Abstract
Genetic control of grass inflorescence architecture is critical given that cereal seeds provide most of the world's food. Seeds are borne on axillary branches, which arise from groups of stem cells in axils of leaves and whose branching patterns dictate most of the variation in plant form. Normal maize (Zea mays) ears are unbranched, and tassels have long branches only at their base. The ramosa2 (ra2) mutant of maize has increased branching with short branches replaced by long, indeterminate ones. ra2 was cloned by chromosome walking and shown to encode a LATERAL ORGAN BOUNDARY domain transcription factor. ra2 is transiently expressed in a group of cells that predicts the position of axillary meristem formation in inflorescences. Expression in different mutant backgrounds places ra2 upstream of other genes that regulate branch formation. The early expression of ra2 suggests that it functions in the patterning of stem cells in axillary meristems. Alignment of ra2-like sequences reveals a grass-specific domain in the C terminus that is not found in Arabidopsis thaliana. The ra2-dm allele suggests this domain is required for transcriptional activation of ra1. The ra2 expression pattern is conserved in rice (Oryza sativa), barley (Hordeum vulgare), sorghum (Sorghum bicolor), and maize, suggesting that ra2 is critical for shaping the initial steps of grass inflorescence architecture.
INTRODUCTION
Plant organogenesis is controlled by meristems, groups of stem cells located at the tips of shoots and roots. Meristems produce cells that give rise to all plant organs but also set aside a population of cells in order to perpetuate themselves. The balance of these two processes, organogenesis and self-perpetuation, guarantees prolonged activity, and such a meristem is said to be indeterminate. The activity of indeterminate meristems gives rise to shoots and branches capable of continued growth. By contrast, determinate meristems, such as those that produce flowers, are consumed after making a certain number of organs (Steeves and Sussex, 1989).
Plant architecture results from activity of the shoot apical meristem, located at the tip of the growing shoot, and axillary meristems, which form in the axils of leaves. During the vegetative phase of growth, leaves are often large and axillary meristems small or dormant. Following environmental and endogenous cues, the shoot apical meristem undergoes a transition to become a reproductive meristem, referred to as an inflorescence meristem. Leaves produced from the inflorescence meristem are often reduced, while axillary meristems become dominant. Flowers, which derive from modified axillary meristems, are borne either directly on the main inflorescence axis or on branches. Variation in the number of branches and their growth potential produces a great diversity of inflorescence morphologies, a striking feature of flowering plants (Weberling, 1989).
Members of the grass family (Poaceae) display a wide array of inflorescence forms that range from the spikes of wheat (Triticum aestivum) and barley (Hordeum vulgare) to the more branched tassels of maize (Zea mays) and sorghum (Sorghum bicolor). The basic unit of grass inflorescence architecture is the spikelet, a compact axillary branch that consists of two bracts subtending one to several reduced flowers (Clifford, 1987). Maize is a monoecious plant that produces male flowers on a terminal tassel and female flowers on lateral ears, which arise in the axils of vegetative leaves. The tassel consists of a main spike with several long, indeterminate branches at the base (Figure 1A), and the ear consists of a single spike with no long branches (Figure 1D). The tassel's main spike and branches, and the entire ear, bear short branches (spikelet pairs) that give rise to two spikelets. In maize, spikelet and spikelet pair meristems are considered determinate because they produce a defined number of structures.
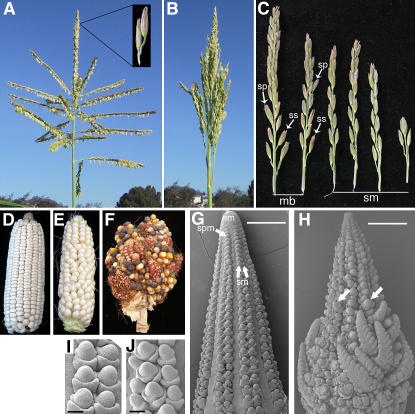
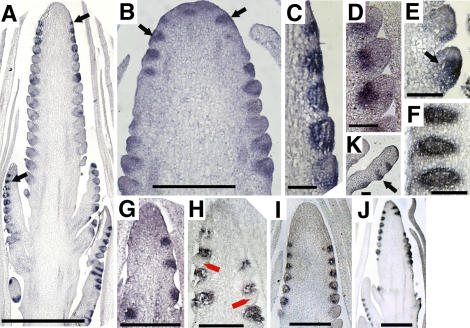
Figure 1.
The ra2 Mutant Phenotype.
(A) Wild-type tassel; inset, a spikelet pair.
(B) ra2-mum4 tassel.
(C) The transition of branch lengths in a ra2 mutant. Mixed branches are formed after main branches (data not shown) and bear single spikelets and spikelet pairs. Spikelet multimers are formed after the mixed branches and consist of more than two single spikelets. mb, mixed branch; sm, spikelet multimer; sp, spikelet pair; ss, single spikelet.
(D) Wild-type ear with well-defined rows of kernels.
(E) ra2-mum4 ear with disorganized rows.
(F) Open-pollinated ra2-dm ear with numerous branches.
(G) to (J) Scanning electron microscopy of developing ears.
(G) Wild-type ear. im, inflorescence meristem; sm, spikelet meristem; spm, spikelet pair meristem.
(H) Scanning electron micrograph of a ra2-R ear showing meristems initiating branches (arrows) and some well-developed branches at the base.
(I) Detail of a wild-type ear row consisting of a pair of spikelet meristems.
(J) Detail of the formation of three spikelet meristems in a ra2-R ear.
Bars = 0.5 mm in (G) and (H) and 50 μm in (I) and (J).
Maize has long been the subject of genetic research, resulting in a large collection of mutants (Neuffer et al., 1997), many of which alter specific stages of inflorescence development (reviewed in McSteen et al., 2000). One group of mutants shows a decreased production of branches and/or spikelet pairs, such as liguleless2 (Walsh and Freeling, 1999), barren stalk1 (ba1) (Gallavotti et al., 2004), barren inflorescence1 (bif1), and bif2 (McSteen and Hake, 2001). The opposite phenotype is observed in the ramosa1 (ra1), ra2, and ra3 mutants, which have tassels with an increased number of long branches as well as branched ears (Vollbrecht et al., 2005), and branched silkless1 and indeterminate spikelet1, which affect determinacy of the spikelet meristem (Chuck et al., 1998, 2002).
Here, we report the positional cloning and characterization of ra2, a recessive mutant first described in 1935 (Emerson et al., 1935). ra2 codes for a putative transcription factor with a LATERAL ORGAN BOUNDARY (LOB) domain that is highly conserved among other plants, including Arabidopsis thaliana, rice (Oryza sativa), barley, and sorghum. ra2 is expressed in the cells that will produce axillary meristems in maize inflorescences, marking the site of meristem initiation. Levels of ra1, which impose determinate fate specifically on spikelet pair meristems, are reduced in ra2 mutants (Vollbrecht et al., 2005), except in the ra2-dm allele that putatively makes a C terminus–only RA2 protein. Based on its expression pattern and the phenotype of loss-of-function mutants, we suggest that ra2 controls multiple aspects of inflorescence architecture by limiting growth of axillary meristems from their inception. Our data suggest that ra2 carries out that function, in part, by promoting ra1 expression in the developing inflorescence.
