Abstract
Bacterial flagellins have been portrayed as a relatively invariant pathogen-associated molecular pattern. We have found within-species, within-pathovar variation for defense-eliciting activity of flagellins among Xanthomonas campestris pv campestris (Xcc) strains. Arabidopsis thaliana FLAGELLIN SENSING2 (FLS2), a transmembrane leucine-rich repeat kinase, confers flagellin responsiveness. The flg22 region was the only Xcc flagellin region responsible for detectable elicitation of Arabidopsis defense responses. A Val-43/Asp polymorphism determined the eliciting/noneliciting nature of Xcc flagellins (structural gene fliC). Arabidopsis detected flagellins carrying Asp-43 or Asn-43 but not Val-43 or Ala-43, and it responded minimally for Glu-43. Wild-type Xcc strains carrying nonrecognized flagellin were more virulent than those carrying a recognized flagellin when infiltrated into Arabidopsis leaf mesophyll, but this correlation was misleading. Isogenic Xcc fliC gene replacement strains expressing eliciting or noneliciting flagellins grew similarly, both in leaf mesophyll and in hydathode/vascular colonization assays. The plant FLS2 genotype also had no detectable effect on disease outcome when previously untreated plants were infected by Xcc. However, resistance against Xcc was enhanced if FLS2-dependent responses were elicited 1 d before Xcc infection. Prior immunization was not required for FLS2-dependent restriction of Pseudomonas syringae pv tomato. We conclude that plant immune systems do not uniformly detect all flagellins of a particular pathogen species and that Xcc can evade Arabidopsis FLS2-mediated defenses unless the FLS2 system has been activated by previous infections.
INTRODUCTION
Bacterial flagellins are a prevalent pathogen-associated molecular pattern (PAMP) that is recognizable by the innate immune systems of plants and animals (Gomez-Gomez and Boller, 2002; Gomez-Gomez, 2004; Nurnberger et al., 2004; Ramos et al., 2004). In Arabidopsis thaliana, flagellin perception and flagellin-elicited defense activation require FLAGELLIN SENSING2 (FLS2), a transmembrane receptor kinase with a leucine-rich repeat (LRR) extracellular domain (Gomez-Gomez and Boller, 2000). FLS2 makes a significant contribution to the resistance of Arabidopsis against virulent Pseudomonas syringae pv tomato (Pst) (Zipfel et al., 2004). Flagellin perception in humans and related animals is mediated by Toll-like receptor5 (TLR5), which is also a transmembrane receptor with a LRR extracellular domain (Hayashi et al., 2001). Direct binding of flagellin by TLR5 has been demonstrated (Mizel et al., 2003; West et al., 2005), and importantly, as this paper went to press, direct binding of flagellin by FLS2 was also reported (Chinchilla et al., 2006). The defenses activated downstream of flagellin perception are receiving increasing attention as a result of recently discovered roles in both basal and race-specific disease resistance in plants and in both innate and adaptive immunity in animals (Asai et al., 2002; Jones and Takemoto, 2004; Kim et al., 2005; Li et al., 2005; Pasare and Medzhitov, 2005).
Flagella play a central role in bacterial biology, enabling bacteria to migrate toward favorable environments and to escape from unfavorable ones (Moens and Vanderleyden, 1996; Otteman and Miller, 1997). Although nonmotile pathogens can still cause disease symptoms, flagellar motility is essential for the overall pathogenicity of bacterial plant pathogens (Panopoulos and Schroth, 1974; Haefele and Lindow, 1987). The filament of a flagellum is a tubular structure made up of 11 protofilaments, which are nearly longitudinal helical arrays of many hundred ∼45-kD flagellin molecules (O'Brien and Bennett, 1972). Although most flagellin is assembled into flagella, flagellin can also leak into the bacterial environment during the construction of flagella (Komoriya et al., 1999), and flagellin is a component of the detritus associated with a bacterial colony. Stray or partially degraded flagellin monomers can be recognized by host cells, inducing defense responses in Drosophila (Lemaitre et al., 1997), mammals (McDermott et al., 2000; Eaves-Pyles et al., 2001a; Sierro et al., 2001), and plants (Felix et al., 1999; Gomez-Gomez et al., 1999).
Different regions of flagellin are recognized by plants and animals. Flg22 and similar peptides, whose sequences are based on a conserved N-terminal domain of flagellins from Pseudomonas aeruginosa and other bacteria, elicit plant defense responses but do not activate the innate immune responses in mammals (Felix et al., 1999; Donnelly and Steiner, 2002). Conversely, the proinflammatory response of mammals to Salmonella flagellin is attributable to other conserved N- and C-terminal regions of flagellin (Eaves-Pyles et al., 2001b; Donnelly and Steiner, 2002; Smith et al., 2003). The recognition of different flagellin domains by plants and animals is not surprising, because FLS2 and TLR5, despite their similarity as transmembrane LRR proteins involved in flagellin perception, are not orthologous proteins. The conserved sites of flagellin that are recognized by animal hosts are essential for bacterial motility (Smith et al., 2003). The flagellin crystal structure and a complete atomic model of the Salmonella flagellar filament are available (Samatey et al., 2001; Yonekura et al., 2003). The conserved N- and C-terminal flagellin domains apparently mediate filament polymerization and are buried within the flagellum structure. The more exposed regions of flagellin exhibit significant variability between bacterial species, and it is interesting that the FLS2- and TLR5-mediated responses of plant and animal innate immune systems have evolved to recognize the buried but widely conserved flagellin domains. However, flagellin domains outside of the flg22 region have been implicated in defense elicitation in other plant–bacteria pathosystems (Taguchi et al., 2003; Takeuchi et al., 2003), and the possibility remains that regions other than flg22 contribute to the induction of innate immunity in Arabidopsis.
Some bacterial species produce a flagellin that is not recognized by host flagellin detection systems. Although the flg22 region of bacterial flagellins is highly conserved, there are a few variable positions, and the plant-associated bacteria Agrobacterium tumefaciens, Sinorhizobium meliloti, and Ralstonia solanacearum diverge at a sufficient number of flg22 residues that peptides based on the flagellins from these species fail to elicit plant responses (Felix et al., 1999; Pfund et al., 2004). Bacteria for which crude flagellin extracts can elicit an FLS2-like response in tomato (Lycopersicon esculentum) cells include Erwinia carotovora, Erwinia chrysanthemi, the P. syringae pathovars glycinea, phaseolicola, tomato, and syringae, P. aeruginosa, P. fluorescens, and Escherichia coli (Felix et al., 1999). An impact of flagellin detection on plant disease resistance has been demonstrated only for Pst strain DC3000 (Zipfel et al., 2004). Crude flagellin extracts from A. tumefaciens, S. meliloti, and the Xanthomonas campestris pathovars vesicatoria, juglandis, and brassica rapa were reported to be inactive for defense elicitation in tomato (Felix et al., 1999), and R. solanacearum strain K60 bacteria are not detected by the Arabidopsis FLS2 system (Pfund et al., 2004). Similarly, some species of animal pathogens produce a flagellin that does not elicit host defenses (Lee et al., 2003; Gewirtz et al., 2004; Andersen-Nissen et al., 2005). Potential effects of within-species flagellin polymorphism on host defense elicitation and disease outcome apparently have not been studied in plant or animal pathogens.
In this study, we focused on host recognition of X. campestris pv campestris (Xcc) flagellin. Xcc is the causal agent of black rot, the most economically significant bacterial disease of crucifers worldwide (Williams, 1980). Xcc is also pathogenic on Arabidopsis (Simpson and Johnson, 1990), making this an attractive model for further study. Xcc is a vascular pathogen that typically enters the plant via hydathodes, specialized pores on the leaf margins of higher plants that connect to the vascular system (Alvarez, 2000). This is in contrast to Pst and P. syringae pv maculicola, which cause leaf disease primarily as a result of mesophyll colonization after entry through stomata or wounds (Schroth et al., 1991). In the xylem, Xcc growth causes darkening of the veins and eventual death of the infected tissue. Many aspects of Arabidopsis–Xcc interactions have been studied (Bent et al., 1992; Lummerzheim et al., 1993; Parker et al., 1993; Buell and Somerville, 1997; Hugouvieux et al., 1998; Godard et al., 2000; Korves and Bergelson, 2003; O'Donnell et al., 2003; Silipo et al., 2005). The extensive progress made by Boller and colleagues concerning Arabidopsis FLS2-mediated flagellin detection (Gomez-Gomez and Boller, 2002; Zipfel et al., 2004) motivated our examination of Arabidopsis–Xcc interactions with respect to flagellin detection. We pursued two complementary goals: examining the structural determinants of flagellin recognition and the effects of flagellin recognition on whole plant–whole pathogen interactions.
RESULTS
Xcc Strain Survey Reveals Variation in Defense Elicitation Activity of Crude Bacterial Extracts
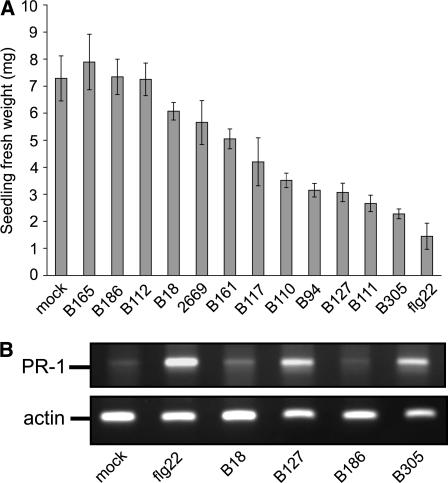
A set of 12 Xcc strains of diverse origin was used to investigate within-pathovar variability in the plant defense elicitation activity of flagellin or other PAMPs. Arabidopsis and other plants activate defense responses such as oxidative burst, PATHOGENESIS-RELATED (PR) gene expression, and callose deposition in response to bacterial flagellin, and seedling growth inhibition has been used as a facile macroscopic phenotype that correlates with the flagellin-induced activation of these defenses (Felix et al., 1999; Gomez-Gomez et al., 1999; Gomez-Gomez and Boller, 2000; Pfund et al., 2004). Growth inhibition is a common phenomenon in Arabidopsis constitutive disease-resistant mutants (Greenberg and Ausubel, 1993; Bowling et al., 1994; Clarke et al., 1998; Yu et al., 1998; Petersen et al., 2000; Maleck et al., 2002). To initiate this study, Arabidopsis accession Columbia (Col-0) seedlings were treated with boiled extracts from the different Xcc strains, because flagellin has been reported to persist in the supernatant through this initial purification step (Felix et al., 1999). We observed (Figure 1A) that boiled extracts from some Xcc strains, such as B94, B127, B111, and B305, were capable of eliciting significant growth inhibition in Arabidopsis seedlings and were therefore termed eliciting Xcc extracts, whereas extracts of other Xcc strains, including B165, B186, and B112, caused little or no reduction in seedling weight after treatment and were termed noneliciting Xcc extracts. Tests in motility agar confirmed that all 12 of these Xcc strains were motile. The level of growth inhibition caused by boiled extracts from strongly eliciting Xcc strains (e.g., Xcc B305) was comparable to the growth inhibition caused by 7.5 μM P. aeruginosa flg22 peptide (Figure 1) (Gomez-Gomez et al., 1999). RT-PCR experiments confirmed that the seedling growth inhibition activity of the bacterial extracts correlated with their ability to induce PR-1 expression (Figure 1B).
Figure 1.
