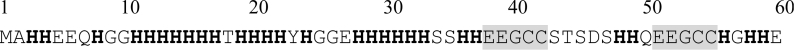Abstract
Hpn is a small cytoplasmic protein found in Helicobacter pylori, which binds Ni2+ ions with moderate affinity. Consisting of 60 amino acids, the protein is rich in histidine (28 residues, 46.7%), as well as glutamate, glycine and serine residues (in total 31.7%), and contains short repeating motifs. In the present study, we report the detailed biophysical characterization of the multimeric status and Ni2+-binding properties of purified recombinant Hpn under physiologically relevant conditions. The protein exists as an equilibration of multimeric forms in solution, with 20-mers (approx. 136 kDa) being the predominant species. Using equilibrium dialysis, ICP-MS (inductively coupled plasma MS) and UV/visible spectroscopy, Hpn was found to bind five Ni2+ ions per monomer at pH 7.4, with a dissociation constant (Kd) of 7.1 μM. Importantly, Ni2+ binding to Hpn is reversible: metal is released either in the presence of a chelating ligand such as EDTA, or at a slightly acidic pH (pH for half dissociation, pH1/2 ∼6.3). Ni2+ binding induces conformational changes within the protein, increasing β-sheet and reducing α-helical content, from 22% to 37%, and 20% to 10% respectively. Growth curves of Escherichia coli BL21(DE3) both with and without the hpn gene performed under Ni2+ pressure clearly implied a role for Hpn to protect the cells from higher concentrations of external metal ions. Similarly, the accumulation of Ni2+ in these cells expressing Hpn from a plasmid was approx. 4-fold higher than in uninduced controls or control cultures that lacked the plasmid. Similarly, levels of Ni2+ in wild-type H. pylori 26695 cells were higher than those in H. pylori hpn-deletion mutant strains. Hpn may potentially serve multiple roles inside the bacterium: storage of Ni2+ ions in a ‘reservoir’; donation of Ni2+ to other proteins; and detoxification via sequestration of excess Ni2+.
Keywords: Helicobacter pylori, histidine-rich protein, Hpn, nickel, storage
Abbreviations: ABC, ATP-binding cassette; Amp, ampicillin; DTT, dithiothreitol; ICP-MS, inductively coupled plasma MS; IPTG, isopropyl β-D-thiogalactoside; LB, Luria–Bertani; LMCT, ligand-to-metal charge transfer; MALDI, matrix-assisted laser-desorption ionization; MT, metallothionein; TTBS, Tris-buffered saline with Tween 20; UV/vis, UV/visible
INTRODUCTION
Helicobacter pylori is a Gram-negative bacterium that colonizes the gastric mucosa of humans [1], and has been shown to be the causative agent of type B gastritis and peptic ulcerations [2–4]. The development of gastric carcinoma and MALT (mucosa-associated lymphoid tissue) lymphoma are also strongly associated with chronic H. pylori infection [5,6]. To colonize and establish infection, H. pylori must produce an assortment of factors to generate a neutral pH in order to survive the extremely acidic environment in the stomach. One of them is a membrane-bound [Ni–Fe] hydrogen-uptake hydrogenase, which permits respiratory-based energy production for the bacteria in the mucosa [7,8]. Another is urease, a dinuclear Ni2+-containing metalloenzyme that accounts for up to 6% of the soluble cellular proteins [9–11], which catalyses the hydrolysis of urea to yield ammonia and carbamate to neutralize the gastric acid, critical for H. pylori's colonization and survival at acidic pH [12].
A constant supply of Ni2+ ions into H. pylori is therefore required for the synthesis and activity of these Ni2+-containing metalloenzymes. To date, members of two general types of Ni2+-specific, membrane-integrated uptake systems have been identified in H. pylori [13]: (i) the multiple-component ABC (ATP-binding cassette) transporters, which are believed to be a four-gene operon designated as abcABCD [14]; and (ii) the nickel–cobalt transporter family, comprising the putatively monomeric NixA protein [15].
However, when excess Ni2+ ions accumulate, they inhibit growth and exhibit toxic effects as a result of interference with normal protein–metal binding and catalysis, as well as the generation of reactive oxygen species [16–18]. The toxicity of metal ions is minimized through the strict control of their intracellular concentrations, which is generally achieved through two processes: regulation of ion transport, i.e. import and efflux from the cell, and sequestration, through the formation of tight complexes with metal-storage proteins [19,20]. As in other bacteria, H. pylori has to strike a delicate balance between the import of Ni2+ ions, their efficient intracellular storage and delivery to Ni2+-dependent metalloenzymes when required.
The completion of the H. pylori genome sequence has revealed the presence of several putative ion-binding proteins and membrane transporters that are involved in the transport and cytoplasmic accumulation of bivalent cations [21]. As mentioned above, integral membrane proteins, such as the NixA and the abcABCD transporters, scavenge Ni2+ ions from the surrounding milieu and import them into the cell. In addition, two proteins with a high affinity for Ni2+ ions have been characterized in H. pylori: the HspA heat-shock protein, a bacterial GroES homologue with a unique histidine-rich C-terminal domain [22], and an unusual protein called Hpn, which is abundant in the cell cytoplasm, accounting for approx. 2% of all protein synthesized [23]. As described previously [23], Hpn consists of 60 amino acids, including 28 histidine residues (Figure 1). The majority of the histidine residues are located within the central part of the protein (from residue 11 to residue 33) and include two separate stretches of six and seven consecutive histidine residues. There are two internal short repeats of Glu-Glu-Gly-Cys-Cys, at positions 38–42 and 51–55, four sets of paired histidine residues (one near the N-terminus, three towards the C-terminus) and also an Xaa-Xaa-His motif at the N-terminus. All these sequence features indicate that this protein would bind metal ions strongly. Mutated strains of H. pylori lacking the hpn gene are four times more sensitive to ranitidine bismuth citrate, a metal-containing drug widely used to treat H. pylori infections, than the wild-type [24,25]. Recently, the H. pylori Ni2+-uptake regulatory protein NikR was found to up-regulate not only the expression of the urease subunit genes, but also the expression of the nixA and hpn genes in the presence of excess Ni2+ [20]. This suggested that H. pylori may respond in a co-ordinated manner to the presence of excess extracellular Ni2+, to allow the optimal import of Ni2+ ions (up-regulation of the nixA gene), optimal incorporation of Ni2+ into other proteins, as well as optimal storage of the free Ni2+ ions (up-regulation of the hpn gene) to minimize any toxic effects due to their accumulation [20].
Figure 1. Amino acid sequence of Hpn from H. pylori.
Histidine residues are in bold, and the two EEGCC pentapeptide repeats are grey-shaded.
In the present paper, we describe the cloning, expression, purification and characterization of the Hpn protein from H. pylori. Most notably, our results indicate that the Hpn protein exists in an equilibration of multimeric states in solution, and binds Ni2+ ions reversibly.
MATERIALS AND METHODS
Bacterial strains and growth conditions
H. pylori 26695 was a gift from Department of Medicine, the University of Hong Kong, and was cultured on Brucella agar (Difco) plates supplemented with 10% defibrinated sheep blood at 37 °C under microaerophilic conditions (5% CO2, 4% O2 and 91% N2), maintained by CampyPak Plus (BBL). Plasmid DNA was maintained in Escherichia coli DH10B. E. coli BL21(DE3) (Stratagene) was used to express the hpn gene. Amp (ampicillin) (100 μg/ml) and IPTG (isopropyl β-D-thiogalactoside) (0.5 mM) were added to LB (Luria–Bertani) broth (USB) when indicated. Chromosomal hpn-deletion mutants were constructed as described previously by electroporating plasmid pHPN2KO (kindly supplied by A. G. Plaut, GRASP Digestive Disease Research Center, Tufts-New England Medical Center, Boston, MA, U.S.A.) into H. pylori 26695 cells in 10% (v/v) glycerol (Bio-Rad gene pulser at 12.5 kV/cm, 25 μF, 600 Ω) [26]. Kanamycin-resistant colonies were isolated, and cell lysates of cultures grown from individual clones were analysed for Hpn production by Western blot analysis, to confirm gene deletion.
DNA techniques
Standard DNA manipulations were performed as described by Maniatis et al. [27], or as recommended by the manufacturers. Plasmid DNA was isolated using the QIAprep Spin mini kit (Qiagen). DNA fragments were purified from 1.0% agarose gels [in 1×TAE (Tris/acetate/EDTA)] with the QIAquick gel extraction kit (Qiagen). PCR was performed on a PerkinElmer 2400 thermal cycler using the Expand High Fidelity PCR System (Roche). Oligonucleotide primers were synthesized by Proligo (Singapore). The hpn gene (HP1427) was amplified from unpurified H. pylori 26695 genomic DNA (from a picked colony) by PCR using primer 1 (5′-TATTCCATGGCTCATATGGCACACCATGAAGAACAGCAC-3′) and primer 2 (5′-TATACTCGAGTTACTCGTGATGCCCGTGGCAGCAACC-3′), which introduce NdeI and XhoI restriction sites (underlined) at the 5′- and 3′-ends of the PCR product respectively. After restriction digestion, the PCR products were ligated into the expression vector pET-32a (Novagen), pre-cut with the same enzymes and gel-purified, to generate the plasmid pET-hpn. The plasmid was sequenced using ABI Prism BigDye (Applied Biosystems) and subsequently transformed into E. coli BL21(DE3) as described previously [27] for protein expression.
