Abstract
Human DSS1 associates with BRCA2, a tumour suppressor protein required for efficient recombinational DNA repair, but the biochemical function of DSS1 is not known. Orthologues of DSS1 are found in organisms such as budding yeast and fission yeast that do not have BRCA2-related proteins, indicating that DSS1 has a physiological role independent of BRCA2. The DSS1 orthologue in Saccharomyces cerevisiae has been shown to associate with the 26 S proteasome and, in the present paper, we report that in the distantly related fission yeast Schizosaccharomyces pombe, Dss1 associates with the 19 S RP (regulatory particle) of the 26 S proteasome. A role for S. pombe Dss1 in proteasome function is supported by three lines of evidence. First, overexpression of two components of the 19 S RP, namely Pad1/Rpn11 and Mts3/Rpn12, rescued the temperature-sensitive growth defect of the dss1 mutant. Secondly, the dss1 mutant showed phenotypes indicative of a defect in proteasome function: growth of the dss1 mutant was inhibited by low concentrations of L-canavanine, an amino acid analogue, and cells of the dss1 mutant accumulated high molecular mass poly-ubiquitylated proteins. Thirdly, synthetic growth defects were found when the dss1 mutation was combined with mutations in other proteasome subunit genes. These findings show that DSS1 has an evolutionarily conserved role as a regulator of proteasome function and suggest that DSS1 may provide a link between BRCA2 and ubiquitin-mediated proteolysis in human cells.
Keywords: BRCA2, DNA repair, DSS1, fission yeast, proteasome, tumour suppressor
Abbreviations: CP, core particle; DTT, dithiothreitol; EMM, Edinburgh minimal medium; GFP, green fluorescent protein; HMW, high molecular weight (‘mass’); MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; RP, regulatory particle; TAP, tandem affinity purification; TEV, tobacco etch virus; Ts, temperature sensitive; WT, wild-type; YES, yeast extract supplemented medium
INTRODUCTION
Human DSS1 was first identified as a gene deleted in patients suffering from a developmental disorder called SHFM (split hand/split foot malformation), which is characterized by missing or fused digits, although whether DSS1 plays a role in the aetiology of this syndrome is uncertain [1]. DSS1 encodes a small acidic protein of 70 amino acid residues with homologues in many, if not all, eukaryotic organisms. Human DSS1 protein was subsequently shown to associate with the product of the breast-cancer susceptibility gene BRCA2 [2], which plays an important role in the recombinational DNA repair in association with RAD51 [3]. Structural studies revealed that DSS1 interacts with a region of BRCA2 involved in binding to single-stranded DNA [4]. However, the biological significance of the DSS1–BRCA2 interaction was uncertain until genetic analysis of DSS1 and BRCA2 homologues in the smut fungus Ustilago maydis revealed that both proteins were required for DNA repair, recombination and genome stability [5,6]. Recent studies in mammalian cells showed that DSS1, like BRCA2, was required for the efficient formation of RAD51-containing nuclear foci in response to DNA damage and that loss of DSS1 expression results in chromosome instability and sensitivity to DNA-damaging agents [7].
The physiological significance of the DSS1–BRCA2 interaction seems clear, but studies in fission yeast (Schizosaccharomyces pombe) and budding yeast (Saccharomyces cerevisiae) indicate that DSS1 has physiological roles that are independent of its association with BRCA2. Orthologues of DSS1 are found in both these yeast species but both organisms appear to lack homologues of BRCA2. Mutants of Sacch. pombe deleted for its dss1+ gene were viable but showed pleiotropic phenotypes including slow growth, cell-cycle delay and temperature sensitivity [2]. The Sacch. cerevisiae DSS1 homologue, known as Sem1, was first identified through genetic interactions with genes encoding components of the exocyst, a multiprotein complex involved in exocytosis [8]. Mutations in Sem1 affect exocytosis, pseudohyphal growth and growth at a high temperature [2,8]. Recently, Sem1 was shown to associate with the 26 S proteasome in Sacch. cerevisiae and to be required for efficient ubiquitin-dependent protein degradation [9–11]. The 26 S proteasome is an ATP-dependent, multisubunit protease containing a proteolytic core, the 20 S CP (core particle), capped at one or both ends by the 19 S RP (regulatory particle), which recognizes ubiquitylated proteins and delivers them to the proteolytic core [12]. In this paper, we present genetic and biochemical evidence suggesting that S. pombe Dss1 associates with and regulates the function of the proteasome. We show that S. pombe Dss1 is important, although not essential, for proteasome function, particularly at high temperature. A surprising finding is that the essential function of Dss1 for growth at high temperature can be bypassed by overexpression of two subunits of the 19 S RP. These observations together with those of other authors show that DSS1 is an evolutionarily conserved regulator of the proteasome.
EXPERIMENTAL
Yeast strains, techniques and expression plasmids
S. pombe strains used in the present study are listed in Table 1. Standard genetic manipulations and growth media [YES (yeast extract supplemented medium) and EMM (Edinburgh minimal medium)] were as described in [13]. For the canavanine sensitivity test, glutamic acid (5 g/l) was used as a nitrogen source in place of ammonium chloride. L-Canavanine sulphate solution (10 mg/ml in sterile water) was added to media after autoclaving. For the expression of N-terminally TAP (tandem affinity purification)-tagged Dss1 (TAP-Dss1), the dss1+ open reading frame was cloned as an NdeI–BamHI fragment into pREP1-NTAP [14]. pREP-based expression plasmids carrying genes encoding proteasome subunits were a gift from Dr Colin Gordon (MRC Human Genetics Unit, Edinburgh, U.K.).
