Abstract
DNA methyltransferase 1 (Dnmt1) has a central role in copying the pattern of DNA methylation after replication which is one manifestation of epigenetic inheritance. With oligonculeotide substrates we show that mouse Dnmt1 has a 30- to 40-fold preference for hemimethylated DNA that is almost lost after addition of fully methylated oligonucleotides. Using long hemimethylated DNA substrates that carry defined methylation patterns and bisulfite analysis of the methylation reaction products, we show a 15-fold preference for hemimethylated CG sites. Dnmt1 moves along the DNA in a random walk methylating hemimethylated substrates with high processivity (>50 sites are visited on average which corresponds to linear diffusion over 6000 bp). The frequency of skipping sites is very low (<0.3%) and there is no detectable flanking sequence preference. CGCTC sites tend to terminate the processive methylation of DNA by Dnmt1. Unmethylated DNA is modified non-processively with a preference for methylation at CCGG sites. We simulate the propagation of methylation patterns using a stochastic model with the specificity of Dnmt1 observed here and conclude that either methylation of several sites is required to propagate the methylation information over several cellular generations or additional epigenetic information must be used.
INTRODUCTION
Epigenetic information is defined as heritable but not encoded in the DNA sequence (1). One form of epigenetic information in human cells is methylation of cytosine residues in DNA at the C5-position at CG sites [reviewed in (2,3)]. DNA methylation is involved in a number of important biological processes, like control of gene expression, imprinting, development, X-chromosome inactivation, genomic integrity and protection of the genome against selfish DNA elements [reviewed in (2–7)]. On the other hand, erroneous DNA methylation causes diseases including cancer [reviewed in (1,8,9)]. The Dnmt1 DNA methyltransferase is responsible for the propagation of the DNA methylation information by maintenance of the DNA methylation pattern after DNA replication [reviewed in (10)]. The enzyme shows a preference for hemimethylated CG sites as they appear after DNA replication. Its activity, therefore, leads to the re-establishment of the same DNA methylation pattern as it was before replication. The accuracy of the transmission of the methylation information to the next cellular generation critically depends on the degree of specificity of Dnmt1 for hemimethylated sites. Surprisingly, this property has not yet been characterized sufficiently, and numbers ranging from 2- to 50-fold were published for oligonucleotide substrates, depending on the substrate, experimental test system and enzyme preparation (11–15). Moreover, many groups have reported that Dnmt1 has reduced specificity in the presence of methylated DNA (11,16–18) which would put into question the whole concept of inheritance of methylation patterns. Other important properties that determine the efficiency of this enzyme in the remethylation of DNA after replication are its processivity and whether it tends to skip hemimethylated target sites or not. Here, we have studied the accuracy, processivity and frequency of skipping sites by Dnmt1 using long hemimethylated DNA molecules that mimic physiological substrates. Our data provide a framework for the understanding of the role of this enzyme in vivo.
MATERIALS AND METHODS
Detailed descriptions of the experiments can be found in supplementary data. In brief, Dnmt1 was purified as described (11,19). Methylation of 30mer oligonucleotide substrates containing one CG site was studied using radioactively labeled AdoMet as described (11). Two different PCR products comprising 634 and 566 bp were amplified from lambda DNA using one 5′-phosphorylated and one standard primer and processed to generate hemimethylated substrates basically as described (19,20). Briefly, they were methylated using either M.SssI or M.HpaII, then the upper strand of the DNA was digested by incubation with λ-exonuclease, which specifically removes the 5′-phosphorylated strand. After a fill in reaction using the same upper primer as before, substrates were obtained that were hemimethylated at all CG or CCGG sites. The substrates were methylated by Dnmt1 in buffer containing 20 mM HEPES, pH 7.0, 1 mM EDTA, 30 mM NaCl, 7% glycerol, 25 µg/ml BSA and 1 mM AdoMet at 37°C and methylation determined by bisulfite analysis performed as described (21–23).