RESULTS
Inflorescences of ra2 Mutants Have Branches with Increased Indeterminacy
In order to analyze the ra2 mutant phenotype, we examined three mutant alleles in defined inbred backgrounds. ra2-R was introgressed into B73 and A188, ra2-dm was found in W22 and further introgressed into that inbred, and ra2-mum4 was introgressed twice into A188. The most consistent tassel phenotype of ra2 mutants is a change in the branch organization of the tassel. Tassels of ra2 mutants have similar numbers of long branches but fewer spikelet pairs. Instead, they have mixed branches that contain both spikelet pairs and single spikelets as well as spikelet multimers, which bear more than two single spikelets (Figure 1C). Although spikelet multimers are also formed in wild-type tassels, they are less frequent and rarely contain more than three spikelets. Thus, a typical ra2 mutant tassel shows an abnormal, gradual transition from long branches to spikelet pairs, with the branches becoming increasingly shorter toward the inflorescence apex (Figure 2, Table 1). Higher orders of branching are also altered in ra2-R, as seen by an increase in number of branches that bear secondary branches and by the total number of secondary branches (Table 2). Finally, for all ra2 mutant alleles, the spikelet pair pedicel is longer, and the angle at which branches are borne with respect to the main inflorescence rachis (Figure 1B) is more acute relative to the wild type (Table 3).
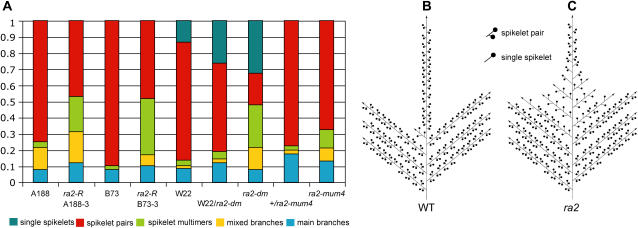
Figure 2.
Tassel Branching Patterns in A188, B73, and W22 Maize Inbred Lines and ra2 Mutants.
(A) The distribution of axillary meristem fates in tassels of ra2 alleles and inbred lines. The area of each class of branch represents its percentage of all tassel branches. Single spikelets were only found in W22 and W22-introgressed families. See Table 1 for error values and significance of a t test.
(B) and (C) A simplified schematic showing the change in branch determinacy imposed on branches of ra2 mutant tassels.
(B) A wild-type tassel, consisting mainly of spikelet pair–bearing branches at the bottom and spikelet pairs directly on the main axis above.
(C) A typical ra2 tassel showing mixed branches and spikelet multimers that gradually become spikelet pairs toward the apex. Branch angle difference is not illustrated.
Table 1.
The Fate of Tassel Primordia Measured in Different ra2 Alleles and Inbred Lines
| Genotype | Total Primordiaa | Main Branchesb | Mixed Branchesb | Spikelet Multimersb | Spikelet Pairsb | Single Spikeletsb,c |
|---|---|---|---|---|---|---|
| A188 | 80.6 ± 9.4 | 0.07 ± 0.02 | 0.13 ± 0.03 | 0.038 ± 0.02 | 0.74 ± 0.03 | – |
| ra2-R, B73-3d | 100.8 ± 16.0 | 0.12 ± 0.06 | 0.19 ± 0.07 | 0.21 ± 0.10 | 0.46 ± 0.09 | – |
| Pe | <0.01 | 0.028 | <0.01 | <0.01 | <0.01 | |
| B73 | 81.1 ± 17.2 | 0.078 ± 0.02 | 0.0017 ± 0.0004 | 0.022 ± 0.02 | 0.9 ± 0.03 | – |
| ra2-R, A188-3f | 68.1 ± 13.7 | 0.1 ± 0.04 | 0.07 ± 0.04 | 0.35 ± 0.10 | 0.4 ± 0.13 | – |
| P | <0.01 | 0.074 | <0.01 | <0.01 | <0.01 | |
| W22/ra2-dm | 99.6 ± 15.5 | 0.12 ± 0.03 | 0.023 ± 0.03 | 0.044 ± 0.03 | 0.54 ± 0.08 | 0.26 ± 0.13 |
| ra2-dm | 73.3 ± 13.6 | 0.078 ± 0.02 | 0.13 ± 0.05 | 0.26 ± 0.08 | 0.19 ± 0.03 | 0.32 ± 0.008 |
| P | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 0.19 |
| +/ra2-mum4 | 99.9 ± 10.7 | 0.17 ± 0.02 | 0.022 ± 0.01 | 0.029 ± 0.01 | 0.77 ± 0.03 | – |
| ra2-mum4 | 81.2 ± 9.9 | 0.13 ± 0.04 | 0.084 ± 0.02 | 0.11 ± 0.06 | 0.67 ± 0.07 | – |
| P | <0.01 | 0.019 | <0.01 | <0.01 | <0.01 |
Total number of primordia on the tassel main axis (average of >12 individuals).
Ratio of the number of primordia that produce a specific kind of lateral organ (main branch, mixed branches, spikelet multimers, spikelet pairs, and single spikelets) to the total number of primordia.
Single spikelets were only found in the W22 inbred line and in ra2-dm segregating families, all of which are in a W22 background.
ra2-R was introgressed into B73 more than three generations.
Significant values of the t test at P = 0.01 are in bold.
ra2-R was introgressed into A188 more than three generations.
Table 2.
Degree of Branching in Inbred Lines and ra2 Alleles
| Genotype | No. of Branched Branches | No. of Secondary Branches |
|---|---|---|
| A188 | 0.4 ± 0.6 | 0.4 ± 0.6 |
| ra2-R, A188-3 | 1.2 ± 1.3 | 1.3 ± 1.4 |
| Pa | 0.038 | 0.023 |
| B73 | 1.5 ± 0.6 | 1.6 ± 0.6 |
| ra2-R, B73-3 | 2.5 ± 0.7 | 7.0 ± 2.2 |
| P | <0.01 | <0.01 |
| +/ra2-dm | 4.1 ± 1.6 | 7.5 ± 1.6 |
| ra2-dm | 5.3 ± 1.5 | 20.3 ± 7.1 |
| P | <0.01 | <0.01 |
| +/ra2-mum4 | 2.2 ± 1.1 | 3.0 ± 1.7 |
| ra2-mum4 | 2.1 ± 0.9 | 3.3 ± 1.6 |
| P | 0.721 | 0.692 |
Significant values of the t test at P = 0.01 are in bold.
Table 3.
Branch Angle and Length of Pedicel in ra2 Compared with Normal Tassels
| Genotype | Tassel Height (cm) | First Tassel Branch Angle | Second Tassel Branch Angle | Pedicel Length (cm)a |
|---|---|---|---|---|
| A188 | 32.6 ± 1.5 | 64.4 ± 26.5 | 80.2 ± 24.4 | 0.5 ± 0.6 |
| ra2-R, A188-3 | 36.5 ± 8.6 | 19.2 ± 16.6 | 24.06 ± 22.1 | 12.0 ± 2.5 |
| Pb | 0.103 | <0.01 | <0.01 | <0.01 |
| B73 | 26.5 ± 4.5 | 5.4 ± 2.5 | 13.3 ± 7.8 | 1.1 ± 0.9 |
| ra2-R, B73-3 | 23.3 ± 1.6 | 4.2 ± 2.2 | 5.6 ± 2.7 | 10.8 ± 3.0 |
| P | 0.017 | 0.148 | <0.01 | <0.01 |
| W22/ra2-dm | 26.3 ± 2.05 | 30.3 ± 15.5 | 36.1 ± 18.1 | 7.8 ± 5.6 |
| ra2-dm | 26.4 ± 2.29 | 11.4 ± 5.3 | 12.4 ± 8.3 | 16.4 ± 3.3 |
| P | 0.83 | <0.01 | <0.01 | <0.01 |
| +/ra2-mum4 | 31.3 ± 2.9 | 60.1 ± 25.2 | 74.3 ± 20.1 | 0.6 ± 1.2 |
| ra2-mum4 | 33.5 ± 3.4 | 14.1 ± 13.5 | 9.3 ± 5.6 | 7.7 ± 4.7 |
| P | 0.094 | <0.01 | <0.01 | <0.01 |
Corresponds to the average length of the pedicels subtending the first five spikelet pairs.