Table 1. S. pombe strains used in the present study.
| Strains | Genotype | Source |
|---|---|---|
| CB4 | h−ade6-M216 leu1-32 | M. Yamamoto (University of Tokyo) |
| CB7 | h+ade6-M210 leu1-32 | M. Yamamoto |
| CB301 | h90dss1::ura4+ade6-M216 leu1-32 ura4-D18 | D. A. Hughes |
| CB309 | h−mts3-1 leu1-32 | C. Gordon |
| CB310 | h−pad1-1 leu1-32 | C. Gordon |
| CB311 | h−pus1::ura4+leu1-32 ura4-D18 | C. Gordon |
| CB312 | h+pad1+::gfp+::ura4+leu1-32 ura4-D18 | C. Gordon |
| CB313 | h+pad1+::gfp+::ura4+dss1::ura4+leu1-32 ura4-D18 | The present study |
Isolation of multicopy suppressors
To construct an S. pombe genomic library lacking the dss1+ gene, genomic DNA was isolated from a dss1Δ mutant strain (CB301) using a genomic DNA extraction kit (Qiagen), the DNA was partially digested with Sau3A and the fragments were inserted into the BamHI site of the pIRT2 vector [15]. A dss1Δ strain was transformed with the library, and transformants were left to grow overnight to recover at 30 °C and then shifted to a restrictive temperature of 35 °C. After 7–10 days, colonies were picked, DNA was prepared from them and plasmids were rescued into Escherichia coli. The plasmids were retested for their ability to suppress the Ts (temperature-sensitive) phenotype of the dss1Δ mutant strain and were then partially sequenced.
Affinity purification from S. pombe
For large-scale TAP, an exponential phase culture of yeast (typically 6.0 litres) was grown in EMM at 30 °C to an A600 of 1.5. Cells were pelleted by centrifugation and washed with 50 mM Tris/HCl (pH 7.5) and 50 mM NaF. The pellet was frozen in liquid nitrogen, placed in a mortar kept on solid CO2, and manually ground with a pestle to a fine powder. The ground powder was collected in a 50 ml screw cap tube and thawed in one pellet volume of ice-cold lysis buffer A containing 10 mM Tris/HCl (pH 8.0), 150 mM NaCl, 2 mM EDTA, 1 mM EGTA, 20 mM NaF, 30 mM β-glycerol phosphate (pH 8.0), 1 mM DTT (dithiothreitol), 0.1% (v/v) Nonidet P40 (or Igepal CA-630) and protease inhibitor cocktail [Complete™ mix (Roche) and 1 mM PMSF]. Lysates were clarified by centrifugation at 15800 g for 20 min at 4 °C and protein concentration was determined with a Bradford assay kit (Bio-Rad). A 200 μl bed volume of IgG–Sepharose 6 Fast Flow beads (Amersham Biosciences), washed in lysis buffer A, was added to the cell extract and the mixture was left to rotate for 90 min at 4 °C. The IgG–Sepharose beads were collected by centrifugation and washed three times with 10 mM Tris/HCl (pH 8.0), 150 mM NaCl and 0.1% Igepal. Protein complexes were cleaved from the beads by digestion of the fusion protein with 100 units of TEV (tobacco etch virus) protease (Invitrogen) in 10 mM Tris/HCl (pH 8.0), 150 mM NaCl, 0.5 mM EDTA, 1 mM DTT and 0.1% Igepal for 2 h at 16 °C. The beads were spun down at 2000 g and the supernatant was collected.
The method described above was modified for affinity purification in the presence or absence of ATP. Ground cells were resuspended in 2 vol. of ice-cold buffer 1 (50 mM Tris/HCl, pH 8.0, 5 mM MgCl2 and 1 mM ATP) with protease inhibitor cocktail. The lysate was clarified twice at 15000 g for 15 min at 4 °C and incubated with IgG–Sepharose beads for 1 h at 4 °C. The beads were washed four times with 10 ml of ice-cold buffer 2 (50 mM Tris/HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2 and 1 mM ATP). To release the bound proteins the beads were boiled in SDS/PAGE sample buffer [50 mM Tris/HCl, pH 6.8, 100 mM DTT, 2%, (w/v), SDS, 10%, (v/v), glycerol and 0.1% (w/v), Bromophenol Blue]. For purification in the absence of ATP, ATP and MgCl2 were omitted from the buffers.
Protein identification by MS
SDS/polyacrylamide gels were stained in 0.25% (w/v) Coomassie Brilliant Blue, 45% (v/v) methanol and 10% (v/v) acetic acid and destained in 25% methanol and 7% acetic acid. Excised bands were rinsed twice in water (10 min each), dehydrated in 50% (v/v) acetonitrile for 10 min, incubated with 50 mM ammonium bicarbonate for 10 min and dehydrated again in 50% acetonitrile. In-gel digestion with 500 ng/ml sequencing-grade modified porcine trypsin (Promega) in 50 mM sodium bicarbonate was performed overnight at 37 °C. Samples for MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS analysis were prepared by mixing a small aliquot of the digest supernatant with an equal volume of a solution of α-cyano-4-hydroxycinnamic acid [10 mg/ml in acetonitrile/0.1% (v/v) trifluoroacetic acid, 1:1, v/v]. Peptide mass ‘fingerprinting’ was performed on a reflectron MALDI–TOF mass spectrometer (Axima-CFR; Kratos Analytical). Mass spectra were internally calibrated with trypsin autolysis peaks [mass/charge (m/z) 842.51 or 2211.10]. Mascot software (Matrix Science) was employed for protein database searching using monoisotopic mass values for each spectrum. Protein identity was based on at least three matching peptides, searching peptide masses allowing for one missed tryptic cleavage with a mass tolerance of 0.2 Da, deriving a molecular mass score with a P<0.05 within the Mascot software. Samples for LC–MS (liquid chromatography/MS) were separated on a 75 μm×15 cm C18 column using a Famos/Switchos/Ultimate system (LC Packings). On-line nanospray ion-trap MS was performed on a Bruker Esquire 3000+ (Bruker Daltonics).
SDS/PAGE analysis and Western blotting
SDS/PAGE was performed using standard methods. Separated proteins were stained with Coomassie Blue or transferred on to a PVDF membrane (Immobilon-P; Millipore). Polyclonal anti-Mts4 antibodies were obtained from Dr Colin Gordon. Polyclonal anti-20 S antibodies, polyclonal anti-ubiquitin antibodies and purified S. cerevisiae 20 S complexes were obtained from Affiniti. Monoclonal anti-α-tubulin antibodies were obtained from Sigma. Processing of membranes was as described previously [16].
