Abstract
In higher plants, leaves initiate in constant spatial and temporal patterns. Although the pattern of leaf initiation is a key element of plant shoot architecture, little is known about how the time interval between initiation events, termed plastochron, is regulated. Here, we present a detailed analysis of plastochron2 (pla2), a rice (Oryza sativa) mutant that exhibits shortened plastochron and precocious maturation of leaves during the vegetative phase and ectopic shoot formation during the reproductive phase. The corresponding PLA2 gene is revealed to be an orthologue of terminal ear1, a maize (Zea mays) gene that encodes a MEI2-like RNA binding protein. PLA2 is expressed predominantly in young leaf primordia. We show that PLA2 normally acts to retard the rate of leaf maturation but does so independently of PLA1, which encodes a member of the P450 family. Based on these analyses, we propose a model in which plastochron is determined by signals from immature leaves that act non-cell-autonomously in the shoot apical meristem to inhibit the initiation of new leaves.
INTRODUCTION
The arrangement of leaves and branches constitutes a major component of plant architecture. Since branches are formed in the axils of leaves, the pattern of leaf initiation has special significance. Leaves are generated from the shoot apical meristem (SAM) in constant spatial (phyllotaxy) and temporal (plastochron) patterns. Thus, knowledge of the regulatory mechanisms of plastochron and phyllotaxy would greatly enhance our understanding of plant architecture. To date, however, neither of these traits has been thoroughly analyzed.
Changes in the timing and the rate of developmental programs, known as heterochrony, can lead to significant morphological changes in organisms and play an important role in evolution (Gould, 1977). In plants, the mechanism of heterochrony or temporal regulation of development is poorly understood, although genes have been identified in several plant species that regulate juvenile–adult phase change (Poethig, 1988; Telfer and Poethig, 1998; Asai et al., 2002). With respect to plastochron, several genes have been identified: PLASTOCHRON1 (PLA1) in rice (Oryza sativa; Miyoshi et al., 2004), terminal ear1 (te1) in maize (Zea mays; Veit et al., 1998), and ALTERED MERISTEM PROGRAM1 (AMP1), PHYTOCHROME B (PHYB), and SERRATE (SE) in Arabidopsis thaliana (Reed et al., 1993; Helliwell et al., 2001; Prigge and Wagner, 2001). pla1, te1, and amp1 show shorter plastochron. By contrast, phyB and se show longer plastochron than the wild type. With each of these genes encoding a distinct class of protein and showing distinct loss-of-function phenotypes, the regulation of plastochron appears complex.
Regulation of phyllotaxy is to some degree independent of plastochron because some plastochron mutants do not exhibit phyllotactic change (Itoh et al., 1998). However, the normal initiation of a leaf primordium requires simultaneous determination of its position and timing. In addition, te1 shows both shortened plastochron and, to a lesser extent, abnormal phyllotaxy (Veit et al., 1998). Accordingly, plastochron and phyllotaxy appear to depend, at least in part, on a common regulatory mechanism.
Two general types of models have been proposed for explaining phyllotactic pattern. The first model, referred to as the inhibitor model (reviewed in Steeves and Sussex, 1989), proposes that existing primordia produce a diffusible inhibitor of leaf formation and that the new primordium can only be initiated at the position where the inhibitor concentration is at a minimum. This model is supported by surgical experiments, in which incisions that separated newly formed leaf primordia from leaf-forming regions of the apex shifted the position of leaf formation toward preexisting leaves (Snow and Snow, 1931). The second model, termed the biophysical model, proposes that physical forces determine phyllotaxy (Green, 1985). This model has received support from experiments in which treatment with expansin, which normally acts to allow cell wall expansion, altered phyllotaxy (Fleming et al., 1997; Pien et al., 2001).
Recently, the auxin transport model has been developed based on the gradients of this plant hormone that are presumed to exist in the shoot apex and the effects that artificial manipulation of auxin levels have on leaf initiation. In this model, each leaf primordium arises at a position where the auxin flows from surrounding regions to create a local concentration maximum (Reinhardt et al., 2000, 2003). Subsequently, the primordium functions as a sink for auxin, thereby depleting auxin from surrounding cells. This model can be considered to be a type of inhibitory field model, since newly formed leaves would inhibit leaf initiation in the neighboring region by reducing auxin content below a critical threshold.
A recent study showed that cytokinin is also involved in phyllotaxy. Maize abphyl1 (abph1) encodes a cytokinin-inducible A-type response regulator, which is thought to act as a negative feedback regulator of cytokinin signaling (Giulini et al., 2004). abphl1 mutations change the normal alternate phyllotaxy (leaves initiated singly on alternate sides of the shoot) to a decussate pattern (successive pairs of leaves initiated at 90° with respect to each other) (Jackson and Hake, 1999). This suggests that cytokinin signaling is involved in leaf initiation through some form of crosstalk with auxin signaling pathways.
As previously discussed, three types of genes whose recessive mutations cause shortened plastochron, including rice PLA1, maize te1, and Arabidopsis AMP1 have been cloned (Chaudhury et al., 1993; Itoh et al., 1998; Veit et al., 1998; Helliwell et al., 2001; Miyoshi et al., 2004). The loss-of-function mutant pla1 shows shortened plastochron and conversion of rachis branches to vegetative shoots, but the phyllotaxy is not modified. pla1 plants have an enlarged SAM that is associated with more frequent cell divisions (Itoh et al., 1998), suggesting that cell division rate may influence plastochron. Supporting this hypothesis, transgenic tobacco (Nicotiana tabacum) plants constitutively expressing cyclin D showed an accelerated rate of cell division and shortened plastochron (Cockcroft et al., 2000). PLA1 encodes a member of the plant-specific subfamily of cytochrome P450 monooxygenases and is expressed in young leaf primordia but not in the SAM (Miyoshi et al., 2004). This suggests that signals from leaf primordia can act non-cell-autonomously to regulate leaf initiation in the SAM. te1 and amp1 show shortened plastochron and abnormal phyllotaxy (Chaudhury et al., 1993; Veit et al., 1998) but not the heterochrony in the reproductive phase observed in pla1. te1 encodes RNA binding protein, while AMP1 encodes Glu carboxypeptidase (Veit et al., 1998; Helliwell et al., 2001). te1 transcripts can be seen in the SAM and young leaf primordia, while AMP1 is expressed in all organs. How these three genes might influence plastochron remains unclear.
We recently isolated several new mutant alleles of a rice gene, termed PLA2, which show shortened plastochron and the conversion of inflorescence branches to vegetative structures. In this article, we describe phenotypes of pla2, the genetic relationship between PLA2 and PLA1, and molecular identification of PLA2. Double mutant phenotypes suggest that PLA2 and PLA1 act independently. By positional cloning, we show that PLA2 encodes MEI2-like RNA binding protein that is likely to be a rice orthologue of te1. Interestingly, despite their similarity, pla2 and te1 show distinct phenotypes, suggesting that differences in the activities of the normal genes may be partly responsible for differences between the shoot architectures of rice and maize. Detailed analyses indicate that the primary function of PLA2 resides in regulating leaf maturation, which in turn plays a major role in regulating plastochron in rice.
