Abstract
The glyoxalase system is a ubiquitous detoxification pathway that protects against cellular damage caused by highly reactive oxoaldehydes such as methylglyoxal which is mainly formed as a by-product of glycolysis. The gene encoding GLOII (glyoxalase II) has been cloned from Leishmania donovani, a protozoan parasite that causes visceral leishmaniasis. DNA sequence analysis revealed an ORF (open reading frame) of ∼888 bp that encodes a putative 295-amino-acid protein with a calculated molecular mass of 32.5 kDa and a predicted pI of 6.0. The sequence identity between human GLOII and LdGLOII (L. donovani GLOII) is only 35%. The ORF is a single-copy gene on a 0.6-Mb chromosome. A ∼38 kDa protein was obtained by heterologous expression of LdGLOII in Escherichia coli, and homogeneous enzyme was obtained after affinity purification. Recombinant L. donovani GLOII showed a marked substrate specificity for trypanothione hemithioacetal over glutathione hemithioacetal. Antiserum against recombinant LdGLOII protein could detect a band of anticipated size ∼32 kDa in promastigote extracts. By overexpressing the GLOII gene in Leishmania donovani using Leishmania expression vector pspαhygroα, we detected elevated expression of GLOII RNA and protein. Overexpression of the GLOII gene will facilitate studies of gene function and its relevance as a chemotherapeutic target. This is the first report on the molecular characterization of glyoxalase II from Leishmania spp. The difference in the substrate specificity of the human and Leishmania donovani glyoxalase II enzyme could be exploited for structure-based drug design of selective inhibitors against the parasite.
Keywords: drug design, glutathione, glyoxalase II, Leishmania donovani, parasite inhibitor, trypanothione
Abbreviations: GLOI, glyoxalase I; GLOII, glyoxalase II; LdGLOII, Leishmania donovani GLOII; Ni-NTA, Ni2+-nitrilotriacetate; ORF, open reading frame; PFGE, pulse-field gradient gel electrophoresis
INTRODUCTION
The glyoxalase system catalyses the conversion of 2-oxoaldehydes into the corresponding 2-hydroxy acids [1–4]. The process involves two consecutive reactions mediated by two enzymes, GLOI (glyoxalase I) (lactoylglutathione lyase, EC 4.4.1.5) and GLOII (glyoxalase II) (hydroxyacylglutathione hydrolase, EC 3.1.2.6). GLOI catalyses the formation of S-D-lactoylglutathione from the hemithioacetal formed non-enzymatically from methylglyoxal and glutathione. GLOII converts S-D-lactoylglutathione into lactate and free glutathione [1–4]. Thus glutathione acts as a cofactor in the overall reaction pathway. The glyoxalase system is present in the cytoplasm of cells and cellular organelles, particularly mitochondria. The two distinct enzymes have been purified from a wide variety of species, including prokaryotes, plants and mammals [5–11]. The widespread distribution suggests that the system fulfils a function of fundamental importance to biological life, and glyoxalase has a distinct role in cell proliferation and maturation [6]. Glyoxalases seem to be ubiquitous, in accord with their proposed role of detoxifying methylglyoxal, which is a physiological substrate, protecting against cellular damage. Methylglyoxal reacts rapidly with both proteins and nucleic acids and thus is both toxic and mutagenic [4,12,13]. Methylglyoxal is produced mainly as a by-product of glycolysis. It is formed mainly by the degradation of triose phosphates, and also by metabolism of ketone bodies, threonine degradation and the fragmentation of glycated proteins [14,15]. GLOI is part of the glyoxalase system present in the cytosol and it prevents the accumulation of these reactive α-oxoaldehydes and thereby suppresses α-oxoaldehyde-mediated glycation reactions [16]. It is a key enzyme of the anti-glycation defence. Genes encoding GLOI and GLOII have been characterized from several species [8–10,17–20].
Leishmania donovani, a flagellated protozoan parasite, is the causative agent of visceral leishmaniasis. Quinquivalent antimonials are the standard first-line treatment for leishmaniasis [21,22], although resistance is a growing problem. The aromatic diamidine pentamidine represents a second line of treatment [23]. Leishmanial patients' refractoriness to existing drugs and the availability of a limited repertoire of drugs has become a rapidly growing problem. Hence, there is an urgent need for the development of new drugs against leishmaniasis. The GLOI activity has been reported in Leishmania braziliensis [24], but very low levels of GLOI and GLOII activity were detected in lysates using glutathione as the substrate [25]. The glyoxalase system of the pathogenic kinetoplastids has been reported recently to be unique, as a consequence of these protozoa possessing an unusual thiol metabolism [25,26]. In these organisms, instead of glutathione, the major low-molecular-mass thiol is trypanothione [N1,N8-bis(glutathionyl)spermidine)] [25]. It has been recently reported that the GLOI system in Leishmania major uses trypanothione as the substitute for glutathione [25]. In Trypanosoma brucei, GLOII strongly prefers thioesters of trypanothione instead of glutathione as substrate [27].
The above findings prompted us to characterize GLOII from Leishmania donovani. In the present paper, we report molecular cloning, expression and characterization of GLOII from Leishmania donovani, the parasite protozoan that is responsible for visceral leishmaniasis. This is the first report on the characterization of GLOII from Leishmania species. The difference in the substrate specificity of the human and Leishmania donovani GLOII suggests that the latter may be a target for antimicrobial therapy.
EXPERIMENTAL
Materials
Trypanothione disulphide was obtained from Bachem. Restriction enzymes and Pfu TaqDNA polymerase were obtained from MBI Fermentas. All other chemicals were of analytical grade and were available commercially.
Parasite and culture conditions
L. donovani AG83 (MHOM/IN/1983/AG83) promastigotes were cultured at 22 °C in modified M199 medium (Sigma) supplemented with 100 units/ml penicillin (Sigma), 100 μg/ml streptomycin (Sigma) and 10% heat inactivated foetal bovine serum (Gibco/BRL, Life Technologies).
Cloning of GLOII gene from L. donovani
An ∼888-bp DNA fragment was amplified from genomic DNA, using a sense primer with a flanking BamHI site (underlined), 5′-CGCGGATCCATGCGCAACTACTGCACA-3′, that coded for the amino acid sequence MRNYCT at positions 1–18, and the antisense primer with a flanking HindIII site (underlined), 5′CCCAAGCTTTCAGTCGCAGGCGTTGT 3′, which corresponded to amino acid residues YNACD including the stop codon, at positions 872–888. PCR was performed in 50 μl reaction volumes containing 100 ng of genomic DNA, 25 pmol each of the gene-specific forward and reverse primers, 200 μM of each dNTP, 2 mM MgCl2 and 5 units of Pfu Taq DNA polymerase. The conditions for PCR were as follows: 94 °C for 10 min, then 35 cycles of 94 °C for 1 min, 57 °C for 45 s and 72 °C for 45 s. Final extension was carried out for 10 min at 72 °C. A single ∼888-bp PCR product was obtained and subcloned into pGEM-T vector (Promega) and subjected to automated sequencing. Sequence analysis was performed by DNAstar, whereas comparisons with other sequences of the database were performed using the search algorithm BLAST [28]. Multiple alignment of amino acid sequences was performed using the ClustalW (http://www.ebi.ac.uk/clustalw/) program. The phylogenetic tree was constructed using PHYLIP style treefile produced by ClustalW. The amplified DNA fragment of ∼888 bp (LdGLOII) was also cloned into the BamHI/HindIII site of pET30a vector (Novagen). The recombinant construct was transformed into the BL21 (DE3) strain of Escherichia coli.