Detection of poly-ubiquitylated proteins
To detect poly-ubiquitylated proteins, cells were lysed in buffer A (described above) supplemented with 50 mM N-ethylmaleimide, an inhibitor of deubiquitylation. Total crude extracts containing equal amounts of proteins were analysed by Western blotting with anti-ubiquitin polyclonal antibody (dilution 1:5000). The membrane was reprobed with mouse monoclonal anti-α-tubulin (dilution 1:5000) as a loading control.
Localization of Pad1/Rpn11–GFP (green fluorescent protein) in live cells
Coverslips were pretreated with polylysine (1 mg/ml) before adding one drop of exponential phase culture grown in YES. Cells were observed and images were captured using a ×63 objective on a Zeiss LSM 510-Meta laser scanning microscope (Zeiss).
RESULTS
Identification of components of the 19 S RP of the proteasome as multicopy suppressors of the dss1Δ mutant
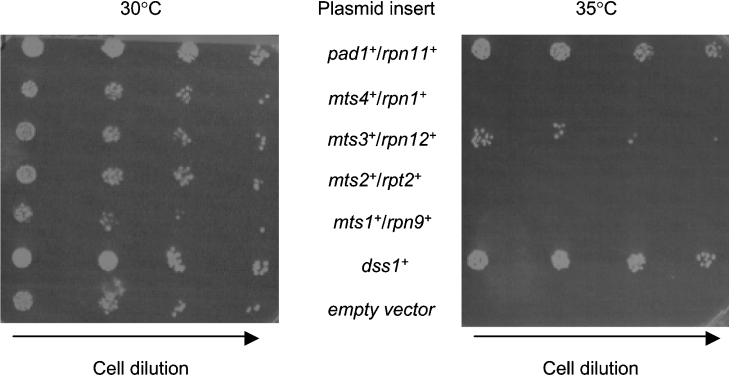
The identification of multicopy suppressors – genes that can suppress a mutant phenotype when expressed from a multicopy plasmid – is a powerful way of identifying genes involved in controlling the same biological process. To understand further the function of Dss1 in S. pombe, we sought to identify multicopy supressors of the Ts phenotype caused by deletion of the dss1+ gene. A genomic library in a multicopy plasmid was constructed with DNA from a strain lacking the dss1+ gene and screened for clones capable of rescuing the dss1Δ Ts phenotype. Two different genomic clones were identified repeatedly and sequence analysis revealed that both clones carried genes encoding components of the 26 S proteasome: one carried the pad1+/rpn11+ gene and the other carried the mts3+/rpn12+ gene. Suppression of the dss1Δ Ts phenotype by expressing just the open reading frame of each of these genes under the control of the nmt1 promoter confirmed that these genes were responsible the suppression (Figure 1).
Figure 1. Overexpression of genes encoding subunits of the 19 S RP suppresses the growth defect of the dss1Δ mutant.
A dss1Δ mutant strain (CB301) was transformed with pREP-based expression plasmids for the genes indicated. Exponential phase cultures of transformants were diluted in a 5-fold series, spotted on to EMM plates and grown at 30 or 35 °C for 7 days.
The isolation of these two genes as multicopy suppressors of dss1Δ suggested a functional relationship between the proteasome and Dss1; both genes encode subunits of the 19 S RP. In Sacch. cerevisiae the 19 S RP contains at least 17 different core proteins and can be dissociated under certain conditions into base and lid subcomplexes [17,18]. The base consists of six ATPases (Rpt1–Rpt6), together with the non-ATPase subunits Rpn1, Rpn2 and Rpn10. The lid consists of the remaining eight non-ATPase subunits. Its components display significant sequence identity with subunits of both the COP9 (constitutively photomorphogenic 9) signalosome complex and the translational initiation factor 3 complex [18]. Both Pad1/Rpn11 and Mts3/Rpn12 are subunits of the lid subcomplex. Mutations in the genes encoding these two subunits were isolated in a screen for mutants that were both resistant to methyl 2-benzimidazolecarbamate, a microtubule inhibitor, and Ts for cell-cycle progression [19,20]. Ts mutations in both genes cause arrest of the cells in mitosis, probably because of a failure to degrade cyclin B/Cdc13 and securin/Cut2. We therefore decided to investigate whether overexpression of other components of the 19 S RP could also rescue the dss1Δ phenotype at 35 °C. Of the five subunits tested, only multicopy pad1+/rpn11+ and mts3+/rpn12+, encoding components of the lid, were able to suppress the Ts phenotype of the dss1Δ mutant strain, suggesting some specificity in the genetic interactions (Figure 1).
Dss1 associates with the 19 S RP of the proteasome in vivo
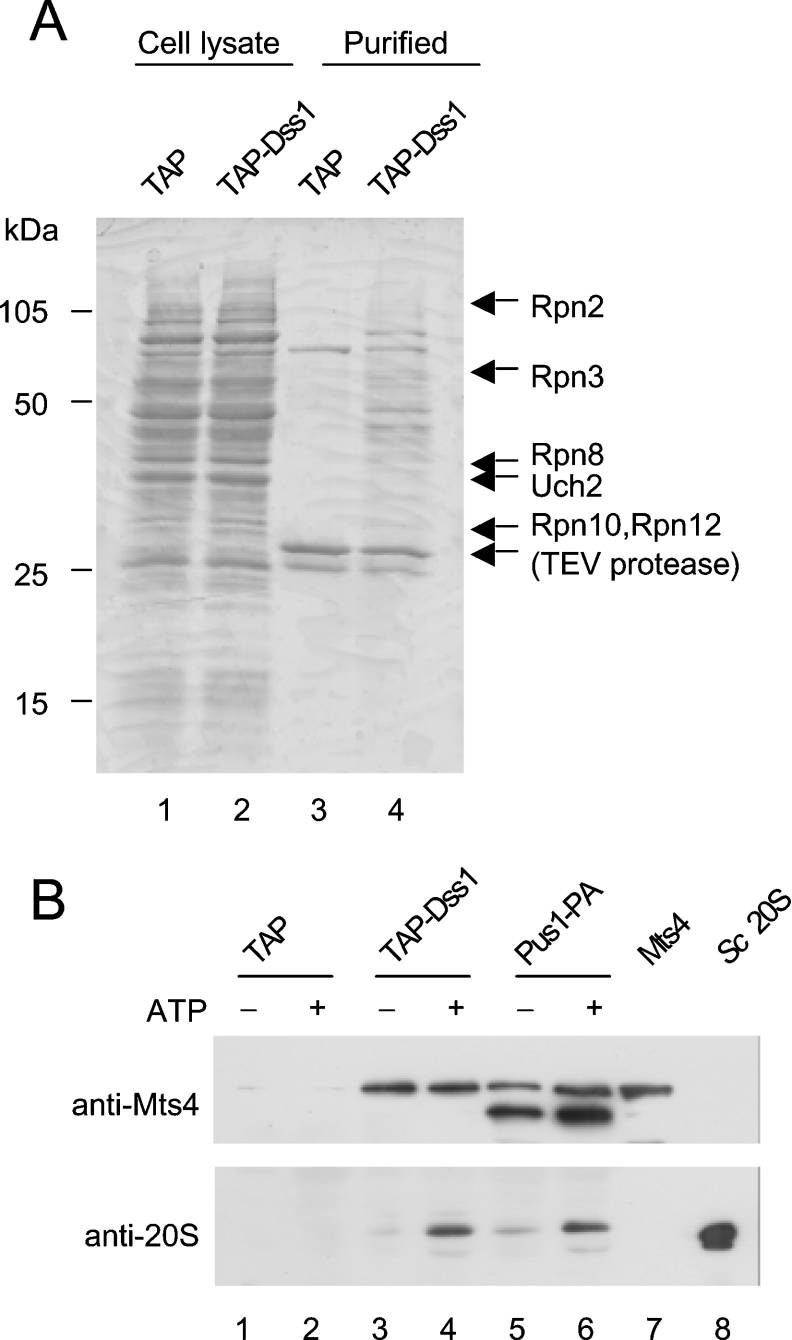
