Abstract
Several transporters belonging to the ABCA subfamily of ABC (ATP-binding cassette) proteins are involved in lipid trafficking. Human ABCA5 and its rat orthologue, rAbca5, represent recently identified subfamily members whose substrate spectrum remains to be defined. The elucidation of (sub)cellular rAbca5 distribution would be expected to provide a basis for optimization of functional analyses. In the present study, we applied in situ hybridization to examine rAbca5 mRNA distribution within sections of rat testis, a tissue expressing high levels of rAbca5 mRNA. We found rAbca5 mRNA to be predominantly expressed in interstitial Leydig cells, which are major sites of testosterone synthesis. To investigate rAbca5 subcellular localization, we constructed expression vectors yielding rAbca5 fused either to EGFP (enhanced green fluorescent protein) or to a peptide bearing the viral V5 epitope. During rAbca5 cDNA cloning, we discovered a splice variant sequence (rAbca5 V20+16), predicted to give rise to a truncated, half-size transporter, which was highly homologous with a human splice variant described by us previously. Quantitative RT (reverse transcription)–PCR demonstrated that the rAbca5 splice variant was expressed in numerous tissues (including testis, brain and lungs), its cDNA amounting to 2.6–11.2% of total rAbca5 cDNA. Transfection of individual rAbca5-EGFP, rAbca5 splice variant-EGFP or transporter-V5 expression plasmids along with organelle marker plasmids into HEK-293 cells (human embryonic kidney 293 cells) revealed that both rAbca5 and splice variant fusion proteins co-localized with marker protein for the Golgi apparatus. Expression of rAbca5 mRNA in Leydig cells, intracellular localization of rAbca5–EGFP/rAbca5–V5 and involvement of rAbca5-related proteins in lipid transport suggest that rAbca5 may participate in intracellular sterol/steroid trafficking.
Keywords: ATP-binding-cassette (ABC) transporter, Golgi, Leydig cell, rat Abca5, real-time PCR, splice variant
Abbreviations: ABC, ATP-binding cassette; DIG, digoxigenin; DsRed, Discosoma sp. fluorescent protein; ECFP, enhanced cyan fluorescent protein; EGFP, enhanced green fluorescent protein; ER, endoplasmic reticulum; 6-FAM, 6-carboxyfluorescein; GAP-43, growth-associated protein 43; HDL, high-density lipoprotein; HEK-293 cells, human embryonic kidney 293 cells; mdr1, multidrug resistance transporter 1; MGB, minor groove binder; ORF, open reading frame; RT, reverse transcription; RTQ PCR, real-time quantitative PCR
INTRODUCTION
ATP-binding-cassette (ABC) proteins have been recognized as key players in physiological transport processes of both prokaryotic and eukaryotic organisms. Most members of the ABC superfamily function as active transporters across biological membranes for a wide variety of substrates. Functional eukaryotic ABC transporter systems comprise two membrane anchor domains and two conserved nucleotide-binding folds. These four regions are assembled either from a single full transporter protein or by homo- or hetero-dimerization of a pair of half-transporters, containing one transmembrane domain and one ATP-binding fold each [1]. The human ABCA subfamily of ABC proteins consists of 12 members representing the full transporter type, of which several (e.g. ABCA1, ABCA2 and ABCA3) appear to be involved in various aspects of lipid homoeostasis. Human ABCA1 has been shown to participate in cellular efflux of phospholipids, leading to a net efflux of cellular cholesterol and contributing to HDL (high-density lipoprotein) generation [2]. Mutations in the genomic locus of ABCA1 are responsible for the phenotype of Tangier disease, which is connected to low plasma concentrations of HDL particles and a high tendency to accumulate atherosclerotic plaques in blood vessels [3]. Overexpression of ABCA2 has been shown to convey resistance to the oestradiol derivative estramustine [4,5]. ABCA3, another ABCA transporter situated at intracellular membranes, has been postulated to contribute to the biogenesis of surfactant lamellar bodies in human lung alveolar type II cells [6]. Surfactant consists of proteins and phospholipids and is necessary to lower surface tension at the gas–liquid interface of the lung. Recently, ABCA3 mutations have been found to be associated with the occurrence of abnormal lamellar bodies and surfactant deficiencies in the lung of newborns [7].
We recently reported the cDNA cloning of human ABCA5 and its rat orthologue, rAbca5 [8]. The human ABCA5 gene belongs to an ABCA cluster of five genes (ABCA5, ABC10, ABCA6, ABCA9 and ABCA8) arranged in a head-to-tail order on human chromosome 17 [9]. A similar cluster of genes (comprising Abca5, Abca6, Abca9, Abca8a and Abca8b) exists in mice [9], indicating the conservation of several gene duplication events between humans and rodents. Although ABCA8 has been implicated in acting as a multi-specific transporter with a substrate spectrum including ochratoxin A, oestradiol-β-glucuronide, taurocholate and leukotriene C4 [10], the substrate spectrum of other protein products of the described ABCA/Abca gene cluster remains to be defined. Analyses of subcellular transporter localization and tissue distribution of transporter expression, however, are expected to provide a basis for optimizing experiments on transporter function.
Northern-blot analyses previously revealed that rAbca5 mRNA was expressed in numerous tissues, including brain and lungs. Most pronounced expression, however, was observed in testis [8]. To specify cell populations accounting for the high rAbca5 mRNA levels in testis [8], we examined rAbca5 transcript expression via in situ hybridization.
During the process of cloning human ABCA5 cDNA, we recently detected a splice variant sequence that we termed ‘ABCA5 V20+16’ [8]. In the present study, we report the characterization of a novel rAbca5 splice variant (rAbca5 V20+16), which is highly homologous with the human ABCA5 V20+16 splice variant. To analyse the relative abundance of the rat splice variant in comparison with total rAbca5 transcripts in rat tissues, we employed an RT (reverse transcription)–RTQ (real-time quantitative) PCR approach. In order to investigate the subcellular distribution of rAbca5 and the rAbca5 splice variant protein product, expression vectors were constructed that comprised rAbca5 cDNA sequences in-frame with the coding sequence for the EGFP (enhanced green fluorescent protein) or a peptide bearing the viral V5 epitope. In HEK-293 cells (human embryonic kidney 293 cells) transiently transfected with individual transporter expression plasmids, the distribution of transporter fusion proteins was observed by fluorescence microscopy and compared with the localization pattern of fluorescent organelle marker proteins.
EXPERIMENTAL
Cell culture
The HEK-293 cell line [11], generously provided by W. Knepel (Department of Molecular Pharmacology, Institute of Pharmacology and Toxicology, University of Goettingen, Goettingen, Germany), and the rat Sertoli cell line SER-W3 [12], kindly provided by B. Oesch-Bartlomowicz and P. Antoniou-Lipfert (Institute of Toxicology, University of Mainz, Mainz, Germany), were maintained in Dulbecco's modified Eagle's medium (Sigma, Munich, Germany) containing 10% (v/v) fetal calf serum (BioWhittacker, Verviers, Belgium) at 37 °C in the presence of 5% CO2. HEK-293 cell line medium was supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin (Biochrom KG, Berlin, Germany). HEK-293 and SER-W3 cultures were split every 72 h in a 1:10 ratio and HEK-293 cells on the day prior to transfection experiments. Primary rat hepatocytes were isolated and cultured as described previously [13].
RNA isolation
Total RNA from tissues of adult male Wistar rats (180–240 g) and from SER-W3 cell cultures was isolated by guanidinium thiocyanate/phenol/chloroform extraction as described in [14].
Non-radioactive in situ hybridization
DIG-11 (digoxigenin 11)-dUTP-labelled antisense and sense cRNA probes were generated by in vitro transcription using the DIG-RNA labelling kit and T3 or T7 polymerase (Roche, Mannheim, Germany). cDNA templates had previously been prepared by a one-step RT–PCR (Invitrogen, Karlsruhe, Germany), using 1 μg of total RNA from rat testis and Abca5-specific oligonucleotide primers, yielding a 232 bp cDNA fragment and containing the T3 and T7 promoter sequences as overhang (primers: RISH.forw, 5′-TAATACGACTCACTATAGGGCTGCCCTAGACAGTCATTC-3′; and RISH.rev, 5′-ATTAACCCTCACTAAAGGGACTCATGGTGCTCACTAGAG-3′). The PCR product, monitored on a 1% agarose gel, was purified using S-300 microspin columns (Amersham Biosciences, Braunschweig, Germany).