RESULTS
Phenotypes of pla2 Mutants
Plastochron and Leaf Size
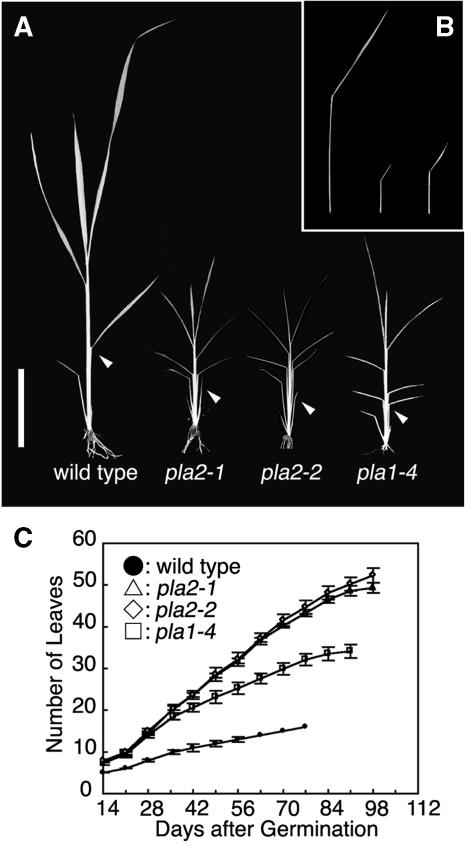
Rice plants normally initiate leaves from the SAM at regular intervals in 1/2 alternate phyllotaxy and, like many grasses, form several juvenile leaves in embryo before dormancy. In both the wild type and pla2, three leaves were present in mature embryos. After germination, both pla2-1 and pla2-2 showed an increased rate of leaf emergence compared with the wild type (Figures 1A and 1C). Since mature wild-type and pla2 embryos have the same number of leaf primordia, the more frequent leaf emergence indicates a shorter plastochron for subsequently formed leaves in the pla2 mutant. Given the similarity of pla2-1 and pla2-2 phenotypes, we chose to focus on pla2-1 in this article. Plastochrons of the wild type and pla2-1 were nearly constant throughout the vegetative phase at 5.0 and 1.8 d, respectively (Figure 1C). For comparison, we examined phenotypes of pla1 mutants, which were reported previously (Itoh et al., 1998; Miyoshi et al., 2004). The plastochron of pla2 is significantly shorter than pla1 (1.8 versus 2.3). Furthermore, in contrast with pla1, in which the angle between the blade and sheath is increased for all leaves, the third leaf of pla2 appears erect due to an irregular blade-sheath boundary (see Supplemental Figure 1 online). The transition of vegetative to reproductive phase was also delayed in pla2-1 and pla2-2. Together with an extended vegetative period, pla2 produced threefold as many leaves as the wild type (49 versus 16 leaves). Assuming that this increase reflects loss of normal PLA2 function, these results suggest that the PLA2 gene normally acts to inhibit leaf initiation. In addition to a shortened plastochron, pla2 plants exhibited significantly smaller leaves than the wild type with respect to the length of blade and sheath and also the width of leaf blade (Figure 1B; see Supplemental Figures 2A to 2C online). As the size of epidermal cells of the third leaf in pla2 was almost comparable to that in the wild type, this size reduction was exclusively due to the reduction in the number of cells (see Supplemental Figure 3 online). The possibility of a causal relationship between leaf size and plastochron is reinforced by the phenotype of pla1, which also shows reduced leaf size and plastochron but not as severe as seen for pla2 (Figure 1B; see Supplemental Figures 2A to 2C online). Thus, there exists a positive correlation between plastochron and leaf size in the wild type, pla1, and pla2.
Figure 1.
Vegetative Phenotypes of Wild-Type, pla2, and pla1 Plants.
(A) Seedlings at 2 weeks after germination. The lamina joint of the third leaf does not bend in pla2-1 and pla2-2, although they do bend in the wild type and pla1-4. Arrowheads indicate the lamina joints of the third leaf. Bar = 5 cm.
(B) Phenotype of the last vegetative leaf (flag leaf) of the wild type, pla2-1, and pla1-4 from left to right. Leaf size is reduced strongly in pla2-1 and pla1-4.
(C) Increase in the number of emerged leaves during development. Closed circles, open triangles, open diamonds, and open squares represent the wild type, pla2-1, pla2-2, and pla1-4, respectively. Bars indicate ± sd.
Precocious Maturation of pla2 Leaves
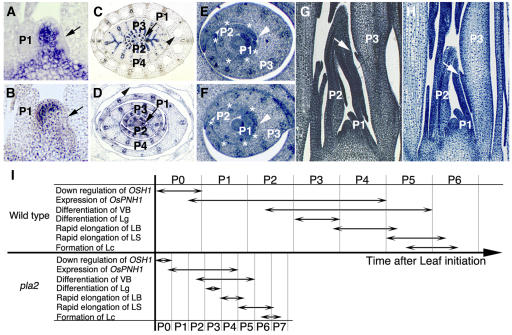
To elucidate the function of PLA2 in leaves, we compared the timing of developmental landmarks for the leaves of wild-type and pla2 plants. To represent different leaves, we used a plastochron numbering (PN) system; P1 represents the youngest primordium, P2 the next youngest, etc. (Itoh et al., 2005). P0 is defined as founder cell stage, which immediately precedes appearance of the primordium bulge on the SAM. In wild-type plants, a series of developmental events can be related to specific PNs. Since the sequence of development for the leaf has been described in detail previously (Itoh et al., 2005), only major events are outlined here. Cells at the P0 stage are recruited as leaf founder cells, an event marked by downregulation of O. sativa HOMEOBOX1 (OSH1) (Figures 2A and 2I). OSH1 is a rice orthologue of maize KNOTTED1 and is expressed in indeterminate cells (Sato et al., 1996). At this stage, O. sativa PNH1, which is a homologue of Arabidopsis PINHEAD/ZWILL and is expressed in the developing vascular bundles and presumptive regions of bundle sheath, is expressed first (Figures 2C and 2I; Nishimura et al., 2002). This expression lasts until P4 stage. Two margins of the leaf primordium overlap and enclose the SAM at P2 stage (Figures 2E and 2I). At this stage, vascular bundles become visible (Figures 2E and 2I). Formation of vascular bundles continues until P5 (Figures 2C and 2I). A protrusion marking the ligule primordium appears at P3 at the boundary between leaf blade and sheath on the adaxial side (Figures 2G and 2I). After differentiation of the ligule primordium, the leaf blade elongates rapidly. This continues from P4 to P5 (Figure 2I). When the leaf blade has nearly stopped elongating, the more basal leaf sheath starts to elongate rapidly until it reaches P6 (Figure 2I). After the leaf sheath elongation has finished, the leaf blade bends away from the shoot axis at its junction with the leaf sheath. At P5, lacunae (air spaces) are formed (Figures 2C and 2I).
Figure 2.
Accelerated Leaf Development in pla2 Plants.
(A), (C), (E), and (G) The wild type.
(B), (D), (F), and (H) pla2-1.
(A) and (B) Expression of OSH1 in the shoot apex. Downregulation of OSH1 is observed in P0 (arrows).
(C) and (D) Expression of Os PNH1 in the shoot apex. The arrows indicate the expression in P1. Lacuna formation is observed in the leaf sheath of P5 in the wild type and P6 in pla2-1 (arrowheads).
(E) and (F) Cross section of shoot apex. Asterisks indicate differentiating procambial strands. The margins of the P2 sheath are overlapping (arrowheads).
(G) and (H) Longitudinal sections of the shoot apex. Arrows indicate the protrusion of ligule primordium.
(I) Schematic representation of the leaf developmental programs in the wild type and pla2. VB, vascular bundle; Lg, ligule; LB, leaf blade; LS, leaf sheath; Lc, lacuna.
In pla2-1, leaves exhibited a normal sequence of PN-related development (Figures 2B, 2D, 2F, 2H, and 2I), except that leaf sheath elongation lasted until P7 (Figure 2I) and lacunae were visible at P6 (Figures 2D and 2I). The downregulation of OSH1 was observed in the presumptive P0 region but not in the presumptive P1 region (Figure 2B). Accordingly, pla2-1 leaves undergo a normal sequence of cell/tissue differentiation. However, since the duration of each PN stage is much shorter in pla2-1 (1.8 d) than in the wild type (5.0 d), these results indicate that pla2-1 leaves mature in a shorter time than wild-type leaves (i.e., pla2-1 leaves mature in just 12.6 d compared with 30 d for wild-type leaves). Therefore, we can conclude that leaf maturation is accelerated in pla2 (Figure 2I). This suggests that PLA2 not only inhibits leaf initiation but also suppresses the progression of programs associated with leaf development.
SAM
Since leaves are initiated on the flank of the SAM, abnormal leaf initiation might be linked to SAM abnormalities. The SAM of pla2 is much larger than that of the wild type (Figures 3A and 3B, Table 1); however, the shape of the SAM, represented by the ratio of the height to the width, was comparable among pla2, pla1, and the wild type (Table 1). Next, to gain insight into whether abnormal leaf initiation was associated with changes in cell division, we performed in situ hybridization experiments with the rice histone H4 gene, which is specifically expressed in the S phase of the cell cycle. The pla2-1 SAM showed an increase in the mean number of cells expressing histone H4 per median longitudinal section of SAM (3.8 in pla2-1 versus 0.7 in the wild type; Figures 3D and 3E, Table 1). This shows that SAM of pla2 has a higher rate of cell division than the wild type, which could compensate for the rapid production of leaf primordia. pla1-4 has an even larger SAM than pla2-2 (Table 1), but the cell division activity was lower than that of pla2-2 (Table 1). This indicates that shortened plastochron is more closely correlated with increased cell divisions than increases in SAM size.
