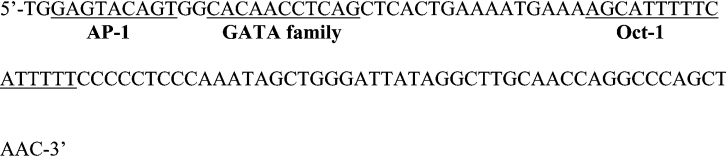Abstract
LOX-1, a receptor for ox-LDL (oxidized low-density lipoprotein), has recently been determined to play a critical role in the progression of atherosclerosis. LOX-1 expression (mRNA and protein) has been shown to be up-regulated by pro-atherogenic stimuli, such as ox-LDL and Ang II (angiotensin II). However, the molecular mechanisms of these up-regulations are unclear. In the present study, we explored LOX-1 transcriptional promoter activation in response to ox-LDL and Ang II. Under basal states, LOX-1 core promoter (LOX-1 −35/+36) was found to be sufficient for its basal activity in HCAECs (human coronary artery endothelial cells). More importantly, we found that ox-LDL (60 μg/ml for 24 h) induced LOX-1 promoter activity significantly and that a 105 bp fragment (between nt −1599 and −1494) was required for this activation. Within this 106 bp fragment, there is a potential binding motif for the transcription factor Oct-1 (octamer-1). By electrophoretic mobility-shift assay, we observed the activation of Oct-1 by ox-LDL. The critical role of Oct-1 in ox-LDL-induced LOX-1 promoter activation was further confirmed by mutagenesis assay. For comparison, we also examined LOX-1 promoter activation in response to Ang II (1 μmol/l for 24 h). Interestingly, another promoter region, between nt −2336 and −1990, was required for Ang II-induced LOX-1 promoter activation. In conclusion, the present study strongly suggests that ox-LDL, by activating Oct-1, induces LOX-1 promoter activation. Furthermore, this study suggests that while ox-LDL and Ang II both induce LOX-1 expression in HCAECs, the underlying mechanisms of promoter activation are different from each other.
Keywords: angiotensin II (Ang II), lectin-like ox-LDL receptor-1 (LOX-1), octamer-1 (Oct-1), oxidized low-density lipoprotein (ox-LDL), oxidative stress, transcription factor
Abbreviations: Ang II, angiotensin II; AP-1, activator protein-1; EMSA, electrophoretic mobility-shift assay; HCAEC, human coronary artery endothelial cell; LDL, low-density lipoprotein; ox-LDL, oxidized LDL; LOX-1, lectin-like ox-LDL receptor-1; NF-κB, nuclear factor κB; Oct-1, octamer-1; PKA, protein kinase A; RAS, renin–angiotensin system; Sp-1, stimulatory protein-1; SV40, simian virus 40
INTRODUCTION
Atherosclerosis is a multifactorial pathological process that affects human health. Dyslipidaemia, particularly increased plasma levels of LDL (low-density lipoprotein), is a major risk factor for atherogenesis [1]. Increasing evidence has demonstrated that oxidatively modified LDL [ox-LDL (oxidized LDL)] plays a more critical role than native LDL in the development of atherosclerosis [2]. Ox-LDL is taken up by monocyte/macrophages and vascular endothelial cells as well as by vascular smooth muscle cells, leading to the formation of lipid-laden foam cells and endothelial dysfunction as well as smooth muscle cell proliferation and migration, the hallmarks of early atherosclerotic lesions [2].
During the early stages of arterial atherogenesis, the endothelial cells are affected by ox-LDL and become dysfunctional. These dysfunctional endothelial cells express adhesion molecules, leading to monocyte/macrophage recruitment. Previous in vitro studies demonstrated that the vascular endothelial cells respond to ox-LDL through a receptor-mediated pathway, which mainly involves the endothelial LOX-1 (lectin-like ox-LDL receptor-1) [3]. Ox-LDL is an important activator of endothelial cells through binding, and then up-regulating the expression of LOX-1, through a positive feedback loop resulting in endothelial cell activation and dysfunction. In fact, studies carried out by our laboratory and others have shown that in vascular endothelial cells, LOX-1 is involved in almost all pro-atherogenic effects of ox-LDL, including the expression of matrix metalloproteinases [4], induction of apoptosis [5], expression of adhesion molecules and resultant monocyte adhesion [6], induction of CD40/CD40L signalling and inflammation [7], and reduced generation of endothelial nitric oxide synthase and release of nitric oxide [8].
In vitro studies have suggested that LOX-1 expression is up-regulated by many pro-atherogenic stimuli, including ox-LDL and the cytokine TNF-α (tumour necrosis factor α) in endothelial cells [9]. Other studies have indicated that Ang II (angiotensin II), a pro-atherogenic risk factor, also up-regulates LOX-1 expression in endothelial cells [10]. The up-regulation of LOX-1 has been confirmed in arterial tissues of atherosclerotic animals [11,12]. Kita [14] and Singh and Mehta [1] have proposed a ‘cross-talk’ between hyperlipidaemia and RAS (renin–angiotensin system) activation in the development of atherosclerosis. This concept of ‘cross-talk’ was confirmed in a recent animal study, which showed that the concurrent blockade of hyperlipidaemia and RAS has a potent synergistic inhibitory effect on LOX-1 expression and arterial atherogenesis in apoE (apolipoprotein E)-deficient mice [12].
These observations, taken together, suggest that LOX-1 plays a critical role in the development of atherosclerosis. This may suggest that increased LOX-1 transcriptional promoter activity may equal increased LOX-1 gene expression and elevated risk of atherosclerosis. Accordingly, decreased LOX-1 promoter activation may reduce the incidence of atherosclerosis and related diseases.
The nucleotide sequence of human LOX-1 promoter has been identified [13]. Although only a few details are known about the transcription factors that modulate LOX-1 gene expression, a number of potential cis-regulatory elements, such as NF-κB (nuclear factor κB) and AP-1 (activator protein-1) binding motifs, are found within the LOX-1 promoter sequence [13]. Nonetheless, human LOX-1 transcriptional promoter has not been extensively examined on the molecular level. In the present study, we analysed ox-LDL-mediated activation of the human LOX-1 promoter in HCAECs (human coronary artery endothelial cells). The purpose of this study was to determine the specific mechanisms for the ox-LDL-induced activation of LOX-1 transcriptional promoter from the basal state. For comparison, we also studied Ang II-induced LOX-1 promoter activation.
MATERIALS AND METHODS
LOX-1 promoter–luciferase reporter plasmid construction
The promoterless luciferase reporter vector (pGL3-Basic) was obtained from Promega (catalogue no. E1751). Three restriction enzyme sites, KpnI, XhoI and BglII, are present upstream of the luciferase-expression gene. Fragments of human LOX-1 promoter (both full-length and deletion mutants) were amplified by PCR, using human genomic DNA as template. The primer pairs and size of PCR products are shown in Table 1. For full-length and 5′ deletion LOX-1 promoters, forward primers were tagged with KpnI restriction site, and reverse primers were tagged with BglII restriction site. As such, KpnI and BglII restriction sites were present on the 5′ and 3′ ends of PCR products, which can be subcloned into pGL3-Basic vector after restriction enzyme digestion followed by T4 DNA ligase-catalysed ligation. For 3′ deletion and finer 5′ or 3′ deletion LOX-1 promoters, forward primers were tagged with KpnI restriction site, and reverse primers were tagged with XhoI restriction site. As such, KpnI and XhoI restriction sites were present on the 5′ and 3′ ends of PCR products. A 96 bp double-stranded DNA was synthesized by Invitrogen, having a KpnI site followed by an XhoI site on the 5′-end and a BglII site on the 3′-end. The nucleotides between XhoI and BglII sites are the same as LOX-1 promoter region from nt −35 to +36, including TATA box and transcription initiation site. Nucleotide number refers to the number of nucleotides upstream of the transcription initiation site. The 96 bp synthetic DNA was first subcloned into pGL3-Basic vector and then 3′ deletion or finer 5′ or 3′ deletion LOX-1 promoters were subcloned into it by restriction enzyme digestion followed by T4 DNA ligase-catalysed ligation.