To investigate whether Dss1 associates with other proteins in vivo, a tagged version of Dss1 was affinity-purified from cells and associated proteins were identified by MS. The method employed was based on the TAP method but only the first purification step – binding of the Protein A units of the tagged protein to IgG-coupled beads – was performed [21]. To do this, a TAP-tagged version of Dss1 (TAP-Dss1) was expressed in a dss1Δ mutant strain and shown to be biologically functional as it rescued the growth defect of the mutant strain. TAP-Dss1 was affinity-purified in a single step by incubation with IgG–Sepharose beads. Dss1 and associated proteins were released from the beads by cleavage of the TAP tag with TEV protease. As a control, the TAP tag itself was expressed in WT (wild-type) cells and purified as described above. The purified samples were separated by SDS/PAGE, stained with Coomassie Blue and the identity of protein bands was inferred following analysis by MS. Gel slices from the TAP- Dss1 purification were processed in parallel with gel slices from the corresponding region of the TAP purification, which served as a negative control. A typical experiment is shown in Figure 2(A). In three independent experiments, a total of 13 proteins were identified that co-purified with TAP-Dss1 but not with TAP (Table 2). Ten of these were core components of the 19 S RP of the 26 S proteasome: three ATPases (Rpt2, Rpt3 and Rpt5) and seven non-ATPases (Rpn2, Rpn3, Rpn5, Rpn7, Rpn8, Rpn10 and Rpn12). Although not all known components of the 19 S RP were identified in these experiments, the presence of both lid and base components indicates that Dss1 associates with the intact 19 S RP. Other proteins in the Dss1 complex were: Uch2, a deubiquitylating enzyme [22]; Cut20/Lid1, a subunit of the APC (anaphase promoting complex)/cyclosome [23,24]; and a homologue of Sacch. cerevisiae Nas6 and human gankyrin [25,26].
Figure 2. Dss1 associates with the proteasome.
(A) TAP tag, expressed in a WT strain (CB4), and TAP-tagged Dss1 (TAP-Dss1), expressed in a dss1Δ mutant strain (CB301), were affinity-purified using IgG–Sepharose. Protein complexes were released from the beads by TEV protease cleavage, separated by SDS/PAGE and stained with Coomassie Blue (lanes 3 and 4). The proteins shown to the right of the gel were identified by MS analysis. Proteins shown in parentheses were present in both the TAP and TAP-Dss1 purifications whereas the others were present only in the TAP-Dss1 purification. (B) TAP or TAP-tagged Dss1 (TAP-Dss1) was expressed in S. pombe cells as described above and affinity-purified using IgG–Sepharose in the presence or absence of ATP. As a control a Protein A-tagged version of Pus1, a component of the 19 S RP, was affinity-purified using the same method. The purified complexes were separated on two SDS/12% polyacrylamide gels and analysed by Western blotting using either anti-Mts4 antibodies (upper panel) or anti-20 S CP antibodies (lower panel). Protein extract from a WT strain overexpressing Mts4 (lane 7) and purified Sacch. cerevisiae 20 S CP (lane 8) were used as controls for the specificity of the antibodies.
Table 2. Proteins identified by MS in the Dss1 complex.
| UniProt accession number | Peptides matched | S. pombe name* | Sacch. cerevisiae homologue | Human homologue |
|---|---|---|---|---|
| O14140 | 4 | Dss1 | Sem1 | DSS1 |
| P33612 | 11 | Mts2/Rpt2 | Rpt2 | S4 |
| O74894 | 3 | Rpt3/SPCC576.10c | Rpt3 | S6b |
| O14126 | 4 | Rpt5/Tbp1/Pam2 | Rpt5 | S6′ |
| O74762 | 16 | Rpn2/SPBC17D11.07c | Rpn2 | S1 |
| O42897 | 11 | Rpn3/SPBC119.01 | Rpn3 | S3 |
| Q9UTM3 | 7 | Rpn5 | Rpn5 | RPN5 |
| Q10335 | 11 | Rpn7/SOBC582.07c | Rpn7 | S10 |
| O74440 | 6 | Rpn8/SPCC1682.10 | Rpn8 | S12 |
| O94444 | 5 | Pus1/Rpn10 | Rpn10 | S5a |
| P50524 | 8 | Mts3/Rpn12 | Rpn12 | S14 |
| Q9UUB6 | 3 | Uch2 | Yuh1 | UCHL5 |
| O42839 | 5 | Cut20/Lid1/Apc4 | Apc4 | APC4 |
| Q10311 | 3 | Nas6/SPAC6C3.08 | Nas6 | Gankyrin |
* For previously hypothetical gene products, proposed names in accordance with the unified nomenclature for proteasome subunits in Sacch. cerevisiae [34] and open reading frame names from the S. pombe genome project are shown.
