Abstract
The Msx1 gene in mice has been proven to be induced by BMP (bone morphogenetic protein) proteins, and three binding sites for SMAD, an intracellular BMP signalling transducer, have already been identified in its promoter. Gel shift analyses were performed and they demonstrated that the consensus found very near the transcription start site, a region designed BP (basal promoter), is functional for binding nuclear proteins from 10.5, 11.5 and 13.5 dpc (days post-coitum) embryos. Notably, this binding occurs only when the SMAD-binding consensus sequence is maintained, suggesting that it is required for the formation of a protein complex over BP. Binding of purified SMAD 1 and SMAD 4 as well as supershift assay with SMAD 1/SMAD 5/SMAD 8 antibody proved that a SMAD protein is present in this complex. Transfection assays in cell cultures with fragments from BP driving the expression of luciferase confirmed that only in the presence of the SMAD consensus site is Msx1 expression activated. A proteomic analysis of the complex components after immunoprecipitation identified several proteins necessary to activate transcription including SMAD 8. Our results suggest that BMP2/BMP4 signalling through SMAD 8 is required for transcriptional activation of the mouse Msx1 gene.
Keywords: bone morphogenetic protein 4 (BMP4) signalling, deletion fragment, Msx1 basal promoter, nuclear protein binding, SMAD consensus, transcriptional activation
Abbreviations: AP1, activator protein 1; BMP, bone morphogenetic protein; BP, basal promoter; CREB, cAMP-response-element-binding protein; CBP, CREB-binding protein; CTF, CCAAT-box binding factor; dpc, days post-coitum; DTT, dithiothreitol; EMSA, electrophoretic mobility-shift assay; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; TBP, TATA box binding protein; TBS, Tris-buffered saline; TGF, transforming growth factor; BetaIG-H3, TGF-β-induced protein IG-H3 precursor; TFA, trifluoroacetic acid; TFIIH, general transcriptional factor II H
INTRODUCTION
Msx1 gene expression in mice occurs at several sites during development, especially sites involved in epithelial–mesenchymal interactions. Prominent expression of the gene is observed in dorsal neural tube, limb buds and derivatives of cranial neural crest from 10.5 to 13.5 dpc (days post-coitum) [1,2]. Furthermore, expression in extraembryonic derivatives, endocardial cells and epithelial cells has also been observed [1,2].
In spite of this large and complex pattern of expression, homozygous knockout mice for Msx1 die at birth exhibiting severe abnormalities limited to the craniofacial region, including a complete cleft of the secondary palate [3,4]. This suggests that the gene is essential for craniofacial bone formation and that, at the other sites where the gene is expressed, its function can be replaced by Msx2, which shows an expression pattern overlapping with that of Msx1 [4–9].
The complex expression pattern of the mouse Msx1 gene during embryogenesis requires a precise mechanism of regulation for its correct spatiotemporal expression. Several groups have been studying the regulation of mouse Msx1 gene over the last 10 years. Song et al. [10] showed that Msx1 forced expression in myoblasts blocks differentiation to myotubes, whereas Woloshin et al. [11] demonstrated that this blockage is due to MyoD synthesis inhibition. These results were strengthened by Thompson-Jaeger and Raghow [12] suggesting that Msx1 expression occurs in undifferentiated and proliferative cells. In 1995, Catron et al. [13] showed that transcriptional repression by Msx1 does not require the homeodomain, whereas Zhang et al. [14] showed the presence of some residues in the N-terminal arm of the protein that seems to be important for transcriptional repression. Kusuoka et al. [15] analysed 1.2 kb sequences upstream of the initiator ATG and suggested the presence of potential cis regulatory elements. Shetty et al. [16] studied the functionality of these elements in C2C12 cells, identifying an SP1 (a transcriptional factor) functional site [15,16]. The same group showed that this SP1 site maps in a region designed minimal promoter, since this region is sufficient to activate Msx1 expression in cell cultures [17]. By studying 5 kb of upstream sequence by transfection assays in C2C12 cell lines, this group proposed that only a 1282 bp region upstream of the transcription starting site is necessary to regulate the expression of the gene. In this region, three E-box and three SP1 sites were found. The minimal promoter contains one of each.
At the same time, our group sequenced the entire 5 kb upstream sequence, and by comparison with the promoter sequences available for other homeobox genes in databases, we could define not only a BP (basal promoter) but also three other boxes where several consensus sequences for transcription factors binding were mapped. The BP was defined by conservation between human and mouse sequences, while the other three boxes showed conservation between genes acting at the same time in the same cells and sharing the same transcription factors [18]. The functionality of these boxes were tested in transfection assays in F12 cells [18].
Using a transgenic approach, MacKenzie et al. [19] and Pereira et al. [20] showed that a 4.9 kb fragment upstream of the translational start site is sufficient to generate the nearly complete expression pattern of the gene [19,20]. Moreover, MacKenzie et al. [19] identified two domains in the Msx1 promoter capable of promoting most of the Msx1 expression pattern independent of the BP: the distal enhancer domain located at −4670 to −4420 and the proximal enhancer located at −2630 to −2553.
A body of evidence indicates that Msx1 can be induced by BMP (bone morphogenetic protein) proteins [21,22]. By analysing the Msx1 gene promoter, three binding sites for SMAD, an intracellular BMP4 signalling regulator, were described by our group [23]. The most distal site is at −3747 bp upstream of the gene. The second one co-localizes with the proximal enhancer, described by MacKenzie et al. [19], and was shown to respond to the enhancer activity [23]. The third one was identified in the region described as BP. This region includes binding sites for SP1, Y-box, NF1.2, JunB, CTF (CCAAT-box binding factor)/CBP (CREB-binding protein, where CREB stands for cAMP-response-element-binding protein), retinoid X receptor, Msx1 and MTI.1 as described by Gonzalez et al. [18].
Recently, Mehra-Chaudhary et al. [24] demonstrated that Msx1 gene transcriptional activation is regulated by a protein complex containing SP1/TBP (TATA box binding protein)/CBP that enhances transcription when bound to the BP. Msx3 protein acts as a potent repressor of Msx1 promoter activity when complexed with histone deacetylase. Msx3-mediated repression can be reversed completely by co-expression of CBP or p300 in a dose-dependent manner [24].
In the present study, we analysed the functionality of the BMP4 response (SMAD) binding site in the BP and the role it plays in activation of transcription. To test for the functionality of this element during basal transcriptional machinery attachment, we carried out EMSA (electrophoretic mobility-shift assay)-analysis with nuclear protein extracts from embryos and fragments of this BP containing or not the SMAD consensus sequence. The results showed that only in the presence of this consensus binding site is a DNA–protein complex formed. Purified SMAD 1 and SMAD 4 attachment was also tested as well as mutagenesis and supershift assays. The results obtained suggested that this element is essential for the formation of a protein complex over BP. To investigate further this issue, transfection assays in P19 cells with fragments containing or not the SMAD consensus site driving the expression of luciferase showed that only in the presence of SMAD-binding site is the Msx1 transcription activated, and the level of this expression increased in the presence of BMP4. Immunoprecipitation of the complex formed showed that several proteins are involved in the activation of Msx1 BP, including BACH1 [an AP1 (activator protein 1)-like protein], TBP-1, TIEG-1 (an SP1-like protein), SMAD 8, MXI1, TFIIH (general transcriptional factor II H), connexin 30 and BetaIG-H3 [TGF-β (transforming growth factor-β)-induced protein IG-H3 precursor].
