Abstract
Programmed frameshifting is one of the translational recoding mechanisms that read the genetic code in alternative ways. This process is generally programmed by signals at defined locations in a specific mRNA. In this study, we report the identification of hepta- and octo-uridine stretches as sole signals for programmed +1 and −1 ribosomal frameshifting during translation of severe acute respiratory syndrome coronavirus (SARS-CoV) ORF 3a variants. SARS-CoV ORF 3a encodes a minor structural protein of 274 amino acids. Over the course of cloning and expression of the gene, a mixed population of clones with six, seven, eight and nine T stretches located 14 nt downstream of the initiation codon was found. In vitro and in vivo expression of clones with six, seven and eight Ts, respectively, showed the detection of the full-length 3a protein. Mutagenesis studies led to the identification of the hepta- and octo-uridine stretches as slippery sequences for efficient frameshifting. Interestingly, no stimulatory elements were found in the sequences upstream or downstream of the slippage site. When the hepta- and octo-uridine stretches were used to replace the original slippery sequence of the SARS-CoV ORF 1a and 1b, efficient frameshift events were observed. Furthermore, the efficiencies of frameshifting mediated by the hepta- and octo-uridine stretches were not affected by mutations introduced into a downstream stem–loop structure that totally abolish the frameshift event mediated by the original slippery sequence of ORF 1a and 1b. Taken together, this study identifies the hepta- and octo-uridine stretches that function as sole elements for efficient +1 and −1 ribosomal frameshift events.
INTRODUCTION
In all organisms, accurate transfer of genetic information is critical for maintaining their genetic traits. During translation, universal decoding rules would be prevailing and guarantee correct decoding of the genetic code. However, cells do evolve various translational recoding mechanisms to interpret the genetic code in alternative ways. Programmed ribosomal frameshifting is one of these recoding mechanisms. This mechanistically diverse process is well characterized in retrotransposons (1,2), bacteria (3–5), insects (6), animals (7) and animal viruses (8–11). Generally, the ribosome can shift its frame during translation elongation either in the forward (3′) or backward (5′) direction, causing +1 or −1 frameshifting. A −1 frameshift event takes place in most cases, such as the gag-pol gene of human immunodeficiency virus 1 (12), the 1a/1b gene of coronavirus infectious bronchitis virus (IBV) (9) and severe acute respiratory syndrome coronavirus (SARS-CoV) (8,13). Examples of +1 frameshifting include GAG3 and POL3 (GAG3-POL3) genes of the retrotransposon Ty3 of yeast (2), the mammalian ornithine decarboxylase antizyme (7) and the thymidine kinase (TK) gene of herpes simplex virus (HSV) (14).
Ribosomal frameshift signals generally contain two elements, a heptanucleotide slippery sequence XXXYYYN (where X = A, G or U and Y = A or U) and a stimulator (10,12). By mutational analysis of the slippery sequence of IBV, Brierley et al. (10) revealed that monotonous runs of Us could give significant levels of frameshifting. Various elements, such as an RNA secondary structure, have been identified as stimulators for efficient frameshifting. However, the HIV slippery sequence could direct efficient frameshifting in mammalian and yeast systems, independent of any secondary structure (15).
SARS-CoV is a group 2 coronavirus. Similar to other coronaviruses, the 5′ unique region of mRNA 1 encodes two large polyproteins, 1a and 1ab. The 1ab polyprotein is translated by a ribosomal frameshifting mechanism, and is a fusion product of ORF 1a and 1b. Through sequence alignment and comparison, a conserved slippery sequence (U UUA AAC) was identified in the 1a/1b region as the slippage site. Evidence provided by Baranov et al. (8) confirmed that a −1 frameshift event did occur during translation of the region. More recently, an atypical RNA pseudoknot and an upstream attenuation signal were shown to regulate this −1 ribosomal frameshift event (8,16). So far, no other frameshifting signal was identified in any other genes from SARS-CoV and other coronaviruses.
In this study, we report the identification of a unique frameshift signal with seven (UUU UUU U) and eight (UUU UUU UU) Us as sole signals for efficient +1 and −1 frameshifting during translation of SARS-CoV ORF 3a variants. SARS-CoV ORF 3a encodes a protein of 274 amino acids. Recent studies demonstrated that 3a protein is a minor structural protein and is associated with the spike protein in virions (17–20). During the course of cloning and expression of ORF 3a, a mixed population of clones with six, seven, eight and nine T stretches located 14 nt downstream of the initiation codon was found. The existence of these ORF 3a variants was confirmed recently by isolation of a heterogeneous population of subgenomic mRNA three transcripts with six, seven, eight and nine Us from the sera of SARS patients (21). Evidence provided here demonstrates that the seven and eight U stretches could function as sole elements for efficient +1 and −1 ribosomal frameshifting during translation of these ORF 3a variants. This report demonstrates a sole requirement of hepta- and octo-uridine as signals for efficient frameshifting during expression of authentic viral genes and reinforces the conclusion drawn by Brierley et al. (10), based on mutagenesis studies, that U stretches in mRNA are particularly slippery compared with other monotonous runs of nucleotides.
MATERIALS AND METHODS
Antibodies
The mouse monoclonal anti-Flag antibody conjugated with HRP, the mouse monoclonal antibody against firefly luciferase and the mouse monoclonal antibody against β-tubulin were obtained from Sigma-Aldrich. The mouse monoclonal antibody against 6× His tag conjugated with HRP was purchased from Santa Cruz Biotechnology Inc. The mouse monoclonal antibody against enhanced green fluorescent protein (EGFP) was obtained from Biomed Diagnostics. The polyclonal antibody against full-length 3a was raised in rabbits.
In vitro transcription and translation
One microgram of each plasmid DNA was transcribed and translated in a total of 50 µl reaction mixture in a TnT coupled in vitro translation system (Promega) in rabbit reticulocyte lysates (RRL), labeled with 50 µCi/ml of [35S]methionine (Amersham Biosciences). The translation products were analyzed on 12% SDS–PAGE and visualized by autoradiography.
