Abstract
Cell motility is a complex biological process, involved in development, inflammation, homeostasis, and pathological processes such as the invasion and metastatic spread of cancer. Here, we describe a genomic screen designed to identify inhibitors of cell migration. A library of 10,996 small interfering RNAs (targeting 5,234 human genes) was screened for their ability to block the migration of a highly motile ovarian carcinoma cell line, SKOV-3, by using a 384-well wound-healing assay coupled with automated microscopy and wound quantification. Two or more small interfering RNAs against four genes, CDK7, DYRK1B, MAP4K4 (NIK/HGK) (MAP4K4, mitogen-activated protein 4 kinase 4), and SCCA-1 (SerpinB3), potently blocked the migration of SKOV-3 cells, concordant with reduced transcript levels. Further studies of the promigratory role of MAP4K4 showed that the knockdown of this transcript inhibited the migration of multiple carcinoma cell lines, indicating a broad role in cell motility and potently suppressed the invasion of SKOV-3 cells in vitro. The effect of MAP4K4 on cellular migration was found to be mediated through c-Jun N-terminal kinase, independent of AP1 activation and downstream transcription. Accordingly, small molecule inhibition of c-Jun N-terminal kinase suppressed SKOV-3 cell migration, underscoring the potential therapeutic utility of mitogen-activated protein kinase pathway inhibition in cancer progression.
Keywords: automated, RNA interference
Cell migration is triggered by a diverse array of cell autonomous and environmental cues, culminating in cycles of microfilament and microtubule reorganization, polarization of the secretory pathway, membrane extension, adhesion to the substratum, and retraction (1, 2). Transduction of promigratory stimuli through one of several key pathways [e.g., the mitogen-activated protein kinase (MAPK) pathway] engages the highly conserved Rho GTPases, Rac1, RhoA, and Cdc42, which integrate these signals with dynamic reorganization of the cytoskeleton (2). Many of the components of these signaling pathways that promote cell migration are commonly activated in tumor cells through mutation or overexpression of promigratory kinases (e.g., Met, Src, and fibroblast and epidermal growth factor receptors), mutation of Ras GTPases, or loss of tumor suppressors, such as the PTEN phosphatase (reviewed in ref 3). Activation of these pathways, and the consequent increase in cell motility, is highly correlated with tissue invasion and distant metastasis (3). Thus, dissecting tumor cell motility is likely to provide important insights into metastatic development.
Cell migration can be monitored by a number of assays in vitro and in vivo, ranging from microscopic observation of movement on a 2D substrate to intravital imaging in rodents (3). One of simplest, termed the wound-healing assay (4), is widely used to monitor the migratory potential of cells, requiring only the introduction of a scratch across the surface of a confluent cell monolayer with a pipette tip, or other blunt instrument, followed by movement of cells across the denuded surface to “heal” the wound. Here, we automated the wound-healing assay to monitor tumor cell migration in 384-well plates by using a custom-built “scratch” device, coupled with automated image capture and quantification of cellular migration. This automated phenotypic screen was used to interrogate the migratory suppression of ≈11,000 small interfering RNAs (siRNAs) in the highly migratory ovarian carcinoma cell line SKOV-3, resulting in the identification and validation of four previously undescribed promigratory genes, including the mitogen-activated kinase, MAP4K4 (also called nck-interacting kinase or hepatocyte progenitor kinase-like/germinal center kinase-like kinase), whose role in cancer cell motility was further characterized.
Results
Development of an Automated Cell Migration Assay.
Measurement of cell migration by the wound-healing assay (4) was automated by using a custom-built, 384-well scratch device, coupled to automated image capture and quantification of wound closure (see Fig. 7 and Supporting Materials and Methods, which are published as supporting information on the PNAS web site). The assay system utilizes a precision-drilled aluminum block into which 384 pipette tips are inserted and fastened. The block is lowered on a static arm into the wells of a 384-well plate containing confluent cell monolayers and shunted ≈3 mm by hydraulic pressure. After the wounds are allowed to close, the cells are fixed and stained with the nuclear stain, DAPI, to enable rapid image capture by automated microscopy.