EXPERIMENTAL
Constructs
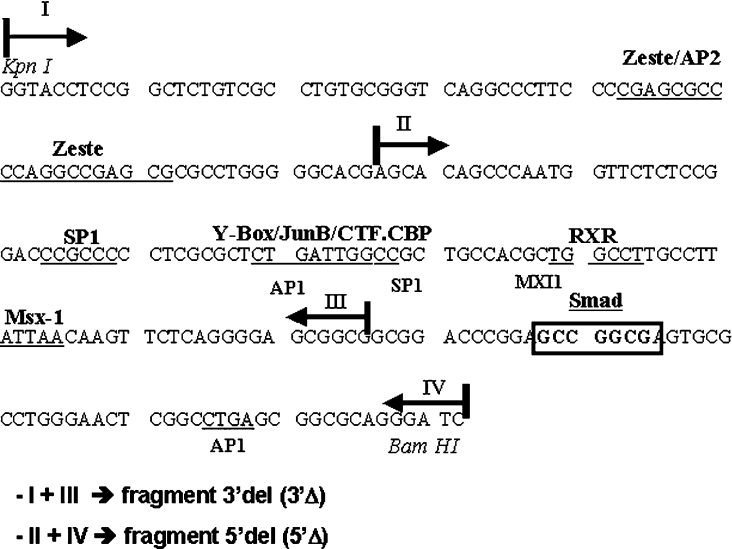
The BP fragment and deletions of BP [5′-deleted (5′Δ) and 3′-deleted (3′Δ)] were obtained by PCR amplification from the construct CS24 [18]. CS24 contains the 4.9 kb Msx1 promoter fragment cloned in pSKT, a modified version of pSK vector [18]. The following primers were used: I (forward), 5′-CCAGGTACCTCCGGCTCTGT-3′; II (reverse), 5′-CAAGAATTCCGCCGCCTCCCCTGAGA-3′; III (forward), 5′-AACGGTACCAGCACAGCCCAATGGTT-3′; and IV (reverse), 5′-TCGAGCGAGCGGGGCCTGGAT-3′. The BP (230 bp) fragment was obtained using restriction enzymes KpnI and BamHI from the construction CS24 [18] and deletion fragments resulted from distinct combinations of forward and reverse primers (Figure 2). PCR products were cloned in the EcoRI and KpnI sites of pSK. These restriction enzyme sites were introduced in the primers used for PCR amplification (underlined). Clones obtained had their sequence confirmed by manual dideoxy DNA sequencing.
Figure 2. Deleted fragments generation scheme.
Subfragments from BP were generated by PCR and cloning. One fragment was deleted in 3′-region (3′Δ) and the other was deleted in 5′-region (5′Δ). Note that the SMAD consensus binding site (boxed sequence) is deleted in the 3′Δ construct.
Embryonic protein extracts
Embryos were obtained from the mouse Balb/c strain. Midday of the day on which plugs were detected was considered as 0.5 dpc. Embryos were dissected and nuclear protein extracts were obtained from head and trunk tissues. The procedure used is a modification of the method described by Dignam et al. [25]. Tissues were suspended in 5 vol. of Buffer A containing 0.3 M sucrose (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2 and 0.1 mM EGTA) and dissociated on a Dounce B homogenizer. Subsequently, 0.4% Nonidet P40 was added followed by another round of homogenization and a 5 min incubation at 4 °C. The material was centrifuged and the pellet was washed twice in Buffer A. Pellets from the last wash were suspended in 2.5 vol. of Buffer C (420 mM NaCl, 20 mM Hepes, pH 7.9, 1.5 mM MgCl2, 0.2 mM EDTA and 25% glycerol) and dissociated. The material was stirred for 45 min at 4 °C and centrifuged. The supernatant was collected and dialysed for 5 h in 50 vol. of Buffer D at 4 °C (20 mM Hepes, pH 7.9, 20% glycerol, 0.1 M KCl and 0.2 mM EDTA). All buffers contained 1 mM DTT (dithiothreitol), 0.5 mM PMSF and 2 μg/ml pepstatin, leupeptin and aprotinin. Protein concentrations were determined by the Bradford method [26].
GST fusion proteins
Constructs containing cDNA of full-length or C-terminally truncated SMAD 1 and 4 proteins cloned at pGEX4T-1 (Amersham Biosciences) were provided by Peter ten Dijke (Ludwig Instruments, Uppsala, Sweden) and have been described by Dennler et al. [27]. Expression of GST fusion proteins was induced in exponentially growing Escherichia coli cultures of the TG1 strain by the addition of 0.2 mM isopropyl β-D-thiogalactoside and incubation at 30 °C for 5 h. Thereafter, fusion proteins were purified according to the manufacturer's (Amersham Biosciences) instructions. SDS/PAGE and Western-blot analyses using anti-goat GST polyclonal antibody (Amersham Biosciences) were performed to determine efficiency of the purification procedure. Extracts were quantified by the Bradford method [26]. For the production of the control GST peptide, the cDNA for SMAD 1 was excised from its corresponding construct by XhoI digestion. Subsequently, the plasmid was religated and transformed into an Epicurean Coli™ XL-1 Blue strain, which was induced to express the GST peptide.
EMSA
Fragments used as probes were obtained by double digesting the constructs described above. Oligonucleotides were synthesized as double-strand (Amitof). Dephosphorylated fragments (1.5–2 pmol) and oligonucleotides (5 pmol) were end-labelled with [γ-32P]ATP and T4 polynucleotide kinase. For binding reactions, 7–10 μg of embryonic extracts or 0.8–1 μg of fusion protein was incubated with 40000–60000 c.p.m. of labelled probe in a binding buffer. The buffer used in reactions with embryonic extracts contained (1×): 10 mM Hepes/KOH (pH 7.9), 60 mM KCl, 1 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 10% glycerol and 2.5 μg of poly(dI-dC)·(dI-dC). A different buffer was used for reactions with fusion proteins (1×): 25 mM Tris/HCl (pH 7.5), 80 mM NaCl, 35 mM KCl, 5 mM MgCl2, 1 mM DTT, 10% glycerol and 1–2.5 μg of poly(dI-dC)·(dI-dC). Reactions were incubated for 40 min at room temperature (25 °C) and then separated by 4.5% native PAGE in 0.5×TBE (1×TBE is 45 mM Tris/borate/1 mM EDTA; run at 4 °C). Initially we performed a dose-dependent competition using 50, 100 and 200 times excess of non-labelled competitors (results not shown) and the dose chosen for experiments was 100 times molar excess. So, for competition reactions, a 100 times molar excess of non-labelled competitor DNA was added 20 min before the addition of the probe. The wild-type oligonucleotide containing only the SMAD consensus binding site and a mutated version of this site was also used as a competitor.
The following oligonucleotides were used: Oligo SMAD wild-type (wt), 5′-GACCCGGAGCCGGCGAGTGCGCC-3′; and Oligo SMAD mutated (mut), 5′-GACCCGGAtCCtGCGAGTGCGCC-3′. The nucleotide changes are shown in boldface and the SMAD element is underlined.
For supershift analysis, protein extracts were incubated for 2 h with 1–2 μg of the antibody at 4 °C either before or after the addition of the probe. The antibodies used were anti-goat IgG polyclonal SMAD 1/SMAD 5/SMAD 8 and anti-rabbit IgG polyclonal E2F-1 (Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.).
Site-specific mutagenesis
To mutate the SMAD-binding site in the 5′-deleted fragment, we used the following primers: III (forward), 5′-AACGGTACCAGCACAGCCCAATGGTT-3′; IV (reverse), 5′-TCGAGCGAGCGGGGCCTGGAT-3′; A (forward), 5′-GACCCGGAtCCtGCGAGTGCGCC-3′; B (reverse), 5′-GGCGCACTCGCaGGaTCCGGGTC-3′. PCR reactions (III+B and IV+A) amplified segments of DNA containing an introduced mutation (using the mutant primers for the SMAD consensus site, primer A or B). After the two products were combined, denatured and allowed to reannel, the DNA polymerase extended the 3′-end of heteroduplexes. After this, the introduced mutation was amplified by using only the outer primers (primers III and IV). The fragment was named 5′Δ mutated fragment and was cloned in pSKT plasmid.