For in situ hybridization of rAbca5 mRNA transcripts, rat testis paraffin sections were mounted on slides coated with 3-aminopropyl-triethoxysilane (Sigma). The slides were allowed to dry at 37 °C overnight. Tissue sections were deparaffinized and rehydrated in serial dilutions of ethanol. Samples were permeabilized using proteinase K (10 μg/ml) for 5 min at 37 °C. Digestion was terminated by washing the samples in a PBS solution (pH 7.4) followed by an acetylation reaction with 0.25% acetic anhydride in 0.1 M Tris/HCl buffer (pH 8.0) for 10 min. Subsequently, the slides were dehydrated in serial dilutions of ethanol and dried at room temperature (20 °C). DIG-labelled riboprobes were diluted in hybridization buffer (Amersham Biosciences). After application of probes, the samples were covered with sterile coverslips and placed on a hot plate at 85 °C for 5 min for probe and target denaturation. Hybridization was performed overnight at 55 °C in a sealed humid chamber containing 50% (v/v) formamide. Non-specific binding or unbound probes were removed by the following post-hybridization washes: 1×SSC (0.15 M NaCl/0.015 M sodium citrate)/0.1% SDS at room temperature (2×5 min), 0.2×SSC/0.1% SDS at 55 °C (3×20 min) and, finally, TBS (Tris-buffered saline) containing 0.1% Tween 20 (TBST; Roche). Signal amplification for detection of rAbca5 mRNA was achieved by the tyramide signal amplification method [15], employing reagents purchased from DuPont–NEN (Boston, MA, U.S.A.). The procedure involved consecutive steps of blocking of non-specific binding, incubation with a horseradish peroxidase-conjugated F(ab)2-antibody fragment against DIG, incubation with biotin–tyramide and, finally, incubation with streptavidin–horseradish peroxidase. Peroxidase activity was visualized by using diaminobenzidine (DAKO ChemMate; DAKO, Hamburg, Germany) as chromogen. For each tissue section, sense riboprobes served as controls.
Identification of a rAbca5 splice variant
The plasmid construct prAbca5-V5, containing the full ORF (open reading frame) of rAbca5 in-frame with the viral V5 epitope sequence, was generated by ligation of the rAbca5 RT–PCR product (obtained from rat testis RNA with the primers: rAbca5.forw, 5′-CTGACCTAGAAAACATGGCTAC-3′; and rAbca5.rev, 5′-GAACACGACTCTGTCTTCCTGC-3′) into the pcDNA3.1/V5-His TOPO vector (Invitrogen). During sequence analysis of the prAbca5-V5 expression plasmid, an rAbca5 full-length variant sequence was identified apart from the major rAbca5 cDNA sequence (EMBL accession no. AJ426052). The variant sequence contained a 16 bp insert at the end of exon 20, as shown previously for a human ABCA5 splice variant [8]. The splice variant sequence was deposited in the EMBL database (accession no. AJ550165) and termed ‘rAbca5 V20+16’.
The plasmid prAbca5 V20+16-V5, containing the full ORF of rAbca5 V20+16 in-frame with the V5 epitope sequence, was generated by ligation of the rAbca5 V20+16 RT–PCR product obtained with the primers Abca5.forw and SV.rev 5′-CACGCTCTCACCAGTAGA-3′ into pcDNA3.1/V5-His TOPO.
Fluorescent protein expression plasmid constructs
The plasmids prAbca5-EGFP and prAbca5 V20+16-EGFP were obtained by digestion with SacI and SacII of the constructs prAbca5-V5 and prAbca5 V20+16-V5 respectively, and ligation of the resulting insert into pEGFP-N1 (BD Biosciences, Palo Alto, CA, U.S.A.). The organelle marker plasmids for co-localization experiments pDsRed-Mito, pDsRed-Golgi and pDsRed-ER (where DsRed stands for Discosoma sp. fluorescent protein and ER for endoplasmic reticulum) were constructed as described previously [16]. The plasmid pDsRed2-Peroxi, yielding a DsRed2 fusion protein targeted to peroxisomes, and the marker plasmid pECFP-Mem, giving rise to a protein containing the N-terminal 20 amino acids of neuromodulin [GAP-43 (growth-associated protein 43)] fused to the N-terminus of ECFP (enhanced cyan fluorescent protein), were purchased from BD Biosciences.
RTQ PCR analysis
Total RNA (1 μg) in 12.5 μl of autoclaved water and 1 μl of dN6 random hexamer primer (Roche) were denatured for 10 min at 70 °C and subsequently subjected to reverse transcription at 42 °C for 60 min in a reaction mixture (30 μl) containing 1× Superscript reverse transcriptase buffer (50 mM Tris/HCl, pH 8.3, 75 mM KCl and 3 mM MgCl2), 12 mM dithiothreitol, 333 μM dNTPs, 50 units of RNase inhibitor (Amersham Biosciences) and 50 units of Superscript reverse transcriptase II (Invitrogen). The resulting cDNA solution was adjusted to a total volume of 100 μl, corresponding to an estimated cDNA concentration of 10 ng/μl, and used in all subsequent RTQ PCR experiments. The relative abundance of rAbca5 V20+16 mRNA (splice variant) with respect to the total amount of rAbca5 (full-length and splice variant) transcripts was determined in total RNA of several rat organs, the rat Sertoli cell line SER-W3 and in freshly isolated primary rat hepatocytes. A PCR amplification target of 198 bp was chosen to contain the 16 bp insertion at the 3′-end. This allowed usage of an identical sense primer in both amplification reactions (RTQ.forw, 5′-AGAAAGCAAATCAGTGCGCTCT-3′). The reverse primer RTQ.rev (5′-CAGTAGAATTTTGAAGCAGCA-3′) was designed to yield total rAbca5 cDNA transcripts, while the reverse primer SVRTQ.rev (5′-TCTGAATCTTCACACGCTCTCAC-3′) was constructed to yield the transcript with the insertion specific to the splice variant. The Taqman probe RTQ1 contained the non-fluorescent quencher MGB (minor groove binder; Applied Biosystems) at the 3′-terminus and 6-FAM (6-carboxyfluorescein) as fluorescence reporter at the 5′-terminus. The probe was designed to hybridize in the centre of the amplification product (RTQ1, 5′-6-FAM-TGCTGTGGTTCCCAT-MGB-3′).
PCR amplification was performed in a 30 μl total reaction volume containing approx. 40 ng of cDNA (10 μl), 15 μl of 2× PCR mastermix including Taq polymerase (Eurogentec), 0.75 μM sense/antisense primer and 250 nM RTQ1 (Applied Biosystems). Triplicate cDNA samples were cycled on the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) with a series of diluted cDNA samples using a full-length rat Abca5 plasmid containing the 16 bp insertion as template to construct a calibration curve as follows: 2 min at 50 °C, 10 min at 95 °C and 49 cycles of 15 s at 95 °C and 60 s at 60 °C. Negative controls without cDNA template were included in each experiment. Data analysis was carried out using ABI SDS 2.1 software. The abundance of transcripts was averaged and calculated as the relative amount of splice variant cDNA transcripts with respect to total rAbca5 cDNA transcripts. Error bars indicate the S.D. for independent experiments.
Transfection and subcellular localization analyses
To investigate the subcellular localization of rat Abca5 and its splice variant protein product, HEK-293 cells were transiently transfected with fusion plasmids, yielding either transporter protein fused to EGFP (prAbca5–EGFP and prAbca5 V20+16–EGFP) or protein tagged with a peptide exhibiting the viral V5 epitope (prAbca5-V5 and prAbca5 V20+16-V5) respectively. Briefly, for analyses of EGFP fusion protein expression, cells were plated on to 60 mm dishes 24 h prior to transfection, and 1 μg of the respective fusion plasmid was transfected into cells with FUGENE 6 (Roche) according to the manufacturer's instructions. In co-transfection experiments, 1 μg of transporter-EGFP fusion plasmid was co-transfected with an organelle marker plasmid (0.25 μg of Golgi marker plasmid; 0.1–0.5 μg of other marker plasmids). The localization of fluorescent proteins was determined microscopically in unfixed cells on a Zeiss fluorescence microscope. Transfections involving V5 epitope expression constructs were performed with cells cultured on 35 mm dishes, employing 0.5 μg of V5 construct, in co-transfection experiments combined with 0.25 μg of pDsRed-Golgi; 48 h after transfection, cells were washed twice with PBS, fixed with 100% methanol for 5 min and washed five times with PBS. Following an incubation in blocking solution (PBS/10% fetal calf serum) for 20 min, cells were incubated for 1 h with an FITC-conjugated antibody raised against the V5 epitope (Invitrogen), diluted 1:500 in blocking solution. Specimens were washed twice with PBS and DsRed-dependent fluorescence of the Golgi marker and FITC-dependent immunofluorescence subsequently observed via fluorescence microscopy. OPENLAB software (Improvision) was applied for image documentation.