Cells and DNA transfection
Cos-7 cells were maintained in DMEM supplemented with 10% fetal bovine serum in the presence of 1% penicillin/streptomycin at 37°C in a 5% CO2 incubator. Semi-confluent Cos-7 cells seeded in 6-well plates were infected with five plaque forming units per cell of the recombinant vaccinia/T7 virus for 2 h followed by transfection of plasmid DNA using the Effectene Transfection reagent (Qiagen). At 24 h post transfection, cells were washed twice with phosphate-buffered saline and lysed in either 200 µl of 2× SDS loading buffer for analysis of the expression or in 500 µl Trizol solution for RNA extraction.
RNA isolation and RT–PCR
Cells transfected with plasmid DNA were lysed in 500 µl Trizol solution. After transferred to fresh tubes, 100 µl of chloroform were added and mixed well by vortex before centrifugation. The upper aqueous phase was precipitated with 2 vol of ethanol at −20°C for 30 min. After centrifugation, the pellets were washed with 70% ethanol and air-dried. The total RNA was resuspended in 20 µl of RNase-free distilled water.
One microgram of total RNA was used as template for reverse transcription in the presence of reverse transcriptase at 42°C for 1 h. One microliter from the reaction was taken out for PCR using the appropriate primers.
Plasmids construction and site-directed mutagenesis
Wild-type SARS 3a cDNA was amplified by PCR and digested with EcoRV and EcoRI. The digested fragment was cloned into EcoRV- and EcoRI-digested pFlag vector, generating pF-3a/6T. The 7T and 8T variants were made by site-directed mutagenesis using the Quikchange™ kit (Stratagene). Point mutations of the seven T stretch in pF-3a/7T were also made by site-directed mutagenesis.
For construction of EGFP-3a fusion constructs and mutants, PCR fragment covering EGFP from pEGFP-C1 (Clontech) was digested with BglII and EcoRV, and cloned into BglII- and EcoRV-digested pSARS-3a/7T, generating pEGFP-3a/7T. Deletions in EGFP and 3a regions were made by overlapping PCR. The PCR fragments were digested with the same set of restriction enzymes and ligated into the same vector.
PCR fragment covering the SARS-CoV ORF 1a and 1b region from nt 12711 to 14110 was digested with EcoRV and EcoRI, and cloned into EcoRV- and EcoRI-digested pFLAG, generating pF-S1ab. Plasmids pF-S1ab/7T and pF-S1ab/8T as well as their corresponding mutants were made by site-directed mutagenesis.
Plasmids pLuc-6T and pLuc-7T were made by insertion of six and seven Ts, respectively, into the luciferase gene at a position 18 nt downstream of the initiator ATG codon. All constructs were confirmed by automated sequencing analysis.
RESULTS
Programmed +1 and −1 frameshifting events occur during translation of SARS-CoV ORF 3a variants
SARS-CoV ORF 3a encodes a minor structural protein of 274 amino acids. Over the course of cloning and expression of the gene, a mixed population of clones with six, seven, eight and nine T stretches located 14 nt downstream of the initiation codon was found. The existence of these ORF 3a variants was confirmed recently in SARS patients (21). To examine the expression of 3a variants, full-length clones covering the 3a and 3b regions with six (pSARS-3a/6T), seven (pSARS-3a/7T) and eight (pSARS-3a/8T) Ts were constructed (Figure 1a). In vitro expression of pSARS-3a/6T in the transcription coupled translation reticulocyte lysate system showed the detection of a dominant product of ∼31 kDa, representing the full-length 3a protein (Figure 1b, lane 1). In addition, a minor band (3a*) of ∼27 kDa was expressed, which represents the initiated product from an internal AUG (22). Interestingly, the 31 kDa full-length 3a protein was also detected when constructs pSARS-3a/7T and pSARS-3a/8T were expressed in the same in vitro expression system (Figure 1b, lanes 2 and 3). The efficiencies of the 31 kDa 3a protein expressed from these two constructs are ∼32 and 20%, respectively, compared with that expressed from pSARS-3a/6T (Figure 1b, lanes 1–3). These results reveal that frameshift events may occur during translation of ORF 3a variants with seven and eight Us.
Figure 1.
Expression of SARS-CoV ORF 3a variants. (a) Diagram of SARS-CoV 3a constructs with six, seven and eight T stretches under the control of T7 promoter. The positions of six, seven and eight T stretches, the C-terminal His tag in constructs pET3a-His/6T, pET3a-His/7T and pET3a-His/8T, and the N-terminal Flag tag in constructs pF-3a/6T, pF-3a/7T and pF-3a/8T, are indicated. (b) Expression of pSARS-3a/6T (lane 1), pSARS-3a/7T (lane 2) and pSARS-3a/8T (lane 3) in vitro in RRL. Polypeptides were labeled with [35S]methionine, separated on SDS–12% polyacrylamide gel and detected by autoradiography. Bands corresponding to the full-length 3a and a minor species 3a* representing an internal initiation product are indicated. Numbers on the left indicate molecular masses in kilodaltons. (c) Expression of pET3a-His/6T (lane 1), pET3a-His/7T (lane 2) and pET3a-His/8T (lane 3) in bacterial cells. Plasmid DNA was transformed into E.coli strain BL-21, and the protein expression was induced by adding 1 mM IPTG. After induction for 2 h, total cell lysates were prepared, resolved on SDS–12% polyacrylamide, and analyzed by western blot with anti-His antibody. Numbers on the left indicate molecular masses in kilodaltons. (d) Expression of pF-3a/6T (lane 2), pF-3a/7T (lane 3) and pF-3a/8T (lane 4) in Cos-7 cells. Cells were infected with the recombinant vaccinia/T7 virus, and transfected with an empty control plasmid (lane 1) and the three Flag-tagged 3a constructs, respectively. At 18 h posttransfection, cells were harvested and lysates prepared. Polypeptides were separated on SDS–12% polyacrylamide and analyzed by western blot with anti-Flag antibody. Numbers on the left indicate molecular masses in kilodaltons.