Immunoprecipitation
Nuclear protein extracts from trunk and head were used in immunoprecipitation assays. First, these extracts were incubated in different tubes with 5′-deleted fragment from BP and the same buffer was used in EMSAs. Reactions were incubated for 40 min at room temperature. After this, 10 μg of primary antibody anti-goat IgG polyclonal SMAD 1/SMAD 5/SMAD 8 (Santa Cruz Biotechnology) was added to the reaction mixture and incubated for 2 h at 4 °C. To complete the reaction, 2 μg of secondary antibody anti-goat IgG (Santa Cruz Biotechnology) was also added and incubated for a further 2 h at 4 °C. After all the probes were centrifuged for 30 min, the pellet was redissolved in 20 μl of 2-mercaptoethanol buffer containing (6×) 0.35 M Tris/HCl (pH 6.8), 10.28% SDS, 36% glycerol, 5% 2-mercaptoethanol and 0.012% Bromophenol Blue. Reactions were boiled for 5 min and then separated by SDS/PAGE (10% polyacrylamide) in 2× Tris/glycine buffer (25 mM Tris/HCl, pH 8.3, 192 mM glycine and 0.1% SDS).
MS analysis
The proteins identified were cut out from the gel and processed for MS according to the following method: non-destructive silver-stained bands were trimmed and treated to remove the silver washing with 1:1 destain solution (30 mM potassium ferricyanide and 100 mM sodium thiosulphate), and the destain solution was removed by washing three times for 15 min each in deionized water. The trimmed gels were transferred to new 1500 μl capped microcentrifuge tubes and washed three times for 15 min each in 50% acetonitrile/25 mM ammonium bicarbonate (pH 8.0); the gels were then soaked in 100% acetonitrile and dried in a SpeedVac for 30 min. After washing and drying, the gel pieces were rehydrated at 4 °C in a digestion buffer (25 mM ammonium bicarbonate, pH 8.0) containing 15 ng/μl enzyme trypsin (Sequence Grade Modified Trypsin, Porcine; Promega) and then incubated overnight at 37 °C. The peptides were extracted from the gel subsequently with 50% acetonitrile/1% TFA (trifluoroacetic acid). Each step was performed for 60 min at 37 °C and all digest extracts were collected and dried in a SpeedVac. Each dried digest was redissolved in 3 μl of 50% acetonitrile/1% TFA. For MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) analysis, 0.5 μl of the redissolved peptides was mixed with fresh α-cyano matrix on a MALDI plate. The spectra were acquired in reflector mode with an optimized method for high resolution in the 800–3000 Da range. Using internal calibration with trypsin peaks T7(842.5099), T4(2211.1046) and close external calibration with Cal Mix 1 (Applied Biosystems), the spectra were processed in a Voyager DE-PRO Biospectrometry workstation from Applied Biosystems. Proteins were identified using the Protein Prospector Search software 4.0.5 (University of California, San Franscisco Mass Spectrometry Facility) to search the databases available in the MS-Fit proteomics tool. The parameters were set to allow 1 miss cleavage with the enzyme trypsin and a peptide mass tolerance of 50–200 p.p.m. Additional protein information was obtained from the SWISS-PROT website.
Western-blot analysis
13.5 dpc nuclear protein extracts from trunk and head were used in the Western-blot assays. For each lane, 30 μg of protein was resolved in an SDS/10% polyacrylamide gel in a Mini Protean 3 system (Bio-Rad) and the gels were blotted on to a Hybond-P membrane (Amersham Biosciences). The membranes were blocked in 5% (w/v) dried milk in TBS (Tris-buffered saline) with 0.05% Tween 20 (TBS-T) for 1 h at room temperature. Incubation with primary antibody was performed at 4 °C overnight and then the membranes were washed five times in TBS-T and incubated with secondary horseradish peroxidase-linked antibodies for 1 h at room temperature. After washing the membranes extensively in TBS-T, antibody binding was detected using ECL® (enhanced chemiluminescence) plus (Amersham Biosciences). Primary antibodies used were: SMAD 1 (T-20) and SMAD 8 (S-20) (Santa Cruz Biotechnology).
Cell line and cell culture
P19 cells, an embryonic carcinoma cell line, were maintained in Dulbecco's modified Eagle's medium (Gibco) supplemented with 7.5% (v/v) newborn calf serum (Sigma) and 2.5% fetal calf serum. Cells were cultivated at 37 °C in 10 cm Petri dishes in a 5% CO2 incubator.
Plasmid constructs for transient transfections assays
The BP fragment and deletions of BP (3′Δ and 5′Δ), already cloned in pSK plasmid, were obtained using restriction enzymes (BP: KpnI and BglII; 3′Δ: KpnI and BglII; 5′Δ: XhoI; and 5′Δ: KpnI), and were inserted into the same restriction sites of pGL2-basic luciferase reporter vector (Promega). The clones obtained had their sequence confirmed by DNA sequencing.
Transient transfection
For the transient assays, P19 cells in a semi-confluent culture were co-transfected using Lipofectin® (Gibco) with 2 μg of each luciferase construct and 50 ng of pRL-CMV plasmids (Promega), according to the manufacturer's (Gibco) instructions. The cells were incubated, 24 h after transfection, with BMP4 (1 and 5 nM) for 5 h in a serum-free medium. After incubation, cells were then washed with PBS and lysed according to Dual Luciferase System protocol (Promega) and luciferase activity was analysed in a TD-20/20 luminometer (Turner Designs). The transfections were performed in triplicate. The results are expressed as relative luciferase activity, calculated as a relationship of luciferase activity to the efficiency of the transfection.
Statistical analysis
Data were analysed by Student's t test for independent samples using the SigmaPlot for Windows (version 4.01) package. Significant differences were indicated for P<0.001.
RESULTS
A SMAD consensus is necessary for nuclear protein binding in the BP
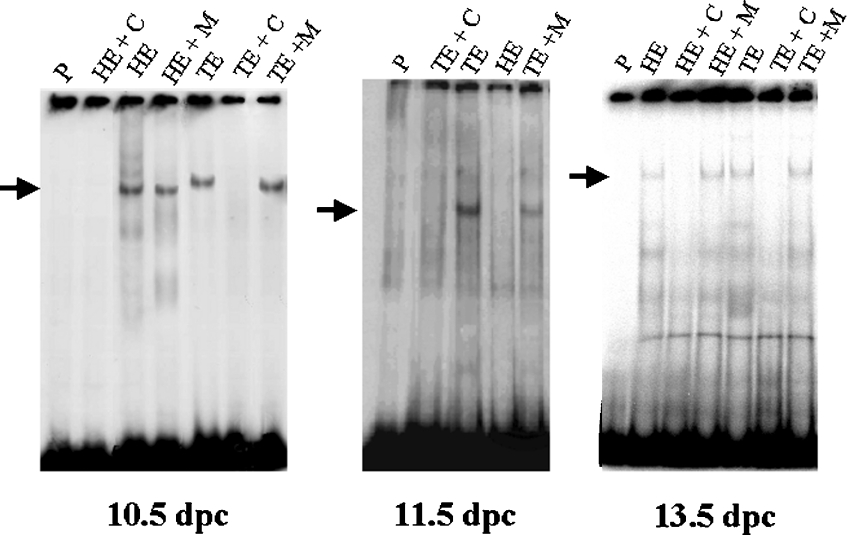
Nuclear protein extracts were obtained using embryos of three distinct ages, encompassing the period of highest and most dynamic Msx1 expression: 10.5, 11.5 and 13.5 dpc. Trunks and heads were treated separately as Msx1 presents a highly distinct temporal expression pattern in these two regions. Extracts were used to investigate binding to the BP domain in EMSA. As controls we used the unlabelled DNA fragment as a competitor as well as a mutated oligonucleotide for the SMAD consensus binding site. We could observe a specific robust binding using 10.5 dpc trunk and head embryo extracts and using 11.5 dpc trunk embryo extract. We also observed binding using 13.5 dpc trunk and head embryo extracts (Figure 1). These results show that the region encompassing the BP domain contains functional binding sites for proteins expressed in 10.5, 11.5 and 13.5 dpc embryos, suggesting the possible capacity of Msx1 BP to attach the transcriptional machinery. Furthermore, the use of a mutated oligo for SMAD consensus as a competitor suggested that one of these sites could be the SMAD consensus binding site.
Figure 1. Gel shift analysis of binding reactions using fragment BP as a probe and head (HE) or trunk (TE) embryo extracts from different ages (10.5, 11.5 and 13.5 dpc).