RESULTS AND DISCUSSION
Accuracy of Dnmt1 for hemimethylated DNA
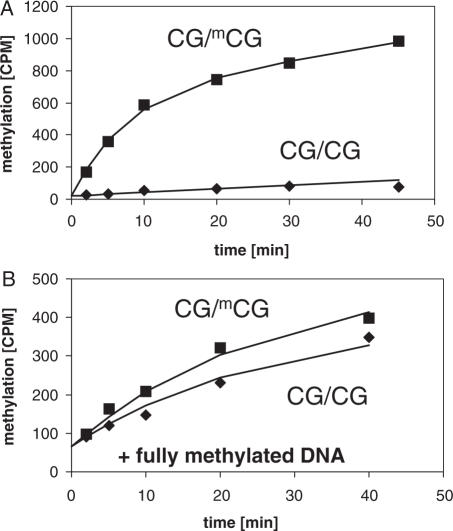
The mouse Dnmt1 enzyme used for these studies is full-length enzyme with an additional His-tag at the N-terminal end. The enzyme was prepared as described (11,19) and is over 90% pure (Supplementary Figure 1). Initially, we have analyzed the specificity of the Dnmt1 enzyme for hemimethylated DNA using 30 bp oligonucleotide substrates containing one centrally positioned CG site. In agreement with previous results (11), we observed a high preference for hemimethylated substrates which is ∼30- to 40- fold in the initial phase of the methylation reaction (Figure 1A). Next, we used hemimethylated DNA substrates to study DNA methylation by Dnmt1 on longer substrates, which is closer to a physiological situation. First, the methylation of substrate 1 (634 bp) hemimethylated at all 54 CG sites was compared with its unmethylated form under identical conditions (Supplementary Table 1). As shown in Figure 2, dense methylation was observed if the hemimethylated substrate was used (average methylation level: 58%), whereas methylation of the unmethylated substrate was only sparse (average methylation level: 4%) which indicates a 15-fold preference of Dnmt1 for the hemimethylated substrate. This ratio is identical to the results obtained with oligonucleotide substrates after 10 min of incubation (Figure 1). Thus, there are no large differences in the specificity of Dnmt1 for hemimethylated substrates if a 30mer oligonucleotide and longer DNAs are compared.
Figure 1.
Methylation of unmethylated and hemimethylated oligonucleotides by Dnmt1. Methylation was analyzed by incorporation of radioactive methyl groups into the DNA.In (A) 1 µM of hemimethylated and unmethylated oligonucleotides (CG30hm and CG30um) and 100 nM enzyme were used. In (B) 1 µM fully methylated oligonucleotide (CG30fm) of the same sequence was added.
Figure 2.
Methylation of unmethylated and hemimethylated long substrates in different reaction tubes. (A) Methylation of unmethylated substrate 1 by Dnmt1. In (B) substrate 1 was prepared in hemimethylated state at all CG sites and used for methylation by Dnmt1. Methylation was analyzed after bisulfite modification of the DNA, PCR amplification, cloning and sequencing of at least 20 individual clones for each DNA strand. In the figure, some examples of the clones obtained and sequenced are shown. Methylated CG sites are shown black, unmethylated white.
Next, we determined the methylation of the hemimethylated and unmethylated oligonucleotide substrates in the presence of an additional fully methylated oligonucleotide and observed an almost complete loss of preference for the hemimethylated target under these conditions (Figure 1B). Since in the cell unmethylated, hemimethylated and fully methylated DNA is present at the same time, this loss of specificity puts into question the whole concept of maintenance methylation. Therefore, we studied the methylation of an unmethylated and hemimethylated long substrate in competition in the same reaction mixture. Equimolar amounts of hemimethylated substrate 1 and unmethylated substrate 2 (566 bp, 44 CG sites) were mixed, incubated with Dnmt1 as described before and their respective methylation pattern was examined by bisulfite analysis (Figure 3A, Supplementary Table 1). We observed average levels of methylation of the hemimethylated and unmethyltated substrates of 57 and 5%, which indicate that the activity and specificity of the enzyme was not changed.
Figure 3.