Western-blot analysis
Buffers were supplemented with Complete protease inhibitor (Roche) and all subsequent steps were carried out at 4 °C. HEK-293 cells were harvested 48 h after transfection, rinsed with TE buffer (20 mM Tris and 1 mM EDTA), suspended in 50 μl of TES buffer (TE buffer with 8.7% sucrose) per 60 mm dish and homogenized at 1400 rev./min. The cell suspension was centrifuged (1000 g for 1 min) and frozen in liquid nitrogen. On thawing at 37 °C, nuclei were removed from the total cellular protein extract by centrifugation (500 g for 5 min) and the post-nuclear supernatant was used for electrophoresis. The protein content was determined as described in [17]. Protein samples (160 μg) were subjected to SDS/PAGE using a stacked gel for separation within the resolving portion of the gel (containing 6% acrylamide in the top stack and 12% acrylamide in the bottom), blotted on to a PVDF membrane by means of the semidry transfer method [13,18] and probed overnight with a 1:750 dilution of a polyclonal antibody raised against the full-length EGFP protein (BD Biosciences) or a 1:1500 dilution of the EGFP monoclonal antibody JL-8 (BD Biosciences). Following incubation with a horseradish peroxidase secondary antibody (Sigma), signals were detected using the Super Signal West Pico kit (Pierce Biotechnology, Rockford, IL, U.S.A.).
RESULTS
In situ hybridization analyses of rat testis and epididymis sections
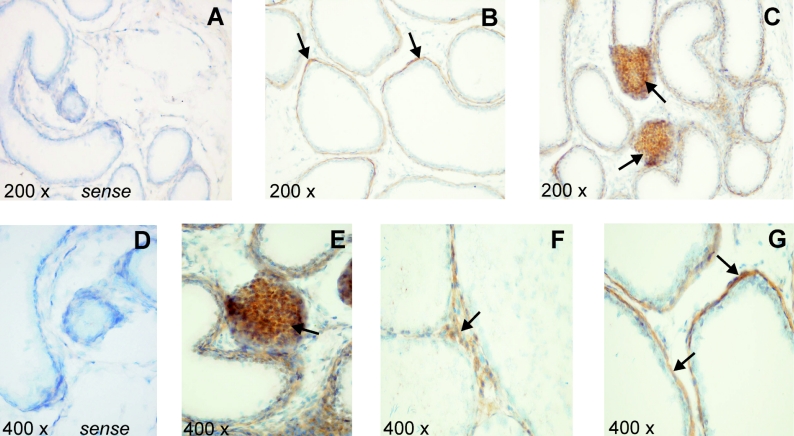
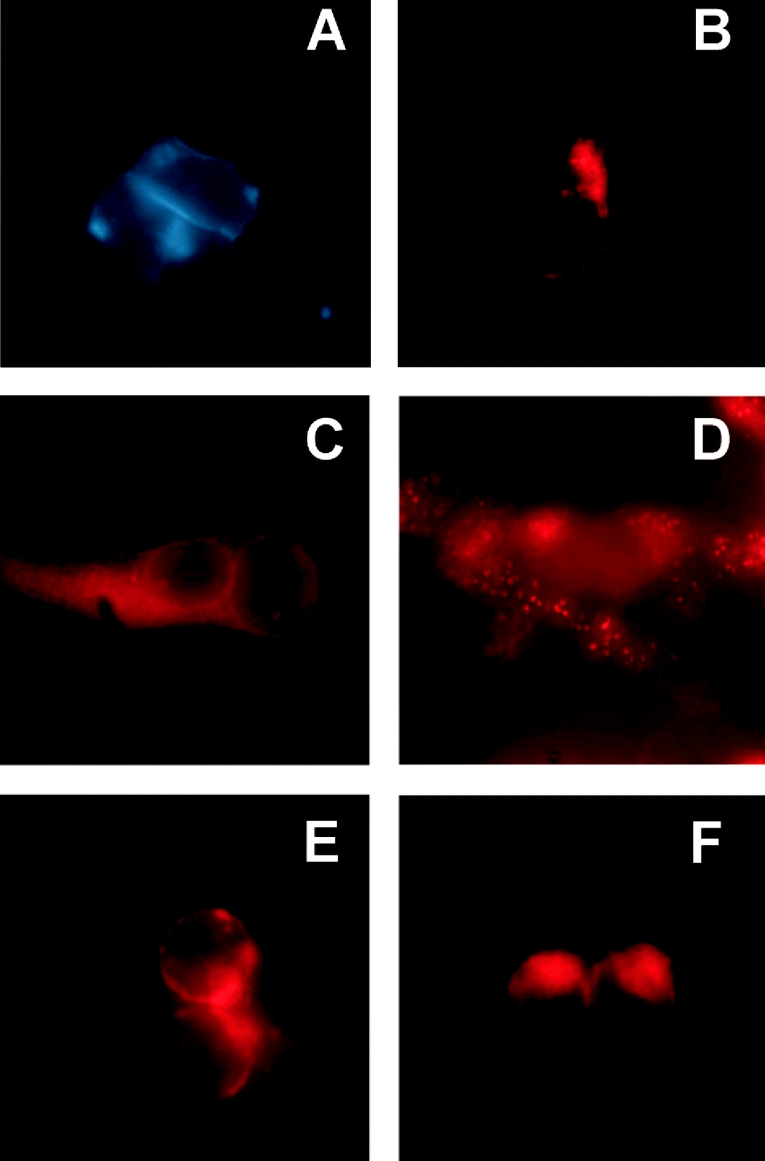
Rat Abca5 mRNA had previously been shown by Northern-blot analyses of rat tissues to be highly expressed in testis [8]. To specify the cell populations responsible for rAbca5 expression, the cellular distribution of rAbca5 transcripts was determined by in situ hybridization of tissue slices of rat testis. Hybridization experiments were performed with a sequence-specific DIG-labelled antisense cRNA probe. Following application of the tyramide signal amplification method, Abca5 mRNA was detected in basal cells of testicular seminiferous tubules and in interstitial cell clusters mainly consisting of Leydig cells (Figure 1). Staining of cells within the seminiferous tubules, e.g. spermatogonia, Sertoli cells, spermatocytes or spermatides, was low in comparison or undetectable. Additionally, in rat epididymis, hybridization signals were detected in the microvilli of the cylindrical epithelium and in fibrocytes of the interstitial region (results not shown).
Figure 1. Expression of rat Abca5 mRNA in testis.
Rat testis paraffin sections were hybridized at 55 °C with the DIG-labelled cRNA antisense probe as described in the Experimental section. (A, D) Results of hybridization with the sense riboprobe (negative control) are shown. (B, C, E–G) Positive hybridization signals (brown) marked with black arrows are shown. Cell nuclei were counterstained with Meyer's haemalaun.
