Abstract
GSTs (glutathione S-transferases; E.C.2.5.1.18) are a supergene family of dimeric multifunctional enzymes that have a major role in detoxification pathways. Using a GST from the mosquito Anopheles dirus (adGSTD4-4), we have characterized the enzymatic and physical properties of Leu-6, Thr-31, Leu-33, Ala-35, Glu-37, Lys-40 and Glu-42. These residues generate two motifs located in the N-terminal domain (domain I) that are functionally conserved across GST classes. The aim of this study was to understand the function of these two motifs. The first motif is a small hydrophobic core in the G-site (glutathione-binding site) wall, and the second motif contains an ionic bridge at the N-terminus of the α2 helix and is also part of the G-site. The mutations in the small hydrophobic core appear to have structural effects, as shown by the thermal stability, refolding rate and intrinsic fluorescence differences. In the Delta class GST, interactions form an ionic bridge motif located at the beginning of the α2 helix. The data suggest that electrostatic interactions in the α2 helix are involved in α-helix stabilization, and disruption of this ionic bridge interaction changes the movement of the α2-helix region, thereby modulating the interaction of the enzyme with substrates. These results show that the small hydrophobic core and ionic bridge have a major impact on structural stabilization, as well as being required to maintain structural conformation of the enzyme. These structural effects are also transmitted to the active site to influence substrate binding and specificity. Therefore changes in the conformation of the G-site wall in the active site appear to be capable of exerting influences on the tertiary structural organization of the whole GST protein.
Keywords: Anopheles dirus, Delta class glutathione S-transferase, mosquito, site-directed mutagenesis
Abbreviations: CDNB, 1-chloro-2,4-dinitrobenzene; DCNB, 1,2-dichloro-4-nitrobenzene; DTT, dithiothreitol; EA, ethacrynic acid; G-site, glutathione-binding site; GST, glutathione S-transferase; PNBC, p-nitrobenzyl chloride; PNPBr, p-nitrophenethyl bromide
INTRODUCTION
Cytosolic GSTs (glutathione transferases; E.C. 2.5.1.18) are a family of dimeric isoenzymes that catalyse the conjugation of glutathione (GSH) to a variety of organic compounds containing an electrophilic centre [1,2], many with carcinogenic and toxic properties [3–7]. This reaction plays an important part in cellular metabolism, transport and subsequent excretion of toxic organic compounds. Other functions of GSTs have been reported, including binding of bilirubin and carcinogens [8,9], isomerization of maleyl acetoacetate [10–12], regulation of the stress kinases [13] and modulation of the ryanodine receptor (a calcium ion channel; [14,15]). A membrane-bound microsomal GST has been well characterized, and seems to be structurally and genetically distinct from the cytosolic enzymes [16]. Despite low sequence homology among GST classes, all of these isoenzymes have very similar tertiary structures, topography of the active site and GSH-binding site [17–19].
The Anopheles dirus mosquito is an important malaria vector in South-East Asia. AdGSTD4-4 is one of four alternatively spliced products from 7.5 kb of the adgst1AS1 gene (An. dirus alternatively spliced GST gene), which has been identified from an An. dirus genomic library [20]. All four spliced products have 45 identical amino acids at the N-terminus, and were named adGST1-1, adGST1-2, adGST1-3 and adGST1-4, according to insect GST nomenclature that is in use [that is, GST-(insect class 1)-(protein 1, 2, 3 and 4) respectively]. However, to be in alignment with a proposed universal GST nomenclature, these were renamed adGSTD1-1, adGSTD2-2, adGSTD3-3 and adGSTD4-4 [21,22]. The subunit number remains the same, since subunits were enumerated as they were initially discovered; ‘D’ refers to GST Delta class and ‘4-4’ refers to the homodimeric isoenzyme. Two available tertiary structures, for adGSTD3-3 and adGSTD4-4, have assisted studies on these proteins [23]. Moreover, GSTs are highly conserved across the insect GST Delta class at the N-terminus (e.g. in An. dirus, An. gambiae, Lucilia cuprina, Musca domestica and Drosophila melanogaster). Therefore, in the present study, characterization of the N-terminal residues in adGSTD4-4 can be used to understand their functions for all insect Delta class GSTs [21,24].
Many hydrophobic residues are found inside a folded protein contributing to tertiary structure as a hydrophobic core. In the G-site (GSH-binding site), there are several such residues (Leu-6, Leu-33 and Ile-52) surrounding the glycine moiety of GSH. In previous studies [25,26], Leu-33 and Ile-52 were shown to impact on structural aspects of the protein, such as folding and stability. In the present paper, we extend our previous studies on the small hydrophobic core in the G-site wall containing Leu-6, Thr-31, Leu-33 and Ile-52.
An induced-fit mechanism apparently conserved across all GST classes occurs through movement of the α2 helix and its flanking regions to modulate G-site affinity for GSH; however, the relative contributions of residues to the flexibility in the α2 helix have not been determined [27–30]. At the N-terminus of the α2 helix, there is an ionic bridge interaction formed by Glu-37, Lys-40 and Glu-42. These residues are on the outside of the protein surface exposed to solvent. This ionic bridge interaction should impact upon movement of part of the active site, in addition to involvement in structural stabilization of the enzyme. Therefore the residues Glu-37, Lys-40 and Glu-42 were studied; furthermore, Ala-35 was changed to arginine in an attempt to increase the ionic bridge interaction in the α2-helix region.
MATERIALS AND METHODS
Site-directed mutagenesis
The mutants were generated using the QuikChange® site-directed mutagenesis protocol (Stratagene). The mutagenic primers used in these experiments were designed based on the sequence of the adGSTD4-4 wild-type gene (GenBank® accession number AF273040). The oligonucleotide primers, each complementary to opposite strands of the vector, were extended during temperature cycling by means of Pfu DNA polymerase, which replicates both plasmid strands with high fidelity. Each mutant was randomly screened by restriction digestion analysis. Mutant plasmids could be distinguished from template by digestion with restriction enzyme corresponding to the restriction recognition site introduced by the mutagenic primers. Then, full-length DNA sequencing in both directions was performed to confirm the engineered clone sequence.
