,ura3-52, ino1(HIS4:LacZ, ura3-52) | Kadaba et al. 2004 |
| Y301 | MATα HIS3 trf4Δ:KANMX4, trm6-504 | Kadaba et al. 2004 |
| Y303 | rrp44-20, ura3-52 | This study |
| Y374 | MATa trm6-504,trf5Δ:KANMX4, gcn2-101, his1-29,ura3-52,ino1(HIS4:LacZ, ura3-52) | This study |
| F8 | MATa ura3-52 ino1 gcn2-1 (HIS4-lacZ, ura3-52) | |
| F22 | MATα his3Δ, leu2Δ, lys2Δ, ura3Δ, trf4Δ: KanMX4 Res gen #11777 | Invitrogen |
| F23 | MATα his3Δ, leu2Δ, lys2Δ, ura3Δ, rrp6Δ: KanMX4 Res gen #16265 | Invitrogen |
| F39 | MATα his3Δ, leu2Δ, lys2Δ, ura3Δ | Invitrogen |
Plasmid construction, trf4 mutant mapping, and site-directed mutagenesis
Plasmids used in this study are shown on Table 2 ▶. Plasmid B288 containing a mutant U6 snRNA gene was created by subcloning a 1.4-kb SphI/PstI fragment containing snr6-59Δ72 from the plasmid PΔBE6, a kind gift from Dr. David Brow (University of Wisconsin, Madison) (Eschenlauer et al. 1993) into SphI/PstI digested YcpLac 111. The high-copy number plasmid containing TRF5 (B232) was created by subcloning a 3.1-kb PstI/BamHI from CB1455 (a generous gift from Dr. Michael Christman, Boston University School of Medicine, Boston, MA) into PstI/BamHI digested YepLac195.
TABLE 2.
Plasmids
| Name | Description | Reference |
| B254 | 3.1-kb SspI/PmeI fragment of TRF4 subcloned from pSup1 into SspI/PmeI YCpLac33 | Kadaba et al. 2004 |
| B256 | 3.1-kb SacI/PstI fragment of TRF4 from B254 into SacI/PstI YEpLac195 | Kadaba et al. 2004 |
| B315 | 3.1-kb TRF4 gene harboring trf4-236 mutation was generated by quick change mutagenesis using B256 as template | This study |
| B316 | KpnI/PstI digested 3.1-kb fragment of trf4-236 from B315 subcloned into KpnI/PstI YCpLac33 | This study |
| pΔBE6 | snr6 Δ59–72 | Eschenlauer et al. 1993 |
| B288 | 1.4-kb SphI/EcoRI fragment of SNR6 subcloned from PΔBE6 into SphI/EcoRI YCpLac111 | This study |
| CB1455 | 3.1-kb PstI/BamHI fragment of TRF5 cloned into PstI/BamHI PRS315 | Dr. Michael Christman |
| B232 | PstI/BamHI 3.1-kb fragment of TRF5 from CB1455 | This study |
| subcloned into PstI/BamHI YEpLac195 | This study | |
| B180 | YCpLac111: CEN4, LEU2 cloning vector | Gietz and Sugino 1988 |
| B181 | YCpLac33: CEN4, URA3 cloning vector | Gietz and Sugino 1988 |
| B184 | YEpLac195: URA3 high copy vector | Gietz and Sugino 1988 |
To map the trf4 point mutants, genomic DNA was isolated from trm6-504/trf4-20–27 mutantsand the TRF4 ORF was amplified by PCR in two independent reactions using primers JA194 and JA193 with Pfu Taq polymerase (Stratagene). The PCR product from each reaction was cloned into the pGEMT vector after generating overhanging adenosines using Taq polymerase (Promega) based on the manufacturer’s specifications. Single bacterial transformants containing trf4 mutant ORFs were screened, and at least two individual plasmids for each mutant were purified and subjected to DNA sequence analysis (MWG Biotech). The DNA sequences were aligned to the TRF4 ORF using the Vector NTI software in order to detect the mutations. The mutation was assigned only if it was found in two or more independently generated clones obtained from the same genomic DNA.
