The often-stated truism that most plant species are resistant to most plant pathogens reflects the many observations that a pathogen isolated from one plant species in most cases cannot infect, reproduce, and cause disease on other distantly related species. What determines pathogen host range is an important and intriguing question in fundamental host–pathogen biology. Many studies have investigated this area with biochemical, pharmacological, and microscopy studies, and now several recent publications (Collins et al., 2003; Lipka et al., 2005), including one in this issue of The Plant Cell (Stein et al., 2006), have focused the power of molecular genetics on the problem with studies of infection (or rather lack of infection) of Arabidopsis thaliana by fungal pathogens of crop plants, particularly powdery mildew species. The new knowledge could have implications for novel strategies for disease control in agriculture.
POWDERY MILDEW INFECTION
Powdery mildews, a large group of Ascomycete fungal species, are obligate biotrophs, meaning that they rely completely on living host plant tissue for survival, and in many cases, each species is specialized to infect a very narrow range of plant species. For example, the powdery mildew fungus Blumeria graminis f. sp hordei (Bgh), which causes a serious disease of barley (Hordeum vulgare), infects only barley and its close relatives. Infection initiates with the germination of conidiospores on the plant leaf surface, followed by the formation of structures called appressoria (a sort of fungal battering ram), from which develop infection hyphae called penetration pegs. These hyphae breach host epidermal cell walls and the infection-induced dome-shaped extensions of the inner surface of the wall, called papillae, probably by physical pressure and enzymatic degradation. The tips of the infection hyphae then expand to form multifingered feeding structures called haustoria that invaginate but don't penetrate the host plasma membrane. This means all nutrients for fungal growth from the host, and potential signal molecules from the pathogen, must cross a double membrane interface (Hückelhoven, 2005; Zhang et al., 2005). The fungal colonies that arise from infection produce the spores for the next cycle of infection, and it is the appearance of these powdery spores on the leaf surface that is the hallmark of successful infection in the experimental systems described below.
GENE-FOR-GENE RESISTANCE AND ADAPTED PATHOGENS
Populations of host species of a biotrophic fungus are frequently highly polymorphic for resistance and susceptibility to isolates of the adapted pathogen species. Resistance is usually associated with the hypersensitive reaction (HR), a localized host cell death response at the infection site that occurs after the fungus has breached the host cell wall and attempted to initiate a haustorium. This type of resistance is mediated by the well-studied gene-for-gene interactions between host resistance (R) genes and pathogen avirulence (Avr) genes. For example, in cultivated barley, >30 different R genes map to the Mla locus, which contains genes encoding nucleotide binding site–leucine-rich repeat (NBS-LRR) proteins that control specific recognition of mildew strains dependent on the corresponding Avr genotype (Zhou et al., 2001).
NONHOST RESISTANCE TO BARLEY MILDEW IN ARABIDOPSIS
In contrast with barley, all individuals of Arabidopsis are resistant to all isolates of Bgh, and this form of resistance, called nonhost resistance, is genetically ill defined (Heath, 2000; Thordal-Christensen, 2003). However, a recent series of articles highlighted here have made significant advances in our understanding of nonhost resistance to mildews. The Arabidopsis–Bgh interaction is particularly well suited for these studies because mildew infection occurs at the leaf surface and is restricted to epidermal cells. Thus, infection attempts are easily visualized by microscopy, and mutant plants with altered responses to infection and reduced resistance to the barley pathogen can be identified. Moreover, at least two species of powdery mildew that cause disease on Arabidopsis are known, and these infections provide comparisons between the steps in infection by mildew species adapted to Arabidopsis and infection by the nonadapted barley mildew.
Nonhost resistance has two phases. During the prehaustorial phase, Bgh spores germinate and form appressoria on the leaf surface of Arabidopsis, cell wall penetration occurs, hyphal growth ceases within the infection-induced papillae at >90% of the infection sites, and haustoria fail to develop (Collins et al., 2003; Assaad et al., 2004). During the post-haustorial phase, haustoria that form at the remaining infection sites become encased in callose and the host cell undergoes the HR. A major difference between gene-for-gene resistance and nonhost resistance to mildews is that the former occurs mainly after haustorium formation, whereas the latter occurs mainly before haustorium formation and mostly is not associated with the HR. However, the HR is a feature in common with the low frequency of nonhost penetrations where initiation of haustoria occurs, leading to the question: How does Bgh cause disease on barley but not on Arabidopsis?