Protein expression and purification
After transformation of the mutant plasmids into Escherichia coli BL21(DE3)pLysS, protein expression was performed. All of the adGSTD4-4 clones were expressed in LB (Luria–Bertani) broth (containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol) and induced with 0.1 mM IPTG (isopropyl β-thiogalactoside) for 3 h at 37 °C. The pellets were collected and kept at −20 °C until used. The expression levels of the protein were determined by SDS/PAGE. The cell pellets from 50 ml of culture were suspended by mixing with 4.8 ml of 50 mM Tris/HCl, pH 7.4, containing 1 mM EDTA, 200 μl of 100 mg/ml lysozyme and 3.6 μl of 1.4 M 2-mercaptoethanol. The suspension was incubated on ice for 20 min, then 50 μl of 1 M DTT (dithiothreitol) was added and the suspension was lysed at 900 lbf/in2 (1 lbf/in2≈6.9 kPa) in a French Press cell. The lysate was then centrifuged at 10000 g at 4 °C for 20 min. The supernatant containing the soluble form of the recombinant enzyme was separated from the pellet. The recombinant adGSTD4-4 mutants and the wild-type protein were purified by using either GSH-affinity chromatography or cation exchange followed by hydrophobic interaction chromatography, as described previously [25]. GSH-affinity chromatography was used according to manufacturer's instructions (Amersham Biociences). A cation-exchange column (SP-XL) followed by hydrophobic interaction chromatography (phenyl-Sepharose column) was used for mutants that could not be purified on the GSH-affinity chromatography column. Briefly, cation-exchange chromatography employed an SP-XL column that was equilibrated with buffer A (20 mM phosphate buffer, pH 7.0, containing 10 mM DTT). After the lysate was applied to the column, the column was washed with buffer A. Then protein was eluted with a linear gradient from 80–500 mM NaCl in buffer A. The major amount of GST enzyme eluted in buffer A containing approx. 250 mM NaCl. The eluted GST fractions were pooled and loaded on to a phenyl-Sepharose column that was equilibrated with buffer A containing 2 M NaCl. Washing steps were performed by using stepwise decreases in salt concentration in buffer A. The GST enzyme was eluted from the phenyl-Sepharose column by using 20% (v/v) ethylene glycol containing 10 mM DTT. The purified enzymes (in 50 mM potassium phosphate, pH 6.5) were stored in 50% (v/v) glycerol at −20 °C until used. Concentrations of the proteins were determined by Bio-Rad protein reagent (Bio-Rad) using BSA as the standard protein, and purity of the proteins was analysed by SDS/PAGE.
Enzymatic characterization
The standard GST assay was performed in 0.1 M potassium phosphate buffer, pH 6.5, in the presence of 3 mM CDNB (1-chloro-2,4-dinitrobenzene) and 10 mM GSH [31]. The rate of conjugation between GSH and CDNB was monitored by measuring continuously the increase in absorbance at 340 nm for 1 min using a SpectraMax 250 apparatus at 25–27 °C. The molar absorption coefficient of 9.6 mM−1·cm−1 was used to convert absorbance into moles [32].
The kinetic experiments were performed as described previously [31]. In brief, CDNB was chosen as the electrophilic substrate for determination of Vmax, Km, kcat and kcat/Km values. The kinetic parameters were determined by varying the CDNB concentration (0.031–3.0 mM) while GSH was held constant at a saturating concentration, and by varying GSH concentrations (0.25–20 mM) at a saturating concentration of CDNB. The initial rate of the enzymatic reaction was measured spectrophotometrically as described for the GST activity assay determination. The steady-state kinetics followed Michaelis–Menten kinetics, except where stated. The maximal velocity (Vmax) and the Michaelis constant (Km) were determined by non-linear regression software analysis (GraphPad Prism 4). The turnover number (or catalytic-centre activity; kcat) and catalytic efficiency (kcat/Km) were calculated on an active-site basis using the subunit molecular mass of each enzyme. The kinetic parameters are the means±S.D. for at least three independent experiments.
The specific activities towards several GST substrates were determined as described previously [31]. All measurements were performed at 25–27 °C in 0.1 M potassium phosphate buffer at either pH 6.5 or 7.5. The GST activities were measured with GSH and five hydrophobic substrates: CDNB, DCNB (1,2-dichloro-4-nitrobenzene), EA (ethacrynic acid), PNPBr (p-nitrophenethyl bromide) and PNBC (p-nitrobenzyl chloride). Specific activities were calculated using the molar absorption coefficient for each substrate [32].
Physical characterization
The enzymes at 0.1 mg/ml in 0.1 M phosphate buffer, pH 6.5, containing 1 mM EDTA and 5 mM DTT were incubated at 45 °C for various times, and the remaining activity was measured in the standard GST assay. Data were plotted as logarithms of the percentage of remaining activity against pre-incubation time. The half-life of the enzyme at 45 °C was calculated from the slope of the plot using the equation: slope=k/2.3, where k=0.693/t1/2.
Enzymes were completely unfolded by incubating 0.5 mg/ml enzymes in unfolding buffer [0.2 M phosphate buffer (pH 7.0)/1 mM EDTA/5 mM dithiothreitol] containing 4 M guanidinium chloride at room temperature (≈25 °C) for 10 min. Unfolded enzymes were allowed to begin refolding by diluting 40-fold in 0.1 M phosphate buffer, pH 6.5, to a final guanidinium chloride concentration of 0.1 M. Appropriate aliquots from this incubation mixture were measured for reactivation by measuring GST activity as a function of time after dilution. GST activity versus time of refolding followed a non-linear regression single-exponential equation [33].
Intrinsic fluorescence of adGSTD4-4 was measured in a single-photon-counting spectrofluorimeter. Excitation was at 295 nm, and emission was scanned from 300–450 nm. Samples contained 0.1 mg/ml GST in 0.1 M potassium phosphate buffer, pH 6.5, and were prepared similarly for wild-type and mutant enzymes. The wavelength that gives the maximum fluorescence intensity (λmax) and the fluorescence intensity at λmax were observed. The experimental data were corrected for both dilution and inner-filter effects, and normalized after background subtraction.
CD measurements in the far-UV region from 200–250 nm were performed with a spectropolarimeter. The enzymes at 0.2 mg/ml in 0.1 M phosphate buffer, pH 6.5, were measured at 25 °C. A cell of 1 cm path length and a 1 nm bandwidth were used. Spectra are the averages of ten scans after subtraction of the average of ten baseline scans.