Table 1. Primer pairs for the amplification of full-length and deletion mutant LOX-1 promoters and the size of PCR products.
| Forward primer | Reverse primer | Size of PCR product (bp) | |
|---|---|---|---|
| LOX-1 (−2336/+36) | 5′-GGGGTACCCACCTACATTATGCAGC-3′ | 5′-GAAGATCTGAGTGAAGCAGTCACGAACTTC-3′ | 2372 |
| LOX-1 (−1999/+36) | 5′-GGGGTACCGAGATCATGCCATTGCAATC-3′ | 5′-GAAGATCTGAGTGAAGCAGTCACGAACTTC-3′ | 2035 |
| LOX-1 (−1500/+36) | 5′-GGGGTACCAGCTAACTTTTGTATTTTTAGTG-3′ | 5′-GAAGATCTGAGTGAAGCAGTCACGAACTTC-3′ | 1536 |
| LOX-1 (−996/+36) | 5′-GGGGTACCGTCTTTGTGACATGAGTCAATC-3′ | 5′-GAAGATCTGAGTGAAGCAGTCACGAACTTC-3′ | 1032 |
| LOX-1 (−498/+36) | 5′-GGGGTACCCAGGACACACCATCGCTGG-3′ | 5′-GAAGATCTGAGTGAAGCAGTCACGAACTTC-3′ | 534 |
| LOX-1 (−2336/−477) | 5′-GGGGTACCCACCTACATTATGCAGC-3′ | 5′-CCGCTCGAGCTTCCAGCGATGGTGTGTCCT-3′ | 1859 |
| LOX-1 (−2336/−1059) | 5′-GGGGTACCCACCTACATTATGCAGC-3′ | 5′-CCGCTCGAGCGCCTTGACAACCTCCTAACCAG-3′ | 1277 |
| LOX-1 (−2336/−1502) | 5′-GGGGTACCCACCTACATTATGCAGC-3′ | 5′-CCGCTCGAGCGGCCTGGTTGCAAGCCTATAATC-3′ | 834 |
| LOX-1 (−2336/−1990) | 5′-GGGGTACCCACCTACATTATGCAGC-3′ | 5′-CCGCTCGAGCGGTGATCTCGGCTCACTGCAAC-3′ | 346 |
| LOX-1 (−1999/−1494) | 5′-GGGGTACCGAGATCATGCCATTGCAATC-3′ | 5′-CCGCTCGAGCGTTAGCTGGGCCTGGTTGC-3′ | 505 |
| LOX-1 (−1846/−1494) | 5′-GGGGTACCGTTCTGGAGATGGATGGTG-3′ | 5′-CCGCTCGAGCGTTAGCTGGGCCTGGTTGC-3′ | 352 |
| LOX-1 (−1794/−1494) | 5′-GGGGTACCTGCCACTGAACAGTACAC-3′ | 5′-CCGCTCGAGCGTTAGCTGGGCCTGGTTGC-3′ | 300 |
| LOX-1 (−1599/−1494) | 5′-GGGGTACCTGGAGTACAGTGGCACAAC-3′ | 5′-CCGCTCGAGCGTTAGCTGGGCCTGGTTGC-3′ | 105 |
| LOX-1 (−1999/−1582) | 5′-GGGGTACCGAGATCATGCCATTGCAATC-3′ | 5′-CCGCTCGAGCTTGTGCCACTGTACTCCAGC-3′ | 417 |
| LOX-1 (−1999/−1775) | 5′-GGGGTACCGAGATCATGCCATTGCAATC-3′ | 5′-CCGCTCGAGCAGGTGTACTGTTCAGTGG-3′ | 224 |
| LOX-1 (−1999/−1876) | 5′-GGGGTACCGAGATCATGCCATTGCAATC-3′ | 5′-CCGCTCGAGCCCACCAAGCAATAACTCCTG-3′ | 123 |
LOX-1 promoter–luciferase construct nomenclature
For ease in writing and understanding, full-length LOX-1 promoter (nt −2336 to +36)–luciferase construct is shortened to ‘LOX-1−2336/+36’. The name of a deletion mutant promoter directly reflects its structure. For example, a 5′ deletion promoter construct, which is deleted from nt −2336 to −2000 and retains sequence from nt −1999 to +36, is named ‘LOX-1−1999/+36’. All 3′ deletion and finer 5′ or 3′ deletion promoters retain the sequence from nt −35 to +36, which is termed ‘LOX-1−35/+36’, and is regarded as a core promoter. A 3′ deletion construct that retains the sequence from nt −2336 to −477 (plus nt −35 to +36) is termed ‘LOX-1−2336/−477’.
DNA sequencing
The nucleotide sequences of full-length and deletion mutant LOX-1 promoters amplified from PCR were verified by dideoxy chain-termination method with appropriate primers, and the procedures were performed in a DNA sequencing laboratory. Nucleotide sequences of all PCR products were analysed by comparison with human LOX-1 promoter sequence found in the GenBank® Nucleotide Sequence Database (accession number AB021922). For all promoters, the consistency was more than 99.8%.
Preparation of ox-LDL
Ox-LDL was prepared as described previously [15]. In brief, human native LDL was obtained from Calbiochem (catalogue no. 437644). It was kept in 50 mmol/l Tris/HCl, 0.15 mol/l NaCl and 2 mmol/l EDTA. After the removal of EDTA by dialysis for 24 h in PBS, native LDL was oxidized by exposure to CuSO4 (5 μmol/l free Cu2+ concentration in PBS) at 37 °C for 24 h. The TBARS (thiobarbituric acid-reacting substances) content of ox-LDL was 10.2±0.48 versus 0.56±0.26 nmol per 100 μg of protein in native LDL, suggesting the oxidation status of LDL.
Cell culture, transient transfection and ox-LDL or Ang II treatment
The initial batch of HCAECs was purchased from Clonetics and cultured in endothelial basal medium as described previously [3]. In brief, cells were cultured to passages 4–5 and seeded in six-well plates. On reaching 80% confluence, 1 μg of LOX-1 promoter–luciferase reporter vector was transfected into HCAECs by FuGENE 6 transfection reagent (Roche catalogue no. 1-814-443) according to the manufacturer's instructions. As an internal control for transfection efficiency, 1 μg of Renilla luciferase vector (pRL-SV40, Promega catalogue no. E2231; SV40 stands for simian virus 40) was co-transfected. The transfection reagent was removed 6 h after transfection, and the transfected HCAECs were cultured in complete culture medium for 36 h. Then, these cells were treated with or without 60 μg/ml ox-LDL for 24 h and harvested for assays mentioned below. In another set of experiments, the transfected HCAECs were treated with or without 1 μmol/l Ang II for 24 h, followed by the same assays. The concentration and incubation time of ox-LDL and Ang II were based on previous studies, showing the maximal effects of them on LOX-1 expression in HCAECs [9,10].
Dual luciferase assay
The luciferase expression/activity was measured by the Dual-Luciferase Report Assay system (Promega catalogue no. E1910), and the luminescence was read by a luminometer (TD-20/20) according to the manufacturer's instructions. This system allows the quantification of activities of both Firefly luciferase (encoded by LOX-1 promoter-pGL3 plasmid construct) and Renilla luciferase (encoded by pRL-SV40 plasmid). The relative values of Firefly luciferase activity were determined by normalizing with Renilla luciferase activity for transfection efficiency.
Computer analysis of the LOX-1 promoter
TRANSFAC database (MatInspector software) was used to search for the potential cis-regulatory elements within human LOX-1 promoter. The threshold was set at 0.88.
EMSA (electrophoretic mobility-shift assay), supershift and competition assays
Nuclear extracts from HCAECs were prepared according to previously published procedures [6]. The 105 bp LOX-1 promoter fragment (between nt −1599 and −1494) was obtained by PCR. Complementary oligonucleotides containing Oct-1 (octamer-1) binding site (5′-TGTCGAATGCAAATCACTAGAA-3′), AP-1 binding site (5′-CGCTTGATGACTCAGCCCGAA-3′) or GATA family binding site (5′-CACTTGATAACAGAAAGTGATAACTCT-3′) were obtained from Invitrogen. All probes were end-labelled with [γ-32P]ATP by T4 polynucleotide kinase, and unincorporated [γ-32P]ATP was removed by a QIAquick Nucleotide Remove kit (Qiagen catalogue no. 28304) according to the manufacturer's instructions. The radiolabelled probes were incubated with nuclear extracts for 30 min at room temperature (24 °C) in 50 mM Tris/HCl buffer (pH 7.5) containing 20% (w/v) glycerol, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM dithiothreitol, 250 mM NaCl and 0.25 mg/ml poly(dI-dC)·(dI-dC). For competition assay, 100-fold excess of non-radiolabelled probes was incubated with nuclear extracts for 10 min before the addition of radiolabelled probes. As negative controls for the competition assay, 100-fold excess of non-radiolabelled probes containing mutant Oct-1 binding site (5′-TGTCGAATGCAAGCCACTAGAA-3′; synthesized by Invitrogen) was used as competitor. For supershift assay, rabbit polyclonal antibody directed to Oct-1 (Santa Cruz Biotechnology catalogue no. sc-232) was used. The experimental method was identical with EMSA except that 1, 2, 3 or 5 μg of anti-Oct-1 antibody was incubated with nuclear extracts on ice for 1 h before the addition of radiolabelled probes. The specificity was verified by conducting a control supershift assay with 5 μg of normal rabbit IgG (Santa Cruz Biotechnology catalogue no. sc-3888), instead of anti-Oct-1 antibody. The DNA–protein complexes were separated by electrophoresis in a 6% (w/v) non-denaturing polyacrylamide gel using 0.25×TBE running buffer (0.0225 M Tris base, 0.0225 M boric acid and 0.0005 M EDTA). The gels were dried for 1 h and exposed to a radiographic film for 24 h at −80 °C.
Site-directed mutagenesis
Site-directed substitution mutation of specific nucleotides within the human LOX-1 promoter region was performed using the In Vitro Site-Directed Mutagenesis system (Promega catalogue no. Q9280) according to the manufacturer's instructions. We first cloned a LOX-1 promoter–luciferase plasmid construct (LOX-1−1599/−1494), containing a potential Oct-1 binding site (5′-ATTTTTCA-3′). To make site-directed substitution mutations within the Oct-1 binding site (from TT to GG), we synthesized a 20-base mutagenic oligonucleotide 5′-AAAAGCATGGTTCATTTTTC-3′. By finishing the mutagenesis method, the original potential Oct-1 binding motif was changed to a mutant binding motif (5′-ATGGTTCA-3′). The mutation was verified by DNA sequencing.