Expression and purification of Hpn
A saturated overnight culture of pET-hpn/E. coli BL21(DE3) was expanded (1:100) in LB medium supplemented with 100 μg/ml Amp, 0.5 mM NiSO4 and 0.5% glucose, and was grown at 37 °C until a D600 of 0.7 was reached. Protein expression was induced by the addition of IPTG to 0.5 mM, and the cultures were then incubated for a further 4 h at 37 °C, before chilling to 4 °C. Cells were harvested by centrifugation at 5000 g for 20 min at 4 °C and were resuspended in 10 ml of ice-cold Buffer A (20 mM sodium phosphate buffer, pH 7.4, 500 mM NaCl, 100 mM imidazole and 1 mM PMSF) per litre of cell culture. Bacteria were ruptured by sonication in the presence of 1% (v/v) Triton X-100. The lysate was centrifuged at 30000 g for 30 min, and the supernatant was filtered through a 0.45-μm-pore-size cellulose acetate syringe filter (Iwaki Glass) before loading on to a NiSO4-impregnated HiTrap Chelating HP column (5 ml; Amersham Biosciences) that had been pre-equilibrated with 10 column vol. of Buffer A. After washing with 10 column vol. of Buffer A, Hpn protein was eluted with Buffer B (20 mM sodium phosphate buffer, pH 7.4, 500 mM NaCl and 500 mM imidazole). The purest fractions, as determined by SDS/PAGE (15% gel), were pooled, concentrated (Centricon YM-3; Millipore) and then purified further by gel-filtration chromatography on Superdex 75 resin (10/300 column; Amersham Biosciences) in buffer appropriate for subsequent use.
Protein analysis
Protein concentrations were determined by BCA (bicinchoninic acid) assay (Bio-Rad) using BSA as the standard. SDS/PAGE was performed according to the method of Maniatis et al. [27], and proteins were visualized with Coomassie Blue R-250.
Immunoblot analysis was used to determine the amount of Hpn present in the various purified Hpn fractions, using a standard curve constructed from Hpn solutions of known concentrations. Fractions were resolved on SDS/15% polyacrylamide gels then transferred on to PVDF membranes (Hybond-P; Amersham Bioscience). After unoccupied sites were blocked with 5% (w/v) non-fat dried milk in TTBS [Tris-buffered saline with Tween 20: 10 mM Tris/HCl, 150 mM NaCl, 0.01% (v/v) Tween 20], membranes were incubated with His·Tag® monoclonal antibody (1:1000 dilution) (Novagen) as recommended by the manufacturers. The membranes were then washed three times with TTBS, incubated with anti-mouse IgG conjugated to horse-radish peroxidase, and developed using the ECL® (enhanced chemiluminescence) Western blotting kit (Amersham Biosciences). Image acquisition and analysis were performed on an ImageScanner (Amersham Biosciences) using ImageMaster 2D Elite software (Amersham Biosciences).
After resolution on an SDS/15% polyacrylamide gel and Coomassie Blue staining, the band corresponding to Hpn was excised and digested in-gel with chymotrypsin (Sigma), with overnight incubation at 37 °C. Peptide fragments were then analysed by MALDI (matrix-assisted laser-desorption ionization)-MS.
To confirm the N-terminal sequence of the recombinant Hpn, the purified protein was subjected to N-terminal analysis by Edman degradation. Aliquots of Hpn (∼5 μg) was subjected to SDS/PAGE, transferred on to a PVDF membrane, and stained with 0.1% Coomassie Blue R-250, before excision and analysis with an automated HPG1000A liquid-phase protein sequencer (Hewlett-Packard).
The free thiolate content of Hpn was determined by the method of Ellman [28]. Briefly, Hpn protein was added to a 1 mM DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)] solution in 20 mM Hepes, pH 7.4, and 100 mM NaCl, to final concentration of 10 μM. After incubation for 2 h at room temperature (25 °C), the amount of generated p-nitrothiolate was determined using the molar absorption coefficient (ϵ412) of 13600 M−1·cm−1 [28].
Determination of the multimerization state of Hpn
To examine the effects of imidazole and DTT (dithiothreitol), as well as Ni2+ binding, on the multimerization state of Hpn, proteins (50 μM) were injected into a Superose 12 (10/300 column) (Amersham Biosciences) gel-filtration column, using running buffers containing 20 mM Hepes, pH 7.4, and 500 mM NaCl, supplemented with 100 or 500 mM imidazole, 5 mM DTT or 250 μM Ni2+ (as NiSO4) when needed. The proteins were equilibrated in the respective running buffer for at least 1 h before injection into the column. The column was calibrated with vitamin B12 (1.355 kDa), lysozyme (14.3 kDa), β-lactoglobulin (35 kDa), BSA (66 kDa), human transferrin (80 kDa) and IgG (150 kDa). Blue Dextrin was used to determine the void volume. Molecular masses were determined by plotting log molecular masses of the standards against the partition coefficient (Kav), where Kav=(Ve−V0)/(Vt−V0), where Ve represents elution volume, V0 is the void volume, and Vt is the total column volume. The aggregation state of recombinant Hpn protein was also analysed by non-denaturing PAGE, according to the method of Coligan et al. [29], calibrated with high-molecular-mass native markers (Amersham Biosciences) and stained using Coomassie Blue R-250.
UV/vis (visible) spectroscopy
Unless otherwise indicated, UV/vis absorption measurements were performed on samples of 50 μM proteins in 20 mM Hepes, pH 7.4, and 100 mM NaCl, using a Varian Cary 3E UV/vis spectrophotometer. Spectra were recorded over the wavelength range 230–650 nm in cuvettes with a pathlength of 1.0 cm.
CD spectroscopy
CD measurements were measured on a Jasco J-720 spectropolarimeter calibrated with a 0.06% (+)-10-camphorsulphonic acid solution (Sigma–Aldrich), using a 0.1-cm-pathlength quartz cell. CD data were obtained for 5 μM Hpn in Tris/HCl buffer, pH 7.4, in both the absence and the presence of 1–5 molar equivalents of Ni2+ (as NiSO4). Instrument optics and sample chamber were continually flushed with 3 litres/min of dry N2 gas. Instrument settings used were: scan range, 190–250 nm; scan rate, 20 nm/min; wavelength step, 0.25 nm; sensitivity, 5 millidegrees; response time, 8 s. The final spectrum, representing an average of six scans, was corrected for the corresponding buffer and smoothed using adjacent averaging or an FFT (fast Fourier transform) filter. Quantitative estimations of the secondary-structure contents were made using the CDPro software package [30].
Influence of Ni2+ and Hpn on the growth of E. coli
(i) E. coli BL21(DE3) cultures, both with and without the plasmid pET-hpn (inoculated from individual colonies), were grown in LB medium (containing 100 μg/ml Amp when required) at 37 °C. Stationary-phase overnight cultures (1 ml) were added to 50 ml of fresh medium supplemented with 500 μM NiSO4, and were incubated for 24 h at 37 °C, with IPTG added where applicable (to a final concentration of 500 μM) when the D600 was approx. 0.6. Cell densities (determined by D600) were measured after 0, 2, 4, 8, 16 and 24 h.
(ii) E. coli BL21(DE3) overnight cultures (as above, 10 μl) were used to inoculate 1 ml of fresh medium containing 0, 50, 100, 200, 500, 600, 700 or 800 μM NiSO4 (as indicated). Cultures were grown for 18 h at 37 °C, with IPTG added where applicable (to 500 μM), before the final cell density (determined by D600) was measured.
Ni2+ accumulation in E. coli
E. coli BL21(DE3) overnight cultures (as above, 10 μl) were added to 1 ml of fresh medium supplemented with 500 μM NiSO4. Cultures were grown for 18 h at 37 °C, with IPTG (1 M) added when required (to a final concentration of 0.5 mM) when the D600 reached approx. 0.6. Cells were pelleted at 4000 g for 10 min and washed three times with PBS, and the cellular Ni2+ content was determined by ICP-MS (inductively coupled plasma MS) (Agilent 7500) with 207Tl as an internal reference.
Ni2+ accumulation in H. pylori
Wild-type or Δhpn mutant H. pylori 26695 cells from fresh plates were resuspended in 10 ml of Brucella broth supplemented with 5% foetal bovine serum and 0.2 mM NiSO4, and were grown for 48 h at 37 °C. Cells were pelleted at 4000 g for 10 min and washed three times with PBS. The cellular Ni2+ content was then determined by ICP-MS similarly. All of the accumulation experiments were carried out with three replicates and an average value was used.