Significant values of the t test at P = 0.01 are in bold.
The female inflorescence of maize, the ear, develops according to a branching program similar to that of the tassel except the ear does not form long branches (Figure 1D). Spikelet pair meristems initiate on the flanks of the ear tip in a regular pattern (Figure 1G) and give rise to two spikelet meristems (Figure 1I). Ears of all three ra2 mutant alleles have disorganized rows of kernels (Figure 1E), and in ra2-R and ra2-dm several long branches are formed (Figure 1F). Spikelet triplets (instead of the typical pair of spikelets) and branches are formed in ra2 mutant ears, reflecting increased indeterminacy of the meristem (Figures 1H and 1J). Ears of the ra2-mum4 allele do not form branches, and the only ear phenotype is row disorganization (Figure 1E). Thus, inflorescence branches of ra2 mutants show additional branching and loss of determinacy in both ear and tassel.
Positional Cloning of ra2
We initially mapped ra2 to a region of chromosome 3 (bin 3.04) between restriction fragment length polymorphism (RFLP) markers asg48 (proximal) and mmp186 (distal). Two flanking markers were used to screen for recombinants among 1069 backcrossed progeny (A188/ra2-R × ra2-dm/ra2-dm). To find additional markers closer to ra2, we assumed conserved synteny between maize and rice and searched for rice genes between the rice asg48 and mmp186 homologs. The rice homologs to maize asg48 and mmp186 are located in two unlinked regions of rice chromosome 1. However, most of the markers in bin 3.04 had homologs in the rice mmp186 region. In addition, all seven maize markers derived from rice genes in the mmp186 region showed linkage to ra2, and some of these were proximal, indicating that the maize region containing asg48 was a small insertion in a region of otherwise completely conserved synteny between rice and maize (Figure 3A).
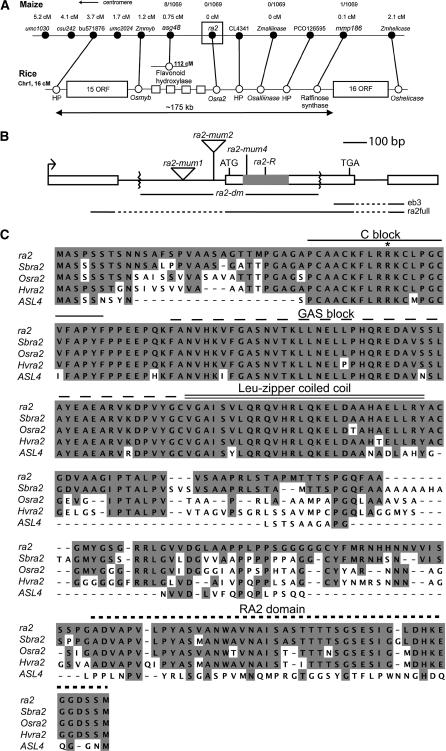
Figure 3.
Cloning and Characterization of ra2.
(A) The genetic map of a portion of maize chromosome 3 (top) is compared with the syntenous physical region of rice chromosome 1 (bottom). Black circles are maize markers/genes. White circles are rice genes and ORFs. The 175-kb segment indicated for the rice physical map corresponds to rice clone AP003339. The rice homolog to maize RFLP marker asg48 is located in an unlinked location of rice chromosome 1, and it is the only nonsyntenic gene of the region shown here between maize and rice. There are four ORFs (white squares) in a 17-kb region between the Osmyb and Osra2 genes for which there are no significantly similar maize sequences. The number and order of rice genes follow their annotation in GenBank but are not drawn to proportion. HP, hypothetical protein.
(B) Structure of ra2 with the LOB domain in gray; diagram begins and ends at transcription initiation and polyadenylation site, respectively. The alleles of ra2 are shown with the ra2-dm deletion indicated by the zigzag marks in intron 1 and exon 2. eb3, the fragment used as a probe for DNA and RNA gel blot hybridizations; ra2full, the probe used for in situ hybridizations.
(C) Alignment of the deduced full-length amino acid sequence of ra2 from maize, sorghum (Sbra2), rice (Osra2), barley (Hvra2), and ASL4 of Arabidopsis. The LOB domain is located near the N terminus and consists of a C-rich domain, a GAS block, and a Leu-zipper domain. Grass ra2 orthologs share a conserved C terminus domain of unknown function. The asterisk indicates the mutation (Arg to His) in the ra2-mum4 allele.
One of the genes tightly linked to the ra2 phenotype (0/1069 recombinants) encoded a protein predicted to carry a LOB domain (Iwakawa et al., 2002; Shuai et al., 2002). We found two maize EST contigs (TC278100 and TC254099) that showed high sequence similarity to the 5′ and 3′ regions of the rice LOB gene and a 2.7-kb genomic contig (MAGI-27324) that spanned the entire gene. This gene is predicted to have two introns and an open reading frame (ORF) in the second exon based on alignment of the EST and genomic contigs (Figure 3B). A 6-kb EcoRI fragment containing ra2 was subcloned from a BAC to identify the entire ORF as well as upstream and downstream sequences. The ORF of ra2 contains a Cys repeat (Cx2Cx6Cx3C), a GAS block, and a Leu-zipper motif (Figure 3C), features conserved in all LOB genes (Iwakawa et al., 2002; Shuai et al., 2002). An additional domain that is conserved in the C terminus of ra2 homologs from grass species is not shared with Arabidopsis.
The maize LOB gene was sequenced from several inbred lines and our ra2 mutant alleles. The ra2-R allele has an 8-bp insertion that introduces a stop codon within the LOB domain. The ra2-dm allele, which arose from a Mutator (Mu) transposon population, has an 1171-bp deletion that eliminates part of the first intron and second exon, including all of the sequence that encodes the LOB domain. The ra2-mum4 allele, which also arose in a Mu population, contains a single base pair mutation in the LOB domain that introduces an Arg-to-His amino acid change in a putative nuclear localization signal (Figures 3B and 3C). The Arg amino acid is conserved in all ra2 orthologs examined (rice, barley, sorghum, and Arabidopsis). Two more alleles were generated by Mu-targeted transposition, ra2-mum1 and ra2-mum2, both of which contain Mu insertions in the first intron (Figure 3B). The molecular nature of the alleles correlates with their phenotype; the two alleles with the strongest ear phenotype have an early stop codon (ra2-R) or a deletion in the LOB domain (ra2-dm), while the weak allele, ra2-mum4, has a single amino acid change in a conserved domain. This collection of independent mutant alleles implies that the maize LOB gene is in fact ra2.
ra2 Is Expressed in the Anlagen of Inflorescence Axillary Meristems
To examine the expression of ra2, hybridization of RNA gel blots from tissues of wild-type and ra2 mutant alleles was performed using a gene-specific probe (eb3 in Figure 3B). ra2 is expressed in ears and tassels of wild-type maize (Figure 4). Wild-type seedlings and roots have no significant expression of ra2. Levels of ra2 transcript are very low in ra2-R tassels, indicating that it may be a null allele. ra2-dm inflorescences make a shorter transcript because of the ∼1.2-kb deletion, and ra2-mum4 mutant tassels have normal levels of ra2 transcript. A weak band of higher molecular weight was observed in some lanes that could be the product of alternative splicing.