The expression of these ORF 3a variants was then tested in bacteria. Three constructs, pET3a-His/6T, pET3a-His/7T and pET3a-His/8T, were made and transformed into Escherichia coli strain BL-21 (Figure 1a). After induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the expression of the His-tagged 3a protein was analyzed by western blot with anti-His antibody. The 32 kDa His-tagged 3a protein was detected in bacterial cells transformed with all three constructs (Figure 1c, lanes 1–3). The efficiencies of the protein expressed from pET3a-His/7T and pET3a-His/8T are ∼35 and 18%, respectively, of that expressed from pET3a-His/6T (Figure 1c).
Examination of the expression of these three ORF 3a variants was finally carried out in mammalian cells. Expression of the Flag-tagged 3a from pF-3a/6T in Cos-7 cells showed very efficient detection of the 32 kDa 3a protein by western blot with anti-Flag antibody (Figure 1d, lane 2). The full-length 3a protein was also detected in cells expressing the two ORF 3a variants with seven (pF-3a/7T) and eight (pF-3a/8T) Ts (Figure 1d, lanes 3 and 4). The expression efficiencies of the full-length 3a protein from these two constructs are ∼28 and 10%, respectively, as compared with that expressed from the construct with six Ts (Figure 1d, lanes 2–4). The detection of the full-length 3a protein expression from these ORF 3a variants in all three expression systems confirms that the hepta- and octo-uridine stretches could direct efficient frameshifting during translation elongation process.
The hepta-uridine stretch functions as a slippery sequence in an ORF 3a variant
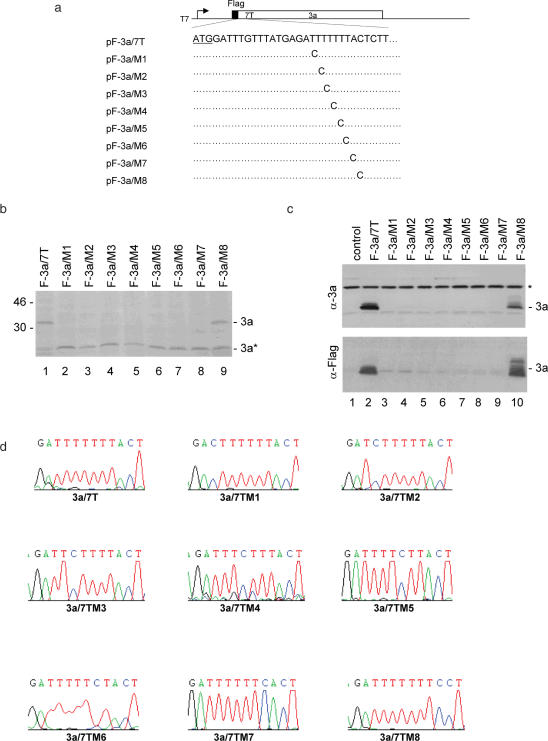
The construct with seven Ts was then used to characterize the slippery sequence. Point mutations of T to C at every individual T position were made, giving rise to pF-3a/7TM1 to M7 (Figure 2a). Studies on HIV frameshifting mechanisms led to the identification of U UUU UUA as a slippery sequence, but mutation of the nucleotide A immediately downstream of the six T stretch did not significantly affect the frameshifting efficiency (15). Accordingly, mutation of the nucleotide A immediately downstream of the seventh T was made, giving rise to pF-3a/7TM8 (Figure 2a). In vitro expression of pF-3a/7T showed, once again, the detection of the Flag-tagged full-length 3a protein (Figure 2b, lane 1). In addition, the internally initiated 3a* product was also detected (Figure 2b, lane 1). However, the full-length 3a expression was significantly decreased to an undetectable level when mutants M1 to M7 were expressed (Figure 2b, lanes 2–8). The detection of similar amounts of 3a* expression from these constructs confirmed that the absence of the full-length 3a protein expression is due to the point mutations introduced into the seven T stretch. Mutation of the nucleotide A immediately downstream of the seven T stretch did not affect the expression of the full-length 3a protein (Figure 2b, lane 9).
Figure 2.
Mutational analysis of the slippery sequence in pF-3a/7T. (a) Diagram of pF-3a/7T and the eight mutants (pF-3a/M1-M8). The initiator ATG codon for 3a is underlined. Also shown are the point mutations introduced into the seven T stretch in each mutant constructs. (b) Expression of pF-3a/7T (lane 1) and mutant constructs (lanes 2–9) in vitro in reticulocyte lysates. Polypeptides were labeled with [35S]methionine, separated on SDS–12% polyacrylamide gel and detected by autoradiography. Bands corresponding to the full-length 3a and a minor species 3a* representing an internal initiation product are indicated. Numbers on the left indicate molecular masses in kilodaltons. (c) Expression of pF-3a/7T and mutant constructs in Cos-7 cells. Cells were infected with the recombinant vaccinia/T7 virus, and transfected with an empty control plasmid (lane 1), pF-3a/7T (lane 2) and the eight mutant constructs (lanes 3–10), respectively. At 18 h posttransfection, cells were harvested and lysates prepared. Polypeptides were separated on SDS–12% polyacrylamide and analyzed by western blot with either anti-3a polyclonal (upper panel) or anti-Flag monoclonal antibodies (lower panel). A background band detected by this antiserum is indicated by asterisk. (d) No deletion of nucleotides or revision of mutations in the seven T regions during transcription of wild type and mutant constructs. Cos-7 cells transfected with pF-3a/7T and the eight mutant constructs, respectively, were lysed by Trizol reagent at 24 h post transfection. Total RNA was extracted from cells expressing and 1 µg of RNA was used for RT–PCR. The RT–PCR products were sequenced by automated nucleotide sequencing. The seven T regions of the RT–PCR products are shown.