Expression and purification procedure
Expression from the construct pET30a-LdGLOII was induced at a D600 of 0.6 with 0.5 mM IPTG (isopropyl β-D-thiogalactoside) (Sigma) at 37 °C for different time periods. Bacteria were then harvested by centrifugation at 5000 g for 15 min, and the cell pellet was resuspended in binding buffer (50 mM sodium phosphate buffer, pH 7.5, 10 mM imidazole, 300 mM sodium chloride, 2 mM PMSF and 30 μl of protease inhibitor cocktail). Lysozyme (100 μg/ml) was added to the cell suspension, which was kept on a rocking platform for 30 min at 4 °C. The resulting cell suspension was sonicated six times for 20 s with 1 min intervals. The lysate was centrifuged at 20000 g for 30 min at 4 °C. The resulting supernatant, which contained the protein, was loaded on to pre-equilibrated Ni-NTA (Ni2+-nitrilotriacetate)–agarose beads (Qiagen). The mixture was kept on a rocking platform for 2 h at 4 °C. It was centrifuged at 400 g for 30 min at 4 °C. The supernatant was discarded and pellet was washed three times with wash buffer (50 mM sodium phosphate buffer, pH 7.5, 50 mM imidazole, 300 mM NaCl, 2 mM PMSF and 30 μl of protease inhibitor cocktail). The protein was eluted with increasing concentrations of imidazole, pH 7.0. The imidazole was removed by dialysis in 20 mM sodium phosphate buffer, pH 7.5. The purified protein was ≥95% pure as judged by SDS/PAGE. The purified protein was divided into 200 μl aliquots and stored at −80 °C.
Cross-linkage of subunits
The recombinant GLOII protein was cross-linked with 0.025% (w/v) glutaraldehyde in PBS (pH 7.0) [29]. The reaction mixture was incubated for 20 min at 37 °C and analysed by SDS/PAGE using a 10% gel with known standards. The protein samples were mixed with an equal volume of loading buffer containing 100 mM Tris/HCl (pH 6.8), 0.4% SDS, 20% (v/v) glycerol and 0.001% (w/v) Bromophenol Blue and subjected to boiling in a water bath for 5 min.
Nucleic acid isolation, PFGE (pulse-field gradient gel electrophoresis) and hybridization analysis
Genomic DNA was isolated from ∼2×109 L. donovani AG83 promastigotes at late exponential phase by standard procedures [30], digested with different restriction endonucleases and subjected to electrophoresis in 0.8% (w/v) agarose gels. The fragments were transferred on to nylon membrane (Amersham Biosciences) and subjected to Southern blot analysis.
Total RNA was isolated from 2×108 L. donovani wild-type promastigotes and from GLOII-overexpressing strain using TRI reagent™ (Sigma). For Northern blot analysis, 15 μg of total RNA was fractionated by denaturing agarose gel electrophoresis and transferred on to nylon membrane following standard procedures.
Leishmania chromosomes were separated by PFGE in which low-melting agarose blocks containing embedded cells (108 exponential phase promastigotes/ml) were electrophoresed in a contour-clamped homogeneous electric field apparatus (CHEF DRIII, Bio-Rad). Saccharomyces cerevisiae chromosomes were used as size markers. PFGE running conditions were as follows: 60 s initial switch time; 120 s final switch time; 24 h run time; 6 V/cm current; 120° including angle. Following the transfer of DNA, RNA and chromosomes on to nylon membranes, the nucleic acids were UV-cross-linked to the membrane in a Stratagene UV cross-linker. Prehybridization was carried out at 65 °C for 4 h in a buffer containing 0.5 M sodium phosphate, 7% (w/v) SDS, 1 mM EDTA, pH 8.0, and 100 μg/ml sheared denatured salmon sperm DNA. The blots were hybridized with denatured [α-32P]dCTP-labelled DNA probe (PCR probe described for the L. donovani GLOII-coding region) at 106 c.p.m./ml, which was labelled by random priming (NEB Blot® Kit, New England Biolabs). Membranes were washed, air-dried and exposed to an imaging plate. The image was developed by PhosphoImager (Fuji FLA-5000) using ImageQuant software (Amersham Biosciences).
GLOII assay
The activity of recombinant purified GLOII was determined at room temperature (25 °C) by measuring the hydrolysis of the thioester in a coupled assay. The assays were performed in a total volume of 0.5 ml of 100 mM Mops buffer (pH 7.2), various concentrations of methylglyoxal (0–600 μM) (Sigma) and 300 μM reduced trypanothione [25,27]. Trypanothione disulphide (1 mM) (Bachem) was reduced with 3 mM dithiothreitol at 60 °C for 20 min before the assay. The resulting reduced trypanothione was used for the assay. The assay mixture was incubated for 10 min followed by the addition of purified recombinant L. donovani GLOI protein. Lactoyltrypanothione was produced by adding 0.3 nmol of recombinant L. donovani GLOI, and the reaction was monitored at 240 nm. At the end of the reaction, 0.3 nmol of recombinant LdGLOII was added, and the rate of decrease in absorbance at 240 nm was measured. Glutathione-dependent GLOII was also measured under similar conditions. The value for Δϵ240 was taken as 6.5 mM−1·cm−1 for S-D-lactoyltrypanothione [27]. The experiment was repeated twice with similar results.
Protein determination
Protein concentration was determined using the method of Bradford [31] using BSA as standard.
Antibody production
The purified recombinant GLOII protein (20 μg) was subcutaneously injected in mice using Freund's complete adjuvant, followed by two booster doses of recombinant GLOII protein (15 μg) with incomplete adjuvant at 2-week intervals to produce the polyclonal antibody against the recombinant GLOII protein. The mice were bled 2 weeks after the second boost, and sera were collected and used for Western blot analysis.
Transfection and overexpression of the GLOII gene in L. donovani
The GLOII ORF (open reading frame) was amplified by PCR using a sense primer with a flanking XbaI site (underlined), 5′-TGCTCTAGAATGCGCAACTACTGCACA-3′, that coded for the amino acid sequence MRNYCT at positions 1–18, and the antisense primer with a flanking HindIII site (underlined), 5′CCCAAGCTT TCAGTCGCAGGCGTTGT 3′, which corresponded to amino acid residues YNACD including the stop codon, at positions 872–888. The amplified DNA fragment (GLOII) was cloned into the XbaI/HindIII site of pspαhygroα Leishmania shuttle vector (kindly provided by Dr Marc Ouellette, Université Laval, Québec, Canada) to create pspαhygroα-LdGLOII containing the hygromycin phosphotransferase gene. The construct (20 μg) was transfected into L. donovani promastigotes by electroporation [2 mm gap cuvettes, 450 V, 500 μF (BTX Electro Cell Manipulator 600)]. Transfectants were selected for resistance with hygromycin (40 μg/ml) as described previously [32].
Western blot analysis
Promastigotes were lysed by sonication on ice four times with a 10 s pulse at 1 min intervals, and cell supernatants were prepared by centrifugation at 20000 g protein (50 μg) from promastigotes was fractionated by SDS/PAGE, and blotted on to nitrocellulose membrane using electrophoretic transfer cell (Bio-Rad). Western Blot analysis was carried out using the ECL® (enhanced chemiluminescence) kit (Amersham Biosciences) according to the manufacturer's protocol. Polyclonal antibody (1:100 dilution) against purified recombinant L. donovani GLOII generated in mice was used for the Western blot analysis. The mouse monoclonal antibody anti-β-tubulin (human) (Santa Cruz) was used as a control. The results shown are from a single experiment typical of at least three giving similar results.