Data analysis
All data represent the means for samples from five independent experiments. Results are expressed as means±S.D. and are analysed with Student's t test. A value of P<0.05 was considered as statistically significant.
RESULTS
Deletion analysis of basal human LOX-1 promoter activity
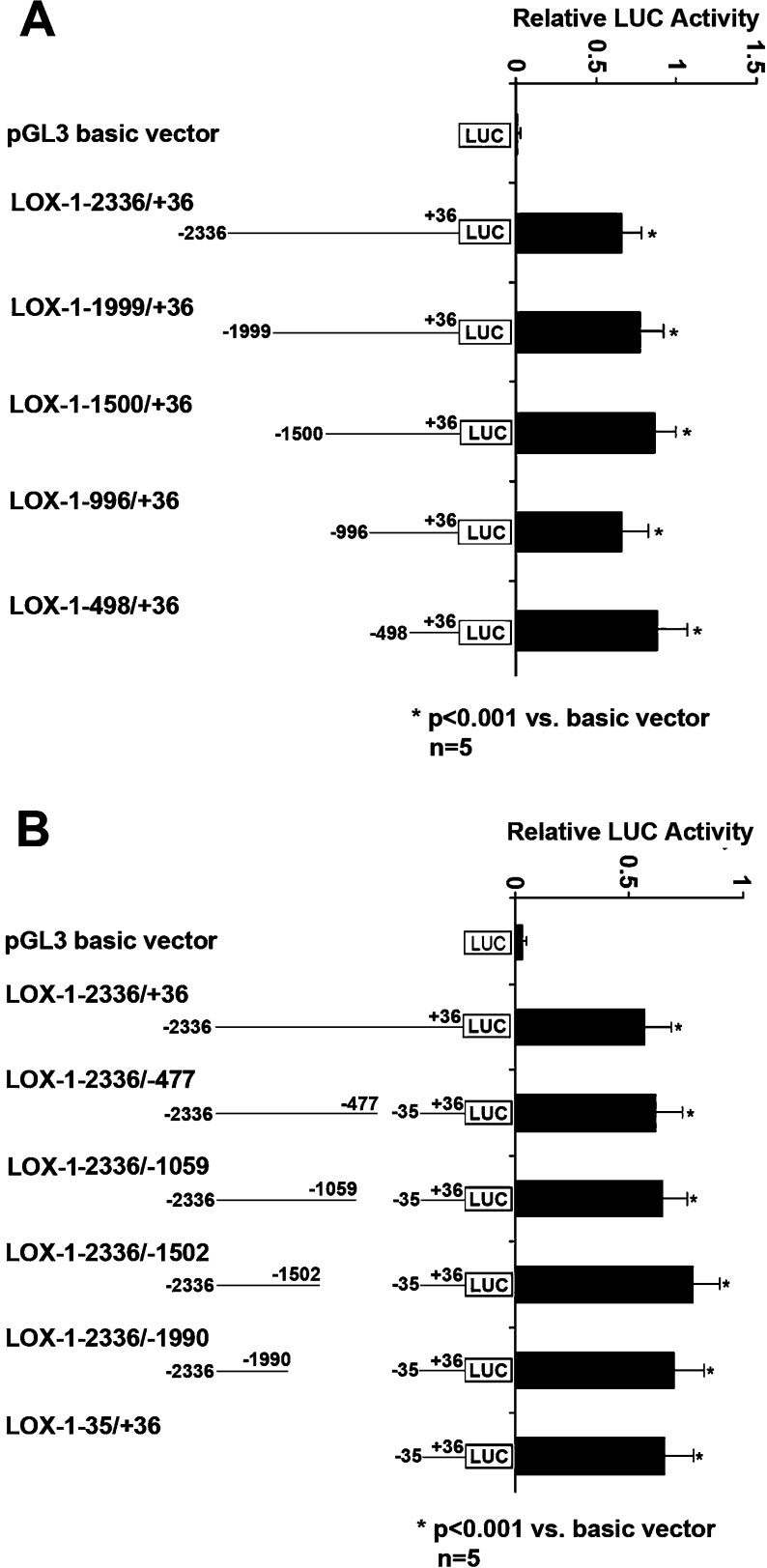
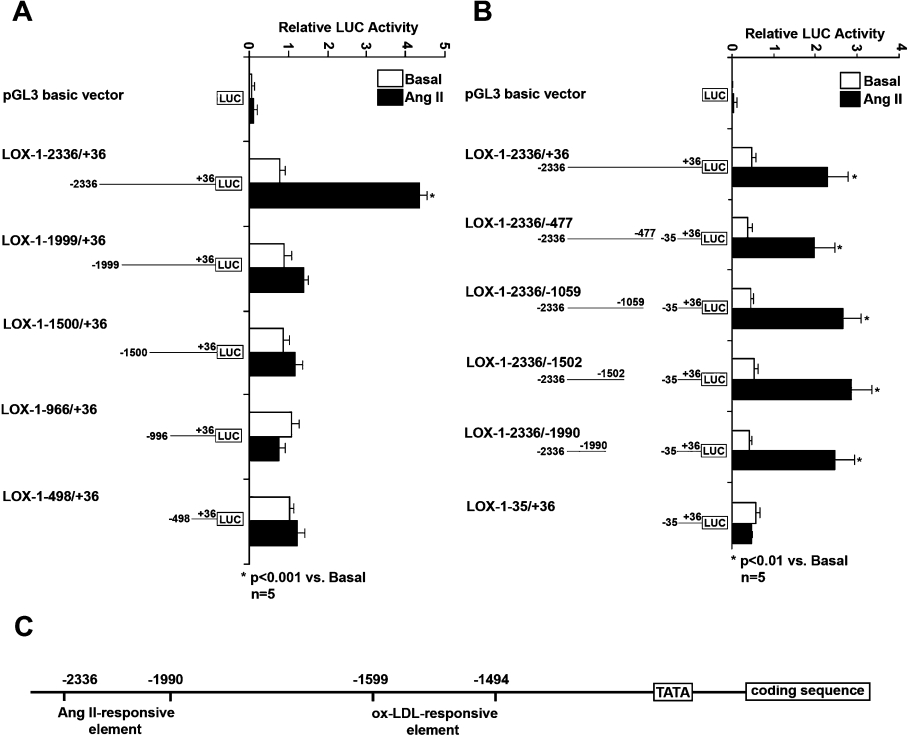
In the basal state, there was no significant difference between fulllength (LOX-1−2336/+36) and 5′ deletion (LOX-1−1999/+36, LOX-1−1500/+36, LOX-1−996/+36 and LOX-1−498/+36) promoter activity (Figure 1A). There was also no significant difference between full-length (LOX-1−2336/+36) and 3′ deletion (LOX-1−2336/–477, LOX-1−2336/−1059, LOX-1−2336/−1502 and LOX-1−2336/−1990) promoter activity. Importantly, LOX-1 core promoter (LOX-1−36/+35) had the same activity as full-length promoter under basal states (Figure 1B).
Figure 1. Analysis of the human LOX-1 promoter under basal states.
The activity of LOX-1 promoters was expressed as the ratio of LOX-1 promoter-mediated firefly LUC (luciferase) activity to Renilla luciferase activity. Results shown are means±S.D. for five independent experiments. (A) The relative activity of full-length LOX-1 promoter (LOX-1−2336/+36) and a series of 5′ deletion promoters (LOX-1−1999/+36, LOX-1−1500/+36, LOX-1−996/+36 and LOX-1−498/+36). (B) The relative activity of full-length LOX-1 promoter and a series of 3′ deletion promoters (LOX-1−2336/−477, LOX-1−2336/−1059, LOX-1−2336/−1502 and LOX-1−2336/−1990) as well as core promoter (LOX-1−36/+35).