Levels of Defense Elicitation Differ among Boiled Extracts from Diverse Strains of Xcc.
(A) Boiled extracts of some but not all strains of Xcc elicit Arabidopsis defense responses, assayed as seedling growth inhibition. Seedling fresh weights (means ± se) are for nine Arabidopsis Col-0 seedlings per treatment, measured after 7 d of growth in 0.5× Murashige and Skoog (MS) medium supplemented with water (mock), 11 μg of flg22, or 11 μg (total protein) of the indicated boiled bacterial extracts.
(B) PR-1 gene expression is activated by flg22 peptide and by boiled extracts from eliciting Xcc strains, but not from noneliciting Xcc strains, consistent with the results shown in (A). Data are for semiquantitative PCR performed on seedling samples taken 24 h after exposure to elicitor extracts. Actin expression was monitored as a control for equivalency between samples.
Phylogenetic Analysis of fliC Genes Predicts Two Distinct Families
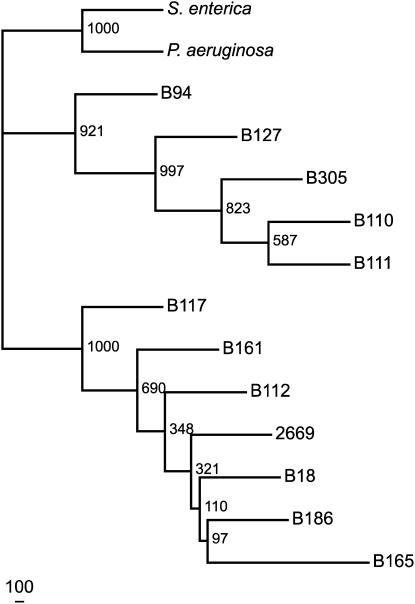
It was shown previously that flagellin is a major determinant of defense elicitation in boiled extracts of some phytopathogenic bacteria (Felix et al., 1999). Therefore, we investigated the hypothesis that flagellin was the causative agent of elicitation in the assays shown in Figure 1 and that the amino acid sequence of flagellin would differ between eliciting and noneliciting strains. Differences in sequence might reveal recognition determinants that are important for plant perception of bacterial flagellins. Using PCR primers based on the published fliC sequence from one Xcc strain (see Methods), we isolated, cloned, and sequenced the flagellin-encoding fliC genes from the 12 different Xcc strains. Derived amino acid sequences from these genes were then compared using nearest-neighbor analysis. As shown in Figure 2, the Xcc FliC products fall into two distinct clades. Several domains across the entire flagellin protein carried sequence differences between the two groups, including the previously characterized flg22 domain (see Supplemental Figure 1 online). Interestingly, the strains that populated the two distinct FliC clades correlated with the two phenotypic classes we observed with regard to elicitation activity of boiled extracts on Arabidopsis Col-0 (Figure 1). For example, the fliC gene products from eliciting Xcc strains B305 and B127 are together in the same clade, distinct from the fliC gene products of noneliciting Xcc strains B18 and B186.
Figure 2.
Phylogenetic Analysis of Xcc fliC Gene Products Predicts Two Distinct Families.
The nearest-neighbor tree was generated by PHYLIP using derived amino acid sequences for full-length flagellins from Xcc strains and the well-studied flagellins from P. aeruginosa and S. enterica serovar Typhimurium as outgroups. Horizontal branch lengths (also presented numerically to the right of each branch) represent bootstrap support values and indicate the number of times out of 1000 trials that the particular branch was predicted.
Variation in FLS2-Dependent Elicitation Activity among Purified Flagellins
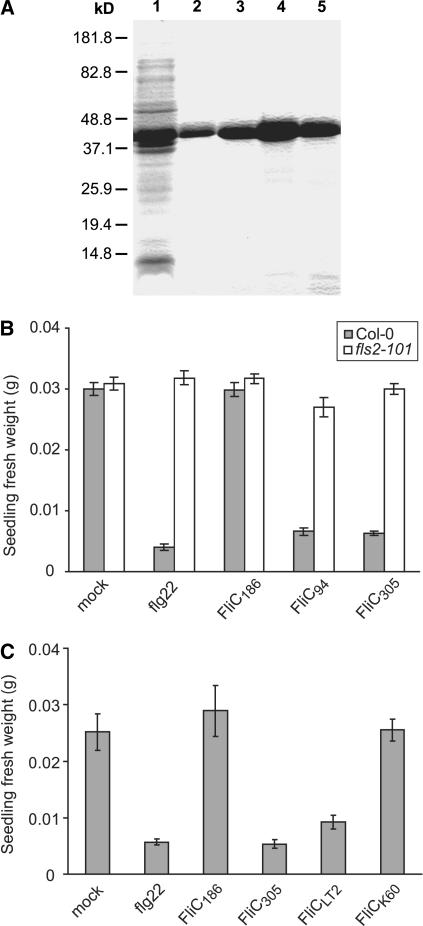
In light of the correlations described above, we sought to investigate the causal relationships between flagellin structure, defense elicitation, and bacterial virulence. Full-length flagellin proteins derived from three representative Xcc strains (B94, B186, and B305) were purified as His6 fusion proteins (Figure 3A). Defense elicitation by these flagellins was then measured using the seedling growth inhibition assay, and the results were consistent with those obtained using crude boiled extracts (Figure 3B). Flagellin from the noneliciting B186 strain (FliC186) was noneliciting on Arabidopsis Col-0 seedlings, whereas flagellins from the eliciting B94 and B305 strains strongly elicited a seedling response. We also tested the ability of these purified flagellins to elicit the Arabidopsis Col-0 fls2-101 mutant line carrying a T-DNA insertion that disrupts FLS2. As with flg22 peptide, the Xcc B94 and B305 flagellins were not perceived by Arabidopsis fls2-101 seedlings (Figure 3B), demonstrating the FLS2-dependence of the response. Flagellin is a very widely studied PAMP and elicitor of innate immunity in plants and animals, and it has previously been shown that some pathogenic bacteria avoid host detection of their flagella by expressing nonrecognized flagellins (Lee et al., 2003; Gewirtz et al., 2004; Andersen-Nissen et al., 2005). These results extend this finding by demonstrating that within a single pathogen taxon (a single species or pathovar), different strains can express different flagellins that vary in their host elicitation activity.
Figure 3.
Elicitation of FLS2-Dependent Plant Responses by Purified His-Tagged Flagellins.
(A) Example of the recombinant His-tagged flagellin samples used in this study. On Coomassie blue–stained SDS-PAGE gels, lane 1 carried E. coli pQE30-fliC total lysate after isopropyl β-d-thiogalactoside induction, and lanes 2 to 5 show four independent samples of His-tagged flagellin after elution from nickel-nitrilotriacetic acid agarose columns.
(B) FLS2-dependent elicitation of Arabidopsis defense responses, assayed as seedling growth inhibition. Seedlings of Arabidopsis Col-0 (shaded bars) and fls2-101 mutant (open bars) were treated with water (mock) as a negative control, 10 μM flg22 peptide as a positive control, or 5 μM purified His-tagged flagellin of Xcc strain B186, B94, or B305. Means ± se are shown.
(C) Arabidopsis Col-0 seedlings were treated with water, 10 μM flg22, or 5 μM purified His-tagged flagellin of Xcc strain B186 or B305, S. enterica serovar Typhimurium strain LT2, or R. solanacearum strain K60. Means ± se are shown.
The elicitation activity of purified flagellin was also determined for two other bacteria whose flagellins have been the subject of past investigation. Full-length His-tagged flagellin derived from the fliC gene of Salmonella enterica serovar Typhimurium strain LT2 inhibited the growth of Arabidopsis Col-0 seedlings (Figure 3C). The crystal structure of this flagellin is available (Samatey et al., 2001). His-tagged flagellin from R. solanacearum strain K60 was noneliciting, as predicted from previous studies (Figure 3C) (Pfund et al., 2004).
The Flg22 Region Carries the Polymorphism Responsible for the Elicitation Activity of Xcc Flagellin
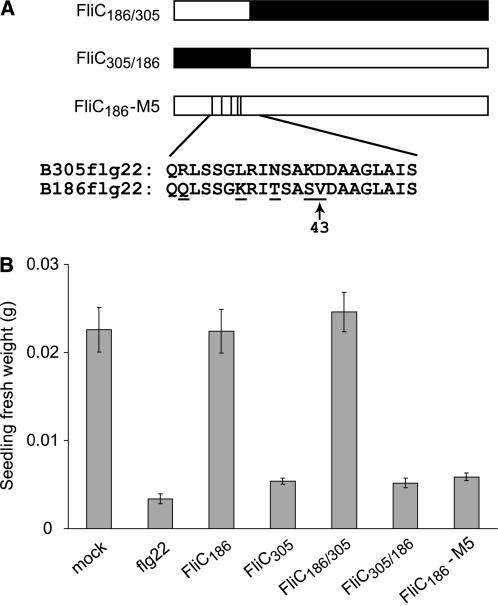
We sought to define the domains of flagellin that elicit defense responses in Arabidopsis. Extensive published studies of the Arabidopsis FLS2-mediated response and similar responses in tomato have focused on elicitation by flg22 peptides (Gomez-Gomez and Boller, 2002; Zipfel et al., 2004) but have left open the possibility that other domains of flagellin also exhibit plant defense-eliciting activity. Flagellin features outside of the flg22 domain are responsible for host elicitation in other plant and animal pathosystems (Che et al., 2000; Eaves-Pyles et al., 2001b; Donnelly and Steiner, 2002; Taguchi et al., 2003; Takeuchi et al., 2003; Murthy et al., 2004). Supplemental Figure 1 online presents an alignment showing all polymorphisms across the derived amino acid sequences of the Xcc flagellins used in this study. To test whether differences in elicitation by Xcc flagellins were attributable to polymorphisms in regions other than the flg22 domain, we constructed hybrid flagellins by swapping segments between the fliC genes of Xcc B305 (eliciting) and B186 (noneliciting), as shown in Figure 4A. The purified, chimeric His6-tagged flagellins were tested for elicitation of plant responses. Like FliC186, FliC186/305 with the N-terminal flg22 region of B186 flagellin did not inhibit the growth of Col-0 seedlings (Figure 4B), suggesting that regions C-terminal to the flg22 domain do not make significant contributions to the elicitation activity of B305 flagellin. By contrast, FliC305/186 with the flg22 region of B305 flagellin strongly elicited seedling growth inhibition (Figure 4). To more precisely locate the eliciting region of B305 flagellin, site-directed mutagenesis was used to create a B186 flagellin that carried B305 sequence only in the flg22 region (a five–amino acid polymorphism; Figure 4A). Purified His6-tagged protein for this FliC186-M5 inhibited the seedling growth of Col-0 as strongly as FliC305, indicating that the flg22 region of Xcc flagellin carries the sole polymorphism responsible for differential elicitation in Arabidopsis (Figure 4B). No contribution to elicitation, as measured by the seedling growth assay, was attributable to flagellin domains other than flg22.
Figure 4.
Use of Purified His-Tagged Domain-Swapped or Region-Mutagenized Flagellin Protein to Identify Elicitation-Active Domains.
(A) Schemes of domain-swap and region-mutagenized flagellins and of the wild-type flg22 regions. Darkened areas of the three constructs represent Xcc B305 flagellin sequences. Amino acid sequences for the flg22 region of two wild-type Xcc flagellins are shown (residues 30 to 51); polymorphic residues within the flg22 region are underlined, and position 43 is marked as a reference point.