RESULTS
Sequence analysis and genomic organization
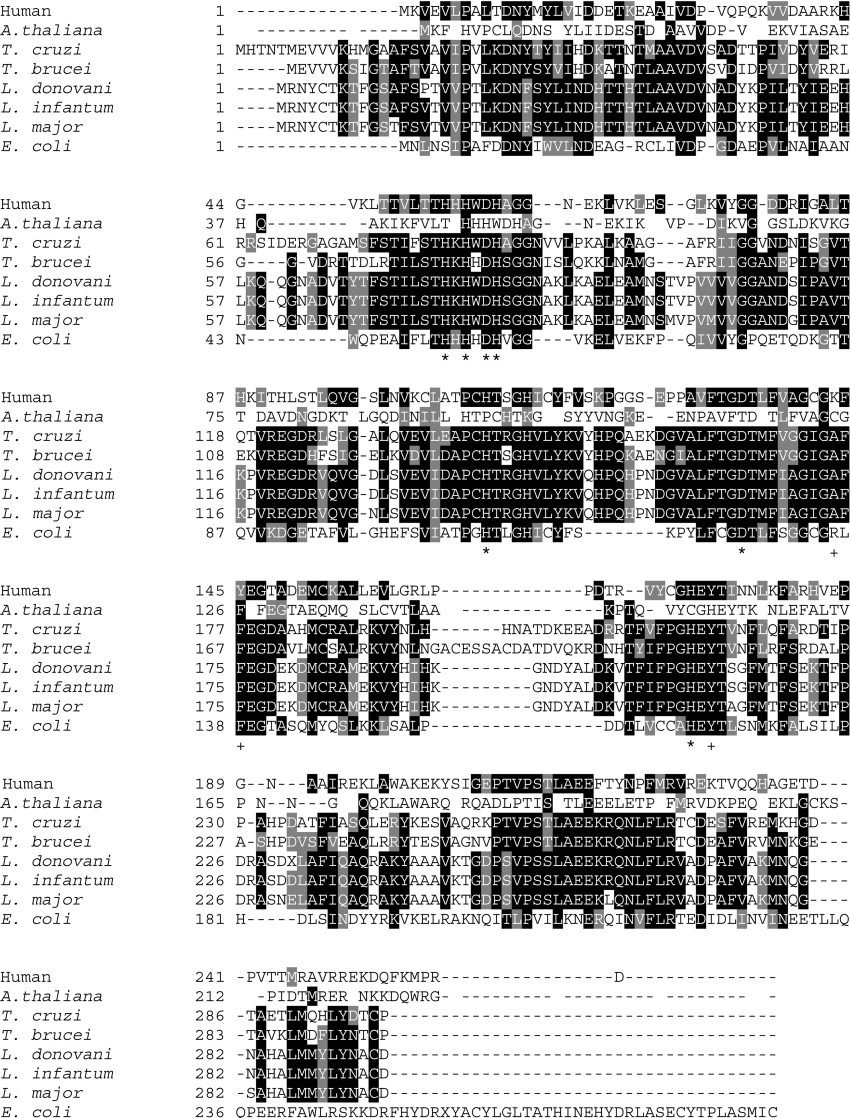
In order to clone the gene encoding GLOII, PCR was performed using specific oligonucleotides whose sequence was based on the Leishmania Genome Sequencing Project of Leishmania infantum (http://www.ebi.ac.uk/parasites/LGN/). Genomic DNA from L. donovani AG83 (MHOM/IN/1983/AG83) promastigotes was used as a template. A single ∼888-bp PCR product was obtained, cloned, and sequenced. A single ORF consisting of ∼888-bp was isolated (LdGLOII gene, GenBank® accession number AY851655) showing 96% identity with L. major trypanothione-dependent GLOII sequence (LmjF12.0220 |||hydroxyacylglutathione hydrolase, putative|Leishmania major|chr 12), 99% identity with L. infantum (LinJ12.0200), 53% identity with Trypanosoma cruzi putative lactoylglutathione lyase-like protein (NCBI accession number AAL96759), 51% identity with T. brucei brucei (NCBI accession number CAD37800), 32% identity with Arabidopsis thaliana (NCBI accession number O24496) and 35% identity with human GLOII (NCBI accession number Q16775). E. coli probable hydroxyacylglutathione hydrolase (NCBI accession number Q47677) shows 28% identical residues (Figure 1).
Figure 1. Multiple sequence alignment of GLOII sequences from L. donovani (AAW52503), L. major (LmjF12.0220; putative), L. infantum (LinJ12.0200), T. cruzi (AAL96759; putative), T. brucei (CAD37800), E. coli (Q47677), human (Q16775) and A. thaliana (O24496) using the ClustalW program.
The amino acids are numbered to the left of the respective sequences. Residues that are identical with or similar to other glyoxalases are indicated in black showing complete identity and grey when they are conserved in at least three sequences. Asterisks (*) indicate amino acids that are responsible for metal binding and+indicates residues that bind to glutathione of human GLOII [34].
The ORF encoded a putative polypeptide of 295 amino acids, with a predicted molecular mass of ∼32.5 kDa, which is very similar to that of L. major (295 amino acids), L. infantum (295 amino acids) and T. cruzi (putative lactoylglutathione lyaselike protein) (299 amino acids) enzymes, but slightly larger than the human (260 amino acids) enzyme (Figure 1). The predicted pI of LdGLOII was determined to be pH 6.0 which is comparable with those of proteins from L. major, L. infantum and T. cruzi. There was only 33% identity between human GLOII and Leishmania donovani GLOII (NCBI accession number AAW52503) sequences (Figure 1). Metal-binding motif THXHXDH, common to all GLOIIs consisting of five histidine and two aspartate residues that link directly to two metal ions [33], is highly conserved in LdGLOII sequence (Figure 1).
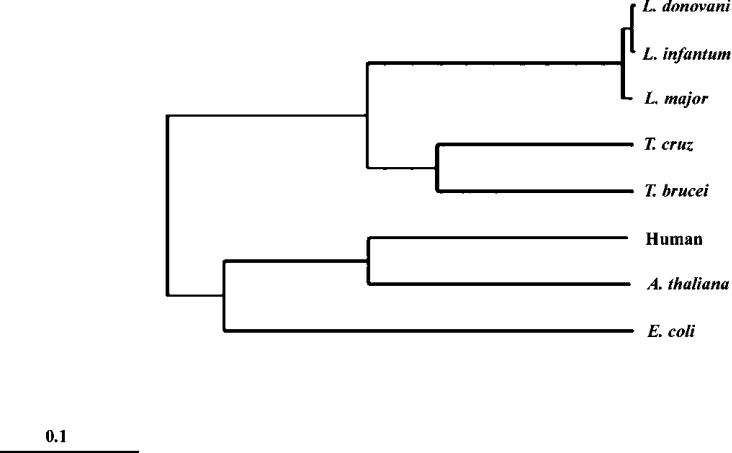
A phylogenetic tree has been constructed (Figure 2) using the LdGLOII sequence and other representative GLOII sequences. The tree indicates the close evolutionary relationship of L. donovani and T. cruzi and T. brucei among the kinetoplastid protozoa. The kinetoplastid GLOII sequences have no similarity with the human enzyme in phylogenetic analysis.