Figure 3.
SAM and Histone H4 Expression in Wild-Type, pla2, and pla1 Plants at 3 Weeks after Germination.
(A) and (D) The wild type.
(B) and (E) pla2-1.
(C) and (F) pla1-4.
(A) to (C) Cleared image of SAMs.
(D) to (F) Expression of histone H4.
Bars = 50 μm.
Table 1.
Phenotypes of the Wild Type, pla2, and pla1 in Vegetative Phase
| Taichung 65 | pla2-1 | pla2-2 | pla1-3 | pla1-4 | |
|---|---|---|---|---|---|
| Plastochron | 5.0 ± 0.0 | 1.8 ± 0.0 | 1.8 ± 0.0 | 3.1 ± 0.1 | 2.3 ± 0.1 |
| SAM width (A) (μm) | 62.9 ± 5.8 | 77.0 ± 8.8 | 72.5 ± 5.5 | 69.5 ± 4.0 | 73.5 ± 3.1 |
| SAM height (B) (μm) | 34.1 ± 3.0 | 43.1 ± 8.2 | 39.9 ± 4.1 | 36.9 ± 3.9 | 40.6 ± 3.2 |
| SAM shape (B/A) | 0.54 ± 0.04 | 0.56 ± 0.07 | 0.55 ± 0.03 | 0.53 ± 0.04 | 0.55 ± 0.05 |
Values indicate the means of 20 experiments ± sd.
Internode Elongation
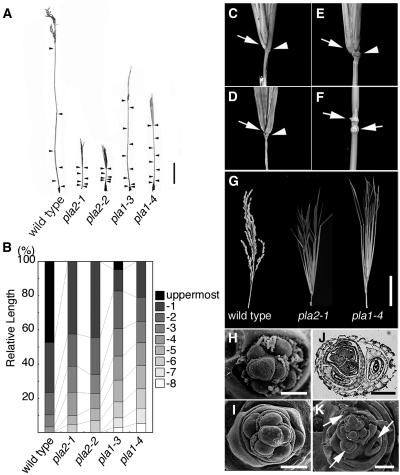
In rice, only the upper four or five internodes elongate substantially at around the vegetative–reproductive transition. Internodes are designated as follows: the internode just below the uppermost one is designated the -1 internode, and the third, fourth, and fifth internodes counted from the top are -2, -3, and -4, respectively (Sunohara et al., 2003). Normally, internode length decreases basipetally in a geometric ratio (i.e., each internode is normally approximately twice as long as the next lower one) (Figures 4A and 4B). Both pla2-1 and pla2-2 mutants were extremely short (<10% of wild-type plant height) and showed an increased number and abnormal pattern of elongated internodes (Figures 4A and 4B). In both pla2-1 and pla2-2, elongation was observed up to the -6 internode, but the uppermost internode did not show substantial elongation (Figures 4C and 4D). Although the geometric pattern of internode elongation was roughly conserved in pla2 (except the uppermost one), the length of each internode was reduced (Figures 4A and 4B). The increase in the number of elongated internodes and the concomitant reduction of each internode length are reminiscent of events occurring in leaves: the increase in the number of leaves and the reduction of leaf size.
Figure 4.
Reproductive Phenotypes of Wild-Type, pla2, and pla1 Plants.
(A) Elongation pattern of internodes. The culm of the wild-type plant is longer than those of pla mutants. Arrowheads indicate nodes.
(B) Schematic representation of the elongation patterns of internodes in the wild type, pla2-1, pla2-2, pla1-3, and pla1-4.
(C) to (E) The nonelongating uppermost internodes of pla2-1, pla2-2, and pla1-4, respectively. Arrows indicate the neck nodes, and arrowheads indicate the flag leaf nodes. The flag leaf is removed in (C) and (D) to clarify the uppermost internode.
(F) Occasional nonelongation of internode in pla1-4. Arrows indicate two successive nodes.
(G) Inflorescences of the wild type, pla2-1, and pla1-4 from left to right. In pla2-1 and pla1-4, vegetative shoots emerge instead of primary branches.
(H) Scanning electron microscopy image of primary branch primordia in the wild type.
(I) Scanning electron microscopy image of primary branch primordia in pla2-1.
(J) Cross section of pla2-1 young inflorescence showing ectopic shoots converted from primary branch primordia.
(K) Scanning electron microscopy image of primary branch primordia in pla1-4. Arrows indicate ectopic shoots.
Bars = 10 cm in (A), 5 cm in (G), 100 μm in (H), (I), and (K), and 200 μm in (J).
The cells of the internodes are somewhat smaller in pla2 than in the wild type (see Supplemental Figure 3 online). By contrast, the number of cells in pla2 internodes was significantly reduced. However, as PLA2 is not expressed in stem (Figure 6C), shortened internodes would not be a direct effect of pla2 mutation but a secondary effect of shortened leaves.
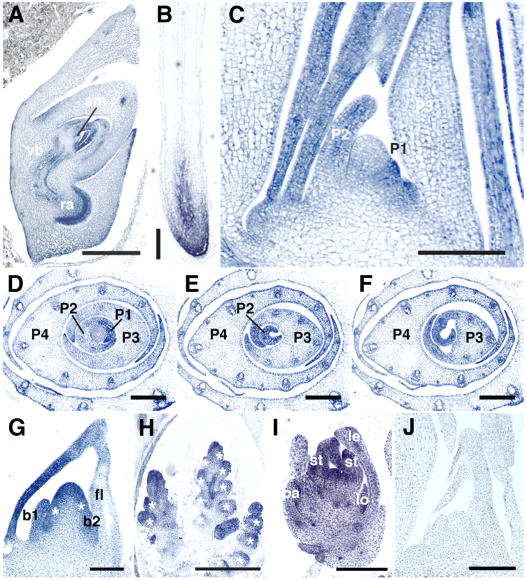
Figure 6.
In Situ Localization of PLA2 Transcripts.
(A) to (I) Antisense probe.
(J) Sense probe.
(A) Longitudinal section of embryo at 10 d after pollination. sm, SAM; vb, vascular bundle; ra, radicle.
(B) Crown root at 1 week after germination.
(C) Longitudinal section of 3-week-old shoot apex.
(D) to (F) Serial transverse sections of 3-week-old shoot apex. Panels are from the bottom to the top. PLA2 signals are strong in marginal regions.
(G) Inflorescence apex at stage In2. Two primary rachis branches (asterisks) are formed. b1 and b2 indicate first and second bracts, respectively.
(H) Inflorescence apex at stage In5. Asterisks indicate spikelet primordia.
(I) Spikelet at stage Sp6. le, lemma; pa, palea; lo, lodicule; st, stamen.
(J) Longitudinal section of shoot apex. No hybridization signals are observed with sense probe.
Bars = 500 μm in (A), 250 μm in (B) and (H), 100 μm in (C), (G), and (J), and 150 μm in (D) to (F) and (I).
The pla1 mutants showed different internode elongation patterns from pla2 and, while dwarf, were significantly taller than pla2 (Figure 4A). Substantial elongation occurred until the -8 internode, but the internode lengths were not in a geometric ratio (Figures 4A and 4B). Frequently, a lower internode was longer than higher ones. The uppermost internode did not elongate in the strong allele pla1-4 (Figure 4E) and elongated only slightly in the mild allele pla1-3. Occasionally some internodes did not elongate, while immediately adjacent upper and lower ones did (Figure 4F).
Inflorescences
After the vegetative–reproductive transition, the rachis meristem of the wild type differentiates bracts (small leaf-like organs associated with floral branches) and branch primordia in 2/5 spiral manner (Figure 4H) and eventually aborts. Normally, bracts remain rudimentary, while the primary rachis branches produce secondary rachis branches and spikelets. In pla2, the reproductive phase starts almost normally, as judged from the formation of flag leaf (the last vegetative leaf morphologically distinguished from other leaves) and bracts. The pla2 inflorescence, however, produced several vegetative shoots with enlarged bracts (Figure 4G) instead of primary branches. A close examination of a young pla2 inflorescence revealed that primary branch primordia were initiated normally in spiral phyllotaxy (Figure 4I) but that several proximal primordia were converted to vegetative shoots (Figure 4J), while more distal primordia aborted. These phenotypes resemble those of pla1 (Figures 4G and 4K) and can be interpreted to result from a failure of certain elements of the vegetative program to terminate properly, leading to their coexpression with reproductive programs of development.