Specificity of DNA–protein complexes is demonstrated by competition reactions using 100 times unlabelled BP (lanes HE+C and TE+C) and by competition reactions using 100 times unlabelled mutant oligonucleotide DNA for the SMAD consensus site (lanes HE+M and TE+M). Each lane indicated as ‘P’ corresponds to migration of the probe alone. The arrows show the specific binding.
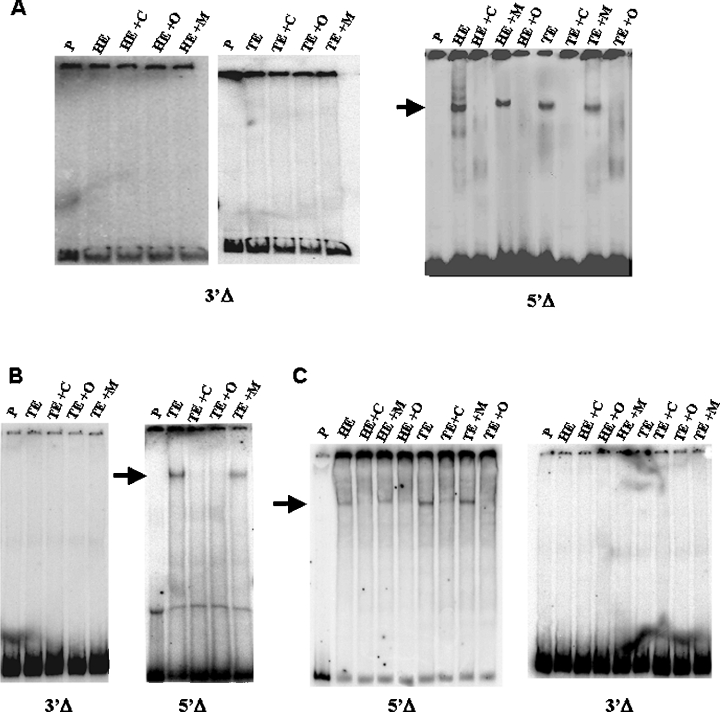
Using the same strategy, we carried out EMSA analysis using two fragments from the BP fragment, one deleting the 5′-region (5′Δ) and the other deleting the 3′-region (3′Δ), both cloned by PCR strategy (Figure 2). These fragments were tested for the binding of embryonic nuclear protein extracts. As shown in Figure 3, the protein–DNA complex formed using 10.5, 11.5 and 13.5 dpc embryo extracts is prevented by deletion of the BP 3′-region (fragment 3′Δ). This deletion eliminates the sequence of the BMP4 response binding site (SMAD consensus binding site). In contrast, 5′Δ fragment retains binding activity, showing that 10.5, 11.5 and 13.5 dpc embryo nuclear proteins recognize only the 5′-containing region of BP (Figure 3). Even in the presence of SP1 and CTF/CBP binding sites, described earlier as responsible for the activation of transcription, we could not observe protein binding in EMSA analysis if the SMAD consensus was not present. Futhermore, the use of the wild-type oligonucleotide for the SMAD consensus binding site as a competitor in 5′Δ fragment EMSA assays abolished protein binding, while the mutated version when used for the same purpose was incapable of eliminating the binding.
Figure 3. EMSA of head (HE) or trunk (TE) nuclear extracts from embryos at different stages of development, using the deleted fragments described in Figure 2.
(A) Gel shift using 10.5 dpc head and trunk nuclear embryo extracts. (B) Gel shift using 11.5 dpc trunk nuclear embryo extract. (C) Gel shift using 13.5 dpc head and trunk nuclear embryo extracts. (A–C) The fragment used as a probe is indicated below each gel. Lanes indicated as ‘P’ represent migration of the probes alone. Competition reactions with 100 times unlabelled probe are indicated as ‘+C’. Competition reactions with 100 times unlabelled wild-type oligonucleotide for the SMAD consensus site are indicated by ‘+O’. Competition reactions with 100 times unlabelled mutated oligonucleotide for the SMAD consensus site are indicated as ‘+M’. The arrow shows the specific binding that occurs only with the 5′Δ fragment.
These results suggest that SMAD-binding site is functional and necessary for the formation of the transcriptional complex in the BP.
Purified SMAD 1 and 4 proteins bind specifically to the SMAD consensus
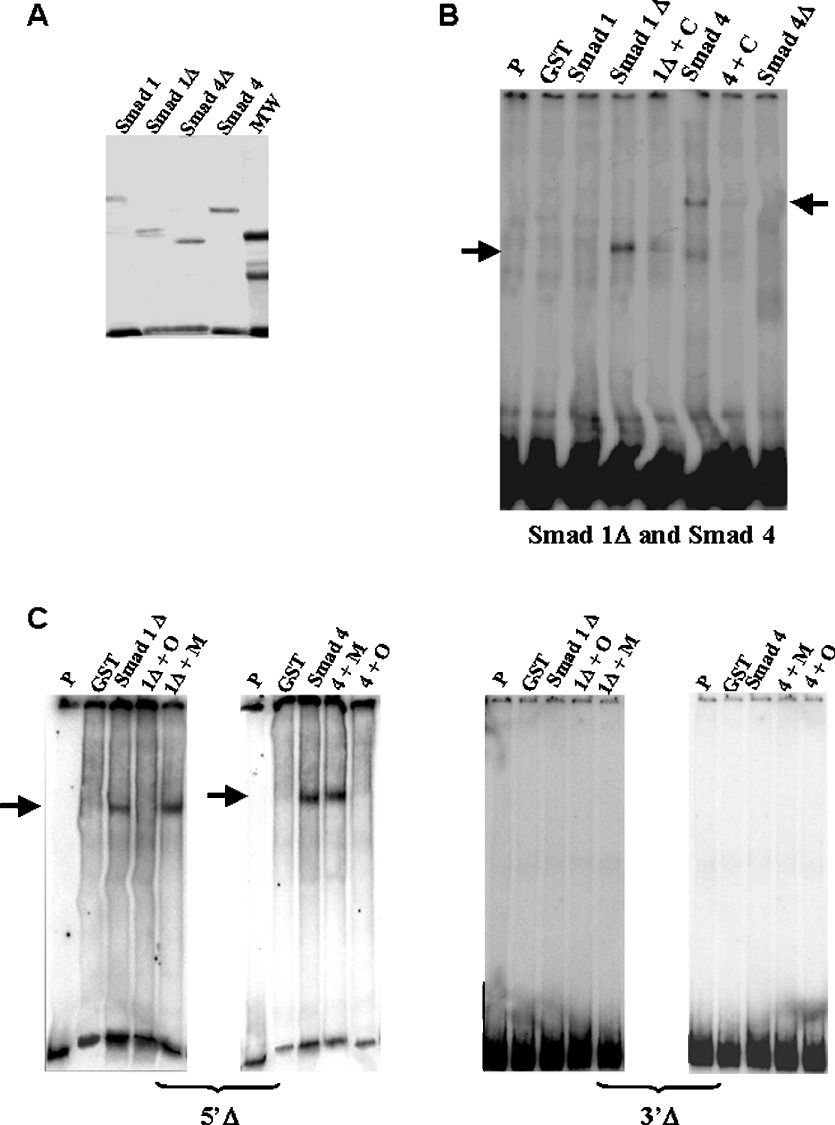
To verify if SMAD proteins bind directly to the SMAD consensus in BP fragment, we used purified GST–SMAD 1 fusion protein. Binding to GST–SMAD 4 was also tested as this protein associates with SMAD 1/SMAD 5/SMAD 8 and binds to DNA [27,28]. The C-terminal region of SMAD proteins consists of a transcription activation domain, called MH2, which has an inhibitory effect on the DNA binding activity of the N-terminal region (MH1) [27,29]. Therefore C-terminally truncated SMAD 1 (SMAD 1Δ) and SMAD 4 (SMAD4Δ) proteins were also produced and tested in our EMSAs (Figure 4A).
Figure 4. Purified SMAD 1 and SMAD 4 bind to BP and 5′Δ and 3′Δ fragments.