Association of Dss1 with the 26 S proteasome is ATP-dependent
No 20 S CP components were identified in the Dss1 complex. However, the complex was purified in the absence of ATP, which is known to affect the association between the 20 and 19 S complexes [27]. It is also known that the association of some proteins with the proteasome is sensitive to ATP [28]. To investigate whether ATP affects the interaction between Dss1 and the proteasome, TAP-Dss1 was purified from cells in the presence or absence of exogenous ATP. Protein complexes were separated by SDS/PAGE and the presence of the 19 S RP and 20 S CP was investigated by Western blotting using antisera raised against Mts4/Rpn1 [29], a 19 S base component, and against the 20 S CP (Figure 2B). Co-purification of Mts4 with Dss1 was unaffected by ATP whereas co-purification of the 20 S CP with Dss1 was markedly reduced in the absence of ATP (Figure 2B, lanes 3 and 4). These results show that Dss1 associates with the 26 S proteasome in an ATP-dependent manner, whereas association of Dss1 with free 19 S RP is unaffected by ATP. The ATP dependence of the Dss1–26 S association may reflect the intrinsic instability of the 20 S–19 S interaction in the absence of ATP. This explanation is consistent with a similar reduction in recovery of the 20 S CP in the absence of ATP found when a tagged version of Pus1/Rpn10 was used to purify the proteasome (Figure 2B, lanes 5 and 6).
Cells lacking Dss1 show phenotypes consistent with a defect in proteasome function
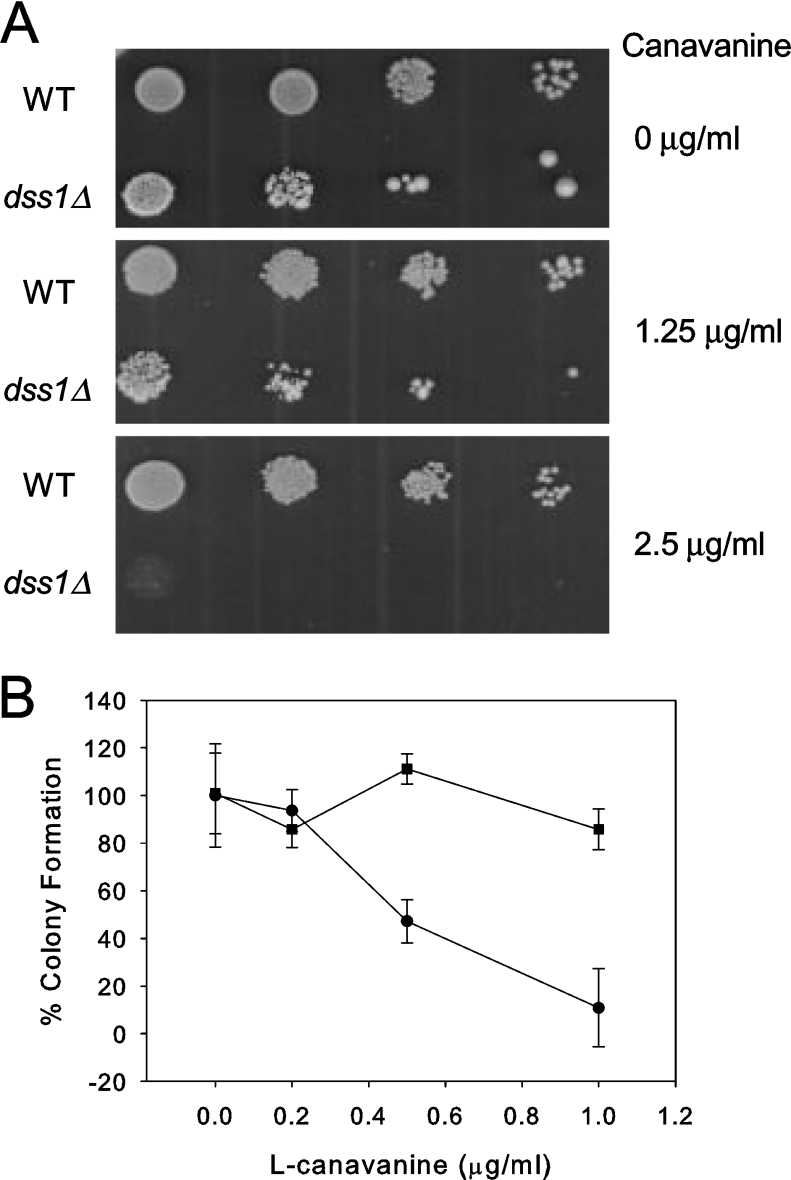
A characteristic phenotype of mutants defective in proteasome function is enhanced sensitivity to canavanine, an arginine analogue. Addition of canavanine to growth media leads to the synthesis of abnormal proteins that are degraded by the proteasome in normal cells; when proteasome activity is compromised cells are more sensitive to the deleterious effects of the abnormal proteins. Mutant dss1Δ cells grew more slowly on media supplemented with 1.25 μg/ml canavanine and failed to grow at all on media with 2.5 μg/ml canavanine (Figure 3A). The plating efficiency of dss1Δ mutant cells on media containing low concentrations of canavanine (0.5–1.0 μg/ml) was significantly reduced relative to WT cells (Figure 3B).
Figure 3. The dss1Δ mutant strain has phenotypes characteristic of a defect in proteasome function.
(A) WT (CB7) and dss1Δ mutant (CB301) cells were diluted in 5-fold series and spotted on to plates containing 0, 1.25, or 2.5 μg/ml L-canavanine as indicated. (B) WT or dss1Δ cells were grown in YES and dilutions plated on EMM plates containing L-canavanine at the indicated concentration and incubated at 29 °C for 10 days. The number of colonies formed in the absence of L-canavanine was taken as 100% plating efficiency. The results are indicated on the graph by the symbol ‘■’ for WT and ‘●’ for the dss1Δ mutant. For each concentration the experiment was performed in triplicate (or in duplicate for the values at 1 μg/ml canavanine) and the mean±S.D. was calculated. The difference in plating efficiency between the strains was statistically significant by the Student's t test at the 0.5 μg/ml (P<0.001) and 1.0 μg/ml (P<0.05) concentrations of L-canavanine.