Expression of these mutant constructs in Cos-7 cells showed similar results to the in vitro expression system. Western blot analysis with anti-3a and anti-Flag antibodies led to the detection of the full-length 3a protein in cells transfected with pF-3a/7T and pF-3a/7TM8, respectively (Figure 2c, lanes 2 and 10). The 3a protein was not detected in cells expressing M1-M7 mutants (Figure 2c, lanes 3–9). These results confirm that mutation of any T in the seven T stretch totally abolishes the frameshift event.
To rule out the possibility that other events, such as RNA editing and transcriptional slippage that might lead to deletion of the extra nucleotide, could account for the expression of the full-length 3a protein from pF-3a/7T, RT–PCR and sequencing analysis of RNA extracted from Cos-7 cells transfected with wild type and mutant pF-3a/7T constructs were performed. The results confirmed that there was no deletion and reversion of the mutations back to wild-type 3a at the mRNA level (Figure 2d).
Sequences upstream and downstream of the hepta-uridine site do not affect the frameshifting efficiency
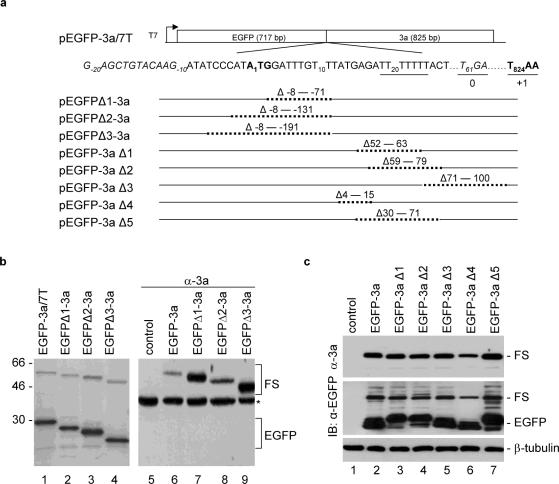
Deletion of sequences upstream and downstream of the slippage site was carried out to search for stimulators that are essential for efficient frameshifting in most of the frameshifting signals identified so far. To facilitate deletion analysis of the upstream sequence, a cDNA fragment covering ORF3a with seven Ts was fused in frame to the 3′ end of the EGFP, giving rise to construct pEGFP-3a/7T (Figure 3a). Expression of this construct was expected to produce a 29 kDa protein, representing the EGFP product terminated at a UGA codon 61 nt downstream of the initiator AUG for 3a (Figure 3a). If frameshifting did occur during translation of this construct, a 55 kDa product terminated at the original UAA codon for 3a and representing a fusion of EGFP and 3a would be expressed (Figure 3a). As expected, expression of this plasmid in RRL in vitro showed the detection of a major 29 kDa protein (Figure 3b, lane 1). In addition, a minor band of ∼55 kDa, representing the frameshifting product, was also detected (Figure 3b, lane 1). The frameshifting efficiency is ∼32%.
Figure 3.
Deletion analysis of sequences upstream and downstream of the slippage site in pEGFP-3a/7T. (a) Diagram showing the structures of pEGFP-3a/7T and eight derivative constructs with deletion at different regions. Sequence covering the fusion region between EGFP and 3a is shown. The initiator AUG for the 3a ORF is indicated in bold, the position of the nucleotide A is designated +1 and the nucleotide positions upstream and downstream of the AUG codon are indicated by minus and plus numbers, respectively. The seven T stretch is underlined, the termination codon TGA for 0 frame is italic and underlined, and the termination codon TAA for +1 frame is underlined and in bold. Also shown are the nucleotides deleted in each construct. (b) Expression of pEGFP-3a/7T (lanes 1 and 6), pEGFPΔ1-3a (lanes 2 and 7), pEGFPΔ2-3a (lanes 3 and 8) and pEGFPΔ3-3a (lanes 4 and 9) in in vitro reticulocyte lysates (lanes 1–4) and in Cos-7 cells (lanes 5–9). The in vitro expressed polypeptides were labeled with [35S]methionine, separated on SDS–12% polyacrylamide gel and detected by autoradiography. Polypeptides expressed in Cos-7 cells were separated on SDS–12% polyacrylamide and analyzed by western blot with anti-3a polyclonal antibodies. Bands corresponding to EGFP, the frameshifting products (FS) and a background band (asterisk) are indicated. Numbers on the left indicate molecular masses in kilodaltons. (c) Expression of pEGFP-3a/7T (lane 2), pEGFP-3aΔ1 (lane 3), pEGFP-3aΔ2 (lane 4) pEGFP-3aΔ3 (lane 5), pEGFP-3aΔ4 (lane 6) and pEGFP-3aΔ5 (lane 7) in Cos-7 cells. Polypeptides expressed were separated on SDS–12% polyacrylamide and analyzed by western blot with either anti-3a, anti-EGFP, or anti-β-tubulin antibodies. Bands corresponding to EGFP, the frameshifting products and -β-tubulin are indicated.
Three deletion constructs with deletion of various upstream sequences were made (Figure 3a). As can be seen, sequences deleted in these constructs are in the EGFP coding region, and are not directly related to the original ORF 3a. Nevertheless, expression of these constructs in vitro showed the detection of a comparable level of frameshifting efficiency (Figure 3b, lanes 2–4). Expression of these constructs in Cos-7 cells showed the detection of the frameshifting product by western blot with polyclonal anti-3a antibodies (Figure 3b, lanes 6–9).
Analysis of the sequences downstream of the seven Us using the MFOLD program predicated two potential stem–loops forming from nt 39 to 56 (starting from AUG for 3a) and from nt 62 to 88. The loop regions could partially base pair with the downstream sequences and form a potential pseudoknot interaction. Deletion of sequences covering this region as well as other sequences upstream and downstream of the seven T stretch was made based on pEGFP-3a/7T. Five constructs (pEGFP-3aΔ1 to pEGFP-3aΔ5), containing deletions of various regions in ORF 3a, were constructed and expressed in Cos-7 cells (Figure 3a). The results obtained showed that these deletions did not have significant effects on the expression of the frameshifting product (Figure 3c, lanes 2–7), suggesting that stimulatory elements may not exist in the surrounding sequences. Taken together, these results demonstrate that the hepta-uridine stretch alone may function as an efficient frameshift signal.