Figure 2. Phylogenetic tree using the amino acid sequences of GLOII from L. donovani and other organisms.
The TreeView program of the ClustalW software was used to create the phylogenetic trees from the multiple alignments.
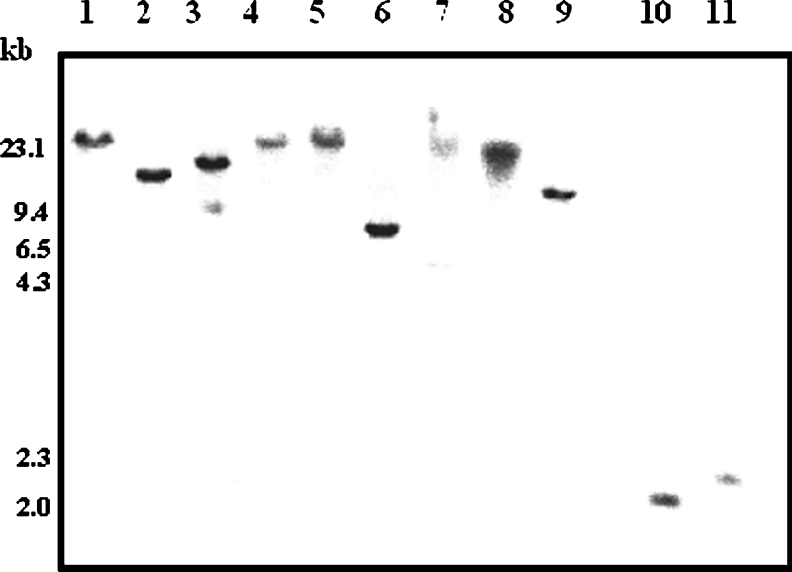
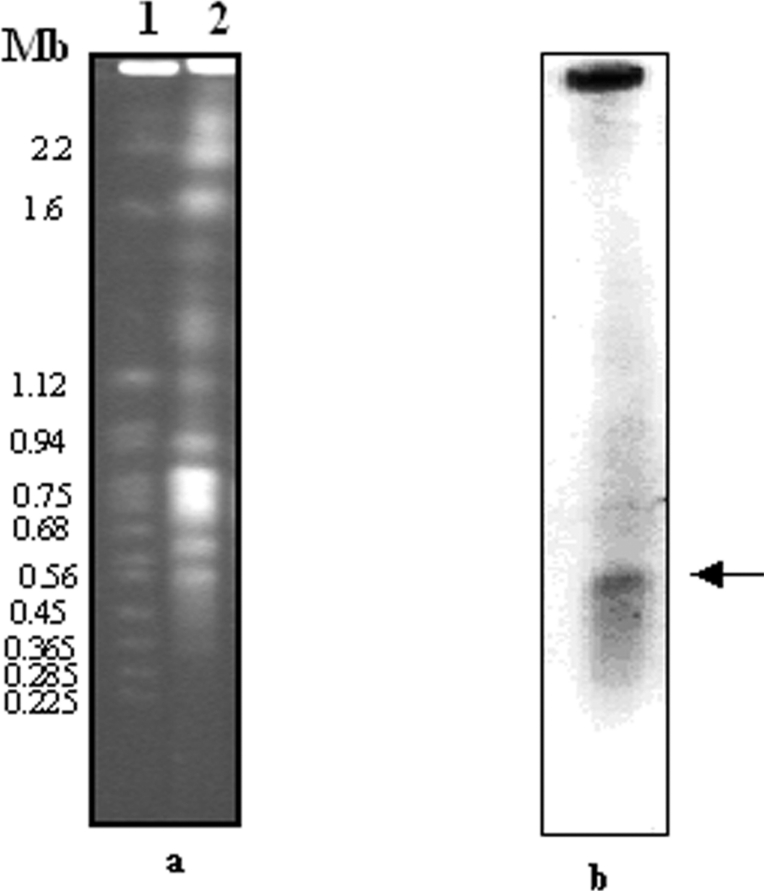
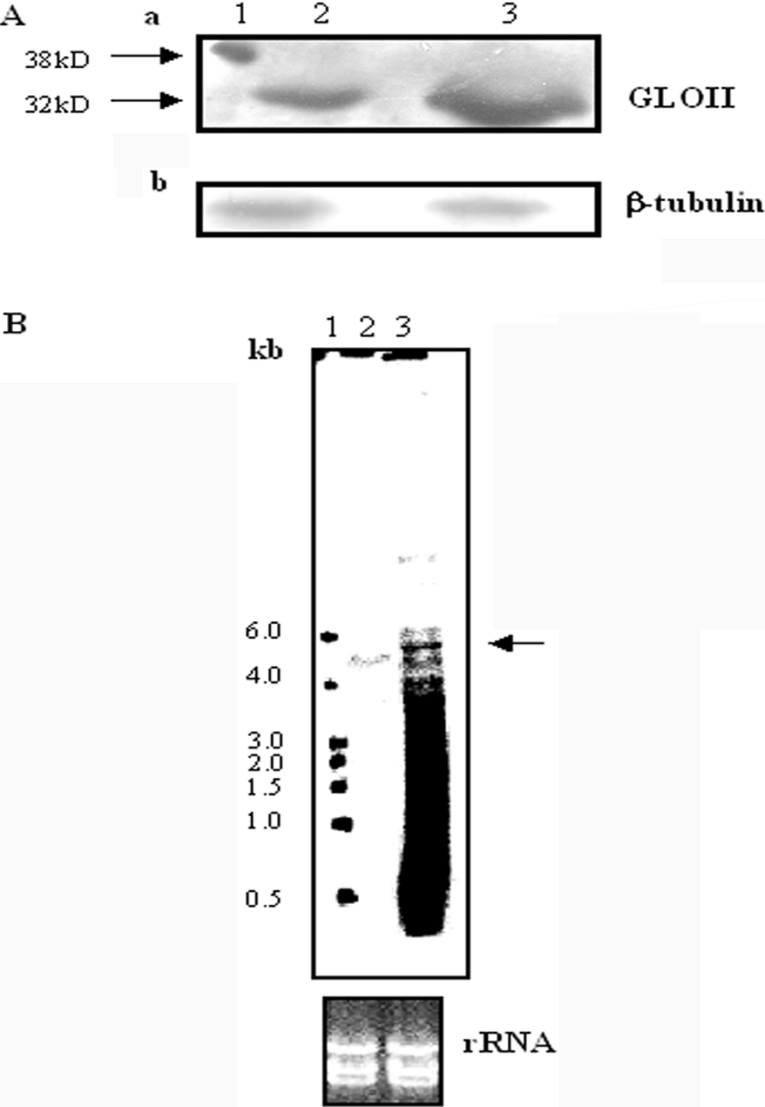
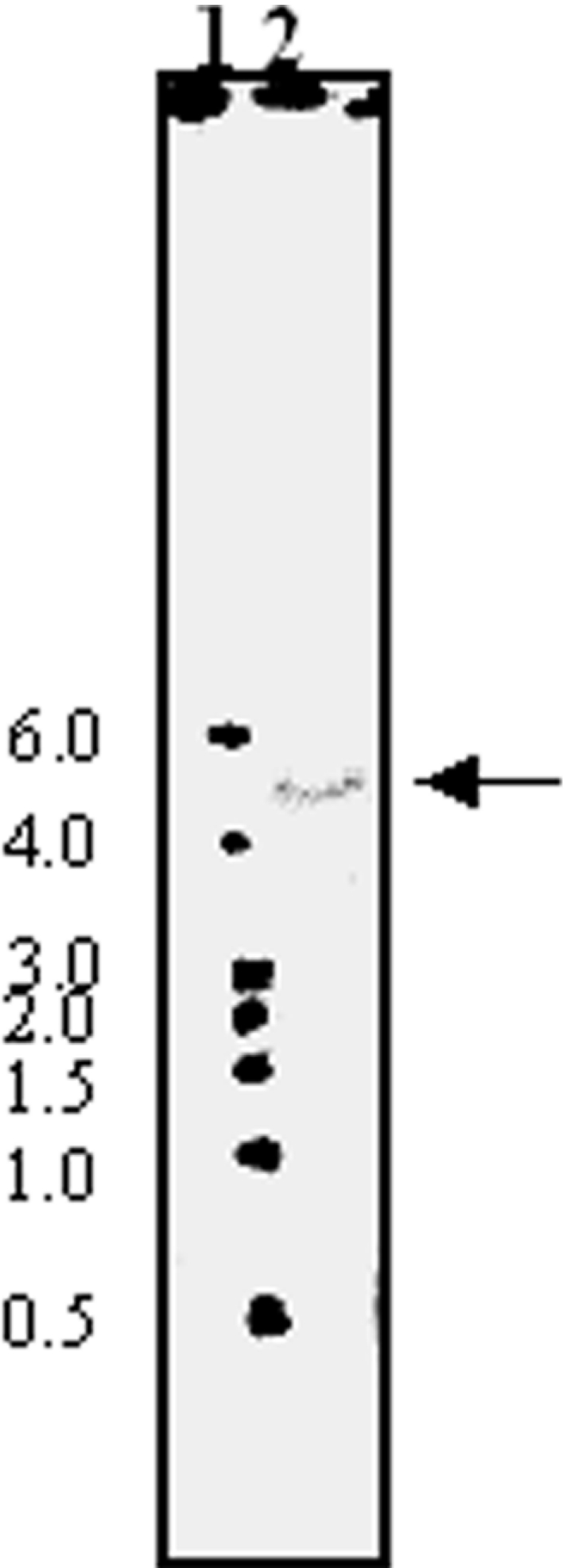
To determine the LdGLOII gene copy number, Southern blot studies were performed as described in the Experimental section using the ∼888-bp PCR product as a probe. A single band was obtained (Figure 3), revealing that it is a single-copy gene. Chromosomal location analysis revealed that LdGLOII gene is placed at a single chromosomal band of 600 kb (Figure 4). These data agree with the Leishmania Genome Sequencing Project findings, according to which GLOII gene has been identified on chromosome 12 (600 kb) in L. major (http://www.genedb.org).
Figure 3. Genomic organization of LdGLOII gene.
Southern blot analysis of LdGLOII gene. Lanes 1–11, restriction digests of L. donovani genomic DNA with BamHI, HindIII, EcoRI, NdeI, XbaI, XhoI, SalI, XmaJI, NotI, BbsI and BglI respectively. The blot was probed with ∼888-bp full-length GLOII gene. DNA molecular-mass marker and the sizes (in kb) are indicated on the left of the Figure.
Figure 4. PFGE analysis of L. donovani indicating chromosomal localization of the GLOII gene.