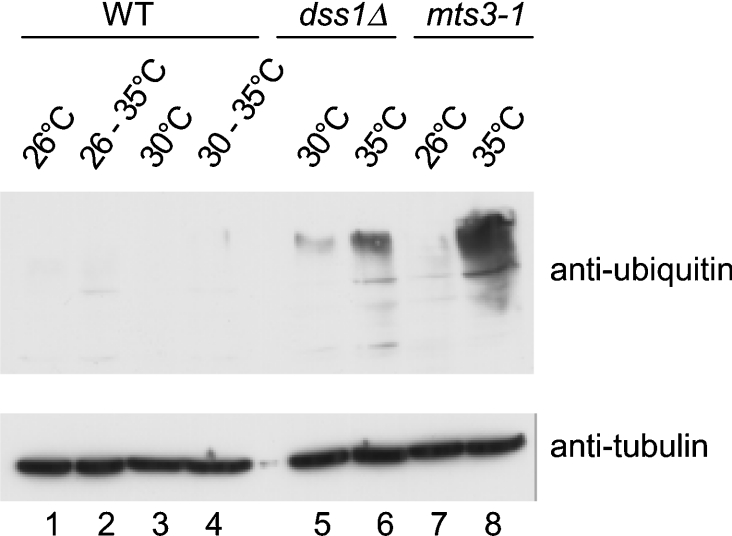
A biochemical phenotype associated with defects in proteasome function is the accumulation of HMW [high molecular weight (‘mass’)] ubiquitin conjugates [20]. To investigate whether the dss1Δ mutant accumulates HMW ubiquitin conjugates, protein extracts made from dss1Δ mutant cells grown at 30 or 35 °C were analysed by Western blotting with anti-ubiquitin antibodies (Figure 4). The result shows that HMW ubiquitylated conjugates were present in the dss1Δ strain at the permissive temperature (30 °C), and at a higher level at the restrictive temperature (35 °C; lanes 5 and 6). A similar, though higher, level of HMW ubiquitin conjugates was seen in a Ts mts3/rpn12 mutant strain at the restrictive temperature (35 °C; lanes 7 and 8). No HMW ubiquitylated conjugates were detected in the WT strain grown at 26, 30 or 35 °C (lanes 1–4). These results show that the dss1Δ mutant has a defect in the processing of ubiquitylated proteins that becomes more severe at higher temperature.
Figure 4. The dss1Δ mutant strain accumulates HMW ubiquitin conjugates.
Exponential phase cultures of WT (CB7), dss1Δ mutant (CB301) and mts3/rpn11 mutant (CB309) strains were grown in YES at the permissive temperature (26 or 30 °C) and a part of each culture was transferred to the restrictive temperature (35 °C) for 4 h. Protein extracts were prepared and analysed by Western blotting with anti-ubiquitin antibodies to detect ubiquitylated conjugates (upper panel) or with anti-α-tubulin as a loading control (lower panel).
To investigate genetic interactions between dss1+ and genes encoding proteasome components, we attempted to construct strains double mutant for dss1 deletion and pus1 (rpn10) deletion or pad1 (rpn11) Ts mutations. The dss1Δ pad1-1Ts double-mutant strain was inviable at 30 °C, a permissive temperature for both single-mutant strains; the spores germinated but formed highly elongated cells that did not divide. The dss1Δ pus1Δ double-mutant strain was viable at 30 °C but grew very slowly, taking 2 weeks to form a small colony (∼1 mm diameter) at 30 °C, and the cells were highly elongated.
Dss1 does not regulate subcellular localization of the proteasome
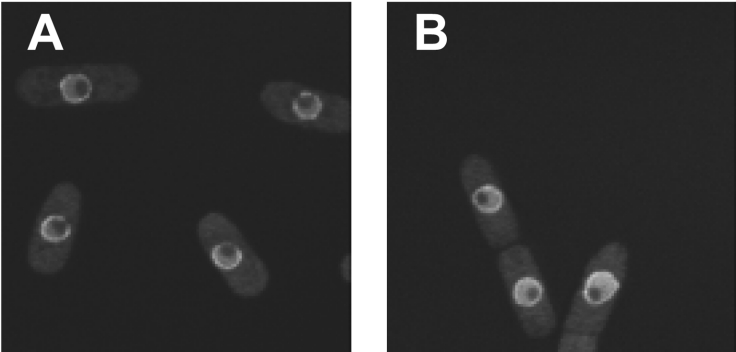
In proliferating cells of S. pombe, the 26 S proteasome is predominantly localized to the inner side of the nuclear membrane [29]. To investigate whether loss of dss1+ function affects the localization of the proteasome, the localization of a Pad1/Rpn11–GFP fusion protein was examined in live WT and dss1Δ mutant cells by confocal fluorescence microscopy. The distribution of Pad1/Rpn11–GFP fusion protein appeared unchanged in dss1Δ cells at 30 °C compared with WT cells (Figure 5). Short incubation (1–2 h) of dss1Δ mutant cells at 35 °C did not affect localization of Pad1/Rpn11–GFP, although prolonged incubation led to severe morphological changes and concomitant changes in Pad1/Rpn11–GFP localization (D. A. Hughes, unpublished work). In conclusion, even complete removal of Dss1 does not detectably affect subcellular localization of the proteasome in morphologically normal cells.
Figure 5. Localization of Pad1/Rpn11–GFP is normal in dss1Δ cells.
Confocal fluorescence images of the cellular localization of the Pad1/Rpn11–GFP fusion protein in live (A) WT (CB312) or (B) dss1Δ mutant (CB313) cells.