MUTATION FOR THE DISSECTION OF NONHOST RESISTANCE: PENETRATION MUTANTS
Microscopic examination of mutated Arabidopsis plants inoculated with Bgh spores has been used to identify mutants with altered responses to infection. More specifically, stains for callose deposition around haustoria or the appearance of increased autofluorescence at infection sites were used to detect mutant plants with increased frequency of haustorial initiation per infection site in rosette leaves. Three genes, PENETRATION1 (PEN1), PEN2, and PEN3, have been identified and cloned (Collins et al., 2003; Lipka et al., 2005; Stein et al., 2006). Single mutants of these three genes have increased frequency of haustorial formation, but no increase in overall susceptibility to Bgh as indicated by colonies producing viable spores.
PEN1 (Collins et al., 2003) encodes a membrane-associated syntaxin containing a SNARE (for soluble N-ethylmaleimide–sensitive factor attachment protein receptor) domain and is a member of a large family of proteins involved in membrane fusion and secretion events. In pen1 mutants, there is reduced inhibition of hyphal development following cell wall penetration, resulting in a sevenfold increase in haustorial initiation. However, the HR occurs, and consequently no fungal colony formation results, which indicates that PEN1 is only one component of a more complex nonhost resistance mechanism that prevents Bgh growth on Arabidopsis. PEN1 functions in resistance through an undefined mechanism involving secretory vesicles. Its expression is induced during Bgh infection, and a functional green fluorescent protein (GFP)–PEN1 fusion is secreted and accumulates at papillae (Assaad et al., 2004; Bhat et al., 2005) that form at the site of infection peg formation. If PEN1 is involved in secretion of building blocks of papillae, its role is partially redundant because electron microscopy analysis of the complete loss-of-function mutant pen1-1 did not find any major detectable alteration in papillae. However, the rate of papilla formation is reduced in Bgh-infected pen1 mutants, and this has been proposed as a potential cause for the breakdown of this early component of nonhost resistance (Assaad et al., 2004).
PEN2 (Lipka et al., 2005) is one of 48 genes encoding predicted glycosyl hydrolases in the Arabidopsis genome. However, the substrates and products of PEN2 activity are presently unknown. pen2 mutants show an increase of Bgh haustoria, similar to pen1 mutants, and additive effects are observed in pen1 pen2 double mutants. Neither mutant has any effect on infection of susceptible genotypes of Arabidopsis by virulent strains of the adapted mildew species Golovinomyces orontii. However, pen2 but not pen1 allows increased haustorium formation and subsequent HR induction in Arabidopsis by the potato pathogen Phytophthora infestans. pen2 also limits growth of Plectosphaerella cucumerina, a necrotrophic pathogen of Arabidopsis. This indicates that the effect of pen2 is broader than that of pen1 (which affects only Bgh infection); together, these results indicate the possibility that PEN1 and PEN2 act in different pathways. In addition, PEN2-GFP functional fusion protein is localized to peroxisomes that move to and accumulate at Bgh penetration sites, consistent with the predicted role of this organelle in delivering an antifungal product to where it is needed to inhibit haustorium development.
PEN3 (Stein et al., 2006) encodes an ATP binding cassette (ABC) transporter protein that was previously annotated as pleiotropic drug resistance-like transporter 8 (PDR8; van den Brule and Smart, 2002). Prior to Bgh infection, functional PEN3-GFP is localized to the plasma membrane and then accumulates at the penetration site during Bgh infection. It also accumulates at the infection sites of the compatible mildew fungus Erysiphe cichoracearum. The frequency of Bgh haustorium formation and subsequent HR also increases in pen3 mutants, and in this respect, they are similar to both pen1 and pen2 mutants (Collins et al., 2003; Stein et al., 2006). Also similar to pen2 mutants, pen3 mutants allow increased haustorium formation by P. infestans and are more susceptible to P. cucumerina. However, while the Arabidopsis mildew pathogen E. cichoracearum infects pen1 and pen2 mutants, it induces extensive salicylic acid–dependent leaf chlorosis and fails to sporulate on pen3 mutants, possibly due to the intracellular accumulation of the cargo of the PDR8 transporter.