RESULTS AND DISCUSSION
Protein expression and purification
In the present study we have characterized Leu-6, Thr-31, Leu-33, Ile-52, Ala-35, Glu-37, Lys-40 and Glu-42, located in domain I in the highly flexible loop region between the β2–α2 or α2–β3 loops. These residues were hypothesized to be involved in structural maintenance or stabilization of the proteins. The mutagenesis study was performed by individually changing each residue chosen to alanine and/or a conservative or non-conservative amino acid. In the present study, Leu-33 mutations generated for a previous report were included to extend that study with further experiments. An additional conservative amino acid replacement, L33I, was generated to confirm the function of Leu-33 in packing of the active site. In addition, Ala-35 was mutated to an arginine in an attempt to increase the ionic bridge interaction in the α2-helix region. All 13 engineered enzymes could be expressed in soluble form at 37 °C in E. coli lysates. Two procedures were utilized for protein purifications: an affinity chromatography on immobilized GSH or sequential purification by ion-exchange chromatography (SP-XL) followed by phenyl-Sepharose chromatography. The latter procedure was used for the engineered enzymes that could not be purified by GSH-affinity chromatography, including L33A, L33F, L33Y and T31A. Purified enzymes gave a single band on SDS/PAGE, with a size of approx. 25 kDa, corresponding to the calculated molecular mass of the GST subunits.
Enzymatic characterization
The kinetic constants for CDNB and GSH show that most of the residue positions only slightly affected enzyme catalysis (Table 1). All of these residues are located in the GSH-binding site, so it is interesting to note that Leu-6 changed the active-site conformation enough to significantly increase Km for the hydrophobic substrate CDNB (3-fold; ANOVA, P<0.001) and decrease the Km for GSH (1.7-fold; ANOVA, P<0.05). GSH steady-state kinetics of the T31A enzyme showed positive co-operativity, with a Hill coefficient of 1.62±0.09 (the Hill coefficient for wild-type enzyme is 0.91±0.02). Positive co-operativity indicates GSH binding in the first active site, then facilitates another GSH binding in the second active site by increasing binding affinity of the vacant binding site [34]. Thr-31 is located in the loop before the α2 helix, and its side chain faces into the G-site in addition to being close to His-38, which interacts directly with GSH. The alanine mutation at Thr-31 must cause a packing rearrangement of the G-site, which increases the Km for GSH 23-fold, as well as generating intersubunit communication between the two active sites. One explanation is that the rearrangement would disturb His-38, which directly interacts with GSH. Mutations of His-38 were shown to have large effects on GSH binding affinity, as well as generating positive co-operativity [25]. The mutation of Ile-52 to alanine caused very large effects on catalysis, as exhibited by a decrease in kcat (1% of wild-type activity) and affinity (14-fold increase in Km), as well as positive co-operativity. The conservative change of Ile-52 to leucine showed smaller effects, including a slight increase in GSH affinity. Mutations of ionic-bridge residues showed only small effects on catalysis, although the GSH binding affinity was affected to a greater extent. Both Glu-37 and Lys-40, located in the middle of the β2–α2 loop, appeared to impact more, suggesting a greater influence on α2-helix movement during induced-fit conformational changes.
Table 1. Steady-state kinetic constants using GSH and CDNB as GST substrates.
The rate of conjugation was monitored continuously by measuring the increase in absorbance at 340 nm for 1 min. The units of Vmax, kcat, Km and kcat/Km are μmol·min−1·mg protein−1, S−1, mM and S−1·mM−1 respectively.
| CDNB | GSH | |||||
|---|---|---|---|---|---|---|
| Enzyme | Vmax | kcat | Km | kcat/Km | Km | kcat/Km |
| Wild-type | 62.5±1.24 | 26.1 | 0.50±0.02 | 51.8 | 0.50±0.10 | 52.1 |
| L33A | 23.5±0.5† | 9.70 | 1.17±0.05† | 8.30 | 8.20±0.43† | 1.20 |
| L33Y | 0.32±0.01† | 0.14 | 0.81±0.12† | 0.17 | 1.10±0.12§ | 0.13 |
| L33F | 1.55±0.03† | 0.65 | 1.30±0.06† | 0.50 | 2.42±0.22† | 0.27 |
| L33I | 53.0±1.71† | 22.2 | 0.73±0.05§ | 30.3 | 0.42±0.01 | 52.8 |
| L6A | 42.3±1.64† | 17.7 | 1.49±0.15† | 11.9 | 0.31±0.01 | 57.1 |
| T31A | 23.3±0.35† | 9.73 | 0.77±0.05‡ | 12.6 | 11.4±0.53*† | 0.85 |
| I52A | 0.630±0.006† | 0.26 | 0.32±0.02 | 0.81 | 7.04±0.05*† | 0.04 |
| I52L | 43.8±1.24† | 18.3 | 0.74±0.02‡ | 24.8 | 0.30±0.04 | 61.1 |
| E37A | 48.5±1.45† | 20.4 | 0.54±0.03 | 37.7 | 1.85±0.09† | 11.0 |
| E37Q | 62.8±0.92 | 26.3 | 0.58±0.05 | 45.3 | 2.26±0.04† | 11.6 |
| K40A | 51.3±1.24† | 21.5 | 0.62±0.04 | 34.6 | 1.14±0.08§ | 18.8 |
| E42A | 63.4±1.95 | 26.5 | 0.57±0.05 | 46.4 | 0.63±0.04 | 42.0 |
| A35R | 53.6±1.18† | 22.4 | 0.52±0.06 | 42.7 | 0.73±0.05 | 30.7 |
* The mutants T31A and I52A showed positive co-operativity upon GSH binding, with Hill coefficients of 1.62±0.09 and 1.49±0.03 respectively.
ANOVA analysis revealed significant differences compared with the wild-type, where indicated: †P<0.001. The absence of a symbol indicates no significant difference compared with wild-type.
ANOVA analysis revealed significant differences compared with the wild-type, where indicated: ‡P<0.01. The absence of a symbol indicates no significant difference compared with wild-type.
ANOVA analysis revealed significant differences compared with the wild-type, where indicated: §P<0.05. The absence of a symbol indicates no significant difference compared with wild-type.
Although the residues studied are in or behind G-site residues, the substrate-specificity results show that these mutations yielded various changes in specificity for several hydrophobic substrates compared with wild-type GST (Table 2). As shown by the kinetic constants, these residues are not critical for enzyme catalysis or substrate binding; however, the mutations influence the enzymes' hydrophobic substrate interaction and catalysis. Compared with wild-type, the mutations at Leu-6, Thr-31 and Ile-52 resulted in decreases in specificity towards several substrates, especially DCNB and PNPBr. The remaining mutations displayed major effects with regard to PNPBr, but specificities were comparable with that of wild-type for CDNB. However, changes in specificity for EA and PNBC as substrates were varied.
Table 2. Substrate specificity changes of the engineered enzymes relative to the wild-type adGSTD4-4.