The hepta- and octo-uridine stretches function as active slippage sites in heterogeneous ORFs
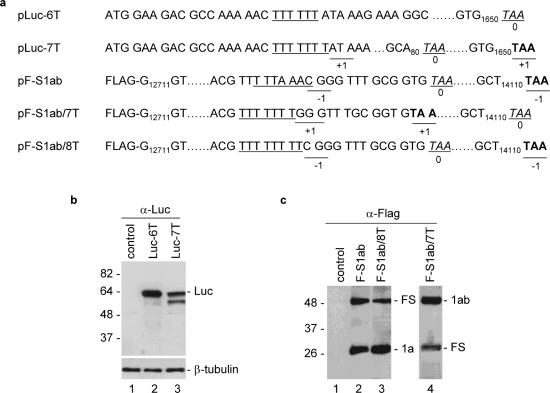
To test further if the hepta- and octo-uridine stretches alone can function as an efficient frameshift signal, the seven and eight Ts were inserted into unrelated ORFs, and their ability to direct frameshifting was analyzed. The six and seven Ts were first inserted into the luciferase gene 14 nt downstream of the initiator AUG, giving rise to constructs pLuc-6T and pLuc-7T (Figure 4a). Expression of these two constructs in Cos-7 cells showed the detection of the 65 kDa full-length luciferase protein in cells expressing pLuc-6T (Figure 4b, lane 2). In addition, a minor band of ∼60 kDa, representing a product probably initiated from an internal AUG codon, was also detected (Figure 4b, lane 2). In cells transfected with pLuc-7T, the full-length 65 kDa luciferase protein was detected, indicating that frameshift event did occur during translation of this construct (Figure 4b, lane 3). Interestingly, the expression of the minor 60 kDa band was increased in cells expressing this constructs (Figure 4b, lane 3). These results indicate that the seven U stretch alone could direct efficient frameshifting.
Figure 4.
Analysis of frameshifting efficiencies mediated by hepta- and octo-uridine stretches in heterogeneous ORFs. (a) Diagram showing the structures of constructs pLuc-6T, pLuc-7T, pF-S1ab, pF-S1ab/7T and pF-S1ab/8T. The slippery sequences (the six T stretch in the case of pLuc-6T) is underlined, the TAA termination codon for 0 frame is italic and underlined, and the TAA termination codon for +1 or −1 frame is underlined and in bold. (b) Expression of pLuc-6T and pLuc-7T in Cos-7 cells. Cells were infected with the recombinant vaccinia/T7 virus, and transfected with an empty control plasmid (lane 1), pLuc-6T (lane 2) and pLuc-7T (lane 3). At 18 h posttransfection, cells were harvested and lysates prepared. Polypeptides were separated on SDS–12% polyacrylamide and analyzed by western blot with anti-luciferase antibodies. Bands corresponding to the full-length luciferase and -β-tubulin are indicated. Numbers on the left indicate molecular masses in kilodaltons. (c) Expression of pF-S1ab, pF-S1ab/7T and pF-S1ab/8T in Cos-7 cells. Cells were infected with the recombinant vaccinia/T7 virus, and transfected with an empty control plasmid (lane 1), pF-S1ab (lane 2), pF-S1ab/7T (lane 3) and pF-S1ab/8T (lane 4). At 18 h posttransfection, cells were harvested and lysates prepared. Polypeptides were separated on SDS–12% polyacrylamide and analyzed by western blot with anti-Flag antibodies. Bands corresponding to 1a (or 1ab) and the frameshifting products are indicated. Numbers on the left indicate molecular masses in kilodaltons.
Replacement of the original slippery sequence of the SARS-CoV ORF 1a/1b with seven and eight U stretches was carried out to test if similar frameshift events could be observed. Plasmid pF-S1ab was constructed by cloning the SARS-CoV 1a/1b region from nt 12711 to 14110 into an expression vector under the control of a T7 promoter (Figure 4a). A Flag tag and a TAA termination codon were added at the 5′ and 3′ ends of the clone, respectively (Figure 4a). As expected, expression of this construct in Cos-7 cells led to the detection of a 25 kDa 1a termination product and a 50 kDa frameshifting product (Figure 4c, lane 2). Replacement of the original slippage site (T TTA AAC) with eight Ts in pF-S1ab/8T produced two products similar to the two proteins produced from the construct with the original slippage site (Figure 4c, lane 3). In construct pF-S1ab/7T, the original slippery sequence was replaced by seven Ts, resulting in the fusion of ORF 1a and 1b (Figure 4a). The termination product therefore contains both the 1a and 1b regions and is similar to the frameshifting product expressed from pF-S1ab and pF-S1ab/8T (Figure 4a). The putative +1 frameshifting product was expected to terminate at the original TAA codon for ORF 1a, which is equivalent to the 1a termination product expressed from pF-S1ab and pF-S1ab/8T (Figure 4a). As expected, expression of this construct resulted in the detection of the 50 kDa 1ab termination product and the 25 kDa frameshifting product (Figure 4c, lane 4).