THE MOLECULAR BASIS OF NONHOST RESISTANCE
What do all of these results tell us about nonhost resistance to mildew in Arabidopsis? First, it is a dynamic process involving organelle movement, secretion processes, membrane changes, and accumulation of three PEN proteins at the infection site (Assaad et al., 2004; Bhat et al., 2005; Lipka et al., 2005; Stein et al., 2006). These processes are potentially activated by infection peg pressure, chemical signals from the pathogen (pathogen-associated molecular patterns [PAMPs]; for example, chitin, a specific constituent of fungal cell walls), or signals resulting from enzymatic degradation of the host cell. Recent studies have highlighted the importance of host receptors for PAMPs in basal resistance (Zipfel and Felix, 2005). Second, nonhost resistance has a complex genetic basis with three genes, PEN1, PEN2, and PEN3, identified so far that are involved in biosynthetic and secretion processes that control prehaustorial resistance. PEN2 and PEN3 probably act in the same pathway because pen3 is epistatic to pen2 in at least one assay (Stein et al., 2006). A plausible model for PEN1 is that it is involved in a vesicle-based secretory pathway, probably specific for nonhost resistance to mildews (Shimada et al., 2006). The cargo of PEN1 and its activity in papillae are as yet undefined. PEN2 is likely involved in peroxisome-based biosynthesis and delivery of one or more unidentified antifungal metabolites to the infection site. Although PEN2 and PEN3 occur in different locations, the fact that pen3 is epistatic to pen2 indicates that the cargo of the membrane-localized ABC transporter protein PDR8 encoded by PEN3 could be the product of PEN2. This product has a general activity and affects infection of several fungal species. Its chemical nature is unknown, although it is presumably antifungal and a cargo molecule of PDR8. Given its broader toxic effects, a simple bioassay approach to its purification should assist its identification. It is unlikely to be the one known antimicrobial small molecule, camelexin, synthesized by Arabidopsis, since the pad3 mutants that are deficient in camelexin biosynthesis do not have a pen phenotype (Stein et al., 2006).
Bgh infections that escape prehaustorial resistance in wild-type and pen mutant plants and progress to form haustoria are mopped up by the HR. Posthaustorial resistance is dependent on genes including PAD4, EDS1, and SAG101 (Lipka et al., 2005; Stein et al., 2006), which are involved in the basal resistance pathways that restrict rampant disease development during compatible host–pathogen interactions and also in some R gene pathways leading to the HR (Parker et al., 1996; Feys et al., 2005). Single mutants of pad4, eds1, and sag101 have little effect on the frequency of Bgh haustoria formation in Arabidopsis. However, where haustoria do form the HR is less frequent, secondary hyphal growth occurs, and microcolonies form. In pen2 sag101 pad4 triple mutants, microcolonies of Bgh with occasional conidiophores and mature spores develop; thus, nonhost resistance is almost completely whittled away (Lipka et al., 2005). This effect is even more evident after inoculation with the pea powdery mildew (Erysiphe pisi). Whereas wild-type plants are completely resistant to this species, the triple mutant sustains sporulating infections similar to those resulting from infection with the Arabidopsis mildew G. orontii (Lipka et al., 2005). A breakdown in nonhost resistance was also observed in the pen3 eds1 double mutant infected with E. pisi (Stein et al., 2006). Thus, nonhost resistance is the sum of pre- and post-haustorial resistance components. The observations of PEN2 association with peroxisomes that move toward the infection site and the involvement of EDS1 in nonhost resistance are consistent with another report that eds1 mutants treated with an actin cytoskeleton inhibitor are more susceptible to the wheat mildew pathogen Blumeria graminis f. sp tritici (Yun et al., 2003). A cytoskeleton-based mechanism that involves vesicle movement and exocytosis and focuses a battery of defense activities at the infection site (and possibly to emerging haustoria) appears to be the basis of nonhost resistance.