Deletion analysis of ox-LDL-induced human LOX-1 promoter activation
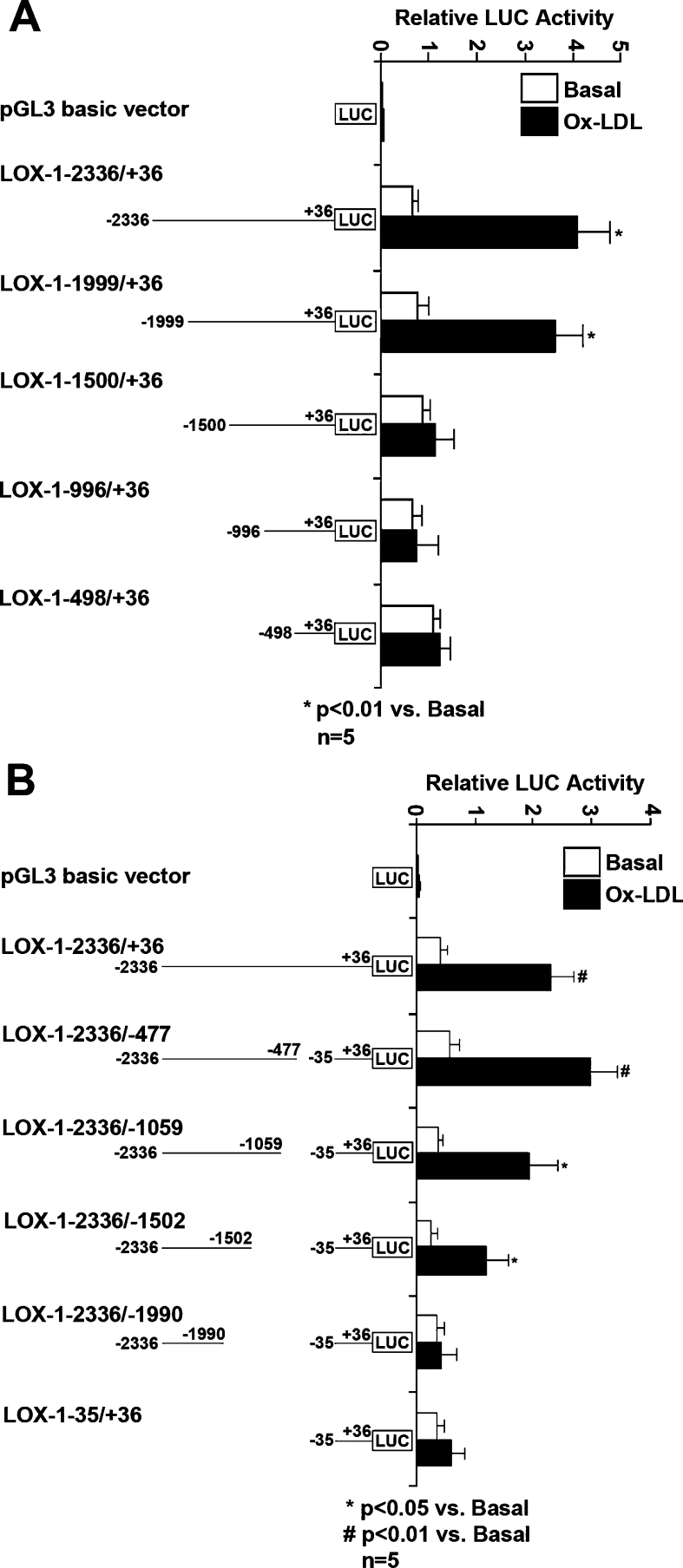
Since the main purpose of the present study was to clarify the mechanism of ox-LDL-induced LOX-1 promoter activation, we treated the LOX-1 promoter–luciferase plasmid-transfected HCAECs with 60 μg/ml ox-LDL for 24 h. Compared with the basal state, we found that only full-length and 5′ deletion LOX-1 promoter (LOX-1−1999/+36) were significantly activated by ox-LDL, while others (LOX-1−1500/+36, LOX-1−996/+36 and LOX-1−498/+36) were not (Figure 2A). For 3′ deletion LOX-1 promoters, most of them (LOX-1−2336/−477, LOX-1−2336/−1059 and LOX-1−2336/−1502) were activated by ox-LDL treatment significantly; however, LOX-1−2336/−1990 was not (Figure 2B). It should be noted that the activity of core promoter (LOX-1−35/+36) itself was not affected by ox-LDL (Figure 2B).
Figure 2. Analysis of human LOX-1 promoter activation induced by ox-LDL.
LOX-1 promoter–luciferase plasmid-transfected HCAECs were treated without (white bar) or with (black bar) 60 μg/ml ox-LDL for 24 h. The promoter activity was expressed as the ratio of LOX-1 promoter-mediated firefly luciferase activity to Renilla luciferase activity. Results shown are means±S.D. for five independent experiments. (A) The relative activity of full-length LOX-1 promoter (LOX-1−2336/+36) and a series of 5′ deletion promoters (LOX-1−1999/+36, LOX-1−1500/+36, LOX-1−996/+36 and LOX-1−498/+36). (B) The relative activity of full-length promoter and a series of 3′ deletion promoters (LOX-1−2336/−477, LOX-1−2336/−1059, LOX-1−2336/−1502 and LOX-1−2336/−1990) as well as core promoter (LOX-1−36/+35).
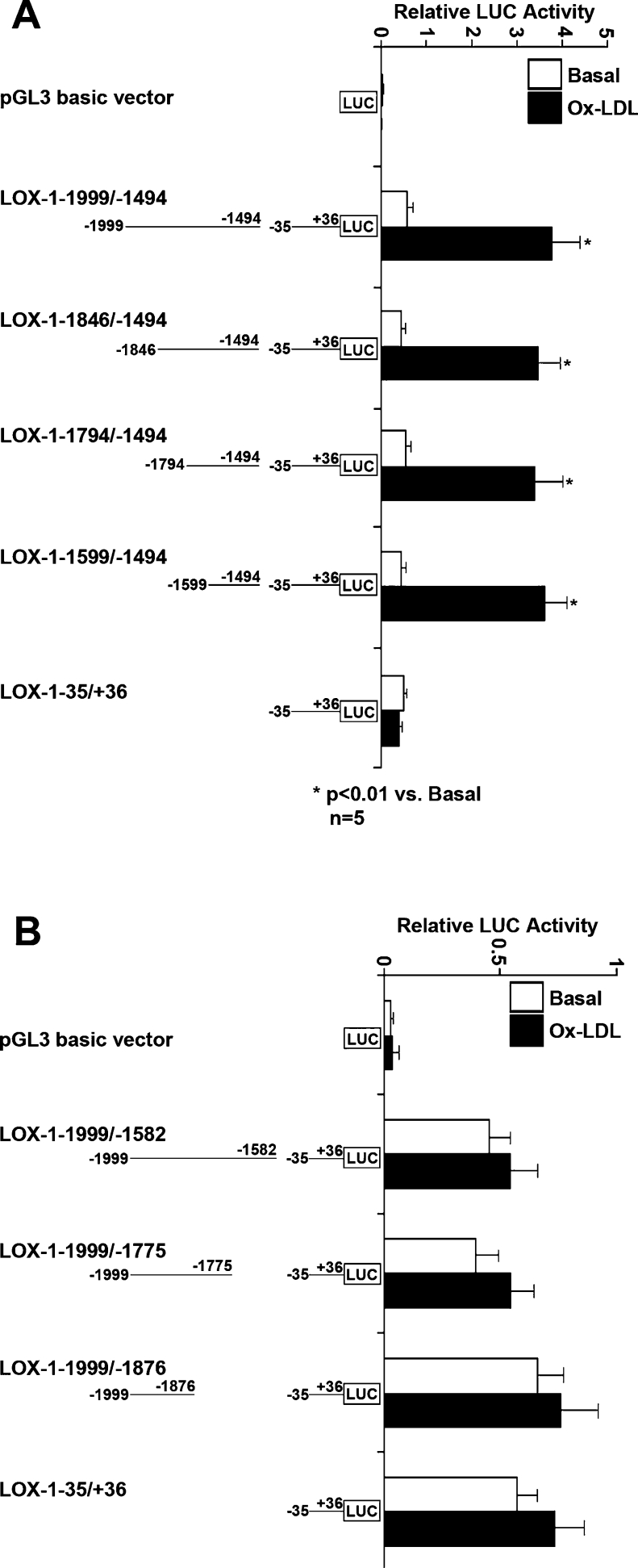
From these experiments, we identified that the LOX-1 promoter region, which responds to ox-LDL, lies between nt −1999 and −1500. Accordingly, we cloned a 506 bp promoter fragment (LOX-1−1999/−1494) containing this region, as well as a series of 5′ and 3′ deletions from this promoter fragment. All these LOX-1 promoter constructs contained an extra nucleotide sequence from nt −35 to +36, serving as the core promoter. After ox-LDL treatment, LOX-1−1999/−1494 and all 5′ deletion promoters (LOX-1−1846/−1494, LOX-1−1794/−1494 and LOX-1−1599/−1494) were activated significantly (Figure 3A). On the other hand, all 3′ deletion promoters (LOX-1−1999/−1582, LOX-1−1999/−1775 and LOX-1−1999/−1876) were barely activated by ox-LDL (Figure 3B). These results suggested that the ox-LDL-responsive cis-regulatory element resides within the 106 bp promoter region, between nt −1599 and −1494.
Figure 3. Further analysis of human LOX-1 promoter activation induced by ox-LDL.
We focused on the promoter region between nt −1999 and −1494. LOX-1 promoter–luciferase plasmid-transfected HCAECs were treated without (white bar) or with (black bar) 60 μg/ml ox-LDL for 24 h. The promoter activity was expressed as the ratio of LOX-1 promoter-mediated firefly luciferase activity to Renilla luciferase activity. Results shown are means±S.D. for five independent experiments. (A) The relative activity of LOX-1 promoter (LOX-1−1999/−1494) and a series of 5′ deletion promoters (LOX-1−1846/−1494, LOX-1−1794/−1494 and LOX-1−1599/−1494) as well as core promoter (LOX-1−36/+35). (B) The relative activity of a series of 3′ deletion promoters (LOX-1−1999/−1582, LOX-1−1999/−1775 and LOX-1−1999/−1876) and core promoter (LOX-1−36/+35).
