The recent cloning of two classical maize mutants, ramosa1 (ra1) by Vollbrecht et al. (2005) and ra2 by Bortiri et al. (2006) (in this issue of The Plant Cell), has identified a pathway that plays a fundamental role in inflorescence architecture in maize. The name ramosa, from the Latin “ramus” meaning “branch,” reflects the phenotype of the ra mutants, which have a highly branched inflorescence. Characterization of orthologs of ra1 and ra2 from other grasses suggests that the ramosa pathway has been involved in the morphological evolution of grass inflorescences (Vollbrecht et al., 2005; Bortiri et al., 2006). This commentary will discuss evolution of the ramosa pathway in the context of some of the concepts of evolution of development (evo-devo).
INFLORESCENCE MORPHOLOGY IN THE GRASSES
The ramosa pathway regulates inflorescence architecture in the grasses. The spikelet is the building block of the grass inflorescence (Figure 1) (Clifford, 1987). The spikelet is a short branch that encloses one or more florets within two leaf-like organs. Grasses differ from each other in their arrangement of branches, spikelets, and florets (Clifford, 1987). Therefore, genes that regulate this arrangement may have been involved in the evolution of the grasses (Kellogg, 2001). The majority of grasses bear spikelets singly. For example, grasses such as rice have many long branches bearing single spikelets (Figure 1C), while barley has a main spike bearing single spikelets (Clifford, 1987). The production of short branches bearing two spikelets is a derived trait of the Andropogoneae, a grass tribe comprising 1000 species, including maize, Sorghum, sugarcane, and Miscanthus (Grass Phylogeny Working Group, 2001; Kellogg, 2001; Mathews et al., 2002). For example, maize has long branches at the base of a main spike with spikelet pairs covering the long branches and main spike (Figure 1A) (McSteen et al., 2000), whereas Sorghum undergoes multiple orders of branching before producing spikelet pairs (Vollbrecht et al., 2005). Recent findings described below show that the ramosa pathway plays a critical role in imposing determinacy on the spikelet pair in the Andropogoneae (Vollbrecht et al., 2005; Bortiri et al., 2006).
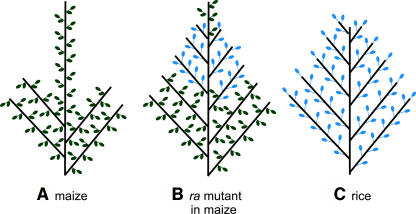
Figure 1.
Simplified Schematic of Inflorescence Morphology in Maize and Rice Compared with the Phenotype of the ra Mutants in Maize.
(A) Maize has long branches at the base of a main spike. Short branches called spikelet pairs cover the main spike and the branches. Note the dramatic switch from long branches to short branches on the main spike.
(B) In the ra1 and ra2 mutants in maize, spikelet pairs on the main spike are converted into long branches bearing single spikelets or mixtures of single spikelets and spikelet pairs. Note that the ra mutants also produce an increased number of long branches bearing spikelet pairs, which have not been drawn for the sake of simplicity.
(C) Rice has many long branches bearing single spikelets.
Thick black lines represent the main spike and the lateral branches, green paired ovals represent paired spikelets, and blue ovals represent single spikelets. The diagram is simplified to illustrate the differences in branching pattern and presence of single versus paired spikelets but does not represent the total number of branches or spikelets.
THE ramosa PATHWAY
The ra1 mutant was first described in 1912 (Gernart, 1912). ra1 mutants affect branching in both the male (tassel) and female (ear) inflorescence (Nickerson and Dale, 1955). In the ra1 tassel, there is a transformation of spikelet pairs (which are in effect short branches) into long branches bearing single or paired spikelets (Vollbrecht et al., 2005) (Figure 1B). The result is a tassel that instead of having a dramatic switch from long branch to short branch (spikelet pair) identity as in normal plants has a gradual switch with the branches getting shorter acropetally. The ear, which is usually unbranched, is highly branched in ra1 mutants, resulting in reduced fertility. It is important to note that ra1 is not required for spikelet pair meristem identity per se, as spikelet pairs do form on branches and at the apical part of the main spike. Rather, ra1 is required for the abrupt switch to determinate growth that occurs when the inflorescence starts to produce spikelet pairs.