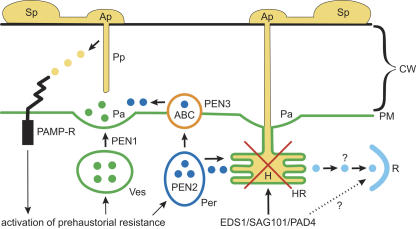
The involvement of the HR and EDS1/PAD4/SAG101, necessary for both basal resistance and resistance mediated by one of two classes NBS-LRR R proteins (Parker et al., 1996; Feys et al., 2005), indicates possible R protein activity in Bgh–Arabidopsis nonhost interactions. However, data counter to this view are provided by Stein et al. (2006). Many Arabidopsis NBS-LRR R genes also require SGT1a and RAR1 for activity (tabulated in Dodds and Schwechheimer, 2002), and Stein et al. (2006) report that whereas pen3 eds1 double mutants show increased epiphytic growth after infection with Bgh and E. pisi and sporulation of E. pisi, pen3 rar1 sgt1a triple mutants do not. So the question of whether R genes are also involved in the post-haustorial phase of nonhost resistance requires further examination. The understanding of nonhost resistance to Bgh in Arabidopsis is summarized in Figure 1.
Figure 1.
Schematic Representation of Arabidopsis Nonhost Resistance to Bgh.
After germination of spores (Sp), formation of appressoria (Ap), and breaching of the host cell wall (CW) by the fungal penetration peg (Pp), ∼90% of penetrations are stopped in cell wall extensions called papillae (Pa). This resistance is dependent on unidentified cargo molecules (green circles) delivered by a PEN1-mediated vesicle (Ves)–based secretion system and on postulated toxin(s) (dark blue circles) synthesized in peroxisomes (Per) in a PEN2-mediated pathway and delivered by a PEN3-encoded ABC transporter in the plasma membrane (PM) to the apoplast and pathogen invasion site. These events are components of prehaustorial resistance. The activation of the dynamic prehaustorial resistance, dependent on cytoskeleton function, may be induced by mechanical or chemical signals resulting from cell wall penetration and/or PAMPs (yellow circles) produced by the pathogen and detected by host PAMP receptors (PAMP-R). Approximately 10% of infections (increased to ∼30 to 90% in pen1, pen2, and pen3 single and double mutants) form haustoria (H) and are stopped (post-haustorial resistance) by basal resistance possibly contributed to by PEN2 pathway products (dark blue circles) and hypersensitive host cell death (HR) dependent on EDS1, SAG101, and PAD4. Because these genes and the HR are also associated with gene-for-gene R genes, it is possible that R proteins could also function to perceive effector molecules (light blue circles) secreted by Bgh haustoria.
WHAT ARE THE REQUIREMENTS FOR MILDEW ADAPTATION?
The articles highlighted here have focused on the nature of Arabidopsis resistance to nonadapted mildews. However, while Bgh fails to infect Arabidopsis, it is adapted to barley. Just the opposite is the case for the Arabidopsis mildew E. cichoracearum. From the pathogen perspective, what is the molecular basis of pathogen adaptation to its host? Evolution of resistance to specific toxins synthesized by the host, for example, the products of the PEN2/PEN3 pathway, is one possibility. Another could involve blunting host basal resistance induced by PAMP perception. Like bacterial pathogens (Abramovitch and Martin, 2004), mildews probably secrete virulence effector molecules to blunt basal resistance, including both pre- and post-haustorial resistance. It is plausible that pathogen effectors are host specific, and those from Bgh could be nonfunctional in Arabidopsis due to failure in their uptake and/or ability to interact with nonhost virulence targets. An analogy is that a key that opens one lock rarely opens another. No Bgh effectors have been described, but evidence for them is provided by sequential inoculation experiments in barley, first with a virulent strain and several hours later with an avirulent strain. Virulent Bgh strains induce susceptibility to host cell penetration and haustorium formation by the normally avirulent strain (Lyngkjaer et al., 2001), consistent with the idea that the first strain delivers effector molecules that induce a susceptible state. Studies of genes expressed in rust haustoria are beginning to provide insights into proteins delivered from these pathogens to infected host cells (Kemen et al., 2005; Catanzariti et al., 2006), and intensive genomics studies of Bgh now in progress could identify mildew effectors (Both et al., 2005; Zhang et al., 2005). Characterization of fungal pathogen effectors and their host targets is now a research priority.