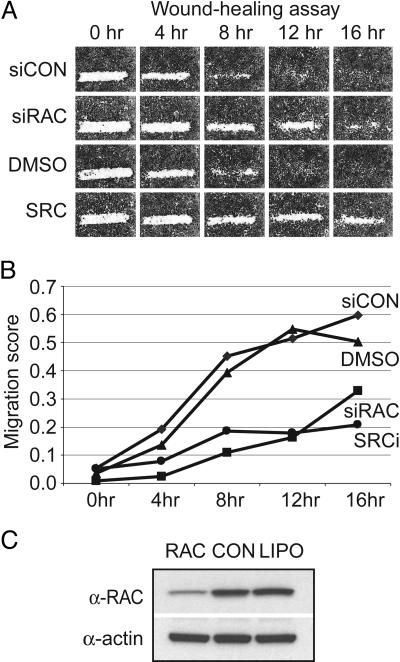
To validate the assay, the temporal migration of a highly motile ovarian carcinoma-derived cell line, SKOV-3, was monitored in the presence of siRNAs, small molecules and appropriate controls. The efficacy of siRNA-mediated migratory inhibition was assessed by using siRNAs homologous to Rac1, one of three Rho GTPases (RhoA, Rac1, and Cdc42) that integrates promigratory signals with dynamic reorganization of the actin cytoskeleton (ref. 2; Fig. 1A). In addition, we tested known small molecule inhibitors of c-Src (SKI-606, ref. 6; and a 2-phenyl-aminoimidazo-[4,5-h]-isoquinolin-9-one, termed compound 43, ref. 7), the activated form of which plays a central role in the motility and invasion of cancer cells, including ovarian (refs. 7 and 8; Fig. 1A). Using a pixel density-based quantitative measure of cell migration (see Supporting Materials and Methods), the addition of the Rac1 siRNA pool or the Src inhibitors retarded SKOV-3 cell migration by 60–70%, respectively, at 12 h after scratching (relative to control wells at the same time points; Fig. 1B). Although migratory inhibition is evident at 8 and 16 h, 12 h represented the largest difference between controls and test siRNAs at a time point where control cells closed the wound. In parallel, we measured cell viability in identically treated 384-well plates by using an ATP-based luminescent assay to monitor potential toxic effects of siRNA transfection and small molecule inhibition on SKOV-3 cells. All screen controls shown in Fig. 1 did not have an appreciable effect on cell viability (luminescent signal was >90% of untreated well signal).
Fig. 1.
Validation of the automated cell motility assay. (A) The temporal (0, 4, 8, 12, and 16 h) migration of SKOV-3 cells in the presence and absence of controls is shown. siCON, FITC-conjugated control siRNA; siRAC, a sequence-specific siRNA targeting RAC; DMSO, dimethyl sulfoxide; SRC, c-Src family kinase inhibitor, compound 43 (5). (B) Quantification of SKOV-3 migration. The degree of scratch closure (“migration score”) was determined by an automated algorithm and plotted as a function of time. Low migration scores reflect migratory inhibition. (C) Western blot demonstrating knockdown of the RAC protein by the RAC-specific siRNA used in A, compared to a control siRNA (CON) and mock transfected cells (LIPO). The same blot reprobed with an anti-actin antibody to demonstrate equal loading is shown below.
The reproducibility of the assay was tested by using a diverse subset of 384 preplated siRNAs targeting 192 genes (two siRNAs per gene plated in duplicate). For these experiments, SKOV-3 cells were reverse transfected on each of three replicate plates, grown to confluency, wounded, and incubated for a further 12 h. After image capture, wells from each of the three replicate plates were scored by the quantitative algorithm (see Supporting Materials and Methods) and the score from each individual well in each of the three replicate runs was compared to the mean well score by using the Pearson correlation coefficient. In each case, r2 was >0.87, demonstrating a high degree of well-to-well consistency (Fig. 8, which is published as supporting information on the PNAS web site).
A Large-Scale siRNA Screen for Inhibitors of SKOV-3 Cell Motility.