Identification of the PLA2 Gene
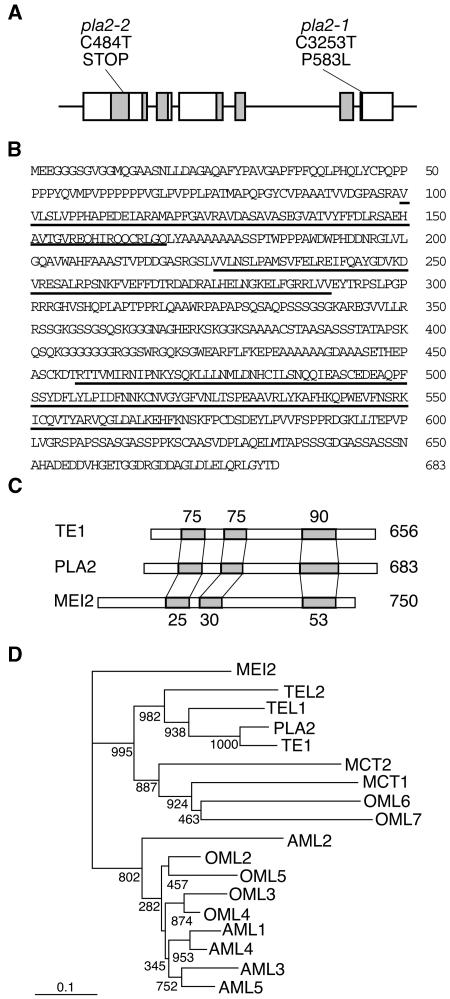
To investigate the molecular function of PLA2, we isolated the PLA2 gene by a map-based cloning strategy. The PLA2 locus is closely linked with the marker R3202 on chromosome 1. Further fine mapping confined the PLA2 locus to a 66-kb region in the P1 artificial chromosome clone P0497A05. This region was predicted to contain nine genes by an annotated rice gene database. Genome sequencing revealed that two independent pla2 alleles had mutations in a putative gene designated P0497A05.11, comprising six exons and five introns (Figure 5A). In pla2-1, a C-to-T single base change causes Pro-to-Leu amino acid substitution in the sixth exon (Figure 5A). In pla2-2, a C-to-T single base change generates a stop codon in the first exon. To clarify whether the candidate gene represented PLA2, we performed a complementation test by introducing a 7.2-kb genomic fragment containing the candidate gene into pla2-2 homozygous plant. As this fragment rescued the mutant phenotypes (see Supplemental Figure 4 online), we concluded that P0497A05.11 is PLA2. We did not observe any additional phenotype in the rescued plants.
Figure 5.
Structures of PLA2 and Related Genes.
(A) Exon/intron structure of the PLA2 gene. Six boxes indicate exons. RNA recognition motifs (RRMs) are shaded. Locations of the two pla2 mutations are indicated: base and amino acid substitutions in exon 6 in pla2-1 and base substitution and non-sense mutation in pla2-2.
(B) Deduced amino acid sequence of PLA2 protein. Underlining indicates RRMs.
(C) Comparison of PLA2, TE1, and MEI2 proteins. RRMs are shaded. Numbers above TE1 and below MEI2 represent amino acid identity between TE1 and PLA2 and MEI2 and PLA2, respectively. Numbers at the right side indicate the number of amino acids.
(D) Phylogenetic tree of MEI2-like RNA binding proteins. Numbers at each branch point indicate bootstrap values. PLA2, OML2, OML3, OML4, OML5, OML6, and OML7 are from O. sativa, TE1 from Z. mays, TEL1, TEL2, AML1, AML2, AML3, AML4, AML5, MCT1, and MCT2 from Arabidopsis, and MEI2 from Schizosaccharomyces pombe.
The predicted PLA2 protein contains three RNA recognition motifs (RRM1, RRM2, and RRM3) and shows high similarity to the maize TE1 and the fission yeast MEI2 proteins (Figures 5B and 5C). The amino acid identities between PLA2 and TE1 in the three RRMs were 75.4, 74.6, and 89.5%, respectively, and those between PLA2 and MEI2 were 24.6, 29.9, and 52.6%, respectively (Figures 5B and 5C).
Anderson et al. (2004) have reported six MEI2-like proteins in rice and nine in Arabidopsis and divided them into two subfamilies: a MEI2-like subfamily and a TE1-like subfamily. PLA2 is identical to OML1 of the TE1-like subfamily. By BLAST search, we have identified an additional MEI2-like protein in rice, OML7, which like OML6, has only the C-terminal RRM. A phylogenetic tree based on a comparison of the highly conserved RRM3 domain indicates that PLA2 is likely to be the orthologue of TE1 (Figure 5D; see Supplemental Figure 5 online).
Expression of the PLA2 Gene
To gain more insight into the biological function of PLA2, we examined the PLA2 expression in detail. We first examined PLA2 expression by RT-PCR analysis. RNA was isolated from 3-week-old vegetative shoot apices, the inflorescence apex, the leaf blade, the leaf sheath, and the root. PLA2 was strongly expressed in shoot apex and inflorescence apex and intermediately in root, while low expression was detected in leaf blade and leaf sheath (see Supplemental Figure 6 online).
To obtain more detailed information on the spatial pattern of PLA2 expression, we performed in situ hybridization experiments. By this method, PLA2 expression was seen throughout the life cycle. In the embryo, transcripts were detected in leaf primordia, vascular bundles, and the radicle (Figure 6A). In the vegetative phase, PLA2 was expressed in crown root apices (Figure 6B) and shoot apices (Figures 6C to 6F). Although Paquet et al. (2005) reported that PLA2/Ostel1/OML1 was expressed in shoot apices but not in roots and leaves, we detected obvious expression in roots, though no mutant phenotypes were apparent in this organ. In the shoot apex, PLA2 expression was first detected in the entire early P1 primordium that later developed into midrib (Figure 6C) and then became extended to the marginal region (Figure 6C). In P2-P4 primordia, the expression was localized to marginal and distal regions and was then downregulated from basal midrib region (Figures 6D to 6F). In older leaves than P4, PLA2 transcripts could not be detected. Almost no or only low levels of PLA2 transcripts could be detected in the SAM (Figure 6C). In the early reproductive phase, PLA2 was expressed in bracts and several external layers of the rachis meristem (Figure 6G), suggesting that at this stage, PLA2 may regulate meristem identity directly. At the later stages, PLA2 was expressed in branch meristems, floral meristems, and floral organs (Figures 6H and 6I). In the control experiment hybridized with sense RNA probe, no hybridization signals were detected (Figure 6J).
Relation between PLA1 and PLA2
Although PLA1 and PLA2 encode unrelated proteins, a member of the P450 family and MEI2-like RNA binding protein, respectively, their loss-of-function mutants show very similar phenotypes. To understand how these genes are related, we assessed whether the expression and function of these genes is interdependent through expression analyses and comparisons of single versus double mutant phenotypes. In a pla2-1 mutant, PLA1 expression was localized normally in the basal abaxial region of young leaf primordia (Figures 7A and 7B). Conversely, PLA2 expression was normal in pla1-4 (Figures 7C and 7D). These results indicate that PLA1 and PLA2 are regulated independently of each other, suggesting that they function in independent pathways.
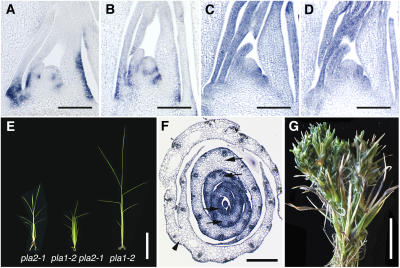
Figure 7.
Relationship between PLA1 and PLA2.
(A) Expression of PLA1 in 1-month-old wild-type shoot apex.
(B) Expression of PLA1 in pla2-1 1-month-old shoot apex.
(C) Expression of PLA2 in 3-week-old shoot apex of the wild type.
(D) Expression of PLA2 in 3-week-old shoot apex of pla1-4.
(E) Seedlings at 2 weeks after germination. pla1-2 pla2-1 initiated more leaves than the pla1-2 or pla2-1 single mutant.
(F) Transverse sections of pla1-2 pla2-1 shoot apex at 1 month after germination. Arrows indicate two successive leaves that are initiated from the same side of the SAM. Arrowhead indicates fusion of the margins of two successive leaves due to shortened plastochron.
(G) Inflorescence of pla1-2 pla2-1. pla1-2 pla2-1 produced a large number of ectopic shoots (cf. with Figure 4G).