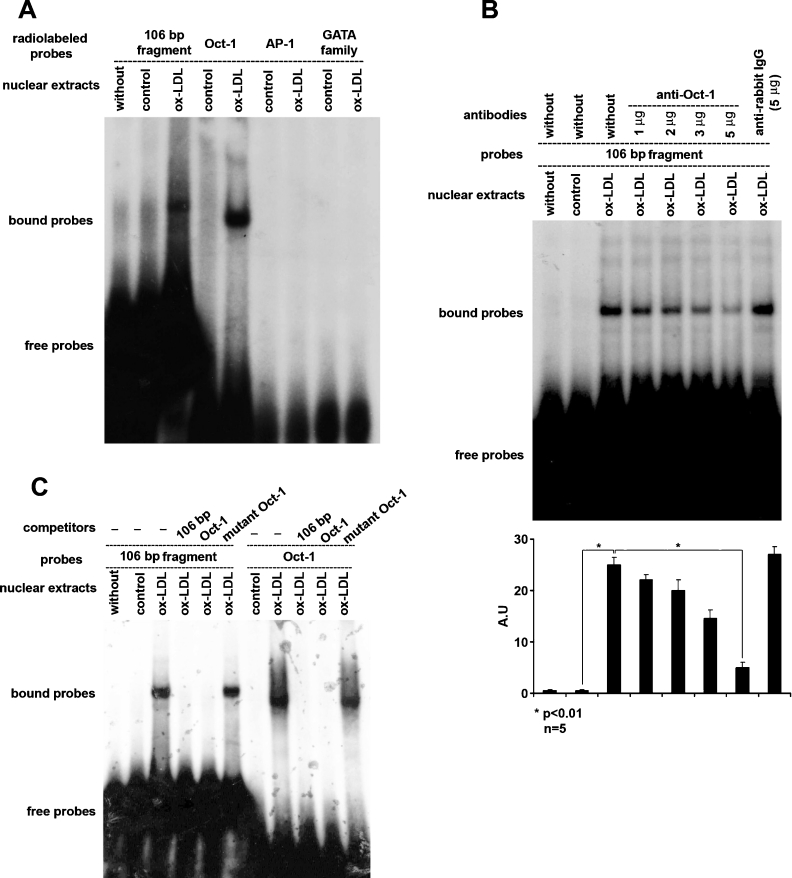
EMSA analysis of the activation of transcription factor Oct-1 by ox-LDL
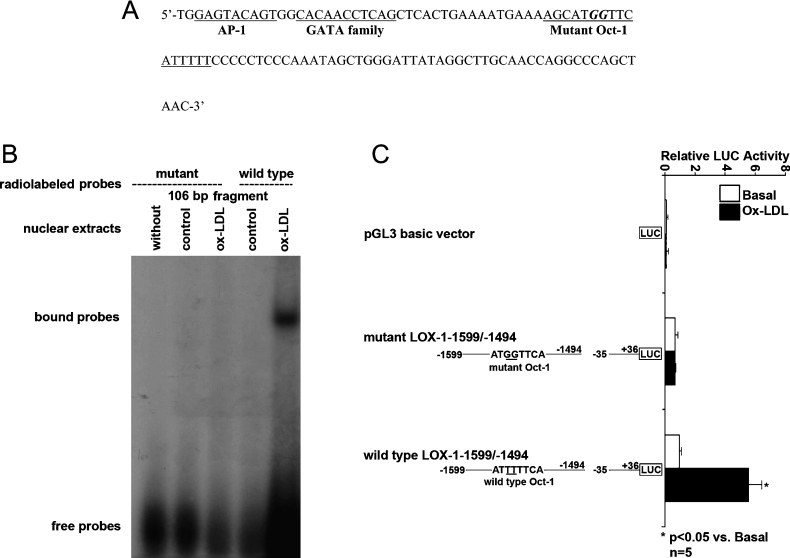
Next, we searched for potential cis-regulatory element(s) within this 106 bp promoter fragment (between nt −1599 and −1494) in the database TRANSFAC, which revealed multiple putative binding sites for transcription factors, including Oct-1, AP-1 and the GATA family (Figure 4).
Figure 4. Nucleotide sequence of human LOX-1 promoter region between nt −1599 and −1494.
Three potential transcription factor (Oct-1, AP-1 and GATA family) binding motifs exist in this region and are underlined.
To determine the transcription factor(s) binding specificity of this 106 bp LOX-1 promoter region and to determine the specific transcription factor(s) responsible for ox-LDL-induced LOX-1 promoter activation, we incubated nuclear extracts from ox-LDL-treated and -untreated HCAECs with radiolabelled 106 bp LOX-1 promoter fragment and performed EMSA. There was no shifted band without ox-LDL treatment. However, a shifted band was observed after ox-LDL treatment, implying the binding of transcription factor(s) (Figure 5A). We also incubated nuclear extracts with radiolabelled oligonucleotides containing putative Oct-1, AP-1 or GATA family binding motifs. There was no shifted band for all probes without ox-LDL treatment. However, after ox-LDL treatment, a shifted band was observed with radiolabelled oligonucleotides containing Oct-1 binding motif, while there was still no shifted band with radiolabelled oligonucleotides containing AP-1 or GATA family binding motifs (Figure 5A).
Figure 5. Analysis of potential cis-regulatory element(s) responsible for ox-LDL-induced LOX-1 promoter activation by EMSA, supershift and competition assays.
(A) Radiolabelled probes containing either 106 bp LOX-1 promoter fragment, putative Oct-1 binding motif, AP-1 binding motif or GATA family binding motif were used respectively. After ox-LDL treatment, shifted band only appeared with probes containing either 106 bp LOX-1 promoter fragment or putative Oct-1 binding motif. (B) In the supershift assay, ox-LDL-induced shifted band was attenuated by the preincubation of nuclear extracts with anti-Oct-1 antibody, but was not affected by normal rabbit IgG. The upper panel is representative of five independent experiments, and the lower panel is the summary of data from these experiments (means±S.D.). (C) In the competition assay, 100-fold excess of non-radiolabelled probes were incubated with nuclear extracts 10 min before the addition of radiolabelled probes. The ox-LDL-induced shifted band was totally blocked by non-radiolabelled competitors, but mutant competitors had no effects.
The binding of Oct-1 to the 106 bp LOX-1 promoter fragment was verified by EMSA–supershift assay using anti-Oct-1 antibody. Although we did not observe a supershifted band as we expected, we found that the preincubation of nuclear extracts with anti-Oct-1 antibody attenuated the ox-LDL-induced shifted band in a concentration-dependent manner. Note that 5 μg of antibody significantly abolished the ox-LDL-induced shifted band. Importantly, rabbit IgG, serving as a control, did not affect ox-LDL-induced shifted band (Figure 5B).
The binding specificity of Oct-1 to LOX-1 promoter was further verified by EMSA-competition assay. We identified that the ox-LDL-induced shifted bands with radiolabelled 106 bp fragment and probes containing Oct-1 binding motif were both blocked by the presence of non-radiolabelled competitors (Figure 5C). However, 100-fold excess of mutant non-radiolabelled probes, with a mutation in Oct-1 binding motif, had no such effect (Figure 5C). We also used 100-fold excess of non-radiolabelled 106 bp LOX-1 promoter fragment as competitors, and observed that the shifted bands with radiolabelled 106 bp fragment and probes containing Oct-1 binding motif were both blocked by the competitors (Figure 5C).
Mutations within the Oct-1 binding motif knock out ox-LDL-induced LOX-1 promoter activation
The role of Oct-1 in ox-LDL-induced activation of human LOX-1 promoter was further addressed by mutational analysis. Two site-directed substitution mutations were made in the potential Oct-1 binding motif within the 106 bp LOX-1 promoter fragment, changing the original Oct-1 binding motif (5′-ATTTTTCA-3′) to a mutant binding motif (5′-ATGGTTCA-3′) (Figure 6A). We found that the mutant 106 bp LOX-1 promoter lost its transcription factor(s) binding ability in response to ox-LDL (Figure 6B). Furthermore, we found that this mutant 106 bp LOX-1 promoter was barely activated by ox-LDL, unlike wild-type 106 bp LOX-1 promoter (Figure 6C).
Figure 6. Mutation analysis of the role of Oct-1 in ox-LDL-induced LOX-1 promoter activation.
(A) Nucleotide sequence of mutant 106 bp LOX-1 promoter fragment. There are two nucleotide mutations (from TT to GG) within Oct-1 binding motif. (B) EMSA showed that the mutant 106 bp LOX-1 promoter fragment lost its transcription factor(s) binding induced by ox-LDL treatment, unlike wild-type 106 bp LOX-1 promoter fragment. The panel is representative of five independent experiments. (C) Mutant 106 bp LOX-1 promoter fragment was not activated by ox-LDL, unlike wild-type 106 bp LOX-1 promoter fragment. Results shown are means±S.D. for five independent experiments.
Analysis of Ang II-induced human LOX-1 promoter activation
The same experimental procedures were employed to identify the nucleotide sequence required for Ang II-induced LOX-1 promoter activation.