Ni2+ binding to Hpn
Binding of Ni2+ to Hpn was determined by equilibrium dialysis. The dialysis tubes (molecular-mass cut-off of 1 kDa), filled with 200 μl of protein suspension (5 μM), were placed into 1000 ml of 20 mM Hepes and 100 mM NaCl, pH 7.4, supplemented with a series of concentrations of Ni2+ from 0 to 100 μM, for overnight incubation at 4 °C. The Ni2+ concentrations inside and outside the dialysis tubes were determined by ICP-MS. Each assay was repeated in triplicate. The results were subjected to Hill analysis for the determination of the stoichiometry and the binding affinity.
Alternatively, 15 molar equivalents of Ni2+ (as NiSO4) were added to 5 μM protein in 20 mM Hepes at pH 7.4. After more than 1 h, the mixture was loaded on to a PD-10 desalting column (Amersham Biosciences) to remove excess Ni2+ ions. The fraction containing the Hpn–Ni2+ complex was collected, and the Ni2+ concentration was determined by ICP-MS.
RESULTS
Expression and purification of Hpn
The hpn gene (HP1427) was amplified by PCR from genomic H. pylori DNA and inserted into pET32a, via the NdeI/XhoI restriction sites, for high-level expression in E. coli. No fusion tag was included as we predicted that the separate stretches of six and seven consecutive histidine residues within the Hpn protein would result in strong binding to Ni2+-charged iminodiacetate-derivatized agarose (HiTrap chelating Sepharose). DNA sequencing revealed that the cloned hpn gene contained several (silent) mutations compared with the published sequence (GenBank® accession number U26361), but the translated product remained unchanged. It was found that adding IPTG to 0.5 mM at a D600 of 0.6–0.8, followed by incubation at 37 °C for 4 h, was optimal for the expression of soluble protein from pET-Hpn in E. coli BL21(DE3). Under these conditions, generally 2–3 mg of soluble Hpn was expressed per litre of cell culture. Hpn bound strongly to the Ni-charged chelating Sepharose resin in the presence of 100 mM imidazole, and could be eluted by 500 mM imidazole. After a second gel-filtration purification step, Hpn was determined to be >95–97% pure by Coomassie Blue staining (Table 1). Upon storage for several days at 4 °C in gel-filtration buffer (which lacked imidazole), the Hpn protein progressively aggregated and precipitated. This could be prevented by the addition of imidazole (500 mM), which increased protein stabilities in solutions to over 1 month (as determined by SDS/PAGE).
Table 1. Purification of expressed Hpn.
The protein content was determined using a BCA protein concentration assay (Bio-Rad). Hpn was quantified from an immunoblot using the His·Tag® monoclonal antibody, normalized against a standard curve of different Hpn concentrations. The percentage Hpn in the fraction was calculated as follows: (mg of Hpn/mg of total protein)×100.
| Fraction | Total protein (mg) | Hpn (mg) | Hpn in fraction (%) | Fold purification |
|---|---|---|---|---|
| Crude extract | 5300 | 2.6 | 0.05 | 1 |
| Crude supernatant | 3279 | 2.5 | 0.08 | 1.6 |
| Ni2+ affinity | 2.7 | 2.3 | 85 | 1700 |
| Gel filtration | 1.5 | 1.4 | >97* | 1940 |
* As determined from a Coomassie Blue-stained gel.
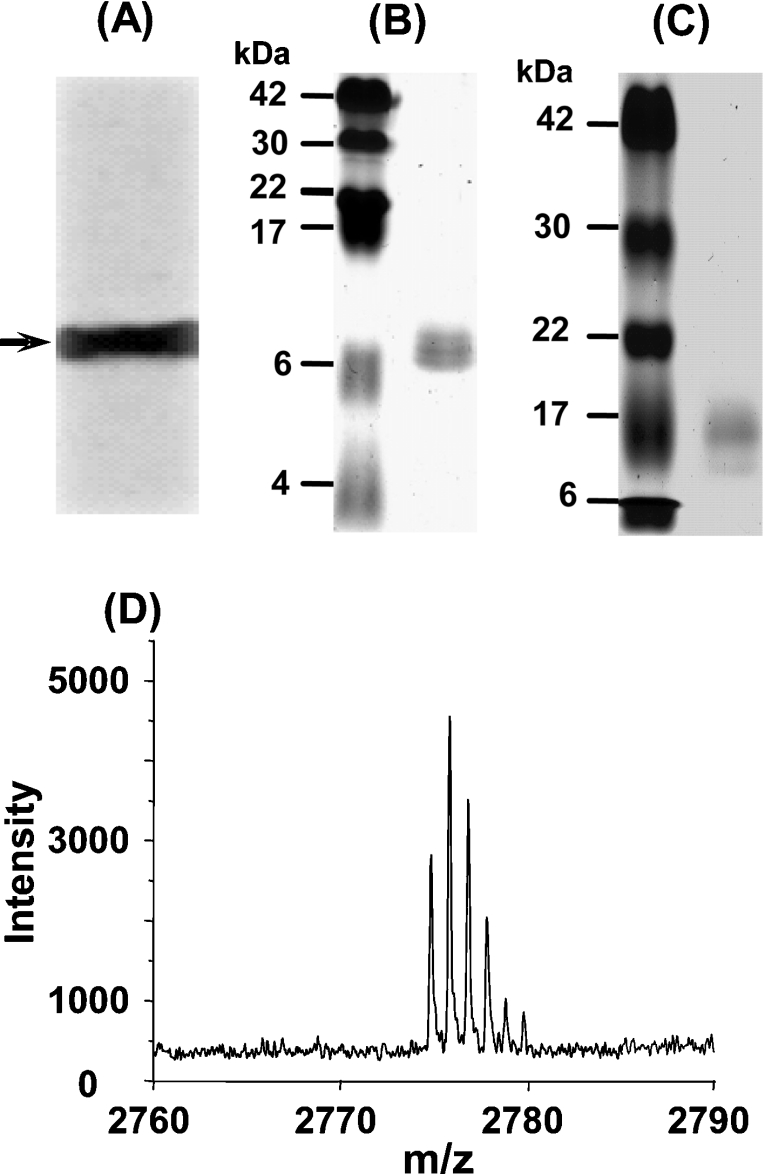
To confirm the identity of the purified recombinant Hpn protein, we performed Western blotting, N-terminal protein sequence analysis and MALDI–TOF (time-of-flight) mass spectrometric analysis. Based on the presence of six and seven consecutive histidine residues in the Hpn protein sequence, we predicted that it would bind with the His·Tag® monoclonal antibody with good affinity. Western blotting experiments confirmed this prediction, with a single intense band appearing on the PVDF membrane (Figure 2A), corresponding to the migratory position of the Hpn protein on a 15% polyacrylamide gel (as a weak band; results not shown).
Figure 2. Analysis of expressed Hpn protein.
(A) Western blotting with His·Tag® antibody. The arrow indicates the position of the recombinant Hpn protein. (B) Purified Hpn subjected to SDS/20% PAGE. The left-hand lane contains the protein markers (sizes are given in kDa), and the right-hand lane contains purified Hpn protein, which migrates to approx. 7 kDa. (C) Purified Hpn subjected to SDS/15% PAGE. The left-hand lane contains the protein markers (sizes are given in kDa), and the right-hand lane contains purified Hpn protein, which migrates to approx. 16 kDa. (D) MALDI-MS spectrum of Hpn showing the major peak at m/z 2773.183 corresponding to the N-terminal fragment of recombinant Hpn.
To perform N-terminal sequencing, Hpn was purified by HiTrap Chelating HP to approx. 95% homogeneity, resolved further by SDS/PAGE, and then electroblotted on to a PVDF membrane to isolate homogeneous protein for N-terminal sequencing. The first seven amino acids were determined to be Ala1-His-His-Glu-Glu-Gln-His7. This revealed that the N-terminal methionine residue had been removed co- or post-translationally in E. coli. MALDI–MS spectrometry of the Hpn digested in-gel with chymotrypsin revealed a major peak at m/z 2773.183 (Figure 2D). This value agrees very well with the mass of m/z 2773.178 for the peptide AHHEEQHGGHHHHHHHTHHHHY of Hpn as predicted by analysis with MS-digest software (http://prospector.ucsf.edu/). Analysis using Ellman's reagent revealed that there were 3.7 thiolate groups per monomer, indicating that four cysteines in Hpn were in reduced form.
Molecular size
As was found previously [23], on standard SDS/10–15% polyacrylamide gels, Hpn migrated with an apparent molecular mass of approx. 16 kDa (Figure 2C), which is considerably larger than its predicted molecular mass of approx. 7 kDa. This is consistent with the properties of other histidine-rich proteins, which also exhibit retarded migration on SDS/polyacrylamide gels [32]. However, when SDS/20% polyacrylamide gels were used, Hpn migrated with an apparent molecular mass of approx. 7 kDa (Figure 2B), which corresponds closely to its predicted monomeric mass (7077 Da).