Mutations that destabilize a downstream stimulator abolish the frameshift event mediated by the original slippery sequence of the SARS-CoV ORF 1a and 1b, but do not affect frameshifting efficiencies mediated by the hepta- and octo-uridine stretches
A pseudoknot structure was shown to be essential for efficient frameshifting during translation of coronavirus ORF 1a and 1b (8,16). In a recent report, Su et al. (16) demonstrated that destabilizing stem II by mutagenesis significantly reduces the frameshifting efficiency occurred at the SARS-CoV 1a and 1b region. Mutations of stem II were then carried out based on pF-S1ab, pF-S1ab/7T and pF-S1ab/8T, giving rise to constructs pF-S1abM, pF-S1ab/7TM and pF-S1ab/8TM, respectively (Figure 5a). The wild type and mutant constructs were then expressed in Cos-7 cells. Expression of pF-S1ab showed, once again, the detection of the 1a termination and 1ab frameshifting products (Figure 5b, lane 1). Expression of pF-S1abM showed the detection of a similar amount of the 1a termination product (Figure 5b, lane 2). However, the 50 kDa frameshifting product was not observed (Figure 5b, lane 2). These results confirm that mutations introduced into stem II abolish the frameshift event. Intriguingly, when pF-S1ab/8T and pF-S1ab/8TM were expressed in Cos-7 cells, equal amounts of termination and frameshifting products were detected from wild type and mutant constructs (Figure 5b, lanes 3–5). Similar results were also obtained when pF-S1ab/7T and pF-S1ab/7TM were expressed (Figure 5b, lanes 6 and 7). These results demonstrate that mutation of a downstream stimulator does not affect the frameshifting efficiencies mediated by the seven and eight U stretches, and reinforce the conclusion that hepta- and octo-uridine stretches are sole elements required for efficient frameshifting.
Figure 5.
Mutational analysis of a downstream stimulator on frameshifting efficiencies mediated by the hepta- and octo-uridine stretches. (a) Diagram showing the slippery sequence and the downstream stem–loop structures of SARS-CoV 1a/1b region. The slippery sequence is underlined and the two stems are indicated. Also indicated are the mutations introduced into stem 2 (in bold). (b) Expression of pF-S1ab (lane 1), pF-S1abM (lane 2), pF-S1ab/7T (lane 6), pF-S1ab/7TM (lane 7), pF-S1ab/8T (lane 3) and pF-S1ab/8T (lane 4) in Cos-7 cells. Cells were infected with the recombinant vaccinia/T7 virus, and transfected with an empty control plasmid (lane 5) and the six constructs, respectively. At 18 h posttransfection, cells were harvested and lysates prepared. Polypeptides were separated on SDS–12% polyacrylamide and analyzed by western blot with anti-Flag antibodies. Bands corresponding to 1a (or 1ab), the frameshifting products (FS) and β-tubulin are indicated. Numbers on the left indicate molecular masses in kilodaltons.
The downstream stimulatory sequence has differential effects on frameshifting efficiencies mediated by the hepta-uridine stretch with mutations at different positions
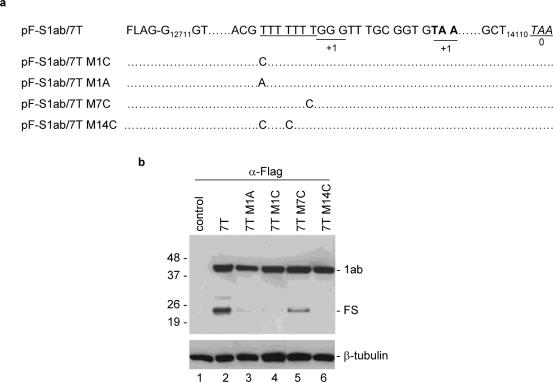
As mutations of any of the seven Us in an ORF3a variant severely reduce the frameshifting efficiency mediated by the seven U slippery sequence, we finally set up to test the effect of a downstream stimulatory element on the frameshift event mediated by the seven T stretch with mutation at different positions. As shown in Figure 6a, mutants containing mutation of the first T to A (pF-slab/7TM1A) and to C (pF-slab/7TM1C), mutation of the seventh T to A (pF-slab/7TM7C), and mutation of the first and fourth Ts to Cs (pF-slab/7TM14C), were made based on pF-slab/7T. Expression of pF-slab/7T showed, once again, the detection of ∼35% of the frameshifting efficiency (Figure 6b, lane 2). Mutation of the first T to either A or C significantly reduces the frameshifting efficiency to ∼1% of wild type (Figure 6b, lanes 3 and 4). Mutation of both the first and fourth Ts to Cs totally abolished the frameshifting (Figure 6b, lane 6). However, mutation of the seventh T to C results in the detection of 15% of frameshifting product (Figure 6b, lane 5). These results confirm that the downstream stimulator has differential effects on the frameshifting efficiencies mediated by the hepta-uridine stretch with mutations at different positions.
Figure 6.
Differential effects of a downstream stimulator on frameshifting efficiencies mediated by wild type and mutant hepta-uridine stretch. (a) Diagram showing the structure of constructs pF-S1ab/7T, pF-S1ab/7TM1C, pF-S1ab/7TM1A, pF-S1ab/7TM7C and pF-S1ab/7TM14C. The seven T stretch is underlined. Also shown are the TAA termination codon for the +1 and 0 frames, and the mutations introduced into the seven T region. (b) Expression of pF-S1ab/7T (lane 2), pF-S1ab/7TM1A (lane 3), pF-S1ab/7TM1C (lane 4), pF-S1ab/7TM7C (lane 5) and pF-S1ab/7TM14C (lane 6) in Cos-7 cells. Cells were infected with the recombinant vaccinia/T7 virus and infected either with an empty control plasmid (lane 1) or each of the five constructs (lanes 2–6). At 18 h posttransfection, cells were harvested and lysates prepared. Polypeptides were separated on SDS–12% polyacrylamide gel and analyzed by western blot with anti-Flag antibodies. Bands corresponding to 1ab, the frameshifting products (FS) and β-tubulin are indicated. Numbers on the left indicate molecular masses in kilodaltons.