Rat Abca5 genomic structure and detection of a rat splice variant
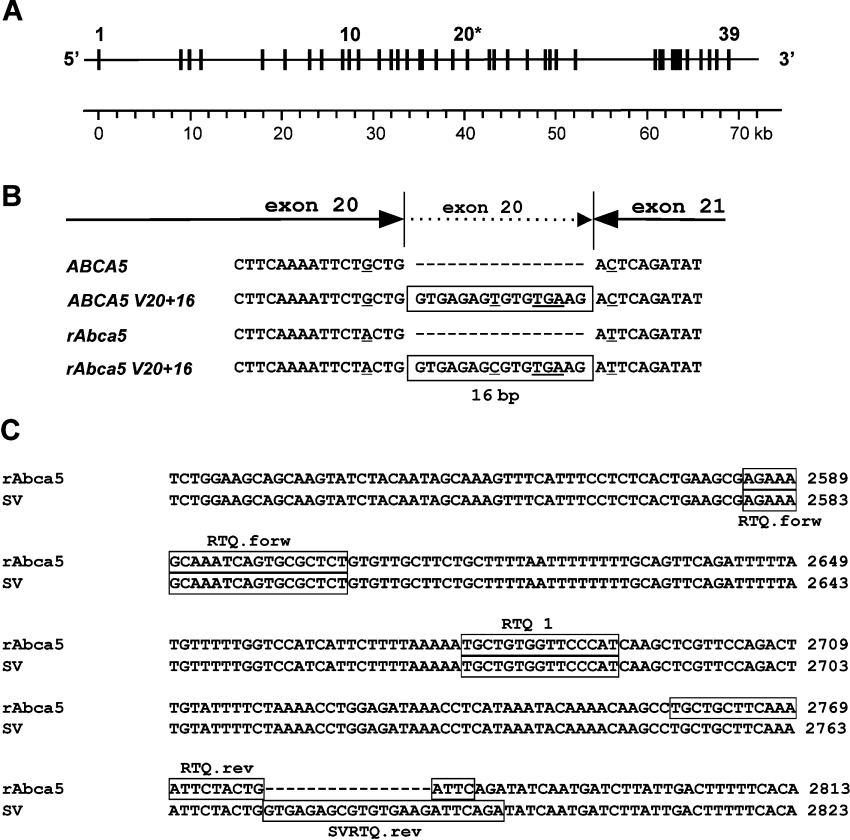
We recently reported the cloning of rat Abca5 cDNA from rat testis RNA [8]. Comparison of the cDNA sequence with the rat genome build 2.1 (the RAT Genome Project) revealed that the translated rat Abca5 sequence was distributed over 38 exons. On further scanning of databases, we identified a 347 bp EST (expressed sequence tag) fragment (GenBank® accession no. CB808508), cloned from female rat pituitary, that showed significant overlap with the 5′-untranslated region of our rat Abca5 cDNA sequence (EMBL accession no. AJ426052). BLAST analyses confirmed that this cDNA fragment originated from a genomic region on rat chromosome 10 in the vicinity of the rAbca5 locus, indicating the existence of an untranslated exon 1 located 8478 bp upstream of the first translated rAbca5 exon described previously [8]. Thus these observations suggest that the rat Abca5 gene consists of 39 exons (Figure 2A), of which exon 2 contains the translation initiation motif AACATGC. This gene structure (39 exons) is in line with the organization of human ABCA5 and murine Abca5 respectively [9].
Figure 2. Detection of a rat Abca5 splice variant.
(A) Structural organization of the rat Abca5 gene on chromosome 10. Exons are numbered in ascending order from the 5′-terminus, starting with the putative first untranslated exon and are depicted as equally sized black boxes. Exon 20, which is extended by 16 bp in the rat Abca5 splice variant rAbca5 V20+16, is marked with an asterisk. A metric scale in corresponding kb is indicated. (B) Enlarged view of the cDNA region in human ABCA5 and rat Abca5 between exons 20 and 21. The conserved 16 bp insertions leading to a premature stop codon in the splice variant of both species are boxed. The premature stop codon and deviating bases are underlined. (C) Design of RTQ PCR assay for selective amplification of rAbca5 V20+16 (EMBL accession no. AJ550165) cDNA. The regions corresponding to the primers and Taqman probe RTQ1 are boxed. The PCR reaction amplifying total rAbca5 and splice variant rAbca5 V20+16 cDNA fragments used primers RTQ.forw and RTQ.rev. To amplify solely the rAbca5 V20+16 cDNA fragment, RTQ.forw and SVRTQ.rev were employed.
For further analyses of subcellular localization of rAbca5, plasmids enabling expression of rAbca5 in transfected cells were constructed. During sequence analyses of rAbca5 expression plasmid clones containing the full ORF of rat Abca5, an alternative rat Abca5 cDNA sequence comprising a 16 bp insert was identified (EMBL accession no. AJ550165). The insert represented a 3′-terminal extension of the exon referred to as exon 20 (Figures 2A and 2B), and was almost identical with the 16 bp insert previously observed for orthologous human ABCA5 [8] (Figure 2B). Since the insert commenced with the canonical intronic splice junction donor motif GT′, it was assumed to result from alternative RNA splicing, leading to partial intron retention. As in the case of the human ABCA5 splice variant, elongation of exon 20 led to introduction of a premature translation stop at 2775 bp with respect to translation initiation, resulting in a putative C-terminally truncated protein of 925 amino acids, exhibiting one complete N-terminal membrane anchor domain and only one nucleotide binding fold. For comparison, the major rAbca5 transcript (EMBL accession no. AJ426052) was expected to encode a protein consisting of 1642 amino acids with two membrane anchor domains and two nucleotide binding folds. Thus a splice variant predicted to give rise to a protein resembling an ABC half-transporter was found to be conserved between human and rat.
Tissue distribution of rat Abca5 splice variant
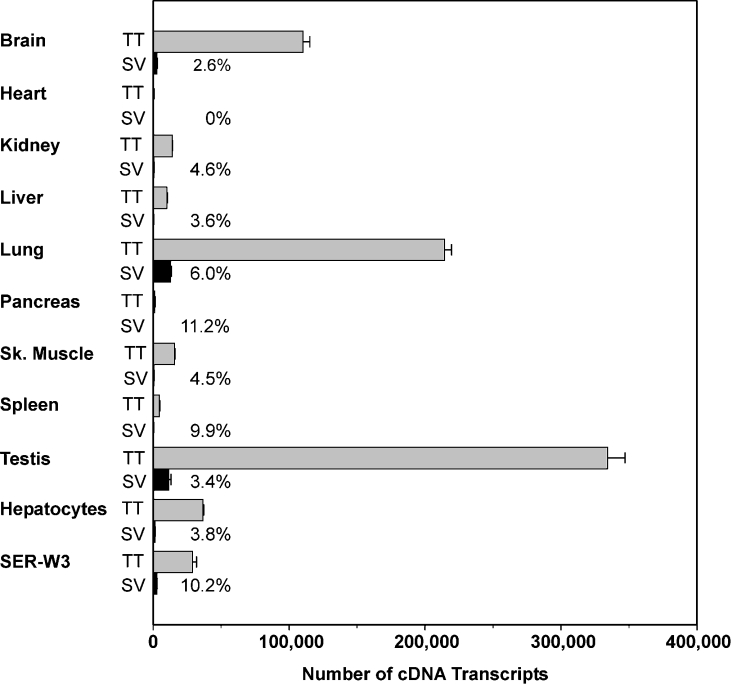
To investigate the tissue distribution of the splice variant rAbca5 V20+16, we carried out an RT–RTQ PCR analysis for nine selected rat organs. As we had previously detected high levels of rAbca5 mRNA in testis and low levels in rat hepatocytes via Northern-blot analyses, we also investigated transcript expression in the rat Sertoli cell line SER-W3 and in freshly isolated primary rat hepatocytes. Expression of the transcript variant was determined with respect to total rAbca5 cDNA transcripts in tissue cDNAs by amplifying a target sequence (of 198 bp for total transcripts and 217 bp for the splice variant; Figure 2C). As shown in Figure 3, expression of the transcript variant was detected for SER-W3 cells, hepatocytes and all organs examined, with the exception of the heart. Relative cDNA amounts varied between 2.6% of the total transporter transcript abundance (rat brain) and 11.2% (pancreas), although general rAbca5 expression was low in pancreas. Total transcript levels for freshly isolated hepatocytes were higher than for liver, indicating that hepatocytes represent a major cell population contributing to liver rAbca5 mRNA expression. Comparison of expression levels for total rAbca5 transcripts among the various organs confirmed results previously obtained through Northern-blot analysis, in which considerable rAbca5 mRNA expression was found in lungs and brain, and the highest expression was observed in rat testis [8].
Figure 3. Quantification of rat Abca5 and rat Abca5 V20+16 cDNA transcripts in nine selected rat organs, freshly isolated rat hepatocytes and a Sertoli cell line (SER-W3).
The total number of cDNA transcripts was determined in 40 ng of reverse transcribed cDNA. The relative amount of splice variant transcript (SV) was calculated with respect to the amount of total rAbca5 cDNA transcripts (TT) and is shown as numbers (%) in the diagram. Error bars indicate the S.D. for experiments performed in triplicate. SV, splice variant.