Methylation of unmethylated and hemimethylated substrate in one reaction. (A) Methylation of unmethylated substrate 1 and hemimethylated substrate 2 by Dnmt1 in one reaction tube. Substrate 2 is hemimethylated at all CG sites in this experiment. (B) Methylation of substrate 2 hemimethylated at all CCGG sites. Methylation was analyzed after bisulfite modification of the DNA, PCR amplification, cloning and sequencing of at least 20 individual clones for each DNA strand. In the figure, some examples of the clones obtained and sequenced are shown. Methylated CG sites are shown black, unmethylated white. In (B) the hemimethylated CCGG sites are shaded gray.
To study the capacity of Dnmt1 to copy an existing methylation pattern, we have prepared substrate 2 hemimethylated at all CCGG sites, but unmethylated at all other CG sites. This DNA was incubated with Dnmt1 and methylation determined as before (Figure 3B, Supplementary Table 1). We observed 61% of methylation at the hemimethylated CCGG sites, while only 5% of the unmethylated CG sites had become modified, such that the preference was ∼12-fold. These results demonstrate that the presence of methylated DNA does not impair specific methylation of longer hemimethylated substrates. The strong allosteric activation and loss of specificity observed with oligonucleotide substrates upon addition of methylated DNA most likely is due to the artificial experimental setup using short oligonucleotide substrates and that is unlikely to affect the maintenance activity of Dnmt1 in the cell.
Processivity of Dnmt1
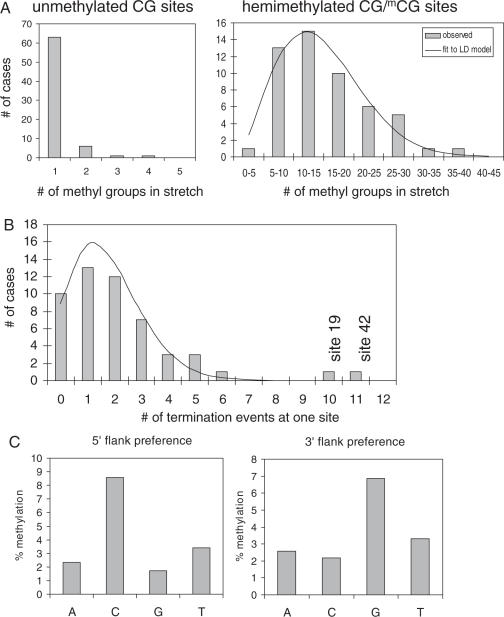
We analyzed the processivity of DNA methylation by Dnmt1 using hemimethylated and unmethylated substrates. Processive methylation is defined by consecutive methylation of one DNA substrate without dissociation of the enzyme from the DNA. Processive methylation of DNA will result in a stretch of methylated sites on the DNA that is not interrupted by unmethylated sites. To determine the number of consecutive methylation events on each substrate, we analyzed the lengths of continuous stretches of methylated sites on each DNA molecule for the experiments shown in Figures 2 and 3A. As shown in Figure 4A, Dnmt1 modifies unmethylated substrates in a distributive reaction, whereas on average 14–16 methyl groups were introduced during one binding event into hemimethylated DNA (average: 16, median value: 14). This number has to be taken as lower limit of processivity, because in many cases the enzyme had reached the end of the substrate molecule during its processive work, where no more methylation is possible. In order to describe the observed length distribution of methylated stretches, we apply the theory of linear diffusion. Since there is no energy input for the movement of Dnmt1 along the DNA, it follows a random walk. As shown in Figure 4A the observed length distribution of methylated stretches is fitted by the random walk model with an average number of 54 diffusional steps under these conditions, if one step is defined as the movement of the enzyme from on CG site to the next. Since the average distance of CG sites on our substrate is 10.6 bp, this means that the enzyme diffuses on the DNA for about 6000 one bp steps. Due to the random nature of the movement back and forth on the DNA, it has a high chance to reach the next methylation site within a distance of ∼80 bp.
Figure 4.
(A) Processivity of the methylation of the unmethylated and hemimethylated CG sites by Dnmt1. The figure displays the distribution of lengths of all stretches of methylated sites for the unmethylated (left) and hemimethylated substrate (right). The line in the right panel indicates a fit to the random walk model with a Pdif of 54. (B) Distribution of the number of termination events of processive methylation of the DNA at the various CG sites. The distribution is roughly fitted by a binomial distribution of the corresponding parameters as indicated by the line, if the two sites 19 and 42 are disregarded. (C) Flanking sequence preference for methylation of unmethylated DNA by Dnmt1.