ra1 was cloned by transposon tagging with Suppressor-mutator and encodes a putative zinc finger transcription factor of the EPF class (Vollbrecht et al., 2005). Putative orthologs of ra1 are found in closely related species in the Andropogoneae, such as Sorghum and Miscanthus. Although transcription factors of the EPF class are found in other species, a clear ortholog of ra1 is not found in rice or Arabidopsis. In maize, ra1 is expressed at the base of the axillary meristems that give rise to the spikelet pair in the inflorescence. There are four types of axillary meristem in the maize inflorescence, named after the structures they produce: the branch meristem, the spikelet pair meristem, the spikelet meristem, and the floral meristem (McSteen et al., 2000). ra1 is not expressed in branch meristems but begins to be expressed as spikelet pair meristems initiate (Vollbrecht et al., 2005). ra1 is not expressed in the spikelet pair meristem itself but is expressed in a region surrounding the base of the meristem. Later, ra1 is expressed at the base of spikelet meristems where it is not known to play a role. It is proposed that ra1 expression imposes determinacy on spikelet pair meristems. The absence of ra1 in spikelet pair meristems of ra1 mutants causes them to become indeterminate producing additional spikelets. Moreover, the absence of ra1 expression in branch meristems of wild-type tassels may allow them to be indeterminate.
There is evidence that ra1 may function by controlling a mobile signal. Characterization of unstable alleles shows that sectors of wild-type tissue can confer a wild-type phenotype over a limited distance (Vollbrecht et al., 2005). Moreover, ra1 is expressed in a boundary expression domain surrounding rather than within the meristem. One possibility is that the RA1 protein itself moves between cells. Another possibility is that ra1 may control a hormonal signal. If a hormone is involved, then a possible candidate is gibberellic acid (GA), which has been implicated in the regulation of tassel branch number in maize. Application of high levels of GA reduces branch number in normal tassels and suppresses the phenotype of ra1 mutant tassels (Nickerson, 1959, 1960). Paradoxically, some of the GA-deficient mutants have fewer tassel branches, and a low level of ectopically applied GA actually promotes tassel branching in these mutants (Evans and Poethig, 1995). Therefore, the role of GA in tassel branching, and its interaction with the ramosa pathway, remains to be resolved. Moreover, other hormones have also been implicated in tassel branching. For example, application of auxin also reduces the number of tassel branches in wild-type plants (Heslop-Harrison, 1960). Thus, the role of hormones in branching of the maize inflorescence may be complex.
The ra2 mutant was first reported in 1935 (Emerson et al., 1935; Hayes, 1939). ra2 mutants have a very similar tassel branching phenotype as ra1, but there are also clear differences. In the tassel, ra2 branches are borne at a more upright angle, and spikelets are borne on elongated pedicels compared with ra1 mutants or the wild ype. Moreover, the ears of ra2 are less severely affected than ra1 (Nickerson and Dale, 1955; Vollbrecht et al., 2005; Bortiri et al., 2006). The cloning of ra2 from maize and related grasses is described in this issue of The Plant Cell (Bortiri et al., 2006). A point of interest is that although ra1 was cloned the traditional way in maize (by transposon tagging), ra2 is one of the first genes to be cloned by map-based cloning in maize. The first published example was the cloning of teosinte glume architecture1 (Wang et al., 2005). Both of these cases made use of colinearity with the rice genome, but walking in maize should become easier when the maize genome sequence is completed (Chandler and Brendel, 2002).
ra2 encodes a putative LOB domain–containing transcription factor (Bortiri et al., 2006). Orthologs of ra2 are found in sorghum, rice, and barley, unlike ra1, which is found in sorghum but not rice (Bortiri et al., 2006). Phylogenetic analysis shows that the closest homolog of ra2 in Arabidopsis is ASYMMETRIC LEAVES2-LIKE4, but the C terminus is completely different in the two genes. ra2 is expressed earlier than ra1 and is in fact one of the earliest genes to be expressed during axillary branching in maize inflorescence development. ra2 is expressed in the anlagen of the branch meristem and spikelet pair meristem and disappears as the meristems grow. ra2 is also expressed when the spikelet meristem initiates. ra2 is conserved in its expression in branches and spikelets of sorghum, barley, and rice, suggesting that it also plays an important role in these species (Bortiri et al., 2006).