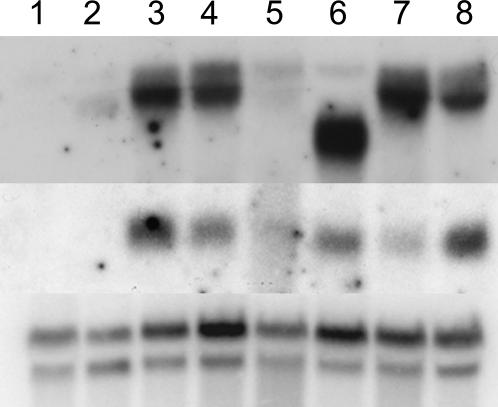
Figure 4.
Tissue Localization of ra2 and ra1 Expression.
RNA gel blot of inbred line B73 seedling (lane 1), B73 root (lane 2), B73 tassel (lane 3), B73 ear (lane 4), ra2-R tassel (lane 5), ra2-dm ear (lane 6), ra2-mum4 tassel (lane 7), and ra1-R ear (lane 8). Top panel is ra2 hybridization, middle is ra1, and bottom is ubiquitin. Note the smaller size transcript in ra2-dm due to the deletion of the LOB domain and that ra1 is expressed in this allele but not the other ra2 alleles.
In situ hybridization was performed to examine the tissue localization of ra2 expression using a full-length ra2 RNA probe (ra2full in Figure 3B). ra2 is expressed in developing tassels in a group of cells that predicts the position of the bract and spikelet pair meristem (Figures 5A and 5C). Closer examination of ra2 expression in young tassels allowed us to determine that ra2 is expressed in the axillary meristem above the inflorescence bracts (Figure 5H). Expression continues transiently as the spikelet pair meristem initiates and then diminishes as the meristem grows. Expression is observed again after the meristem enlarges, at the position where the spikelet pair meristem branches to produce the spikelet meristem (Figure 5E). ra2 expression in ears is similarly observed in the anlagen of spikelet pair meristems (Figures 5B and 5F). The same expression pattern was observed with the gene-specific eb3 probe in wild-type tassels (Figure 5I). In both the tassel and ear, expression is not seen in the branching that results in floral meristems. As a negative control, we performed the same procedure on similar tissues using a sense probe and saw no signal (data not shown). Antisense probe on ra2-R tassels showed much weaker expression than the wild type (data not shown).
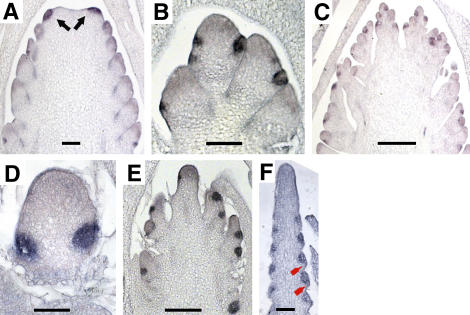
Figure 5.
ra2 Expression Marks the Position of Inflorescence Axillary Meristems.
(A) to (C) and (E) to (I) ra2 expression in wild-type tissue.
(A) Tassel.
(B) Ear.
(C) Longitudinal section of a tassel tip showing ra2 expression predicting the position of spikelet pair meristems.
(D) Detail of ra1 expression in spikelet pair meristems of a tassel.
(E) Detail of ra2 expression in the anlagen of spikelet meristem.
(F) Front view of a row of spikelet pair meristems in an ear.
(G) Expression in a tassel primordium that is just initiating branch meristems.
(H) Hybridization of a young tassel tip showing expression of ra2 in axillary meristem anlagen above the bract primordia (red arrows).
(I) Tip of a tassel of similar stage of development as in (A) showing hybridization with a gene-specific probe.
(J) and (K) ra2 expression in ra mutants: ra3 tassel (J); ra1 tassel branch (K).
Bars = 0.5 mm (A), (B), and (G) to (J) and 50 μm in (C) to (F) and (K).
We also performed in situ hybridization to determine if ra2 is expressed in the axillary meristems that produce long branches of the tassel. Because these are the first meristems to form in the tassel, it was necessary to collect meristems that were transitioning from vegetative to reproductive. Transition meristems can be distinguished from vegetative meristems by an increased height (Bonnett, 1940). In shoot meristems of 4-week-old seedlings, ra2 is expressed at the site of long branch meristem initiation (Figure 5G). No expression was detected in the vegetative meristem (data not shown). In summary, ra2 marks the position of all nonfloral axillary meristems in the developing inflorescence, which includes long branch, spikelet pair, and spikelet meristems.
ra2 Is Required for Normal ra1 Expression
ra1, ra2, and ra3 mutants have similar branching phenotypes except that ra2 has upright branches (Neuffer et al., 1997; Vollbrecht et al., 2005). In order to determine whether ra2 expression was dependent on ra1 or ra3, we performed in situ hybridization on developing tassels. We observed the same pattern of expression of ra2 in tassels of ra1-R (Figure 5K) and ra3-R mutants (Figure 5J), suggesting that ra2 is not dependent on either ra1 or ra3. To find out whether ra2 and ra1 expression overlap, we performed in situ hybridization of ra1 in wild-type tassels using the probe described by Vollbrecht et al. (2005). The onset of ra1 expression was observed at the base of spikelet pair meristems (Figure 5D), clearly overlapping with ra2 expression. We also performed RNA gel blot analysis of ra1 expression in a ra2 mutant and vice versa. As shown in Figure 4 (top panel, lane 8), ra2 expression is not greatly affected in ra1 mutants. By contrast, Vollbrecht et al. (2005) reported that ra1 expression is significantly reduced in ra2-R mutants. Indeed, our RNA gel blots show that ra1 is significantly reduced in ra2-R and ra2-mum4 mutants (Figure 4, middle panel, lanes 5 to 7). However, ra1 is still expressed in ears of ra2-dm (Figure 4, middle panel, lane 6), which putatively makes a truncated protein consisting of only the last 59 amino acids (Figure 3B). This result would suggest that the C-terminal domain of RA2 is sufficient for ra1 transcript accumulation. The hypothesis that ra2 is required for ra1 expression fits with the RNA gel blot analysis as well as the timing of expression; ra2 is expressed earlier than ra1, and their expression patterns overlap briefly (Vollbrecht et al., 2005).
The Pattern of ra2 Expression Is Conserved among Grasses
The coding sequence of ra2 is highly conserved among rice, barley, sorghum, and maize (Figure 3C). In order to examine the variation in ra2 expression among grasses, in situ hybridizations were performed. Sorghum is more closely related to maize and also has spikelet pairs; however, the primary axis of the sorghum inflorescence bears indeterminate branches, and spikelet pairs are borne on branches of second and third order. Rice, a distant relative of maize, makes inflorescences that branch once to make second-order branches that bear multiple single spikelets. In contrast with those grasses, barley produces inflorescences without branches, and the single spikelets are borne directly on the main axis. Similar to the expression pattern in maize, ra2 is expressed transiently in each anlagen of multiple axillary meristem types of sorghum inflorescences, including all indeterminate branches and spikelet pairs (Figures 6A to 6C), but not in floral meristems. ra2 is similarly expressed in the anlagen of primary branch meristems and spikelet meristems of rice inflorescences (Figures 6D and 6E), while in barley, ra2 is expressed only in the spikelet meristem anlagen on the main axis of the inflorescence (Figure 6F). These results demonstrate that both the sequence and expression pattern of ra2 are conserved and suggest an important role for ra2 in all grasses.