The substrates were used at the following concentrations: CDNB, 3 mM; DCNB, 1 mM; PNPBr, 0.1 mM; PNBC, 0.1 mM; and EA, 0.2 mM. The reactions were performed at a constant GSH concentration.
| Substrate specificity (μmol·min−1·mg protein−1) | ||||||
|---|---|---|---|---|---|---|
| Enzyme | Substrate… | CDNB | DCNB | EA | PNPBr | PNBC |
| Wild-type | 52.5±0.52 | 0.035±0.006 | 0.286±0.062 | 0.074±0.012 | 0.064±0.002* | |
| L33A | 18.3±0.3* | 0.031±0.015 | 0.332±0.061 | 0.019±0.004* | 0.024±0.004* | |
| L33Y | 0.253±0.001* | <0.0016* | 0.092±0.006* | <0.006* | <0.007* | |
| L33F | 1.023±0.018* | <0.0016* | 0.059±0.007* | <0.006* | <0.007* | |
| L33I | 45.4±0.37* | 0.047±0.002‡ | 0.247±0.006 | 0.007±0.001* | 0.044±0.002* | |
| L6A | 28.0±0.53* | 0.0053±0.0003* | 0.380±0.004 | 0.017±0.002* | 0.031±0.002* | |
| T31A | 17.5±0.33* | 0.0059±0.0006* | 0.186±0.008† | <0.002* | 0.039±0.004* | |
| I52A | 0.511±0.010* | <0.001* | 0.307±0.023 | <0.004* | 0.011±0.001* | |
| I52L | 35.4±0.36* | 0.025±0.001 | 0.373±0.005 | 0.018±0.001* | 0.036±0.003* | |
| E37A | 39.1±0.74* | 0.055±0.005† | 0.135±0.010* | <0.008* | 0.122±0.006* | |
| E37Q | 47.8±0.22* | 0.053±0.001† | 0.431±0.026* | 0.033±0.004* | 0.062±0.002 | |
| K40A | 37.7±0.16* | 0.037±0.001 | 0.110±0.004* | 0.038±0.003* | 0.080±0.002* | |
| E42A | 55.1±0.40‡ | 0.059±0.001* | 0.24±0.02 | 0.016±0.001* | 0.045±0.002* | |
| A35R | 44.9±0.56* | 0.056±0.003* | 0.37±0.04 | 0.020±0.001* | 0.041±0.002* | |
ANOVA analysis revealed significant differences compared with the wild-type, where indicated: *P<0.001. The absence of a symbol indicates no significant difference compared with wild-type.
ANOVA analysis revealed significant differences compared with the wild-type, where indicated: †P<0.01. The absence of a symbol indicates no significant difference compared with wild-type.
ANOVA analysis revealed significant differences compared with the wild-type, where indicated: ‡P<0.05. The absence of a symbol indicates no significant difference compared with wild-type.
Both GSH- and hydrophobic-substrate-binding sites are located in different parts of the same active-site pocket. Therefore differences in the mutant enzymes appear to occur from changes in active-site conformation that disturb orientation of the substrates, as found in the wild-type enzyme. In addition, binding modes and orientations of the various substrates in the active site appear to be unique, as shown by dissimilar effects on specificity of the mutant enzymes.
Physical characterization
The half-life at 45 °C of the mutant enzymes compared with adGSTD4-4 wild-type showed that the mutants decreased in stability, except for T31A, which displayed a slight increase (Table 3). L6A decreased enzyme stability approx. 9-fold, showing a major impact on structural maintenance. The mutation of ionic-bridge residues (Glu-37, Lys-40 and Glu-42) to alanine and the mutation of Ala-35 to arginine also resulted in decreases in enzyme stability, suggesting that the mutations cause a rearrangement of the α2-helix, owing to disruption of the ionic bridge interaction. The replacement of positively charged Glu-37 with a similarly sized, polar glutamine residue decreased enzyme stability 2.3-fold, supporting the idea of charge distribution involvement in structural stabilization of the enzyme. However, the attempt to increase charge distribution with the A35R mutation decreased the half-life of the enzyme 1.7-fold. This suggests that residues in the β2–α2-helix loop appear to affect helix dipole moment, which impacts upon structural stabilization of the enzyme.
Table 3. Thermal stability of wild-type and engineered adGSTD4-4 mutants at 45 °C.
The remaining GST activity was measured after incubating the enzyme at various time points at 45 °C.
| Enzyme | Half-life (t1/2) at 45 °C (min) |
|---|---|
| Wild-type | 15.3±0.31 |
| L33A | 45.3±1.56* |
| L33Y | 71.7±1.71* |
| L33F | 212±17.9* |
| L33I | 30.4±1.5* |
| L6A | 1.73±0.01* |
| T31A | 22.7±1.77* |
| I52A | 9.58±0.65* |
| I52L | 18.1±1.33 |
| E37A | 1.73±0.04* |
| E37Q | 6.54±0.54* |
| K40A | 3.94±0.20* |
| E42A | 8.44±0.48* |
| A35R | 9.01±0.54* |
ANOVA analysis revealed a significant difference compared with the wild-type enzyme, where indicated (*P<0.001); the absence of an asterisk indicates no significant difference compared with wild-type (in these experiments, exclusively the I52L mutant).
AdGSTD4-4 became completely unfolded after incubation for 10 min in 4 M guanidinium chloride, as monitored by intrinsic fluorescence and CD spectroscopy. Refolding rates and percentages of recovered activities of wild-type and mutant enzymes were determined (Table 4). L6A showed a decreased refolding rate (2.5-fold compared with wild-type), suggesting the initial folding process was disturbed, which also slightly reduced the activity recovered as well as decreasing the enzyme stability, as described above. L33A showed an increased refolding rate of 1.6-fold and 3-fold greater stability than the wild-type enzyme, demonstrating that its final conformation is more stable. However, the mutants L33I and I52L showed only small effects on the refolding process. The ionic bridge mutations (Glu-37, Lys-40, Glu-42 and Ala-35) all decreased enzyme stability in the range 1.8- to 8.9-fold, although the refolding rates were altered differently. These data suggest that the changes in the refolding rates occur from differential ‘flexing’ of the engineered enzyme. An interesting observation for the E37A enzyme was that it possessed a slightly increased refolding rate of 1.3-fold, as well as a 1.8-fold increase in recovered activity, even though the enzyme had an 8.9-fold decreased stability (Table 3). This suggests that this residue position contributes to structural maintenance, but not to initial folding events. Therefore the ionic bridge motif appears to have a function in structural maintenance and modulation of α2-helix movement, as well as a minor effect on folding.