Evidence from both expression data and double mutant analysis suggests that ra2 acts upstream of ra1 (Vollbrecht et al., 2005; Bortiri et al., 2006). As discussed above, ra2 mutants have a tassel phenotype very similar to ra1, but the ear is less severely affected. Double mutants between ra2 and a weak allele of ra1 have a greatly enhanced ear phenotype, suggesting that ra2 and ra1 act in the same pathway. Convincing evidence was provided by RNA gel blot analysis and RNA in situ hybridization experiments, which showed that ra1 expression is reduced in ra2 mutants (Vollbrecht et al., 2005; Bortiri et al., 2006). Therefore, ra2 is proposed to regulate ra1. Although it has not been shown that their interaction is direct, there is some evidence that ra2 may be involved in transcriptional activation of ra1 (Bortiri et al., 2006). However, as ra1 is not expressed everywhere that ra2 is expressed, there must be additional factors involved in the regulation of ra1.
EVOLUTION OF THE ramosa PATHWAY
The central premise of evo-devo is that changes in the overall morphology of organisms can be traced to early changes in development and in particular to changes in genes controlling development (Carroll et al., 2005). As transcription factors control many developmental processes, it is common to find that diversification of morphology between closely related organisms has involved changes in (1) how transcription factors are regulated or (2) how transcription factors interact with their target genes (Doebley and Lukens, 1998; Carroll et al., 2005). Another powerful mechanism in evo-devo is (3) co-option, whereby transcription factors are co-opted for a new purpose in a different species (Carroll et al., 2005). I discuss the evolution of the ramosa pathway in the context of some of these major mechanisms of evo-devo. I refer the reader to additional recent reviews for other examples of evo-devo in plants (Friedman et al., 2004; Kellogg, 2004; Irish and Litt, 2005; Piazza et al., 2005).
Changes in How Transcription Factors Are Regulated
A common mechanism of evo-devo is changes in the regulation of genes controlling development (Carroll et al., 2005). For example, spectacular changes in the body plans of animals (arthropods and vertebrates) correlate with shifts in the expression pattern of Hox genes (Averof and Patel, 1997; Cohn and Tickle, 1999). Also, striking changes in plant architecture correlate with changes in the level of teosinte branched1 expression during the domestication of maize from teosinte (Doebley et al., 1997).
Characterization of ra1 expression within the Andropogoneae shows that changes in ra1 expression correlate with changes in morphology (Vollbrecht et al., 2005). Miscanthus has a similar morphology to maize in that long branches and spikelet pairs are formed. However, in Miscanthus, there are more long branches relative to maize, and this correlates with a delay in the onset of ra1 expression. Sorghum is highly branched relative to maize and produces spikelet pairs only after multiple rounds of branching. This phenotype correlates with a delay in peak ra1 expression compared with maize and Miscanthus. Thus, expression of ra1 is correlated with the imposition of determinate spikelet pair meristem identity in Miscanthus and Sorghum. Hence, ra1 is likely playing a similar role in these species (Vollbrecht et al., 2005).
Although changes in the expression of ra1 correlate with changes in morphology in the Andropogoneae, it is challenging to determine whether a gene is truly changed in its timing when the experiments are done in different species. Vollbrecht and coworkers performed an in-depth analysis comparing expression data to morphological data at different stages of development in the three species. This is similar to the challenge of experiments that have been done in mice, chicks, and fish, whereby changes in the timing of Hox gene expression were correlated with differences in body plans using markers of vertebrate development as a guide (Burke et al., 1995; Anand et al., 2003). One way of getting around the issue of timing (or developmental stage) when comparing gene expression in different organisms is to perform transgenic experiments of the type that are used routinely in animal studies (Belting et al., 1998; Anand et al., 2003; Carroll et al., 2005). For example, the sorghum ra1 promoter could be linked to a reporter gene and transformed into maize to determine if its expression truly occurs later than the native maize ra1 gene. These experiments would also answer the question of whether the changes in the timing of ra1 expression in sorghum are due to changes in the sorghum promoter or trans-acting factors upstream of the promoter. If the activity of the sorghum promoter was delayed in maize, then this would imply that changes had occurred in the cis-regulatory region of ra1 in sorghum, opening the door to experiments to determine the exact molecular basis of the change in cis-regulation. A starting point, which has been used in both animals and plants, would be to compare the regulatory regions of ra1 from different species to identify conserved noncoding sequences potentially important in function (Shashikant et al., 1998; Kaplinsky et al., 2002).
Many evo-devo stories start with a correlation between morphology and gene expression pattern (Bharathan et al., 2002). Whether changes in gene expression are causative of morphological change requires further testing, again, usually with transgenic experiments (Kim et al., 2003). Frequently, evo-devo stories in both plants and animals end here due to limitations in the ability to transform nonmodel organisms. A great advantage of the grasses is that many, including rice, sorghum, maize, and barley, can be transformed. Whether changes in the timing of ra1 expression are causative of the evolutionary change in morphology in these species could be tested in further studies. For example, can expression of the maize ra1 gene in sorghum decrease branching? The results of these experiments would be very exciting because evidence of causation is rare in evo-devo.