(B) Elicitation of Arabidopsis defense responses, assayed as seedling growth inhibition. Arabidopsis Col-0 seedlings were treated with water (mock), 10 μM flg22, 5 μM purified His-tagged flagellin of Xcc strain B186 or B305, or the hybrid flagellins described for (A). Means ± se are shown.
A Single Amino Acid Polymorphism That Is Critical for Elicitation Activity
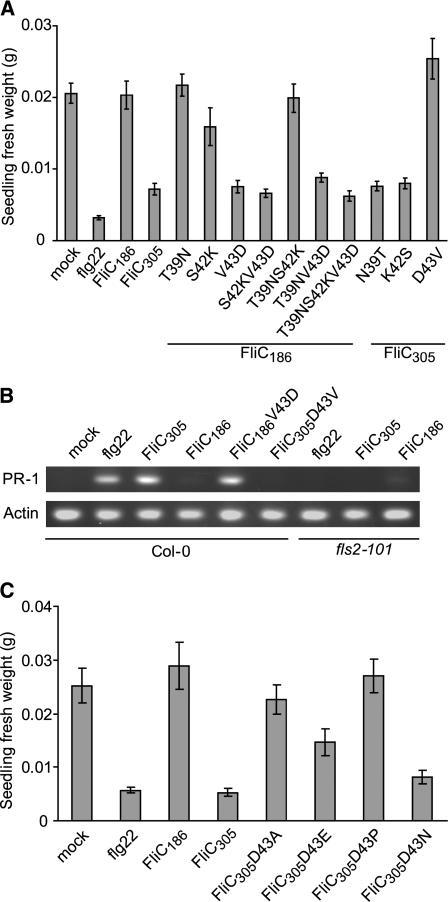
Five amino acid positions are polymorphic between the flagellin flg22 domains from eliciting Xcc strains such as B305 and those of nonelicitors such as B186. Three of these residues are within the 15–amino acid flg15 peptide that is sufficient for plant elicitation (Felix et al., 1999; Meindl et al., 2000). To identify specific residues that determine elicitation activity on Arabidopsis Col-0, we used site-directed mutagenesis to alter B186 and B305 flagellins at these positions. Position 43 was identified as the key polymorphic determinant of activity. Seedling inhibition assays using purified full-length His-tagged flagellin protein (Figure 5A) showed that FliC186V43D had the same eliciting activity as FliC305, whereas the reciprocal FliC305D43V had no eliciting activity, similar to FliC186. By contrast, the B186/B305 residue swaps at amino acid positions 39 and 42 had minimal effects on elicitation activity. The experimental data from double- and triple-swap FliC186(T39N S42K), FliC186(T39N V43D), FliC186(S42K V43D), and FliC186(T39N S42K V43D) corroborated that the V43D substitution identifies the polymorphism critical for flagellin eliciting activity (Figure 5A). We used PR-1 gene expression as a marker to verify these defense elicitation results. PR-1 mRNA expression was significantly induced in Arabidopsis seedlings by flg22 peptide treatment, as well as by the treatments with FliC305 and FliC186V43D, whereas FliC305D43V had lost the ability to elicit PR-1 gene expression (Figure 5B).
Figure 5.
Identification and Dissection of Amino Acid Residue 43 as the Key Polymorphic Determinant of Xcc Flagellin for Arabidopsis Defense Elicitation.
(A) Defense elicitation activity of purified His-tagged flagellins carrying one, two, or three amino acid changes as noted, with defense elicitation assayed as seedling growth inhibition. Arabidopsis Col-0 seedlings were treated with water (mock), 10 μM flg22, or 5 μM of the designated purified His-tagged flagellin. Means ± se are shown.
(B) PR-1 gene expression activated by flg22 peptide or by purified His-tagged flagellins that carry the Asp-43 residue, consistent with the results shown in (A). Data are for semiquantitative PCR performed on Arabidopsis Col-0 or fls2-101 mutant seedling samples taken 24 h after exposure to elicitor. Actin expression was monitored as a control.
(C) Defense elicitation activity of purified His-tagged flagellins carrying different amino acids at Xcc flagellin residue 43, assayed as for (A). Means ± se are shown.
Further amino acid change experiments were performed to study the charge and structural features of amino acid position 43 that determine perception of Xcc flagellin by Arabidopsis. Asp-43 was replaced with Ala, Pro, Asn, and Glu. Purified His-tagged FliC305D43E protein retained an acidic side chain at position 43 yet lost substantial elicitation activity, whereas FliC305D43N traded the acidic side chain for an amide side chain with similar overall structure yet exhibited little if any loss of elicitation activity (Figure 5C). Arabidopsis was almost completely nonresponsive not only to FliC305D43P but also to FliC305D43A (Figure 5C). Hence, the structural features at flagellin amino acid position 43 that allow recognition by Arabidopsis are strictly constrained.
Isogenic Xcc Strains That Express Eliciting or Noneliciting Flagellin Exhibit Similar Virulence on Arabidopsis
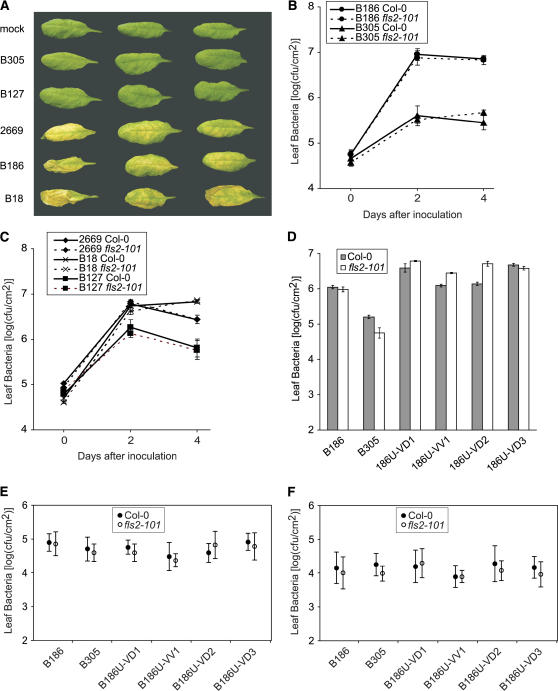
The contribution of flagellin recognition to overall disease resistance against Xcc was examined. We initially hypothesized that Xcc strains whose extracts were more strongly defense-eliciting on a particular plant host (Figure 1), and that expressed a flagellin with elicitation activity on that host (Figure 3), would exhibit reduced virulence on that host relative to strains that were noneliciting. Leaves of 4-week-old Arabidopsis Col-0 plants were inoculated by vacuum infiltration with five representative Xcc strains. We observed faint chlorosis after 3 d in the plants inoculated with Xcc B18, B186, and 2669 (strains that produced noneliciting extracts; Figure 1). The severity of chlorosis increased over time, producing symptoms typical of virulent Xcc infiltrated into Arabidopsis leaf mesophyll (Figure 6A) (Simpson and Johnson, 1990; Parker et al., 1993; Buell and Somerville, 1997). By contrast, no disease symptoms were visible in Col-0 plants inoculated with the eliciting Xcc strains B127 and B305, even after 2 weeks. Bacterial growth within Arabidopsis Col-0 was consistent with these visible differences in disease symptoms; populations of B305 and B127 were 50- to 100-fold lower than those of the virulent strains 4 d after leaf mesophyll inoculation (Figures 6B and 6C). Although B127 also lacked virulence on a Brassica rapa and a Brassica oleracea accession, the other four Xcc strains caused typical black rot disease on these plants (see Methods; data not shown), confirming that B305 in particular did not exhibit a generic loss of virulence. Hence, the defense-eliciting activity of boiled extracts and flagellins from the surveyed Xcc strains correlated with a decrease in virulence of those same strains on Arabidopsis Col-0. Note, however, that these correlations do not precisely implicate flagellin as the causal agent of reduced virulence. The experiments represented in Figures 6B and 6C also included Arabidopsis Col-0 fls2-101 mutant plants, and those results suggested that flagellin was not the causal agent of the observed differences in virulence. Loss of the FLS2 flagellin detection system did not make the plants more susceptible (Figures 6B and 6C).
Figure 6.
Growth of Xcc Strains, Including Isogenic Xcc B186 Strains, Expressing Eliciting or Noneliciting Flagellins in Arabidopsis Wild-Type and fls2-101 Leaves.
(A) Decreased virulence of eliciting Xcc strains. Three representative leaves for each treatment are shown from 4-week-old Arabidopsis Col-0 plants 6 d after vacuum infiltration with 106 cfu/mL of the indicated bacteria.
(B) and (C) Similar reduced growth of eliciting Xcc strains on Arabidopsis Col-0 and fls2-101. Plant rosettes were vacuum infiltrated with 106 cfu/mL of the indicated Xcc strains, and leaf samples were surface-sterilized before maceration and dilution plating to assess bacterial populations in the leaf interior. Data are presented as means ± se.
(D) Populations of isogenic Xcc strains carrying eliciting or noneliciting flagellins after inoculation by vacuum infiltration. Leaf samples were taken 3 d after vacuum infiltration of Arabidopsis Col-0 (shaded bars) or fls2-101 (open bars) with 106 cfu/mL of the indicated Xcc strains. Strains B186U-VD1, B186U-VD2, and B186U-VD3 are independent Xcc B186 derivatives carrying precise gene replacements with fliC186V43D integrated at the fliC locus. B186U-VV1 is derived from the same sacB-fliC186V43D integration event that yielded B186U-VD1 but is a resolution product that carries the original (wild-type) fliC186 locus. Data are means ± se for one experiment; similar results were obtained in two separate experiments.
(E) and (F) Populations of isogenic Xcc strains carrying eliciting or noneliciting flagellins after inoculation by spraying a bacterial suspension onto plants grown under constant temperature and humidity (E) or onto plants undergoing leaf guttation (F). A suspension of ∼109 cfu/mL Xcc bacteria in 0.04% Silwet L-77 was misted onto rosette leaves of intact plants, and internal leaf bacterial populations were determined 4 d after inoculation. The graphs report analysis of variance (means ± 95% confidence intervals) for pooled data from three independent experiments.
To further investigate any causal role of flagellin perception in disease resistance, we first reproduced the finding that Arabidopsis FLS2 contributes to plant defense against Pst DC3000 (Zipfel et al., 2004). As predicted, modest but significant reductions in bacterial populations and disease symptom severity were observed after spray inoculation of wild-type (FLS2+) plants compared with fls2-101 plants (see Supplemental Figure 2 online). These experiments and those of Zipfel et al. (2004) used the presence or absence of a functional Arabidopsis FLS2 to postulate contributions of flagellin recognition to disease outcome; analogous experiments in which Pst flagellin expression is manipulated directly have not been reported.