The native molecular mass of the protein in the presence of imidazole, DTT and Ni2+ was estimated by gel-filtration chromatography. As shown in the elution profile (Figure 3A), in the presence of 100 mM imidazole, a large fraction of the purified recombinant Hpn formed a high-molecular-mass aggregate of >500 kDa (eluted at approx. 8.1 ml). There were six other major species observable, corresponding to molecular masses of 55, 34, 26, 20, 14 and 7 kDa, suggesting that Hpn was present in a range of multimeric forms. However, when DTT was added to the Hpn solution to a final concentration of 5 mM, before loading on to the Superose 12 column, the relative amount of the high-molecular-mass aggregate (eluted at approx. 8.1 ml) decreased significantly (Figure 3B), and the majority of the protein eluted with an apparent molecular mass of 136 kDa (eluted at approx. 10.4 ml), with two very minor peaks at 55 and 26 kDa. When the eluted fraction corresponding to 136 kDa was re-injected into the gel-filtration column under the same conditions, a similar elution profile was observed (results not shown), suggesting that an equilibrium exists between multimeric forms. When the amount of imidazole was increased to 500 mM in the presence of 5 mM DTT, this fraction (approx. 136 kDa) become the dominant species with little high-molecular-mass aggregate (approx. 8.1 ml of eluate) (Figure 3C). When analysed on a SDS/15% polyacrylamide gel, all of the separately collected fractions migrated with an similar molecular mass of approx. 16 kDa (as in Figure 2C), showing that they are the same protein with different multimerization states.
Figure 3. Analysis of Hpn multimeric state using gel-filtration chromatography.
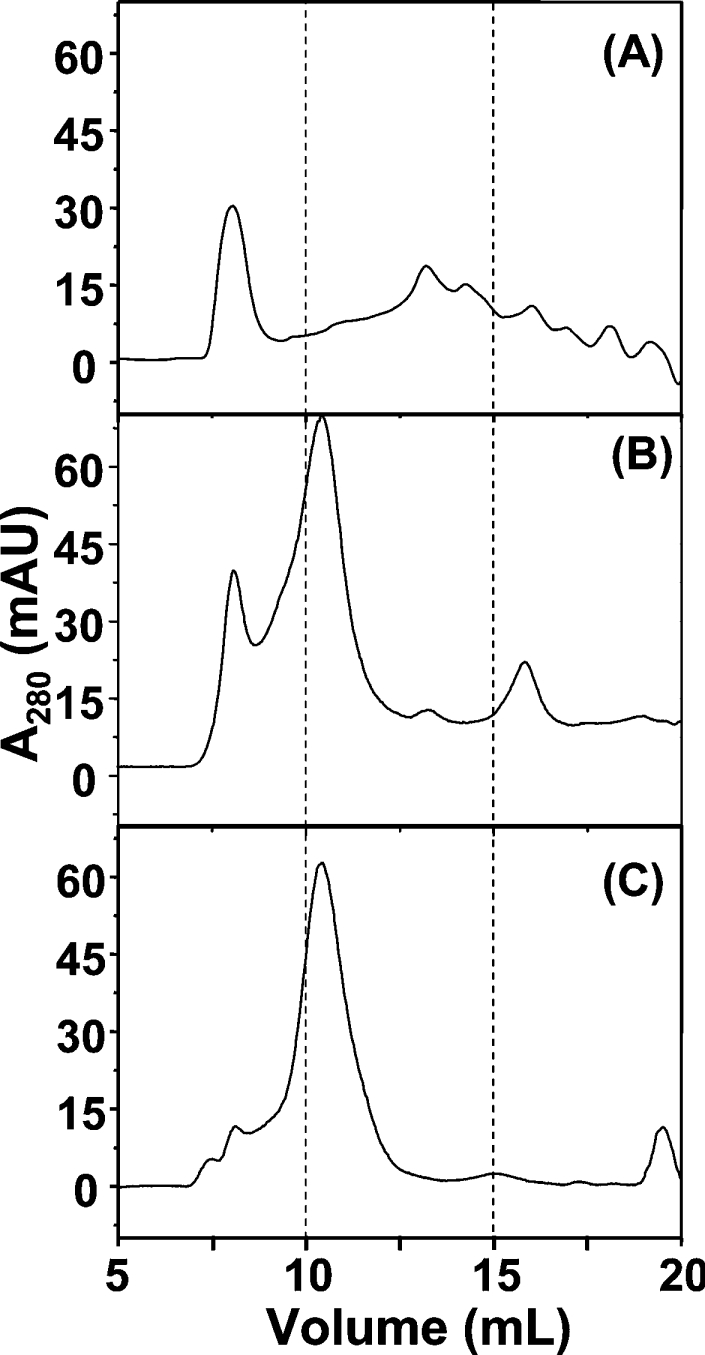
Elution profile (monitoring at 280 nm) of Hpn after gel filtration on Superose 12 (10/300) column, using running buffers containing 20 mM Hepes, pH 7.4, and 500 mM NaCl, supplemented with (A) 100 mM imidazole, (B) 100 mM imidazole plus 5 mM DTT, or (C) 500 mM imidazole plus 5 mM DTT. Proteins were equilibrated in the respective running buffer for at least 1 h immediately before analysis.
Ni2+–Hpn interaction
As shown in Figure 3, Hpn was present mainly as a 136 kDa multimer in buffers containing 500 mM imidazole and 5 mM DTT; otherwise, it will aggregate to over 500 kDa (Supplementary Figure S1 at http://www.BiochemJ.org/bj/393/bj3930285add.htm). It is impossible to investigate the Ni2+ binding in the presence of high concentrations of imidazole and DTT because of their competitive metal-binding capabilities. Therefore we studied the ‘effective’ Ni2+-binding mode under physiologically relevant conditions (without imidazole). The choice of these experimental conditions was supported further by the findings that Hpn (2% of the total proteins of H. pylori) was absent in the two-dimensional gels of H. pylori cellular protein fractions (R. Ge, Q.-Y. He, J.-D. Huang and H. Sun, unpublished work), indicating that Hpn was unlikely to be present as a monomer inside the H. pylori cells.
The binding of Ni2+ with apo-Hpn was studied by ICP-MS. It was found that freshly prepared Hpn contained fewer than one Ni2+ ion per ten Hpn molecules (results not shown). To a solution of Hpn (50 μM, in 20 mM Hepes buffer at pH 7.4), 15 molar equivalents of Ni2+ (as NiSO4) were added, then the excess Ni2+ was removed by desalting on a PD-10 column. The Hpn-bound Ni2+ was quantified by comparing the amount of Ni2+ present in the protein fraction with a standard curve (results not shown). This revealed that 5.1±0.2 (n=3, mean±S.D.) Ni2+ ions were present per Hpn monomer. In order to examine whether free cysteine residues in Hpn were involved in the Ni2+ binding, the Ni2+-bound Hpn was subjected to the method of Ellman [28], and the number of free thiolate groups was determined to be 0.4, indicating that almost all the four cysteine residues took part in the Ni2+ binding.
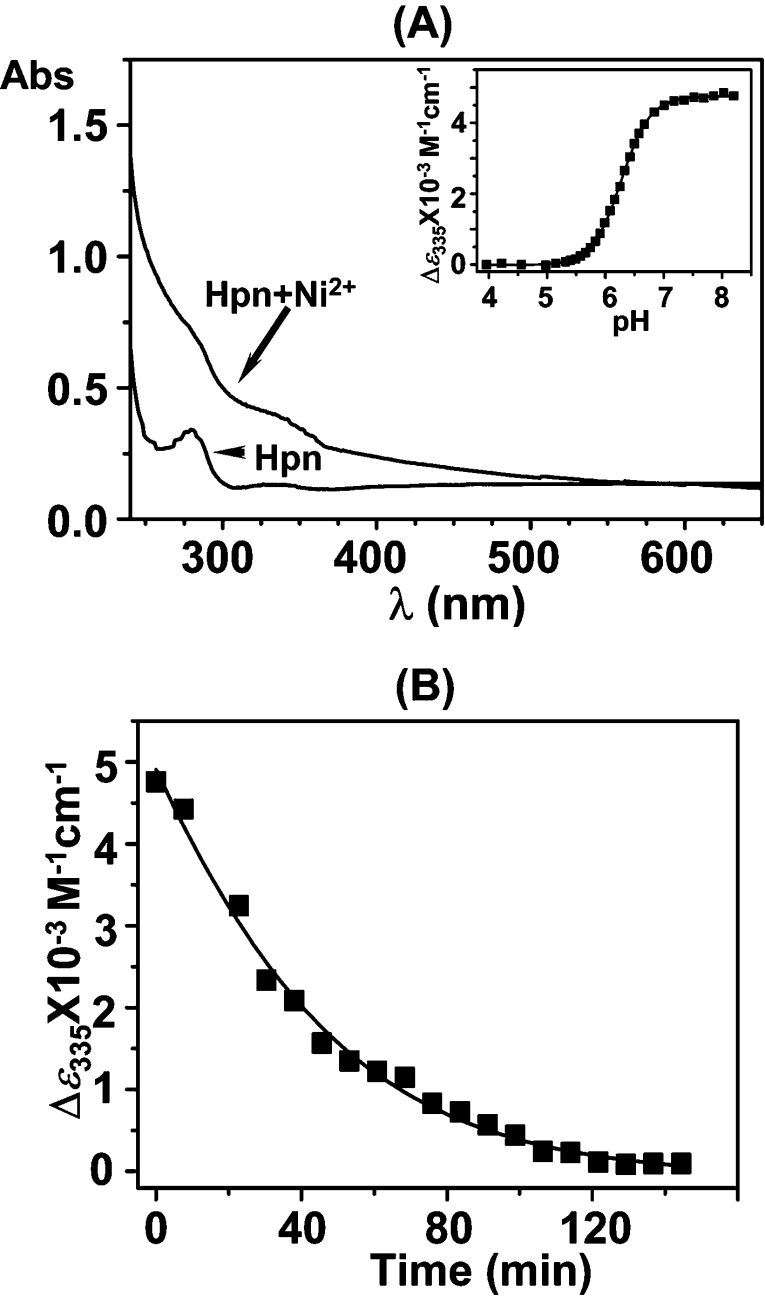
UV/vis spectroscopy was used to characterize further the Ni2+–Hpn interaction. Complexation of Ni2+ to proteins normally leads to the production of new absorptions in the visible region due to LMCT (ligand-to-metal charge transfer) transitions [33]. Consequently, these new absorption peaks can be readily used to monitor protein–metal binding. The UV/vis absorption spectrum of a 50 μM Hpn solution was recorded (Figure 4A). After the sequential addition of different molar equivalents of Ni2+ to the purified protein solution, two shoulder peaks gradually appeared, centred at approx. 286 and 335 nm, indicative of Ni2+ binding (Figure 4A). The band at 335 nm was attributed to a ligand [e.g. N(His)]→Ni2+ LMCT transition [34]. A plot of Δϵ286 against the molar ratio (r) of Ni2+ (as NiSO4) to the apo-protein is also shown in Supplementary Figure S2 (http://www.BiochemJ.org/bj/393/bj3930285add.htm). It can be seen that with the increase of r, the absorption at 286 nm (and 335 nm) increases in intensity and reaches a plateau at r≈4.8±0.2, which suggests that five Ni2+ ions bind strongly per Hpn monomer.