(a) Chromosomes of S. cerevisiae and L. donovani separated and visualized with ethidium bromide before blotting. (b) The same blot probed with LdGLOII gene probe. The arrow shows the hybridizing band of L. donovani. Sizes are given in Mb.
Northern blotting of total L. donovani RNA and PCR-generated ∼888-bp gene probe revealed a single transcript of ∼ 5 kb (Figure 5). The presence of a single RNA band in the corresponding Northern blot analysis indicated further the existence of a single encoding gene.
Figure 5. Expression analysis of LdGLOII gene.
Northern blot analysis of total RNA from L. donovani promastigotes (exponential phase culture). Lane 1; molecular-mass markers (sizes in kb), lane 2; total RNA from L. donovani AG83 probed with a ∼888-bp full-length GLOII gene.
Overexpression and purification of full-length LdGLOII enzyme in E. coli
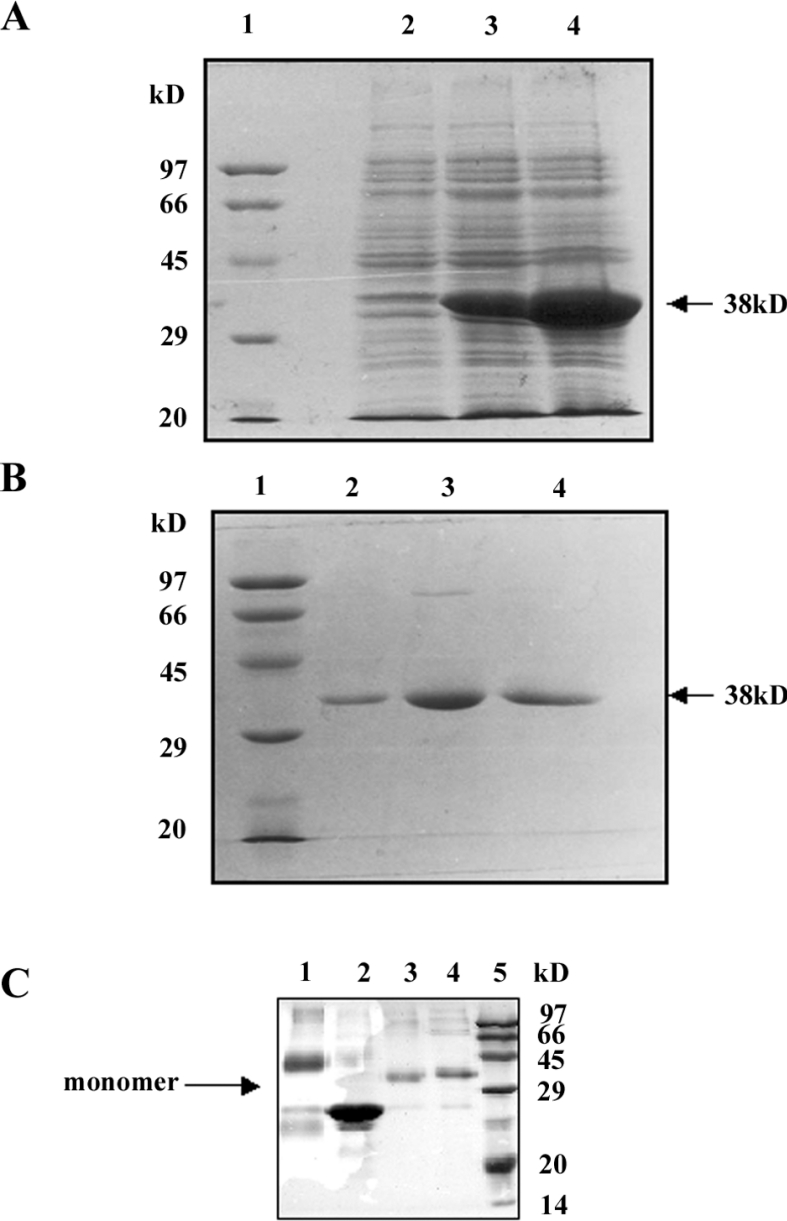
In order to characterize the recombinant protein, the encoding LdGLOII sequence was cloned in-frame in a pET30a expression vector with its own start ATG codon. The resultant pET30a-LdGLOII construct was transformed into E. coli and protein overexpression was induced as described in the Experimental section. A protein with a molecular mass that matched the estimated ∼ 38 kDa according to the amino acid composition of LdGLOII with a His6 tag and an S-tag present at its N-terminal end was induced (Figure 6A). The recombinant protein was purified on an Ni-NTA affinity chromatography column (Figure 6B). Purification of His-tagged LdGLOII by metal-affinity chromatography yielded ∼5 mg of pure protein from a 1-litre bacterial culture.