Table 4. Refolding rate constants and percentages of recovered activity from the reversible refolding experiment for the wild-type adGSTD4-4 enzyme and its engineered mutants.
The proteins were denatured initially with 4 M guanidinium chloride at room temperature for 10 min. Refolding rate constants (kref) were determined by non-linear regression analysis using a single-exponential equation (GraphPad Prism 4).
| Refolding experiment | ||
|---|---|---|
| Enzyme | Refolding rate constant (min−1) | % recovery |
| Wild-type | 0.54±0.02 | 20.7±0.67 |
| L33A | 0.85±0.05* | 18.6±0.55 |
| L33Y | nd | nd |
| L33F | nd | nd |
| L33I | 0.53±0.01 | 14.7±0.76* |
| L6A | 0.22±0.02* | 14.9±0.77* |
| T31A | 0.41±0.02* | 10.1±0.41* |
| I52A | nd | nd |
| I52L | 0.67±0.02* | 20.1±0.61 |
| E37A | 0.69±0.05* | 37.7±1.49* |
| E37Q | 0.74±0.03* | 14.0±0.32* |
| K40A | 0.26±0.02* | 14.2±0.95* |
| E42A | 0.32±0.02* | 31.5±1.44* |
| A35R | 0.40±0.01‡ | 29.6±0.64† |
ANOVA analysis revealed significant differences compared with the wild-type, where indicated: *P<0.001. The absence of a symbol indicates no significant difference compared with the wild-type. nd, not determined (low activity precluded measurement).
ANOVA analysis revealed significant differences compared with the wild-type, where indicated: †P<0.01. The absence of a symbol indicates no significant difference compared with the wild-type. nd, not determined (low activity precluded measurement).
ANOVA analysis revealed significant differences compared with the wild-type, where indicated: ‡P<0.05. The absence of a symbol indicates no significant difference compared with the wild-type. nd, not determined (low activity precluded measurement).
The intrinsic fluorescence property of tryptophan in proteins is modulated by various interactions in the environment around the tryptophan residue. Therefore changes in λmax and the fluorescence intensity of the intrinsic fluorescence spectrum can suggest that changes have occurred in the local tertiary structure of the protein. There are two tryptophan residues present in adGSTD4-4 (Trp-64 and Trp-191). Trp-64 has the side chain exposed to solvent, located in the subunit interface at the base of the GSH-binding site, whereas Trp-191 is buried in the interior of domain II. Therefore this technique can be used to monitor active-site conformation indirectly. The same protein concentration of wild-type and mutant enzymes was used for comparison of the maximum emission wavelengths (λmax) and fluorescence intensities (Table 5). The λmax of intrinsic tryptophan fluorescence for the mutants was approximately the same as that for the wild-type (333±1 nm), except for I52A, which displayed a red-shifted spectrum with a λmax at 339 nm. Therefore I52A affected tertiary structure at the active site, causing a change in polarity in the tryptophan environment. In addition, there were differences in intensity of fluorescence between wild-type and several mutant proteins, indicating a limited conformational alteration in the active site of the final structure. The charged residue mutations (Glu-37, Lys-40 and Glu-42) displayed decreased fluorescence intensity. Located at the C-terminal end of the β4-sheet, Trp-64 is in proximity with the α2-helix from which the ND2 side-chain nitrogen of Asn-47 extends 3.07 Å (1 Å=0.1 nm) to have a cation–π interaction with the aromatic ring of Trp-64. Disruption of the ionic bridge interaction would most likely increase flexibility of the α2 helix, thereby disrupting this interaction and increasing solvent exposure of Trp-64, with a concomitant decrease in fluorescence. In addition, a rearrangement of the active-site topology appears to occur for the G-site-residue mutations Leu-6, Thr-31, Leu-33 and Ile-52, which affects orientation of Trp-64, as well as of its neighbouring residues, which also may contribute to changes in fluorescence.
Table 5. The maximum emission wavelength (λmax) and intrinsic fluorescence intensity at λmax of tryptophan fluorescence of adGSTD4-4 and the engineered enzymes.
The excitation wavelength (λex) was set at 295 nm, and emission was scanned from 300–450 nm. Samples (n=3) contained 0.1 mg/ml protein in 0.1 M potassium phosphate buffer, pH 6.5. The percentage intensities compared with wild-type enzyme were measured at fluorescence λmax averaged over three scans, corrected for dilution and inner-filter effects.
| Intrinsic fluorescence (λex 295 nm) | ||
|---|---|---|
| Enzyme | λmax (nm) | % intensity |
| Wild-type | 333±0 | 100 |
| L33A | 334±0 | 60.8* |
| L33Y | 334±0 | 51.6* |
| L33F | 334±0 | 47.9* |
| L33I | 333±0 | 109.1 |
| L6A | 334±0 | 138.6* |
| T31A | 334±0 | 99.1 |
| I52A | 339±0 | 98.8 |
| I52L | 333±0 | 115.5* |
| E37A | 333±0 | 66.8* |
| E37Q | 334±0 | 107.7 |
| K40A | 334±0 | 68.4* |
| E42A | 333±0 | 75.5* |
| A35R | 334±0 | 102.1 |
ANOVA analysis revealed a significant difference compared with the wild-type enzyme, where indicated (*P<0.001); the absence of an asterisk indicates no significant difference compared with the wild-type.
CD spectroscopy was performed to determine whether secondary structure content of the proteins had changed. CD spectroscopy showed two ‘troughs’ at 210 and 222 nm, owing to the high helical content of the proteins. All the proteins had a similar far-UV CD spectrum (results not shown). This shows that, although some mutations affect active-site conformation, the overall secondary structure content of the enzymes has not been altered.