Figure 4. Binding of Ni2+ to recombinant Hpn as determined by UV/vis absorption spectra.
(A) UV/vis absorption spectra of Hpn (50 μM) in 20 mM Hepes, pH 7.4, and 100 mM NaCl (lower trace) and after addition of 5 molar equivalents of Ni2+ (as NiSO4) (upper trace). Inset, pH titration of Ni2+-saturated Hpn, monitoring the putative Ni2+–protein LMCT at 335 nm. (B) Kinetics of Ni2+ release from Hpn in the presence of 120 molar equivalents of EDTA at pH 6.8 and 298 K, as indicated by the absorption at 335 nm.
We investigated how the intensity of the absorption at 335 nm altered as the pH of a Ni2+-saturated Hpn solution was systematically varied. As can be seen in the inset of Figure 4(A), there were no significant changes in the absorption at 335 nm at pH values greater than approx. 6.8. This indicated that the amount of Ni2+ bound to the protein remained relatively stable above this pH value. However, upon lowering the pH of the solution from approx. pH 6.8 to 5.8 (by the careful addition of 1 M HCl), the intensity of the absorption at 335 nm decreased markedly. At pH values lower than approx. 5.0, the absorption at 335 nm remained essentially unchanged. The pH for half dissociation of Ni2+ from the protein complex (pH½) was determined to be 6.3±0.1.
A competition binding assay between Hpn and EDTA for free Ni2+ was performed analogously. Upon the addition of a large excess of EDTA (120 molar equivalents) to a Ni2+-saturated Hpn solution at pH 6.8, the absorption at 335 nm (characteristic of an LMCT) decreased exponentially with a half-life of 41±1 min (Figure 4B). However, this peak was restored to original levels upon the addition of excess Ni2+ ions, demonstrating that Ni2+ binding is reversible (results not shown).
To determine the Ni2+-binding affinity and capacity of the protein, the pure protein was dialysed against 20 mM Hepes buffer (pH 7.4) with different amount of Ni2+. As shown in Figure 5, at the concentration used (5 μM), the binding of Ni2+ to Hpn reached saturation at 40 μM, and the protein was able to bind to 4.8±0.1 Ni2+ ions per monomer in a slightly co-operative fashion (Hill coefficient of approx. 1.7). The dissociation constant (Kd) was calculated to be 7.1 μM.
Figure 5. Binding of Ni2+ to recombinant Hpn as determined by equilibrium dialysis.
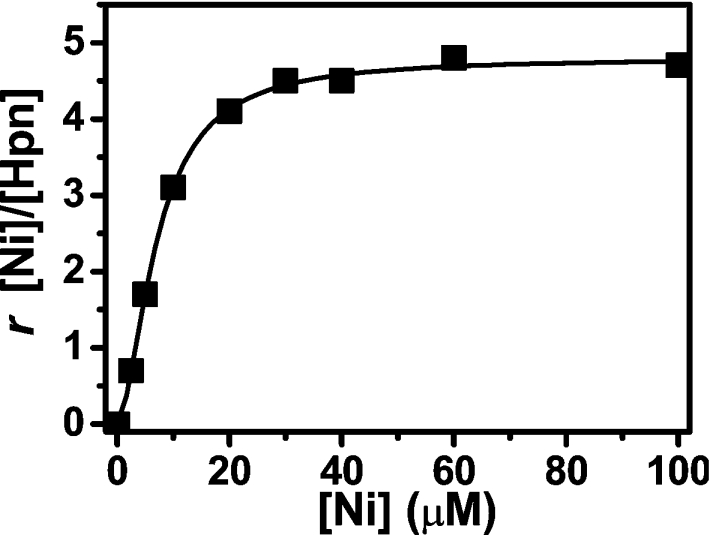
Hpn (5 μM, 200 μl) was dialysed overnight at 4 °C against 1000 ml of 20 mM Hepes, pH 7.4, and 100 mM NaCl containing 0, 2, 5, 10, 20, 30, 40, 60 or 100 μM NiSO4. Hpn-bound Ni2+ was determined by ICP-MS (subtracting Ni2+ concentrations inside and outside the dialysis tubing). Shown is a Hill plot of the molar ratio (r) of bound Ni2+ to Hpn protein against concentration of NiSO4 in the dialysis buffer. Each point represents the average value of three independent experiments.
The CD spectrum of Hpn in 20 mM Tris/HCl at pH 7.4 (Figure 6) is characterized by a negative peak with a minimum at approx. 217 nm and a positive peak with a maximum at 197 nm, attributed to the presence of a mixture of α-helix, β-sheet, turn and unordered form. Upon the addition of different amounts of molar equivalents of Ni2+ ions (as NiSO4) to the apo-protein solution, the negative peak at 217 nm increased gradually, indicative of a conformational change within the protein induced by the metals. Quantitative analysis using CDPro [30] indicated that α-helix content was reduced from approx. 20% to 10%, whereas the β-structure increased from 22% to 37% after addition of 5 molar equivalents of Ni2+.
Figure 6. Binding of Ni2+ to recombinant Hpn as determined by CD.
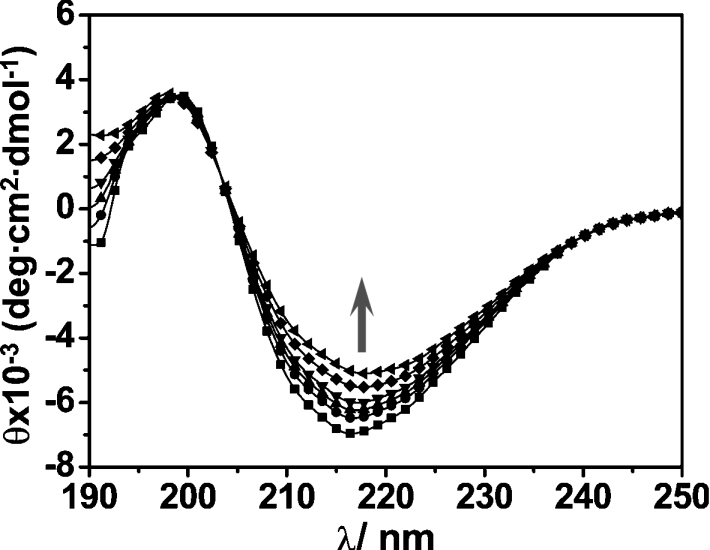
CD data were obtained for 5 μM Hpn in Tris/HCl buffer, pH 7.4, in both the absence and the presence of 1–5 molar equivalents of Ni2+ (as NiSO4). The arrow indicates the increases in ellipticity values (approx. 217 nm) as the protein was titrated with 0–5 molar equivalents of Ni2+.