Figure 6. Overexpression and purification of LdGLOII protein.
(A) Coomassie Blue staining of SDS/PAGE gel showing overexpression of full-length LdGLOII protein in E. coli. The pET30a-bacterial extract before induction (lane 2) and after induction (lanes 3 and 4) at 1 and 4 h respectively with 0.5 mM IPTG (isopropyl β-D-thiogalactoside). The arrow shows the induced recombinant GLOII protein. Broad-range protein molecular-mass marker (Bio-Rad) was used to identify the size (in kDa) of the recombinant protein (Lane 1). (B) Purification of the GLOII protein on Ni-NTA affinity resin. Lane 1, broad-range molecular-mass marker (Bio-Rad). Lanes 2–4 are eluted fractions showing purified GLOII protein from the affinity column: lane 2, fraction eluted with 150 mM imidazole, lane 3, fraction eluted with 80 mM imidazole and lane 4, fraction eluted with 60 mM imidazole. (C) Analysis of subunit structure of GLOII protein. Protein samples were run on a 10% SDS/polyacrylamide gel. Lane 1 shows recombinant GLOI cross-linked with 0.025% glutaraldehyde showing a band size of ∼46 kDa (dimer), lane 2 shows recombinant GLOI (23 kDa) without glutaraldehyde, lane 3 shows recombinant GLOII cross-linked with 0.025% glutaraldehyde, and lane 4 shows recombinant GLOII without glutaraldehyde. A band size of ∼38 kDa representing GLOII was observed in lanes 3 and 4. Lane 5, broad-range protein molecular-mass markers (Bio-Rad). Sizes in kDa are indicated on the right of the Figure.
In order to determine the number of subunits in the recombinant GLOII, the homogeneous protein was cross-linked with the bifunctional reagent glutaraldehyde (0.025%) before electrophoresis on a 10% polyacrylamide gel in the presence of SDS (Figure 6C). Lane 1 shows recombinant GLOI cross-linked with 0.025% glutaraldehyde showing a band size of ∼ 46 kDa (dimer). The results in lane 2 show recombinant GLOI (23 kDa) without glutaraldehyde. Lane 3 shows recombinant GLOII cross-linked with 0.025% glutaraldehyde. Lane 4 shows recombinant GLOII without glutaraldehyde. A band size of ∼38 kDa representing GLOII was observed in lanes 3 and 4. These results show that the predominant species observed on SDS/PAGE corresponded to the molecular mass of a dimer in the case of GLOI in L. donovani, whereas GLOII corresponded to a monomer. Lysozyme (14.4 kDa) from chicken-egg white, a known monomer when cross-linked with the bifunctional reagent glutaraldehyde on electrophoresis on a 10% polyacrylamide gel in the presence of SDS, appeared as a band of ∼14 kDa (results not shown).
Recombinant GLOII was used to raise polyclonal antibody in BALB/c mice as described in the Experimental section. The antiserum recognized a ∼38 kDa fusion protein on a Western blot of purified recombinant LdGLOII fusion protein (20 ng) (Figure 7A). In a Western blot using size-fractionated parasite protein, the antiserum could detect a band of anticipated LdGLOII size ∼32 kDa in promastigote extracts (200 μg), which is in agreement with the value calculated from the predicted sequence (Figure 7A). Our attempts to analyse GLOII protein in the mastigote extracts (200 μg) with the same polyclonal antibody failed. This could be due to the nature of the antiserum.
Figure 7. Analysis of (A) protein and (B) RNA from promastigotes of wild-type L. donovani (WT) and GLOII overexpressor.
(A, a) Recombinant GLOII protein (rGLOII) (20 ng) was used as a control. Polyclonal antiserum against LdGLOII generated in mouse was used to detect GLOII protein in fractionated extracts obtained from cultures harvested after 48 h of growth. Lane 1, 20 ng of rGLOII protein, lane 2, 200 μg of total cell lysate of wild-type AG83 promastigotes, lane 3, 200 μg of total cell lysate from GLOII overexpressor. (A, b) The same blot was reprobed with antibody against β-tubulin protein to normalize the loading on to each lane of the gel. (B) Total RNA was isolated from exponential phase culture of L. donovani wild-type promastigotes (lane 2) and GLOII overexpressor (lane 3). Lane 1, molecular-mass markers. Total RNA was electrophoresed, transferred on to a membrane and probed with a ∼888-bp full-length GLOII gene as described in the Experimental section. An ethidium bromide-stained RNA gel showing equal loading of the RNA is shown. The results shown are from a single experiment typical of at least three giving similar results.
Overexpression of GLOII in L. donovani
To evaluate the consequences of GLOII overexpression, GLOII protein was measured in wild-type and GLOII-overexpressing L. donovani. Western blot analysis of wild-type and GLOII-overexpressing cell extract demonstrated a significant increase in GLOII protein in the overexpressors (Figure 7A). Northern blot analysis of wild-type and GLOII-overexpressing cells showed overexpression of GLOII transcript (Figure 7B).
LdGLOII activity
The kinetic parameters of recombinant LdGLOII were determined for S-D-lactoyltrypanothione as a substrate in a coupled reaction as mentioned in the Experimental section. LdGLOII hydrolysed S-D-lactoyltrypanothione with a Vmax value of 0.14 μmol·min−1·mg−1 and a Km value of 0.039 mM (Table 1). LdGLOII showed a high level of specificity for S-D-lactoyltrypanothione and little activity with S-D-lactoylglutathione. The Km of 187 μM has been reported for the human GLOII with S-D-lactoylglutathione [11]. The preference for trypanothione hemithioacetal over glutathione hemithioacetal by LdGLOII could be due to the absence of three conserved basic residues, namely Arg-249, Lys-252 and Lys-143, which are known to be involved in complexing with the substrate in human GLOII [34]. These residues are highly conserved in all GLOII studied so far, except for kinetoplastids [27].
Table 1. Comparison of kinetic parameters of recombinant L. donovani and human GLOII.
The values of human enzyme are taken from [11]. ND, not detected.
| L. donovani | |||
|---|---|---|---|
| Parameter | Km (μM) | Vmax (μmol·min−1·mg−1) | Human Km (μM) |
| S-D-lactoyltrypanothione | 39 | 0.14 | |
| S-D-lactoylglutathione | ND | ND | 187 |
DISCUSSION
All organisms require systems to shield them from chemical stress, such as the antioxidant enzymes that detoxify endogenous oxidants and the enzymes that metabolize exogenous toxins. However, endogenous toxins such as the reactive α-oxoaldehyde, methylglyoxal, are also by-products of metabolism [4]. The glyoxalase system is a ubiquitous detoxification pathway that protects against cellular damage caused by methylglyoxal. The glutathione-dependent glyoxalase system is present in all organisms analysed so far [35,36]. Recently, Irsch and Krauth-Siegel [27] characterized the GLOII from T. brucei. It is the first GLOII that does not use glutathione as cofactor. Trypanothione, a unique thiol in the kinetoplastid organisms, replaces glutathione in the glyoxalase system of the African trypanosome T. brucei.