To determine the contribution of flagellin perception to plant defense against Xcc, we tested plant fls2 mutants and also constructed and tested isogenic Xcc B186 strains that carry eliciting or noneliciting flagellins. The isogenic strains were constructed by unmarked replacement of the chromosomal fliC gene and carried either fliC186 (wild type, noneliciting) or fliC186V43D (eliciting) at the original fliC locus. These strains retained similar motility on motility agar (see Methods; data not shown). As shown in Figures 6D and 6E, no reproducible differences were observed in the virulence of these Xcc B186 fliC isogenic strains on Arabidopsis Col-0 leaves, in multiple experiments in which plants were inoculated by vacuum infiltration or by spray inoculation (as in Zipfel et al., 2004). However, in agricultural settings, Xcc infects leaves primarily by vascular colonization through hydathodes rather than by mesophyll colonization through stomates (Alvarez, 2000). Therefore, a hydathode invasion assay that more closely mimics this natural mode of infection was used, and multiple repetitions of this assay also revealed no significant differences in the virulence of the Xcc B186 fliC isogenic strains on Arabidopsis Col-0 leaves (Figure 6F). No differences in the visible disease symptoms caused by the fliC-isogenic Xcc strains were detected in any of the experiments described above (data not shown). In addition, by any of the three inoculation methods, none of the strains achieved significantly different population levels (Figure 6) or disease symptoms on fls2-101 plants compared with wild-type Col-0. It remains possible that some other infection assay condition might reveal an effect, but these results suggest that possession of a defense-eliciting flagellin does not significantly constrain the growth of virulent Xcc in Arabidopsis leaves.
Wild-type Xcc B305 strains were included in these experiments and yielded additional interesting results. As noted above, Xcc B305 grew significantly less well than Xcc B186 in Arabidopsis vacuum infiltration experiments (Figures 6B and 6D). However, B305 grew similarly on Col-0 or fls2-101 plants. This finding reinforces the suggestion that the reduced virulence of Xcc strains such as B305 is not attributable to their production of an eliciting flagellin. It was further intriguing that, after spraying or hydathode inoculation of Arabidopsis, Xcc B305 and B186 grew to similar population levels (Figures 6E and 6F). The reproducible differences in the growth of B305 relative to B186 after vacuum or syringe infiltration into leaves were not observed after spraying or hydathode inoculation. This suggests that some Xcc B305 defense-eliciting compounds other than flagellin are more efficiently made, introduced by infiltration, or detected in leaf mesophyll as opposed to leaf vascular tissue. Alternatively, defense suppression by Xcc B305 may proceed more effectively in vascular tissue.
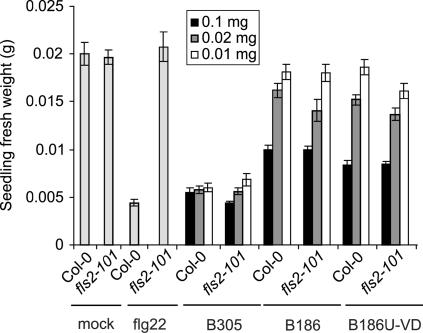
The defense elicititation by Xcc boiled extracts was briefly reexamined in light of the observation that flagellin type (eliciting or noneliciting) had no detectable effect on infection outcomes. When crude boiled extracts from the isogenic, motile Xcc B186 strains expressing eliciting or noneliciting flagellins were tested, they caused indistinguishable defense elicitation on Arabidopsis Col-0 (Figure 7). Elicitation of Col-0 plants compared with fls2-101 plants was indistinguishable for extracts from any of the tested strains (Figure 7). These results indicate that defense elicitors other than flagellin are present in these extracts and that they mask the elicitation activity of flagellin as a result of their greater abundance and/or stronger specific activity. Dose–response analysis suggested that Xcc B305 produces more of these other elicitors than Xcc B186, or a more active form (Figure 7).
Figure 7.
Defense Elicitation by flg22 Peptide or by Boiled Xcc Extracts Applied at Different Concentrations to Arabidopsis Col-0 and fls2-101 Plants.
Defense responses were assayed as seedling growth inhibition. Fresh weights (means ± se) are for 10 seedlings per treatment, measured after 10 d of growth in 0.5× MS medium supplemented with water (mock), 10 μM flg22, or the indicated amounts of total protein for boiled bacterial extracts. Xcc B186U-VD strains express fliC186V43D in place of wild-type fliC as a result of unmarked gene replacement at the fliC locus.
Pretreatment with Flagellin Restricts the Growth of Pst DC3000 and Xcc B186
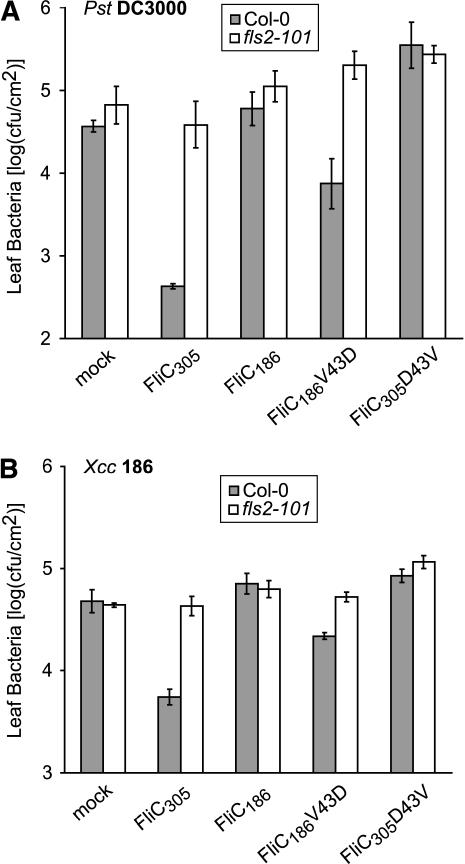
Because Xcc B186 and B186 fliC186V43D strains all grew similarly on Arabidopsis Col-0 and fls2-101 mutant plants, it was of interest to further investigate whether flagellin-elicited responses could restrict the growth of Xcc in Arabidopsis. We treated leaves with purified wild-type or mutant Xcc flagellin proteins and then inoculated the same leaves with Xcc B186 or Pst DC3000 bacteria 1 d later. As shown in Figure 8A, in leaves pretreated with FliC305 and FliC186V43D, the growth of Pst DC3000 was reduced in Col-0 wild-type leaves but not in the fls2-101 mutant. These findings with Pst are consistent with published data for plants treated with flg22 peptide (Zipfel et al., 2004). Significantly, treatment of leaves with eliciting flagellins also restricted the growth of Xcc B186 (Figure 8B). As expected, pretreatment with the noneliciting FliC186 or FliC305D43V flagellins did not reduce the growth of either bacterial strain (Figures 8A and 8B). These experiments demonstrate that although Xcc strains that express an eliciting flagellin are not constrained by the host FLS2 system in previously noninfected plants (Figure 6), a preceding flagellin-mediated elicitation of FLS2-dependent responses can constrain the growth of Xcc in Arabidopsis (Figure 8B).
Figure 8.
Restriction of Pst and Xcc Growth as a Result of Leaf Elicitation with Purified His6-Tagged Flagellin Protein.
(A) Leaf populations of Pst DC3000 (means ± se) after 2 d of growth on leaves that were exposed to purified flagellin 24 h before inoculation with bacteria. Arabidopsis wild-type Col-0 (shaded bars) and fls2-101 (open bars) leaves were treated by syringe infiltration with water (mock) or with 1 μM of the His-tagged flagellins FliC305, FliC186, FliC186V43D, or FliC305D43V, then inoculated the next day with 105 cfu/mL Pst DC3000 by syringe infiltration.
(B) Leaf populations of Xcc after 2 d of growth on leaves that were exposed to purified flagellins 24 h before inoculation with 5 × 105 cfu/mL Xcc B186, as in (A).
DISCUSSION
PAMP Receptor or R Gene?
The findings of this study challenge past portrayals of flagellin as a relatively stable PAMP. Previous studies had reported that crude flagellin preparations (boiled extracts) from many bacterial species can elicit defense-associated responses in plants but that flagellins from R. solanacearum, S. meliloti, and A. tumefaciens did not elicit these responses (Felix et al., 1999; Gomez-Gomez et al., 1999; Pfund et al., 2004). This parallels findings from the study of mammalian innate immune systems, in which the flagellins of numerous bacteria are recognized via TLR5 but certain pathogen species produce flagellins that escape detection (Lee et al., 2003; Gewirtz et al., 2004; Andersen-Nissen et al., 2005). We now report within-species polymorphism for the host elicitation activity of flagellins, with different strains of the same bacterial pathovar producing recognizable or nonrecognizable versions of this host defense elicitor.
This finding suggests some reexamination of our conceptions of PAMPs and avirulence genes. This subject has substantial practical relevance because a major issue facing farmers and plant breeders is the variable efficacy of plant R genes as pathogen populations shift toward individuals that lack a recognized version of the corresponding avirulence gene product (Baker et al., 1997; Simmonds and Smartt, 1999). PAMPs have been characterized as relatively invariant molecular markers that a eukaryotic host can use to recognize the presence of many different potentially pathogenic species (Medzhitov and Janeway, 2000; Akira et al., 2001; Dangl and Jones, 2001; Gomez-Gomez and Boller, 2002; Nurnberger et al., 2004). Avirulence genes of plant pathogens, on the other hand, encode defense-eliciting products that often vary among different strains of a single pathogen species (Leach and White, 1996; Dangl and Jones, 2001). In the last decade, it has become clear that many avirulence genes are dispensable, often encoding factors that contribute to pathogen virulence but with pathogen strains that lack a particular avirulence gene being fairly common (Leach and White, 1996; Kjemtrup et al., 2000). PAMPs, as typically conceived, encode indispensable molecules such as flagellin that are conserved across many species and that a pathogen cannot simply delete from its repertoire. However, plant viruses provide useful examples of avirulence genes that, like PAMPs, are indispensable for pathogen viability. A virus capsid protein or replicase is an essential component, and variation for these avirulence genes is often possible only at the level of polymorphisms that change a single amino acid in the gene product (Culver and Dawson, 1989; Cruz and Baulcombe, 1993; Li et al., 1999; Palanichelvam and Schoelz, 2002; Jenner et al., 2003; Kobayashi and Hohn, 2004). It remains appropriate to call flagellin a PAMP given its very widespread and indispensable presence among bacterial pathogens and its activity as an elicitor of host immune responses. However, our data show that this PAMP can exist as a relatively variable compound, not unlike many avirulence gene products.
Findings about FLS2 and flagellin also offer an instructive push to broaden our conception of R genes. FLS2 is like numerous R genes in that it encodes an LRR-containing product that controls pathogen recognition and defense activation in plants. Admittedly, FLS2 is more like Toll-like receptors of mammalian innate immunity than plant R gene products, in that the FLS2-mediated defense response is only mildly effective at slowing pathogen infection and the response does not involve the hypersensitive response programmed cell death that is often associated with R gene–mediated defenses (Akira et al., 2001; Dangl and Jones, 2001; Gomez-Gomez and Boller, 2002). However, there are examples of other R genes for which the hypersensitive response is not a required part of their activity and of R gene alleles that are only moderately effective at slowing pathogen multiplication (McIntosh et al., 1995; Greenberg and Yao, 2004). R genes were historically defined by the presence of a detectable gene-for-gene genetic interaction with a pathogen, but as research progresses, R genes are being discovered and used without clear knowledge of the corresponding pathogen virulence/avirulence. And FLS2, it is now clear, can encounter pathogen species that are polymorphic for expression of the eliciting flagellin molecule. FLS2 might seem less like a typical R gene in that it acts against multiple pathogen species, whereas many R genes are known to act against only one species, but specificity for only a single pathogen species is not a defining trait of R genes (Rossi et al., 1998; Cooley et al., 2000; Xiao et al., 2001). Lastly, FLS2 is like many R genes in that functional alleles are often not present in healthy plant populations, whereas altered function of mammalian Toll-like receptors is rare and is associated with significant immune disorders (Abreu and Arditi, 2004; Netea et al., 2004). Some Arabidopsis accessions isolated from wild populations lack a functional FLS2 (Gomez-Gomez and Boller, 2002; M. Dunning and A.F. Bent, unpublished results), including the widely studied and reasonably immunocompetent Wassilewskija (Ws-0) accession. Overall, it seems increasingly appropriate to think of FLS2 as another type of plant R gene.