Figure 6.
ra2 Transiently Marks the Position of Axillary Inflorescence Meristems in Sorghum, Rice, and Barley.
(A) to (C) Sorghum.
(A) Expression in first-order branch primordia.
(B) Expression in second-order branch primordia.
(C) Expression in spikelet pair meristem primordia.
(D) and (E) Rice.
(D) Expression in primary branch primordia.
(E) Expression in spikelet meristem primordia.
(F) Expression in the anlagen of a spikelet meristem of a barley inflorescence. Notice that the expression is above the bracts (red arrows).
Bars = 250 μm.
DISCUSSION
ra2 mutants produce inflorescence branches with increased indeterminacy or meristematic potential. We cloned ra2 by positional cloning and determined that it encodes a LOB domain protein. Multiple alleles provided proof that the gene was cloned. ra2 is expressed early in the inflorescence in cells that predict the position of axillary meristem initiation. The expression in different mutant backgrounds places ra2 upstream of other branch regulators. Conservation of sequence and expression pattern in other grass species suggests that ra2 plays a critical role in the architecture of all grass inflorescences.
Positional Cloning of ra2 Demonstrates That It Encodes a LOB Domain Protein
We undertook positional cloning to characterize the ra2 gene, given the recent developments in maize genomic tools (Rabinowicz et al., 1999; Palmer et al., 2003; Yuan et al., 2003) and sequencing of the rice genome (Sasaki et al., 2002; International Rice Genome Sequencing Project, 2005). The strategy relies on the hypothesis that the relative order of genes is conserved among distantly related grass species (Moore et al., 1995; Song et al., 2002; Klein et al., 2003). We were fortunate to find ra2 using a relatively small mapping population (1069) thanks to the synteny in the ra2 region, a large number of alleles, and the presence of a ra2 ortholog in rice. One of our closest flanking markers, however, was not located in this region in rice. As sequencing of the maize genome progresses, the ability to quickly identify the boundaries of syntenic regions should facilitate more rapid progress in positional cloning.
ra2 encodes a protein with a LOB domain, named after the enhancer trap pattern of the first gene isolated in this family (Shuai et al., 2002). ra2 has highest sequence similarity to LOB/ASL4 and is most likely its ortholog, based on results from a phylogenetic analysis that included all Arabidopsis LOB domain genes (see Supplemental Figure 1 online). Forty-two members of this plant-specific family are found in Arabidopsis. The rice genome contains 20 genes with 40% or more conserved amino acids in the LOB domain when compared with ASL4. Our searches have identified 12 unique genes in maize; however, maize has not yet been fully sequenced. ASL4 loss of function mutants in Arabidopsis show no phenotype (Shuai et al., 2002), and ASL4 expression, at the boundary of lateral organs long after the organs have been initiated, differs from ra2. Recent reports show that some LOB genes are important in root morphogenesis in Arabidopsis (Okushima et al., 2005) and rice (Inukai et al., 2005; Liu et al., 2005). Ectopic expression of certain LOB genes causes repression of knotted1-like (knox) genes, whose products promote stem cell proliferation (Lin et al., 2003; Chalfun-Junior et al., 2005). The opposite effect, increased expression of knox genes, has been reported in the LOB gene mutant asymmetric leaves2 (Ori et al., 2000; Semiarti et al., 2001). Our RNA gel blots and in situ hybridization, however, showed no difference in the levels of knotted1 transcript or expression pattern between wild-type and ra2 mutant inflorescences, nor did we find differences in the expression of other knox genes (gn1, rs1, and lg3) when measured by RT-PCR (see Supplemental Figure 2 online).
LOB domain proteins of Arabidopsis and rice localize to the nucleus and are thus thought to function in transcriptional control (Iwakawa et al., 2002; Liu et al., 2005). The LOB domain contains a putative nuclear localization signal. The ra2-mum4 allele introduces an Arg-to-His amino acid change in the nuclear localization signal in support of the notion that RA2 plays a role in the nucleus. In addition to the LOB domain that is conserved across plants, the grass species share a second C-terminal domain. Experiments in yeast indicate that the LOB domain of ARL1, a rice gene required for adventitious root formation, may function as a transcriptional repressor (Liu et al., 2005), whereas the C-terminal portion may act as an activator. Our ra2-dm allele contains a deletion that eliminates the LOB domain but can presumably make an in-frame protein consisting of the last 59 amino acids in the C terminus. This allele has a severe phenotype but may retain some of its function as discussed later.
ra Mutants in Maize Affect the Determinacy of Branches
Processes that regulate axillary meristem formation are beginning to be understood thanks to a number of mutants that either fail to make branches or make too many. Mutants of the tomato lateral suppressor (ls) gene fail to produce vegetative branches, and their inflorescences have fewer flowers (Schumacher et al., 1999). las, the Arabidopsis ortholog of ls, is required for normal development of axillary meristems during the vegetative phase but does not play as significant a role in the inflorescence (Greb et al., 2003). The function of ls seems to be conserved in monocots because mutations in the rice ortholog, monoculm1, significantly decrease tiller production (Li et al., 2003). The bHLH protein encoded by ba1 in maize is required for all axillary branches, including ear and basal vegetative branches (Gallavotti et al., 2004). A related protein, LAX, shares a similar function in rice (Komatsu et al., 2003). No obvious orthologs are found in Arabidopsis.
An interesting group of mutants links the plant hormone auxin to formation of axillary meristems. These include pin-formed1 (pin1), pinoid1 (pid1), and monopteros1, all of which are deficient in production of lateral organs, and auxin-resistant1, which has increased shoot branching (reviewed in McSteen and Leyser, 2005). PIN proteins are transporter-like membrane proteins that are thought to function in polar auxin transport (Galweiler et al., 1998). Localization of PIN1 protein within the cell appears to be an early indicator of lateral organ initiation (Reinhardt et al., 2003). PID1 is thought to be important in this localization, as high levels of PID activity lead to the apical localization of PIN, while low levels lead to the basal localization of PIN (Friml et al., 2004).
Many mutants with increased branching occur because of increased bud outgrowth, as in max, dad, and ramosus mutants in Arabidopsis, petunia (Petunia hybrida), and pea (Pisum sativum), respectively (McSteen and Leyser, 2005). Increased bud outgrowth occurs through a novel signaling molecule identified by analysis of the MAX genes (Booker et al., 2005). The phenotype is similar to the teosinte branched1 mutant in maize (Doebley et al., 1997), which is encoded by a bHLH protein. tb1 mutants have more branches arising from vegetative nodes and make secondary and tertiary branches (Hubbard et al., 2002). TB1 functions not only to repress axillary bud initiation but also axillary branch length.