Mutations in TRF4 ORF were created by a PCR-based Quick Change XL Site-directed mutagenesis kit (Stratagene) in plasmid (B256) as the template using the manufacturer’s specifications. Two primers containing the desired mutation were designed so that they hybridized to the same sequence on the opposite strands. The mutations introduced an AfeI restriction site that was used to screen for the desired changes in B315 (hctrf4-236). The presence of the desired mutations was confirmed by DNA sequence analysis of the area mutagenized. A single-copy number plasmid carrying trf4-236 was generated by subcloning a 3.1-kb KpnI/PstI restriction fragment from B315, trf4-236 into KpnI/PstI digested YCpLac33 to create B316.
RNA isolation, Northern blotting, purification of poly(A) RNA, and cDNA cloning
Total RNA was isolated as described by Kohrer and Domdey (1990). RNAs were separated using a 10 cm × 14 cm × 0.8 mm 6% polyacrylamide-bis-acrylamide 8 M urea gel in 1× TBE buffer (Maniatis et al. 1982) for 1 h at 450 V. The separated RNA was transferred to a HyBond N+ membrane (Pal Biosciences) in the cold at 14 V for 3 h in 0.5× TBE unless otherwise stated. Blots were probed with radiolabeled deoxyoligonucleotides (0.1 pmol) in 5 mL of hybridization buffer (0.25M Na2HPO4, 1 mM EDTA, 1% BSA, 7% SDS). Probes were radiolabeled by incubating 10 pmol of deoxyoligonucleotide with γ-32P-ATP > 5000 Ci/mmol (Amersham Pharmacia) and T4 polynucleotide kinase (New England Biolabs) at 37°C for 1 h. Unincorporated γ-P32 ATP was separated from radiolabeled oligonucleotide by centrifugation through a 0.7-mL G-25 microspin column containing water for 1 min at 2500g. Sequences of all oligodeoxynucleotides used in this study can be found in Table 3 ▶.
TABLE 3.
Oligonucleotides
| Oligo | Target | Sequence |
| JA11 | *tRNAi Met | TCGGTTTCGATCCGAGGACATCAGGGTTATGA |
| JA99 | *5S rRNA | TCGCGTATGGTCACCCACTACA |
| JA133 | *5S rRNA | TGGTAGATATGGCCGCAACC |
| JA134 | *5S rRNA | AGATTGCAGCACCTGAGTTTC |
| JA151 | *tRNACAALeu | TGGTTGCTAAGAGATTCGAAC |
| JA48 | *5′ IMT3 | ACGGCGCTTAACTTTTATG |
| JA66 | *5′ IMT4 | GGCGCTTAGCCAACTTG |
| JA72 | *5′ IMT2 | GGCGCTGCTAAATCATGAG |
| JA382 | *5′ IMT1 | CACGGCGCTGCTAATCTTGCAATGCCTCT |
| JA114 | *tRNAUAUILe | TGCTCGAGGTGGGGWTTGAACCCACGACGG |
| JA15 | *tRNAeMet | TGCTCCAGGGGAGGTTCGAACTCTCGACC |
| JA275 | trf4-236 | CCGATTTGTATTTGCCTGGTAGCGCTATTGCTTGCGTGGTAACGAGCG |
| JA276 | trf4-236 | CGCTCGTTACCACGCAAGCAATAGCGCTACCAGGCAAATACAAATCGG |
| JA311 | oligo d(T) adaptor | GGCCACGCGTCGACTACTTTTTTTTTTTTTTTTT |
| JA310 | Adaptor | GGCCACGCGTCGACTAC |
| JA150 | tRNAiMet | TGGCGCAGTGGAAGCGC |
| JA193 | TRF4 | GATGTACAGTTCAGTGCATC |
| JA194 | TRF4 | GTGTGAATAAGCAAGGGAAC |
| JA346 | 5′ TRF5 deletion | TTATTTTTCAAATAAACAAACGAGGGCGGAGTTTATTGGGTCGTCCGTACGCTGCAGGTCGAC |
| JA347 | 3′ TRF5 deletion | AAGAGCCTGGCCTTTAGAGAGCCAGTAGTCCCTTCTCGTTTGAGCATCGATGAATTCGAGCTC |
*Deoxyoligonucleotides used in Northern hybridization.