The nice fit of the observed lengths' of continuously methylated stretches by the random walk model indicates that the data presented here are consistent with a random walk of Dnmt1 on the DNA. The methylation profiles are not in agreement with a model proposing a preferential movement of the enzyme in one direction, because then one would expect an accumulation of methylation at one end of the substrates, which is not observed.
Frequency of skipping target sites by Dnmt1
We checked the probability of Dnmt1 to miss target sites during the processive move on hemimethylated DNA. In this case, one would expect to observe two modified sites separated by one or two unmodified sites. Altogether on the hemimethylated substrates, we observed 85 stretches of more than two modified sites next to each other (>1000 methylated sites in total). Within these only three gaps of a size of one or two sites (indicative of the enzyme skipping one or two sites) were detected. This result demonstrates that Dnmt1 has a very low probability to skip a hemimethylated target site (P < 0.3%). Together with the result that Dnmt1 does not change its target strand during processive methylation of DNA (19), it indicates that Dnmt1 keeps intimate contact to the DNA for the duration of the diffusional walk.
Our data can be compared with results of Vilkaitis et al. (2005) who performed a similar experiment using CG hemimethylated substrates and bisulfite sequencing (24). However, they report much shorter average lengths of the processive methylation (median value: 5). In addition, within 22 runs of methylated sites they reported 15 gaps of one or two sites, indicating a much higher skipping rate of the enzyme than observed here. One reason for the reduced processivity and increased skipping rate could be in the preparation and purification of the enzyme or the hemimethylated substrate or the DNA sequence of the substrates. Vilkaitis et al. (2005) worked with a truncated Dnmt1 that had an N-terminal deletion of 290 amino acid residues. However, the truncation is unlikely to be of great influence, since in additional experiments they did not observe differences between the processivity of the full-length and truncated Dnmt1. In addition, it is possible that slight variations between the buffer conditions of both studies are responsible for the differences, because changes in the salt content have been shown previously to influence the processivity of DNA interacting enzymes (25,26).
Termination of processive methylation
We observed that in the experiments with hemimethylated DNA many stretches of methylated DNA terminated at the sites 19 (10 cases) and 42 (11 cases) (Figure 4B). It is not conceivable that these sites represent start sites for the DNA methylation by Dnmt1, because due to the random movement of Dnmt1 backwards and forwards on the DNA, methylation takes place on the 3′ and 5′ side of the original start site such that the original start site most likely will be situated somewhere in the center of the stretch of completely methylated DNA.
Consequently, the sites 19 and 42 represent termination sites that constitute barriers to the continuation of processive DNA methylation by Dnmt1. To understand this observation, we first investigated whether a large distance of these sites to their next neighbor sites could be due to the preferential termination of methylation. In our substrate, the CG sites are separated by up to 36 bp. Within this distance, there is no correlation detectable between the distances of CG sites to their neighbor sites and the termination frequency (data not shown), which indicates that the dwell time of Dnmt1 on the DNA is sufficiently long to allow for traveling this distance in most cases. Next, we analyzed for statistical significance: The observed distribution of termination events at all sites (except for the 10 and 11 events at sites 19 and 42) was roughly fitted by a binomial distribution indicating no strong deviation from statistics for most sites. However, the probability of finding two sites with more than nine termination events in this distribution was very low (P = 1.4 × 10−4), which suggests that these sites play a special role for Dnmt1. Interestingly, both sites are flanked by the same CTC trinucleotide sequence on their 3′ side. There was no sequence similarity on the 5′ side of the CG sequence and there were no additional CGCTC sites in our substrate. This result suggests that CGCTC sites function as termination site for DNA methylation by Dnmt1 either by promoting dissociation of the enzyme from the DNA or by forming a very tight interaction that prevents continuation of linear diffusion. Further experiments will show if this sequence also functions in other substrates and which mechanism is operative. It should be noticed that approaching a CGCTC site not necessarily led to termination of processive DNA methylation as indicated by the finding that several continuously methylated stretches span over the CGCTC sites.