In the present paper, we have sought to understand the function of two features in domain I of adGSTD4-4 that have been found in all insect Delta class GSTs: a small hydrophobic core in the G-site and an ionic bridge at the N-terminus of the α2-helix. The available structure of adGSTD4-4 shows that Leu-6, Thr-31, Leu-33 and Ile-52 form part of the wall in the GSH-binding site (Figure 1A). The mutation of Leu-6 leads to a structural perturbation in the G-site, resulting in altered substrate specificity, decreased stability, as well as modulation of the refolding of the enzyme. From a previous study [25], increasing hydrophobicity of the residue in the Leu-33 position increased enzyme stability 3- to 14-fold, although the mutants lost enzyme activity (0.9–62% compared with wild type). In the present study, Leu-33 mutations altered intrinsic fluorescence intensity when compared with wild-type enzyme, suggesting changes in topology of the active site. The change in structural conformation and active-site topology of the Leu-33 mutations was confirmed by a trypsin proteolytic experiment: the mutation of Leu-33 to alanine increased the proteolytic rate almost 2-fold compared with the wild-type (results not shown).The conservative mutation of Leu-33 to isoleucine resulted in small effects on specificity and catalysis, the most notable of which was a 10-fold decrease for PNBr. L33I displayed only a 2-fold increase in half-life, with all other physical properties being similar to those of the wild-type. These data support the idea that the role of Leu-33 is to contribute to the small hydrophobic core for structural maintenance. Previously, Ile-52 had been characterized in adGSTD3-3 [26]. In the tertiary structures of adGSTD3-3 (PDB identification no. 1JLV) and adGSTD4-4 (PDB identification no. 1JLW), the location of Ile-52 is identical in both structures. A mutation of Ile-52 to alanine showed a decrease in enzyme stability of approx. 5.5-fold, a 24-fold increase in the Km for GSH and a 2-fold decrease in kcat [26]. For adGSTD4-4, the I52A mutant showed different effects, with a 1.6-fold decrease in stability, a 14-fold increase in the Km for GSH and a 100-fold decrease in kcat. The conservative replacement of Ile-52 with leucine showed smaller effects in enzyme properties, and the I52L mutant was very similar to the wild-type enzyme in terms of its physical properties. These data show that Ile-52 has roles in structural integrity and maintenance, as well as in the initial folding of the protein. Therefore it may be surmised that the small hydrophobic core formed by Leu-6, Leu-33 and Ile-52 at the G-site wall plays a critical role in packing and stabilization of the active site into an appropriate conformation, and this packing also impacts upon tertiary structure of the whole protein. A study of available crystal structures of other GST classes shows several hydrophobic residues clustered in the G-site wall in a similar fashion. For example, in the human GST isoenzyme hGSTP1-1 (PDB identification no. 11GS), these residues are Phe-8, Trp-38 and Leu-52, whereas in maize GST-I (PDB identification no. 1AXD) they are Phe-35, Ile-33 and Val-54. These residues are located in the G-site wall, and therefore are probably involved in packing of the active-site conformation as well.
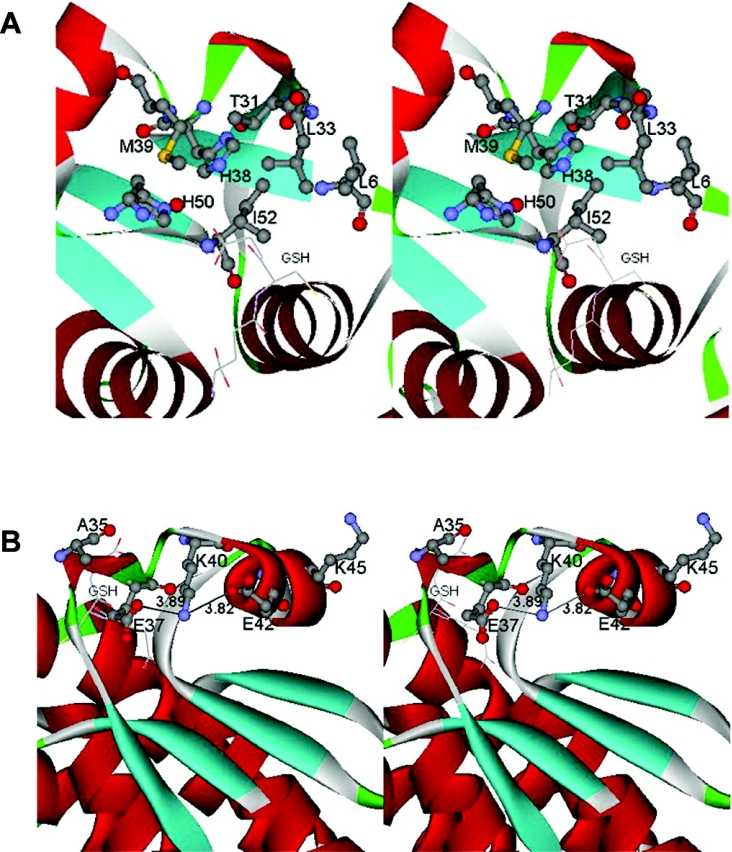
Figure 1. Stereo views of domain I region characterized in this study.
(A) Stereo view of the active-site pocket of adGSTD4-4. Leu-6, Leu-33, Thr-31 and Ile-52 form part of the G-site wall. The hydrophobic residues Leu-6, Leu-33 and Ile-52 form a small hydrophobic cluster in the G-site. GSH in the active site is shown in stick form. (B) Stereo view of the charged residues located near the α2 helix of adGSTD4-4. The ionic bridge motif formed by the charged residues (Glu-37, Lys-40 and Glu-42) is exposed to the solvent, located at the N-terminus of the α2 helix. GSH in the active site is shown in stick form. Both parts of the Figure were created with Accelrys DS ViewerPro 5.0.
Several studies show α2 helices of Delta, Theta, Sigma and Pi GSTs to have high conformational flexibility and to display the highest temperature factors of the whole protein [29,35,36]. In insect Delta class GSTs, there is a unique ionic bridge motif exposed to solvent on the α2 helix consisting of three charged residues: Glu-37, Lys-40 and Glu-42 (Figure 1B). The data suggest that this feature influences the flexibility of the α2 helix and its flanking region in Delta class GSTs. In the present study, the alanine mutations of Glu-37, Lys-40 and Glu-42 in adGSTD4-4 showed structural effects in the domain I region, as shown by decreased thermal stability and changes in refolding rates and intrinsic fluorescence intensities. The apparent changes in α2-helix movements resulting from mutations of the charged residues also appear to modulate interaction of the enzymes with substrates, as shown by changes in substrate specificity. The charged residues and ionic bridge motif in this region appear to play a role in maintenance of a stable conformation, as well as in influencing refolding of the enzymes. The attempt to increase the ionic-bridge interaction by constructing the A35R mutant did not result in any improvement in kinetic properties or enzyme stability, although it did appear to have some effects on substrate specificity. This may be because the side chain of arginine in A35R is not in the correct orientation for interacting with the other residues. In hGSTP1-1, there is an ion pair formed by Cys-47 and Lys-54 at the end of the α2 helix. The disruption of the electrostatic interaction between Cys-47 and Lys-54 causes an increased mobility of the α2 helix, local structural changes, lowered GSH-binding affinity and strong positive co-operativity towards GSH [37,38]. Therefore electrostatic interactions in the α2 helix appear to be involved in α-helix stabilization for GSTs in general. In cephalopod Sigma class GSTS1-1, charged residues at each end of the α2 helix (Asp-37 and Lys-42 at the N- and C-termini of the α2 helix respectively) have been described to interact with the helix dipole, and thereby stabilize the helix [39]. This feature is also found in adGSTD4-4 (residues Glu-37 and Lys-45), as well as in the blood fluke (Schistosoma japonicum) GST, Sj26GST, a GST of 26 kDa (PDB identification no. 1M9A; residues Glu-37 and Lys-44). These charged residues would appear therefore to interact with the helix dipole, and contribute to stabilizing the α2 helix.