(A) Gel of purified recombinant proteins SMAD 1, SMAD 1Δ, SMAD 4 and SMAD 4Δ. MW, molecular mass standards. (B) Gel shift analysis of complexes between GST–SMAD fusion proteins and BP fragment. The protein used is indicated above each lane. Lanes indicated as ‘P’ and ‘GST’ show migration of the probe alone and reaction using GST peptide respectively. We observed binding reactions involving SMAD 1ΔMH2 (called SMAD 1Δ) and SMAD 4 (arrows). Competition reactions are indicated by the symbol ‘+C’ and correspond to addition of 100 times unlabelled BP. (C) Gel shift analysis of purified GST–SMAD 1ΔMH2 and SMAD 4 fusion protein complexes with the 5′Δ and 3′Δ fragments. The fragment used as a probe is indicated below each gel. Lanes indicated as ‘P’ and ‘GST’ show migration of the probe alone and reaction using GST peptide respectively. Competition reactions with 100 times unlabelled wild-type oligonucleotide for the SMAD consensus site are indicated as ‘+O’ and competition reactions with 100 times unlabelled mutated oligonucleotide for the SMAD consensus site are indicated as ‘+M’. The arrows show the specific binding.
As shown in Figure 4(B), SMAD 1Δ and SMAD 4 bind directly to the BP. Binding was observed only using full-length SMAD 4 and MH2-truncated SMAD 1 (SMAD 1Δ). The presence of two bands in the EMSA may be due to oligomerization, either between SMAD proteins or the GST peptide present in the fusion [27]. A two-band binding profile has been previously observed [27,30].
Purified SMAD 1Δ and SMAD 4 were also tested for the binding with 5′-deleted (5′Δ) and 3′-deleted (3′Δ) fragments (Figures 4C and 4D). Binding was observed only when the fragment used retained the SMAD consensus (5′Δ).
These results suggest that the SMAD consensus sequence located at the BP recognizes SMAD proteins such as SMAD 1Δ and SMAD 4.
SMAD proteins are present in 13.5 dpc embryo nuclear extracts and participate in the protein complex formed with a SMAD consensus oligonucleotide
In order to test for the presence of SMAD proteins in embryonic nuclear extracts, the 23 bp oligonucleotide containing only the SMAD consensus binding site flanked by sequences from the BP but devoid of any other motif was used in EMSAs with 13.5 dpc nuclear protein extract. As observed in Figure 5(A), the oligonucleotide was able to bind proteins in 13.5 dpc head and trunk extracts and the use of the oligo as a competitor abolished the binding.
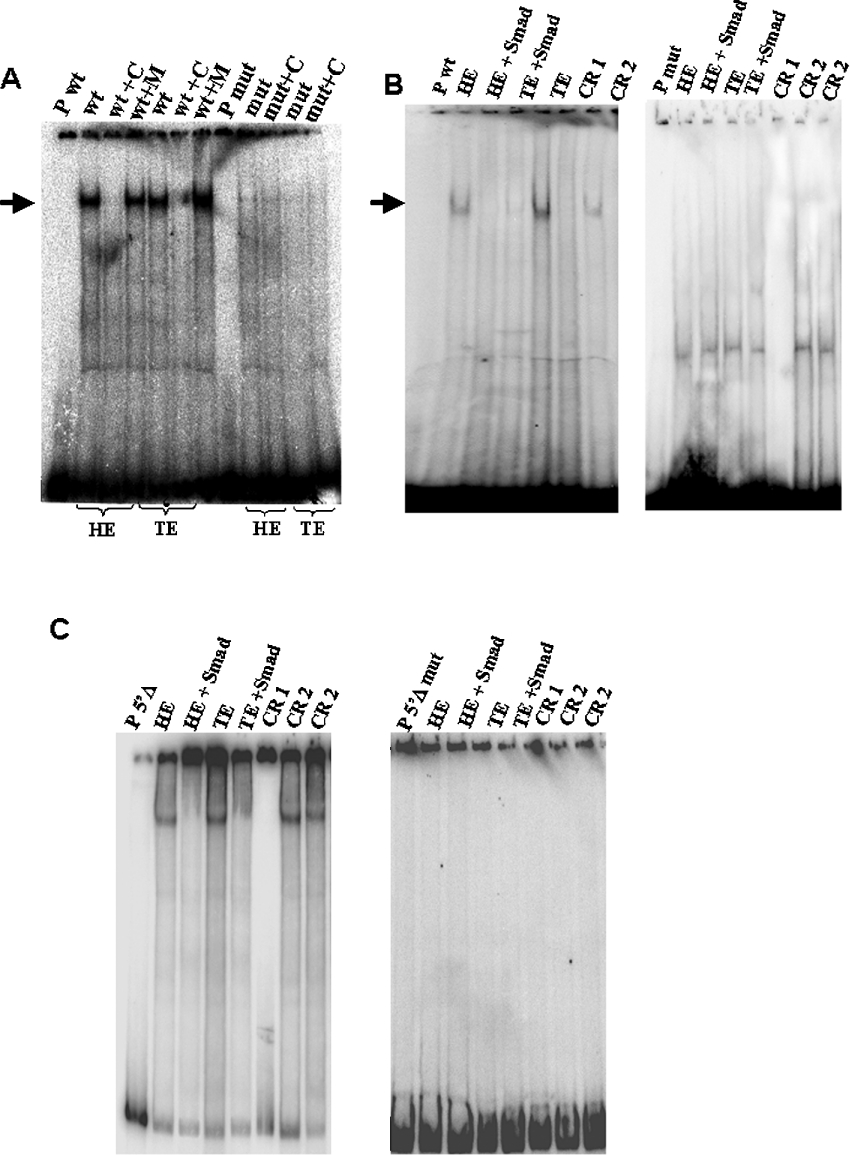
Figure 5. SMAD proteins are present in 13.5 dpc embryo nuclear extracts and participate in the protein complex formed with a SMAD consensus oligonucleotide.
(A) EMSA performed with head (HE) and trunk (TE) nuclear extracts from 13.5 dpc embryos and the wild-type (wt) or mutant (mut) oligonucleotides for the SMAD consensus binding site. Lanes indicated as ‘P wt’ and ‘P mut’ show migration of the probe alone. Competition reactions are indicated by the symbol ‘+C’ and correspond to the addition of 100 times unlabelled wild-type oligonucleotide. Competition reactions with 100 times unlabelled mutated oligonucleotide are indicated as ‘+M’. The arrows show specific binding confirming that mutations in the SMAD consensus site eliminate binding. (B) Supershift analysis of binding between head and trunk 13.5 dpc embryo nuclear extract and wild-type and mutated oligonucleotide. Lanes indicated as ‘P wt’ and ‘P mut’ show migration of the probe alone. Lanes indicated as HE and TE correspond to incubation of the probe with protein extract from head and trunk respectively. The addition of the SMAD 1/SMAD 5/SMAD 8 antibody in binding reactions with protein extract from head and trunk is analysed in lanes indicated as ‘head+SMAD’ and ‘trunk+SMAD’ respectively. Reaction indicated as CR 1 corresponds to a control reaction without nuclear proteins and CR 2 corresponds to a control reaction using an unspecific antibody (anti-P53). (C) Supershift analysis of binding between head and trunk 13.5 dpc embryo nuclear extract and 5′Δ fragment and 5′Δ mutated fragment. Lanes indicated as ‘P 5′Δ’ and ‘P 5′Δmut’ show migration of the probe alone. Lanes indicated as head and trunk correspond to incubation of the probe with protein extract from head and trunk respectively. The addition of the SMAD 1/SMAD 5/SMAD 8 antibody in binding reactions with protein extract from head and trunk is analysed in lanes indicated as ‘HE+SMAD’ and ‘TE+SMAD’ respectively. Reaction indicated as CR 1 correspond to a control reaction without nuclear proteins and CR 2 corresponds to a control reaction using an unspecific antibody (anti-P53).
To demonstrate further that this binding is due to the SMAD consensus, we performed EMSA analysis with the mutated version of the oligo and 13.5 dpc embryo nuclear protein extracts as a competitor for the binding to the wild-type oligonucleotide. As observed in Figure 5(A), the introduction of mutations in the SMAD consensus site is sufficient to abolish binding to the oligo, thus confirming the specificity of the binding.