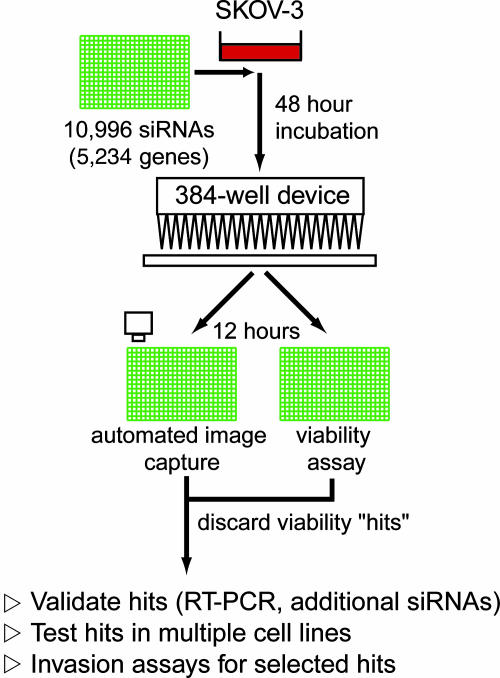
We used a preplated library of 10,996 siRNAs, targeting 5,234 genes (described in Supporting Materials and Methods) to identify inhibitors of cellular motility in SKOV-3 cells (Fig. 2). Cells were reverse transfected as described above and incubated for 48 h before wounding. The screen was performed in duplicate (≈22,000 wells) and quantitatively scored. Measurement of cell viability was performed in a set of duplicate siRNA library plates, and the luminescence of each well was compared to the normalized mean well intensity of each 384-well plate. To eliminate siRNAs that induced cell growth arrest or cell death and would not migrate as a consequence, we adopted a viability cutoff score of 0.9 (10% deviation from the plate mean), below which siRNAs were disregarded.
Fig. 2.
Schematic of the SKOV-3 siRNA screen and followup.
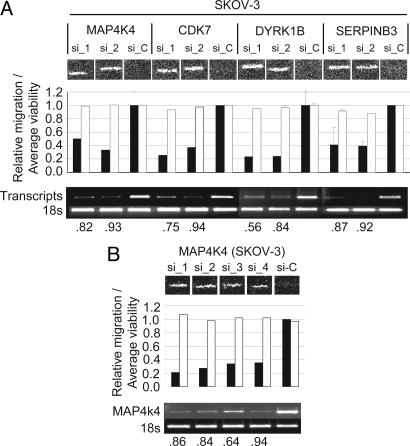
The top ≈5% of wells in which SKOV-3 cells migrated the least (n = 532), were chosen for further analysis (Table 1, which is published as supporting information on the PNAS web site), based on a statistical review of the screen (see Supporting Materials and Methods). Because of the significant potential for off-target effects when considering the phenotypic effects of single siRNAs, we focused only on those transcripts targeted by at least two independent siRNA sequences (n = 22), with the assumption that a similar phenotypic effect observed with two siRNAs would be less likely to occur by chance (Table 2, which is published as supporting information on the PNAS web site). To test this assumption, we resynthesized the siRNAs from the library sequences and monitored transcript knockdown by semiquantitative RT-PCR in parallel with migratory inhibition. Of the 46 siRNAs targeting 22 genes (20 genes targeted by 2 siRNAs and 2 genes targeted by 3 siRNAs), 36 (78%; 17 unique genes) yielded migratory phenotypes similar to those observed in the primary screen. In contrast to the high degree of concordance at a phenotypic level, independent siRNAs against only 4 of the 22 genes; MAP4K4, DYRK1B, CDK7, and SerpinB3, exhibited similar reductions in transcript levels, consistent with phenotypic inhibition (Fig. 3A).
Fig. 3.
Identification and validation of four promigratory genes. (A) Validation of migratory inhibition by phenotypic and transcriptional analysis. The migratory inhibition elicited by two independent siRNA duplexes targeting four genes, MAP4K4 (NM_004834), CDK7 (NM_001799), DYRK1B (NM_004714), and SERPINB3 (NM_006919), is shown compared to control siRNA and quantified by the automated algorithm (black bars, migration score; white bars, relative cellular viability). RT-PCR analysis is shown for each transcript, and the relative transcriptional knockdown was quantified by using imagej (National Institutes of Health). The extent of transcript knockdown is shown as a ratio to control transfected cells beneath the gel picture. (B) Four unique siRNA duplexes (three from the primary screen and one additional sequence) have similar effects on the migration of SKOV-3 cells, reducing motility by up to 75%, with a corresponding knockdown of MAP4K4 transcript ranging from 64% to 94%.