In coevolved host–pathogen interactions, effectors (Avr gene products) are recognized by host R proteins. Pathogen survival depends on polymorphisms in pathogen Avr genes and host R genes (gene-for-gene interactions) so that the pathogen is able to reproduce in at least some interactions. Single gene differences between formae speciales of B. graminis (individuals of the same pathogen species taxonomically separated by their strict adaptation to different but closely related host species) indicate that Avr and R gene equivalents also determine nonhost resistance at this taxonomic level (Tosa, 1989). Similarly, single pathogen genes that encode small secreted proteins, such as PWL2, determine the host range of strains of the blast fungus Magnaporthe grisea on different grass species (Kang et al., 1995; Sweigard et al., 1995). These data again suggest R gene–like functions in nonhost resistance in these interactions. However, whether R/Avr gene–like systems function in interactions where host and nonhost plants have a wider taxonomic separation (e.g., barley and Arabidopsis) is unknown and indeed may not be required in nonhost resistance. In the absence of effector activity, PAMP signal flux alone may be sufficient for induction of the HR by nonadapted pathogen penetration. Thus, the HR in Bgh–pen mutant interactions may be due to the inactivity rather than direct recognition of Bgh effectors. A similar situation has been described in flax mutants that constitutively express resistance responses and where normally compatible flax rust infection (no gene-for-gene interaction) is stopped with the HR (Howles et al., 2005).
Nonprotein small (e.g., pathogen-encoded toxins) and large molecules could also act as virulence effectors. Since proteins are transported from biotrophic fungi to plants, it is plausible that small RNAs derived from RNA interference (RNAi)–like pathways in the pathogen could be as well. These hypothetical pathogen-derived RNAi molecules could silence host genes involved in basal resistance, and coupled with the sequence specificity of the gene silencing process, the restricted host ranges of fungal species could derive partly from nucleotide sequence mismatches of RNAi and target mRNA sequences in nonhost interactions. Indeed, profiling of expression of host genes during mildew infection of barley indicates downregulation of defense genes at the time of haustorial formation during compatible mildew infections (Caldo et al., 2004), and the Bgh transcript-related Neurospora quelling gene qde-3 is reported to increase in abundance during host infection (Zhang et al., 2005).
In summary, post-haustorial resistance of Arabidopsis to Bgh involving the HR could be because (1) Bgh effectors are recognized or (2) Bgh effectors don't work in Arabidopsis, and the HR is a consequence of high signal flux through PAMP-induced basal resistance pathways. The latter does not require direct recognition of Bgh effectors by the nonhost Arabidopsis. These are important issues for further investigation.
CAN COMPONENTS OF NONHOST RESISTANCE BE ENGINEERED TO CONTROL ADAPTED PATHOGENS?
Finally, can information about nonhost resistance in Arabidopsis lead to development of new sources of disease resistance in crop plants? For example, could components of nonhost resistance from Arabidopsis be used to protect barley crops? If nonadapted pathogens are indeed hypersensitive to nonhost antifungal toxins, the cloning of complete gene pathways for prehaustorial resistance (antifungal toxin synthesis and delivery mechanisms if needed; the PEN2/PEN3 pathway) from Arabidopsis and their transfer to barley is a strategy that can be tested. The toxic effects of these products extend beyond mildew species (Lipka et al., 2005; Stein et al., 2006), so this approach might be broadly effective against a range of fungal pathogens.