DISCUSSION
We report here the identification of two unique frameshift events with seven (UUU UUU U) and eight (UUU UUU UU) Us as sole signals for efficient +1 and −1 frameshifting during translation of SARS-CoV ORF 3a variants containing seven and eight U stretches located 14 nt downstream of the initiation codon. Expression of clones with six, seven and eight Ts in an in vitro expression system, bacteria and in intact cells showed detection of the full-length 3a protein. To study the mechanisms that control the expression of the full-length 3a protein from clones with seven and eight Ts, site-directed mutagenesis was carried out based on the construct with seven Ts (TTT TTT TA). The results showed that mutation of any T in the TTT TTT TA significantly reduced the expression of the full-length 3a protein. However, mutation of the nucleotide A immediately downstream of the seven T stretch did not affect the 3a expression, suggesting that the full-length 3a protein is expressed by a +1/−2 frameshifting mechanism and the UUU UUU U is the slippage site. Attempts were subsequently made to identify upstream and downstream regulatory elements by deletion analysis. Neither downstream stem–loop structures and pseudoknots, nor upstream sequences with stimulatory function were identified, suggesting that the hepta- and octo-uridine stretches could function as sole elements for efficient ribosomal frameshifting. This possibility was further confirmed by insertion of seven Ts into the coding region of luciferase gene and by replacement of the slippery sequence of SARS-CoV ORF 1a and 1b with seven and eight Ts. Moreover, mutations introduced into stem II of the SARS-CoV ORF 1b region totally abolished frameshifting mediated by the original slippery sequence of SARS-CoV ORF 1a and 1b, but did not affect frameshifting mediated by the seven or eight U stretches. Taken together, this study demonstrates that the hepta- and octo-uridine stretches are sole elements for efficient programmed ribosomal frameshift events.
Expression of the full-length 3a protein from the construct with seven Ts (with a single-nucleotide insertion) requires +1/−2 frameshifting at the slippage site (UUU UUU U). At present, we do not know exactly whether +1 or −2 frameshifting occurs. Attempts were made to purify the frameshifting products from various expression systems and to determine the precise sequence across the slippage site, but without success. Generally, +1 frameshifting is considered the main event taking place at the slippage site since the phenylalanine tRNAGAA could form perfect pairing in the +1 frame with UUU, but only one base pairing could form in the −2 frame with GAU. Another interesting question is whether the putative +1 frameshifting at the seven U stretch requires ribosomal slippage at both P and A sites. Mutations introduced into the slippery sequence in pF-3a/M1, M2, M3 and M4 would make imperfect pairs at the P site if +1 frameshifting occurs, resulting in the decrease of the frameshifting efficiency. In mutant pF-3a/M5, M6 and M7, perfect base pairs were maintained at the P site in the event of +1 frameshifting. As the expression of the full-length 3a protein was significantly reduced when these mutant constructs were expressed, it suggests that re-pairing at the A site was not successful, resulting in the reduction of the frameshifting efficiency. Based on these observations, it would be reasonable to conclude that a double slippage mechanism is responsible for this frameshift event, and exact base pairing is required. At present, however, we cannot rule out the possibility that a slippage-independent mechanism, such as GAG3-POL3 genes of yeast Ty3 that are dependent on slow decoding of certain codons, while not involving peptidyl-tRNA slippage (2,23), and the mammalian ornithine decarboxylase antizyme frameshift event in which a slippage was not necessary as shown by mutagenesis studies (7), could account for this frameshift event.
Similarly, detection of the full-length 3a protein from the construct with eight Ts (with the insertion of two Ts) would require a −1 frameshift event at the slippage site (UUU UUU UU). Interestingly, considerably less amounts of the full-length product in all three expression systems were detected from this construct than that from the construct with seven Ts. It suggests that +1 frameshifting is much more efficient than −1 frameshifting at this position. However, when the original slippery sequence of SARS-CoV ORF 1a and 1b was replaced by seven and eight U stretches, very similar or even higher frameshifting efficiency was consistently observed from the construct with eight Ts. Mutations that destroyed the downstream stimulatory structure did not alter/reverse the frameshifting efficiency of the two constructs, suggesting that either additional sequences located in the region or long range RNA interaction may favor −1 frameshifting.
In addition to the slippery sequence, the majority of the known frameshifting signals also contain a stimulatory element. In some cases, a secondary structure or tertiary interaction, such as a pseudoknot structure downstream of the slippage site, could significantly increase the frameshifting efficiency. In other cases, sequence specific elements, such as the Shine–Dalgarno (24,25) or sequences partially complementary with yeast 18S rRNA (26), are required. Furthermore, space between the slippage site and the stimulatory element, especially the downstream secondary structure, also plays a critical regulatory role in the frameshifting efficiency (24). Although certain stem–loop structures and potential pseudoknot interaction downstream of the slippage site in ORF 3a variants may be formed, deletion of sequence within 100 nt downstream of the U stretch showed that these potential structural elements did not affect the seven U stretch-mediated frameshifting, indicating that either long-range RNA interaction may play a role or stimulatory elements may be dispensable for these frameshifting events. However, the fact that frameshifting was still efficient in other ORF context, such as the luciferase gene, argues against the first possibility. This conclusion was reinforced by the observation that mutation of stem II in the SARS-CoV 1a/1b region totally abolished the frameshift event mediated by the original slippery sequence of ORF 1a and 1b, but rendered no effect on the frameshift event mediated either by the seven Us or the eight Us. Previously, a G-rich sequence was reported to induce net +1 frameshift in HSV TK gene (14). In this case, the frameshifting efficiency was not augmented by downstream structures and ribosomal pausing (14). A recent report also showed that ribosomal pausing had little correlation with the frameshifting efficiency (27).
Another mechanism that may account for the expression of full-length 3a protein from ORF3a variants with seven and eight Us is transcriptional slippage. During transcription of homopolymeric DNA templates, DNA-dependent RNA polymerase could slip and insert non-templated nucleotide(s) into the growing RNA chain. Examples include the expression of dnaX in Thermus thermophilus (28) and bacterial IS elements (29). In this study, we show no deletion or revision of the mutations in the seven T regions during transcription of wild type and mutant constructs, arguing against the possibility that transcriptional slippage may play a role in the expression of the ORF3a variant with seven Us. More systematic cloning, sequencing and expression of constructs with seven and eight Ts are under way to address this issue further.