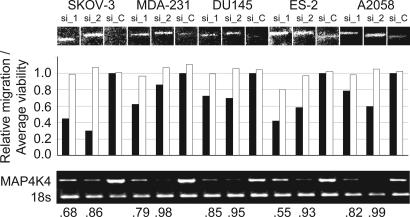
We next tested the effect of inhibiting these four transcripts in other migratory carcinoma cells from different anatomic origins, ES-2 (ovarian), MDA-MB-231 (breast), A2058 (melanoma), and DU-145 (prostate), to assess whether the effects of transcript reduction were unique to the SKOV-3 cells used in the primary screen or reflect more general effects on the migration of cancer cells. RNA interference-mediated knockdown of MAP4K4 (Figs. 3B and 4) and CDK7 (data not shown) affected the migration of all cell types tested. In contrast, inhibition of DYRK1B and SerpinB3 affected the motility of SKOV-3 and only two of four of the other cell lines tested (data not shown).
Fig. 4.
Inhibition of MAP4K4 affects the motility of multiple carcinoma cell lines. Shown are the affects of the two most potent MAP4K4 siRNAs on the motility of SKOV-3, MDA-MB-231 (MDA-231), DU-145, ES-2, and A2058. A graphical representation of migratory inhibition relative to control siRNA is shown above RT-PCR analysis of the MAP4K4 transcript in each of the cell lines.
Suppression of MAP4K4 Affects Cell Motility and Invasion.
We further characterized the role of the MAPK, MAP4K4, in cancer cell motility for the following reasons: First, siRNAs targeting MAP4K4 variably retarded the migration of all motile carcinoma cells tested, suggesting a central role for this kinase in cell migration. Second, MAP4K4-deficient mice exhibit specific defects in the migration of neural crest cells during early development (8), supporting a physiological role in cell migration. Third, overexpression of a rare cancer-associated splice variant of MAP4K4 in rat intestinal epithelial cells was reported to increase cellular invasiveness in the presence of hepatocyte growth factor/scatter factor, suggesting a role in cancer development (9).
Fig. 3B illustrates the quantitative effects of four independent siRNAs targeting the MAP4K4 gene (three from the primary screen and one additional siRNA, hereafter termed si_1–si_4; sequences are listed in Supporting Materials and Methods) on SKOV-3 migration. The migratory inhibition, which ranged from 50% to 66% relative to control siRNA-transfected cells, was consistent with the degree of transcript knockdown determined by RT-PCR (Fig. 3B), which ranged from 64% to 94%. The effect of the two most potent siRNAs (si_1 and si_2) on other highly motile carcinoma cells, and their associated transcriptional inhibition, are depicted in Fig. 4. Migratory inhibition is evident in all four cell lines relative to control siRNA (si_c), with variable potency. We assume that the variability reflects the following: (i) variable transfection efficiencies, (ii) differences in the relative expression of the gene, (iii) differences in signaling pathway dynamics in the different cell types, and (iv) the effects of variable cellular densities on the ability of the automated scoring algorithm to comparably score different cell types relative to SKOV-3.
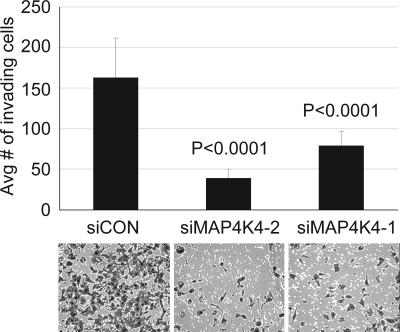
Because of the strong relationship between increased cancer cell motility, tissue invasion, and metastasis (3), we next asked whether transient MAP4K4 knockdown could affect cell invasion. SKOV-3 cells were transfected by using si_1, si_2, or a control scrambled siRNA, and invasion was monitored by using a matrigel-coated (Boyden) chamber assay. Invasion was inhibited by 76% and 52% with si_2 and si_1, respectively, relative to control transfected cells (Fig. 5).
Fig. 5.
Down-regulation of MAP4K4 decreases SKOV-3 cell invasion in vitro. Cells transiently transfected with siRNAs were subjected to Boyden chamber assays of cell invasion through matrigel. The data were collected from five individual consecutive fields of view (×10 objective) of each of four replicate invasion chambers. Representative photomicrographs are shown beneath the graph.
MAP4K4 Signals Through c-Jun N-Terminal Kinase, Independent of AP1 Activation and Downstream Transcription.