Expression of rAbca5–EGFP and rAbca5 V20+16–EGFP fusion proteins in HEK-293 cells as detected by immunoblot analyses
The ORFs of both rat Abca5 and the splice variant rAbca5 V20+16 were used to construct expression plasmids containing the coding region for the fluorescent tag EGFP at the 3′-terminus of the respective ORF. HEK-293 cells were used for the expression of the resulting fluorescent fusion proteins. To investigate whether continuous transporter–EGFP fusion proteins were generated, immunoblot analysis was performed. Cells were homogenized 48 h after transfection, and the homogenates were centrifuged for sedimentation of the nuclear fraction. The post-nuclear supernatants were subjected to PAGE, Western transfer and development of immunoreactive protein bands with different primary antibodies raised against EGFP. As shown in Figure 4, the polyclonal primary antibody reacted with EGFP, which exhibited the expected molecular mass of approx. 30 kDa, in samples of cells transfected with pEGFP-N1 as a positive control. In addition, protein bands in the range 195–230 kDa were detected in samples of prAbca5–EGFP-transfected cells, approximately corresponding to an expected electrophoretic mobility of the rAbca5 full transporter (186 kDa) fused to EGFP (∼30 kDa; Figure 4). The appearance of multiple immunoreactive bands is indicative of post-transcriptional modification and the observed smear of immunoreactive protein, although focused around 200 and 220 kDa, is typical for glycosylated proteins, e.g. for the P-glycoprotein (MDR) subgroup of ABC transporters [13]. Nevertheless, further experiments are required to specify the nature of possible post-translational modifications of the rAbca5 protein.
Figure 4. Expressions of EGFP, rAbca5–EGFP fusion protein and rAbca5 splice variant fusion protein in transiently transfected HEK-293 cells (Western-blot analysis).
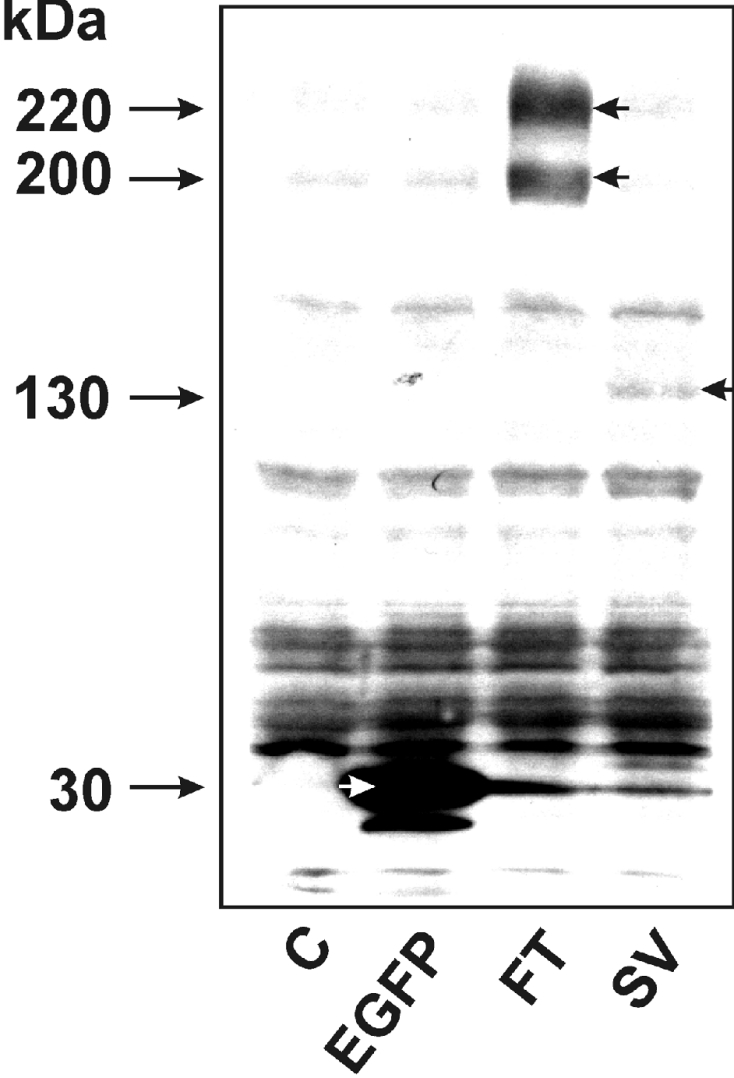
Homogenates of HEK-293 cells were prepared 48 h after transfection with 1 μg of pEGFP-N1 or an rAbca5–EGFP fusion plasmid. Following sedimentation of nuclei, the post-nuclear supernatant was subjected to immunoblot analysis as outlined in the Experimental section, applying 160 μg of protein per lane. EGFP and EGFP fusion proteins were detected with a polyclonal antibody (BD Biosciences) raised against the full-length EGFP protein. The samples were derived from the following cells: C, untransfected control cells; EGFP, transfected with pEGFP-N1 plasmid; FT, transfected with prAbca5–EGFP; SV, transfected with prAbca5 V20+16–EGFP. Short arrows denote immunoreactive EGFP (white arrow) and transporter–EGFP fusion proteins (black arrows).
In homogenates of cells transfected with the prAbca5 V20+16–EGFP construct, a specific protein band was detected at approx. 130–135 kDa (Figure 4). Since the calculated molecular mass of the rAbca5 splice variant protein would amount to 104 kDa, the observed electrophoretic mobility of the immunoreactive protein would be consistent with the size of the rAbca5 splice variant–EGFP fusion protein. The development of Western blots with a monoclonal primary antibody raised against EGFP (JL-8) yielded the same immunoreactive bands for EGFP, rAbca5–EGFP and the splice variant fusion protein as the polyclonal antibody raised against EGFP (results not shown). Thus these results support the conclusion that the fluorescence observed in HEK-293 cells transfected with the transporter-EGFP expression plasmids was due to the expression of intact rAbca5–EGFP or rAbca5 splice variant–EGFP fusion protein respectively.