Branching of the maize ra mutants differs in that branches with limited growth potential are transformed into indeterminate branches. In this regard, they are more similar to floricaula and leafy mutants of Antirrhinum and Arabidopsis, respectively, which change the determinate floral meristem into an inflorescence-like meristem (Coen et al., 1990; Weigel et al., 1992). In ra2 mutants, short branches that normally make spikelet pairs are converted into longer branches that make multiple spikelets. Long branches at the base of the tassel also have increased secondary branching. In the ear, the initiation of more than two spikelet meristems per row causes disorganization of the rows, and in ra2-R and ra2-dm, some of the spikelet meristems acquire an indeterminate fate and become long branches. Thus, the mutant phenotype indicates that one function of ra2 is to restrict branch growth and to establish determinacy on the spikelet pair meristems. In addition to a change in branch determinacy, ra2 mutant tassels also have upright branches. These two features, branch angle and branch length, may be coordinately regulated. For example, the brevipedicellus mutation of Arabidopsis results in downward, shorter pedicels compared with the upright, long pedicels of the wild type (Douglas et al., 2002; Smith and Hake, 2003).
ra2 Predicts the Position of Branch Anlagen in the Inflorescence
Maize inflorescences offer an ideal system to follow the ontogeny of axillary meristem formation. The developmental series seen in tassel or ear primordia provides a view of multiple stages of axillary meristem initiation and growth. Scanning electron microscopy shows that the first sign of axillary growth is the presence of a bract, a modified leaf. Spikelet pair meristems form in axils of bracts. ra2 expression is first seen prior to any visible bump on the surface of the inflorescence, suggesting that it may mark the site of both bract and spikelet pair meristem initiation. As the bract grows out, but before the axillary meristem develops, ra2 expression is restricted to a group of cells in the axil of the bract in maize (Figure 5H) and barley (Figure 6F). Thus, ra2 may initially define both bract and axillary meristem anlagen, but as the bract grows, it becomes restricted to just the cells that are destined to be the axillary meristem.
The ra2 expression pattern is highly conserved among grasses with widely different inflorescence architecture (i.e., maize, sorghum, barley, and rice). These cereals represent monophyletic groups that account for most of the species of Poaceae (Grass Phylogeny Working Group, 2001), suggesting that ra2 may play a similar role in inflorescence development of all grasses. This hypothesis is supported by the finding that ra2 maps to a quantitative trait locus for branch number in sorghum, a closely related species to maize with a more branched inflorescence (P. Brown and S. Kresovich, personal communication). Thus, we predict that a loss of function in rice should cause increased branch growth or indeterminacy. Availability of the ra2 gene sequence will enable quantitative trait loci analyses in other cereals and association analysis in maize to collectively better define the role of ra2 in quantitative variation in the inflorescence (Thornsberry et al., 2001).
Regulation of ra1 by ra2
Considering the expression patterns of ra1 and ra2, we propose a model to explain how these genes function to regulate branch architecture in cereal species. ra1 is likely to be downstream of ra2 based on maize data, including a reduction of ra1 expression in a ra2 mutant and timing of expression. ra2 appears to be independent of ra3 based on our in situ results. ra2 is expressed in the anlagen of the bract and meristem early in inflorescence development, while ra1 is expressed at a later stage at the base of emerging spikelet pair meristems. The expression patterns of ra1 and ra2 overlap briefly and then ra2 expression disappears in the zone where ra1 is expressed. Maize tassels have both determinate (spikelet pair) and indeterminate branches. Although ra2 is transiently expressed in indeterminate branch anlagen, ra1 is not (Vollbrecht et al., 2005). This result suggests that other factors required for normal expression of ra1, besides ra2, are missing in these first formed branch anlagen. The inability to promote ra1 expression in these branch anlagen likely explains their indeterminacy. Interestingly, ra1 is not found in rice, where spikelets form in multimers on indeterminate branches. These results suggest that in maize and sorghum, ra2 promotes spikelet pair meristem fate through its regulation of ra1. In grasses where ra1 is not present, spikelet pair meristems do not form. These grasses still form spikelets and have defined patterns of determinate and indeterminate branches. We hypothesize that ra2 imposes determinacy in these species through other targets.
Although our data suggest that ra1 is downstream of ra2, it is clear there are additional targets for ra2. ra2 mutant tassel branches are upright, which is not a feature of ra1 mutants. The ra1 ra2 double mutants have a synergistic phenotype with increased branching (Vollbrecht et al., 2005), suggesting that both proteins are likely to interact with additional components. Finally, ra1 is still expressed in the ra2-dm allele, which has a severe phenotype. ra2-dm has an in-frame deletion of the LOB domain, leaving the grass-specific conserved C-terminal domain. These results suggest that the domain is sufficient for ra1 expression, but not sufficient for full RA2 activity. Further experiments will determine the exact nature of their interactions.
Conclusion
The final shape of a ra2 mutant tassel is reminiscent of a typical raceme, which has branches that get progressively shorter toward the apex (Weberling, 1989), a pattern established through apical dominance. ra2 reinforces apical dominance by abruptly imposing a determinate fate on lateral organs, causing the switch from long branches to spikelet pairs. In maize, and likely in related grasses like sorghum, this function is performed, at least in part, by ra1. The conserved expression pattern and sequence of ra2 in maize, sorghum, barley, and rice suggest that it plays an important role in the organization of axillary meristems in all cereals. However, its downstream targets or the timing of activation may be different across grass species, resulting in varying degrees of branch growth that produce a wide array of inflorescence morphologies.
METHODS
Phenotype
To characterize inflorescence architecture of inbred lines and introgressed alleles, the ratio of each type of tassel branch borne on the main axis of the inflorescence was calculated. The number of main branches, branches with more than two single spikelets (spikelet multimers), branches with both single spikelets and spikelet pairs (mixed branches), and spikelet pairs along the main inflorescence axis was counted. The number of each type of branch was divided by the total to obtain the ratio. The tassel height, angle of the first two branches, and the length of the pedicels subtending the first five spikelet pairs were also measured.
Marker Development and Recombinant Screen
Markers umc1030, an SSR marker proximal to ra2, and PCR_mmp186 were used to screen for recombinants among 1069 seedlings of backcrossed progenies (A188/ra2-R × ra2-dm/ra2-dm). DNA minipreps were prepared from discs of leaf tissue from 2-week-old seedlings cut into Eppendorf tubes and extracted as described (Konieczny and Ausubel, 1993) except that samples were not frozen with liquid nitrogen. Recombinant seedlings were allowed to grow to score the phenotype of tassel and ear.
The initial mapping stage was done using markers listed in the IBM neighbors map. In order to find more closely linked markers to ra2, BLAST searches in various maize (Zea mays) EST and GSS databases (http://maizegdb.org, http://magi.plantgenome.iastate.edu, and http://maize.tigr.org) were performed with rice (Oryza sativa) genes that were linked to the rice mmp186 homolog as queries. Maize genes that showed high similarity to the rice queries were sequenced to search for polymorphisms among the alleles involved in our mapping populations. Cleaved-amplified polymorphisms or single nucleotide polymorphisms were used. Some PCR products were used as RFLP probes.
Physical Maps
The online FPC maps available at the Arizona Genomics Institute (http://www.genome.arizona.edu/) website were used to find an asg48 BAC contig. To obtain BACs for the distal region to ra2, the Clemson University Genomics Institute (CUGI) B73 BAC library was screened with the RFLP probe mmp186 and the eb3 probe (Figure 3B). BACs were obtained from CUGI, and their correspondence to markers was confirmed by PCR and DNA gel blot hybridization.