Effect of Ni2+ on the growth of E. coli
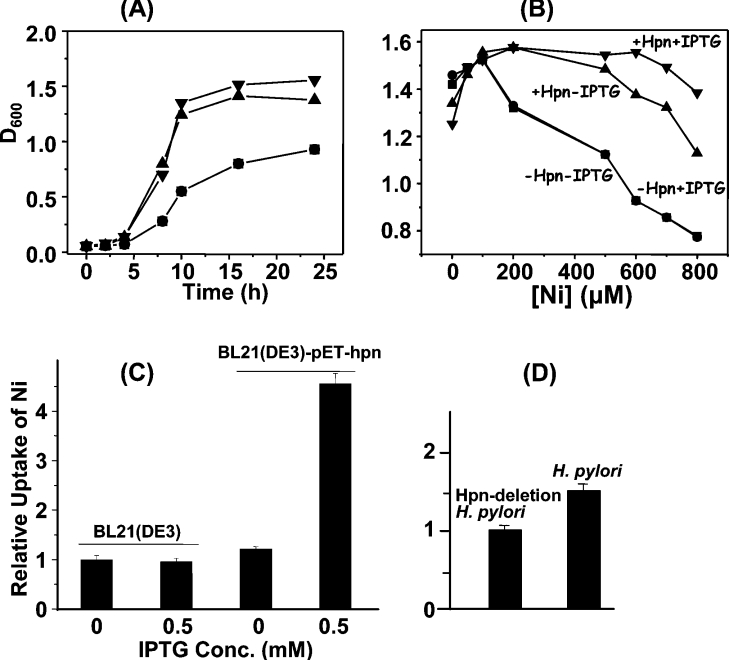
E. coli BL21(DE3) cells with and without the plasmid pET-hpn were grown in LB medium supplemented with different amounts of NiSO4. The growth at 37 °C in the presence of 500 μM Ni2+, as a representative, was followed by the measurement of D600 values (Figure 7A). The D600 values for the E. coli BL21(DE3) cells with hpn were about twice those of cells without hpn after 8 h. In addition, the D600 values for the E. coli BL21(DE3) cells with hpn were slightly higher upon addition of IPTG than those without (Figure 7B). In contrast with the very similar D600 values for the E. coli BL21(DE3) cells with hpn grown at [Ni2+]≤500 μM, the D600 values for the E. coli BL21(DE3) cells without hpn fell significantly at [Ni2+]≥200 μM. More importantly, the difference in D600 values (overnight culture) for E. coli BL21(DE3) cells harbouring hpn became more evident with Ni2+ concentrations increased to 500 μM or over with and without IPTG, i.e. the D600 values were higher for those cells with IPTG than for those without.
Figure 7. Effects of Ni2+ on growth of E. coli and Ni2+ accumulation in E. coli and in H. pylori.
(A) Growth curves of freshly inoculated cultures of E. coli BL21(DE3) both with and without the hpn gene (pET-hpn) in LB medium containing Ni2+ (as NiSO4, 500 μM). (B) The influence of Hpn expression on the growth of E. coli BL21(DE3) cells cultured in LB medium supplemented with NiSO4 at 0, 50, 100, 200, 500 or 800 μM. ▼, +pET-hpn, with IPTG induction (0.5 mM); ▲, +pET-hpn, no IPTG induction; ●, −pET-hpn, with IPTG induction (0.5 mM); ■, −pET-hpn, no IPTG induction. (C) E. coli BL21(DE3) cells were cultured in LB medium supplemented with 500 μM NiSO4, with or without the plasmid pET-Hpn, and with or without IPTG induction of Hpn expression. (D) The wild-type and Hpn-deleted H. pylori 26695 cells were cultured in fresh Brucella broth supplemented with 5% foetal bovine serum and 200 μM NiSO4. The Ni2+ content associated with the cell pellet was determined using ICP-MS.
Cellular Ni2+ accumulation
To comply with the conditions used in the expression of Hpn, the cells treated with 0.5 mM Ni2+ were pelleted and washed three times with PBS, and the cellular Ni2+ content was determined by ICP-MS. In E. coli BL21(DE3) harbouring pET-hpn where Hpn expression had been induced with IPTG, the Ni2+ ion content was found to be approx. 4-fold higher than in uninduced cells, or in cells that did not contain pET-hpn (Figure 7C). The cellular accumulation of Ni2+ was then investigated in cultures of H. pylori 26695 which contained a wild-type copy of the gene, and in constructed strains where the hpn gene had been deleted. The deletion of the hpn gene was confirmed by PCR (Supplementary Figure S3 at http://www.BiochemJ.org/bj/393/bj3930285add.htm), and by the disappearance of the Hpn protein from cell lysates, using Western blot analysis, probing with the His·Tag® monoclonal antibody. After 48 h of culture in medium containing 0.2 mM NiSO4, ICP-MS analysis indicated that intracellular levels of Ni2+ were 1.5-fold higher in the wild-type strain than in the Δhpn strain (Figure 7D). In addition, the accumulation of Ni2+ in H. pylori 26695 is at least 2-fold greater than that in E. coli, on a gram-for-gram basis (wet cell mass).
DISCUSSION
The transition metal Ni2+ is essential for anaerobic metabolism in micro-organisms, including the medically important Gram-negative bacterium H. pylori. However, too much ‘free’ or unbound Ni2+ can be toxic and therefore intracellular concentrations must be regulated tightly [10,13]. Initial studies have indicated that the small cytoplasmic Hpn protein may play a pivotal role in Ni2+ homoeostasis, binding or transport [23], and Hpn was named to emphasize its origins in H. pylori and its affinity for Ni2+ [23]. Although the specific amino acids essential for metal binding remain to be established, several conserved histidine, glutamate and aspartate residues stand out as potential metal ligands. All four cysteine residues of Hpn were found to be involved in Ni2+ binding. It is possible that the two neighbouring double cysteines seem to be possible to take part in the co-ordination with the same Ni2+ ion. The motif Asp-Xaa-His-His-Xaa-Xaa-Glu (or DXHHXXE) is a potential metal-binding site, and is conserved in approx. 300 proteins listed in the SwissProt database ([35]; http://www.expasy.org/sprot/). A significant proportion of these proteins are histidine-rich and have been implicated in metal binding, such as UreE [36], IGPD (imidazole glycerol-phosphate dehydratase) [37], HspA and HypB [38,46]. Another conserved pentapeptide motif: His-Glu-Xaa-Xaa-His (or HEXXH) appears regularly in protein sequences, especially in metallopeptidases. Roughly half of all metallopeptidases characterized to date contain this motif, which has been implicated in metal binding in X-ray crystal structures [39]. Low-stringency homologous sequence searching using various BLAST programs [40] failed to find any meaningful protein matches to Hpn, the only ‘hits’ containing ubiquitous short histidine-rich motifs (results not shown). Consequently, the structural fold and organization of Hpn cannot presently be inferred by homology.
The hpn gene was cloned into a pET32 vector, and was successfully expressed (untagged) in a predominantly soluble form in E. coli. Approx. 77% of the amino acid residues in Hpn are predicted to be >16% solvent-exposed using PredictProtein ([41]; http://cubic.bioc.columbia.edu/predictprotein/), indicating that the protein should be highly hydrophilic. Denaturing conditions were not required for Hpn binding to Ni2+-impregnated Sepharose resin, suggesting that enough contiguous histidine residues are solvent-exposed under the conditions used to be readily available either for chelation with the immobilized metal ions, or possibly for protein–protein interactions. This is congruent with the predicted hydrophilicity of the histidine-rich regions by PredictProtein (results not shown).
After purifying milligram quantities of recombinant Hpn, we were able to investigate in detail its multimeric form, conformation and Ni-binding properties in vitro. On native gels, Hpn migrated as a single band with an apparent molecular mass of >500 kDa (Supplementary Figure S1 at http://www.BiochemJ.org/bj/393/bj3930285add.htm). However, it eluted as a mixture of species after gel-filtration chromatography on a Superose 12 column in a buffer containing 20 mM Hepes, pH 7.4, 500 mM NaCl and 100 mM imidazole buffer (Figure 3A). Upon addition of 5 mM DTT to the running buffer, the relative amounts of the high-molecular-mass aggregate dramatically decreased, indicative of the presence of an intermolecular oxidation. The protein fractions with apparent molecular masses of approx. 55 and approx. 136 kDa correspond to a ‘dimer’ and ‘pentamer’ of tetramers respectively. It is possible that the 20 (±2)-mer Hpn multimer may be the physiologically relevant form of the protein in vivo. In the presence of higher amounts of imidazole, the relative amount of the high-molecular-mass aggregate decreased further, indicating that imidazole plays a role in the prevention of protein precipitation and restoring the preferred multimerization state.
A number of other histidine-rich proteins have been reported to exist as multimers (Supplementary Table S1 at http://www.BiochemJ.org/bj/393/bj3930285add.htm) [32,37,42–46]. Based on these data, it seems that the higher the percentage of histidine in a metal-binding protein, the greater the propensity of the protein to form higher-order multimers. One explanation for this is that histidine residues located on different protein monomers might facilitate protein multimerization in some way, possibly through their tendency to align or ‘stack’ on top of one another. It is conceivable that the disappearance of the low-molecular-mass Hpn band (as determined by SDS/PAGE) after storage for several days in buffers lacking imidazole may be due to the formation of a high-molecular-mass aggregate that is large enough to precipitate. Conversely, when present in concentrations of no less than 500 mM, imidazole in solution may locate between the protein monomers, inhibiting intermolecular histidine stacking, consequently stabilizing the protein. However, we cannot exclude the possibility that other biophysical processes are behind multimer formation/disassociation.