Compared with the basal state, the full-length LOX-1 promoter (LOX-1−2336/+36) was activated by Ang II treatment (1 μmol/l for 24 h), while all 5′ deletion LOX-1 promoters (LOX-1−1999/+36, LOX-1−1500/+36, LOX-1−996/+36 and LOX-1−498/+36) were not (Figure 7A). Meanwhile, all 3′ deletion LOX-1 promoters (LOX-1−2336/−477, LOX-1−2336/−1059, LOX-1−2336/−1502 and LOX-1−2336/−1990) were activated by Ang II. Again, the core promoter (LOX-1−36/+35) was not activated by Ang II (Figure 7B). Accordingly, we narrowed the LOX-1 promoter region, which responds to Ang II, to a 347 bp fragment, between nt −2336 and −1990. Note that this region (between nt −2336 and −1990) is different from the region (between nt −1599 and −1494) that is required for ox-LDL-induced LOX-1 promoter activation (Figure 7C).
Figure 7. Analysis of LOX-1 promoter activation in response to Ang II.
(A, B) Full-length and 5′ and 3′ deletion LOX-1 promoter–luciferase constructs were transfected into HCAECs, followed by the treatment with 1 μmol/l Ang II for 24 h. The promoter activities were measured and the means±S.D. for five independent experiments are shown. (C) A diagram showing that different promoter regions are required for LOX-1 promoter activation in response to ox-LDL and Ang II.
DISCUSSION
The present study has demonstrated that the nucleotide sequence required for basal human LOX-1 promoter activity is localized in the region between nt −36 and +35. More importantly, this study showed that the cis-regulatory element, which is responsible for ox-LDL-induced LOX-1 promoter activation, resides in the region between nt −1599 and −1494. Furthermore, we identified that a binding motif for the transcription factor Oct-1 is present within this region and that Oct-1 binding appears to correlate with ox-LDL-induced LOX-1 promoter activation. On the other hand, we found that the nucleotide sequence required for Ang II-induced LOX-1 promoter activation localizes in the region between nt −2336 and −1990, suggesting that different mechanisms are involved in ox-LDL- and Ang II-induced LOX-1 promoter activation.
Analysis of basal human LOX-1 promoter activity
Several in vitro studies have demonstrated that under basal states, LOX-1 gene expression (mRNA and protein) is very low in HCAECs [3–6]. The present study, together with others [13,16], provides solid evidence that LOX-1 promoter is constitutively active, but at a relative low level, and that it is activated by ox-LDL and Ang II (Figures 2A and 7A).
In order to identify the specific nucleotide sequence for basal promoter activity, we compared the activity of full-length human LOX-1 promoter and a series of 5′ and 3′ LOX-1 promoter deletion mutants and found that the core promoter (LOX-1−36/+35) has the same activity as full-length LOX-1 promoter (Figure 1B). Accordingly, we propose that LOX-1 core promoter is required and sufficient for basal LOX-1 promoter activity as well as basal gene expression. Within this core promoter are the transcription initiation site and TATA box (TATTTAAA) located at nt −29. Besides the TATA box, a CAAT box (CCAAT) was identified at nt −99, upstream of the core promoter [13]. The presence of these elements suggests that LOX-1 is not a housekeeping gene but an inducible gene. The requirement of the core promoter for basal LOX-1 promoter activity is somewhat inconsistent with two previously published studies [13,16]. One study showed that the cis-regulatory elements required for basal LOX-1 promoter activity exist in two regions. One is from nt −150 to −90 containing GC box and CAAT box, and the other is from nt −50 to +30 containing TATA box [13]. Another study showed that it is the region between nt −150 and −81 that is important for basal LOX-1 promoter activity [16]. Both studies were carried out in HeLa cells, a cervical carcinoma cell line, and not the primary vascular endothelial cells that were used in the present study. Different cells may respond differently to the same stimulus [17]. On the other hand, the transfection of primary endothelial cells with plasmid is more difficult than that of HeLa cells. For HCAECs, the transfection efficiency is approx. 10–25%, but for HeLa cells, the transfection efficiency could be as high as 90%. As such, the discrepancy between present study and studies by others may be explained by the use of different cell lines. Furthermore, it has been widely acknowledged that TATA box, CAAT box and GC box are all potential targets for Sp-1 (stimulatory protein-1), a ubiquitous and constitutively active transcription factor that binds to the promoters of many genes [18]. Based on our present findings (requirement of TATA box for basal LOX-1 promoter activity) and the previous ones (requirement of CAAT and GC boxes for basal LOX-1 promoter activity), we believe that transcription factor Sp-1 may play an important role in basal LOX-1 promoter activity and gene expression. However, this needs to be confirmed by additional studies.
Analysis of ox-LDL- and Ang II-induced LOX-1 promoter activation
Increasing evidence has suggested that ox-LDL/hyperlipidaemia and Ang II/RAS activation are two major stimuli for LOX-1 expression and atherosclerosis [1,3,9–11]. However, it is not known if the regulation of LOX-1 gene expression is transcriptional or translational or both. Although LOX-1 mRNA has been shown to be up-regulated by ox-LDL [9] and Ang II [10], it is not clear whether this effect is due to the stabilization of LOX-1 mRNA or increased transcription of LOX-1 mRNA. Our study shows that the human LOX-1 promoter is activated by ox-LDL and Ang II. Our main purpose was to identify further the specific cis-regulatory element(s) and associated transcription factor(s), which are responsible for ox-LDL-induced LOX-1 promoter activation.
We first compared ox-LDL-induced activation of full-length human LOX-1 promoter and a series of 5′ and 3′ LOX-1 promoter deletion mutants. The results (Figure 2) suggested that the nucleotide sequence that responds to ox-LDL lies in the promoter region between nt −1999 and −1500. After ox-LDL treatment, the LOX-1 promoter (LOX-1−1999/−1494) containing only this region had the same activity as full-length LOX-1 promoter (Figures 2A and 3A).
In order to determine the specific cis-regulatory element(s) responsible for ox-LDL-induced LOX-1 promoter activation, we further analysed the LOX-1 promoter (LOX-1−1999/−1494) by comparing a series of 5′ and 3′ deletion mutants. The results (Figure 3) suggested the localization of ox-LDL-responsive cis-regulatory element(s) in the region between nt −1599 and −1494, which is a 106 bp fragment. This 106 bp promoter fragment, like full-length LOX-1 promoter, can be significantly activated by ox-LDL (Figures 2A and 6C). However, it is to be noted that the LOX-1 core promoter (LOX-1−35/+36), containing TATA box as well as transcription initiation site, is indispensable for ox-LDL-induced LOX-1 promoter activation, because all LOX-1 promoter constructs without the core promoter cannot be activated by ox-LDL (results not shown). We postulate that, after ox-LDL treatment, there is a conformational change within the LOX-1 promoter. It is induced by transcription factor(s) and/or cofactor(s) binding, bringing the region between nt −1599 and −1494 to the proximity of core promoter. This conformational change and the interaction among transcription factors lead to the activation of LOX-1 promoter.
Previous studies have demonstrated that both ox-LDL and Ang II stimulate intracellular oxidative stress and induce the expression of LOX-1 in endothelial cells [19]. Furthermore, pretreatment of endothelial cells with antioxidants attenuates ox-LDL- and Ang II-induced LOX-1 expression [19], implying that ox-LDL and Ang II stimulate LOX-1 expression possibly through the same signalling pathway, which is associated with oxidative stress. As such, we hypothesized that Ang II activates LOX-1 promoter through the same promoter region as ox-LDL. To examine this hypothesis, we used the same methodology to examine the promoter region responsible for Ang II-induced LOX-1 promoter activation. Unexpectedly, we found that another region, between nt −2336 and −1990, is required for LOX-1 promoter activation in response to Ang II. There could be two explanations for this observation. First, ox-LDL and Ang II induce intracellular oxidative stress, which then induces the activation of the same transcription factor. The binding motif for this specific transcription factor is present within the region between nt −1599 and −1494 (required for ox-LDL-induced promoter activation), and within the region between nt −2336 and −1990 (required for Ang II-induced promoter activation). After ox-LDL or Ang II treatment, the activation of this specific transcription factor causes the LOX-1 promoter activation followed by LOX-1 gene expression. The other possibility is that oxidative stress in response to ox-LDL and Ang II may induce the activation of different transcription factors. The binding motif for one exists in the region between nt −1599 and −1494, and the binding motif for the other exists in the region between nt −2336 and −1990. Accordingly, although ox-LDL and Ang II cause the same initial signalling, different downstream steps seem to be involved in ox-LDL- and Ang II-induced LOX-1 promoter activation. To examine these two possibilities, further experiments are required to examine the activation of potential transcription factors in response to ox-LDL and Ang II in endothelial cells.
Central role of Oct-1 in ox-LDL-induced LOX-1 promoter activation
Computer analysis showed that within the 106 bp promoter region, which is required for ox-LDL-induced LOX-1 promoter activation, there are at least three potential transcription factor binding motifs, including Oct-1 binding motif and AP-1 binding motif as well as GATA family binding motif (Figure 4). Although the presence of these transcription factor binding motifs within the LOX-1 promoter has already been suggested by previous studies [13,16,20], it is merely based on computer analysis. Furthermore, ox-LDL has been shown to activate transcription factors including NF-κB and AP-1 in endothelial cells [19]; however, there is no solid evidence presented to clearly support their roles in ox-LDL-induced LOX-1 promoter activation and LOX-1 gene expression.