Changes in How Transcription Factors Interact with Their Target Genes
A few evo-devo case studies have shown that changes in the coding regions of transcription factors can alter their interaction with downstream targets (Galant and Carroll, 2002; Ronshaugen et al., 2002; Maizel et al., 2005). This approach requires determining the direct target genes of transcription factors, which is very powerful for understanding the evolution of developmental pathways. A recent article beautifully illustrates this approach. LEAFY (LFY) regulates flower development in Arabidopsis, but the function of LFY in other species differs (Maizel et al., 2005). In an elegant series of microarray experiments, Maizel and coworkers showed that alterations in the DNA binding domain of LFY in flowering plants have resulted in LFY having different targets in different species. Therefore, evo-devo in the LFY pathway has involved changes in the coding region of LFY.
ra1 and ra2 also have differences in their coding regions compared with gene family members in Arabidopsis. ra1 has an invariant amino acid change in the zinc finger DNA binding domain compared with EPF zinc fingers in rice and Arabidopsis (Vollbrecht et al., 2005). Moreover, all grass ra2 genes have a specific C-terminal putative activation domain that is absent from LOB genes in Arabidopsis (Bortiri et al., 2006). Therefore, members of the LOB and EPF gene families in the grasses may have different targets and, hence, different functions from their eudicot relatives. Identification of the direct target genes of the ramosa pathway will be important in unraveling the evolution of this pathway.
Changes in the targets of transcription factors may also be caused by changes in the cis-regulatory regions of their target genes (Carroll et al., 2005). An excellent example from plants is provided by the CYCLOIDEA (CYC) pathway. CYC is a TCP domain–containing transcription factor that regulates floral asymmetry in Antirrhinum by altering growth of organs in the upper region of the floral meristem (Luo et al., 1996). RADIALIS (RAD), a MYB domain–containing transcription factor, is a direct downstream target of CYC (Costa et al., 2005). Evolution of the interaction was analyzed in Arabidopsis, which has CYC and RAD homologs but has radially symmetrical flowers. Costa and coworkers showed that expression of Antirrhinum CYC in Arabidopsis cannot induce the Arabidopsis RAD-like genes because they do not have CYC binding sites in their regulatory regions. Therefore, evolution of the CYC pathway has involved changes in the target gene RAD and in particular changes in the cis-regulatory regions of RAD. Whether evolution of the ramosa pathway involves a direct interaction between ra2 and ra1 remains to be determined.
Co-Option
A common mechanism in evo-devo is co-option of transcription factors for different purposes in different tissues (Carroll et al., 2005). ra1 plays a major role in spikelet pair meristem determinacy in the Andropogoneae but is not present in rice, which, correspondingly, does not have spikelet pairs. Maize and rice last shared a common ancestor ∼50 million years ago near the base of the grass radiation, leading to a crucial question: Was ra1 co-opted in the Andropogoneae or lost in rice? Answering this question will require identifying additional ra1 homologs within the Andropogoneae, within the Oryzeae, and most importantly from a species basal to both lineages to determine if ra1 was present prior to the divergence of the two groups. If ra1 was lost in a lineage leading to rice, what were the steps that led to its loss? Was it initially no longer expressed and then became a pseudogene? If ra1 is specific to the Andropogoneae, was the gene co-opted in the lineage leading to Andropogoneae for the evolution of the spikelet pair meristem? In the example described above, it was proposed that RAD had been co-opted in the Antirrhinum lineage through addition of CYC binding sites to the regulatory regions of RAD (Costa et al., 2005). In order to address the mechanism by which ra1 may have been co-opted, further analysis of its upstream regulator(s) and regulatory sequences are required. In animals, co-option of multiple target genes and entire signaling pathways has been demonstrated (Weatherbee et al., 1998; Keys et al., 1999). Therefore, co-option sometimes requires changes to the cis-regulatory elements of multiple target genes, again emphasizing the importance of identifying the targets in the rest of the pathway.