Bars = 100 μm in (A) to (D), 5 cm in (E), 200 μm in (F), and 2.5 cm in (G).
To elucidate genetic interaction, we made pla1-2 pla2-1 double mutants. As was the case for single pla1 and pla2 mutants, no abnormalities were apparent in embryos of pla1-2 pla2-1. After germination, pla1-2 pla2-1 initiated leaves more rapidly than either single mutant (Figure 7E): the plastochron, 1.1 d, was much shorter than those of pla1-2 (2.8 d) and pla2-1 (1.8 d). The pla1-2 pla2-1 SAM was larger than wild-type SAM but smaller than each single mutant SAM (data not shown). pla1-2 pla2-1 also showed aberrant phyllotaxy. Cross sections revealed that in pla1-2 pla2-1, two successive leaf primordia were frequently formed on the same side of the SAM or with a divergence angle deviated from 180° (Figure 7F). In addition, two successive leaves could sometimes be seen fused on their margins (Figure 7F), probably due to an extremely short plastochron. None of the internodes in the double mutants showed substantial elongation (data not shown). In the reproductive phase, pla1-2 pla2-1 produced many more ectopic shoots than pla1-2 and pla2-1 (Figure 7G), despite pla1-2 being a weak allele that produces both normal inflorescence branches and a small number of ectopic shoots.
The independent regulation of gene expression and more severe phenotypes of the double mutants compared with the single mutants suggest that PLA1 and PLA2 act in the independent pathways that both contribute redundantly to the regulation of plastochron and the duration of vegetative phase.
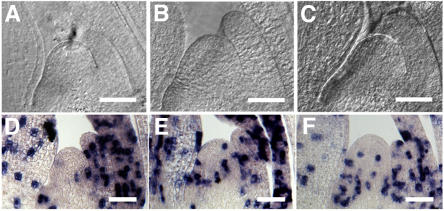
Expression Profiles of PLA2 and te1 Differ
Since rice PLA2 and maize te1 are orthologous and loss-of-function mutants for both genes are available, it was of interest to make functional comparisons of these genes. With respect to gene expression, transcripts of both genes accumulate in the marginal and distal regions of young leaf primordia but appear downregulated in the basal central region (Figures 6C, 8A, and 8B; Veit et al., 1998). However, significant expression differences are observed in early stages of leaf development. PLA2 transcripts are first observed in the central region of early P1 primordia and then extend toward the marginal region (Figure 6C), while te1 is first expressed in semicircular bands whose arms include the incipient marginal region of P0 primordia but do not include the central region (Figures 8A and 8B; Veit et al., 1998). In summary, te1 is expressed earlier than PLA2 (P0 versus P1); the expression extends from primordia margins to bracket the central region (in contrast with central-to-marginal progression in PLA2). A further difference in expression is observed in the reproductive phase, where strong PLA2 expression is seen in bracts and inflorescence meristems (Figure 6G), in contrast with the te1 inflorescence, where no transcripts were detected in RNA gel blot analysis (Veit et al., 1998). More recently, quite low levels of transcripts were detected in young tassel and ear only by a more sensitive competitive RT-PCR (D. Jeffares, unpublished data). These results suggest that te1 expression is significantly lower during the reproductive phase than is seen for PLA2.
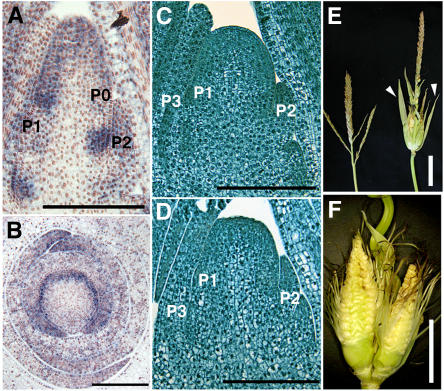
Figure 8.
Phenotypes of te1.
(A) In situ localization of te1 transcripts in longitudinal section of late vegetative shoot apex.
(B) In situ localization of te1 transcripts in transverse section of late vegetative shoot apex.
(C) Wild-type shoot apex.
(D) te1 shoot apex.
(E) Tassels of the wild type (right) and te1 (left). Arrowheads indicate elongated husk leaves.
(F) Enlarged view of ectopic ears in the tassel of te1. Husk leaves are removed.
Bars = 200 μm in (A) to (D), 5 cm in (E), and 2.5 cm in (F).
Mutant phenotypes also differ between pla2 and te1 mutants. While both pla2 and te1 initiate small leaves more rapidly than the wild type, the extent of plastochron reduction is larger in pla2 (65%) than te1 (35%) (Table 1; see Supplemental Table 1 online). The divergence angle between successive leaves in pla2 is normal (180°) but is sometimes variable in te1 (Veit et al., 1998). While pla2 had large but normally shaped SAM, te1 had a reduced misshapen SAM (Figures 8C and 8D; see Supplemental Table 1 online). Both mutants were dwarf but differed in internode elongation. Although dwarf, the relative pattern of internode elongation seen for pla2 was nearly normal (Figure 4B), while te1 showed irregular patterns of internode elongation (Veit et al., 1998). The most conspicuous differences were detected in the reproductive phase. Although both mutants developed large bracts (Figures 4G and 8E), the fates of subtended organs were quite different (Figures 4G and 8F). In pla2, primary branches were converted to vegetative shoots, while in te1, the normally male (stamen bearing) tassel branches were converted to female (kernel bearing) branches (Figure 8F). In some inbred backgrounds, this feminization was much less common (B. Veit, unpublished data).
DISCUSSION
In this article, we describe pla2 mutants that show a combination of phenotypes related to the formation of leaves, including shortened plastochron, the reduction of leaf size due to precocious maturation, and the conversion of primary reproductive branches to vegetative shoots. While these phenotypes are very similar to those previously reported for pla1 (Miyoshi et al., 2004), the results presented here suggest that the two genes normally function in distinct regulatory pathways. We show that PLA2 encodes RNA binding protein that is unrelated to PLA1, a member of the P450 family. The transcriptional regulation of each gene is independent of the activity of the other. Finally, double mutants show synergistic phenotypes not seen in either single mutant. Taken together, these results provide support for a model in which the rate of leaf initiation, or plastochron, is directly coupled to the rate at which previously formed leaves mature.
PLA2 Encodes a MEI2-Like RNA Binding Protein
Our genetic and molecular analysis indicates that PLA2 is the rice orthologue of te1, which like PLA2, encodes a MEI2-like RNA binding protein and shares a similar loss-of-function phenotype. MEI2-like proteins are named for their similarity to MEI2, which in S. pombe plays a crucial role in the transition to meiosis. Members of this family share a characteristic ribonucleoprotein-type RRM at their C terminus. In S. pombe, this RRM forms a complex with meiRNA, a noncoding RNA encoded by SME2 locus (Watanabe and Yamamoto, 1994). The formation of this complex is essential for meiosis I but is not required for premeiotic DNA synthesis (Yamashita et al., 1998), which is thought to involve complexes with as yet undefined RNAs.
Although the high degree of conservation between the C-terminal RRMs of PLA2 and MEI2 suggested that they might bind similar target RNA(s), we could find nothing similar to SME2 in rice. Other possible interactions remain to be tested. Recently, it was reported that signaling via small RNAs, such as small-interfering RNA and microRNA, is involved in many developmental processes in plants (Kidner and Martienssen, 2005), with some related to heterochrony (Bollman et al., 2003; Hunter et al., 2003; Achard et al., 2004; Peragine et al., 2004). This idea is supported by the result that overexpression of miR156b caused shortened plastochron in Arabidopsis (Schwab et al., 2005). Thus, it is possible that PLA2 binds to small RNAs and is involved in an RNA interference–mediated process. In any case, detection of the target RNAs of PLA2 should provide useful insights into molecular mechanisms of leaf development.