We examined the role of these transcription factors by performing EMSA, supershift and competition assays and found that only Oct-1, but not AP-1 or GATA family, was activated by ox-LDL in HCAECs (Figure 5). In the supershift assay, although we did not observe a supershifted band after preincubation with anti-Oct-1 antibody, we found that the ox-LDL-induced shifted band was attenuated by the presence of the antibody in a concentration-dependent manner. This may be explained by the interaction between Oct-1 and its antibody affecting its DNA binding affinity. Subsequently, the role of Oct-1 in ox-LDL-induced LOX-1 promoter activation was further confirmed by mutagenesis assay (Figures 6B and 6C).
Oct-1 belongs to a transcription factor family, POU domain family, which is composed of members that share a common domain called POU domain. There are five classes of POU domain family proteins, of which Oct-1 is ubiquitously expressed and functions as either a positive or a negative transcriptional regulator of a wide range of cellular genes, including housekeeping genes and tissue-specific genes. These genes encode products participating in a variety of cellular developmental processes such as cell division, differentiation, specification and survival. The regulation of these genes is dependent on the interaction between Oct-1 and the octamer motifs (ATTTGCAT or its variants) present in the promoter or enhancer of these genes [21,22]. Like other transcription factors, Western blotting and immunoprecipitation analyses revealed that the phosphorylation/dephosphorylation of Oct-1 is a common mechanism that regulates the binding of itself to octamer motifs and the activation of target gene transcription. Previous studies have shown that Oct-1 is phosphorylated in cell-cycle-dependent, cell-type-dependent, protein kinase-dependent and sequence-dependent manners [23]. For instance, PKA (protein kinase A)-mediated phosphorylation of Oct-1 inhibits its DNA-binding activity, while PKC- and CK2 (protein kinase CK2)-mediated phosphorylation of Oct-1 enhances its DNA-binding activity [23]. Furthermore, in addition to protein kinases, oxidation/reduction might be another mechanism whereby the phosphorylation of Oct-1 and Oct-1-mediated transcription of a specific gene is regulated [23]. As discussed earlier, ox-LDL induces intracellular oxidative stress. As such, we postulate that the following steps are involved in ox-LDL-induced LOX-1 expression: ox-LDL→oxidative stress→Oct-1 phosphorylation→Oct-1 binding to DNA→target gene (LOX-1) transcription. In addition to phosphorylation, on activation, Oct-1 has been demonstrated to regulate, positively or negatively, a variety of tissue-specific genes by recruiting tissue-specific cofactors [22]. In vascular endothelial cells, Oct-1 negatively regulates von Willebrand factor and VCAM-1 (vascular cell adhesion molecule-1) gene expression [17], whereas it positively regulates fgl2/fibroleukin [24], endothelial cell-specific TIE2 [25] and inducible nitric oxide synthase gene expression [26]. As a positively acting transcription factor, Oct-1 has been shown to enhance gene expression through interaction with a variety of DNA-binding protein partners, basal transcription factors or tissue-specific coactivators. Sp-1 family members and NF-κB have been shown to interact with Oct-1 in regulating gene expression [24–26]. We found that LOX-1 core promoter and promoter region containing Oct-1 binding motif are both required for ox-LDL-induced LOX-1 promoter activation. Interestingly, as mentioned earlier, the TATA box within the LOX-1 core promoter is a potential binding site for Sp-1. We, therefore, postulate that Sp-1 might be constitutively recruited to LOX-1 core promoter under basal states. After ox-LDL treatment, the binding of Oct-1 to LOX-1 promoter and the interaction between Oct-1 and Sp-1 facilitate LOX-1 promoter activation and gene expression.
In HCAECs, NF-κB is activated by ox-LDL and has been shown to be involved in the ox-LDL-induced up-regulation of several genes [6,27]. Interestingly, we did not find the correlation between NF-κB and ox-LDL-induced LOX-1 promoter activation in the present study. Although we observed NF-κB activation by ox-LDL (results not shown), its binding motif is not present within the 106 bp LOX-1 promoter region. More importantly, in the EMSA, the ox-LDL-induced shifted band with radiolabelled 106 bp probes was not affected by non-radiolabelled probes containing NF-κB binding motif (results not shown). These observations point against a major role for NF-κB in ox-LDL-induced LOX-1 promoter activation. Nonetheless, we cannot totally exclude its role in this process. In contrast, a previous computer analysis study suggested a potential binding motif for NF-κB within the LOX-1 promoter between nt −2336 and −1990 [13], which is required for Ang II-induced LOX-1 promoter activation. Consistent with this report, we previously showed the activation of NF-κB by Ang II [19]. Although we do not have direct evidence suggesting a role for NF-κB in Ang II-induced LOX-1 promoter activation so far, we are currently studying the correlation between Ang II, NF-κB and LOX-1 promoter activation. In general, different transcription factors seem to be involved in LOX-1 promoter activation in response to ox-LDL and Ang II. Considering the fact that there is a ‘cross-talk’ between ox-LDL and Ang II in inducing LOX-1 expression and atherosclerosis, it is highly possible that an interaction between Oct-1 and NF-κB exists and that NF-κB enhances the activity of Oct-1, particularly when both stimuli (ox-LDL and Ang II) coexist.
Besides NF-κB, ox-LDL also activates several other transcription factors in vascular endothelial cells, such as STAT (signal transducer and activators of transcription), ATF-2 (activating transcription factor 2), ELK-1 (ets-like gene-1), CREB (cAMP-response-element-binding protein) and PPAR (peroxisome-proliferator-activated receptor) [28–30]. However, their binding motifs are not present within the 106 bp LOX-1 promoter region. The role of AP-1 in ox-LDL-treated HCAECs is also controversial [6,31]. In the present study, we did not find the activation of AP-1 by ox-LDL (Figure 5A), although a potential binding motif for AP-1 is present within LOX-1 promoter.
In the present study, the binding of Oct-1, AP-1 and the GATA family to DNA motifs was not detected under basal states (Figure 5A). We believe that these transcription factors are constitutively active in HCAECs, but to a low extent. In the EMSA, we used 2 μg of nuclear extracts. The small amount of nuclear extracts may explain the lack of transcription factor binding detected under basal states.
Since the binding of transcription factor(s) to the 106 bp LOX-1 promoter fragment was totally blocked by non-radiolabelled probes containing putative Oct-1 binding motif (Figure 5C) and the mutant 106 bp LOX-1 promoter (without Oct-1 binding) totally lost its activation in response to ox-LDL (Figure 6C), we conclude that Oct-1 is the key transcription factor that is required for ox-LDL-induced LOX-1 promoter activation. However, the role of Oct-1 in ox-LDL-induced LOX-1 promoter activation and gene expression needs to be further studied. Studies need to be performed to determine (i) whether ox-LDL treatment increases Oct-1 mRNA or protein expression and/or protein phosphorylation, (ii) whether ox-LDL induces the translocation of Oct-1 from the cytoplasm to the nucleus and (iii) whether other transcription factors or cofactors also contribute to ox-LDL-induced activation of LOX-1 promoter.
Summary
In the present study, human LOX-1 promoter activity was studied in HCAECs. We found that the nucleotide sequence for LOX-1 basal promoter activity resides within the region between nt −36 and +35. More importantly, we identified that a 105 bp region of the LOX-1 promoter, from nt −1599 to −1494, along with the core promoter, is required for ox-LDL-induced LOX-1 promoter activation. Within this region there is an Oct-1 binding motif that binds Oct-1. Mutation of this motif disrupts Oct-1 binding and eliminates ox-LDL-induced activation of the LOX-1 promoter. These results strongly suggest that Oct-1 plays an important role in ox-LDL-induced LOX-1 promoter activation in HCAECs. In addition, the present study suggests that the promoter region for Ang II-induced LOX-1 promoter activation is different from the region required for ox-LDL-induced LOX-1 promoter activation.
Acknowledgments
This work was supported by a Scientist Development grant from the American Heart Association and a Merit Review grant from the Department of Veterans' Affairs.