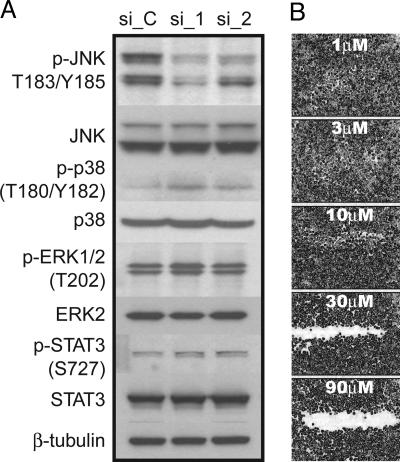
To address the underlying signaling mechanism(s) through which MAP4K4 mediates its effects on cell migration, we asked whether MAP4K4 signaling converges on one or more of the key MAPK transduction pathways, i.e., c-Jun N-terminal kinase (JNK), p38, or Erk MAPK (10). Knockdown of MAP4K4 by two independent siRNAs had no appreciable effect on the phosphorylation of Erk1/2 (Fig. 6), consistent with a lack of migratory inhibition observed by using the MEK inhibitor, U0126 (11), or siRNAs specific to the MEK1 kinase (data not shown). Similarly, cells transfected with MAP4K4 siRNAs did not show a decrease in detectable levels of phosphorylated p38 MAPK, which were low in SKOV-3 cells. In contrast, phosphorylation of JNK was significantly decreased in MAP4K4 siRNA transfected cells, consistent with reports showing that MAP4K4 can phosphorylate JNK in vitro (12). That JNK plays a significant role in mediating MAP4K4 signaling was demonstrated by inhibition of SKOV-3 migration with the JNK-specific inhibitor, SP600125 (Fig. 7). Migration was potently inhibited at concentrations of 30 μM and above without any significant effect on the viability of the cells. This effect on SKOV-3 migration is consistent with the effect of SP600125 on JNK-dependent cellular migration in NBT-II bladder tumor cells, MDA-MB-231 cells, fish keratinocytes (50 μM) (13), and fibroblasts (20 μM) (14). These results, together with the observation that siRNAs targeting Rac1 inhibit SKOV-3 migration (Fig. 1), tentatively suggest a constitutively active Rac1-MEKK1-JNK pathway in SKOV-3 cells. However, additional work is required to substantiate whether the major effect is mediated via JNK and whether constitutively activated JNK can rescue the migration of cells with diminished MAP4K4.
Fig. 6.
Decrease in JNK phosphorylation after siRNA-mediated knockdown of MAP4K4. (A) Western blots of SKOV-3 total protein lysates collected 48 h after transfection with control (si_C) or MAP4K4-specific siRNAs (si_1 and si_2). Blots were probed with phospho-specific antibodies (p) and antibodies recognizing total JNK, p38, ERK, and STAT-3. (B) Scratch migration assay of SKOV-3 cells exposed to increasing concentrations of the JNK inhibitor, SP600125.
We next asked whether the effect of JNK on SKOV-3 migration was mediated through activation of AP1 transcription. To first address this question, the expression profile of cells transiently transfected with two MAP4K4 siRNAs (si_1 and si_2) was assessed by oligonucleotide microarray hybridization on a 22,500-member array. We did not observe modulation of any known AP1 target genes; indeed, the only significantly altered probe sets when compared to control cells were those homologous to the MAP4K4 transcript, strongly indicating a lack of AP1-mediated transcription. Basal c-Jun protein and phosphoprotein levels were essentially undetectable by Western blot in SKOV-3 cells (data not shown), further confirming the lack of AP1 activation.
MAP4K4 has also been reported to activate a number of other proteins, including kinases within the JAK/STAT pathway. Because JAK/STAT signaling is highly active in high grade ovarian carcinomas and has been shown to be important for SKOV-3 cell motility by RNA interference-mediated knockdown (15), we examined the phosphorylation of STAT-3 in the presence or absence of MAP4K4 siRNAs. No significant changes were observed (Fig. 7), further supporting a major role for JNK in mediating SKOV-3 cell motility.
Discussion
Epithelial tumor progression depends on the sequential acquisition of multiple cellular traits, including the dissolution of cell–cell contacts and increased motility, enabling the invasion of adjacent tissues and, ultimately, the colonization of distant body sites. To address the underlying molecular mechanisms that promote cancer cell motility, we developed an automated 384-well wound-healing assay, coupled with automated microscopy and quantification.