Poly (A)+ RNA was purified using the same protocol described in Kadaba et al. (2004) except that poly(A)+ RNA was isolated from 200-mL cultures using 0.05 g Oligo (dT) cellulose.
For the creation of cDNAs of tRNAiMet, 10 μg of total RNA were treated with RNase-free DNase for 20 min at 37°C. The DNase-treated RNA was phenol-extracted and precipitated with ethanol at −20°C for 1 h. The RNA was pelleted by centrifugation, washed with 70% ethanol, and air-dried. One microgram of this RNA was combined with 50 pmol of an oligo d(T) primer (JA311), 1 mM dNTP and incubated at 80°C for 10 min, then immediately placed on ice for 5 min. Reactions were started by the addition of reverse transcriptase buffer (1× final conc.), four units of RNasin (Promega), and 20 units of M-MLV reverse transcriptase (Promega), and incubation at 42°C was carried out for 1.5 h. Two microliters of the completed cDNA reaction were combined with 2.5 μL of each primer (JA312) 20 pmol/μL, and 5′ primer (JA150) 20 pmol/μL, 2 μL dNTP (200 mM) and adjusted to a final volume of 20 μL with dH2O, after which it was incubated at 95°C for 5 min. Immediately after preincubation, 5 μL of 10× Taq DNA polymerase buffer A (Promega), 5 μL of MgCl2 (25 mM), and 0.5 μL of Taq DNA polymerase, 5 U/μL (Promega), and dH2O were added to a final volume of 50 μL. The PCR reaction was carried out under the following conditions: 95°C for 30 sec, 55°C for 40 sec, and 72°C for 75 sec for 30 cycles followed by final elongation at 72°C for 7 min. The PCR product was gel-purified (Qiagen) and ligated to pGEMT-Easy vector (Promega) using T4 DNA ligase, and the ligation was used to transform DH10B electromax competent cells (Invitrogen). Colony hybridization was performed in order to identify positive colonies carrying tRNAiMet clones with 32P-labeled JA11 probe specific to tRNAiMet according to the procedure described by Maniatis et al. (1982). Plasmid DNA from the positive colonies was analyzed by DNA sequencing (MWG) using the primer JA294. The DNA sequences were aligned to the DNA sequence of the IMT genes obtained from the Saccharomyces genome database using the Vector NTI software program (Invitrogen).
Acknowledgments
We thank Drs. Michael Christman, David Brow, and Roy Long for generous gifts of reagents. We thank members of the Anderson laboratory, Roy Long, and John LaCava for critical reading of the manuscript. This work was supported by a research grant from the National Institutes of Health (GM069949-01A1).
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2305406.
REFERENCES
- Allmang, C., Kufel, J., Chanfreau, G., Mitchell, P., Petfalski, E., and Tollervey, D. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18: 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J., Phan, L., Cuesta, R., Carlson, B.A., Pak, M., Asano, K., Bjork, G.R., Tamame, M., and Hinnebusch, A.G. 1998. The essential Gcd10p–Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes & Dev. 12: 3650–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L. and Koonin, E.V. 1999. DNA polymerase β-like nucleoti-dyltransferase superfamily: Identification of three new families, classification and evolutionary history. Nucleic Acids Res. 27: 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnez, J.G. and Moras, D. 1999. Transfer RNA. In Oxford handbook of nucleic acid structure (ed. S. Neidle), pp. 603–651. Oxford University Press, Oxford, UK.