In the present study of adGSTD4-4, we examined the roles of conserved residues in the N-terminal domain (domain I) that are involved in a small hydrophobic core in the G-site wall and an ionic bridge motif in the N-terminus of the α2 helix. The results have shown that both the small hydrophobic core and the ionic bridge motif can have a major impact on structural stabilization, as well as being required to maintain the structural conformation of the enzyme. The packing of the small hydrophobic core is important for structural integrity and, as part of the G-site wall, directly affects GSH binding. These packing effects in the active site appear not only to affect the G-site, but are transmitted through rearrangement of the active-site residues to the adjacent H-site, thereby impacting on specificity as well as catalysis.
Acknowledgments
This work was funded by the TRF (Thailand Research Fund). A.V. was supported by a Royal Golden Jubilee Ph.D. Research Scholarship.
References
- 1.Jakoby W. B., Habig W. H. Glutathione transferases. In: Jakoby W. B., editor. Enzymatic Basis of Detoxication, vol. 2. New York: Academic Press; 1980. pp. 63–94. [Google Scholar]
- 2.Hayes J. D., Flanagan J. U., Jowsey I. R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong R. N. Structure, catalytic mechanism, and evolution of the glutathione transferases. Chem. Res. Toxicol. 1997;10:2–18. doi: 10.1021/tx960072x. [DOI] [PubMed] [Google Scholar]
- 4.Mannervik B., Funk M., Frank H., Seidel A. Glutathione S-transferase A1-1-catalysed conjugation of bay and fjord region diol epoxides of polycyclic aromatic hydrocarbons with glutathione. Carcinogenesis. 1996;17:1491–1498. doi: 10.1093/carcin/17.7.1491. [DOI] [PubMed] [Google Scholar]
- 5.Mannervik B., Danielson U. H. Glutathione transferases – structure and catalytic activity. CRC Crit. Rev. Biochem. 1988;23:283–337. doi: 10.3109/10409238809088226. [DOI] [PubMed] [Google Scholar]
- 6.Wilce M. C. J., Parker M. W. Structure and function of glutathione S-transferases. Biochim. Biophys. Acta. 1994;1205:1–18. doi: 10.1016/0167-4838(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 7.Ketterer B. A bird's eye view of the glutathione transferase field. Chem. Biol. Interact. 2001;138:27–42. doi: 10.1016/s0009-2797(01)00277-0. [DOI] [PubMed] [Google Scholar]
- 8.Litwack G., Ketterer B., Arias I. M. Ligandin: a hepatic protein which binds steroids, bilirubin, carcinogens and a number of exogenous organic anions. Nature (London) 1971;234:466–467. doi: 10.1038/234466a0. [DOI] [PubMed] [Google Scholar]
- 9.Hayes J. D., Pulford D. J. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. CRC Crit. Rev. Biochem. Mol. Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 10.Board P., Baker R. T., Chelvanayagam G., Jermiin L. S. Zeta, a novel class of glutathione transferases in a range of species from plants to humans. Biochem. J. 1997;328:929–935. doi: 10.1042/bj3280929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polekhina G., Board P. G., Blackburn A. C., Parker M. W. Crystal structure of maleylacetoacetate isomerase/glutathione transferase zeta reveals the molecular basis for its remarkable catalytic promiscuity. Biochemistry. 2001;40:1567–1576. doi: 10.1021/bi002249z. [DOI] [PubMed] [Google Scholar]
- 12.Board P. G., Taylor M. C., Coggan M., Parker M. W., Lantum H. B., Anders M. W. Clarification of the role of key active site residues of glutathione transferase Zeta/maleylacetoacetate isomerase by a new spectrophotometric technique. Biochem. J. 2003;374:731–737. doi: 10.1042/BJ20030625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adler V., Yin Z., Fuchs S. Y., Benezra M., Rosario L., Tew K. D., Pincus M. R., Sardana M., Henderson C. J., Wolf C. R., et al. Regulation of JNK signaling by GSTp. EMBO J. 1999;18:1321–1334. doi: 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dulhunty A., Gage P., Curtis S., Chelvanayagam G., Board P. The glutathione transferase structural family includes a nuclear chloride channel and a ryanodine receptor calcium release channel modulator. J. Biol. Chem. 2001;276:3319–3323. doi: 10.1074/jbc.M007874200. [DOI] [PubMed] [Google Scholar]
- 15.Board P. G., Coggan M., Chelvanayagam G., Easteal S., Jermiin L. S., Schulte G. K., Danley D. E., Hoth L. R., Griffor M. C., Kamath A. V., et al. Identification, characterization, and crystal structure of the omega class glutathione transferases. J. Biol. Chem. 2000;275:24798–24806. doi: 10.1074/jbc.M001706200. [DOI] [PubMed] [Google Scholar]
- 16.Jakobsson P.-J., Morgenstern R., Mancini J., Ford-Hutchinson A., Persson B. Common structural features of MAPEG – a widespread superfamily of membrane associated proteins with highly divergent functions in eicosanoid and glutathione metabolism. Protein Sci. 1999;8:689–692. doi: 10.1110/ps.8.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirr H., Reinemer P., Huber R. X-ray crystal structures of cytosolic glutathione S-transferases. Implications for protein architecture, substrate recognition and catalytic function. Eur. J. Biochem. 1994;220:645–661. doi: 10.1111/j.1432-1033.1994.tb18666.x. [DOI] [PubMed] [Google Scholar]
- 18.Wilce M. C. J., Board P. G., Feil S. C., Parker M. W. Crystal structure of a theta-class glutathione transferase. EMBO J. 1995;14:2133–2143. doi: 10.1002/j.1460-2075.1995.tb07207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossjohn J., McKinstry W. J., Oakley A. J., Verger D., Flanagan J., Chelvanayagam G., Tan K.-L., Board P. G., Parker M. W. Human theta class glutathione transferase: the crystal structure reveals a sulfate-binding pocket within a buried active site. Structure. 1998;6:309–322. doi: 10.1016/s0969-2126(98)00034-3. [DOI] [PubMed] [Google Scholar]