Elicitation-Active Sites of Flagellin
This study dissected the entire flagellin protein to identify defense-eliciting structural elements. The FLS2/flagellin system has been the focus of much recent study, but most of these studies have used flg22 peptides and have not attended to the possibility that other flagellin domains might contribute to flagellin perception by the plant. Within the flg22 domain, past studies that noted differing elicitation abilities between flagellins of different species have not identified the naturally occurring amino acid polymorphisms that control the FLS2-mediated defense activation in Arabidopsis. We used purified full-length flagellins to determine that domains other than flg22 contribute very little to the elicitation of FLS2-dependent or FLS2-independent defense responses in Arabidopsis. Alignment analysis of Xcc FliC derived amino acid sequences revealed major divergence across several different domains in addition to the flg22 region, but domain swaps showed no contribution to defense elicitation by domains other than flg22. We subsequently identified the specific polymorphism responsible for the variation in elicitation activity. Domain swaps and site-directed mutagenesis together showed that across all observed Xcc flagellin polymorphisms, the amino acid 43 polymorphism determined elicitation activity. The D43V change was always part of the swaps that caused the loss of elicitation ability of Xcc B305 flagellin, and importantly, the reciprocal V43D substitution converted noneliciting Xcc B186 flagellin to an eliciting flagellin. Bacterial motility was not detectably altered by these flagellin sequence interconversions.
Although only the flg22 region of the purified Xcc flagellins was found to harbor elicitation activity on Arabidopsis, we cannot rule out the possibility that other flagellin domains or posttranslational modifications serve as elicitors of defense responses on other plants. In the flagellins of Salmonella dublin, E. coli, and Salmonella muenchen, conserved N- and C-terminal domains distinct from the flg22 domain have been implicated in the activation of animal TLR5-mediated inflammatory responses (Eaves-Pyles et al., 2001b; Donnelly and Steiner, 2002; Murthy et al., 2004). Flagellins from Pseudomonas avenae, P. syringae pv glycinea, and Pst have also been reported to induce plant cell death in nonhost rice (Oryza sativa) and tobacco (Nicotiana tabacum) plants, and the defense elicitation activity of flagellin from P. syringae pv glycinea has been attributed to domains other than the flg22 region (Che et al., 2000; Taguchi et al., 2003; Takeuchi et al., 2003). Flagellin glycosylation apparently can also serve as a specific determinant of compatibility between phytopathogenic bacteria and plants (Takeuchi et al., 2003). However, our findings reinforce the focus on the flg22 domain for studies of Arabidopsis flagellin perception. Other amino acids within the flg22 domain and across the entire flagellin protein may influence overall structure, potentially affecting localization within the plant and precise positioning of the flg22 domain with respect to its plant receptor, but the amino acid at position 43 clearly plays a significant role in plant defense elicitation.
Our studies of flagellin position 43 suggest that structural aspects beyond acidic charge determine elicitation activity. Replacing the charged Asp residue with the similarly charged yet structurally different Glu residue resulted in a relatively complete loss of activity, whereas replacement of Asp with structurally similar but nonacidic Asn allowed the retention of significant activity. Interestingly, the smaller, less sterically hindered Ala residue also showed loss of activity, indicating that hydrogen bonding of Asp-43 may play a central role in receptor elicitation. Not surprisingly, the gross structural changes caused by a Pro at position 43 produced a flagellin with no elicitation activity.
The work described here was conducted to lay the groundwork for future structure–function research on pathogen recognition by plants, in expectation that the plant flagellin receptor would be identified. FLS2 was the predicted receptor based on earlier studies that showed, for example, that Arabidopsis membrane preparations exhibited specific binding of bioactive flg peptides (and not inactive analogs), with this binding lost in preparations from plants carrying mutations in FLS2 (Bauer et al., 2001). It had also been shown that TLR5, the transmembrane protein with an extracellular LRR that mediates mammalian responsiveness to flagellin, directly binds flagellin (Mizel et al., 2003; West et al., 2005). It remained possible that FLS2 may instead sense flagellin indirectly by monitoring changes that flagellin elicits in other host molecules, as occurs with those R gene products that sense Avr proteins via a guard mechanism (Kooman-Gersmann et al., 1996; Van der Biezen and Jones, 1998; Quirino and Bent, 2003; Nurnberger et al., 2004). However, the very recent report by Chinchilla and colleagues (2006) provides strong evidence that the flg22 peptide is in fact bound directly and specifically by FLS2 and that FLS2 determines the specificity of flg22 perception in planta. Hence, this study on Xcc flagellins seems likely to have defined bioactive residues that determine elicitation activity during physical interaction with FLS2.
Organism-Level Effects of Flagellin Perception
Recently, it was reported that plant perception of Pst flagellins could restrict the invasion of bacteria at an early step after Pst strain DC3000 bacteria were sprayed onto Arabidopsis leaves but that the flagellin detection system was ineffective when vacuum infiltration was used to introduce bacteria directly into the leaf mesophyll (Zipfel et al., 2004). We reproduced these results. Presumably, Pst is restricted at an early step during infection that is bypassed by vacuum infiltration. This may be a population/neighborhood-level restriction, wherein early invading bacteria are present in low quantities and elicit mild FLS2-mediated defenses that restrict subsequent invaders in the same or adjacent host cells. Vacuum infiltration may foster an infection that is sufficiently synchronous to tip the balance in favor of the pathogen, with essentially all inoculated Pst able to enter and initiate a type III secretion-mediated suppression of defenses (Alfano and Collmer, 2004) before the host can use FLS2 to muster a more effective defense response.
We investigated whether the plant perception of Xcc flagellin limits infection by Xcc. Early results with wild-type Xcc strains showed a correlation between the absence of a recognized flagellin and the relative virulence on Arabidopsis Col-0, but this comparison among genetically diverse strains was apparently misleading. Isogenic Xcc strains expressing eliciting and noneliciting flagellins as a result of precise gene replacement at the fliC locus did not exhibit detectable differences in growth or virulence on plants. Bacteria were applied to leaves using vacuum infiltration, spray, or hydathode invasion methods, hydathode invasion being the primary infection route of Xcc in natural settings. The experiments suggested that possession of an eliciting flagellin is not sufficient to restrict the growth of otherwise virulent Xcc in Arabidopsis leaves. Experiments with Brassica oleracea TO1434 also did not reveal differences in virulence between these fliC-isogenic Xcc strains (W. Sun and A.F. Bent, unpublished results). The reduced growth of Xcc strains such as B305 may instead be attributable to the production of defense elicitors other than flagellin (Figures 1, 6, and 7; see discussion below) or to the production of type III secreted effectors (Alfano and Collmer, 2004) that are less effective at defense suppression within Arabidopsis than those made by members of the Xcc B186 clade.
There are a number of possible explanations for the observation that elicitation-active flagellins do not reduce the virulence of the Xcc strains making those flagellins. Xcc bacteria may not produce enough flagellin monomers in leaves to sufficiently induce host defense responses. All Xcc strains used in this study were motile in standard motility assays, but the speed of Xcc spread on motility plates was much slower than that of Pst. Although the results were for bacteria taken from rich culture media rather than from plants, light microscopy also revealed that the percentage of motile Xcc bacteria (∼1 to 3%) was much lower that that of mobile Pst bacteria (∼50%; data not shown). Xcc may also retain more intact flagella and exude fewer flagellin monomers than Pst. The potential benefit of reducing the levels of free flagellin monomers is suggested by our observation that treatment of plants with purified eliciting Xcc flagellin caused the subsequent growth of Xcc B186 to be restricted, whereas Xcc B186 expressing an eliciting flagellin but with no exogenous flagellin added did not exhibit this reduced growth in plants. Another factor contributing to the differential effectiveness of FLS2 systems against Pst as opposed to Xcc may be that Pst typically invades through stomates and colonizes the leaf mesophyll, whereas Xcc typically invades through hydathodes and colonizes the xylem (Schroth et al., 1991; Alvarez, 2000). If FLS2 systems are less sensitive or less effective in the leaf vasculature relative to the mesophyll, this raises the possibility that the hydathode/vascular invasion strategy may benefit Xcc in part by allowing for the avoidance of host flagellin detection systems. It is also possible that Xcc does not need its flagellin once it is positioned at a hydathode and that it sheds its flagellin at an early stage of infection to prevent recognition. For several bacterial pathogens, motility is an important virulence trait that is nevertheless dispensable for virulence if the pathogen is directly inoculated into the host (Panopoulos and Schroth, 1974; Haefele and Lindow, 1987; Tans-Kersten et al., 2001; Pfund et al., 2004). Lastly, Xcc effectors such as those transported by type III secretion may simply be better at suppressing and/or overcoming FLS2-mediated responses than are the Pst effectors. These possibilities all represent testable hypotheses for future investigation. Regardless of the mechanism, our results provide evidence that the presence of FLS2-eliciting flagellins is of variable relevance in the overall plant defense response against different bacterial pathogens.
This finding regarding flagellin does not imply that Xcc bacteria entirely escape detection by plant basal defense systems. For example, lipopolysaccharides, harpins, and cold shock proteins of X. campestris have been implicated as general elicitors of plant defense responses (Newman et al., 1995; Meyer et al., 2001; Felix and Boller, 2003; Kim et al., 2004; Silipo et al., 2005). Our results reveal that flagellin detection systems are apparently circumvented by Xcc bacteria, but plant–microbe interactions involve multiple layers of defense and counterdefense, and other elicitors of basal defenses remain that may limit plant colonization by Xcc.
A broader point is brought up by the results showing that pretreatment with Xcc flagellins can elicit FLS2-mediated responses that immunize the plant and reduce the severity of subsequent Xcc infection. Depending on the microbial populations and environmental conditions involved, the most significant contribution of FLS2-mediated defenses in some natural settings may be to reduce the severity of subsequent infections by the same pathogen, or by different pathogens.
Together, these data show that within a single pathogen species, flagellin is not necessarily a stable eliciting molecule and that the contribution of flagellin perception to plant disease resistance can vary significantly across different host–pathogen combinations. Future studies using diverse Brassica hosts and environmental settings may provide further insight into why plants can or cannot use flagellin perception to restrict the growth of Xcc pathogens. It will also be valuable to couple the available structure–function data for flagellin elicitation activity with similar information on the host flagellin receptor, to gain detailed knowledge of the recognition events that mediate flagellin perception and plant defense activation.
METHODS
The data in each figure are from representative experiments that were repeated at least twice.
Plant Materials and Bacterial Strains
Arabidopsis thaliana ecotype Col-0 and fls2-101 were used in this study. The fls2-101 line is homozygous for a T-DNA insertion that was initially characterized at the Salk knockout facility (JP71.4 G09), with the homozygous mutant line subsequently isolated and confirmed by DNA gel blot analysis and growth inhibition assay (Pfund et al., 2004). Plants were typically grown at 22°C under 9-h photoperiods at 100 to 150 μE·m−2·s−1 in controlled environment chambers. Xanthomonas campestris pv campestris strains were provided by Al Poplawsky and Wesley Chun (University of Idaho), except for strain 2669, which was from Robert Stall (University of Florida). Xcc and Pseudomonas syringae pv tomato were typically grown in NYG liquid or NYGA solidified medium (5 g/L bactopeptone, 3 g/L yeast extract, and 20 mL/L glycerol, with 15 g/L agar for solid medium).