To demonstrate that BMP-regulated SMADs are present in the complex formed between 13.5 dpc embryo proteins and the SMAD oligonucleotide or a mutated version of the oligo, we carried out supershift assays using human SMAD 1/SMAD 5/SMAD 8 antibody. We also performed supershift assays using human SMAD 1/SMAD 5/SMAD 8 antibody and 5′Δ fragment or 5′Δ mutated fragment. Addition of SMAD antibody to the reaction induced a supershift in the EMSA assay when the oligo or the 5′Δ fragment was used, showing that SMAD proteins are present in the complex formed with the SMAD consensus (Figures 5B and 5C).
BMP4 signalling is necessary to activate Msx1 transcription
To verify if the SMAD consensus binding site is necessary to the transcriptional activation of Msx1 gene, we performed transient transfection assays using P19 cells and constructs containing or not the SMAD consensus binding site. The BP fragment and the two deleting fragments (5′Δ and 3′Δ) were cloned into the pGL2-basic luciferase reporter vector (Promega) (BP, 5′Δ and 3′Δ respectively). These three constructions were transfected into P19 cells in the presence or absence of BMP4 factor in the medium and the luciferase activity was measured using the luciferase assay system (Promega) in a luminometer.
As we observed in Figure 6, both constructions containing the SMAD consensus site (constructions BP and 5′Δ) showed a significant induction of luciferase activity and in the presence of BMP4, the activity was increased. In contrast, the levels of luciferase when we transfected the construction containing the 3′Δ fragment (that do not contain the SMAD consensus site) were minimal and also did not respond to BMP4 induction.
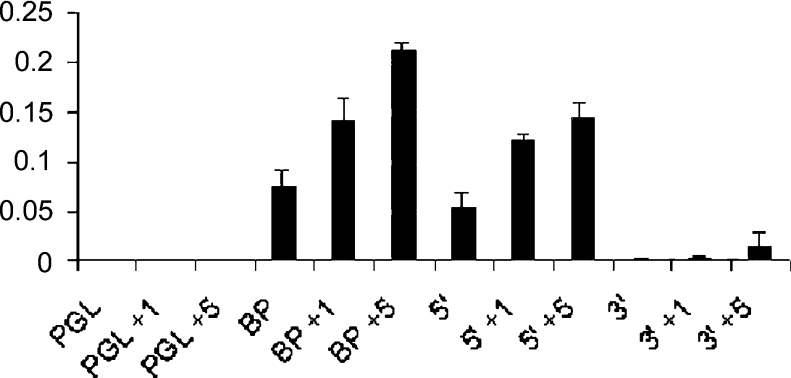
Figure 6. P19 cells were transiently transfected using the reporter plasmid PGL-basic2 (PGL – containing the luciferase gene) alone or using the constructions BP, 5′Δ and 3′Δ.
The cells were treated or not, 24 h after transfection, with 1 nM (PGL+1) or 5 nM (PGL+5) BMP4 for 5 h. Whole cell lysates were prepared and analysed for luciferase activity. The histogram shows induction of luciferase activity when we used both constructions containing the SMAD consensus binding site (BP and 5′Δ) and this activity is increased in the presence of 1 and 5 nM BMP4 (BP+1 nM, BP+5 nM, 5′Δ+1 nM and 5′Δ+5 nM). In contrast, the levels of luciferase using the construction without the SMAD consensus binding site (3′Δ) were minimal even in the presence of BMP4 (3′Δ+1 nM and 3′Δ+5 nM). PGL, PGL+1 nM and PGL+5 nM indicate the reporter plasmid alone, untreated with BMP and treated with 1 and 5 nM BMP4 respectively. These data are representative with P value <0.001.
These results suggest that SMAD consensus binding site is necessary to activate Msx1 transcription and that it is specifically activated by BMP4 signals.
SMAD 8 participates in the complex formed between BP and nuclear proteins
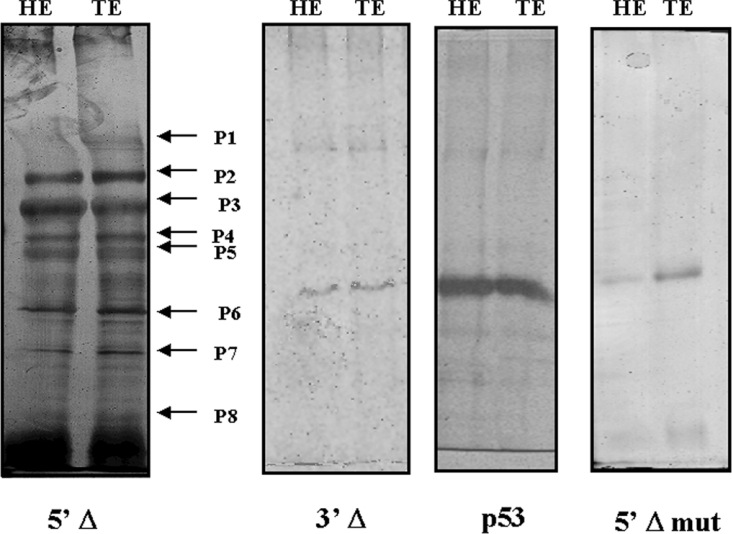
To identify proteins that participate in the complex formed with the SMAD consensus binding site, we carried out immunoprecipitation assays using anti-SMAD 1/SMAD 5/SMAD 8 antibody in binding reactions with 13.5 dpc head and trunk embryo nuclear proteins and the 5′Δ fragment. After immunoprecipitation, the protein complex was separated by gel electrophoresis (SDS/PAGE). The immunoprecipitated complex showed apparently the same composition when we used nuclear protein extracts from head or trunk (Figure 7). Immunoprecipitation assays was also done using 3′Δ fragment or 5′Δ mutated fragment, but a protein complex was not formed (Figure 7).
Figure 7. SDS/polyacrylamide gel of immunoprecipitation products.
Lanes named HE and TE correspond to reactions using 13.5 dpc embryo nuclear protein extracts from head and trunk respectively with anti-SMAD 1/SMAD 5/SMAD 8 antibody. The fragment used as a probe [5′-deleted fragment (5′Δ), 5′Δ mutated fragment (5′Δ mut) and 3′Δ fragment (3′Δ)] is indicated below each gel. The immunoprecipitated complex showed apparently the same composition using nuclear protein extracts from head or trunk. The arrows P1–P8 in the gel using 5′Δ fragment indicate the protein bands analysed by MS. The gel using 3′Δ fragment and 5′Δ mutated fragment did not show any immunoprecipitated protein. A control immunoprecipitation using an unspecific antibody (anti-P53) also did not show any protein.
Each identified protein band was excised from the gel and prepared for MS analysis by trypsin digestion. Eight different components of the complex were identified by MALDI–TOF analysis using Protein Prospector Search software 4.0.5 and they are listed in Table 1.
Table 1. Data bank (InterProScan, Motif Scan in a Protein Sequence and ExPASy) analysis of the proteins present in the complex formed over BP.
| Spot number | Accesion number | Name | Description | pI | Molecular mass (Da) |
|---|---|---|---|---|---|
| P1 | P49135 | XPB_MOUSE | ATP-dependent 3′–5′-DNA helicase, a component of the core-TFIIH basal transcription factor, involved in nucleotide excision repair of DNA and, when complexed to CAK (CDK activating kinase where CDK stands for cyclin-dependent kinase), in RNA transcription by RNA polymerase II. Acts by opening DNA either around the RNA transcription start site or the DNA damage. | 6.8 | 89127 |
| P2 | P97302 | BAC1_MOUSE | Transcriptional regulator that acts as repressor or activator. Binds in vitro to NF-E2 (nuclear transcription factor erythroid 2) binding sites. Plays important roles in co-ordinating transcription activation and repression by MAFK (musculoaponeurotic fibrosarcoma protein K). | 4.9 | 81375 |
| P3 | Q15582 | BGH3_HUMAN | BetaIG-H3. | 7.6 | 74682 |
| P4 | O89091 | TIEG-1_MOUSE | Transcriptional repressor involved in the regulation of cell growth. Inhibits cell growth. | 9.0 | 51756 |
| P5 | O88685 | PRSA_MOUSE | TBP-1. The 26 S protease is involved in the ATP-dependent degradation of ubiquitinated proteins. The regulatory (or ATPase) complex confers ATP dependence and substrate specificity to the 26 S complex (by similarity). | 5.1 | 49493 |
| P6 | Q9JIW5 | SMA9_MOUSE | Mother against decapentaplegic Smad 8. Transcriptional modulator activated by BMP type 1 receptor kinase. Is a receptor-regulated Smad (R-Smad). | 8.7 | 48314 |
| P7 | P70689 | CXB6_MOUSE | Connexin 30 | 8.7 | 30367 |
| P8 | P50540 | MXI1_MOUSE | Transcriptional repressor. MXI1 binds with MAX to form a sequence-specific DNA-binding protein complex that recognizes the core sequence 5′-CAC[GA]TG-3′. MXI1 thus antagonizes MYC transcriptional activity by competing for MAX. | 6.8 | 25978 |
Interestingly, one of the proteins found was SMAD 8. Other SMADs were not detected in the complex formed over the BP in this assay although their presence in the cells cannot be discarded.