PLA2 Regulates the Rate of Leaf Maturation
Our analysis of the loss-of-function mutant phenotypes indicates that PLA2 is a positive regulator of leaf size and a negative regulator of leaf initiation, although Paquet et al. (2005) have suggested, based on a more limited analysis of in situ hybridization data, that PLA2/Ostel1/OML1 is a positive regulator of leaf initiation. Our analysis, however, shows that in the absence of PLA2 function, small leaves develop as a result of precocious maturation (i.e., accelerated cell/tissue differentiation). Thus, PLA2 would normally negatively regulate the rate of leaf maturation. This regulation of leaf maturation could be viewed as the primary function of PLA2, considering that PLA2 expression is predominantly localized to leaf primordia (Figure 6). In this respect, a positive correlation between leaf size and plastochron is suggestive. We propose that the shortened plastochron of pla2 is an indirect consequence of precocious leaf maturation and, as such, resembles pla1, which shows similar phenotypes and which is also expressed in leaf primordia but not in the SAM (Itoh et al., 1998; Miyoshi et al., 2004). In summary, analyses of pla2 and pla1 provide two independent examples that suggest that the rate of leaf maturation plays a significant role in regulating the rate of leaf initiation.
It is significant that expression domains in leaf primordia differ between PLA2 and PLA1. PLA2 is expressed strongly in marginal and distal regions (Figure 6), whereas PLA1 expression is observed in the proximal abaxial region (Miyoshi et al., 2004). It is known that leaf maturation proceeds from the distal to proximal region (Freeling, 1992; Muehlbauer et al., 1997). Thus, PLA2 may be involved in the maturation of distal tissues, while PLA1 is more involved in maturation of proximal tissues. Since pla1 pla2 double mutants exhibit much shorter plastochron than either mutant, PLA2 and PLA1 appear to have complementary functions in leaf maturation that together exert a strong effect on leaf plastochron.
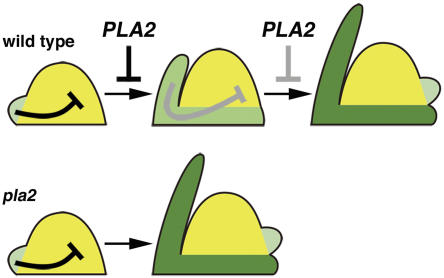
It remains unclear how leaf maturation could affect plastochron. To date, almost no studies have focused specifically on the temporal regulation of leaf initiation but instead have considered factors that influence spatial regulation to give a characteristic leaf arrangement, or phyllotaxy. Several types of models have been proposed to explain this patterning, including the inhibitor, biophysical, and auxin transport models (Green, 1985; Steeves and Sussex, 1989; Reinhardt et al., 2003). Although the proposed mechanisms differ between models, they all maintain that young leaf primordia inhibit new primordium formation in a proximity-dependent manner. We propose that these schemes may also explain temporal regulation of leaf initiation (i.e., preexisting leaf primordia inhibit the precocious initiation of the next leaf). Based on data presented here, in which the precocious maturation of pla2 and pla1 leaves is associated with a reduced plastochron, we propose a model that explains how PLA2 regulates plastochron (Figure 9). In this model, we postulate that leaves lose their inhibitory activity as they mature. Thus, in pla2, the inhibitory activity rapidly declines due to precocious maturation, resulting in more rapid leaf initiation. It remains for future study to determine the nature of this inhibitory signal and to understand its relationship to leaf maturation.
Figure 9.
Model of Plastochron Regulation.
Leaf initiation is inhibited by the preexisting immature leaf primordia. PLA2 limits the rate of leaf development; as leaf development proceeds, the inhibitory effect becomes weakened and finally allows the next leaf to be initiated. In pla2, leaves mature precociously, and the inhibitory effect is cancelled sooner than in the wild type.
PLA2 Ensures Normal Termination of Vegetative Development
Both pla2 and pla1 show heterochronic traits, including the delayed transition to reproductive phase and conversion of rachis branches to vegetative shoots, suggesting that these genes are required for normal termination of the vegetative phase. In maize, dominant heterochronic mutants Teopod1 (Tp1), Tp2, and Tp3 show a similar phenotype (Poethig, 1988; Dudley and Poethig, 1991, 1993; Bassiri et al., 1992) in which branches of tassel are converted into shoots. The viviparous8 (vp8) mutations enhance the conversion in Tp1, Tp2, and Tp3 backgrounds, although vp8 never produces tassel shoots by itself (Evans and Poethig, 1997). Since these maize genes would have different molecular functions from PLA2, many genes appear to be involved in the termination of the vegetative program. Interestingly, vp8 mutants show shortened plastochron, although Tp mutants do not. This suggests that vp8 may play a similar role as PLA1 and PLA2. As Tp mutants are dominant, tp genes may positively regulate vegetative programs. Unfortunately, these maize genes have not been cloned, and we do not have orthologous mutants in rice. However, one expectation is that tp homologues of rice are negatively regulated by PLA2, and they are expressed ectopically in the pla2 mutant, where they promote the ectopic expression of vegetative programs.
A failure in the normal termination of the vegetative phase in pla2 and pla1 might also account for overgrowth of bracts that normally remain vestigial (Figure 4; Itoh et al., 1998), a phenotype that is also seen in Tp mutants of maize. Since both PLA2 and PLA1 are expressed strongly in bracts (Figure 6; Miyoshi et al., 2004), it is natural to speculate that normal bracts suppress the expression of the vegetative program in inflorescence. In the absence of PLA2 or PLA1 activity, bracts might adopt a pattern of development more similar to the much larger vegetative leaf. While vegetative leaves would mature more rapidly in the absence of PLA activity, they would still attain a larger size than normal bracts.
In contrast with PLA1, which is expressed only in bracts, PLA2 is expressed in the upper region of the inflorescence meristem and branch meristems as well as in bracts. This indicates that PLA2 may have some functions in addition to the inhibition of bract development. The inflorescence phenotype, however, does not differ between pla2 and pla1 except for the severity. It is reported that some MEI2-like genes in Arabidopsis are expressed in inflorescence meristems (Anderson et al., 2004). It may be that MEI2 homologue(s) other than PLA2 in rice are expressed in inflorescence meristems, where they function redundantly.
Plastochron Is More Closely Correlated to Cell Division Than SAM Size
We have already reported that for pla1, a shortened plastochron is correlated with an enlargement of the SAM and an increased frequency of cell divisions (Itoh et al., 1998). Our observations on pla2 offer further support for this correlation but suggest that SAM size may be less significant. For example, although the SAM of pla2-2 is not as large as that of pla1-4, it has a shorter plastochron. Similarly, the SAM of the pla1-2 pla2-1 double mutant, which has a smaller SAM than either single mutant, has the shortest plastochron. The maize equivalent of pla2, te1, also shows a reduced SAM compared with the wild type but also has a shortened plastochron. Ikeda et al. (2005) have recently reported that aberrant panicle organization1 of rice shows slightly shortened plastochron, but the SAM size is not modified. By contrast, Cockcroft et al. (2000) reported that higher cell division activity associated with constitutive expression of cyclin D in tobacco shortened plastochron without altering the SAM size. These results suggest that a reduced plastochron is more tightly linked to higher rates of cell division than to an increase in the size of the SAM. However, it should be noted that since no or only low levels of PLA1 and PLA2 transcripts are observed in SAM, their effects on cell divisions in the SAM are likely to be indirect. We propose that precocious maturation of leaves leads to a premature reduction of signal(s) that normally limits cell division in the SAM.
Functional Diversification of PLA2-Like Activities
In addition to delaying the maturation of leaves, PLA2 and closely related genes appear to encompass additional activities as deduced from phenotypic differences. These differences could be explained in several ways. One explanation would suppose that the genes regulate distinct downstream targets either by complexing with distinct target RNAs or, alternatively, through the formation of similar complexes that activate distinct pathways in maize and rice. A second explanation would attribute functional differences to the overlapping but nonidentical expression patterns of the two genes. Such expression differences have been described among several FLORICAULA (FLO)/LEAFY (LFY) orthologues in grasses, including rice RFL, maize Zea FLO/LFY1 (zfl1) and zfl2, and Lolium temlentum LFY (Kyozuka et al., 1998; Gocal et al., 2001; Bomblies et al., 2003), and may be responsible for clear differences in the inflorescence architecture between these species.
Our analysis of expression differences seen for PLA2-like genes might largely account for changes in apparent activity during development and between species. During vegetative development, both PLA2 and te1 affect plastochron, with both genes normally acting to extend plastochron. However, the reduction of plastochron associated with loss of function is relatively modest in te1 compared with pla2. More obvious differences are associated with the SAM, which is reduced and misshapen in te1 compared with the enlarged but relatively normal shaped SAM of pla2. Finally, the extent to which internode development is affected differs, with more shortened internodes observed in the te1 mutant. We propose that all of these differences can be related to observed differences in expression between the PLA2 versus te1 gene, where the ratio of expression in the SAM versus leaf is significantly higher for te1. By this model, PLA2-like genes would act to delay maturation processes not just in developing leaves, but in other tissues in which they are expressed. Higher expression of te1 in the SAM would promote proliferation of cells and thus enhance the sizes of the SAM and associated internodes. A broader activity for PLA2-like genes is also supported by the expression patterns of related genes in Arabidopsis, which are specifically expressed in the apical meristem regions of the shoot and root that contain pluripotent stem cells (Anderson et al., 2004).