- Ballantyne, S., Daniel Jr., D.L., and Wickens, M. 1997. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol. Biol. Cell 8: 1633–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavappa, R. and Sigler, P.B. 1991. The 3 Å crystal structure of yeast initiator tRNA: Functional implications in initiator/elongator discrimination. EMBO J. 10: 3105–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow, D.A. 1987. In vitro transcripts of a yeast variant 5 S rRNA gene exhibit alterations in 3′-end processing and protein binding. J. Biol. Chem. 262: 13959–13965. [PubMed] [Google Scholar]
- Butler, J.S. 2002. The yin and yang of the exosome. Trends Cell. Biol. 12: 90–96. [DOI] [PubMed] [Google Scholar]
- Calvo, O., Cuesta, R., Anderson, J., Gutierrez, N., Garcia-Barrio, M.T., Hinnebusch, A.G., and Tamame, M. 1999. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4167–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell Jr., F.E. and Setzer, D.R. 1992. Transcription termination by RNA polymerase III: Uncoupling of polymerase release from termination signal recognition. Mol. Cell. Biol. 12: 2260–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakshusmathi, G., Kim, S.D., Rubinson, D.A., and Wolin, S.L. 2003. A La protein requirement for efficient pre-tRNA folding. EMBO J. 22: 6562–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzarelli, N.R., Gerrard, S.P., Schlissel, M., Brown, D.D., and Bogenhagen, D.F. 1983. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell 34: 829–835. [DOI] [PubMed] [Google Scholar]
- Dickson, K.S., Thompson, S.R., Gray, N.K., and Wickens, M. 2001. Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J. Biol. Chem. 276: 41810–41816. [DOI] [PubMed] [Google Scholar]
- Edmonds, M. 2002. A history of poly A sequences: From formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 71: 285–389. [DOI] [PubMed] [Google Scholar]
- Eschenlauer, J.B., Kaiser, M.W., Gerlach, V.L., and Brow, D.A. 1993. Architecture of a yeast U6 RNA gene promoter. Mol. Cell. Biol. 13: 3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D. and Sugino, A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., Szweykowska-Kulinska, Z., Motorin, Y., Fasiolo, F., and Simos, G. 1997. Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: A review. Biochimie 79: 293–302. [DOI] [PubMed] [Google Scholar]
- Hopper, A.K. and Phizicky, E.M. 2003. tRNA transfers to the limelight. Genes & Dev. 17: 162–180. [DOI] [PubMed] [Google Scholar]
- Hu, J., Xu, D., Schappert, K., Xu, Y., and Friesen, J.D. 1995. Mutational analysis of Saccharomyces cerevisiae U4 small nuclear RNA identifies functionally important domains. Mol. Cell. Biol. 15: 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, H.Q., Motorin, Y., Jin, Y.X., and Grosjean, H. 1997. Pleiotropic effects of intron removal on base modification pattern of yeast tRNAPhe: An in vitro study. Nucleic Acids Res. 25: 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba, S., Krueger, A., Trice, T., Krecic, A.M., Hinnebusch, A.G., and Anderson, J. 2004. Nuclear surveillance and degradation of hypo-modified initiator tRNAMet in S. cerevisiae. Genes & Dev. 18: 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H., Suddath, F.L., Quigley, G.J., McPherson, A., Sussman, J.L., Wang, A.H., Seeman, N.C., and Rich, A. 1974. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185: 435–440. [DOI] [PubMed] [Google Scholar]
- Kohrer, K. and Domdey, H. 1990. Preparation of high molecular weight RNA. Methods Enzymol. 194: 398–405. [DOI] [PubMed] [Google Scholar]
- Kuai, L., Fang, F., Butler, J.S., and Sherman, F. 2004. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 101: 8581–8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava, J., Houseley, J., Saveanu, C., Petfalski, E., Thompson, E., Jacquier, A., and Tollervey, D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724. [DOI] [PubMed] [Google Scholar]