Seedling Inhibition Assays
Seedling inhibition assays were performed as described by Pfund et al. (2004). Typically, 10 5-d-old Arabidopsis seedlings per treatment were transferred to a 24-well plate (one seedling per well), with each well carrying 400 μL of 0.5× MS salts and 10 μM flg22 peptide or ∼5 μM (80 μg/mL) purified His-tagged flagellin protein (as noted); after 10 to 14 d of further growth, each seedling was briefly blotted dry and weighed. Preparation of boiled bacterial extracts was also as described by Pfund et al. (2004).
Cloning, Sequencing, and Phylogenetic Analysis of fliC Genes
The fliC loci including flanking sequence were cloned using PCR primers designed from the Xcc (ATCC33913) genomic sequence (da Silva et al., 2002). The primers were 5′-GACTTGAGCGTGGTCATG-3′ and 5′-TTCAGCAGATGCAGTCG-3′. The 1.7-kb products containing fliC and ∼250 bp of flanking sequence on each end were blunt-cloned into pCR-BluntII-TOPO (Invitrogen), and the nucleotide sequence was determined by sequencing three independent clones generated from independent PCR runs. Derived amino acid sequences were then generated and aligned using ClustalX (Thompson et al., 1997). Phylogenetic analysis using the alignment shown in Supplemental Figure 3 online was conducted by nearest-neighbor analysis using the PHYLIP package (version 3.6; Felsenstein, 1989) with 1000 bootstrap trials; the final phylogeny shown in Figure 2 is the consensus of the 1000 generated trees.
Expression and Purification of His-Tagged Flagellin Proteins
His-tagged FliC proteins were prepared from amplified fliC PCR products subcloned into the pQE30 vector (Qiagen) essentially according to the supplier's instructions. Full-length open reading frames of Xcc fliC were isolated using the forward primer 5′-AAAGGATCCATGGCACAGGTAATCAACACC-3′ and the reverse primer 5′-TTTAAGCTTTACTGCAGCAGGCTCAGCACG-3′. The resultant product was cleaved by BamHI and HindIII and subcloned into the pQE30 vector. Escherichia coli XL1-Blue cells were transformed with the plasmids and selected at 37°C on Luria-Bertani plates containing 100 μg/mL ampicillin. A single colony was placed into 2 mL of Luria-Bertani medium containing 100 μg/mL ampicillin and grown overnight at 37°C. A 50-fold dilution was made, and the culture was grown at 37°C until it reached a density of OD600 = 0.5 to 0.7. Isopropyl β-d-thiogalactoside was then added to a final concentration of 1 mM to induce the expression of flagellin proteins. After 4 to 5 h of incubation, the cells were collected by centrifugation and stored at −80°C until use. Cells were lysed by sonication in buffer B (7 M urea, 100 mM NaH2PO4, 10 mM Tris-Cl, and 5 mM imidazole, pH 8.0), and supernatants were then loaded onto nickel-nitrilotriacetic acid agarose Superflow columns (Qiagen), which were subsequently washed with buffer B and buffer C (7 M urea, 100 mM NaH2PO4, 10 mM Tris-Cl, and 5 mM imidazole, pH 6.3). The bound His6-tagged proteins were then eluted with buffer E (8 M urea, 100 mM NaH2PO4, and 10 mM Tris-Cl, pH 4.5) and dialyzed extensively in PBS (pH 7.4). The concentration of proteins was determined using the BCA protein assay kit (Pierce). The purity of flagellin proteins was verified by SDS-PAGE gel staining with Coomassie Brilliant Blue G 250.
Domain Swapping between Xcc B186 and B305 Flagellin Proteins
Domain swaps were generated by PCR of fliC alleles using splicing by overlap extension (Horton et al., 1989). The 5′ fragments of the B186 and B305 fliC gene were amplified using PCR primers 5′-TACAACGGCGACCAGACCCAG-3′ and 5′-TTCGACCATCGCGCCTTCGG-3′, and the 3′ fragments were amplified using PCR primers 5′-AACGACGGCATCTCGCTGGC-3′ and 5′-AAAAGCTTATGCGGTGAGGTTGCCCT-3′. The gel-purified PCR products with a 48-bp overlap were added together into a new PCR without primers, then after 12 cycles of self-extension (94°C for 2 min, followed by 12 cycles of 94°C for 1 min, 55°C for 30 s, and 72°C for 4 min), the primers 5′-TACAACGGCGACCAGACCCAG-3′ and 5′-AAAAGCTTATGCGGTGAGGTTGCCCT-3′ were added and the PCR was continued using the same parameters for 30 more cycles. The resulting PCR products were gel-purified and subcloned into pCR-BluntII-TOPO vector (Invitrogen) and then into pQE30. Note that there were multiple options for locating the precise junction of this domain swap, because there are no polymorphisms between B186 and B305 flagellins between amino acids 43 (within the flg22 region) and 87.
Site-Directed Mutagenesis
Point mutations were generated by circular PCR with DpnI digestion to eliminate background wild-type plasmid pQE30-fliC. Briefly, two synthetic oligonucleotide primers containing the desired mutation, each complementary to opposite strands of the same target sequence, were extended during temperature cycling by PfuTurbo DNA polymerase (Stratagene). After cycling, the products were treated with DpnI to specifically digest the methylated parental DNA template. The resultant products, which included double-stranded annealed plasmids with a strand break at the 5′ end of each primer, were then transformed into XL1-Blue supercompetent cells. The resultant mutated plasmid constructs were verified by sequencing. For substitution of the flg22 block from B305 flagellin into the B186 flagellin, an adapted directed mutagenesis approach was used. A pair of primers, 5′-CAGGCCGCGGATCTGCGTGGTGAAGCGCTCGGAGATTGCC-3′ and 5′-ACAGTTCGAGCATGGCGCTGAGCATCCAGCGTCTGTCCTC-3′, was used to amplify the nucleotide sequences corresponding to the flg22-encoding region of B305 fliC. The product was gel-purified, and the denatured strands were used as mutation primers for pQE30-B186fliC mutagenesis as described above.
Construction of fliC Gene Replacement Strains of Xcc B186
The full-length fliC coding region and ∼1 kb of upstream genomic sequence were amplified using Pfu polymerase with the forward primer 5′-AAATCTAGAACAACGCGACCAGACC-3′ and the reverse primer 5′-AAAACTCGAGATGCGGTGAGGTTGCCC-3′ with XbaI and XhoI restriction sites, respectively. The resultant PCR product was subcloned into pCR-BluntII-TOPO vector. Mutations at amino acid residue Val-43 were introduced by site-directed mutagenesis as described above. XbaI fragments with mutated fliC genes were cloned into the pUFR80 suicide vector (courtesy of Dean Gabriel, University of Florida), a suicide vector that allows the generation of precise unmarked chromosomal gene replacements in Gram-negative bacteria (Ried and Collmer, 1987). pUFR80 is a pUC119 derivative (it will not replicate in Xcc) that carries an NPTII gene for kanamycin selection and a sacB gene that confers sucrose toxicity. pUFR80+fliC products were electroporated into Xcc B186 and subjected to kanamycin selection. For electroporation, Xcc cells were washed in 10% glycerol and resuspended to ∼1 × 1011 colony-forming units (cfu)/mL. Cells were incubated with 1 to 2 μg of DNA and pulsed in a 1-mm cuvette (FisherBiotech) at 1.75 kV (White and Gonzalez, 1995). Cells were grown for 2 to ∼4 h in NYG and subsequently transferred to NYGA with 50 μg/mL kanamycin. After integrants had been selected, single clones were picked and cultured in NYG medium overnight with no kanamycin. The cultures were then spread onto NYGA plates with 5% sucrose to select sucrose-insensitive clones. The fliC genotype of kanamycin-sensitive/sucrose-insensitive Xcc colonies was determined by sequencing PCR products from genomic DNA of these strains (template DNA isolated using the genomic DNA isolation kit; Promega).
Bacterial Motility Assays
Small samples of freshly grown bacterial cells taken from NYGA medium were stabbed onto swarm agar plates (10 g/L tryptone and 0.3% noble agar) (Boles and McCarter, 2000) and incubated at 28°C. Colony spread caused by swimming motility was differentiated from spread attributable to population increase by the presence of a thin leading edge of bacteria extending out from the colony within rather than on the surface of the swarm agar plates. By light microscopy, flagella-mediated motility was revealed by brief rapid movements of individual bacteria in a straight line for many micrometers, in contrast with twitching and tumbling movements.
Virulence Assays and Bacterial Growth Assays
All strains were grown at 28°C on NYGA medium. For vacuum infiltration, a fresh culture of bacteria was scraped off the plate, resuspended in 10 mM MgCl2 to 106 cfu/mL with 0.005% Silwet L-77 (OSi Specialties), and vacuum-infiltrated into leaves. Three days after infection, leaf discs from four different leaves were combined and ground in 10 mM MgCl2 with a microcentrifuge tube plastic pestle, with triplicate replication (12 leaves total). The samples were vortex-mixed, diluted 1:10 serially, and plated on NYGA solid medium, supplemented with rifampicin in the case of Pst DC3000, and viable colonies were counted after 2 d of growth at 28°C. Alternatively, samples were taken at 0, 2, and 4 d after infiltration and surface-sterilized in 70% ethanol for 45 s, then rinsed three times with sterile distilled water before grinding in 10 mM MgCl2. For syringe inoculation, a fresh culture of bacteria was scraped off the plate, resuspended in sterile water to 105 cfu/mL (for Pst DC3000) and to 5 × 105 cfu/mL (for Xcc B186), and infiltrated into leaves by syringe infiltration using a 1-cc plastic syringe with no needle. For spray inoculation, Pst was resuspended in 10 mM MgCl2/0.04% Silwet L-77 at 5 × 108 cfu/mL, Xcc was used at 109 cfu/mL, and bacterial populations were determined using leaf disc samples as described above, with surface-sterilization before grinding. For hydathode inoculation, 3-week-old Arabidopsis Col-0 and fls2-101 mutant plants were placed in a dew chamber (wall, 10°C; water, 28°C; and air, 22°C) for 4 to 5 h until guttation drops formed at the edges of the leaves. Bacteria at ∼1 × 109 cfu/mL in 10 mM MgCl2/0.04% Silwet L-77 were then gently misted onto leaves, and plants were returned to their normal growth chamber and covered with transparent domes to maintain increased humidity for 2 d. To assess bacterial populations in hydathode-infected leaves, four entire leaves of similar size were removed 4 d after inoculation, surface-sterilized with 15% H2O2 for 5 min, washed three times with sterile distilled water, and ground in 10 mM MgCl2, and extracts were serially diluted on NYGA plates as described above. Triplicate samples were tested (12 leaves per treatment). Virulence assays on Brassica rapa accession C1-22 and Brassica oleracea accession TO1434 (courtesy of T. Osborn, University of Wisconsin–Madison) were conducted using a toothpick carrying Xcc bacteria from an NYGA plate to stab the midvein of seedling leaves on soil-grown plants, or separately by misting of an Xcc suspension onto guttating seedling leaves of soil-grown plants, or separately by syringe inoculation of bacteria into leaf mesophyll. One-way analysis of variance was performed using Minitab (release 14).