We have found that Hpn can bind up to five Ni2+ ions per monomer under physiologically relevant conditions (i.e. at pH 7.4). More importantly, this binding is reversible, as revealed by both pH titration experiments and competition binding experiments with EDTA (Figure 4). This result raises the intriguing possibility that Hpn releases bound Ni2+ ions when the environmental pH decreases to approx. 6.8, a physiological pH value. The moderate affinity (Kd of approx. 7.1 μM) of Hpn for Ni2+ is comparable with that of HypB (Kd of approx. 2.3 μM), a protein that has been suggested to store Ni2+ in its N-terminal histidine-rich region (16 histidine residues out of 54 residues) in Bradyrhizobium japonicum [46]. In this manner, H. pylori may buffer itself against a potentially damaging fall in the environmental pH. As shown in a previous report [23], there appears to be no direct interaction between Hpn and urease, since hpn gene deletion has little affect on the urease activity. It is possible that Hpn may donate Ni2+ to a chaperone protein (e.g. HspA) with higher affinity, which would transport the Ni2+ ions to other proteins and ultimately to a Ni2+-containing enzyme [21]. The presence of other functions of the protein is also possible and needs to be studied further.
Interestingly, Ni2+ release from the protein appeared to occur in a single process, although one may have expected five micro-steps (generating micro pKa values). This may be due to the high kinetic liability of Ni2+ binding. A similar kinetic profile has been observed for Zn2+ (and Cd2+) release from MT (metallothionein; Zn7-MT), i.e. no more than two steps were observed [47]. A facile Ni2+-to-ligand exchange mechanism would facilitate intraprotein metal transfer, possibly assisting rapid metal trafficking to and from other proteins or enzymes.
The appearance of absorption peaks at 286 and 335 nm upon the addition of Ni2+ to apo-Hpn indicates that the Ni2+ is probably co-ordinated to nitrogen (and thiolate) centres, the majority presumably from the imidazole moieties of histidine residues. Similar absorptions have been observed in other Ni2+-binding proteins such as NikR, a protein known to bind to Ni2+ via three histidine nitrogens and one cysteine [33,48]. These UV/vis data also indicated that Ni2+ co-ordinates to ligands in either a 4-co-ordinated diamagnetic square-planar or a distorted square-planar geometry [10,34], consistent with silent EPR signals of the Ni2+-saturated Hpn (results not shown), which suggests that the Ni2+ ions may be in low-spin states. Further studies are required to firmly establish the co-ordination geometry of Ni2+-bound Hpn.
Similar to the situation in other metal-binding proteins, CD spectroscopy revealed that the binding of Ni2+ ions induced a protein conformational change in Hpn, with the α-helical content reduced from approx. 20% to 10%, and β-sheet content increased from approx. 22% to 37% (Figure 6). We are currently investigating the biological and biophysical significance of this change in protein structure.
In E. coli BL21(DE3) cells where the expression of the plasmid-based hpn gene had been induced, clearly Ni2+ had less influence on the cell growth than in those cells without hpn, based on the D600 values (Figures 7A and 7B). The higher D600 values for E. coli BL21(DE3) cells with the hpn gene in the presence of IPTG than those in the absence of IPTG indicated that Hpn may have a protective role against excess Ni2+ (and probably other heavy metals) in the environment. A similar pattern was also observed in H. pylori previously [25]. These data are in agreement with Ni2+ accumulation by ICP-MS. Ni2+ accumulation from the extracellular milieu was approx. 4-fold higher than in uninduced cultures, or in cells that did not contain the plasmid (Figure 7C). An analogous Ni2+-accumulation pattern was observed in H. pylori cultures under similar conditions. As the transmembrane metal transport machineries in E. coli and H. pylori are quite distinct, this suggests that Hpn may promote the gradual intracellular accumulation of Ni2+ through a ‘passive’ (equilibrium-driven) sequestration mechanism. The slight difference in Ni2+ accumulation between H. pylori and H. pylori with the hpn gene deleted also suggests that some other proteins, such as Hpn-like protein, play a similar role in cells.
In view of its short length (60 amino acids) and extremely high content of potential metal-binding residues, Hpn shares some similarities with MT, a zinc-storage protein rich in cysteine residues [47]. MT has been known to play a fundamental role in the metabolism of zinc and copper under various physiological conditions and its binding capability to metals is comparable with that of Hpn towards Ni2+. Moreover, MT also appears to play a role in sequestering toxic metals such as Cd2+ and Hg2+. It will be of great interest to see whether future work reveals any structural or functional similarities between these two important metal-binding proteins. With its large multimeric quaternary structure, possibly based around an arrangement of tetramers, Hpn may be regarded as a ‘ferritin-like’ Ni2+ protein, which regulates levels of Ni2+ within the bacterium.
In conclusion, the 60-amino-acid exceptionally histidine-rich Hpn protein from H. pylori binds tightly and reversibly to Ni2+ ions in a pH-dependent manner. Hpn binds approx. five Ni2+ ions per monomer with a Kd of 7.1 μM, and adopts a variety of multimeric structures in solution, with the 20 (±2)-mer form predominating under mildly reducing conditions. The growth and accumulation experiments performed under Ni2+ pressure clearly imply a role for Hpn to protect the cells from higher concentrations of external metal ions. Given its high intracellular concentration in H. pylori (approx. 2% of total protein), our data are consistent with Hpn functioning as an intracellular ‘reservoir’ of Ni2+ ions, playing a key role in bacterial Ni2+ homoeostasis.
Online Data
Acknowledgments
This work was supported by the Research Grants Council of Hong Kong (to H. Z. S., HKU703904P); the Research Fund for the Control of Infectious Diseases (to J. D. H., 01030182 and 02040192); the Area of Excellence Scheme of the University Grants Committee and the University of Hong Kong. Dr Harry Huaxiang Xia (Faculty of Medicine, The University of Hong Kong) is acknowledged for the gift of H. pylori 26695 and Miss Qing Gu for the culture of H. pylori. We thank Professor A. G. Plaut (GRASP Digestive Disease Research Center at the Tufts-New England Medical Center) for the hpn-deletion plasmid pHPN2KO, and Professor Sunny I. Chan (Caltech and Academic Sinica, Taipei, Taiwan) and Professor Dean E. Wilcox (Dartmouth College, Hanover, NH, U.S.A.) for helpful comments.