We queried a custom library of 10,996 synthetic siRNAs targeting 5,234 transcripts to identify genes involved in the migration of SKOV-3 cells, a highly motile cell line derived from the ascites of a patient with metastatic ovarian adenocarcinoma. Although we identified 532 siRNAs with highly significant effects on migration (i.e., the highest ranked 5%; see Supporting Materials and Methods), which included many genes known to play a role in cell motility, we chose to examine the 22 genes targeted by at least two siRNAs, because of the significant potential for off-target phenotypic effects elicited by a single siRNA. Evaluation of resynthesized siRNAs targeting these 22 genes showed a relatively high degree of phenotypic consistency (74%), but a much lower consistency in terms of correlated transcriptional knockdown and migratory suppression of both siRNAs. Four genes, CDK7, MAP4K4, DYRK1B, and SerpinB3, were validated by these criteria. Given the apparently high off-target rate, we anticipate that future screens should encompass additional siRNAs against each target or secondary screens designed to interrogate the subgroup of identified hits (532 in this case) with additional, independent siRNAs.
We chose to further examine the role of MAP4K4 in cancer cell motility primarily because the protein is involved developmental cell migration (7) and is reported to augment cellular motility and invasion of rat intestinal epithelial cells in the presence of hepatocyte growth factor (8). Cumulatively, our data show that MAP4K4 is involved in the motility of multiple cancer cell lines, signaling upstream of JNK, and that inhibition of MAP4K4 results in suppression of invasion in vitro. Activation of JNK by MAP4K4 has been shown previously; however, it is reported to be indirect, acting through TAK1, MKK4, and MKK7 (11). However, we have been unable to detect appreciable basal levels of phospho-MKK4 or phospho-MKK7 (data not shown), suggesting that MAP4K4 may activate JNK through other kinases, at least in this cell type. Downstream of JNK, we have shown that AP1-dependent transcription is not involved in SKOV-3 cell migration; however, the substrates through which MAP4K4-JNK mediates its effects are not yet defined. Multiple direct targets of JNK phosphorylation have been described (reviewed in ref. 9), including the microtubule-associated proteins, MAP1B, DCX, and MAP2, and proteins involved in actin dynamics, such as Spir, and paxillin, which plays a key role in adhesion dynamics. Although all of these proteins are candidates for being targeted by JNK in SKOV-3 cells, a detailed examination of their possible role is beyond the scope of this report. The mouse homolog of MAP4K4, Nik, is reported to bind β1-integrin and associate with actin filaments (16). This observation may reflect a need to have MAP4K4 in close proximity to phosphorylation targets at points of substrate contact or that MAP4K4 also plays a structural role in cell movement.
Involvement of MAP4K4 in cell invasion has been suggested by one report in which rat intestinal cells overexpressing the protein were significantly more invasive in the presence of hepatocyte growth factor than parental rat intestinal epithelial cells (9). The same report also documented a striking complexity of MAP4K4 isoforms as well as an increased expression in tumor cells and primary tumors. Our own analysis has identified several splice variants of the gene in SKOV-3 cells, the dominant form of which is most similar to transcript variant 1 in the National Center for Biotechnology Information databases (data not shown). However, we have been unable to verify over-expression of MAP4K4 in primary tumor samples, either in our databases (http://symatlas.gnf.org) or the extensive tumor data sets available at Oncomine (www.oncomine.org). Notably, the expression of MAP4K4 in the data sets housed at Oncomine is measured both by cDNA and oligonucleotide microarrays, representing several different probes homologous to the gene. Thus, we suggest that MAP4K4 activity is triggered either by overactivity of an upstream kinase or by a constitutive autocrine loop that involves receptors signaling into the Rac1-MEKK1-MAP4K4-JNK pathway.
In summary, we developed a fully automated approach to interrogate the molecular basis of cell motility. We exemplified this approach by screening a large siRNA library and identifying several genes with promigratory activity. One of these genes, MAP4K4, appears to play a central role in cell migration and invasion in vitro and most likely does so through the JNK pathway, independent of AP1-mediated transcription.
Materials and Methods
Cell Culture.
Cells were obtained from the American Type Culture Collection (ATCC) and cultured according to ATCC recommendations. Media was supplemented with 10% Australian-sourced FBS/100 units/ml penicillin/100 μg/ml streptomycin/2 mM l-glutamine. RPMI medium 1640 was additionally supplemented with 1 mM sodium pyruvate/10 mM Hepes buffer.
Automated Wound-Healing Assay Device.