DISCUSSION
In this paper, we present biochemical and genetic evidence showing that Dss1 associates with the 19 S RP of the proteasome and is required for efficient ubiquitin-mediated proteolysis. In biochemical purification studies, Dss1 co-purified with the 19 S RP in the absence of ATP and associated with the intact 26 S proteasome in the presence of ATP (Figure 2). In this respect, Dss1 differs from the ‘proteasome interacting proteins’ described by Verma et al. [28], which associate with 19 S (and 26 S) complexes only in the absence of ATP. The association of Dss1 with the 19 S RP in S. pombe is similar to the reported association of Sacch. cerevisiae Sem1 with the 19 S RP [9–11]. From our studies, we cannot say whether Dss1 is a core subunit of the 26 S proteasome or an accessory protein present in non-stoichiometric amounts. In Sacch. cerevisiae, Sem1 efficiently co-purifies with the proteasome, suggesting that it may be a stoichiometric subunit; however, upon gel filtration of cell extracts, most of the Sem1 protein is found in fractions not containing proteasomes [10].
A role for Dss1 in proteasome function is supported by analysis of the phenotypes of a mutant defective in dss1+ function. Like other mutants defective in proteasome function, the dss1Δ mutant strain was supersensitive to canavanine, an amino acid analogue that causes the production of abnormal proteins (Figure 3), and accumulated HMW ubiquitin conjugates, indicating a defect in the turnover of ubiquitylated proteins (Figure 4). This accumulation of HMW ubiquitin conjugates was significant at the permissive temperature for the mutant (30 °C), but was more pronounced at a higher temperature (35 °C) that prevents continued proliferation of the mutant strain. Furthermore, synthetic growth defects were observed when the dss1Δ mutation was combined with mutations in other proteasome subunits. Similar biochemical and genetic observations have been made with the sem1 mutant in Sacch. cerevisiae [9–11]. Unlike most of the genes encoding core subunits of the proteasome in S. pombe, dss1+ is not an essential gene and therefore cannot be essential for proteasome function. However, the results presented here argue that dss1+ function is important for efficient functioning of the proteasome, particularly at higher temperatures.
The molecular function of Dss1 and the role of its interaction with the proteasome remain to be established. The interaction with the proteasome must be important because the Ts growth defect of the dss1Δ mutant was rescued by overexpression of two lid components of the 19 S RP (Figure 1). Multicopy suppression of proteasome subunit mutants by other proteasome subunits has been reported previously [30], but the situation we report is unusual in that overexpression of the proteasome subunits can rescue a complete gene deletion of the dss1+ gene and not just a Ts missense or protein-truncating mutation. It was reported that in the Sacch. cerevisiae sem1 mutant strain, proteasomes were less stable in vitro than Sem1-containing proteasomes, possibly due to decreased stability of the lid or decreased affinity between the lid and base substructures of the 19 S RP [10,11]. The suppression of the Ts growth defect of the S. pombe dss1Δ mutant by overexpression of two lid components is suggestive of a role for Dss1 in the stability of the lid. Dss1 does not appear to be required for the correct subcellular localization of the proteasome, although subtle effects could be missed by the technique we have used (Figure 5) and we were able to purify proteolytically active 26 S proteasomes from dss1Δ mutant cells (L. Jossé and D. A. Hughes, unpublished work).
DSS1 is of interest in the fields of cancer biology and DNA repair because of its interaction with the tumour suppressor protein BRCA2. We have shown in a previous study that human DSS1 can functionally replace S. pombe Dss1 [2] and more recent evidence indicates that human DSS1 can associate with the proteasome [9,11]. These observations and those in the present paper raise the interesting possibility that DSS1 may be involved in linking BRCA2 with the proteasome. BRCA2 is implicated in the control of protein ubiquitylation by its association with BRCA1, BARD1 and other proteins in an E3 ubiquitin ligase complex called BRCC (BRCA1–BRCA2-containing complex) [31] and has also been implicated in the Fanconi anaemia pathway that controls mono-ubiquitylation of the FANCD2 protein [32]. It will be of considerable interest to determine whether human DSS1 mediates an interaction between BRCA2 and the proteasome. Alternatively, DSS1 may function independent of the proteasome. A recent study in S. pombe showed that Dss1 is required for efficient export of mRNA from the nucleus to cytoplasm and interacts directly with several components of the nuclear export machinery [33]. DSS1 certainly has a surprisingly diverse set of functions for such a small protein.
Acknowledgments
We thank Dr Colin Gordon for generously providing materials and invaluable advice, Steve Aves for the pIRT2 plasmid, Chris Storey for analysis of samples by MS, Chi Tang for generous help with confocal microscopy and Alan Ashworth (Institute of Cancer Research, London, U.K.) for communicating results before publication. I. M. S. P. was supported by an undergraduate PRODEP III scholarship (Ministry of Education, Portugal). This work was supported by a grant from the Association for International Cancer Research to D. A. H.