During reproductive development, the relatively strong expression of PLA2 compared with te1 correlates well with differences in mutant phenotypes. During this phase, pla2 mutants form vegetative branches bearing normal leaves in place of inflorescence branches. By contrast, relatively minor changes are seen in te1 mutants, where some feminization of basal branches is seen. Other aspects of inflorescence development are relatively normal, including the formation of a relatively dense array of floral branches. Thus, it would appear that expression differences can largely account for the functional diversification between PLA2 and te1.
In this article, we have documented a clear role for PLA2 in regulating the maturation of leaf primordia that in turn is clearly linked to the processes that regulate the formation of new leaves. Our molecular characterization of PLA2 indicates that it effects this regulation through an RNA binding protein that is expressed in young leaf primordia. Thus, the primary function of PLA2 is to regulate the rate of leaf maturation. PLA2 also has an important function in the suppression of the vegetative program in the reproductive phase. Our analysis also provides key insights into the nature of signals that regulate the initiation of leaves. Although previous studies have shown that leaf initiation can be inhibited by preexisting leaves in a proximity-dependent fashion, our study shows that the maturation state of the leaf would have a strong influence on this inhibition. Further analyses focused on the nature of these maturation-dependent signals should afford valuable insights into the regulation of plant development.
METHODS
Plants and Plastochron Measurement
We identified two single-gene recessive mutants from an M2 population of rice (Oryza sativa) cv Taichung 65 mutagenized with N-methyl-N-nitrosourea that showed a short plastochron like pla1. Allelism test revealed that the two mutants, designated pla2-1 and pla2-2, were allelic to each other but distinct from PLA1. We also used pla1-3 and pla1-4 mutants derived from cv Taichung 65 (Miyoshi et al., 2004). pla1-4 is the most severe allele among four pla1 alleles. Mutant and wild-type plants were grown in pots or in paddy fields under natural conditions. Transgenic plants were grown in a biohazard greenhouse at 30°C in the day and 25°C at night.
When every new leaf blade had emerged completely from the sheath of the previous leaf, five plants at the same developmental stage were sampled, and the number of leaves was counted for each plant. At any stage of the vegetative phase, the number of immature leaves before emergence was nearly constant in both pla2 and pla1 (four or five leaves) and wild-type plants (three or four leaves). This indicates that leaf emergence rate corresponds to leaf initiation rate. Thus, we consider that leaf emergence rate is equal to leaf initiation rate (plastochron).
For generating double mutants between pla1 and pla2, we crossed PLA1-2/pla1-2 heterozygous plants with pollen of PLA2-1/pla2-1. pla1-2 pla2-1 plants were identified by phenotypic characterization and molecular genotyping.
Developmental stages of rice inflorescence were represented according to the system of Ikeda et al. (2004).
The standard reference allele te1-1 that had been introgressed into a B73 background that had been grown under standard field conditions was used for comparisons with maize (Zea mays).
Paraffin Sectioning and SAM Measurement
Shoot apices of the wild type and pla2 were fixed in FAA (formaldehyde:glacial acetic acid:ethanol [1:1:18]) for 24 h at 4°C and then dehydrated in a graded ethanol series. Dehydrated samples in 100% ethanol were replaced with xylene and embedded in Paraplast plus (Oxford Labware). Microtome sections (8 μm thick) were stained with Delafield's hematoxyline and observed with a light microscope. For measurement of SAM sizes, dehydrated shoot apices were cleared in the benzyl-benzoate-four-and-a-half fluid devised by Herr (1982). Cleared shoot apices were observed with a microscope (IMT-2; Olympus) equipped with Nomarski differential interference contrast optics. The width of the shoot apex was measured just above the P1 leaf primordium insertion, and the height of the shoot apex was given as the shortest distance from the line used for measuring the width to the tip of the apex.
For maize, shoot apices were infiltrated and sectioned as described by Jackson et al. (1994).
Scanning Electron Microscopy
Dehydrated samples in 100% ethanol were infiltrated with 3-methyl-butyl-acetate, critical point dried, sputter coated with platinum, and observed under a scanning electron microscope (S-4000; Hitachi) at an accelerating voltage of 10 kV.
In Situ Hybridization
Paraffin sections were prepared as described above except that 8-μm-thick microtome sections were applied to slide glasses coated with APS (Matsunami Glasses). Digoxigenin-labeled antisense and sense probes were prepared from the full-length cDNAs of histone H4, PLA1, and PLA2. In situ hybridization and immunological detection of the hybridization signals were performed as described by Kouchi and Hata (1993), except that hybridizations were performed at 55°C for PLA1 and PLA2 because of the high GC contents of these genes. For te1, in situ hybridization was performed as described by Jackson et al. (1994) except that hybridizations were performed at 60 to 65°C because of the high GC content.
RNA Isolation and Analysis
Total RNA was extracted from 100 mg of tissue (shoot apices, leaves, and inflorescence apices) using TRIZOL reagent (Invitrogen). After RNase-free DNase (Takara) treatment, 2.5 μg of RNA was reverse transcribed using oligo(dT) primer and SuperScript III (Invitrogen). PCR was performed for 30 cycles at 94°C for 45 s, 58°C for 30 s, and 72°C for 30 s using 5′-ACAAGGCGTTCCACAAGCAACC-3′ and 5′-GGCGCTGTCATGAGCTCCTG-3′ for PLA2 and 5′-TCCATCTTGGCATCTCTCAG-3′ and 5′-GTACCCGCATCAGGCATCTG-3′ for ACTIN.
Map-Based Cloning
F2 populations of PLA2/pla2-1 (subsp japonica) and cv Kasalath (subsp indica) were used as mapping populations. Using multiple restriction fragment length polymorphism, sequence-tagged sites, and cleaved-amplified polymorphic sequence markers, the PLA2 locus was confined to a 66-kb region on the P1 artificial chromosome clone P0497A05. For complementation tests, the 7.2-kb SpeI fragment, including the PLA2 candidate and 1.8 kb direct upstream of initiation codon, was cloned into binary vector and introduced into pla2-2 homozygotes by the Agrobacterium tumefaciens–mediated transformation method (Hiei et al., 1994).
Phylogenetic Analysis
Multiple sequence alignments were performed using ClustalX (Thompson et al., 1997) and BOXSHADE (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html) programs. The phylogenetic tree was constructed based on the well-conserved RRM3 domain by the neighbor-joining method using ClustalX and TreeView (Page, 1996) programs. The numbers at the branching points indicate the times that each branch topology was found during bootstrap analysis (n = 1000).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AB244276 to AB244282 (PLA2 and OML2 to OML7, respectively), CAA15822 (MEI2), AAK29419 (te1), NP_568946 (At5g61960, AML1), NP_973674 (At2g42890, AML2), NP_193546 (At4g18120, AML3), NP_196346 (At5g07290, AML4), NP_849727 (At1g29400, AML5), NP_189242 (At3g26120, TEL1), NP_176943 (At1g67770, TEL2), NP_174902 (At1g37140, MCT1), and NP_196410 (At5g07930, MCT2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Boundary between Leaf Blade and Sheath.
Supplemental Figure 2. Changes of Leaf Size in Wild-Type, pla2, and pla1 Plants.
Supplemental Figure 3. Epidermal Cells of the Third Leaves and Cells of -2 Internodes.
Supplemental Figure 4. Complementation Test of the pla2 Mutation.
Supplemental Figure 5. Amino Acid Sequence Alignment of MEI2-Like Proteins, Which Are Used to Construct the Phylogenetic Tree.
Supplemental Figure 6. RT-PCR Analysis of the PLA2 mRNA Expression in Different Tissues.
Supplemental Table 1. Plastochron and SAM in the Wild Type and te1.
Supplementary Material
Acknowledgments
We thank Makoto Matsuoka (Nagoya University, Nagoya, Japan) for kindly providing OSH1 and histone H4 cDNA clones and Momoyo Ito and Kyoko Ikeda (University of Tokyo) for technical advice. We also thank Noboru Washizu, Ken-Ichiro Ichikawa, Shizue Nakata, and Hiroshi Kimura (University of Tokyo) for their assistance in cultivating rice plants at the Experimental Farm of the University of Tokyo. This work is supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (14036206 and 16208002 to Y.N.) and by Marsden Fund grants to B.V.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yasuo Nagato (anagato@mail.ecc.u-tokyo.ac.jp).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037622.