The automated device consists of a machined aluminum block into which 384 holes are drilled wide enough to accommodate 12.5-μl pipette tips (Matrix Technologies, catalog no. 5302, Hudson, NH). Sterilized pipette tips are inserted into the block and suspended on a vertical tracking arm. 384-well clear bottom tissue culture plates (Greiner Bio-One, Frickenhausen, Germany; see below) are placed on a level platform below the aluminum block. Upon initiation, the aluminum block is lowered to a point at which the pipette tips touch the bottom of each of the 384 wells (after plate-specific calibration). With the pipette tip block engaged, the platform is shifted ≈3 mm (well diameter = 3.70 mm) by hydraulic pressure, resulting in uniform “scratches” in each of the 384 well plates (see also Supporting Materials and Methods).
Wound-Healing Assays.
siRNAs were purchased from Dharmacon (Lafayette, CO) or Qiagen (Valencia, CA) and prepared and dispensed into 384-well plates as described in ref. 17. Motile cells were plated at high density (4,000–5,000 cells per well) in 384-well clear bottom tissue culture plates and cultured for up to 24 h. For 384-well siRNA transfections, cells were added directly to a siRNA/transfection reagent mixture, resulting in a “reverse transfection” as described in ref. 17 and in detail in Supporting Materials and Methods. Compounds were added onto the cell layers 12 h before scratching at a final concentration of 0.5% DMSO for all conditions. Media was changed in all experiments at 24 h after transfection. Assay plates were fitted with metal low-evaporation covers and incubated at 37°C, 5% CO2 in humidified tissue culture incubators. All liquid dispensing steps were performed by using a Multidrop 384 dispenser (Titertek, Huntsville, AL), and all liquid aspiration steps were done by using an EMBLA plate washer (Molecular Devices). Confluent cell layers were scratched by using the automated device. Plates were reincubated to allow wound closure and then fixed with formaldehyde (Sigma-Aldrich) at a final concentration of 3.7%. Cells were washed two times with PBS, permeabilized with Triton X-100 (Sigma-Aldrich), and stained with the nuclear stain DAPI (Molecular Probes). Wells were further washed twice with PBS (supplemented with 0.9 mM calcium/0.5 mM magnesium) by using the EMBLA plate washer. Each well of the 384-well plate was photographed by a fluorescent microscope retooled by Q3DM, Inc. (San Diego) to automate image capture. A ×4 objective lens was used to visualize a majority of the space of each well within one field of view.
Quantitative Scoring Method for Cell Migration.
The derivation and use of the automated scoring algorithm to assess cell migration, and the statistical treatment of the screen data are described in detail in Fig. 9, which is published as supporting information on the PNAS web site.
Cell Viability.
Cells were plated into an identical set of assay plates and processed identically to the sister wound closure plates, up to the point of plate scratching. Viability was measured by using Cell Titer Glo (Promega) according to the manufacturer's protocol. The mean luminescent intensity of each plate was calculated, and the percent of the plate mean was calculated for each well. siRNAs or molecules that resulted in a percent mean of <90% were considered to negatively effect viability and were eliminated from further followup.
Cell Invasion.
Invasion was measured by using an 8-μm pore, 24-well format matrigel-coated transwell chamber assay (Becton Dickinson Discovery Labware, Bedford, MA) according to the manufacturer's recommended conditions (see also Supporting Materials and Methods). Membranes were photographed by using brightfield and pictures of five consecutive vertical fields of view were taken by using the ×10 objective lens. The number of cells in each field of view was counted manually. For all conditions, a minimum of four replicate chambers were assayed.
RT-PCR.
siRNA-treated cells were lysed in situ and RNA were isolated by using the Qiagen RNeasy kit. cDNA was prepared by using the Thermoscript RT-PCR system (Invitrogen). PCR was carried out for 26 cycles by using an annealing temperature of 60°C. The PCR buffer and Taq polymerase were from the PCR SuperMix High Fidelity reaction mixture (Invitrogen). 18S PCR primers were from the QuantumRNA Classic 18S kit (Ambion, Austin, TX). Gene-specific primers for MAP4K4 were 5′-GTGGTCAATGTGAATCCTACC-3′ for the forward primer, and 5′-CCAAGTGACCAGTTTCCACAG-3′ for the reverse primer.
Western Blotting.