- 20.Pongjaroenkit S., Jirajaroenrat K., Boonchauy C., Chanama U., Leetachewa S., Prapanthadara L., Ketterman A. J. Genomic organization and putative promoters of highly conserved glutathione S-transferases originating by alternative splicing in Anopheles dirus. Insect Biochem. Mol. Biol. 2001;31:75–85. doi: 10.1016/s0965-1748(00)00107-7. [DOI] [PubMed] [Google Scholar]
- 21.Chelvanayagam G., Parker M. W., Board P. G. Fly fishing for GSTs: a unified nomenclature for mammalian and insect glutathione transferases. Chem. Biol. Interact. 2001;133:256–260. [Google Scholar]
- 22.Wongsantichon J., Harnnoi T., Ketterman A. J. A sensitive core region in the structure of glutathione S-transferases. Biochem. J. 2003;373:759–765. doi: 10.1042/BJ20030394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oakley A. J., Harnnoi T., Udomsinprasert R., Jirajaroenrat K., Ketterman A. J., Wilce M. C. J. The crystal structures of glutathione S-transferases isozymes 1-3 and 1-4 from Anopheles dirus species B. Protein Sci. 2001;10:2176–2185. doi: 10.1110/ps.ps.21201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mannervik B., Awasthi Y. C., Board P. G., Hayes J. D., Di Ilio C., Ketterer B., Listowsky I., Morgenstern R., Muramatsu M., Pearson W. R., et al. Nomenclature for human glutathione transferases. Biochem. J. 1992;282:305–306. doi: 10.1042/bj2820305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vararattanavech A., Ketterman A. Multiple roles of glutathione binding-site residues of glutathione S-transferase. Protein Pept. Lett. 2003;10:441–448. doi: 10.2174/0929866033478654. [DOI] [PubMed] [Google Scholar]
- 26.Winayanuwattikun P., Ketterman A. J. Catalytic and structural contributions for glutathione binding residues in a delta class glutathione S-transferase. Biochem. J. 2004;382:751–757. doi: 10.1042/BJ20040697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricci G., Caccuri A. M., Lo Bello M., Rosato N., Mei G., Nicotra M., Chiessi E., Mazzetti A. P., Federici G. Structural flexibility modulates the activity of human glutathione transferase P1-1. Role of helix 2 flexibility in the catalytic mechanism. J. Biol. Chem. 1996;271:16187–16192. doi: 10.1074/jbc.271.27.16187. [DOI] [PubMed] [Google Scholar]
- 28.Stella L., Caccuri A. M., Rosato N., Nicotra M., Lo Bello M., De Matteis F., Mazzetti A. P., Federici G., Ricci G. Flexibility of helix 2 in the human glutathione transferase P1-1. Time-resolved fluorescence spectroscopy. J. Biol. Chem. 1998;273:23267–23273. doi: 10.1074/jbc.273.36.23267. [DOI] [PubMed] [Google Scholar]
- 29.Stella L., Nicotra M., Ricci G., Rosato N., Di Iorio E. E. Molecular dynamics simulations of human glutathione transferase P1-1: analysis of the induced-fit mechanism by GSH binding. Proteins. 1999;37:1–9. [PubMed] [Google Scholar]
- 30.Labrou N. E., Mello L. V., Clonis Y. D. Functional and structural roles of the glutathione-binding residues in maize (Zea mays) glutathione S-transferase I. Biochem. J. 2001;358:101–110. doi: 10.1042/0264-6021:3580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jirajaroenrat K., Pongjaroenkit S., Krittanai C., Prapanthadara L., Ketterman A. J. Heterologous expression and characterization of alternatively spliced glutathione S-transferases from a single Anopheles gene. Insect Biochem. Mol. Biol. 2001;31:867–875. doi: 10.1016/s0965-1748(01)00032-7. [DOI] [PubMed] [Google Scholar]
- 32.Habig W. H., Pabst M. J., Jakoby W. B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- 33.Stenberg G., Dragani B., Cocco R., Mannervik B., Aceto A. A conserved “hydrophobic staple motif” plays a crucial role in the refolding of human glutathione transferase P1-1. J. Biol. Chem. 2000;275:10421–10428. doi: 10.1074/jbc.275.14.10421. [DOI] [PubMed] [Google Scholar]
- 34.Segel I. H. New York: John Wiley & Sons, Inc.; 1993. Enzyme kinetics. Behavior and analysis of rapid equilibrium and steady-state enzyme systems. [Google Scholar]
- 35.Ji X., Von Rosenvinge E. C., Johnson W. W., Tomarev S. I., Paitigorsky J., Armstrong R. N., Gilliland G. L. Three-dimensional structure, catalytic properties, and evolution of a sigma class glutathione transferase from squid, a progenitor of the lens S-crystallins of cephalopods. Biochemistry. 1995;34:5317–5328. doi: 10.1021/bi00016a003. [DOI] [PubMed] [Google Scholar]
- 36.Oakley A. J., Lo Bello M., Ricci G., Federici G., Parker M. W. Evidence for an induced-fit mechanism operating in Pi class glutathione transferases. Biochemistry. 1998;37:9912–9917. doi: 10.1021/bi980323w. [DOI] [PubMed] [Google Scholar]
- 37.Ricci G., Lo Bello M., Caccuri A. M., Pastore A., Nuccetelli M., Parker M. W., Federici G. Site-directed mutagenesis of human glutathione transferase P1-1. Mutation of Cys-47 induces a positive cooperativity in glutathione transferase P1–1. J. Biol. Chem. 1995;270:1243–1248. doi: 10.1074/jbc.270.3.1243. [DOI] [PubMed] [Google Scholar]
- 38.Lo Bello M., Battistoni A., Mazzetti A. P., Board P. G., Muramatsu M., Federici G., Ricci G. Site-directed mutagenesis of human glutathione transferase P1-1. Spectral, kinetic, and structural properties of Cys-47 and Lys-54 mutants. J. Biol. Chem. 1995;270:1249–1253. [PubMed] [Google Scholar]
- 39.Stevens J. M., Armstrong R. N., Dirr H. W. Electrostatic interactions affecting the active site of class Sigma glutathione S-transferase. Biochem. J. 2000;347:193–197. [PMC free article] [PubMed] [Google Scholar]