- Lee, Y. and Nazar, R.N. 1997. Ribosomal 5 S rRNA maturation in Saccharomyces cerevisiae. J. Biol. Chem. 272: 15206–15212. [DOI] [PubMed] [Google Scholar]
- Li, Z., Reimers, S., Pandit, S., and Deutscher, M.P. 2002. RNA quality control: Degradation of defective transfer RNA. EMBO J. 21: 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie 3rd, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E.F., and Sambrook, J. 1982. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- McMahon, M.E., Stamenkovich, D., and Petes, T.D. 1984. Tandemly arranged variant 5S ribosomal RNA genes in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 12: 8001–8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P. 2001. Purification of yeast exosome. Methods Enzymol. 342: 356–364. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M., and Tollervey, D. 1997. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466. [DOI] [PubMed] [Google Scholar]
- Moras, D., Comarmond, M.B., Fischer, J., Weiss, R., Thierry, J.C., Ebel, J.P., and Giege, R. 1980. Crystal structure of yeast tRNAAsp. Nature 288: 669–674. [DOI] [PubMed] [Google Scholar]
- O’Connor, J.P. and Peebles, C.L. 1991. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol. Cell. Biol. 11: 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, P.W., Bellatin, J.A., and Lockheart, A. 1983. Altered maturation of sequences at the 3′ terminus of 5S gene transcripts in a Saccharomyces cerevisiae mutant that lacks a RNA processing endonuclease. EMBO J. 2: 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, P.W., Lockheart, A., and Bellatin, J. 1987. The effects of protein synthesis inhibition, and of mutations rna1.1 and rna82.1, on the synthesis of small RNAs in yeast. FEBS Lett. 214: 143–148. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16: 272–278. [DOI] [PubMed] [Google Scholar]
- Raijmakers, R., Schilders, G., and Pruijn, G.J. 2004. The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur. J. Cell Biol. 83: 175–183. [DOI] [PubMed] [Google Scholar]
- Sherman, F., Fink, G.R., and Lawrence, C.W. 1974. Methods of yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Stefano, J.E. 1984. Purified lupus antigen La recognizes an oligoridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36: 145–154. [DOI] [PubMed] [Google Scholar]
- Tekamp, P.A., Garcea, R.L., and Rutter, W.J. 1980. Transcription and in vitro processing of yeast 5 S rRNA. J. Biol. Chem. 255: 9501–9506. [PubMed] [Google Scholar]
- van Hoof, A., Lennertz, P., and Parker, R. 2000a. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J. 19: 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000b. Yeast exosome mutants accumulate 3′-extended poly-adenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova, S., Wolf, J., Martin, G., Blank, D., Dettwiler, S., Friedlein, A., Langen, H., Keith, G., and Keller, W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Pawel-Rammingen, U., Åström, S., and Byström, A.S. 1992. Mutational analysis of conserved positions potentially important for initiator tRNA function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle, E. and Ruegsegger, U. 1999. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 23: 277–295. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Castano, I.B., De Las Penas, A., Adams, C., and Christman, M.F. 2000. Pol κ: A DNA polymerase required for sister chromatid cohesion [see comments]. Science 289: 774–779. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Castano, I.B., Adams, C., Vu, C., Fitzhugh, D., and Christman, M.F. 2002. Structure/function analysis of the Saccharomyces cerevisiae Trf4/Pol σ DNA polymerase. Genetics 160: 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers, F., Rougemaille, M., Badis, G., Rousselle, J.C., Dufour, M.E., Boulay, J., Regnault, B., Devaux, F., Namane, A., Seraphin, B., et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737. [DOI] [PubMed] [Google Scholar]