Subcellular localization of rAbca5–EGFP and rAbca5–V5 fusion proteins
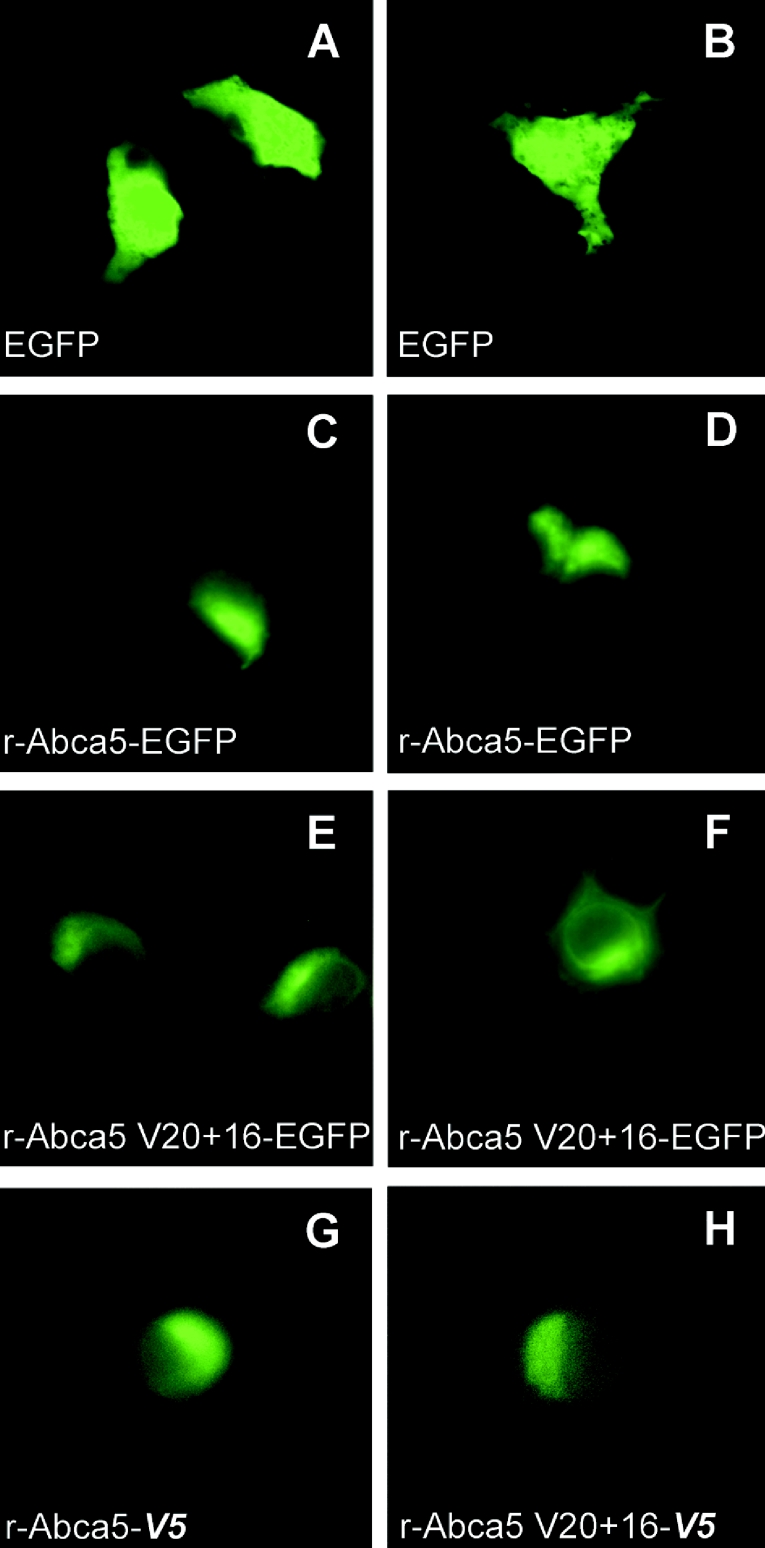
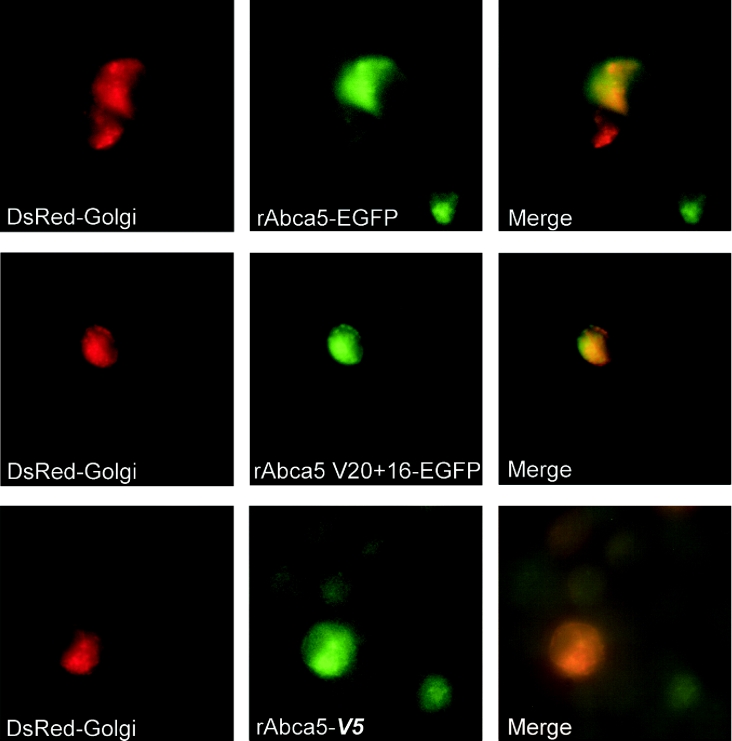
The expression of green fluorescent rat Abca5 and splice variant rAbca5 V20+16 fusion proteins was monitored in transiently transfected HEK-293 cells via fluorescence microscopy. In cells observed for a period of up to 72 h (time course not shown), an expression maximum of EGFP-tagged proteins was reached within 48 h. The fluorescence of cells transfected for control purposes with the pEGFP-N1 plasmid was diffuse (Figures 5A and 5B), highlighting the shape of entire cells. This localization is in accordance with the expected cytoplasmic distribution of EGFP, and could clearly be distinguished from rAbca5–EGFP or rAbca5 splice variant–EGFP fusion protein-dependent fluorescence. Rat Abca5–EGFP distribution was focused in a rim-shaped fashion asymmetrically around the nucleus, forming condensed caps at one end of the nucleus (Figures 5C and 5D). Although expression of the tagged splice variant protein was less pronounced than that of the full transporter fusion protein, the pattern of distribution of the rAbca5 V20+16–EGFP fusion protein revealed marked similarities to localization patterns observed for the full transporter fusion protein (Figures 5E and 5F). The distribution of V5 epitope-tagged rAbca5 and splice variant proteins (bearing a C-terminal V5 epitope) was determined by immunofluorescence using an FITC-conjugated antibody directed against the V5 epitope. V5-tagged fusion protein localization was consistent with the localization of the EGFP-tagged transporter proteins (Figures 5G and 5H). Tagged fusion protein distribution was compared with the localization of fluorescent organelle marker proteins expressed in HEK-293 cells. As evident from Figures 5 and 6, localization of EGFP-tagged rAbca5 proteins resembled the distribution of the red fluorescent marker for the Golgi system. Indeed, in HEK-293 cells co-transfected with the Golgi marker expression plasmid and either of the expression plasmids for EGFP-tagged rAbca5, an overlap of red and green fluorescence was observed, indicative of co-localization in individual cells in which expression of both fluorescent proteins occurred (Figure 7). Accordingly, localization of V5-tagged rAbca5 and V5-tagged splice variant also overlapped with Golgi marker distribution (as shown for rAbca5–V5 in Figure 7). Thus these data support the conclusion that, in transfected HEK-293 cells, rAbca5 and rAbca5 splice variant fusion proteins are localized primarily in an intracellular compartment, most likely representing the Golgi system.
Figure 5. Expressions of rAbca5–EGFP, rAbca5 splice variant–EGFP and V5 epitope-tagged transporter fusion proteins in transiently transfected HEK-293 cells (magnification, ×630).
(A–F) HEK-293 cells 48 h after transfection with 1 μg of individual plasmid per 60 mm culture dish. Cells were transfected with pEGFP-N1 alone (A, B), prAbca5–EGFP (C, D) or prAbca5 V20+16–EGFP (E, F). (G, H) HEK-293 cells 48 h after transfection with 0.5 μg of individual plasmid per 3.5 mm culture dish, transporter expression being probed with an FITC-labelled antibody raised against V5. Cells were transfected with prAbca5-V5 (G) or with prAbca5 V20+16-V5 (H).
Figure 6. Expression of organelle marker proteins in transiently transfected HEK-293 cells (magnification, ×630).
Images show HEK-293 cells after transfection with 0.1–0.5 μg of marker expression plasmid per 60 mm culture dish. (A) Cells transfected with pECFP-Mem (0.1 μg), giving rise to a protein containing the N-terminal 20 amino acids of neuromodulin (GAP-43) fused to the N-terminus of ECFP (plasma membrane and intracellular distribution), 72 h after transfection. (B) Cells transfected with pDsRed-Mito (0.25 μg), yielding a protein containing the mitochondrial targeting sequence of cytochrome c oxidase subunit VIII, fused to DsRed, 48 h after transfection. (C) Cells transfected with pDsRed-Endo (0.5 μg), yielding a protein containing the ER-targeting sequence of calreticulin, fused to DsRed, 48 h after transfection. (D) Cells transfected with pDsRed2-Peroxi (0.25 μg), leading to a protein of DsRed2, fused to the peroxisomal targeting signal 1 (SKL), 72 h after transfection. (E, F) Cells transfected with pDsRed-Golgi (0.25 μg), expressing a protein exhibiting the Golgi-targeting sequence within the N-terminus of β-1,4-galatosyltransferase, fused to DsRed, 48 h (E) and 24 h (F) after transfection.
Figure 7. Co-localization of rAbca5 fusion proteins with Golgi marker (magnification, ×630).