For post-haustorial resistance, the cloning and transfer of putative Arabidopsis receptors (R genes) for nonadapted crop plant pathogens is also possible. However, so far no R gene equivalents of the NBS-LRR type for nonadapted fungal pathogens have been identified in Arabidopsis, although data from mildew and rice blast studies indicate R gene determination of formae speciales distinctions by grass relatives of barley and rice (Tosa, 1989; Kang et al., 1995; Sweigard et al.,1995). These resistances used as transgenes would not necessarily be more durable than classical single gene-for-gene type resistances that rapidly select virulent variants of previously avirulent fungi. Furthermore, as discussed above, post-haustorial resistance to pathogens from more distantly related hosts may be based on nonspecific basal resistance mechanisms present in all species. If nonhost resistance is due to deficiencies (e.g., inappropriate effectors) of the nonadapted pathogen, a route to novel resistance might be interspecific transfer of the virulence targets of effectors, such as components of a nonhost basal resistance response. These transferred components might be less susceptible to suppression by the adapted pathogen than the corresponding host machinery.
There is also evidence that machinery for prehaustorial nonhost resistance can be activated by mutation in a host plant and provide resistance against an adapted pathogen. The best example is the Mlo system in barley, which negatively affects a prehaustorial mildew resistance dependent on the Ror2 gene, the barley homolog of PEN1 (Collins et al., 2003). Recessive mutants of Mlo in both barley and Arabidopsis are resistant to adapted mildew species (Panstruga, 2005), which indicates that this form of resistance cannot be negated by adapted pathogen effectors. However, whereas mlo mutations are ineffective against rusts (Jorgensen, 1992) and mlo mutant barley is more susceptible than the wild type to the rice blast pathogen M. grisea (Jarosch et al., 1999), the PEN1 pathway in Arabidopsis appears to be specific against mildew species (Shimada et al., 2006). A challenge for molecular genetics will be to identify genes analogous to Mlo for combating these pathogens through activation of prehaustorial defenses.
References
- Abramovitch, R.B., and Martin, G.B. (2004). Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 7 356–364. [DOI] [PubMed] [Google Scholar]
- Assaad, F.F., Qiu, J., Youngs, H., Ehrhardt, D., Zimmerli, L., Kalde, M., Wanner, G., Peck, S.C., Edwards, H., Ramonell, K., Somerville, C.R., and Thordal-Christensen, H. (2004). The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol. Biol. Cell 15 5118–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, R.A., Miklis, M., Schmelzer, E., Schulze-Lefert, P., and Panstruga, R. (2005). Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. USA 102 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both, M., Csukai, M., Stumpf, M.P., and Spanu, P.D. (2005). Gene expression profiles of Blumeria graminis indicate dynamic changes to primary metabolism during development of an obligate biotrophic pathogen. Plant Cell 17 2107–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldo, R.A., Nettleton, D., and Wise, R.P. (2004). Interaction-dependent gene expression in Mla-specified response to barley powdery mildew. Plant Cell 16 2514–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti, A., Dodds, P.N., Lawrence, G.J., Ayliffe, M.A., and Ellis, J.G. (2006). Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, N.C., Thordal-Christensen, H., Lipka, V., Bau, S., Kombrink, E., Qui, J.L., Huckelhoven, R., Stein, M., Freialdenoven, A., Somerville, S.C., and Schulze-Lefert, P. (2003). Snare-protein-mediated disease resistance at the plant cell wall. Nature 425 973–007. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., and Schwechheimer, C. (2002). A breakdown in defense signaling. Plant Cell 14 (suppl.), S5–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., Wiermer, M., Bhat, R.A., Moisan, L.J., Medina-Escobar, N., Neu, C., Cabral, A., and Parker, J.E. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, M.C. (2000). Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3 315–319. [DOI] [PubMed] [Google Scholar]