Sequencing of Alleles and ra2 Characterization
The ra2 ORF of the inbred lines B73, A188, W22, and Mo17 as well as the alleles ra2-R and ra2-mum4 were PCR amplified and sequenced with primers full_Zm_LOB_F (5′-CCTAATCTTCTCCCTCTCCTCGTG-3′) and full_Zm_LOB_R (5′-CCTTCCTTCTTTCACCGACTCC-3′). To identify the site of Mu insertions in alleles ra2-mum1 and ra2-mum2, a Mu_out primer (5′-AGAGAAGCCAACGCCWCGCCTCYATTTCGTC-3′) in combination with 5′_Zm_LOB_F (5′-CCCTCCCCTTTCTATTTCCATTG-3′) and 5′_Zm_LOB_R (5′-TGGCGAACTTCTGCGGCTC-3′) or 5′_insitu_R (5′-TGGCGAACTTCTGCGGCTC-3′) was used. The ra2-dm allele was amplified and sequenced with primers 5′_Zm_LOB_F and full_Zm_LOB_R.
To obtain a full genomic sequence, a ra2-containing BAC (b0097N14) was restricted with EcoRI, and DNA gel blot hybridization with the eb3 probe was performed. The hybridizing band was gel extracted, purified with a QIAquick column (Qiagen), and cloned into an EcoRI-linearized pBluescript SK+ (Stratagene) plasmid for sequencing.
The ra2 transcript ends were amplified and sequenced using a tassel cDNA library. PCR fragments were generated with T7 and the internal primer EB3_R (5′-CGTCCTGCCTTACGCTTCCG-3′) to obtain the 3′ end. Primer pAD_GAL4 (5′-AAAAAGAGATCGAATTAGGATCCTCTG-3′) and the internal primer 5_end (5′-CAATGGAAATAGAAAGGGGAGGG-3′) were used for the 5′ end. All PCR products were cloned into a pGEM-T Easy (Promega) vector for sequencing.
Gene Expression
RNA from inflorescences measuring between 0.5 and 1 cm was extracted using the Trizol (Invitrogen) method, and poly(A)+ was prepared using DyNAbeads (Dynal Biotech). Approximately 4 μg of poly(A)+ was loaded in each lane of a formaldehyde denaturing gel, and the blotted membrane was hybridized with the eb3 probe. The same membrane was stripped and hybridized with a ra1 probe. As a control to estimate the relative amount of RNA in the lanes, the membrane was reprobed with a polyubiquitin fragment.
Five micrograms of DNase-treated total RNA were used for cDNA synthesis, and first strand cDNA was made using a SuperScript III kit (Invitrogen) following the manufacturer's directions with an oligo(dT)20 primer. One-thirtieth of the reaction volume was used as template for amplifications, and actin was used as a control. Twenty-six cycles were used for all genes except for actin, which was amplified with 32 cycles. Primers used for amplifications were gn1_F (5′-CGGTGTCGTCCTCGTCCTTC-3′), gn1_R (5′-TCAGCCTCTCCAGCAGGTCC-3′), rs1_F (5′-TTCAGAGACGGAGAAGATTGC-3′), rs1_R (5′-CAGGAGAACTACAAGCCATGC-3′), actin_F (5′-AAGTACCCGATTGAGCATGG-3′), and actin_R (5′-GATGGAGTTGTACGTGGCCT-3′). The lg3 primers were as published before (Schneeberger et al., 1998). Levels of kn1 transcript were estimated by hybridization to a blotted membrane with ∼10 μg of total RNA per lane from ra2 alleles and wild-type tissue.
Scanning Electron Microscopy
Maize tissue for scanning electron microscopy was fixed in FAA (50% ethanol, 5% acetic acid, 3.7% formaldehyde, and 0.5% Triton) for 1 h at 4°C and dehydrated in an ethanol series to 100% ethanol. The samples were critical point dried, sputter coated with gold palladium for 45 s, and viewed on a Hitachi S-4700 scanning electron microscope at an accelerating voltage of 5 kV.
Searches for Other LOB Genes and Phylogenetic Analysis
We searched for other maize LOB genes in The Institute for Genomic Research EST database (http://tigrblast.tigr.org/tgi/) and the Iowa State University genomic sequences database (http://maize.ece.iastate.edu/). Rice LOB genes were identified using the Gramene database (www.gramene.org). We selected nonredundant sequences and aligned them as amino acids using ClustalX (Thompson et al., 1997). A Bayesian phylogenetic analysis was conducted using MrBayes version 3.0b4 (Huelsenbeck and Ronquist, 2001) on the Clustal-aligned amino acid data set of the LOB domains of all Arabidopsis thaliana LOB genes and the rice and maize LOB genes found in the databases listed above. The default settings were used for initial conditions, and four Markov chain Monte Carlo chains were allowed to run for 3 million generations. Trees from the Bayesian analysis were summarized in PAUP* 4.0b10 (Swofford, 2001) after discarding the first 50,000 generations.
In Situ Hybridizations
The protocol of Jackson (1992) was used with the following modifications. Immature inflorescences were fixed in FAA for 1 h. To make ra2 RNA probes, fragments were amplified from a tassel library and the PCR products were cloned into the pGEM-T Easy plasmid. Probes for barley (Hordeum vulgare), sorghum (Sorghum bicolor), and rice in situ hybridization were amplified using cDNA from immature inflorescences as template and consisted of the entire ORF. Plasmids with the appropriate orientation were linearized, and an RNA probe was synthesized using a DIG RNA labeling mix (Roche Diagnostics). Hybridization and incubations with antibody and detection solution were done by placing two slides together, face to face, with 200 μL of the corresponding solution. Hybridization was performed overnight at 55°C, and the slides were then washed twice in 0.2× SSC for 30 min each at 55°C and twice in 0.5 M NaCl, 10 mM Tris, and 1 mM EDTA for 5 min at 37°C and RNase treated for 20 min at 37°C. Detection was performed by incubating the slides for 1 h with Anti-DIG antibody (Roche Diagnostics) diluted 1/500 in buffer A (1% BSA, 100 mM Tris, pH 7.5, 150 mM NaCl, and 0.3% Triton X-100). Slides were then washed four times for 20 min each at room temperature with buffer A and once with detection buffer (0.1 M Tris, pH 9.5, 0.1 M NaCl, and 50 mM MgCl2). Final detection was performed by adding 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium (Roche Diagnostics) in detection buffer, and the reaction was allowed to proceed for 24 to 48 h. Negative controls consisted of hybridizing a sense probe on wild-type tissue and an antisense probe on ra2-R tissue. To rule out the possibility that the expression pattern observed with the full-length probe was due to cross-hybridization with other maize LOB genes, we repeated the experiment with an antisense eb3 probe on wild-type tassels.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ327701 (maize), DQ327702 (barley), and DQ327703 (sorghum).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogeny of LOB Genes.
Supplemental Figure 2. Expression of knox Genes in ra2 Mutant Inflorescences.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Science Foundation (DBI-0110189) to S.H., R.M., and T.R. and by the USDA–Agricultural Research Service to S.H. E.B. was supported by National Science Foundation Grant DBI-0310210. We thank Paula McSteen for discovering the ra2-mum4 allele, Don McCarty and the UniformMu National Science Foundation project for allowing us to screen their Mu lines, Tracy Yamawaki for the initial ra2 mapping, Delilah Wood for use of the scanning electron microscopy facility, Devin O'Connor for help with ra1 in situ analysis, and Jennifer Fletcher for reviewing the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sarah Hake (maizesh@nature.berkeley.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.039032.