Depending of the sequence context, CGCTC sites also function as binding site of the CTCF insulator protein (CCCTC) (27), which is involved in the control of imprinting and X-inactivation (28). One role of CTCF is to separate methylated and unmethylated parts of the DNA. On the basis of our data one could speculate that CGCTC sites could result in insulation of DNA methylation by a 2-fold effect: either by recruiting CTCF protein or in a CTCF independent way by preventing that Dnmt1 moves to the adjacent DNA. Both modes of action would result in the inhibition of spreading of DNA methylation into unmethylated regions of the DNA.
Flanking sequence preferences of Dnmt1 for methylation of unmethylated CG sites
In the experiments using unmethylated substrates, we observed 83 methylation events altogether. Since the number of methylation events did not suffice for a simultaneous statistical analysis with respect to next base pair on the 5′ and 3′ side, we studied 5′ and 3′ flanking preferences separately (Figure 4C). Out data show that Dnmt1 prefers to methylate DNA in a CCGG context which is methylated at ∼10 times higher frequency than all other sides. This conclusion is also illustrated by the observation that on the unmethylated substrates >40% of all methylation events occurred at CCGG sites although these represented only 10% of all target sites. We realized that by chance we observed a flanking preference on the unmethylated DNA that is identical to the sequence used for hemimethylation in the experiment shown in Figure 3B. However, the experiments shown in Figures 2 and 3A were carried out completely independently from that shown in Figure 3B such that there was no change of a partially methylation of the unmodified substrates.
There is no preference for flanking sequences reported for the catalytic activity of Dnmt1 so far. Our finding agrees to results by Flynn et al. (29) who reported increased binding of Dnmt1 to GCGG and CCGC sites. In contrast, the CCGG flanking sequence is disfavored by Dnmt3a and Dnmt3b (30). The difference in the flanking sequence preferences of Dnmt1 and Dnmt3a and 3b might allow in the future to determine whether aberrant de novo methylation was due to malfunction of Dnmt1 or Dnmt3a and 3b.
Implications for the propagation of methylation patterns in vivo
DNA methylation carries information that is used for control of gene expression. One of the most fascinating questions regarding DNA methylation is to understand how methylation patterns are copied after DNA replication to achieve a stable transmission of epigenetic information to next cellular generations. This function has been attributed to Dnmt1, which can re-establish a methylation pattern after DNA replication on the daughter strands of the DNA, because it prefers methylation at hemimethylated DNA sites. Errors in the propagation of methylation patterns are due to imperfect maintenance methylation as well as de novo methylation at previously unmethylated sites. In order to explore the accuracy of this mechanism we have analyzed the specificity of Dnmt1 for hemimethylated sites. Using long hemimethylated substrates, we show that Dnmt1 prefers hemimethylated sites ∼15-fold over unmethylated, irrespective of the presence of other sites in vicinity. This level of fidelity is compatible with in vivo data, which indicate that methylation is maintained with an accuracy of ∼95% and de novo methylation occurs with ∼5% (31–33), suggesting an ∼20-fold preference for methylation of hemimethylated sites after DNA replication. This preference of Dnmt1 sets an upper limit of accuracy for the propagation of punctual patterns of DNA methylation.