References
- 1.Crackower M. A., Scherer S. W., Rommens J. M., Hui C. C., Poorkaj P., Soder S., Cobben J. M., Hudgins L., Evans J. P., Tsui L. C. Characterization of the split hand/split foot malformation locus SHFM1 at 7q21.3-q22.1 and analysis of a candidate gene for its expression during limb development. Hum. Mol. Genet. 1996;5:571–579. doi: 10.1093/hmg/5.5.571. [DOI] [PubMed] [Google Scholar]
- 2.Marston N. J., Richards W. J., Hughes D., Bertwistle D., Marshall C. J., Ashworth A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol. Cell. Biol. 1999;19:4633–4642. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venkitaraman A. R. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell (Cambridge, Mass.) 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 4.Yang H., Jeffrey P. D., Miller J., Kinnucan E., Sun Y., Thoma N. H., Zheng N., Chen P. L., Lee W. H., Pavletich N. P. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 5.Kojic M., Kostrub C. F., Buchman A. R., Holloman W. K. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell. 2002;10:683–691. doi: 10.1016/s1097-2765(02)00632-9. [DOI] [PubMed] [Google Scholar]
- 6.Kojic M., Yang H., Kostrub C. F., Pavletich N. P., Holloman W. K. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol. Cell. 2003;12:1043–1049. doi: 10.1016/s1097-2765(03)00367-8. [DOI] [PubMed] [Google Scholar]
- 7.Gudmundsdottir K., Lord C. J., Witt E., Tutt A. N., Ashworth A. DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep. 2004;5:989–993. doi: 10.1038/sj.embor.7400255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jantti J., Lahdenranta J., Olkkonen V. M., Soderlund H., Keranen S. SEM1, a homologue of the split hand/split foot malformation candidate gene Dss1, regulates exocytosis and pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. U.S.A. 1999;96:909–914. doi: 10.1073/pnas.96.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krogan N. J., Lam M. H., Fillingham J., Keogh M. C., Gebbia M., Li J., Datta N., Cagney G., Buratowski S., Emili A., et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell. 2004;16:1027–1034. doi: 10.1016/j.molcel.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 10.Funakoshi M., Li X., Velichutina I., Hochstrasser M., Kobayashi H. Sem1, the yeast ortholog of a human BRCA2-binding protein, is a component of the proteasome regulatory particle that enhances proteasome stability. J. Cell Sci. 2004;117:6447–6454. doi: 10.1242/jcs.01575. [DOI] [PubMed] [Google Scholar]
- 11.Sone T., Saeki Y., Toh-e A., Yokosawa H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:28807–28816. doi: 10.1074/jbc.M403165200. [DOI] [PubMed] [Google Scholar]
- 12.Hartmann-Petersen R., Seeger M., Gordon C. Transferring substrates to the 26 S proteasome. Trends Biochem. Sci. 2003;28:26–31. doi: 10.1016/s0968-0004(02)00002-6. [DOI] [PubMed] [Google Scholar]
- 13.Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 14.Tasto J. J., Carnahan R. H., McDonald W. H., Gould K. L. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- 15.Hindley J., Phear G., Stein M., Beach D. Sucl+ encodes a predicted 13-kilodalton protein that is essential for cell viability and is directly involved in the division cycle of Schizosaccharomyces pombe. Mol. Cell. Biol. 1987;7:504–511. doi: 10.1128/mcb.7.1.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodcock S. A., Hughes D. A. p120 Ras GTPase-activating protein associates with fibroblast growth factor receptors in Drosophila. Biochem. J. 2004;380:767–774. doi: 10.1042/BJ20031848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glickman M. H., Rubin D. M., Fried V. A., Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glickman M. H., Rubin D. M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V. A., Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell (Cambridge, Mass.) 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson C. R., Wallace M., Seeger M., Dubiel W., Gordon C. Mts4, a non-ATPase subunit of the 26 S protease in fission yeast is essential for mitosis and interacts directly with the ATPase subunit Mts2. J. Biol. Chem. 1997;272:25768–25777. doi: 10.1074/jbc.272.41.25768. [DOI] [PubMed] [Google Scholar]
- 20.Gordon C., McGurk G., Dillon P., Rosen C., Hastie N. D. Defective mitosis due to a mutation in the gene for a fission yeast 26 S protease subunit. Nature (London) 1993;366:355–357. doi: 10.1038/366355a0. [DOI] [PubMed] [Google Scholar]
- 21.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 22.Li T., Naqvi N. I., Yang H., Teo T. S. Identification of a 26 S proteasome-associated UCH in fission yeast. Biochem. Biophys. Res. Commun. 2000;272:270–275. doi: 10.1006/bbrc.2000.2767. [DOI] [PubMed] [Google Scholar]
- 23.Berry L. D., Feoktistova A., Wright M. D., Gould K. L. The Schizosaccharomyces pombe dim1+ gene interacts with the anaphase-promoting complex or cyclosome (APC/C) component lid1+ and is required for APC/C function. Mol. Cell. Biol. 1999;19:2535–2546. doi: 10.1128/mcb.19.4.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamashita Y. M., Nakaseko Y., Kumada K., Nakagawa T., Yanagida M. Fission yeast APC/cyclosome subunits, Cut20/Apc4 and Cut23/Apc8, in regulating metaphase-anaphase progression and cellular stress responses. Genes Cells. 1999;4:445–463. doi: 10.1046/j.1365-2443.1999.00274.x. [DOI] [PubMed] [Google Scholar]
- 25.Hori T., Kato S., Saeki M., DeMartino G. N., Slaughter C. A., Takeuchi J., Toh-e A., Tanaka K. cDNA cloning and functional analysis of p28 (Nas6p) and p40.5 (Nas7p), two novel regulatory subunits of the 26 S proteasome. Gene. 1998;216:113–122. doi: 10.1016/s0378-1119(98)00309-6. [DOI] [PubMed] [Google Scholar]
- 26.Higashitsuji H., Itoh K., Nagao T., Dawson S., Nonoguchi K., Kido T., Mayer R. J., Arii S., Fujita J. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat. Med. 2000;6:96–99. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- 27.Hendil K. B., Hartmann-Petersen R., Tanaka K. 26 S proteasomes function as stable entities. J. Mol. Biol. 2002;315:627–636. doi: 10.1006/jmbi.2001.5285. [DOI] [PubMed] [Google Scholar]
- 28.Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R. J. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkinson C. R., Wallace M., Morphew M., Perry P., Allshire R., Javerzat J. P., McIntosh J. R., Gordon C. Localization of the 26 S proteasome during mitosis and meiosis in fission yeast. EMBO J. 1998;17:6465–6476. doi: 10.1093/emboj/17.22.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkinson C. R., Penney M., McGurk G., Wallace M., Gordon C. The 26 S proteasome of the fission yeast Schizosaccharomyces pombe. Philos. Trans. R. Soc. London B Biol. Sci. 1999;354:1523–1532. doi: 10.1098/rstb.1999.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong Y., Hakimi M. A., Chen X., Kumaraswamy E., Cooch N. S., Godwin A. K., Shiekhattar R. Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell. 2003;12:1087–1099. doi: 10.1016/s1097-2765(03)00424-6. [DOI] [PubMed] [Google Scholar]
- 32.Howlett N. G., Taniguchi T., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G., et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 33.Thakurta A. G., Gopal G., Yoon J. H., Kozak L., Dhar R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. EMBO J. 2005;24:2512–2513. doi: 10.1038/sj.emboj.7600713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finley D., Tanaka K., Mann C., Feldmann H., Hochstrasser M., Vierstra R., Johnston S., Hampton R., Haber J., McCusker J., et al. Unified nomenclature for subunits of the Saccharomyces cerevisiae proteasome regulatory particle. Trends Biochem. Sci. 1998;23:244–245. doi: 10.1016/s0968-0004(98)01222-5. [DOI] [PubMed] [Google Scholar]