References
- Bonnett, O.T. (1940). Development of the staminate and pistillate inflorescences of sweet corn. J. Agric. Res. 60 25–33. [Google Scholar]
- Booker, J., Sieberer, T., Wright, W., Williamson, L., Willett, B., Stirnberg, P., Turnbull, C., Srinivasan, M., Goddard, P., and Leyser, O. (2005). MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 8 443–449. [DOI] [PubMed] [Google Scholar]
- Chalfun-Junior, A., Franken, J., Mes, J.J., Marsch-Martinez, N., Pereira, A., and Angenent, G.C. (2005). ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal-distal patterning in Arabidopsis petals. Plant Mol. Biol. 57 559–575. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Meeley, R., and Hake, S. (1998). The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 12 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Muszynski, M., Kellogg, E., Hake, S., and Schmidt, R.J. (2002). The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298 1238–1241. [DOI] [PubMed] [Google Scholar]
- Clifford, H.T. (1987). Spikelet and floral morphology. In Grass Systematics and Evolution, T.R. Soderstrom, K.W. Hilu, C.S. Campbell, and M.E. Barkworth, eds (Washington, DC: Smithsonian Institution Press), pp. 21–30.
- Coen, E.S., Romero, J.M., Doyle, S., Elliot, R., Murphy, G., and Carpenter, R. (1990). FLORICAULA: A homeotic gene required for flower development in Antirrhinum majus. Cell 63 1311–1322. [DOI] [PubMed] [Google Scholar]
- Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386 485–488. [DOI] [PubMed] [Google Scholar]
- Douglas, S.J., Chuck, G., Dengler, R.E., Pelecanda, L., and Riggs, C.D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, R.A., Beadle, G.W., and Fraser, A.C. (1935). A summary of linkage studies in maize. Cornell Univ. Agric. Exo. Sta. Mem. 180 1–83. [Google Scholar]
- Friml, J., et al. (2004). A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306 862–865. [DOI] [PubMed] [Google Scholar]
- Gallavotti, A., Zhao, Q., Kyozuka, J., Meeley, R.B., Ritter, M.K., Doebley, J.F., Pe, M.E., and Schmidt, R.J. (2004). The role of barren stalk1 in the architecture of maize. Nature 432 630–635. [DOI] [PubMed] [Google Scholar]
- Galweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282 2226–2230. [DOI] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group (2001). Phylogeny and subfamilial classification of the Poaceae. Ann. Mo. Bot. Gard. 88 373–457. [Google Scholar]
- Greb, T., Clarenz, O., Schafer, E., Muller, D., Herrero, R., Schmitz, G., and Theres, K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, L., McSteen, P., Doebley, J., and Hake, S. (2002). Expression patterns and mutant phenotypes of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162 1927–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J.P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17 754–755. [DOI] [PubMed] [Google Scholar]
- Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Shibata, Y., Gomi, K., Umemura, I., Hasegawa, Y., Ashikari, M., Kitano, H., and Matsuoka, M. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43 467–478. [DOI] [PubMed] [Google Scholar]
- Jackson, D. (1992). In situ hybridization in plants. In Plant Molecular Pathology: A Practical Approach, S.J. Gurr, M.J. McPherson, and D.J. Bowles, eds (Oxford, UK: Oxford University Press), pp. 163–174.
- Klein, P.E., Klein, R.R., Vrebalov, J., and Mullet, J.E. (2003). Sequence-based alignment of sorghum chromosome 3 and rice chromosome 1 reveals extensive conservation of gene order and one major chromosomal rearrangement. Plant J. 34 605–621. [DOI] [PubMed] [Google Scholar]
- Komatsu, K., Maekawa, M., Ujiie, S., Satake, Y., Furutani, I., Okamoto, H., Shimamoto, K., and Kyozuka, J. (2003). LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 100 11765–11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Li, X., et al. (2003). Control of tillering in rice. Nature 422 618–621. [DOI] [PubMed] [Google Scholar]
- Lin, W.C., Shuai, B., and Springer, P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Wang, S., Yu, X., Yu, J., He, X., Zhang, S., Shou, H., and Wu, P. (2005). ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 43 47–56. [DOI] [PubMed] [Google Scholar]
- McSteen, P., and Hake, S. (2001). barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128 2881–2891. [DOI] [PubMed] [Google Scholar]
- McSteen, P., Laudencia-Chingcuanco, D., and Colasanti, J. (2000). A floret by any other name: Control of meristem identity in maize. Trends Plant Sci. 5 61–66. [DOI] [PubMed] [Google Scholar]
- McSteen, P., and Leyser, O. (2005). Shoot branching. Annu. Rev. Plant Biol. 56 353–374. [DOI] [PubMed] [Google Scholar]
- Moore, G., Devos, K.M., Wang, Z., and Gale, M.D. (1995). Cereal genome evolution. Grasses, line up and form a circle. Curr. Biol. 7 737–739. [DOI] [PubMed] [Google Scholar]
- Neuffer, M.G., Coe, E.H., and Wessler, S.R. (1997). Mutants of Maize. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Okushima, Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Palmer, L.E., Rabinowicz, P.D., O'Shaughnessy, A.L., Balija, V.S., Nascimento, L.U., Dike, S., de la Bastide, M., Martienssen, R.A., and McCombie, W.R. (2003). Maize genome sequencing by methylation filtration. Science 302 2115–2117. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Rabinowicz, P.D., Schutz, K., Dedhia, N., Yordan, C., Parnell, L.D., Stein, L., McCombie, W.R., and Martienssen, R.A. (1999). Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat. Genet. 23 305–308. [DOI] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., et al. (2002). The genome sequence and structure of rice chromosome 1. Nature 420 312–316. [DOI] [PubMed] [Google Scholar]
- Schneeberger, R., Tsiantis, M., Freeling, M., and Langdale, J.A. (1998). The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125 2857–2865. [DOI] [PubMed] [Google Scholar]
- Schumacher, K., Schmitt, T., Rossberg, M., Schmitz, G., and Theres, K. (1999). The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 96 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Pena, C.G., and Springer, P.S. (2002). The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M.S., and Hake, S. (2003). The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., Llaca, V., and Messing, J. (2002). Mosaic organization of orthologous sequences in grass genomes. Genome Res. 12 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves, T.A., and Sussex, I.M. 1989. Patterns in Plant Development. (Cambridge, UK: Cambridge University Press).
- Swofford, D.L. (2001). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. (Sunderland, MA: Sinauer Associates).
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTALX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry, J.M., Goodman, M.M., Doebley, J., Kresovich, S., Nielsen, D., and Buckler, E.S. (2001). Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28 286–289. [DOI] [PubMed] [Google Scholar]
- Vollbrecht, E., Springer, P.S., Goh, L., Buckler, E.S., and Martienssen, R. (2005). Architecture of floral branch systems in maize and related grasses. Nature 436 1119–1126. [DOI] [PubMed] [Google Scholar]
- Walsh, J., and Freeling, M. (1999). The liguleless2 gene of maize functions during the transition from the vegetative to the reproductive shoot apex. Plant J. 19 489–495. [DOI] [PubMed] [Google Scholar]
- Weberling, F. (1989). Morphology of Flowers and Inflorescences. (Cambridge, UK: Cambridge University Press).
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859. [DOI] [PubMed] [Google Scholar]
- Yuan, Y., SanMiguel, P.J., and Bennetzen, J.L. (2003). High-Cot sequence analysis of the maize genome. Plant J. 49 249–255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.