Conclusions
The cloning of ra1 and ra2 from maize has identified a pathway regulating inflorescence morphology. Cloning of the putative orthologs of ra1 and ra2 from other species indicates that evolution of this pathway may have been involved in the evolution of inflorescence morphology. The grasses are a premier model system for evo-devo studies: there is tremendous diversity in inflorescence morphology, the phylogeny is well understood, and many species are transformable so hypotheses can be tested. Moreover, maize in particular is an excellent model system for studying selection as it was domesticated from its wild ancestor teosinte a mere 10,000 years ago (Doebley, 2004). Indeed, ra1 was shown to have been a target of selection during maize domestication or improvement (Vollbrecht et al., 2005). Future studies to address the role of the ramosa pathway within maize, within the Andropogoneae, and within the grasses will be important in understanding the evolution of the grasses and furthermore will provide an understanding of the mechanisms of evo-devo.
Acknowledgments
I thank Sarah Hake and Esteban Bortiri for communication of results prior to publication, Esteban Bortiri, David Braun, Sarah Hake, Erik Vollbrecht, members of the McSteen lab, and The Plant Cell reviewers for suggestions on the manuscript, and Toby Kellogg, Simon Malcomber, and Andrew Doust for discussions on inflorescence diversity in the grasses. I apologize to the authors of other great stories in evo-devo whose work I couldn't cite due to space limitations.
References
- Anand, S., Wang, W.C.H., Powell, D.R., Bolanowski, S.A., Zhang, J., Ledje, C., Pawashe, A.B., Amemiya, C.T., and Shashikant, C.S. (2003). Divergence of Hoxc8 early enhancer parallels diverged axial morphologies between mammals and fishes. Proc. Natl. Acad. Sci. USA 100 15666–15669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averof, M., and Patel, N.H. (1997). Crustacean appendage evolution associated with changes in Hox gene expression. Nature 388 682–686. [DOI] [PubMed] [Google Scholar]
- Belting, H.-G., Shashikant, C.S., and Ruddle, F.H. (1998). Modification of expression and cis-regulation of Hoxc8 in the evolution of diverged axial morphology. Proc. Natl. Acad. Sci. USA 95 2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathan, G., Goliber, T.E., Moore, C., Kessler, S., Pham, T., and Sinha, N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296 1858–1860. [DOI] [PubMed] [Google Scholar]
- Bortiri, E., Chuck, G., Vollbrecht, E., Rocheford, T., Martienssen, R., and Hake, S. (2006). ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, A., Nelson, C., Morgan, B., and Tabin, C. (1995). Hox genes and the evolution of vertebrate axial morphology. Development 121 333–346. [DOI] [PubMed] [Google Scholar]
- Carroll, S.B., Grenier, J.K., and Weatherbee, S.D. (2005). From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. (Oxford: Blackwell Publishing).
- Chandler, V.L., and Brendel, V. (2002). The maize genome sequencing project. Plant Physiol. 130 1594–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford, H.T. (1987). Spikelet and floral morphology. In Grass Systematics and Evolution, T.R. Soderstrom, K.W. Hilu, C.S. Campbell, and M.E. Barkworth, eds (Washington, D.C.: Smithsonion Institution Press), pp. 21–30.
- Cohn, M.J., and Tickle, C. (1999). Developmental basis of limblessness and axial patterning in snakes. Nature 399 474–479. [DOI] [PubMed] [Google Scholar]
- Costa, M.M.R., Fox, S., Hanna, A.I., Baxter, C., and Coen, E. (2005). Evolution of regulatory interactions controlling floral asymmetry. Development 132 5093–5101. [DOI] [PubMed] [Google Scholar]
- Doebley, J. (2004). The genetics of maize evolution. Annu. Rev. Genet. 38 37–59. [DOI] [PubMed] [Google Scholar]
- Doebley, J., and Lukens, L. (1998). Transcriptional regulators and the evolution of plant form. Plant Cell 10 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386 485–488. [DOI] [PubMed] [Google Scholar]
- Emerson, R.A., Beadle, G.W., and Fraser, A.C. (1935). A summary of linkage studies in maize. Cornell Univ. Agric. Exp. Stn. Memoir 180 1–83. [Google Scholar]
- Evans, M., and Poethig, R.S. (1995). Gibberellins promote vegetative phase change and reproductive maturity in maize. Plant Physiol. 108 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, W.E., Moore, R.C., and Purugganan, M.D. (2004). The evolution of plant development. Am. J. Bot. 91 1726–1741. [DOI] [PubMed] [Google Scholar]