HEK-293 cells were co-transfected with either transporter-EGFP expression plasmid (prAbca5–EGFP or prAbca5 V20+16–EGFP) and pDsRed-Golgi or, alternatively, with the plasmid prAbca5-V5 (expressing V5 epitope-tagged rAbca5) and pDsRed-Golgi, as outlined in the Experimental section. Expression of fusion proteins was documented separately after 24 h (full rAbca5–EGFP), 36 h (rAbca5 V20+16–EGFP) and 48 h (full rAbca5–V5) respectively and merged using Openlab software (Improvision).
DISCUSSION
Characterization of the rAbca5 V20+16 splice variant
Transcripts of 40–60% of all human genes have been estimated to undergo alternative splicing, which may contribute to complexity of the functional spectrum of resulting protein products [19]. Alternative splicing has been described for genes belonging to the same cluster as human ABCA5 (for ABCA6 [20], ABCA9 [21] and ABCA10 [22]), and thus appears to be prevalent among members of this ABCA gene subgroup. Nevertheless, elucidation of the functional impact of splice variant protein products remains a future challenge. We have demonstrated that the ABCA5/rAbca5 splice variant V20+16, formerly described for human ABCA5 [8], is conserved between human and rat species. In the present study, quantitative RT–PCR analyses revealed that the rAbca5 splice variant was expressed in most of the rat tissues examined, its cDNA constituting a proportion of total rAbca5 cDNA transcripts between 2.6 and 11.2%. These results suggest that the rAbca5 splice variant is not simply the result of low-frequency erroneous splicing events, but that the splice variant protein product fulfils a physiological function in numerous tissues. Since a stop codon is introduced by elongation of exon 20 in the splice variant, the putative translation product would represent a protein truncated at the C-terminus, lacking most of the second half of the major full transporter. As the second nucleotide-binding domain and most of the second membrane anchor domain would be missing, the splice variant protein product would be expected to resemble ABC half-transporters in structure. Four protein domains (two transmembrane domains and two nucleotide binding folds) are required for ABC transporter systems to be functional [1]; thus the splice variant protein product would be predicted not to act as a functional transporter in the monomer constellation. However, various ABC half-transporters belonging to other ABC subfamilies, e.g. the ABCB or ABCG subfamilies, are known to form functional transporter complexes by hetero- or homo-dimerization [1], and this principle might also apply to the rAbca5 variant protein.
A question that was raised in the course of our studies was whether the truncated C-terminus of the rAbca5 splice variant protein might affect subcellular distribution. Following transfection of HEK-293 cells with expression plasmids for rAbca5–EGFP or rAbca5 splice variant–EGFP fusion proteins, the distribution of EGFP-tagged transporter proteins was observed via fluorescence microscopy of unfixed cell cultures. Hence, the splice variant product rAbca5 V20+16–EGFP represented the first example of a C-terminally truncated ABCA/Abca splice protein product to be visibly expressed in cell culture. The EGFP-tagged rAbca5 splice variant protein exhibited a localization pattern resembling that of the EGFP-tagged full transporter protein and co-localized with a fluorescent Golgi marker protein. Since the possibility cannot be fully ruled out that a particular protein/peptide tag might alter the localization pattern of a fusion protein as compared with the native, untagged protein, we conducted further experiments involving expression in HEK-293 cells of alternatively V5-tagged transporter proteins, bearing the viral V5 epitope within a peptide considerably shorter than the EGFP sequence. The distribution of rAbca5–V5 and rAbca5 V20+16–V5 fusion proteins, as demonstrated via immunofluorescence of fixed cells, was in accordance with Golgi marker localization, thus corroborating the results obtained with EGFP-tagged transporter proteins. Although detection of the endogenous untagged transporter proteins via immunohistochemistry would be required to confirm rat transporter subcellular localization in the natural tissue context, our results indicate that both rAbca5 and the splice variant protein reside in membranes of the same intracellular compartment, probably constituting the Golgi system. Provided that proximity between the full-transporter and the splice variant protein exists, interaction between the half-transporter and the major full-length transporter may be possible. Questions relating to whether the rAbca5 V20+16 splice variant protein might fulfil a role in regulation of full-transporter function or whether it might convey substrate translocation as part of a half-transporter dimer, however, require further investigation.
Implications of rAbca5 localization analyses
Many ABC proteins of the full transporter type, e.g. ABCA1, are primarily localized at the plasma membrane. Several full transporters of the ABCA subfamily, however, have been found to reside at intracellular organelle membranes. ABCA3, for example, which is involved in the biogenesis of surfactant, has been shown to be situated in vesicle membranes and lamellar bodies of alveolar type II cells [6]. Human ABCA2, a related transporter demonstrated to confer resistance to the oestradiol derivative estramustine, is highly expressed in brain and peripheral blood leucocytes and is targeted to lysosomes [4,5]. Rat Abca2 is localized to lysosomal membranes and in part to the Golgi apparatus in rat oligodendrocytes [23]. The present study provides evidence for an intracellular localization of rAbca5 in a compartment most likely representing the Golgi apparatus.
Investigation of rAbca5 mRNA expression, either by Northern-blot analyses [8] or by RT–RTQ PCR, revealed high expression in testis. To pinpoint cell populations in testis contributing to this rAbca5 mRNA expression, we performed in situ hybridization. The results indicated that rAbca5 mRNA is expressed in basal cells and in interstitial cell clusters that consist mainly of Leydig cells.
Leydig cells are known to process cholesterol and synthesize essential steroid hormones such as testosterone. Cholesterol acts as a precursor for steroid synthesis. The first steps of cholesterol-dependent steroid hormone synthesis take place in the mitochondria, whereas the final conversion into testosterone occurs at the ER. Testosterone is not only crucial for spermatogenesis within the seminiferous tubules, but also promotes development and maintenance of male secondary sex characteristics, which requires transport of the hormone from the site of synthesis to the seminiferous tubules and into the bloodstream respectively. The intracellular localization of rAbca5, presumably at the Golgi apparatus, and pronounced expression in Leydig cells, indicate that this transporter may play a role in intracellular sterol/steroid trafficking. Rat Abca5-dependent processes may be of relevance in cholesterol distribution within the Leydig cells, steroid hormone release or in transcellular transport of steroid hormones. Insight has been gained on uptake, metabolism, cellular redistribution and efflux of cholesterol [24], although many aspects of sterol transport and of steroid translocation within and through membranes remain to be defined. A contribution of membrane-situated ABC transporters to sterol trafficking has been recognized lately. In particular, the half-transporters ABCG5 and ABCG8, forming a functional dimer, are now known to limit absorption of plant sterols and cholesterol in the intestine [1,25], and ABCA1 has been characterized in mediating apolipoprotein A-I-associated cellular export of cholesterol and phospholipids from cells, presumably by acting as a phospholipid flippase [1–3]. ABCA1 is also expressed in Sertoli cells of the testis and participation of ABCA1 in regulation of male fertility, in particular in lipid efflux from Sertoli cells, has been suggested by studies involving Abca1−/− mice [26].
Intriguingly, a similar pattern of mRNA expression in rat testis as described here for rAbca5 has previously been demonstrated for the distantly related rat mdr1b (multidrug resistance transporter 1b; Abcb1b): Leydig cells, peritubular cells and Sertoli cells were shown to be sites of mdr1b mRNA production [27]. Several functional implications of this expression pattern may be inferred for mdr1 proteins, which are predominantly situated in the plasma membrane, including a protective role of mdr1 proteins as functional constituents of the blood–testis barrier, e.g. in prevention of diffusion of toxic substances into the seminiferous tubules. mdr1b mRNA expression in Leydig cells, however, may point to participation of this transporter in steroid hormone secretion. Indeed, several reports provide evidence that steroids, e.g. corticosteroids or oestrogens, may function as substrates of mdr1-type transporters [28,29].
In summary, human ABCA5 and rat Abca5 represent novel ABC transporters for which the exact substrate spectrum remains to be clarified. Knowledge of distinct cell populations expressing this transporter and of subcellular transporter distribution provide a basis for optimization of future functional experiments. Due to pronounced expression of rAbca5 mRNA in Leydig cells of rat testis, however, and its homology to other ABCA transporters involved in aspects of lipid transport, a role of rAbca5 in intracellular sterol/steroid trafficking, probably complementary to other ABC transporters involved in lipid homoeostasis, may be anticipated.
Acknowledgments
We thank S. Blume, O. Walter and Gudrun Rüdell for their expert technical assistance, B. Oesch-Bartlomowicz and P. Antoniou-Lipfert for the gift of SER-W3 cells and W. Knepel for providing HEK-293 cells. F.P. is indebted to the Studienstiftung des Deutschen Volkes for a Ph.D. scholarship.