In order to confirm the presence of SMAD 8 and other SMAD as SMAD 1 in 13.5 dpc nuclear extracts from trunk and head, we performed a Western-blot assay using SMAD 1 and 8 antibodies. Our results showed that both proteins are present in the extracts although in different concentrations (Figure 8).
Figure 8. Western-blot analysis using 13.5 dpc embryo nuclear protein extracts from head (HE) and trunk (TE) with anti-SMAD 1 or anti-SMAD 8 antibodies.
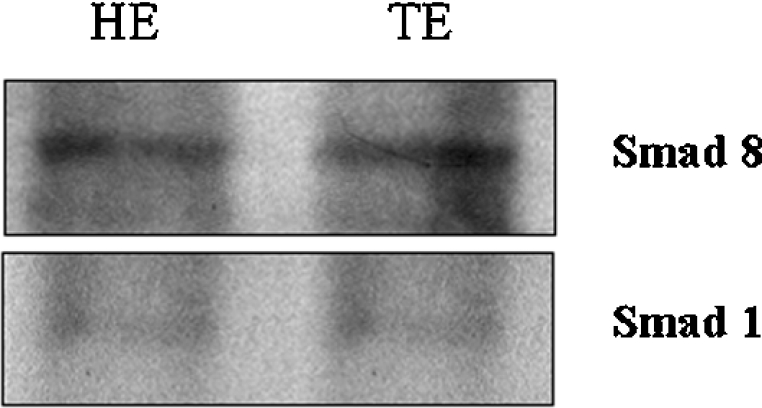
The extract and the antibody used are indicated in each gel. The bands showed the presence of each protein in the extracts but in different concentrations.
DISCUSSION
The mouse Msx1 gene is orthologous to the Drosophila msh gene and can be induced by BMP proteins. These proteins are members of the TGF-β superfamily of signalling factors and act by association to serine/threonine kinase receptors in the target cell, which then activate SMAD proteins. SMAD 1/SMAD 5/SMAD 8 are specifically activated by BMP2/BMP4 signals. The presence of three elements similar to the consensus established for Mad, the Drosophila melanogaster relative of SMAD 1/SMAD 5/SMAD 8, in the promoter of the Msx1 gene was described by our group [23]. These proteins recognize the motif 5′-GCCGnCG-3′ and we showed that, in mice, the same motif was capable of being recognized by SMAD proteins at least in a region characterized as PED (proximal enhancer domain) [19].
In the present study, to investigate further these binding sites, we tested the functionality of the SMAD consensus binding site located in the BP using EMSA analysis with nuclear protein extracts from 10.5–13.5 dpc embryos. Protein extracts were obtained using embryos of three distinct ages, encompassing the period of highest and most dynamic Msx1 expression: 10.5, 11.5 and 13.5 dpc. Trunk and head were treated separately as Msx1 has a highly distinct temporal expression pattern in these two regions. From 10.5 to 11.5 dpc, Msx1 is observed in lateral surface ectoderm, lateral mesoderm and the neuroepithelium that will form the dorsal part of the neural tube, including the region that gives rise to the migratory neural crest cells. At 13.5 dpc, Msx1 displays a more restricted expression pattern, being expressed in mandibular and maxillary regions, tooth buds, digits, mammary glands, internal optic cup and hair follicles [1,4]. Indeed, using alternative deletions from the BP in EMSA analysis, we could observe that only the fragment that contains the SMAD consensus binding site exhibits binding activity with nuclear protein extracts of different ages. Moreover, with the 11.5 dpc protein extract, only the trunk extract was capable of binding to this fragment. These results indicate that a DNA–protein complex can be formed only in the presence of the SMAD consensus.
The minimal promoter for this gene was described by Takahashi et al. [17] and shown to contain an E-box and a SP1 consensus site. More recently, Mehra-Chaudhary et al. [24] proposed that this minimal promoter consisted of SP1/TBP/CBP consensus binding sites. However, from our results, we can suggest that the presence of these consensus binding sites by itself is not sufficient to catalyse basal transcriptional machinery complex since promoter fragments containing only the SP1/TBP/CBP recognition sequences are not capable of attaching any nuclear protein. On the contrary, DNA fragments containing these sequences plus the SMAD consensus binding site were shown to be able to complex with nuclear proteins.
To test the capacity of this region to ligate SMAD proteins, we used purified SMAD 1 and 4 proteins in EMSAs. Specific binding was observed using SMAD 1Δ and SMAD 4 purified proteins, suggesting that the SMAD consensus site can recognize SMAD proteins. Moreover, mutational analysis of this site showed that when the consensus was mutated, binding of nuclear proteins was prevented, confirming the necessity of the SMAD consensus site to induce the formation of a protein complex over the BP. The presence of a SMAD protein in this complex was verified by supershift assay using anti-SMAD 1/SMAD 5/SMAD 8 antibody.
Moreover, in transient transfection assays we confirmed that the transcription of the Msx1 gene occurs only in the presence of the SMAD consensus binding site. Furthermore, this transcription was highly induced by the presence of BMP4.
Altogether, these results confirm that the SMAD consensus, homologous with the MAD motif, is also functional within the BP, as suggested by the high degree of evolutionary conservation for this consensus binding site in Msh/Msx genes.
We also identified some of the proteins that are present in the complex formed over the BP using an immunoprecipitation assay and a proteomic analysis. Eight different proteins were identified. One of them was TFIIH, necessary for the basal apparatus to initiate transcription. Other proteins identified were: BAC-1, an AP1-like protein; TIEG-1, an SP1-like protein; TBP-1; connexin 30, which, despite being a transmembrane protein, can have fragments occurring in the nucleus as has been already described for connexin 43 [31]; MXI1, a transcriptional repressor that antagonizes Myc transcriptional activity; BetaIG-H3, a TGF-β-induced IG-H3 protein precursor; and SMAD 8. Other proteins can also participate in this complex although not detected in our experiments. This can be the case of SMAD 4 that was not identified in our approach but is probably involved in the complex. A Western-blot analysis showed that SMAD 1, as well as SMAD 8, is present in 13.5 dpc nuclear extracts although SMAD 1 presents a lower concentration. This result can indicate that Sma8 is preferentially recruited to the protein complex formed over BP. Nevertheless, with this study, we can propose that SMAD 8 is the protein that transduces BMP4 signalling for the mouse Msx1 gene at least at the studied stages. Moreover, SMAD 8 attachment to the BP is also necessary to nucleate the formation of a protein complex that will give rise to the basal transcriptional machinery. In addition, the SP1/TBP/CBP consensus is probably recognized by proteins like BACH1, TIEG-1 and TBP1 and needs the attachment of other proteins like MXI1 at a very close position.
Acknowledgments
We thank Lilian Ayres Sá and Terezinha Maria Castro Silva for technical assistance, Dr Ulisses Gazos Lopes (Laboratório de Parasitologia Molecular, Instituto de Biofísica Carlos Chagas Filho, Universidade Federal do Rio de Janeiro) for a critical revision of this paper and Rio de Janeiro Proteomic Network for the MS facilities. This work was supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and FAPERJ (Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro).