References
- 1.Singh B. M., Mehta J. L. Interactions between the renin-angiotensin system and dyslipidemia: relevance in the therapy of hypertension and coronary heart disease. Arch. Intern. Med. 2003;163:1296–1304. doi: 10.1001/archinte.163.11.1296. [DOI] [PubMed] [Google Scholar]
- 2.Naderi G. A., Asgary S., Shirvany H. Anti-oxidant effect of flavonoids on the susceptibility of LDL oxidation. Mol. Cell. Biochem. 2003;246:193–196. [PubMed] [Google Scholar]
- 3.Mehta J. L., Li D. Identification and autoregulation of receptor for OX-LDL in cultured human coronary artery endothelial cells. Biochem. Biophys. Res. Commun. 1998;248:511–514. doi: 10.1006/bbrc.1998.9004. [DOI] [PubMed] [Google Scholar]
- 4.Li D., Liu L., Chen H., Sawamura T., Ranganathan S., Mehta J. L. LOX-1 mediates oxidized low-density lipoprotein-induced expression of matrix metalloproteinases in human coronary artery endothelial cells. Circulation. 2003;107:612–617. doi: 10.1161/01.cir.0000047276.52039.fb. [DOI] [PubMed] [Google Scholar]
- 5.Chen J., Mehta J. L., Haider N., Li D. Role of caspases in ox-LDL-induced apoptotic cascade in human coronary artery endothelial cells. Circ. Res. 2004;94:370–376. doi: 10.1161/01.RES.0000113782.07824.BE. [DOI] [PubMed] [Google Scholar]
- 6.Li D., Chen H., Romeo F., Sawamura T., Mehta J. L. Statins modulate oxidized low-density lipoprotein-mediated adhesion molecule expression in human coronary artery endothelial cells: role of LOX-1. J. Pharmacol. Exp. Ther. 2002;302:601–605. doi: 10.1124/jpet.102.034959. [DOI] [PubMed] [Google Scholar]
- 7.Li D., Liu L., Chen H., Mehta J. L. LOX-1, an oxidized LDL endothelial receptor, induces CD40/CD40L signaling in human coronary artery endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2003;23:816–821. doi: 10.1161/01.ATV.0000066685.13434.FA. [DOI] [PubMed] [Google Scholar]
- 8.Mehta J. L., Li D. Y., Chen H. J., Joseph J., Romeo F. Inhibition of LOX-1 by statins may relate to upregulation of eNOS. Biochem. Biophys. Res. Commun. 2001;289:857–861. doi: 10.1006/bbrc.2001.6070. [DOI] [PubMed] [Google Scholar]
- 9.Li D., Mehta J. L. Upregulation of endothelial receptor for oxidized LDL (LOX-1) by oxidized LDL and implications in apoptosis of human coronary artery endothelial cells: evidence from use of antisense LOX-1 mRNA and chemical inhibitors. Arterioscler. Thromb. Vasc. Biol. 2000;20:1116–1122. doi: 10.1161/01.atv.20.4.1116. [DOI] [PubMed] [Google Scholar]
- 10.Li D. Y., Zhang Y. C., Philips M. I., Sawamura T., Mehta J. L. Upregulation of endothelial receptor for oxidized low-density lipoprotein (LOX-1) in cultured human coronary artery endothelial cells by angiotensin II type-1 receptor activation. Circ. Res. 1999;84:1043–1049. doi: 10.1161/01.res.84.9.1043. [DOI] [PubMed] [Google Scholar]
- 11.Chen H., Li D., Sawamura T., Inoue K., Mehta J. L. Upregulation of LOX-1 expression in aorta of hypercholesterolemic rabbits: modulation by losartan. Biochem. Biophys. Res. Commun. 2000;276:1100–1104. doi: 10.1006/bbrc.2000.3532. [DOI] [PubMed] [Google Scholar]
- 12.Chen J., Li D., Schaefer R., Mehta J. L. Cross-talk between dyslipidemia and rennin–angiotensin system and the role of LOX-1 and MAPK in atherogenesis: studies with the combined use of rosuvastatin and candesartan. Atherosclerosis. 2005 doi: 10.1016/j.atherosclerosis.2005.04.016. in the press. [DOI] [PubMed] [Google Scholar]
- 13.Aoyama T., Sawamura T., Furutani Y., Matsuoka R., Fujiwara H., Masaki T. Structure and chromosomal assignment of the human lectin-like oxidized low-density-lipoprotein receptor-1 (LOX-1) gene. Biochem. J. 1999;39:177–184. [PMC free article] [PubMed] [Google Scholar]
- 14.Kita T. LOX-1, a possible clue to the missing link between hypertension and atherogenesis. Circ. Res. 1999;84:1113–1115. doi: 10.1161/01.res.84.9.1113. [DOI] [PubMed] [Google Scholar]
- 15.Li D., Yang B., Mehta J. L. Ox-LDL induces apoptosis in human coronary artery endothelial cells: role of PKC, PTK, bcl-2, and Fas. Am. J. Physiol. 1998;275:H568–H576. doi: 10.1152/ajpheart.1998.275.2.H568. [DOI] [PubMed] [Google Scholar]
- 16.Yamanaka S., Zhang X. Y., Miura K., Kim S., Iwao H. The human gene encoding the lectin-type oxidized LDL receptor (OLR1) is a novel member of the natural killer gene complex with a unique expression profile. Genomics. 1998;54:191–199. doi: 10.1006/geno.1998.5561. [DOI] [PubMed] [Google Scholar]
- 17.Schwachtgen J. L., Remacle J. E., Janel N., Brys R., Huylebroeck D., Meyer D., Kerbiriou-Nabias D. Oct-1 is involved in the transcriptional repression of the von Willebrand factor gene promoter. Blood. 1998;92:1247–1258. [PubMed] [Google Scholar]
- 18.Osborne T. F., Gil G., Goldstein J. L., Brown M. S. Operator constitutive mutation of 3-hydroxy-3-methylglutaryl coenzyme A reductase promoter abolishes protein binding to sterol regulatory element. J. Biol. Chem. 1988;263:3380–3387. [PubMed] [Google Scholar]
- 19.Mehta J. L., Hu B., Chen J., Li D. Pioglitazone inhibits LOX-1 expression in human coronary artery endothelial cells by reducing intracellular superoxide radical generation. Arterioscler. Thromb. Vasc. Biol. 2003;23:2203–2208. doi: 10.1161/01.ATV.0000094411.98127.5F. [DOI] [PubMed] [Google Scholar]
- 20.Nagase M., Abe J., Takahashi K., Ando J., Hirose S., Fujita T. Genomic organization and regulation of expression of the lectin-like oxidized low-density lipoprotein receptor (LOX-1) gene. J. Biol. Chem. 1998;273:33702–33707. doi: 10.1074/jbc.273.50.33702. [DOI] [PubMed] [Google Scholar]
- 21.Ortego M., Hernandez A. G., Bustos C., Tunon J., Egido J. 3-Hydroxy-3-methylglutaryl coenzyme A reductase inhibitors increase the binding activity and nuclear level of Oct-1 in mononuclear cells. Eur. J. Pharmacol. 2002;448:113–121. doi: 10.1016/s0014-2999(02)01938-6. [DOI] [PubMed] [Google Scholar]
- 22.Zhao F. Q., Zheng Y., Dong B., Oka T. Cloning, genomic organization, expression, and effect on beta-casein promoter activity of a novel isoform of the mouse Oct-1 transcription factor. Gene. 2004;326:175–187. doi: 10.1016/j.gene.2003.10.023. [DOI] [PubMed] [Google Scholar]
- 23.Grenfell S. J., Latchman D. S., Thomas N. S. Oct-1 and Oct-2 DNA-binding site specificity is regulated in vitro by different kinases. Biochem. J. 1996;315:889–893. doi: 10.1042/bj3150889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu M., Leibowitz J. L., Clark D. A., Mendicino M., Ning Q., Levy G. A. Gene transcription of fgl2 in endothelial cells is controlled by Ets-1 and Oct-1 and requires the presence of both Sp1 and Sp3. Eur. J. Biochem. 2003;270:2274–2286. doi: 10.1046/j.1432-1033.2003.03595.x. [DOI] [PubMed] [Google Scholar]
- 25.Fadel B. M., Boutet S. C., Quertermous T. Octamer-dependent in vivo expression of the endothelial cell-specific TIE2 gene. J. Biol. Chem. 1999;274:20376–20383. doi: 10.1074/jbc.274.29.20376. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y. M., Ko C. B., Park Y. P., Kim Y. J., Paik S. G. Octamer motif is required for the NF-κB-mediated induction of the inducible nitric oxide synthase gene expression in RAW 264.7 macrophages. Mol. Cell. 1999;9:99–109. [PubMed] [Google Scholar]
- 27.Li D., Saldeen T., Romeo F., Mehta J. L. Oxidized LDL upregulates angiotensin II type 1 receptor expression in cultured human coronary artery endothelial cells: the potential role of transcription factor NF-κB. Circulation. 2000;102:1970–1976. doi: 10.1161/01.cir.102.16.1970. [DOI] [PubMed] [Google Scholar]
- 28.Maziere C., Conte M. A., Maziere J. C. Activation of JAK2 by the oxidative stress generated with oxidized low-density lipoprotein. Free Radical Biol. Med. 2001;31:1334–1340. doi: 10.1016/s0891-5849(01)00649-9. [DOI] [PubMed] [Google Scholar]
- 29.Napoli C., Quehenberger O., De Nigris F., Abete P., Glass C. K., Palinski W. Mildly oxidized low density lipoprotein activates multiple apoptotic signaling pathways in human coronary cells. FASEB J. 2000;14:1996–2007. doi: 10.1096/fj.99-0986com. [DOI] [PubMed] [Google Scholar]
- 30.Hayashida K., Kume N., Minami M., Inui-Hayashida A., Mukai E., Toyohara M., Kita T. Peroxisome proliferator-activated receptor alpha ligands activate transcription of lectin-like oxidized low density lipoprotein receptor-1 gene through GC box motif. Biochem. Biophys. Res. Commun. 2004;323:1116–1123. doi: 10.1016/j.bbrc.2004.08.193. [DOI] [PubMed] [Google Scholar]
- 31.Cho S., Hazama M., Urata Y., Goto S., Horiuchi S., Sumikawa K., Kondo T. Protective role of glutathione synthesis in response to oxidized low density lipoprotein in human vascular endothelial cells. Free Radical Biol. Med. 1999;26:589–602. doi: 10.1016/s0891-5849(98)00232-9. [DOI] [PubMed] [Google Scholar]