To date, GLOII sequences have been reported in several species, including prokaryotes, plants, mammals, yeast and bacteria [8–11]. The present paper describes the molecular cloning and characterization of LdGLOII. The gene of GLOII is located on a 600-bp chromosome. Comparison of the GLOII from L. donovani and that of L. major, L. infantum and T. cruzi protein showed 51% identity with T. cruzi lactoylglutathione lyase-like protein, 96% identity with L. major GLOII and 99% identity with L. infantum GLOII protein. There was 32% identity with A. thaliana, 35% homology with human GLOII and 28% identity with E. coli probable hydroxyacylglutathione hydrolase. Phylogenetic tree analysis showed a close evolutionary relationship of L. donovani and T. cruzi and T. brucei among the kinetoplastid protozoa. From an evolutionary perspective, it is interesting to note that the kinetoplastid GLOII had very little similarity with human and A. thaliana enzyme in phylogenetic analysis, thereby ruling out a common evolutionary origin of the two enzymes.
The enzyme is trypanothione-dependent rather than glutathione-dependent. GLOII strongly prefers thioesters of trypanothione instead of glutathione as substrate. LdGLOII showed a high level of specificity for trypanothione hemithioacetal and little activity with glutathione hemithioacetal.
Sequence analysis of LdGLOII showed the conservation of metal-binding motif THXHXDH [33]. This metal-binding motif, along with two histidine residues and an aspartate residue interacts directly with two metal ions and is highly conserved in all glyoxalases studied so far [34]. It has been reported previously that storage of T. brucei GLOII causes a simultaneous decrease of kcat and Km [27]. A similar observation was made with GLOII of L. donovani. As in A. thaliana, the reason for this could also be the loss of metal as described for the A. thaliana enzyme [11].
The molecular mass of the enzyme was calculated as 32500 Da, higher than that of the human enzyme (22861 Da) and A. thaliana (28791 Da). GLOII from L. donovani with a predicted pI of 6.0 is more acidic than the human enzyme (pI 8.5). Western blot analysis of whole-cell lysates of promastigotes of L. donovani using the polyclonal antibody against GLOII enzyme, shows a single band of approx. 32 kDa, and the same antibody recognized the recombinant protein of the expected size of the GLOII–His6 tag fusion protein of approx. 38 kDa. Cross-linking studies established that the recombinant GLOII is a monomer.
Overexpression of GLOII in transfectants was observed both by Northern and Western blot analysis. The difference observed between the mRNA and protein levels expressed in the transfected cells could be due to post-transcriptional regulation which seems to be the mechanism of choice for gene expression in Leishmania and other lower organisms [37].
The exact biological role of the ubiquitous glyoxalase system is not known. It appears that one of the main roles of the glyoxalase system is glutathione-based detoxification of methylglyoxal. Characterization of the LdGLOII gene and expression of the protein will facilitate studies of the structural and functional aspects of the enzyme.
The glyoxalase system has received a considerable amount of attention as a possible anti-malarial target and for its possible anti-trypanosomal activity [38,39]. Inhibitors of GLOI and GLOII have been shown to inhibit the growth of tumour cells [15]. The three-dimensional structure of the human GLOII bound to analogues of the substrate revealed three conserved basic residues, Arg-249, Lys-252 and Lys-143, that are in close proximity and show specificity to glutathione [34]. The glycine carboxylate of the substrate glutathione interacts directly with the side chains of Arg-249 and Lys-252 [34]. These critical residues are not conserved in LdGLOII. The substrate for GLOII in Leishmania is trypanothione thioesters. These thioesters carry an overall positive charge because of the spermidine moiety, whereas glutathione thioesters carry a negative charge. The active site of the mammalian enzyme therefore cannot accommodate positively charged trypanothione thioesters. A difference in the substrate specificity of the human and LdGLOII enzyme renders the glyoxalase system an attractive target for antileishmanial chemotherapy. Finally, in view of the uniqueness of LdGLOII enzyme, it could be exploited for structure-based drug designing of selective inhibitors against the parasite.
Acknowledgments
A.M. is supported by a Senior Research Fellowship from the Council of Scientific and Industrial Research, India. P.K.P. is supported by a grant from the University Grants Commission, New Delhi, India.
References
- 1.Thornalley P. J. The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 1990;269:1–11. doi: 10.1042/bj2690001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vender Jagat D. L. Glutathione: chemical, biochemical and medical aspects. In: Dolphin D., Avramovic O., Poulson R., editors. Coenzymes and Cofactors, vol. IIIA. New York: John Wiley and Sons; 1989. pp. 597–641. [Google Scholar]
- 3.Racker E. The mechanism of action of glyoxalase. J. Biol. Chem. 1951;190:685–696. [PubMed] [Google Scholar]
- 4.Thornalley P. J. Pharmacology of methylglyoxal: formation, modification of proteins and nucleic acids, and enzymatic detoxification-a role in pathogenesis and antiproliferative chemotherapy. Gen. Pharmacol. 1996;27:565–573. doi: 10.1016/0306-3623(95)02054-3. [DOI] [PubMed] [Google Scholar]
- 5.Kimura A., Inoue Y. Glyoxalase I in micro-organisms: molecular characteristics, genetics and biochemical regulation. Biochem. Soc. Trans. 1993;21:518–522. doi: 10.1042/bst0210518. [DOI] [PubMed] [Google Scholar]
- 6.Deswal R., Chakaravarty T. N., Sopory S. K. The glyoxalase system in higher plants: regulation in growth and differentiation. Biochem. Soc. Trans. 1993;21:527–530. doi: 10.1042/bst0210527. [DOI] [PubMed] [Google Scholar]
- 7.Marmstal E., Aronsson A. C., Mannervik B. Comparison of glyoxalase I purified from yeast (Saccharomyces cerevisiae) with the enzyme from mammalian sources. Biochem. J. 1979;183:23–30. doi: 10.1042/bj1830023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maiti M. K., Krishnasamy S., Owen H. A., Makaroff C. A. Molecular characterization of glyoxalase II from Arabidopsis thaliana. Plant Mol. Biol. 1997;35:471–481. doi: 10.1023/a:1005891123344. [DOI] [PubMed] [Google Scholar]
- 9.Ridderstrom M., Saccucci F., Hellman U., Bergman T., Principato G., Mannervik B. Molecular cloning, heterologous expression and characterization of human glyoxalase II. J. Biol. Chem. 1996;271:319–323. doi: 10.1074/jbc.271.1.319. [DOI] [PubMed] [Google Scholar]