References
- 1.Marshall B. J., Warren J. R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;i:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Marshall B. J. Helicobacter pylori. Am. J. Gastroenterol. 1994;89:S116–S128. [PubMed] [Google Scholar]
- 3.Blaser M. J. Gastric Campylobacter-like organisms, gastritis, and peptic ulcer disease. Gastroenterology. 1987;93:371–383. doi: 10.1016/0016-5085(87)91028-6. [DOI] [PubMed] [Google Scholar]
- 4.Blaser M. J. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J. Infect. Dis. 1990;161:626–633. doi: 10.1093/infdis/161.4.626. [DOI] [PubMed] [Google Scholar]
- 5.Forman D., Newell D. G., Fullerton F., Yarnell J. W., Stacey A. R., Wald N., Sitas F. Association between infection with Helicobacter pylori and risk of gastric cancer: evidence from a prospective investigation. Br. Med. J. 1991;302:1302–1305. doi: 10.1136/bmj.302.6788.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsonnet J., Hansen S., Rodriguez L., Gelb A. B., Warnke R. A., Jellum E., Orentreich N., Vogelman J. H., Friedman G. D. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 1994;330:1267–1271. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 7.Maier R. J., Fu C., Gilbert J., Moshiri F., Olson J., Plaut A. G. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol. Lett. 1996;141:71–76. doi: 10.1111/j.1574-6968.1996.tb08365.x. [DOI] [PubMed] [Google Scholar]
- 8.Olson J. W., Maier R. J. Molecular hydrogen as an energy source for Helicobacter pylori. Science. 2002;298:1788–1790. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- 9.Hu L. T., Mobley H. L. Purification and N-terminal analysis of urease from Helicobacter pylori. Infect. Immun. 1990;58:992–998. doi: 10.1128/iai.58.4.992-998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulrooney S. B., Hausinger R. P. Nickel uptake and utilization by microorganisms. FEMS Microbiol. Rev. 2003;27:239–261. doi: 10.1016/S0168-6445(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 11.Dunn B. E., Campbell G. P., Perez-Perez G. I., Blaser M. J. Purification and characterization of urease from Helicobacter pylori. J. Biol. Chem. 1990;265:9464–9469. [PubMed] [Google Scholar]
- 12.McGee D. J., Mobley H. L. Mechanisms of Helicobacter pylori infection: bacterial factors. Curr. Top. Microbiol. Immunol. 1999;241:155–180. doi: 10.1007/978-3-642-60013-5_9. [DOI] [PubMed] [Google Scholar]
- 13.Eitinger T., Mandrand-Berthelot M. A. Nickel transport systems in microorganisms. Arch. Microbiol. 2000;173:1–9. doi: 10.1007/s002030050001. [DOI] [PubMed] [Google Scholar]
- 14.Hendricks J. K., Mobley H. L. Helicobacter pylori ABC transporter: effect of allelic exchange mutagenesis on urease activity. J. Bacteriol. 1997;179:5892–5902. doi: 10.1128/jb.179.18.5892-5902.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauerfeind P., Garner R. M., Mobley L. T. Allelic exchange mutagenesis of nixA in Helicobacter pylori results in reduced nickel transport and urease activity. Infect. Immun. 1996;64:2877–2880. doi: 10.1128/iai.64.7.2877-2880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misra M., Olinski R., Dizdaroglu M., Kasprzak K. S. Enhancement by L-histidine of nickel(II)-induced DNA–protein cross-linking and oxidative DNA base damage in the rat kidney. Chem. Res. Toxicol. 1993;6:33–37. doi: 10.1021/tx00031a005. [DOI] [PubMed] [Google Scholar]
- 17.Chakrabarti S. K., Bai C., Subramanian K. S. DNA–protein crosslinks induced by nickel compounds in isolated rat lymphocytes: role of reactive oxygen species and specific amino acids. Toxicol. Appl. Pharmacol. 2001;170:153–165. doi: 10.1006/taap.2000.9097. [DOI] [PubMed] [Google Scholar]
- 18.Kawanishi S., Inoue S., Oikawa S., Yamashita N., Toyokuni S., Kawanishi M., Nishino K. Oxidative DNA damage in cultured cells and rat lungs by carcinogenic nickel compounds. Free Radical Biol. Med. 2001;31:108–116. doi: 10.1016/s0891-5849(01)00558-5. [DOI] [PubMed] [Google Scholar]
- 19.Bruins M. R., Kapil S., Oehme F. W. Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 2000;45:198–207. doi: 10.1006/eesa.1999.1860. [DOI] [PubMed] [Google Scholar]
- 20.Contreras M., Thiberge J. M., Mandrand-Berthelot M. A., Labigne A. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol. Microbiol. 2003;49:947–963. doi: 10.1046/j.1365-2958.2003.03621.x. [DOI] [PubMed] [Google Scholar]
- 21.Tomb J.-F., White O., Kerlavage A. R., Clayton R. A., Sutton G. G., Fleischmann R. D., Ketchum K. A., Klenk H. P., Gill S., Dougherty B. A., et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 22.Kansau I., Raymond J., Bingen E., Courcoux P., Kalach N., Bergeret M., Braimi N., Dupont C., Labigne A. Genotyping of Helicobacter pylori isolates by sequencing of PCR products and comparison with the RAPD technique. Res. Microbiol. 1996;147:661–669. doi: 10.1016/0923-2508(96)84023-x. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert J. V., Ramakrishna J., Sunderman F. W., Jr, Wright A., Plaut A. G. Protein Hpn: cloning and characterization of a histidine-rich metal-binding polypeptide in Helicobacter pylori and Helicobacter mustelae. Infect. Immun. 1995;63:2682–2688. doi: 10.1128/iai.63.7.2682-2688.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H., Zhang L., Szeto K. Y. Bismuth in medicine. Met. Ions Biol. Syst. 2004;41:333–378. [PubMed] [Google Scholar]
- 25.Mobley H. L., Garner R. M., Chippendale G. R., Gilbert J. V., Kane A. V., Plaut A. G. Role of Hpn and NixA of Helicobacter pylori in susceptibility and resistance to bismuth and other metal ions. Helicobacter. 1999;4:162–169. doi: 10.1046/j.1523-5378.1999.99286.x. [DOI] [PubMed] [Google Scholar]
- 26.Ge Z., Taylor D. E. H. pylori DNA transformation by natural competence and electroporation. In: Clayton C. L., Mobley H. L. T., editors. Helicobacter pylori Protocols. Totowa: Humana Press Inc.; 1997. pp. 145–152. [DOI] [PubMed] [Google Scholar]
- 27.Maniatis T., Fritsch E. F., Sambrook J. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1982. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- 28.Ellman G. L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 29.Coligan J. E., Dunn B. M., Ploegh H. L., Speicher D. W., Wingfield P. T. New York: John Wiley & Sons Inc.; 2001. Current Protocols in Protein Science. [Google Scholar]
- 30.Sreerama N., Woody R. W. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 31. Reference deleted.
- 32.Watt R. K., Ludden P. W. The identification, purification, and characterization of CooJ: a nickel-binding protein that is co-regulated with the Ni-containing CO dehydrogenase from Rhodospirillum rubrum. J. Biol. Chem. 1998;273:10019–10025. doi: 10.1074/jbc.273.16.10019. [DOI] [PubMed] [Google Scholar]
- 33.Chivers P. T., Sauer R. T. NikR repressor: high-affinity nickel binding to the C-terminal domain regulates binding to operator DNA. Chem. Biol. 2003;9:1141–1148. doi: 10.1016/s1074-5521(02)00241-7. [DOI] [PubMed] [Google Scholar]
- 34.Lever A. B. P. 2nd edn. New York: Elsevier Science; 1984. Inorganic Electronic Spectroscopy. [Google Scholar]
- 35.Boeckmann B., Bairoch A., Apweiler R., Blatter M.-C., Estreicher A., Gasteiger E., Martin M. J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Remaut H., Safarov N., Ciurli S., Van Beeumen J. Structural basis for Ni2+ transport and assembly of the urease active site by the metallochaperone UreE from Bacillus pasteurii. J. Biol. Chem. 2001;276:49365–49370. doi: 10.1074/jbc.M108304200. [DOI] [PubMed] [Google Scholar]
- 37.Sinha S. C., Chaudhuri B. N., Burgner J. W., Yakovleva G., Davisson V. J., Smith J. L. Crystal structure of imidazole glycerol-phosphate dehydratase: duplication of an unusual fold. J. Biol. Chem. 2004;279:15491–15498. doi: 10.1074/jbc.M312733200. [DOI] [PubMed] [Google Scholar]
- 38.Rey L., Murillo J., Hernando Y., Hidalgo E., Cabrera E., Imperial J., Ruiz-Argueso T. Molecular analysis of a microaerobically induced operon required for hydrogenase synthesis in Rhizobium leguminosarum biovar viciae. Mol. Microbiol. 1993;8:471–481. doi: 10.1111/j.1365-2958.1993.tb01591.x. [DOI] [PubMed] [Google Scholar]
- 39.Rawlings N. D., Barrett A. J. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 40.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rost B., Liu J. The PredictProtein server. Nucleic Acids Res. 2003;31:3300–3304. doi: 10.1093/nar/gkg508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hattan S. J., Laue T. M., Chasteen N. D. Purification and characterization of a novel calcium-binding protein from the extrapallial fluid of the mollusc, Mytilus edulis. J. Biol. Chem. 2001;276:4461–4468. doi: 10.1074/jbc.M006803200. [DOI] [PubMed] [Google Scholar]
- 43.Suk J. Y., Kim Y. S., Park W. J. HRC (histidine-rich Ca2+ binding protein) residues in the lumen of sarcoplasmic reticulum as a multimer. Biochem. Biophys. Res. Commun. 1999;263:667–671. doi: 10.1006/bbrc.1999.1432. [DOI] [PubMed] [Google Scholar]
- 44.Lee M. H., Pankratz H. S., Wang S., Scott R. A., Finnegan M. G., Johnson M. K., Ippolito J. A., Christianson D. W., Hausinger R. P. Purification and characterization of Klebsiella aerogenes UreE protein: a nickel-binding protein that functions in urease metallocenter assembly. Protein Sci. 1993;2:1042–1052. doi: 10.1002/pro.5560020617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sriwanthana B., Island M. D., Maneval D., Mobley H. L. Single-step purification of Proteus mirabilis urease accessory protein UreE, a protein with a naturally occurring histidine tail, by nickel chelate affinity chromatography. J. Bacteriol. 1994;176:6836–6841. doi: 10.1128/jb.176.22.6836-6841.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu C., Olson J. W., Maier R. J. HypB protein of Bradyrhizobium japonicum is a metal-binding GTPase capable of binding 18 divalent nickel ions per dimer. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2333–2337. doi: 10.1073/pnas.92.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasler D. W., Jesen L. T., Zerbo O., Winge D. R., Vašák M. Effect of the two conserved prolines of human growth inhibitory factor (metallothionein-3) on its biological activity and structure fluctuation: comparison with a mutant protein. Biochemistry. 2000;39:14567–14575. doi: 10.1021/bi001569f. [DOI] [PubMed] [Google Scholar]
- 48.Schreiter E. R., Sintchak M. D., Guo Y., Chivers P. T., Sauer R. T., Drennan C. L. Crystal structure of the nickel-responsive transcription factor NikR. Nat. Struct. Biol. 2003;10:794–799. doi: 10.1038/nsb985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.