Flagellin Pretreatment
Treatments with His6-tagged flagellin were performed by syringe infiltration using a 1-cc plastic syringe with no needle to introduce 1 μM flagellin solution into the leaves. One day later, the same flagellin-treated leaves were inoculated with 105 cfu/mL Pst DC3000 or 5 × 105 cfu/mL Xcc B186 by syringe infiltration, and 2 d after that, bacterial populations were measured by dilution plating of homogenized leaf samples as described above. For each treatment, two leaves from each of six to eight plant replicates were used. Each experiment was repeated at least twice.
Semiquantitative RT-PCR
Six-day-old seedlings were treated with 5 μM His6-tagged Xcc B305 and B186 wild-type and mutant flagellins or 5 μM flg22 peptide in 0.5× MS medium for 24 h and then collected for RNA isolation. Total RNA was isolated using Trizol reagent according to the provided protocol (Invitrogen). cDNA was synthesized by reverse transcriptase Superscript III (Invitrogen) using 5 μg of total RNA as template. Two microliters of cDNA product from reverse transcription reactions was used as template for PCR performed with Taq polymerase (Promega) for 40 cycles at 51°C. The primer pairs 5′-GTAGGTGCTCTTGTTCTTCCC-3′/5′-CACATAATTCCCACGAGGATC-3′ and 5′-AGGTTCTGTTCCAGCCATC-3′/5′-TTAGAAGCATTTCCTGTGAAC-3′ were used to track PR-1 and actin gene expression, respectively. PCR products were separated by gel electrophoresis and stained with ethidium bromide before viewing.
Accession Numbers
The Arabidopsis Genome Initiative number for FLS2 is At5g46330.1. DNA sequence data for the Xcc fliC genes can be found in the GenBank data library under accession numbers DQ356455 to DQ356466.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ClustalX Alignment of Xcc Flagellin-Derived Amino Acid Sequences from the 12 Xcc Strains Used in This Study.
Supplemental Figure 2. Restriction of the Growth of Pst DC3000 in Arabidopsis Attributable to FLS2-Mediated Responses.
Supplemental Figure 3. ClustalX Alignment of Xcc Flagellin-Derived Amino Acid Sequences from 12 Different Xcc Strains as Well as Salmonella enterica serovar Typhimurium and Pseudomonas aeruginosa.
Supplementary Material
Acknowledgments
We thank Al Poplawsky and Wesley Chun (University of Idaho) and Robert Stall (University of Florida) for the Xcc strains; Dean Gabriel (University of Florida) for providing the sacB vector before publication; Jorge Escalante (University of Wisconsin–Madison) for genomic DNA from S. enterica serovar Typhimurium; Frances Yap for assistance with protein purification; Jian Yao for help with motility assays and statistics; and Ruth Genger and Lori Adams-Phillips for critical reading of the manuscript. C.P. was a Department of Energy Energy Biosciences Research Fellow of the Life Sciences Research Foundation. This project was also supported by Department of Energy Grant DE-FG02-02ER15342 to A.F.B.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Andrew F. Bent (afb@plantpath.wisc.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.037648.
References
- Abreu, M.T., and Arditi, M. (2004). Innate immunity and Toll-like receptors: Clinical implications of basic science research. J. Pediatr. 144 421–429. [DOI] [PubMed] [Google Scholar]
- Akira, S., Takeda, K., and Kaisho, T. (2001). Toll-like receptors: Critical proteins linking innate and acquired immunity. Nat. Immunol. 2 675–680. [DOI] [PubMed] [Google Scholar]
- Alfano, J.R., and Collmer, A. (2004). Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42 385–414. [DOI] [PubMed] [Google Scholar]
- Alvarez, A.M. (2000). Black rot of crucifers. In Mechanisms of Resistance to Plant Diseases, A. Slusarenko, R.S.S. Fraser, and L.C. van Loon, eds (Dordrecht, The Netherlands: Kluwer), pp. 21–52.
- Andersen-Nissen, E., Smith, K.D., Strobe, K.L., Rassoulian Barrett, S.L., Cookson, B.T., Logan, S.M., and Aderem, A. (2005). Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. USA 102 9247–9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interactions. Science 276 726–733. [DOI] [PubMed] [Google Scholar]
- Bauer, Z., Gomez-Gomez, L., Boller, T., and Felix, G. (2001). Sensitivity of different ecotypes and mutants of Arabidopsis thaliana toward the bacterial elicitor flagellin correlates with the presence of receptor-binding sites. J. Biol. Chem. 276 45669–45676. [DOI] [PubMed] [Google Scholar]
- Bent, A., Innes, R., Ecker, J., and Staskawicz, B. (1992). Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol. Plant Microbe Interact. 5 372–378. [DOI] [PubMed] [Google Scholar]
- Boles, B.R., and McCarter, L.L. (2000). Insertional inactivation of genes encoding components of the sodium-type flagellar motor and switch of Vibrio parahaemolyticus. J. Bacteriol. 182 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell, C.R., and Somerville, S.C. (1997). Use of Arabidopsis recombination lines reveals a monogenic and a novel digenic resistance mechanism to Xanthomonas campestris pv campestris. Plant J. 12 21–29. [DOI] [PubMed] [Google Scholar]
- Che, F.S., Nakajima, Y., Tanaka, N., Iwano, M., Yoshida, T., Takayama, S., Kadota, I., and Isogai, A. (2000). Flagellin from an incompatible strain of Pseudomonas avenae induces a resistance response in cultured rice cells. J. Biol. Chem. 275 32347–32356. [DOI] [PubMed] [Google Scholar]
- Chinchilla, D., Bauer, Z., Regenass, M., Boller, T., and Felix, G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, M.B., Pathirana, S., Wu, H.J., Kachroo, P., and Klessig, D.F. (2000). Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz, S.S., and Baulcombe, D.C. (1993). Molecular analysis of potato virus X isolates in relation to the potato hypersensitivity gene Nx. Mol. Plant Microbe Interact. 6 707–714. [DOI] [PubMed] [Google Scholar]
- Culver, J.N., and Dawson, W.O. (1989). Tobacco mosaic virus coat protein: An elicitor of the hypersensitive reaction but not required for the development of mosaic symptoms in Nicotiana sylvestris. Virology 173 755–758. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- da Silva, A.C., et al. (2002). Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417 459–463. [DOI] [PubMed] [Google Scholar]
- Donnelly, M.A., and Steiner, T.S. (2002). Two nonadjacent regions in enteroaggregative Escherichia coli flagellin are required for activation of Toll-like receptor 5. J. Biol. Chem. 277 40456–40461. [DOI] [PubMed] [Google Scholar]
- Eaves-Pyles, T.D., Murthy, K., Liaudet, L., Virag, L., Ross, G., Soriano, F.G., Szabo, C., and Salzman, A.L. (2001. a). Flagellin, a novel mediator of Salmonella-induced epithelial activation and systemic inflammation: IκBα degradation, induction of nitric oxide synthase, induction of proinflammatory mediators, and cardiovascular dysfunction. J. Immunol. 166 1248–1260. [DOI] [PubMed] [Google Scholar]
- Eaves-Pyles, T.D., Wong, H.R., Odoms, K., and Pyles, R.B. (2001. b). Salmonella flagellin-dependent proinflammatory responses are localized to the conserved amino and carboxyl regions of the protein. J. Immunol. 167 7009–7016. [DOI] [PubMed] [Google Scholar]
- Felix, G., and Boller, T. (2003). Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J. Biol. Chem. 278 6201–6208. [DOI] [PubMed] [Google Scholar]
- Felix, G., Duran, J.D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18 165–176. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). PHYLIP—Phylogeny inference package (version 3.2). Cladistics 5 164–166. [Google Scholar]
- Gewirtz, A.T., Yu, Y., Krishna, U.S., Israel, D.A., Lyons, S.L., and Peek, R.M., Jr. (2004). Helicobacter pylori flagellin evades Toll-like receptor 5-mediated innate immunity. J. Infect. Dis. 189 1914–1920. [DOI] [PubMed] [Google Scholar]
- Godard, F., Lummerzheim, M., Saindrenan, P., Balague, C., and Roby, D. (2000). hxc2, an Arabidopsis mutant with an altered hypersensitive response to Xanthomonas campestris pv. campestris. Plant J. 24 749–761. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez, L. (2004). Plant perception systems for pathogen recognition and defence. Mol. Immunol. 41 1055–1062. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5 1003–1011. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez, L., and Boller, T. (2002). Flagellin perception: A paradigm for innate immunity. Trends Plant Sci. 7 251–256. [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez, L., Felix, G., and Boller, T. (1999). A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 18 277–284. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., and Ausubel, F.M. (1993). Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J. 4 327–341. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., and Yao, N. (2004). The role and regulation of programmed cell death in plant-pathogen interactions. Cell. Microbiol. 6 201–211. [DOI] [PubMed] [Google Scholar]
- Haefele, D.M., and Lindow, S.E. (1987). Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae. Appl. Environ. Microbiol. 53 2528–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, F., Smith, K.D., Ozinsky, A., Hawn, T.R., Yi, E.C., Goodlett, D.R., Eng, J.K., Akirak, S., Underhill, D.M., and Aderem, A. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410 1099–1103. [DOI] [PubMed] [Google Scholar]
- Horton, R.M., Hunt, H.D., Ho, S.N., Pullen, J.K., and Pease, L.R. (1989). Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene 77 61–68. [DOI] [PubMed] [Google Scholar]
- Hugouvieux, V., Barber, C.E., and Daniels, M.J. (1998). Entry of Xanthomonas campestris pv. campestris into hydathodes of Arabidopsis thaliana leaves: A system for studying early infection events in bacterial pathogenesis. Mol. Plant Microbe Interact. 11 537–543. [DOI] [PubMed] [Google Scholar]
- Jenner, C.E., Wang, X., Tomimura, K., Ohshima, K., Ponz, F., and Walsh, J.A. (2003). The dual role of the potyvirus P3 protein of Turnip mosaic virus as a symptom and avirulence determinant in brassicas. Mol. Plant Microbe Interact. 16 777–784. [DOI] [PubMed] [Google Scholar]
- Jones, D.A., and Takemoto, D. (2004). Plant innate immunity—direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 16 48–62. [DOI] [PubMed] [Google Scholar]
- Kim, J.G., Jeon, E., Oh, J., Moon, J.S., and Hwang, I. (2004). Mutational analysis of Xanthomonas harpin HpaG identifies a key functional region that elicits the hypersensitive response in nonhost plants. J. Bacteriol. 186 6239–6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.G., da Cunha, L., McFall, A.J., Belkhadir, Y., DebRoy, S., Dangl, J.L., and Mackey, D. (2005). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121 749–759. [DOI] [PubMed] [Google Scholar]
- Kjemtrup, S., Nimchuk, Z., and Dangl, J.L. (2000). Effector proteins of phytopathogenic bacteria: Bifunctional signals in virulence and host recognition. Curr. Opin. Microbiol. 3 73–78. [DOI] [PubMed] [Google Scholar]
- Kobayashi, K., and Hohn, T. (2004). The avirulence domain of Cauliflower mosaic virus transactivator/viroplasmin is a determinant of viral virulence in susceptible hosts. Mol. Plant Microbe Interact. 17 475–483. [DOI] [PubMed] [Google Scholar]
- Komoriya, K., Shibano, N., Higano, T., Azuma, N., Yamaguchi, S., and Aizawa, S.I. (1999). Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34 767–779. [DOI] [PubMed] [Google Scholar]
- Kooman-Gersmann, M., Honee, G., Bonnema, G., and De Wit, P.J.G.M. (1996). A high-affinity binding site for the AVR9 peptide elicitor of Cladosporium fulvum is present on plasma membranes of tomato and other solanaceous plants. Plant Cell 8 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korves, T.M., and Bergelson, J. (2003). A developmental response to pathogen infection in Arabidopsis. Plant Physiol. 133 339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, J.E., and White, F.F. (1996). Bacterial avirulence genes. Annu. Rev. Phytopathol. 34 153–179. [DOI] [PubMed] [Google Scholar]
- Lee, S.K., Stack, A., Katzowitsch, E., Aizawa, S.I., Suerbaum, S., and Josenhans, C. (2003). Helicobacter pylori flagellins have very low intrinsic activity to stimulate human gastric epithelial cells via TLR5. Microbes Infect. 5 1345–1356. [DOI] [PubMed] [Google Scholar]
- Lemaitre, B., Reichhart, J.-M., and Hoffmann, J.A. (1997). Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 94 14614–14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.W., Lucy, A.P., Guo, H.S., Li, W.X., Ji, L.H., Wong, S.M., and Ding, S.W. (1999). Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 18 2683–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Lin, H., Zhang, W., Zou, Y., Zhang, J., Tang, X., and Zhou, J.M. (2005). Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA 102 12990–12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lummerzheim, M., de Oliveira, D., Castresana, C., Miguens, F.C., Louzasa, E., Roby, D., Van Montagu, M., and Timmerman, B. (1993). Identification of compatible and incompatible interactions between Arabidopsis thaliana and Xanthomonas campestris pv campestris and characterization of the hypersensitive response. Mol. Plant Microbe Interact. 6 532–544. [Google Scholar]
- Maleck, K., Neuenschwander, U., Cade, R.M., Dietrich, R.A., Dangl, J.L., and Ryals, J.A. (2002). Isolation and characterization of broad-spectrum disease-resistant Arabidopsis mutants. Genetics 160 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott, P.F., Ciacci-Woolwine, F., Snipes, J.A., and Mizel, S.B. (2000). High affinity interaction between Gram-negative flagellin and a cell surface polypeptide results in human monocyte activation. Infect. Immun. 68 5525–5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh, R.A., Wellings, C.R., and Park, R.F. (1995). Wheat Rusts: An Atlas of Resistance Genes. (Dordrecht, The Netherlands: CSIRO Australia and Kluwer Academic Publishers).