Acknowledgments
This work was supported by the Agency for Science Technology and Research, Singapore, and by grant R-154-000-212-112 from the National University of Singapore. Funding to pay the Open Access publication charges for this article was provided by the Agency for Science Technology and Research, Singapore.
Conflict of interest statement. None declared.
REFERENCES
- 1.Belcourt M.F., Farabaugh P.J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farabaugh P.J., Zhao H., Vimaladithan A. A novel programmed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell. 1993;74:93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blinkowa A.L., Walker Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III γ subunit from within the τ subunit reading frame. Nucleic Acids Res. 1990;18:1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craigen W.J., Caskey C.T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- 5.Gurvich O., Baranov P.V., Zhou J., Hammer A.W., Gesteland R., Atkins J.F. Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. EMBO J. 2003;22:5941–5950. doi: 10.1093/emboj/cdg561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckenbach A., Robson S., Crozier R.H. Single nucleotide +1 frameshifts in an apparently functional mitochondrial cytochrome b gene in ants of the genus Polyrhachis. J. Mol. Evol. 2005;60:141–152. doi: 10.1007/s00239-004-0178-5. [DOI] [PubMed] [Google Scholar]
- 7.Matsufuji S., Matsufuji T., Miyazaki Y., Murakami Y., Atkins J.F., Gesteland R.F., Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baranov P.V., Henderson C.M., Anderson C., Gesteland R., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brierley I., Jenner A.J., Inglis S.C. Mutational analysis of the ‘slippery-sequence’ component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacks T., Madhani H.D., Masiarz F., Varmus H. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacks T., Power M.D., Masiarz F., Luciw P., Barr P.J., Varmus H. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 13.Dos Ramos F., Carrasco M., Doyle T., Brierley I. Programmed −1 ribosomal frameshifting in the SARS coronavirus. Biochem. Soc. Trans. 2004;32:1081–1083. doi: 10.1042/BST0321081. [DOI] [PubMed] [Google Scholar]
- 14.Horsburgh B., Kollmus H., Hauser H., Coen D. Translational recoding induced by G-rich mRNA sequences that form unusual structures. Cell. 1996;86:949–959. doi: 10.1016/S0092-8674(00)80170-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson W., Braddock M., Adams S., Rathjen P., Kingsman S., Kingsman A. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988;55:1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- 16.Su M.C., Chang C.T., Chu C.H., Tsai C.H., Chang K.Y. An atypical RNA pseudoknot stimulator and an upstream attenuation signal for −1 ribosomal frameshifting of SARS coronavirus. Nucleic Acids Res. 2005;33:4265–4275. doi: 10.1093/nar/gki731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito N., Mossel E.C., Narayanan K., Popov V.L., Huang C., Inoue T., Peters C.J., Makino S. Severe acute respiratory syndrome coronavirus 3a protein is a viral structural protein. J. Virol. 2005;79:3182–3186. doi: 10.1128/JVI.79.5.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen S., Lin P.S., Chao Y.C., Zhang A., Yang X., Lim S.G., Hong W., Tan Y.J. The severe acute respiratory syndrome coronavirus 3a is a novel structural protein. Biochem. Biophys. Res. Commun. 2005;330:286–292. doi: 10.1016/j.bbrc.2005.02.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan X., Li J., Shan Y., Yang Z., Zhao Z., Chen B., Yao Z., Dong B., Wang S., Chen J., et al. Subcellular localization and membrane association of SARS-CoV 3a protein. Virus Res. 2005;109:191–202. doi: 10.1016/j.virusres.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng R., Yang R.F., Shi M.D., Jiang M.R., Xie Y.H., Ruan H.Q., Jiang X.S., Shi L., Zhou H., Zhang L., et al. Characterization of the 3a protein of SARS-associated coronavirus in infected Vero E6 cells and SARS patients. J. Mol. Biol. 2004;341:271–279. doi: 10.1016/j.jmb.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan T.H., Barkham T., Fielding B.C., Chou C.F., Shen S., Lim S.G., Hong W., Tan Y.J. Genetic lesions within the 3a gene of SARS-CoV. Virol J. 2005;2:51. doi: 10.1186/1743-422X-2-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Y.J., Teng E., Shen S., Tan T.H., Goh P.Y., Fielding B.C., Ooi E.E., Tan H.C., Lim S.G., Hong W. A novel severe acute respiratory syndrome coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 2004;78:6723–6734. doi: 10.1128/JVI.78.13.6723-6734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farabaugh P.J. Programmed translational frameshifting. Annu. Rev. Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 24.Larsen B., Wills N.M., Gesteland R.F., Atkins J.F. rRNA-mRNA base pairing stimulates a programmed -1 ribosomal frameshift. J. Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larsen B., Peden J., Matsufuji S., Matsufuji T., Brady K., Maldonado R., Wills N.M., Fayet O., Atkins J.F., Gesteland R.F. Upstream stimulators for recoding. Biochem. Cell Biol. 1995;73:1123–1129. doi: 10.1139/o95-121. [DOI] [PubMed] [Google Scholar]
- 26.Li Z., Stahl G., Farabaugh P.J. Programmed +1 frameshifting stimulated by complementarity between a downstream mRNA sequence and an error-correcting region of rRNA. RNA. 2001;7:275–284. doi: 10.1017/s135583820100190x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kontos H., Napthine S., Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsen B., Wills N.M., Nelson C., Atkins J.F., Gesteland R.F. Nonlinearity in genetic decoding: homologous DNA replicase genes use alternatives of transcriptional slippage or translational frameshifting. Proc. Natl Acad. Sci. USA. 2000;97:1683–1688. doi: 10.1073/pnas.97.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baranov P.V., Hammer A.W., Zhou J., Gesteland R.F., Atkins J.F. Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 2005;6:R25. doi: 10.1186/gb-2005-6-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]