Protein isolation, SDS/PAGE and Western blotting were all carried out by using standard techniques. Antibodies sources are described in Supporting Materials and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Quinn Deveraux, Christopher Stroh, and Venkat Reddy for critically reading the manuscript and Drs. Pedro Aza-Blanc, Serge Batalov, and Sumit Chanda, Mr. Abel Gutierez, and Mr. Paul deJesus for siRNA design, library construction, assay development, and advice.
Abbreviations
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- siRNA
small interfering RNA
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Lauffenburger D. A., Horwitz A. F. Cell. 1996;84:359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- 2.Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons J. T., Horwitz A. R. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 3.Sahai E. Curr. Opin. Genet. Dev. 2005;15:87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Todaro G. J., Lazar G. K., Green H. J. Cell. Physiol. 1965;66:325–333. doi: 10.1002/jcp.1030660310. [DOI] [PubMed] [Google Scholar]
- 5.Goldberg D. R., Butz T., Cardozo M. G., Eckner R. J., Hammach A., Huang J., Jakes S., Kapadia S., Kashem M., Lukas S., et al. J. Med. Chem. 2003;46:1337–1349. doi: 10.1021/jm020446l. [DOI] [PubMed] [Google Scholar]
- 6.Golas J. M., Lucas J., Etienne C., Golas J., Discafani C., Sridharan L., Boghaert E., Arndt K., Ye F., Boschelli D. H., et al. Cancer Res. 2005;65:5358–5364. doi: 10.1158/0008-5472.CAN-04-2484. [DOI] [PubMed] [Google Scholar]
- 7.Wiener J. R., Windham T. C., Estrella V. C., Parikh N. U., Thall P. F., Deavers M. T., Bast R. C., Mills G. B., Gallick G. E. Gynecol. Oncol. 2003;88:73–79. doi: 10.1006/gyno.2002.6851. [DOI] [PubMed] [Google Scholar]
- 8.Xue Y., Wang X., Li Z., Gotoh N., Chapman D., Skolnik E. Y. Development (Cambridge, U.K.) 2001;128:1559–1572. doi: 10.1242/dev.128.9.1559. [DOI] [PubMed] [Google Scholar]
- 9.Wright J. H., Wang X., Manning G., LaMere B. J., Le P., Zhu S., Khatry D., Flanagan P. M., Buckley S. D., Whyte D. B., et al. Mol. Cell. Biol. 2003;23:2068–2082. doi: 10.1128/MCB.23.6.2068-2082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C., Jacobson K., Schaller M. D. J. Cell Sci. 2004;117:4619–4628. doi: 10.1242/jcs.01481. [DOI] [PubMed] [Google Scholar]
- 11.Favata M. F., Horiuchi K. Y., Manos E. J., Daulerio A. J., Stradley D. A., Feeser W. S., Van Dyk D. E., Pitts W. J., Earl R. A., Hobbs F., et al. J. Biol. Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 12.Yao Z., Zhou G., Wang X. S., Brown A., Diener K., Gan H., Tan T. H. J. Biol. Chem. 1999;274:2118–2125. doi: 10.1074/jbc.274.4.2118. [DOI] [PubMed] [Google Scholar]
- 13.Huang C., Rajfur Z., Borchers C., Schaller M. D., Jacobson K. Nature. 2003;424:219–223. doi: 10.1038/nature01745. [DOI] [PubMed] [Google Scholar]
- 14.Javelaud D., Laboureau J., Gabison E., Verrecchia F., Mauviel A. J. Biol. Chem. 2003;278:24624–24628. doi: 10.1074/jbc.M301942200. [DOI] [PubMed] [Google Scholar]
- 15.Burke W. M., Jin X., Lin H. J., Huang M., Liu R., Reynolds R. K., Lin J. Oncogene. 2001;20:7925–7934. doi: 10.1038/sj.onc.1204990. [DOI] [PubMed] [Google Scholar]
- 16.Poinat P., De Arcangelis A., Sookhareea S., Zhu X., Hedgecock E. M., Labouesse M., Georges-Labouesse E. Curr. Biol. 2002;12:622–631. doi: 10.1016/s0960-9822(02)00764-9. [DOI] [PubMed] [Google Scholar]
- 17.Aza-Blanc P., Cooper C. L., Wagner K., Batalov S., Deveraux Q. L., Cooke M. P. Mol. Cell. 2003;12:627–637. doi: 10.1016/s1097-2765(03)00348-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.