References
- 1.Dean M., Hamon Y., Chimini G. The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res. 2001;42:1007–1017. [PubMed] [Google Scholar]
- 2.Wang N., Silver D. L., Costet P., Tall A. R. Specific binding of ApoA-I, enhanced cholesterol efflux, and altered plasma membrane morphology in cells expressing ABC1. J. Biol. Chem. 2000;275:33053–33058. doi: 10.1074/jbc.M005438200. [DOI] [PubMed] [Google Scholar]
- 3.Bodzioch M., Orso E., Klucken J., Langmann T., Böttcher A., Diederich W., Drobnik W., Barlage S., Büchler C., Porsch-Ozcurumez M., et al. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 1999;22:347–351. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- 4.Vulevic B., Chen Z., Boyd J. T., Davis W., Walsh E. S., Belinsky M. G., Tew K. D. Cloning and characterization of human adenosine 5′-triphosphate-binding cassette, sub-family A transporter 2 (ABCA2) Cancer Res. 2001;61:3339–3347. [PubMed] [Google Scholar]
- 5.Ile K. E., Davis W., Jr, Boyd J. T., Soulika A. M., Tew K. D. Identification of a novel first exon of the human ABCA2 transporter gene encoding a unique N-terminus. Biochim. Biophys. Acta. 2004;1678:22–32. doi: 10.1016/j.bbaexp.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Yamano G., Funahashi H., Kawanami O., Zhao L. X., Ban N., Uchida Y., Morohoshi T., Ogawa J., Shioda S., Inagaki N. ABCA3 is a lamellar body membrane protein in human lung alveolar type II cells. FEBS Lett. 2001;508:221–225. doi: 10.1016/s0014-5793(01)03056-3. [DOI] [PubMed] [Google Scholar]
- 7.Shulenin S., Nogee L. M., Annilo T., Wert S. E., Whitsett J. A., Dean M. ABCA3 gene mutations in newborns with fatal surfactant deficiency. N. Engl. J. Med. 2004;350:1296–1303. doi: 10.1056/NEJMoa032178. [DOI] [PubMed] [Google Scholar]
- 8.Petry F., Kotthaus A., Hirsch-Ernst K. I. Cloning of human and rat ABCA5/Abca5 and detection of a human splice variant. Biochem. Biophys. Res. Commun. 2003;300:343–350. doi: 10.1016/s0006-291x(02)02827-9. [DOI] [PubMed] [Google Scholar]
- 9.Annilo T., Chen Z., Shulenin S., Dean M. Evolutionary analysis of a cluster of ATP-binding cassette (ABC) genes. Mamm. Genome. 2003;14:7–20. doi: 10.1007/s00335-002-2229-9. [DOI] [PubMed] [Google Scholar]
- 10.Tsuruoka S., Ishibashi K., Yamamoto H., Wakaumi M., Suzuki M., Schwartz G. J., Imai M., Fujimura A. Functional analysis of ABCA8, a new drug transporter. Biochem. Biophys. Res. Commun. 2002;298:41–45. doi: 10.1016/s0006-291x(02)02389-6. [DOI] [PubMed] [Google Scholar]
- 11.Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 12.Pogan F., Masson M. T., Lagelle F., Charuel C. Establishment of a rat Sertoli cell line that displays the morphological and some of the functional characteristics of the native cell. Cell Biol. Toxicol. 1997;13:453–463. doi: 10.1023/a:1007475928452. [DOI] [PubMed] [Google Scholar]
- 13.Ziemann C., Bürkle A., Kahl G. F., Hirsch-Ernst K. I. Reactive oxygen species participate in mdr1b mRNA and P-glycoprotein overexpression in primary rat hepatocyte cultures. Carcinogenesis. 1999;20:407–414. doi: 10.1093/carcin/20.3.407. [DOI] [PubMed] [Google Scholar]
- 14.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Speel E. J., Saremaslani P., Roth J., Hopman A. H., Komminoth P. Improved mRNA in situ hybridization on formaldehyde-fixed and paraffin-embedded tissue using signal amplification with different haptenized tyramides. Histochem. Cell Biol. 1998;110:571–577. doi: 10.1007/s004180050319. [DOI] [PubMed] [Google Scholar]
- 16.Liu Q., Berchner-Pfannschmidt U., Möller U., Brecht M., Wotzlaw C., Acker H., Jungermann K., Kietzmann T. A Fenton reaction at the endoplasmic reticulum is involved in the redox control of hypoxia-inducible gene expression. Proc. Natl. Acad. Sci. U.S.A. 2004;12:4302–4307. doi: 10.1073/pnas.0400265101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowry O. H., Rosebrough N. J., Farr A. C., Randall R. J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 18.Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 19.Modrek B., Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 20.Kaminski W. E., Wenzel J. J., Piehler A., Langmann T., Schmitz G. ABCA6, a novel A subclass ABC transporter. Biochem. Biophys. Res. Commun. 2001;285:1295–1301. doi: 10.1006/bbrc.2001.5326. [DOI] [PubMed] [Google Scholar]
- 21.Piehler A., Kaminski W. E., Wenzel J. J., Langmann T., Schmitz G. Molecular structure of a novel cholesterol-responsive A subclass ABC transporter, ABCA9. Biochem. Biophys. Res. Commun. 2002;295:408–416. doi: 10.1016/s0006-291x(02)00659-9. [DOI] [PubMed] [Google Scholar]
- 22.Wenzel J. J., Kaminski W. E., Piehler A., Heimerl S., Langmann T., Schmitz G. ABCA10, a novel cholesterol-regulated ABCA6-like ABC transporter. Biochem. Biophys. Res. Commun. 2003;306:1089–1098. doi: 10.1016/s0006-291x(03)01097-0. [DOI] [PubMed] [Google Scholar]
- 23.Zhou C., Zhao L., Inagaki N., Guan J., Nakajo S., Hirabayashi T., Kikuyama S., Shioda S. Atp-binding cassette transporter ABC2/ABCA2 in the rat brain: a novel mammalian lysosome-associated membrane protein and a specific marker for oligodendrocytes but not for myelin sheaths. J. Neurosci. 2001;21:849–857. doi: 10.1523/JNEUROSCI.21-03-00849.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soccio R. E., Breslow J. L. Intracellular cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 2004;24:1150–1160. doi: 10.1161/01.ATV.0000131264.66417.d5. [DOI] [PubMed] [Google Scholar]
- 25.Berge K. E., Tian H., Graf G. A., Yu L., Grishin N. V., Schultz J., Kwiterovich P., Shan B., Barnes R., Hobbs H. H. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science. 2000;290:1771–1775. doi: 10.1126/science.290.5497.1771. [DOI] [PubMed] [Google Scholar]
- 26.Selva D. M., Hirsch-Reinshagen V., Burgess B., Zhou S., Chan J., McIsaac S., Hayden M. R., Hammond G. L., Vogl A. W., Wellington C. L. The ATP-binding cassette transporter ABCA1 mediates lipid efflux from Sertoli cells and influences male fertility. J. Lipid Res. 2004;45:1040–1050. doi: 10.1194/jlr.M400007-JLR200. [DOI] [PubMed] [Google Scholar]
- 27.Melaine N., Liénard M.-O., Dorval I., Le Goascogne C., Lejeune H., Jégou B. Multidrug resistance genes and P-glycoprotein in the testis of the rat, mouse, guinea pig, and human. Biol. Reprod. 2002;67:1699–1707. doi: 10.1095/biolreprod.102.003558. [DOI] [PubMed] [Google Scholar]
- 28.Ueda K., Okamura N., Hirai M., Tanigawara Y., Saeki T., Kioka N., Komano T., Hori R. Human P-glycoprotein transports cortisol, aldosterone, and dexamethasone, but not progesterone. J. Biol. Chem. 1992;267:24248–24252. [PubMed] [Google Scholar]
- 29.Kim W. Y., Benet L. Z. P-glycoprotein (P-gp/MDR1)-mediated efflux of sex-steroid hormones and modulation of P-gp expression in vitro. Pharm. Res. 2004;21:1284–1293. doi: 10.1023/b:pham.0000033017.52484.81. [DOI] [PubMed] [Google Scholar]