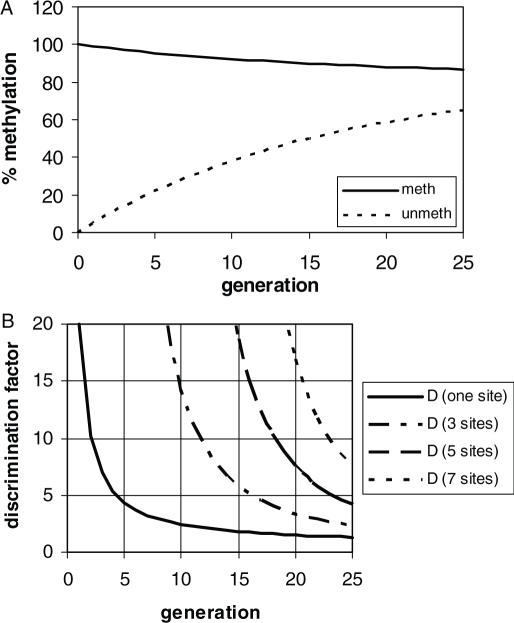
To simulate the process of the propagation of a DNA methylation pattern, we apply the model of stochastic maintenance of DNA methylation (31,33) and started with two populations of DNA molecules, one methylated at a certain site and the other not, and followed the average methylation of both populations over several generations (Figure 5). The information content of the methylation pattern is given by the ratio of the probabilities to find the site still methylated after N generations in the first population over finding it methylated in the second population after the same time (discrimination factor). If we assume a discrimination factor of 10 is required for transmission of the epigenetic information, we observe that the signal is lost already after three generations, if a single CG site is considered (Figure 5). However, if the biological system makes use of the combined reading of the methylation state of several CG sites, our simulation suggests that combination of 5 sites would allow to propagate a signal over 18 cellular generations and 7 sites suffice to propagate the signal over 23 generations which is already close to the typical somatic lifetime of many cells. This observation could explain why methylation of more than one CG site is required for strong gene inhibition and targeting of methyl cytosine binding proteins to methylated DNA. On the other hand, in the light of these data, it is not surprising that loss of methylation information occurs during aging and development. In the cell the specificity of maintenance methylation could be increased by local differences in the rate of methylation of unmodified CG which could be mediated by other chromatin modifications like histone 3 at lysine 9 methylation. Such mechanism could make use of Dnmt1 or one of the Dnmt3 enzymes and combine the effects of the various epigenetic systems into self enforcing epigenetic circuits whose state can be transmitted stably.
Figure 5.
Simulation of the average methylation level of an unmethylated and hemimethylated population of DNA molecules during several generations. For the simulation an average level of maintenance methylation of 95% and a 20-fold preference for hemimethylated DNA was used. In (A) the average methylation levels of the hemimethylated and unmethylated populations at one CG site are shown, in (B) the discrimination factor (defined as the ratio of the average methylation levels of both populations) is shown if one, three, five or seven CG sites are concerned.
Supplementary Material
Acknowledgments
This work has been supported by the DFG priority program Epigenetics (JE 252/4) and the BMBF NGFN2 (SMP Epigenetics, 01GR0497) BioFuture, and BioChance programs. Funding to pay for the open access publication charges for this article was provided by International University Bremen.
Conflict of interest statement. None declared.
REFERENCES
- 1.Egger G., Liang G., Aparicio A., Jones P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–293. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 4.Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 5.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature Rev. Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 6.Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 7.Reik W., Dean W., Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg A.P., Tycko B. The history of cancer epigenetics. Nature Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 9.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nature Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 10.Hermann A., Gowher H., Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol. Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PubMed] [Google Scholar]
- 11.Fatemi M., Hermann A., Pradhan S., Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J. Mol. Biol. 2001;309:1189–1199. doi: 10.1006/jmbi.2001.4709. [DOI] [PubMed] [Google Scholar]
- 12.Tollefsbol T.O., Hutchison C.A., III Mammalian DNA (cytosine-5-)-methyltransferase expressed in Escherichia coli, purified and characterized. J. Biol. Chem. 1995;270:18543–18550. doi: 10.1074/jbc.270.31.18543. [DOI] [PubMed] [Google Scholar]