- Galant, R., and Carroll, S.B. (2002). Evolution of a transcriptional repression domain in an insect Hox protein. Nature 415 910–913. [DOI] [PubMed] [Google Scholar]
- Gernart, W. (1912). A new subspecies of Zea mays L. Am. Nat. 46 616–622. [Google Scholar]
- Grass Phylogeny Working Group (2001). Phylogeny and subfamilial classification of the grasses (Poaceae). Ann. Mo. Bot. Gard. 88 373–457. [Google Scholar]
- Hayes, H. (1939). Recent linkage studies in maize IV ramosa ear-2 (ra2). Genetics 24 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison, J.S. (1960). The experimental control of sexuality and inflorescence architecture in Zea mays L. Proc. Linn. Soc. Lond. 172 108–123. [Google Scholar]
- Irish, V.F., and Litt, A. (2005). Flower development and evolution: Gene duplication, diversification and redeployment. Curr. Opin. Genet. Dev. 15 454–460. [DOI] [PubMed] [Google Scholar]
- Kaplinsky, N.J., Braun, D.M., Penterman, J., Goff, S.A., and Freeling, M. (2002). Utility and distribution of conserved noncoding sequences in the grasses. Proc. Natl. Acad. Sci. USA 99 6147–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, E.A. (2001). Evolutionary history of the grasses. Plant Physiol. 125 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, E.A. (2004). Evolution of developmental traits. Curr. Opin. Plant Biol. 7 92–98. [DOI] [PubMed] [Google Scholar]
- Keys, D.N., Lewis, D.L., Selegue, J.E., Pearson, B.J., Goodrich, L.V., Johnson, R.L., Gates, J., Scott, M.P., and Carroll, S.B. (1999). Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science 283 532–534. [DOI] [PubMed] [Google Scholar]
- Kim, M., McCormick, S., Timmermans, M., and Sinha, N. (2003). The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 424 438–443. [DOI] [PubMed] [Google Scholar]
- Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383 794–799. [DOI] [PubMed] [Google Scholar]
- Maizel, A., Busch, M.A., Tanahashi, T., Perkovic, J., Kato, M., Hasebe, M., and Weigel, D. (2005). The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308 260–263. [DOI] [PubMed] [Google Scholar]
- Mathews, S., Spangler, R.E., Mason-Gamer, R.J., and Kellogg, E.A. (2002). Phylogeny of Andropogoneae inferred from phytochrome B, GBSSI, and NDHF. Int. J. Plant Sci. 163 441–450. [Google Scholar]
- McSteen, P., Laudencia-Chingcuanco, D., and Colasanti, J. (2000). A floret by any other name: Control of meristem identity in maize. Trends Plant Sci. 5 61–66. [DOI] [PubMed] [Google Scholar]
- Nickerson, N.H. (1959). Sustained treatment with gibberellic acid of five different kinds of maize. Ann. Mo. Bot. Gard. 46 19–37. [Google Scholar]
- Nickerson, N.H. (1960). Studies involving sustained treatment of maize with gibberellic acid II: Responses of plants carrying certain tassel-modifying genes. Ann. Mo. Bot. Gard. 47 243–261. [Google Scholar]
- Nickerson, N.H., and Dale, E.E. (1955). Tassel modifications in Zea mays. Ann. Mo. Bot. Gard. 42 195–211. [Google Scholar]
- Piazza, P., Jasinski, S., and Tsiantis, M. (2005). Evolution of leaf developmental mechanisms. New Phytol. 167 693–710. [DOI] [PubMed] [Google Scholar]
- Ronshaugen, M., McGinnis, N., and McGinnis, W. (2002). Hox protein mutation and macroevolution of the insect body plan. Nature 415 914–917. [DOI] [PubMed] [Google Scholar]
- Shashikant, C.S., Kim, C.B., Borbely, M.A., Wang, W.C.H., and Ruddle, F.H. (1998). Comparative studies on mammalian Hoxc8 early enhancer sequence reveal a baleen whale-specific deletion of a cis-acting element. Proc. Natl. Acad. Sci. USA 95 15446–15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht, E., Springer, P.S., Goh, L., Buckler IV, E.S., and Martienssen, R. (2005). Architecture of floral branch systems in maize and related grasses. Nature 436 1119–1126. [DOI] [PubMed] [Google Scholar]
- Wang, H., Nussbaum-Wagler, T., Li, B., Zhao, Q., Vigouroux, Y., Faller, M., Bomblies, K., Lukens, L., and Doebley, J.F. (2005). The origin of the naked grains of maize. Nature 436 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherbee, S.D., Halder, G., Kim, J., Hudson, A., and Carroll, S. (1998). Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 12 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]