- 10.Bito A., Haider M., Hadler I., Breitenbach M. Identification and phenotypic analysis of two glyoxalase II encoding genes from Saccharomyces cerevisiae, GLO2 and GLO4, and intracellular localization of the corresponding proteins. J. Biol. Chem. 1997;272:21509–21519. doi: 10.1074/jbc.272.34.21509. [DOI] [PubMed] [Google Scholar]
- 11.Ridderstrom M., Mannervik B. Molecular cloning and characterization of thioesterase glyoxalase II from Arabidopsis thaliana. Biochem. J. 1997;322:449–454. doi: 10.1042/bj3220449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lo T. W., Westwood M. E., McLellan A. C., Selwood T., Thornalley P. J. Binding and modification of proteins by methylglyoxal under physiological conditions: a kinetic and mechanistic study with Nα-acetylarginine, Nα-acetylcysteine, and Nα-acetyllysine, and bovine serum albumin. J. Biol. Chem. 1994;269:32299–32305. [PubMed] [Google Scholar]
- 13.Marinari U. M., Ferro M., Sciaba L., Finollo R., Bassi A. M., Brambilla G. DNA-damaging activity of biotic and xenobiotic aldehydes in Chinese hamster ovary cells. Cell Biochem. Funct. 1984;2:243–248. doi: 10.1002/cbf.290020411. [DOI] [PubMed] [Google Scholar]
- 14.Thornalley P. J. The glyoxalase system in health and disease. Mol. Aspects Med. 1993;14:287–371. doi: 10.1016/0098-2997(93)90002-u. [DOI] [PubMed] [Google Scholar]
- 15.Thornalley P. J. Glutathione-dependent detoxification of α-oxoaldehydes by the glyoxalase system: involvement in disease mechanisms and antiproliferative activity of glyoxalase I inhibitors. Chem. Biol. Interact. 1998;111–112:137–151. doi: 10.1016/s0009-2797(97)00157-9. [DOI] [PubMed] [Google Scholar]
- 16.Carrington S. J., Douglas K. T. The glyoxalase enigma – the biological consequences of a ubiquitous enzyme. IRCS Med. Sci. 1986;14:763–768. [Google Scholar]
- 17.Rhee H. I., Sato N., Murata K., Kimura A. Nucleotide sequence of the glyoxalase I gene Pseudomonas putida. Agric. Biol. Chem. 1988;52:2243–2246. [Google Scholar]
- 18.Ranganathan S., Walsh E. S., Godwin A. K., Tew K. D. Cloning and characterization of human colon glyoxalase-I. J. Biol. Chem. 1993;268:5661–5667. [PubMed] [Google Scholar]
- 19.Kim N. S., Umezawa Y., Ohmura S., Kato S. Human glyoxalase I. cDNA cloning, expression, and sequence similarity to glyoxalase I from Pseudomonas putida. J. Biol. Chem. 1993;268:11217–11221. [PubMed] [Google Scholar]
- 20.Espartero J., Sanchez-Aguayo I., Pardo J. M. Molecular characterization of glyoxalase-I from a higher plant; upregulation by stress. Plant Mol. Biol. 1995;29:1223–1233. doi: 10.1007/BF00020464. [DOI] [PubMed] [Google Scholar]
- 21.Aggarwal P., Handa R., Singh S., Wali J. P. Kala-azar: new development in diagnosis and treatment. Indian J. Pediatr. 1999;66:63–71. doi: 10.1007/BF02752355. [DOI] [PubMed] [Google Scholar]
- 22.Berman J. D. Treatment of New World cutaneous and mucosalleishmaniases. Clin. Dermatol. 1996;14:519–522. doi: 10.1016/0738-081x(96)00048-x. [DOI] [PubMed] [Google Scholar]
- 23.Amato V., Amato J., Nicodemo A., Uip D., Amato-Neto V., Durate M. Treatment of mucocutaneous leishmaniasis with pentamidine isothionate. Ann. Dermatol. Venereol. 1998;125:492–495. [PubMed] [Google Scholar]
- 24.Darling T. N., Blum J. J. D-Lactate production by Leishmania braziliensis through the glyoxalase pathway. Mol. Biochem. Parasitol. 1988;28:121–128. doi: 10.1016/0166-6851(88)90059-x. [DOI] [PubMed] [Google Scholar]
- 25.Vickers T. J., Greig N., Fairlamb A. H. A trypanothione-dependent glyoxalase I with a prokaryotic ancestry in Leishmania major. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13186–13191. doi: 10.1073/pnas.0402918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sousa Silva M., Ferreira A. E. N., Tomás A. M., Cordeiro C., Ponces Freire A. Quantitative assessment of the glyoxalase pathway in Leishmania infantum as a therapeutic target by modelling and computer simulation. FEBS J. 2005;272:1–11. doi: 10.1111/j.1742-4658.2005.04632.x. [DOI] [PubMed] [Google Scholar]
- 27.Irsch T., Krauth-Siegel R. L. Glyoxalase II of African trypanosomes is trypanothione-dependent. J. Biol. Chem. 2004;279:22209–22217. doi: 10.1074/jbc.M401240200. [DOI] [PubMed] [Google Scholar]
- 28.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C. D., Abdelal A. T. The gdhB gene of Pseudomonas aeruginosa encodes an arginine-inducible NAD+-dependent glutamate dehydrogenase which is subject to allosteric regulation. J. Bacteriol. 2001;183:490–499. doi: 10.1128/JB.183.2.490-499.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellofato V., Cross G. A. Expression of a bacterial gene in a trypanosomatid protozoan. Science. 1989;244:1167–1169. doi: 10.1126/science.2499047. [DOI] [PubMed] [Google Scholar]
- 31.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 32.Papadopoulou B., Roy G., Ouellette M. A novel antifolate resistance gene on the amplified H circle of Leishmania. EMBO J. 1992;11:3601–3608. doi: 10.1002/j.1460-2075.1992.tb05444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zang T. M., Hollman D. A., Crawford P. A., Crowder M. W., Makaroff C. A. Arabidopsis glyoxalase II contains a zinc/iron binuclear metal center that is essential for substrate binding and catalysis. J. Biol. Chem. 2001;276:4788–4795. doi: 10.1074/jbc.M005090200. [DOI] [PubMed] [Google Scholar]
- 34.Cameron A. D., Ridderstrom M., Olin B., Mannervik B. Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue. Structure. 1999;7:1067–1078. doi: 10.1016/s0969-2126(99)80174-9. [DOI] [PubMed] [Google Scholar]
- 35.Jerzykowski T., Winter R., Matuszewski W., Piskorska D. A re-evaluation of studies on the distribution of glyoxalases in animal and tumour tissues. Int. J. Biochem. 1978;9:853–860. doi: 10.1016/0020-711x(78)90036-8. [DOI] [PubMed] [Google Scholar]
- 36.Ghoshal K., Banerjee A. B., Ray S. Methylglyoxal-catabolizing enzymes of Leishmania donovani promastigotes. Mol. Biochem. Parasitol. 1989;35:21–29. doi: 10.1016/0166-6851(89)90138-2. [DOI] [PubMed] [Google Scholar]
- 37.Clayton C. E. Life without transcriptional control? From fly to man and back again. EMBO J. 2002;21:1881–1888. doi: 10.1093/emboj/21.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thornalley P. J., Strath M., Wilson R. J. Antimalarial activity in vitro of the glyoxalase I inhibitor diester, S-p-bromobenzylglutathione diethyl ester. Biochem. Pharmacol. 1994;47:418–420. doi: 10.1016/0006-2952(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 39.D'silva C., Daunes S., Rock P., Yardley V., Croft S. L. Structure–activity study on the in vitro antiprotozoal activity of glutathione derivatives. J. Med. Chem. 2000;43:2072–2078. doi: 10.1021/jm990259w. [DOI] [PubMed] [Google Scholar]