- Medzhitov, R., and Janeway, C., Jr. (2000). Innate immune recognition: Mechanisms and pathways. Immunol. Rev. 173 89–97. [DOI] [PubMed] [Google Scholar]
- Meindl, T., Boller, T., and Felix, G. (2000). The bacterial elicitor flagellin activates its receptor in tomato cells according to the address-message concept. Plant Cell 12 1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, A., Puhler, A., and Niehaus, K. (2001). The lipopolysaccharides of the phytopathogen Xanthomonas campestris pv. campestris induce an oxidative burst reaction in cell cultures of Nicotiana tabacum. Planta 213 214–222. [DOI] [PubMed] [Google Scholar]
- Mizel, S.B., West, A.P., and Hantgan, R.R. (2003). Identification of a sequence in human Toll-like receptor 5 required for the binding of Gram-negative flagellin. J. Biol. Chem. 278 23624–23629. [DOI] [PubMed] [Google Scholar]
- Moens, S., and Vanderleyden, J. (1996). Functions of bacterial flagella. Crit. Rev. Microbiol. 22 67–100. [DOI] [PubMed] [Google Scholar]
- Murthy, K.G., Deb, A., Goonesekera, S., Szabo, C., and Salzman, A.L. (2004). Identification of conserved domains in Salmonella muenchen flagellin that are essential for its ability to activate TLR5 and to induce an inflammatory response in vitro. J. Biol. Chem. 279 5667–5675. [DOI] [PubMed] [Google Scholar]
- Netea, M.G., van der Graaf, C., Van der Meer, J.W., and Kullberg, B.J. (2004). Toll-like receptors and the host defense against microbial pathogens: Bringing specificity to the innate-immune system. J. Leukoc. Biol. 75 749–755. [DOI] [PubMed] [Google Scholar]
- Newman, M.A., Daniels, M.J., and Dow, J.M. (1995). Lipopolysaccharide from Xanthomonas campestris induces defense-related gene expression in Brassica campestris. Mol. Plant Microbe Interact. 8 778–780. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol. Rev. 198 249–266. [DOI] [PubMed] [Google Scholar]
- O'Brien, E.J., and Bennett, P.M. (1972). Structure of straight flagella from a mutant Salmonella. J. Mol. Biol. 70 133–152. [DOI] [PubMed] [Google Scholar]
- O'Donnell, P.J., Schmelz, E.A., Moussatche, P., Lund, S.T., Jones, J.B., and Klee, H.J. (2003). Susceptible to intolerance—a range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J. 33 245–257. [DOI] [PubMed] [Google Scholar]
- Otteman, K.M., and Miller, J.F. (1997). Roles for motility in bacterial-host interactions. Mol. Microbiol. 24 1109–1117. [DOI] [PubMed] [Google Scholar]
- Palanichelvam, K., and Schoelz, J.E. (2002). A comparative analysis of the avirulence and translational transactivator functions of gene VI of Cauliflower mosaic virus. Virology 293 225–233. [DOI] [PubMed] [Google Scholar]
- Panopoulos, N.J., and Schroth, M.N. (1974). Role of flagellar motility in invasion of bean leaves by Pseudomonas phaseolicola. Phytopathology 64 1389–1397. [Google Scholar]
- Parker, J.E., Barber, C.E., Fan, M.-J., and Daniels, M.J. (1993). Interaction of Xanthomonas campestris with Arabidopsis thaliana: Characterization of a gene from X. c. pv. raphani that confers avirulence to most A. thaliana accessions. Mol. Plant Microbe Interact. 6 216–224. [DOI] [PubMed] [Google Scholar]
- Pasare, C., and Medzhitov, R. (2005). Toll-like receptors: Linking innate and adaptive immunity. Adv. Exp. Med. Biol. 560 11–18. [DOI] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103 1111–1120. [DOI] [PubMed] [Google Scholar]
- Pfund, C., Tans-Kersten, J., Dunning, F.M., Alonso, J.M., Ecker, J.R., Allen, C., and Bent, A.F. (2004). Flagellin is not a major defense elicitor in Ralstonia solanacearum cells or extracts applied to Arabidopsis thaliana. Mol. Plant Microbe Interact. 17 696–706. [DOI] [PubMed] [Google Scholar]
- Quirino, B.F., and Bent, A.F. (2003). Deciphering host resistance and pathogen virulence: The Arabidopsis/Pseudomonas interaction as a model. Mol. Plant Pathol. 4 517–530. [DOI] [PubMed] [Google Scholar]
- Ramos, H.C., Rumbo, M., and Sirard, J.C. (2004). Bacterial flagellins: Mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 12 509–517. [DOI] [PubMed] [Google Scholar]
- Ried, J.L., and Collmer, A. (1987). An nptI-sacB-sacR cartridge for constructing directed, unmarked mutations in Gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57 239–246. [DOI] [PubMed] [Google Scholar]
- Rossi, M., Goggin, F.L., Milligan, S.B., Kaloshian, I., Ullman, D.E., and Williamson, V.M. (1998). The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 95 9750–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatey, F.A., Imada, K., Nagashima, S., Vonderviszt, F., Kumasaka, T., Yamamoto, M., and Namba, K. (2001). Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410 331–337. [DOI] [PubMed] [Google Scholar]
- Schroth, M.N., Hildebrand, D.C., and Panopoulos, N. (1991). Phytopathogenic pseudomonads and related plant-associated pseudomonads. In The Prokaryotes, 2nd ed. A. Balows, H.G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer, eds (New York: Springer Verlag), pp. 3104–3131.
- Sierro, F., Dubois, B., Coste, A., Kaiserlian, D., Kraehenbuhl, J.P., and Sirard, J.C. (2001). Flagellin stimulation of intestinal epithelial cells triggers CCL20-mediated migration of dendritic cells. Proc. Natl. Acad. Sci. USA 98 13722–13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silipo, A., Molinaro, A., Sturiale, L., Dow, J.M., Erbs, G., Lanzetta, R., Newman, M.A., and Parrilli, M. (2005). The elicitation of plant innate immunity by lipooligosaccharide of Xanthomonas campestris. J. Biol. Chem. 280 33660–33668. [DOI] [PubMed] [Google Scholar]
- Simmonds, N.W., and Smartt, J. (1999). Principles of Crop Improvement. (Oxford, UK: Blackwell Science).
- Simpson, R.B., and Johnson, L.J. (1990). Arabidopsis thaliana as a host for Xanthomonas campestris pv campestris. Mol. Plant Microbe Interact. 3 233–237. [Google Scholar]
- Smith, K.D., Andersen-Nissen, E., Hayashi, F., Strobe, K., Bergman, M.A., Barrett, S.L.R., Cookson, B.T., and Aderem, A. (2003). Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4 1247–1253. [DOI] [PubMed] [Google Scholar]
- Taguchi, F., Shimizu, R., Nakajima, R., Toyoda, K., Shiraishi, T., and Ichinose, Y. (2003). Differential effects of flagellins from Pseudomonas syringae pvs. tabaci, tomato and glycinea on plant defense response. Plant Physiol. Biochem. 41 165–174. [Google Scholar]
- Takeuchi, K., Taguchi, F., Inagaki, Y., Toyoda, K., Shiraishi, T., and Ichinose, Y. (2003). Flagellin glycosylation island in Pseudomonas syringae pv. glycinea and its role in host specificity. J. Bacteriol. 185 6658–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tans-Kersten, J., Huang, H., and Allen, C. (2001). Ralstonia solanacearum needs motility for invasive virulence on tomato. J. Bacteriol. 183 3597–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Biezen, E.A., and Jones, J.D. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23 454–456. [DOI] [PubMed] [Google Scholar]
- West, A.P., Dancho, B.A., and Mizel, S.B. (2005). Gangliosides inhibit flagellin signaling in the absence of an effect on flagellin binding to Toll-like receptor 5. J. Biol. Chem. 280 9482–9488. [DOI] [PubMed] [Google Scholar]
- White, T.J., and Gonzalez, C.F. (1995). Electroporation of Xanthomonas. In Electroporation Protocols for Microorganisms, J.A. Nickoloff, ed (Totowa, NJ: Humana Press), pp. 135–141.
- Williams, P.H. (1980). Black rot: A continuing threat to world crucifers. Plant Dis. 64 736–742. [Google Scholar]
- Xiao, S., Ellwood, S., Calis, O., Patrick, E., Li, T., Coleman, M., and Turner, J.G. (2001). Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291 118–120. [DOI] [PubMed] [Google Scholar]
- Yonekura, K., Maki-Yonekura, S., and Namba, K. (2003). Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424 643–650. [DOI] [PubMed] [Google Scholar]
- Yu, I.C., Parker, J., and Bent, A.F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. USA 95 7819–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E.J., Jones, J.D.G., Felix, G., and Boller, T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.