References
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131 3357–3365. [DOI] [PubMed] [Google Scholar]
- Anderson, G.H., Alvarez, N.D., Gilman, C., Jeffares, D.C., Trainor, V.C., Hanson, M.R., and Veit, B. (2004). Diversification of genes encoding mei2-like RNA binding proteins in plants. Plant Mol. Biol. 54 653–670. [DOI] [PubMed] [Google Scholar]
- Asai, K., Satoh, N., Sasaki, H., Satoh, H., and Nagato, Y. (2002). A rice heterochronic mutant, mori1, is defective in the juvenile-adult phase change. Development 129 265–273. [DOI] [PubMed] [Google Scholar]
- Bassiri, A., Irish, E.E., and Poethig, R.S. (1992). Heterochronic effects of Teopod 2 on the growth and photosensitivity of the maize shoot. Plant Cell 4 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollman, K.M., Aukerman, M.J., Park, M.Y., Hunter, C., Berardini, T.Z., and Poethig, R.S. (2003). HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130 1493–1504. [DOI] [PubMed] [Google Scholar]
- Bomblies, K., Wang, R.L., Ambrose, B.A., Schmidt, R.J., Meeley, R.B., and Doebley, J. (2003). Duplicate FLORICAULA/LEAFY homologs zfl1 and zfl2 control inflorescence architecture and flower patterning in maize. Development 130 2385–2395. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Letham, S., Craig, S., and Dennis, E.S. (1993). amp1 - A mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J. 4 907–916. [Google Scholar]
- Cockcroft, C.E., den Boer, B.G., Healy, J.M., and Murray, J.A. (2000). Cyclin D control of growth rate in plants. Nature 405 575–579. [DOI] [PubMed] [Google Scholar]
- Dudley, M., and Poethig, R.S. (1991). The effect of a heterochronic mutation, Teopod2, on the cell lineage of the maize shoot. Development 111 733–739. [DOI] [PubMed] [Google Scholar]
- Dudley, M., and Poethig, R.S. (1993). The heterochronic Teopod1 and Teopod2 mutations of maize are expressed non-cell-autonomously. Genetics 133 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, M.M.S., and Poethig, R.S. (1997). The viviparous8 mutation delays vegetative phase change and accelerates the rate of seedling growth in maize. Plant J. 12 769–779. [Google Scholar]
- Fleming, A.J., McQueen-Mason, S., Mandel, T., and Kuhlemeier, C. (1997). Induction of leaf primordia by the cell wall protein expansin. Science 276 1415–1418. [Google Scholar]
- Freeling, M. (1992). A conceptual framework for maize leaf development. Dev. Biol. 153 44–58. [DOI] [PubMed] [Google Scholar]
- Giulini, A., Wang, J., and Jackson, D. (2004). Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 430 1031–1034. [DOI] [PubMed] [Google Scholar]
- Gocal, G.F., King, R.W., Blundell, C.A., Schwartz, O.M., Andersen, C.H., and Weigel, D. (2001). Evolution of floral meristem identity genes. Analysis of Lolium temulentum genes related to APETALA1 and LEAFY of Arabidopsis. Plant Physiol. 125 1788–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, S.J. (1977). Ontogeny and Phylogeny. (Cambridge, MA: Belknap Press of Harvard University Press).
- Green, P.B. (1985). Surface of the shoot apex: A reinforcement-field theory for phyllotaxis. J. Cell Sci. Suppl. 2 181–201. [DOI] [PubMed] [Google Scholar]
- Helliwell, C.A., Chin-Atkins, A.N., Wilson, I.W., Chapple, R., Dennis, E.S., and Chaudhury, A. (2001). The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr, J.M., Jr. (1982). An analysis of methods for permanently mounting ovules cleared in four-and-a-half type clearing fluids. Stain Technol. 57 161–169. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hunter, C., Sun, H., and Poethig, R.S. (2003). The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13 1734–1739. [DOI] [PubMed] [Google Scholar]
- Ikeda, K., Nagasawa, N., and Nagato, Y. (2005). ABERRANT PANICLE ORGANIZATION1 temporally regulates meristem identity in rice. Dev. Biol. 282 349–360. [DOI] [PubMed] [Google Scholar]
- Ikeda, K., Sunohara, H., and Nagato, Y. (2004). Developmental course of inflorescence and spikelet in rice. Breed. Sci. 54 147–156. [Google Scholar]
- Itoh, J., Nonomura, K., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., Kitano, H., and Nagato, Y. (2005). Rice plant development: From zygote to spikelet. Plant Cell Physiol. 46 23–47. [DOI] [PubMed] [Google Scholar]
- Itoh, J.I., Hasegawa, A., Kitano, H., and Nagato, Y. (1998). A recessive heterochronic mutation, plastochron1, shortens the plastochron and elongates the vegetative phase in rice. Plant Cell 10 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D., and Hake, S. (1999). Control of phyllotaxy in maize by the abphyl1 gene. Development 126 315–323. [DOI] [PubMed] [Google Scholar]
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize Knotted1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413. [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2005). The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 8 38–44. [DOI] [PubMed] [Google Scholar]
- Kouchi, H., and Hata, S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238 106–119. [DOI] [PubMed] [Google Scholar]
- Kyozuka, J., Konishi, S., Nemoto, K., Izawa, T., and Shimamoto, K. (1998). Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc. Natl. Acad. Sci. USA 95 1979–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, K., Ahn, B.O., Kawakatsu, T., Ito, Y., Itoh, J., Nagato, Y., and Kurata, N. (2004). PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc. Natl. Acad. Sci. USA 101 875–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlbauer, G.J., Fowler, J.E., and Freeling, M. (1997). Sectors expressing the homeobox gene liguleless3 implicate a time-dependent mechanism for cell fate acquisition along the proximal-distal axis of the maize leaf. Development 124 5097–5106. [DOI] [PubMed] [Google Scholar]
- Nishimura, A., Ito, M., Kamiya, N., Sato, Y., and Matsuoka, M. (2002). OsPNH1 regulates leaf development and maintenance of the shoot apical meristem in rice. Plant J. 30 189–201. [DOI] [PubMed] [Google Scholar]
- Page, R.D. (1996). TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Paquet, N., Bernadet, M., Morin, H., Traas, J., Dron, M., and Charon, C. (2005). Expression patterns of TEL genes in Poaceae suggest a conserved association with cell differentiation. J. Exp. Bot. 56 1605–1614. [DOI] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien, S., Wyrzykowska, J., McQueen-Mason, S., Smart, C., and Fleming, A. (2001). Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc. Natl. Acad. Sci. USA 98 11812–11817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig, R.S. (1988). Heterochronic mutations affecting shoot development in maize. Genetics 119 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge, M.J., and Wagner, D.R. (2001). The Arabidopsis serrate gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13 1263–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Mandel, T., and Kuhlemeier, C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Sato, Y., Hong, S.K., Tagiri, A., Kitano, H., Yamamoto, N., Nagato, Y., and Matsuoka, M. (1996). A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proc. Natl. Acad. Sci. USA 93 8117–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab, R., Palatnik, J.F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8 517–527. [DOI] [PubMed] [Google Scholar]
- Snow, M., and Snow, R. (1931). Experiments on phyllotaxis. I. The effect of isolating a primordium. Philos. Trans. R. Soc. Lond. B 221 1–43. [Google Scholar]
- Steeves, T.A., and Sussex, I.M. (1989). Patterns in Plant Development. (Cambridge, UK: Cambridge University Press).
- Sunohara, H., Satoh, H., and Nagato, Y. (2003). Mutations in panicle development affect culm elongation in rice. Breed. Sci. 53 109–117. [Google Scholar]
- Telfer, A., and Poethig, R.S. (1998). HASTY: A gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125 1889–1898. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit, B., Briggs, S.P., Schmidt, R.J., Yanofsky, M.F., and Hake, S. (1998). Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature 393 166–168. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., and Yamamoto, M. (1994). S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78 487–498. [DOI] [PubMed] [Google Scholar]
- Yamashita, A., Watanabe, Y., Nukina, N., and Yamamoto, M. (1998). RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 95 115–123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.