- Howles, P., Lawrence, G.J., Finnegan, J., McFadden, H., Ayliffe, M.A., Dodds, P.N., and Ellis, J.G. (2005). Auto-active alleles of the flax L6 rust resistance gene induce non race-specific rust resistance associated with the hypersensitive response. Mol. Plant Microbe Interact. 18 570–582. [DOI] [PubMed] [Google Scholar]
- Hückelhoven, R. (2005). Powdery mildew susceptibility and biotrophic infection strategies. FEMS Microbiol. Lett. 245 9–17. [DOI] [PubMed] [Google Scholar]
- Jarosch, B., Kogel, K.-H., and Schaffrath, U. (1999). The ambivalence of the barley Mlo locus: Mutations conferring resistance against powdery mildew (Blumeria grammis f. sp. hordei) enhance susceptibility to the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 12 508–514. [Google Scholar]
- Jorgensen, J.H. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63 141–152. [Google Scholar]
- Kang, S., Sweigard, J.A., and Valent, B. (1995). The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 8 939–948. [DOI] [PubMed] [Google Scholar]
- Kemen, E., Kemen, A., Rafiqi, M., Hempel, U., Mendgen, K., Hahn, M., and Voegele, R.T. (2005). Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant Microbe Interact. 18 1130–1139. [DOI] [PubMed] [Google Scholar]
- Lipka, V., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183. [DOI] [PubMed] [Google Scholar]
- Lyngkjaer, M.F., Carver, T.L.W., and Zeyen, R.J. (2001). Virulent Blumeria graminis infection induces penetration susceptibility and suppresses race-specific hypersensitive resistance against avirulent attack in Mla1-barley. Physiol. Mol. Plant Pathol. 59 243–256. [Google Scholar]
- Panstruga, R. (2005). Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 33 389–392. [DOI] [PubMed] [Google Scholar]
- Parker, J.E., Holub, E.B., Frost, L.N., Falk, A., Gunn, N.D., and Daniels, M.J. (1996). Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8 2033–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, C., Lipka, V., O'Connell, R., Okuno, T., Schulze-Lefert, P., and Takano, Y. (2006). Non-host resistance in Arabidopsis–Colletotrichum interactions acts at the cell periphery and requires actin filament function. Mol. Plant Microbe Interact. 19 270–279. [DOI] [PubMed]
- Stein, M., Dittgen, J., Sánchez-Rodriguez, C., Hou, B.-H., Molina, A., Schulze-Lefert, P., Lipka, V., and Somerville, S. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter plants by direct penetration. Plant Cell 18 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard, J.A., Carroll, A.M., Kang, S., Farrall, L., Chumley, F.G., and Valent, B. (1995). Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell 7 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen, H. (2003). Fresh insight into processes of nonhost resistance. Curr. Opin. Plant Biol. 6 351–357. [DOI] [PubMed] [Google Scholar]
- Tosa, Y. (1989). Evidence on wheat for gene-for-gene relationship between formae speciales of Erysiphe graminis and genera of gramineous plants. Genome 32 918–924. [Google Scholar]
- van den Brule, S., and Smart, C.C. (2002). The plant PDR family of ABC transporters. Planta 216 95–106. [DOI] [PubMed] [Google Scholar]
- Yun, B., Atkinson, H.A., Gaborit, C., Greenland, A., Read, N.D., Pallas, J.A., and Loake, G.J. (2003). Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non-host resistance in Arabidopsis against wheat powdery mildew. Plant J. 34 768–777. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Henderson, C., Perfect, E., Carver, T.L.W., Thomas, B.J., Skamnioti, P., and Gurr, S.J. (2005). Review of genes and genomes, needles and haystacks: Blumeria graminis and functionality. Mol. Plant Pathol. 6 561–575. [DOI] [PubMed] [Google Scholar]
- Zhou, F., Kurth, J., Wei, F., Elliott, C., Valè, G., Yahiaoui, N., Keller, B., Somerville, S., Wise, R., and Schulze-Lefert, P. (2001). Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C., and Felix, G. (2005). Plants and animals: A different taste for microbes? Curr. Opin. Plant Biol. 8 353–360. [DOI] [PubMed] [Google Scholar]