References
- 1.Robert B., Sassoon D., Jacq B., Gehring W., Buckingham M. Hox-7, a mouse homeobox gene with a novel pattern of expression during embryogenesis. EMBO J. 1989;8:91–100. doi: 10.1002/j.1460-2075.1989.tb03352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill R. E., Jones P. F., Rees A. R., Sime C. M., Justice M. J., Copeland N. G., Jenkins A., Graham E., Davidson D. R. A new family of mouse homeobox-containing genes:molecular structure, chromosomal location, and developmental expression of Hox-7-1. Genes Dev. 1989;3:26–37. doi: 10.1101/gad.3.1.26. [DOI] [PubMed] [Google Scholar]
- 3.Satokata I., Maas R. Msx-1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 4.Houzelstein D., Cohen A., Buckingham M. E., Robert B. Insertional mutation of the mouse Msx-1 homeobox gene by an nLacZ reporter gene. Mech. Dev. 1997;65:123–133. doi: 10.1016/s0925-4773(97)00065-8. [DOI] [PubMed] [Google Scholar]
- 5.Davidson D. R., Hill R. E. Msh-like genes: a family of homeobox genes with wide ranging expression during vertebrate development. Semin. Dev. Biol. 1991;2:405–412. [Google Scholar]
- 6.Davidson D. The functional and evolution of Msx genes: pointers and paradoxes. Trends Genet. 1995;11:405–411. doi: 10.1016/s0168-9525(00)89124-6. [DOI] [PubMed] [Google Scholar]
- 7.MacKenzie A., Ferguson M. W. J., Sharpe P. T. Expression patterns of the homeobox gene, Hox-8, in the mouse embryo suggest a role in specifying tooth initiation and shape. Development. 1992;115:403–420. doi: 10.1242/dev.115.2.403. [DOI] [PubMed] [Google Scholar]
- 8.Catron K. M., Wang H., Hu G., Shen M. M., Abate-Shen C. Comparison of Msx-1 and Msx-2 suggests a molecular basis for functional redundancy. Mech. Dev. 1996;55:185–199. doi: 10.1016/0925-4773(96)00503-5. [DOI] [PubMed] [Google Scholar]
- 9.Monaghan A. P., Davidson D. R., Sime C., Graham E., Baldock R., Bhattacharya S. S., Hill R. E. The Msh-like homeobox genes define domains in the developing vertebrate eye. Development. 1991;112:1053–1061. doi: 10.1242/dev.112.4.1053. [DOI] [PubMed] [Google Scholar]
- 10.Song K., Wang Y., Sassoon D. Expression of Hox-7 in myoblasts results in inhibition of differentiation and induces cell transformation. Nature (London) 1992;360:477–481. doi: 10.1038/360477a0. [DOI] [PubMed] [Google Scholar]
- 11.Woloshin P., Song K., Degnin C., Killary A. M., Goldhamer D. J., Sassoon D., Thayer M. J. Msx-1 inhibits MyoD expression in fibroblast 10T1/2 cell hybrids. Cell (Cambridge, Mass.) 1995;82:611–620. doi: 10.1016/0092-8674(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 12.Thompson-Jaegar S., Raghow R. Exogenous expression of Msx-1 renders myoblasts refractory to differentiation into myotubes and elicits enhancer biosynthesis of four unique mRNAs. Mol. Cell. Biochem. 2000;208:63–69. doi: 10.1023/a:1007069317131. [DOI] [PubMed] [Google Scholar]
- 13.Catron K. M., Zhang H., Marshall S. C., Inostroza J. A., Wilson J. M., Abate C. Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. Mol. Cell. Biol. 1995;15:861–871. doi: 10.1128/mcb.15.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H., Catron K. M., Abate-Shen C. A role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TATA binding protein interaction and transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1764–1769. doi: 10.1073/pnas.93.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kusuoka M., Takahashi T., Gurom C., Raghow R. Murine homeobox-containing gene, Msx-1 analysis of genomic organization, promoter structure and potential autoregulatory cis-acting elements. Genomics. 1994;21:85–91. doi: 10.1006/geno.1994.1228. [DOI] [PubMed] [Google Scholar]
- 16.Shetty S., Takahashi T., Matsui H., Ayengar R., Raghow R. Transcriptional autorepression of Msx-1 gene is mediated by interactions of Msx-1 protein with a multi-protein transcriptional complex containing TATA-binding protein, SP1 and cAMP-response-element-binding protein-binding protein (CBP/p300) Biochem. J. 1999;339:751–758. [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi T., Guron C., Shetty S., Matsui H., Raghow R. A minimal murine Msx-1 gene promoter. J. Biol. Chem. 1997;272:22667–22678. doi: 10.1074/jbc.272.36.22667. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez S. M., Ferland L. H., Robert B., Abdelhay E. Structural and functional analysis of mouse Msx-1 gene promoter: sequence conservation with human MSX1 promoter points at potential regulatory elements. DNA Cell Biol. 1998;17:561–572. doi: 10.1089/dna.1998.17.561. [DOI] [PubMed] [Google Scholar]
- 19.MacKenzie A., Purdie L., Davidson D., Collinson M., Hill R. Two enhancers domains control early aspects of the complex expression pattern of Msx-1. Mech. Dev. 1997;62:29–40. doi: 10.1016/s0925-4773(96)00646-6. [DOI] [PubMed] [Google Scholar]
- 20.Pereira M., Houzelstein D., Vinci G., Cohen A., Abdelhay E., Robert B. Analysis of the mouse Msx-1 gene promoter using embryonic stem cell-mediated transgenesis. Transgenics. 1999;2:403–416. [Google Scholar]
- 21.Jones C. M., Lyons K. M., Hogan B. L. Involvement of bone morphogenetic protein-4 (BMP-4) and Vgr-1 in morphogenesis and neurogenesis in the mouse. Development. 1991;111:531–542. doi: 10.1242/dev.111.2.531. [DOI] [PubMed] [Google Scholar]
- 22.Lyons K. M., Hogan B. L., Robertson E. J. Colocalization of BMP 7 and BMP 2 RNAs suggests that these factors cooperatively mediate tissue interactions during murine development. Mech. Dev. 1995;50:71–83. doi: 10.1016/0925-4773(94)00326-i. [DOI] [PubMed] [Google Scholar]
- 23.Martinez C. E. A., Binato R., Gonzalez S., Pereira M., Benoit R., Abdelhay E. Characterization of Smad motif similar to Drosophila Mad in mouse Msx-1 promoter. Biochem. Biophys. Res. Commun. 2002;291:655–662. doi: 10.1006/bbrc.2002.6502. [DOI] [PubMed] [Google Scholar]
- 24.Mehra-Chaudhary R., Matsui H., Ragow R. Msx-3 protein recruits histone deacetylase to down-regulate the msx-1 promoter. Biochem. J. 2001;353:13–22. [PMC free article] [PubMed] [Google Scholar]
- 25.Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1488. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 27.Dennler S., Itoh S., Vivien D., Dijke P., Huet S., Gauthier J.-M. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishida W., Hamamoto T., Kusanagi K., Yagi K., Kawabata M., Takehara K., Sampath T., Kato M., Miyazono K. Smad 6 is a Smad1/5-induced Smad inhibitor. J. Biol. Chem. 2000;275:6075–6079. doi: 10.1074/jbc.275.9.6075. [DOI] [PubMed] [Google Scholar]
- 29.Kim J., Johnson K., Chen H.-J., Carroll S., Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature (London) 1997;388:304–307. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- 30.Kusanagi K., Inoue H., Ishidou Y., Mishima H. K., Kawabata M., Miyazono K. Characterization of a bone morphogenetic protein-responsive smad-binding element. Mol. Biol. Cell. 2000;11:555–565. doi: 10.1091/mbc.11.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dang X., Doble B. W., Kardami E. The carboxy-tail of connexin-43 localizes to the nucleus and inhibits cell growth. Mol. Cell. Biochem. 2003;242:35–38. [PubMed] [Google Scholar]