- 13.Tollefsbol T.O., Hutchison C.A., III Control of methylation spreading in synthetic DNA sequences by the murine DNA methyltransferase. J. Mol. Biol. 1997;269:494–504. doi: 10.1006/jmbi.1997.1064. [DOI] [PubMed] [Google Scholar]
- 14.Flynn J., Glickman J.F., Reich N.O. Murine DNA cytosine-C5 methyltransferase: pre-steady- and steady-state kinetic analysis with regulatory DNA sequences. Biochemistry. 1996;35:7308–7315. doi: 10.1021/bi9600512. [DOI] [PubMed] [Google Scholar]
- 15.Pradhan S., Bacolla A., Wells R.D., Roberts R.J. Recombinant human DNA (cytosine-5) methyltransferase. I. Expression, purification, and comparison of de novo and maintenance methylation. J. Biol. Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- 16.Fatemi M., Hermann A., Gowher H., Jeltsch A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur. J. Biochem. 2002;269:4981–4984. doi: 10.1046/j.1432-1033.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- 17.Christman J.K., Sheikhnejad G., Marasco C.J., Sufrin J.R. 5-Methyl-2′-deoxycytidine in single-stranded DNA can act in cis to signal de novo DNA methylation. Proc. Natl Acad. Sci. USA. 1995;92:7347–7351. doi: 10.1073/pnas.92.16.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bacolla A., Pradhan S., Roberts R.J., Wells R.D. Recombinant human DNA (cytosine-5) methyltransferase. II. Steady-state kinetics reveal allosteric activation by methylated dna. J. Biol. Chem. 1999;274:33011–33019. doi: 10.1074/jbc.274.46.33011. [DOI] [PubMed] [Google Scholar]
- 19.Hermann A., Goyal R., Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 20.Roth M., Jeltsch A. Biotin-avidin microplate assay for the quantitative analysis of enzymatic methylation of DNA by DNA methyltransferases. Biol. Chem. 2000;381:269–272. doi: 10.1515/BC.2000.035. [DOI] [PubMed] [Google Scholar]
- 21.Clark S.J., Harrison J., Paul C.L., Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W., Molloy P.L., Paul C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warnecke P.M., Stirzaker C., Song J., Grunau C., Melki J.R., Clark S.J. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002;27:101–107. doi: 10.1016/s1046-2023(02)00060-9. [DOI] [PubMed] [Google Scholar]
- 24.Vilkaitis G., Suetake I., Klimasauskas S., Tajima S. Processive methylation of hemimethylated CpG sites by mouse Dnmt1 DNA methyltransferase. J. Biol. Chem. 2005;280:64–72. doi: 10.1074/jbc.M411126200. [DOI] [PubMed] [Google Scholar]
- 25.Ehbrecht H.J., Pingoud A., Urbanke C., Maass G., Gualerzi C. Linear diffusion of restriction endonucleases on DNA. J. Biol. Chem. 1985;260:6160–6166. [PubMed] [Google Scholar]
- 26.Jeltsch A., Pingoud A. Kinetic characterization of linear diffusion of the restriction endonuclease EcoRV on DNA. Biochemistry. 1998;37:2160–2169. doi: 10.1021/bi9719206. [DOI] [PubMed] [Google Scholar]
- 27.Filippova G.N., Fagerlie S., Klenova E.M., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reik W., Lewis A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nature Rev. Genet. 2005;6:403–410. doi: 10.1038/nrg1602. [DOI] [PubMed] [Google Scholar]
- 29.Flynn J., Azzam R., Reich N. DNA binding discrimination of the murine DNA cytosine-C5 methyltransferase. J. Mol. Biol. 1998;279:101–116. doi: 10.1006/jmbi.1998.1761. [DOI] [PubMed] [Google Scholar]
- 30.Handa V., Jeltsch A. Profound flanking sequence preference of Dnmt3a and Dnmt3b mammalian DNA methyltransferases shape the human epigenome. J. Mol. Biol. 2005;348:1103–1112. doi: 10.1016/j.jmb.2005.02.044. [DOI] [PubMed] [Google Scholar]
- 31.Pfeifer G.P., Steigerwald S.D., Hansen R.S., Gartler S.M., Riggs A.D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc. Natl Acad. Sci. USA. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laird C.D., Pleasant N.D., Clark A.D., Sneeden J.L., Hassan K.M., Manley N.C., Vary J.C., Jr, Morgan T., Hansen R.S., Stoger R. Hairpin-bisulfite PCR: assessing epigenetic methylation patterns on complementary strands of individual DNA molecules. Proc. Natl Acad. Sci. USA. 2004;101:204–209. doi: 10.1073/pnas.2536758100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riggs A.D., Xiong Z. Methylation and epigenetic fidelity. Proc. Natl Acad. Sci. USA. 2004;101:4–5. doi: 10.1073/pnas.0307781100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.